Abstract
Bardet–Biedl syndrome (BBS) is a rare autosomal recessive ciliopathy characterized by retinitis pigmentosa (RP), truncal obesity, cognitive impairment, hypogonadism in men, polydactyly, and renal abnormalities with severe renal dysfunction. Twenty-two causative genes have already been reported for this disorder. In this study, we identified two unrelated Japanese patients with clinical diagnoses of BBS associated with compound heterozygous SCLT1 mutation. Patient 1 was a 10-year-old girl, and patient 2 was a 22-year-old man. Both the patients showed severe renal dysfunction in childhood, RP, mild intellectual disability, short stature, and truncal obesity, without oral aberrations and polydactyly. Patient 2 also had hypogonadism. We identified two missense variants in SCLT1, c.[1218G > A] and [1631A > G], in both the patients by next-generation sequencing. Subsequent cDNA analysis revealed that c.1218G > A affected exon 14 skipping in SCLT1. To date, SCLT1 has been reported as the causative gene of oral–facial–digital syndrome type IX, and Senior–Løken syndrome. The phenotypes of both the present patients were compatible with BBS. These results highlight SCLT1 as an additional candidate for BBS phenotype in an autosomal recessive manner.
Electronic supplementary material
The online version of this article (10.1007/s13730-020-00472-y) contains supplementary material, which is available to authorized users.
Keywords: Bardet–biedl syndrome, Ciliopathy, SCLT1, Next-generation sequencing
Introduction
Bardet–Biedl syndrome (BBS) is a rare genetic disorder, an autosomal recessive ciliopathy, characterized by retinitis pigmentosa (RP), truncal obesity, cognitive impairment, hypogonadism in men, polydactyly, and renal abnormalities [1]. BBS has an occurrence of approximately 1/100,000 in North America and Europe [2]. The Online Mendelian Inheritance in Man (https://www.omim.org/) has reported 22 causative genes for BBS. Although the genes that have been associated with BBS so far are inherited in an autosomal or digenic recessive manner, we recently reported an OFD1 (gene responsible for causing oral–facial–digital syndrome type I in women) [3] aberration in a man with BBS phenotype that was inherited in an X-linked manner [4]. A renal abnormality in BBS is recognized as renal dysplasia or nephronophthisis (NPHP) [1], and renal dysfunction is observed in 53–82% of BBS patients [1]. Renal symptoms begin in childhood with polyuria, polydipsia, and nephrogenic anemia, and reach end-stage renal disease (ESRD). Therefore, renal abnormalities are the most important reasons for mortality in BBS patients.
Recently, the sodium channel and clathrin linker 1 gene (SCLT1, 4q28.2) has been reported to be the causative gene for oral–facial–digital syndrome type IX (OFD9) [5], and Senior–Løken syndrome (SLSN) [6]. SCLT1 encodes an adaptor protein associated with the sodium voltage-gated channel alpha subunit 10 and clathrin [7], and SCLT1 was identified as one of the component proteins of the distal appendages of centrioles, which was proposed to anchor cilia to the plasma membrane [8]. However, few patients with SCLT1 aberrations have been reported. Here, we report two unrelated patients clinically diagnosed with BBS, who carry the same compound heterozygous SCLT1 mutations.
Case reports
Patient 1
Patient 1 was a 10-year-old Japanese girl with non-consanguineous parents. The patient had no significant family history (Fig. 1a). Her gestational age was 39 weeks 2 days and birth weight was 2700 g. She had intellectual disability, autism, and motor developmental delay (head control 3–4 months old, turn over 9 months old) because of which she had recently received special education support. She also had visual disability due to RP, which was diagnosed by flat electroretinogram. The patient was first detected with mild proteinuria (urine protein/creatinine ratio 0.36) at the age of 10 through an annual school urinalysis, which revealed severe renal dysfunction (serum creatinine was 5.6 mg/dL, estimated glomerular filtration rate was 8 mL/min/1.73 m2) and hyperuricemia (9.3 mg/dL). Her urine β2-microglobulin was > 10,000 μg/dL. Renal echography revealed bilateral hyperechogenicity, ambiguous cortico-medullary junction, and small renal cysts. Renal biopsy was not performed. Her height was 121.0 cm (− 2.84 SD) and body weight was 28.6 kg (obesity ratio = 27.8%). The oral cavity of the patient showed no apparent abnormalities.
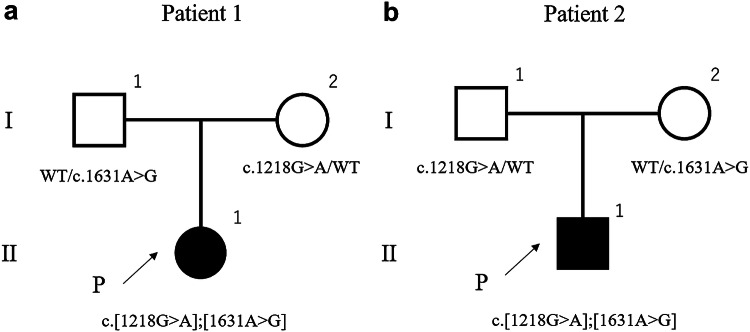
Fig. 1.
Pedigrees of the two families. The patients had no siblings, and their parents were non-consanguineous
Patient 2
Patient 2 was a 23-year-old Japanese man with non-consanguineous parents, with no family history of BBS (Fig. 1b). His gestational age was 41 weeks and 1 day and birth weight was 3442 g. At the age of 1 year, he was diagnosed with hepatic fibrosis after analysis of a liver biopsy. The patient had nystagmus and visual impairment, and was diagnosed with RP at the age of 3 years by an ophthalmologist. His renal function had deteriorated, and the patient was initiated on peritoneal dialysis at the age of 11 years. At the time of kidney transplantation, his urine β2-microglobulin was 4519 μg/dL and N-acetylglucosaminidase was 3.9 U/L. The patient had received living-donor kidney transplant from his father at the age of 13 years. The patient also showed other abnormalities, such as micropenis, mild cognitive impairment, short stature (156.8 cm, − 2.4 SD), and obesity (obesity ratio = 57.3%). The oral cavity of the patient showed no apparent abnormalities. These symptoms indicated BBS in the patient, even though polydactyly was not observed.
Genetic analysis
Genetic analysis was performed after obtaining written informed consent from both the patients and their parents. This study was approved by the Institutional Review Board of Kobe University School of Medicine (IRB approval number 86 and 301). Genomic DNA was extracted from peripheral blood leukocytes of the patients using the Quick Gene Mini 80 system (Wako Pure Chemical Industries, Ltd., Tokyo, Japan). Targeted sequencing using next-generation sequencing (NGS) was conducted for 159 genes (Online Resource 1) associated with congenital anomalies of the kidney, the urinary tract, and NPHP-related ciliopathies. NGS samples were prepared using the HaloPlex HS target enrichment system kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions for the MiSeq platform (Illumina, San Diego, CA, USA). The mutations detected by NGS were confirmed by standard Sanger direct sequencing using the 3130 Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA).
For total RNA extraction from leukocytes, blood, RiboPure RNA Purification Kit (Thermo Fisher Scientific), and RNA stabilization agent (RNAlater; Thermo Fisher Scientific) were used. Total RNA was reverse-transcribed into cDNA using EcoDry Premix (Double Primed; Takara Bio USA, Inc., Mountain View, CA, USA). We performed nested PCR using different pairs of SCLT1 primers: for exon 14 first forward primer: 5′-GTGACAATCCAAGAAGCCAACC-3′, and reverse: 5′-TGAACGCTCTGAAACCAGGA-3′, and second forward primer: 5′-AGGAGAAGCAAAAAGAAGAAGACA-3′, and reverse: 5′-TCTAGTTCTTCTTCCACTGCTTT-3′. The products were electrophoretically fractionated; the gel band was sliced out, and cDNA was extracted using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany). The cDNA was sequenced using a dye terminator cycle sequencing kit and an automatic DNA sequencer as mentioned above.
Pathogenicity predictions were performed in accordance with the American College of Medical Genetics (ACMG) guidelines [9]. Several websites, including CADD [10], PROVEAN [11], SIFT [12], PolyPhen-2 [13], and Mutation Taster [14], were also used to predict variant pathogenicity. We also determined GERP ++ [15] and phyloP [16] conservation scores. Human Splicing Finder (HSF) was used for the prediction of exon skipping [17].
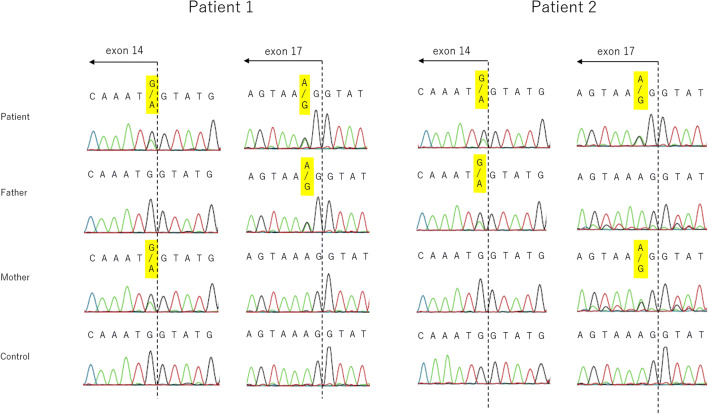
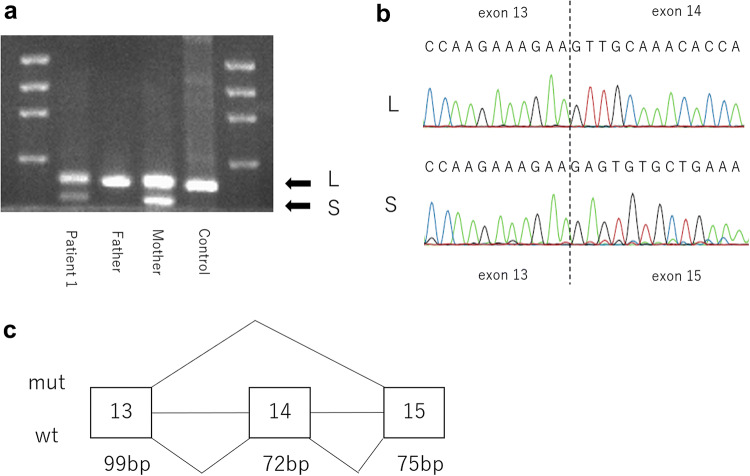
Both patient 1 and patient 2 displayed two identical compound heterozygous SCLT1 mutations: NM_144643.4: c.1218G > A, p.Met406Ile, and c.1631A > G, p.Lys544Arg (Fig. 2). We identified these variants in the parents (both mother and father) of both these patients. For the missense variants, the CADD score was 24.9 for c.1218G > A and 24.4 for c.1631A > G. PROVEAN was neutral and SIFT was deleterious for both the mutants, PP2 was benign for c.1218G > A and possibly damaging for c.1631A > G, and Mutation Taster showed it to be damaging in the case of both the variants (Table 1). The GERP ++ scores were 4.49 and 5.31, and Phylop scores were 3.984 and 5.589 for c.1218G > A and c.1631A > G, respectively. HSF predicted potential alteration in splicing for c.1218G > A, but did not predict any effect on splicing for c.1631A > G. According to ACMG guidelines, both the missense variants met uncertain significance criteria (PM2 and PP3). Because these results could not determine pathogenicity in the patients, we performed cDNA analysis for c.1218G > A. The variants were found to be c.1218G > A in exons 14 adjacent to the introns. We identified exon 14 skipping by cDNA analysis, and the skipping was found to result in in-frame deletion (72 bp) (Fig. 3).
Fig. 2.
Direct sequencing of SCLT1 mutations in the two families. Both patients had two identical mutations in exon 14 and 17. The mutation in exon 14 was derived from the mother in patient 1 and from the father in patient 2
Table 1.
The evaluation of two missense variants in the study
| CADD score | PROVEAN | SIFT | PolyPhen2 | MutationTaster | Human Splicing Finder | GERP ++ | phyloP | HGMD | ClinVar | gnomAD | dbSNP | HGVD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.1218G > A | 24.9 | − 1.33 | Deleterious | Benign | Disease causing | Potential alteration of splicing | 4.49 | 3.984 | – | – | – | – | – |
| c.1631A > G | 24.4 | − 1.28 | Deleterious | Possibly damaging | Disease causing | Probably no impact on splicing | 5.31 | 5.589 | – | – | 0.000836% | rs762215370 | 0.12% |
CADD combined annotation dependent depletion, GERP genomic evolutionary rate profiling, gnomAD genome aggregation database, HGMD the human gene mutation database, HGVD human genetic variation database, PROVEAN protein variation effect analyzer, SIFT sorting intolerant from tolerant
Fig. 3.
The splicing aberrations in SCLT1 at exons 14 in patient 1. Electrophoresis revealed two DNA bands (a), and exon 14 skipping of the shorter band (b). This results in a 72-bp deletion (c). L longer band, S shorter band
Discussion
BBS is a clinically recognizable disorder owing to its specific symptoms. The clinical diagnosis of BBS according to Beales et al. [18] is as follows: patients should display at least four features from the six primary features: rod-cone dystrophy (almost synonymous with RP), polydactyly, obesity, learning disabilities, hypogonadism in males, and renal anomalies. In the current study, patient 1 had RP, obesity, intellectual disability, and renal dysfunction, and patient 2 had RP, obesity, intellectual disability, renal dysfunction, and hypogonadism. Therefore, we clinically diagnosed them with BBS. In patient 1, renal abnormality was compatible with NPHP based on inspection and clinical findings. In patient 2, although the detailed clinical course before ESRD was unknown, the renal dysfunction in the patient might have been due to NPHP. The clinical symptoms of BBS and its related ciliopathies, including SLSN, Joubert syndrome, Meckel syndrome, and Alström syndrome, show an overlap [19]. These diseases are comprehensively called NPHP-related ciliopathies (NPHP-RC). These diseases are likely to result in an ESRD due to NPHP. The onset time at which the renal replacement therapy is required in NPHP patients may vary depending on the causative genes; in patients with SCLT1 mutation, it is likely to be in the second decade. We analyzed most of the NPHP-RC genes in the patients by NGS (Online Resource 1), and were able to identify SCLT1 as the gene responsible for BBS. Comprehensive analysis using NGS is a useful strategy for patients with any NPHP-RC.
Patients with SCLT1 mutations are extremely rare. The first patient with SCLT1 aberration reported by Adly et al. [5] presented a severe midline cleft lip, microcephaly, bilateral coloboma, abnormal genitalia, and congenital heart disease, but polydactyly was not observed; he was diagnosed with OFD9. A Japanese girl reported by Katagiri et al. [6] showed RP and NPHP, resulting in the diagnosis of SLSN. In addition, she showed mild intellectual disability and obesity, but no polydactyly was observed. The clinical manifestations of our patients and those of the patient reported by Katagiri et al. [6] are similar. Polydactyly is seen in most patients with BBS, except in patients with BBS due to SDCCAG8 aberrations. Although the SCLT1 knockout mice exhibited polydactyly [20], no patients with polydactyly and SCLT1 aberrations has been reported, thus far. Therefore, humans with SCLT1 aberrations may have normal digits.
Renal diseases are common in BBS patients. Katagiri et al. reported that the patient showed severe renal disorder during childhood [6]. The patient reported by Adly et al. died at the age of 3 months; therefore, details regarding renal diseases of this patient are unknown [5]. Patients with SCLT1 aberrations may have exacerbated renal dysfunction. Maddirevula et al. recently reported two siblings with congenital pan hypopituitarism [21]. However, they did not show any syndromic features, including developmental delay. To establish the clinical manifestations of SCLT1 aberrations, further studies are required.
Although the pathogenicity of the two missense variants described in this study remains controversial, we believe that these variants are pathogenic. The c.1218G > A mutation is a novel variant and was predicted to be pathogenic in some in silico analyses (SIFT, Mutation Taster) but not in others (PROVEAN, PP2). This variant was located within exon 14, near intron 14. It was predicted using HSF that this variant would affect exon skipping, and we indeed identified a splicing aberration in exon 14 skipping by cDNA analysis (Fig. 3). This exon skipping was the same as that in the case of the patient reported by Katagiri et al. [6]. The c.1631A > G mutation was registered in dbSNP as rs762215370 and its allele frequency in the Japanese population was reported to be 0.12%. This variant was located within exon 17, near intron 17; therefore, it might affect exon skipping. Although HSF predicted it to have no effect on splicing, Katagiri et al. have reported that this variant might induce the skipping of SCLT1 exon 17 or exon 17 and 18 and could induce a 6-bp deletion or could have no effect on exon skipping or deletion. Except for the 6-bp deletion, these skipping events might occur in healthy individuals [6]. In addition, the existence of three patients (two patients reported by us and one by Katagiri et al.) with similar phenotypes and the same genotype strongly suggests that these mutations are pathological.
The reason behind the occurrence of identical mutations (c.1218G > A and c.1631A > G) in the two unrelated patients in this study is currently unknown. To the best of our knowledge, the two families were unrelated. The c.1218G > A mutation has not been registered in the gnomAD browser (https://gnomad.broadinstitute.org/), Integrative Japanese Genome Variation Database (iJGVD, https://ijgvd.megabank.tohoku.ac.jp/), or Human Genetic Variation Database (HGVD, https://www.hgvd.genome.med.kyoto-u.ac.jp/); however, c.1631A > G showed very low allele frequency in gnomAD and HGVD (0.00082% and 0.12%, respectively). Similar to that in the patients in this study, the patient reported by Katagiri et al. [6] also possessed the c.1631A > G mutation. Further studies are needed to clarify the genetic distribution of NPHP-RC in the Japanese population.
In conclusion, we report about two unrelated BBS patients with identical SCLT1 aberrations. Based on our investigations, we suggest that SCLT1 is the gene responsible for causing BBS without polydactyly. Although our study did not include whole exome or whole genome sequencing by NGS, the findings could contribute to a greater understanding of NPHP-RC patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank all the study participants and their families. We are profoundly grateful to Mrs. Yoshimi Nozu (Kobe University) for her excellent technical assistance. We would like to thank Editage (www.editage.jp) for English language editing. This work was supported by the Health Labor Sciences Research Grant for the Research on Measures for Intractable Diseases (H24-nanchi-ippan-041 to K.I.; H29-nanchi-ippan-039 to N.M.), and the Japan Society for the Promotion of Science (KAKENHI Grant number JP15K09261 and 18K08243 to N.M.).
Compliance with ethical standards
Conflict of interest
Kazumoto Iijima received funding support from Daiichi Sankyo Co., Ltd., Zenyaku Kogyo Co., Ltd. and Astellas Pharma, Inc.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee at which the studies were conducted (IRB approval number 86 and 301) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Forsythe E, Beales PL. Bardet-Biedl Syndrome. In: Adam MP, Ardinger HH, Pagon RA, editors. GeneReviews®. Seattle: University of Washington; 2003. pp. 1993–2019. [PubMed] [Google Scholar]
- 2.Forsythe E, Kenny J, Bacchelli C, Beales PL. Managing Bardet-Biedl syndrome-now and in the future. Front Pediatr. 2018;6:23. doi: 10.3389/fped.2018.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feather SA, Woolf AS, Donnai D, Malcolm S, Winter RM. The oral-facial-digital syndrome type 1 (OFD1), a cause of polycystic kidney disease and associated malformations, maps to Xp22.2-Xp22.3. Hum Mol Genet. 1997;6(7):1163–1167. doi: 10.1093/hmg/6.7.1163. [DOI] [PubMed] [Google Scholar]
- 4.Sakakibara N, Morisada N, Nozu K, Nagatani K, Ohta T, Shimizu J, et al. Clinical spectrum of male patients with OFD1 mutations. J Hum Genet. 2019;64(1):3–9. doi: 10.1038/s10038-018-0532-x. [DOI] [PubMed] [Google Scholar]
- 5.Adly N, Alhashem A, Ammari A, Alkuraya FS. Ciliary genes TBC1D32/C6orf170 and SCLT1 are mutated in patients with OFD type IX. Hum Mutat. 2014;35(1):36–40. doi: 10.1002/humu.22477. [DOI] [PubMed] [Google Scholar]
- 6.Katagiri S, Hayashi T, Yoshitake K, Murai N, Matsui Z, Kubo H, et al. Compound heterozygous splice site variants in the SCLT1 gene highlight an additional candidate locus for Senior-Løken syndrome. Sci Rep. 2018;8(1):16733. doi: 10.1038/s41598-018-35152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Cummins TR, Tyrrell L, Black JA, Waxman SG, Dib-Hajj SD. CAP-1A is a novel linker that binds clathrin and the voltage-gated sodium channel Na(v)1.8. Mol Cell Neurosci. 2005;28(4):636–649. doi: 10.1016/j.mcn.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, et al. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013;27(2):163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, On behalf of the ACMG Laboratory Quality Assurance Committee et al. Standards and guidelines for the interpretation of sequence variants: a Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 13.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. Mutation Taster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 15.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet. 1999;36(6):437–446. [PMC free article] [PubMed] [Google Scholar]
- 19.Braun DA, Hildebrandt F. Ciliopathies. Cold Spring Harb Perspect Biol. 2017; 9(3). [DOI] [PMC free article] [PubMed]
- 20.Li J, Lu D, Liu H, Williams BO, Overbeek PA, Lee B, et al. Sclt1 deficiency causes cystic kidney by activating ERK and STAT3 signaling. Hum Mol Genet. 2017;26(15):2949–2960. doi: 10.1093/hmg/ddx183. [DOI] [PubMed] [Google Scholar]
- 21.Maddirevula S, Alzahrani F, Al-Owain M, Al Muhaizea MA, Kayyali HR, AlHashem A, et al. Autozygome and high throughput confirmation of disease genes candidacy. Genet Med. 2019;21(3):736–742. doi: 10.1038/s41436-018-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.