Abstract
The role of our gut microbiota in health and disease is largely attributed to the collective metabolic activities of the inhabitant microbes. A system-level framework of the microbial community structure, mediated through metabolite transport, would provide important insights into the complex microbe-microbe and host-microbe chemical interactions. This framework, if adaptable to both mouse and human systems, would be useful for mechanistic interpretations of the vast amounts of experimental data from gut microbiomes in murine animal models, whether humanized or not. Here, we constructed a literature-curated, interspecies network of the mammalian gut microbiota for mouse and human hosts, called NJC19. This network is an extensive data resource, encompassing 838 microbial species (766 bacteria, 53 archaea, and 19 eukaryotes) and 6 host cell types, interacting through 8,224 small-molecule transport and macromolecule degradation events. Moreover, we compiled 912 negative associations between organisms and metabolic compounds that are not transportable or degradable by those organisms. Our network may facilitate experimental and computational endeavors for the mechanistic investigations of host-associated microbial communities.
Subject terms: Biochemical networks, Microbial ecology, Literature mining, Microbiome
| Measurement(s) | metabolic process • transport • macromolecule catabolic process • gut microbiome measurement |
| Technology Type(s) | digital curation • Phylogenetic Analysis |
| Factor Type(s) | Species of microorganisms • Type of host cells |
| Sample Characteristic - Organism | Homo sapiens • Mus musculus • Bacteria • Archaea • Eukaryota |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.12336164
Background & Summary
The mammalian intestinal tract is colonized by various microorganisms, called the gut microbiota or microbiome1–3. Recent advances in metagenomics have revealed that alterations in the human gut microbiota are implicated in a number of disorders, such as obesity, inflammatory bowel disease, colorectal cancer, and diabetes4–7. At the center of the gut microbiota functions are the various interactions between microbes and their interplay with the host environment2,6,8. Microbes degrade diet-derived and host-derived chemical substances, and release the degradation products to other members of the community. The microbial transport of nutrients and metabolic byproducts gives rise to competition for resources and cooperative relationships via metabolic cross-feeding2,8. The metabolites secreted by the microbes are absorbed by host tissues, and translate into beneficial or detrimental mediators of host physiology6,9. As a result, such microbe-microbe and microbe-host interactions form a complex ecological network in the gut environment10.
In the microbiome research, one common practice for reconstructing metabolite-mediated microbial networks is to combine the entire biochemical reactions inferred from annotated metagenomes11,12. This method, by its nature, does not delineate biochemical reactions to the species from which they originate, making it difficult to elucidate interspecies interactions. On the other hand, there exist previous works on the modeling of diverse interspecies interactions explicitly mediated by metabolites that are transported (imported or exported) by individual microbial species13,14. Yet, these works are based on error-prone, automated identification protocols for transportable metabolites, which are possibly inaccurate to some degrees. There are ongoing computational efforts towards biologically realistic microbial interactions, by using manually curated, constraint-based metabolic models or relatively simple kinetic models15,16. Nevertheless, most of these models are far from the scale of diversity seen in the gut community, which typically comprises hundreds of different microbial species. Notably, this scale of microbial diversity has been recently captured by constraint-based metabolic models with semi-automatic model reconstructions17, but they still exhibit limited biological accuracies18–20.
Recently, we have constructed an extensive, literature-curated interspecies metabolic interaction network of the human gut microbiota, NJS16, which represents another system-level framework for gut microbiota analysis10. This network is primarily based on biological knowledge and experimental evidence documented in the literature. The network NJS16 encompasses >4,000 small-molecule transport and macromolecule degradation events of >500 bacterial and archaeal species and 3 human cell types. Although NJS16 is useful to explore the microbial community inside the human gut, mechanistic studies in the microbiome research field have been mainly conducted on animal models, rather than on human subjects, due to the technical and regulatory limitations on human experimentation21,22. Regarding animal models, physiological, anatomical, and genetic similarities between humans and mice, as well as massively accumulated knowledge of mouse genetics, have facilitated the use of murine models, to elucidate causality and mechanisms of host-microbiota interactions4,7,23. In this regard, a phylogenetic extension of NJS16 to murine gut microbes would be useful for the system-level mechanistic exploration of gut microbiota functions using murine animal models.
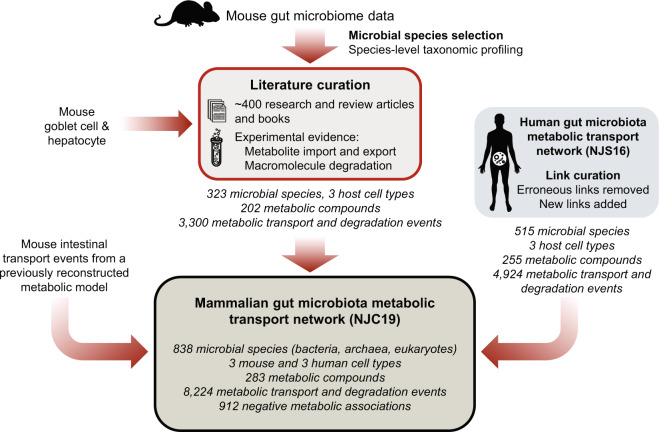
Here, we present a literature-curated, interspecies metabolic interaction network of the microbiota associated with the mouse and human gut, NJC19. To our knowledge, NJC19 represents the largest ever, literature-based network data resource for the mammalian gut microbiota, as a compilation of information from 769 research and review articles and textbooks (Fig. 1). This network is an advancement from our previous network, NJS16, which is limited to the human gut microbiota10. Specifically, NJC19 greatly expands the diversity of microbial species and host cells to those relevant to the mouse gut environments, and even covers a certain range of eukaryotic microbes that were completely missing in the predecessor NJS16. Therefore, NJC19 serves as a global network template, adaptable to the gut microbiota of either a mouse, human, or humanized mouse. Moreover, not only does NJC19 incorporate metabolite transport and macromolecule degradation events of the microbiota, but it also provides literature-annotated, negative information of which metabolic compounds are not able to be transported or degraded by the organisms. Such negative information would be useful to curate computational microbial models, such as constraint-based metabolic models, which can include false-positive transport reactions from automatic genome annotations.
Fig. 1.
Construction of the mammalian (mouse and human) gut microbiota interaction network NJC19. The flow chart of the network construction is presented. NJC19 is mainly built upon literature-curated, metabolic information of the mouse gut microbiota, combined with the revised version of NJS16 that represents the human gut microbiota interaction network.
We expect our network NJC19 to be a useful template for the mechanistic interpretation of various microbiome data from murine and human experiments.
Methods
Collection of mouse microbiome data and taxonomic identification for NJC19 construction
We aimed to construct a large-scale network for the mammalian gut microbiota that comprises microbial species populating the mouse and human gut. Figure 1 provides the overview of our network construction procedure. To construct the network, we started by collecting raw shotgun metagenome and 16S rRNA gene sequence data from fecal and cecal samples of laboratory and wild-caught mice from seven different studies3,24–29, as detailed in Online-only Table 1. It is noteworthy that the inclusion of the data from wild-caught mice3 allows the coverage of diverse microbial communities associated with natural murine lifestyles. The species-level taxonomic profiling of the shotgun metagenome sequence data was performed using the MetaPhlAn v2.0 software, which utilizes clade-specific marker sequences to identify microbial taxa30. When using MetaPhlAn v2.0, the “sensitive-local” mapping option was selected. For the taxonomic profiling of 16S rRNA gene sequence data, we used the open-reference OTU picking workflow of QIIME v1.8.0 with Greengenes v13_8_pp reference files31, and then selected species-level microbial taxa from the results. Among all species detected from the metagenome and 16S rRNA gene sequence data, priority for the collection of metabolic information (see below) was given to species absent in our previous network, NJS1610. In the case of the metagenome sequence data, the number of the detected species was rather excessive for our further processing; therefore, among those species, we only considered the species inhabiting ≥90% of the metagenome samples (with the relative abundance ≥0.001%) in each study. We found that the genera of these selected species account for the vast majority [89.6 ± 4.3% (avg. ± s.d.)] of the total microbial abundances in the metagenome samples. In addition, we manually considered some relevant species, such as Citrobacter rodentium32 (Online-only Table 2–3).
Online-only Table 1.
Sources of mouse microbiome samples for microbial species identification for NJC19 construction.
| - Shotgun metagenome sequence data | ||||
|---|---|---|---|---|
| Reference | Mouse strains | Sample source | No. of samples | Data source |
| Wang J, Linnenbrink M, Künzel S, Fernandes R, Nadeau M-J, Rosenstiel P et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci U S A. 2014;111:E2703–10. | wild-caught mice | stool | 26 | Kindly provided by the authors' group |
| Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV et al. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature. 2014;514:638–41. | (B6.129X1-Fut2tm1Sdo/J) mice backcrossed greater than 7 generations to BALB/c | stool | 12 | GSE60874 |
| Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–81. | C57BL/6 (TLR5−/− and wild-type) | cecum | 10 | mgp6393 |
| Langille MG, Meehan CJ, Koenig JE, Dhanani AS, Rose RA, Howlett SE et al. Microbial shifts in the aging mouse gut. Microbiome. 2014;2:50. | C57BL/6 | stool | 21 | mgp3907 |
| Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–17. | BALB/c T-bet-/- Rag2-/- | stool | 6 | mgp6698 |
| - 16S rRNA gene sequence data | ||||
| Reference | Mouse strains | Sample source | No. of samples | Data source |
| Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proceedings of the National Academy of Sciences. 2010;107:18933–8. | C57BL/6 and ICR-derived HR intercross line | stool | 645 | UNL Core for Applied Genomics and Ecology |
| Wang J, Linnenbrink M, Künzel S, Fernandes R, Nadeau M-J, Rosenstiel P et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci U S A. 2014;111:E2703–10. | wild-caught mice | stool | 66 | ERP004395 |
| Linnenbrink M, Wang J, Hardouin EA, Künzel S, Metzler D, Baines JF. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol. 2013;22:1904–16. | house mice | cecum | 201 | ERP001970 |
| Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV et al. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature. 2014;514:638–41. | (B6.129X1-Fut2tm1Sdo/J) mice backcrossed greater than 7 generations to BALB/c. | stool | 14 | mgp10494 |
| Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–17. | BALB/c T-bet−/−, Rag2−/− | stool | 154 | mgp6698 |
Online-only Table 2.
List of literature sources of metabolic information used for NJC19 construction.
| Ref. # | First author | Year | Title |
|---|---|---|---|
| 1 | Fontes | 2010 | Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates |
| 2 | Lynd | 2002 | Microbial cellulose utilization: fundamentals and biotechnology |
| 3 | Patel | 1980 | Isolation and Characterization of an Anaerobic, Cellulolytic Microorganism, Acetivibrio cellulolyticus gen. nov., sp. nov. |
| 4 | Illeghems | 2013 | Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem |
| 5 | Oren | 2002 | Halophilic Microorganisms and their Environments (Chapter 4) |
| 6 | Lazarev | 2011 | Complete Genome and Proteome of Acholeplasma laidlawii |
| 7 | Kay | 2001 | Recurrent achromobacter piechaudii bacteremia in a patient with hematological malignancy |
| 8 | Kiredjian | 1986 | Alcaligenes piechaudii, a New Species from Human Clinical Specimens and the Environment |
| 9 | Reverdy | 1984 | Nosocomial colonization and infection by Achromobacter xylosoxidans |
| 10 | Rogosa | 1969 | Acidaminococcus gen. n., Acidaminococcus fermentans sp. n., Anaerobic Gram-negative Diplococci Using Amino Acids as the Sole Energy Source for Growth |
| 11 | Chang | 2010 | Complete genome sequence of Acidaminococcus fermentans type strain (VR4T) |
| 12 | Eschenlauer | 2002 | Ammonia Production by Ruminal Microorganisms and Enumeration, Isolation, and Characterization of Bacteria Capable of Growth on Peptides and Amino Acids from the Sheep Rumen |
| 13 | Jumas-Bilak | 2007 | Acidaminococcus intestini sp. nov., isolated from human clinical samples |
| 14 | Clark | 1995 | Acidimicrobium ferrooxidans gen. nov., sp. nov.: mixed-culture ferrous iron oxidation with Sulfobacillus species |
| 15 | Kuseel | 1999 | Microbial Reduction of Fe(III) in Acidic Sediments: Isolation of Acidiphilium cryptum JF-5 Capable of Coupling the Reduction of Fe(III) to the Oxidation of Glucose |
| 16 | Zhou | 2007 | Isolation of a strain of Acidithiobacillus caldus and its role in bioleaching of chalcopyrite |
| 17 | Ko | 2013 | The role of Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans in arsenic bioleaching from soil |
| 18 | Willems | 1992 | Transfer of Several Phytopathogenic Pseudomonas Species to Acidovorax as Acidovorax avenae subsp. avenae subsp. nov., comb. nov., Acidovorax avenae subsp. citrulli, Acidovorax avenae subsp. cattleyae, and Acidovorax konjaci |
| 19 | Guettler | 1999 | Actinobacillus succinogenes sp. nov., a novel succinic-acid producing strain from the bovine rumen |
| 20 | Bruhlmann | 1994 | Pectinolytic Enzymes from Actinomycetes for the Degumming of Ramie Bast Fibers |
| 21 | Bruns | 2003 | Aeromicrobium marinum sp. nov., an abundant pelagic bacterium isolated from the German Wadden Sea |
| 22 | Abbott | 2002 | The Genus Aeromonas: Biochemical Characteristics, Atypical Reactions, and Phenotypic Identification Schemes |
| 23 | Siebers | 2005 | Unusual pathways and enzymes of central carbohydrate metabolism in archaea |
| 24 | Stacy | 2014 | Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection |
| 25 | Todar | - | Todars Online Textbook of Bacteriology |
| 26 | Dagorn | 2013 | Effect of GABA, a Bacterial Metabolite, on Pseudomonas fluorescens Surface Properties and Cytotoxicity |
| 27 | Derrien | 2004 | Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacteria |
| 28 | Derrien | 2010 | Mucin-bacterial interactions in the human oral cavity and digestive tract |
| 29 | Killer | 2011 | Fermentation of mucin by bifidobacteria from rectal samples of humans and rectal and intestinal samples of animals |
| 30 | Tailford | 2015 | Mucin glycan foraging in the human gut microbiome |
| 31 | Ze | 2013 | Some are more equal than others: the role of "keystone" species in the degradation of recalcitrant substrates |
| 32 | Rautio | 2003 | Reclassification of Bacteroides putredinis (Weinberg et al., 1937) in a New Genus Alistipes gen. nov., as Alistipes putredinis comb. nov., and Description of Alistipes finegoldii sp. nov., from Human Sources |
| 33 | Song | 2006 | Alistipes onderdonkii sp. nov. and Alistipes shahii sp. nov., of human origin |
| 34 | Sieber | 2012 | Genomic insights into syntrophy: The paradigm for anaerobic metabolic cooperation |
| 35 | Ezaki | 2001 | Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus |
| 36 | Falony | 2006 | Cross-feeding between bifidobacterium longum BB536 and acetate-convertin, butyrate-producing colon bacteria during growth on oligofructose |
| 37 | Flint | 2007 | Interactions and competition within the microbial community of the human colon: links between diet and health |
| 38 | Louis | 2009 | Diversity, metabolism and microbial ecology of butyrate-producing bacteria from large intestine |
| 39 | Macfarlane | 2012 | Bacteria, colonic fermentation, and gastrointestinal health |
| 40 | Pryde | 2002 | The microbiology of butyrate formation in the human colon |
| 41 | Sato | 2008 | Isolation of lactate-utilizing butyrate-producing bacteria from human feces andin vivo administration ofAnaerostipes caccae strain L2 and galacto-oligosaccharides in a rat model |
| 42 | Scott | 2013 | Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro |
| 43 | Belenguer | 2006 | Two Routes of Metabolic Cross-Feeding between Bifidobacterium adolescentis and Butyrate-Producing Anaerobes from the Human Gut |
| 44 | Belenguer | 2007 | Impact of pH on Lactate Formation and Utilization by Human Fecal Microbial Communities |
| 45 | Charrier | 2006 | A novel class of coa-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria |
| 46 | Duncan | 2004 | Lactate-Utilizing Bacteria, Isolated from Human Feces, That Produce Butyrate as a Major Fermentation Product |
| 47 | Lawson | 2004 | Anaerotruncus colihominis gen. nov., sp. nov., from human faeces |
| 48 | Drake | 2008 | Old acetogens, new light |
| 49 | Haba | 2000 | Isolation of lipase-secreting bacteria by deploying used frying oil as selective substrate |
| 50 | Shields | 2013 | Efficacy of a Marine Bacterial Nuclease against Biofilm Forming Microorganisms Isolated from Chronic Rhinosinusitis |
| 51 | Willerding | 2011 | Lipase Activity among Bacteria Isolated from Amazonian Soils |
| 52 | Balestrazzi | 2007 | Nuclease-producing bacteria in soil cultivated with herbicide resistant transgenic white poplars |
| 53 | Bentley | 1982 | Biosynthesis of Vitamin K (Menaquinone) in Bacteria |
| 54 | LeBlanc | 2011 | B-group vitamin production by lactic acid bacteria - Current knowledge and potential applications |
| 55 | Leviton | 1952 | Microbiological Synthesis of Vitamin B12 by Propionic Acid Bacteria |
| 56 | Martens | 2002 | Microbial production of vitamin B12 |
| 57 | Rodionov | 2003 | Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes |
| 58 | Degrassi | 1997 | Purification and Characterization of an Acetyl Xylan Esterase from Bacillus pumilus |
| 59 | Giannella | 1971 | Vitamin B12 uptake by intestinal microorganisms: mechanism and relevance to syndromies of intestinal bacterial growth |
| 60 | Saxena | 2003 | Purification strategies for microbial lipases |
| 61 | Heinken | 2013 | Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut |
| 62 | Hermann | 2003 | Industrial production of amino acids by coryneform bacteria |
| 63 | LeBlanc | 2013 | Bacteria as vitamin suppliers to their host: a gut microbiota perspective |
| 64 | Meyers | 1996 | Lipase production by lactic acid bacteria and activity on butter oil |
| 65 | Rodionov | 2009 | A novel class of modular transporters for vitamins in prokaryotes |
| 66 | Shimizu | 2008 | Vitamins and Related Compounds: Microbial Production, in Biotechnology: Special Processes |
| 67 | Takeno | 2007 | Anaerobic growth and potential for amino acid production by nitrate respiration in Corynebacterium glutamicum |
| 68 | Thompson | 2012 | Metabolism of sugars by genetically diverse species of oral Leptotrichia |
| 69 | Berstenhorst | 2009 | Vitamins and Vitamin-like Compounds: Microbial Production |
| 70 | Burke | 1982 | Bacillus subtilis Extracellular Nuclease Production Associated with the spoOH Sporulation Locus |
| 71 | Burkholder | 1942 | Synthesis of vitamins by intestinal bacteria |
| 72 | Koropatkin | 2012 | How glycan metabolism shapes the human gut microbiota |
| 73 | Macfarlane | 2005 | Colonization of Mucin by Human Intestinal Bacteria and Establishment of Biofilm Communities in a Two-Stage Continuous Culture System |
| 74 | McNulty | 2011 | The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins |
| 75 | Sonnenburg | 2010 | Specificity of Polysaccharide Use in Intestinal Bacteroides Species Determines Diet-Induced Microbiota Alterations |
| 76 | Cuskin | 2015 | Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism |
| 77 | Chassard | 2010 | The cellulose-degrading microbial community of the human gut varies according to the presence or absence of methanogens |
| 78 | Flint | 2012 | Microbial degradation of complex carbohydrates in the gut |
| 79 | Flint | 2008 | Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis |
| 80 | Shah | 1989 | Proposal To Restrict the Genus Bacteroides (Castellani and Chalmers) to Bacteroides fragilis and Closely Related Species |
| 81 | Krieg | 2010 | Bergey's Manual of Systematic Bacteriology, Volume 4: The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes |
| 82 | Deguchi | 1992 | Nutritional Requirements in Multiple Auxotrophic Lactic Acid Bacteria: Genetic Lesions Affecting Amino Acid Biosynthetic Pathways in Lactococcus lactis, Enterococcus faecium, and Pediococcus acidilactici |
| 83 | Nishiyama | 2009 | Bacteroides graminisolvens sp. nov., a xylanolytic anaerobe isolated from a methanogenic reactor treating cattle waste |
| 84 | Holdeman | 1974 | New Genus, Coprococcus, Twelve New Species, and Emended Descriptions of Four Previously Described Species of Bacteria from Human Feces |
| 85 | Macy | 1979 | The Biology of Gastrointestinal Bacteroides |
| 86 | Salyers | 1977 | Fermentation of Mucin and Plant Polysaccharides by Strains of Bacteroides from the Human Colon |
| 87 | Dodd | 2010 | Transcriptomic Analyses of Xylan Degradation by Prevotella bryantii and Insights into Energy Acquisition by Xylanolytic Bacteroidetes |
| 88 | Macfarlane | 1992 | Synthesis and Release of Proteases by Bacteroides fragilis |
| 89 | Macfarlane | 1991 | Formation of glycoprotein degrading enzymes by Bacteroides fragilis |
| 90 | Macfarlane | 1986 | Protein Degradation by Human Intestinal Bacteria |
| 91 | McBain | 1998 | Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites |
| 92 | Ridlon | 2005 | Bile salt biotransformations by human intestinal bacteria |
| 93 | Schink | 1987 | Pathway of propionate formation from ethanol in Pelobacter propionicus |
| 94 | Fukiya | 2009 | Conversion of cholic acid and chenodeoxycholic acid into their 7-oxo derivatives by Bacteroides intestinalis AM-1 isolated from human feces |
| 95 | Hatamoto | 2014 | Bacteroides luti sp. nov., an anaerobic, cellulolytic and xylanolytic bacterium isolated from methanogenic sludge |
| 96 | Martens | 2011 | Recognition and Degradation of Plant Cell Wall Polysaccharides by Two Human Gut Symbionts |
| 97 | Goodman | 2009 | Identifying Genetic Determinants Needed to Establish a Human Gut Symbiont in Its Habitat |
| 98 | Smith | 1996 | Studies on Amine Production in the Human Colon: Enumeration of Amine forming Bacteria and Physiological Effects of Carbohydrate and pH |
| 99 | Rakoff-Nahoum | 2014 | An Ecological Network of Polysaccharide Utilization among Human Intestinal Symbionts |
| 100 | Jenkins | 1982 | Differences in susceptibilities of species of the Bacteroides fragilis group to several beta-lactam antibiotics: indole production as an indicator of resistance. |
| 101 | Endo | 2012 | Comparison of Fructooligosaccharide Utilization by Lactobacillus and Bacteroides Species |
| 102 | Cato | 1976 | Reinstatement of Species Rank for Bacteroides fragilis, B. ovatus, B. distasonis, B. thetaiotaomicron, and B. vulgatus: Designation of Neotype Strains for Bacteroides fragilis (Veillon and Zuber) Castellani and Chalmers and Bacteroides thetaiotaomicron (Distaso) Castellani and Chalmers |
| 103 | Macfarlane | 2003 | Regulation of short-chain fatty acid production |
| 104 | Cooke | 2006 | Newly identified vitamin K-producing bacteria isolated from the neonatal faecal flora |
| 105 | Johnson | 1986 | Bacteroides caccae sp. nov., Bacteroides merdae sp. nov., and Bacteroides stercoris sp. nov. Isolated from Human Feces |
| 106 | Mahowald | 2009 | Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla |
| 107 | Musso | 2011 | Interaction between gut microbiota and host metabolism predisposing to obesity and diabetes |
| 108 | Payne | 2012 | Gut microbial adaptation to dietary consumption of fructose, artificial sweeteners and sugar alcohols |
| 109 | Rey | 2013 | Metabolic niche of a prominent sufate-reducing human gut bacterium |
| 110 | Rey | 2010 | Dissecting the in vivo metabolic potential of two human gut acetogens |
| 111 | Samuel | 2006 | A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism |
| 112 | Samuel | 2007 | Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut |
| 113 | Shoaie | 2013 | Understanding the interactions between bacteria in the human gut through metabolic modeling |
| 114 | Smith | 1998 | Enumeration of amino acid fermenting bacteria in the human large intestine: effects of pH and starch on peptide metabolsim and dissimilation of amino acids |
| 115 | Sonnenburg | 2006 | A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism |
| 116 | Sonnenburg | 2005 | Glycan Foraging in Vivo by an Intestine-Adapted Bacterial Symbiont |
| 117 | Ze | 2012 | Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon |
| 118 | Backhed | 2005 | Host-bacterial mutualism in the human intestine |
| 119 | Blaut | 2013 | Ecology and physiology of the intestinal tract |
| 120 | Chassard | 2008 | Bacteroides xylanisolvens sp. nov., a xylan-degrading bacterium isolated from human faeces |
| 121 | Degnan | 2014 | Human Gut Microbes Use Multiple Transporters to Distinguish Vitamin B12 Analogs and Compete in the Gut |
| 122 | Fischbach | 2011 | Eating for two: How metabolism establishes interspecies interactions in the gut |
| 123 | Gibson | 2004 | Dietary modulation of the human colonic microbiota: updating the concept of prebiotics |
| 124 | Kayahara | 1994 | Δ22-β-Muricholic acid in monoassociated rats and conventional ratsacid in monoassociated rats and conventional rats |
| 125 | Conly | 1993 | The absorption and bioactivity of bacterially synthesized menaquinones |
| 126 | Pokusaeva | 2011 | Cellodextrin Utilization by Bifidobacterium breve UCC2003 |
| 127 | Pompei | 2007 | Folate production by bifidobacteria as a potential probiotic property p-aminobenzoic acid? |
| 128 | Ramirez-Farias | 2009 | Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii |
| 129 | Rossi | 2011 | Folate production by probiotic bacteria |
| 130 | Rossi | 2005 | Fermentation of Fructooligosaccharides and Inulin by Bifidobacteria: a Comparative Study of Pure and Fecal Cultures |
| 131 | Rossi | 2010 | Bifidobacteria: Genomics and Molecular Aspects (Chapter 6. Probiotic properties of bifidobacteria) |
| 132 | Salyers | 1977 | Fermentation of Mucins and Plant Polysaccharides by Anaerobic Bacteria from the Human Colon |
| 133 | Tanaka | 1999 | Screening of lactic acid bacteria for bile salt hydrolase activity |
| 134 | Vernazza | 2005 | Carbohydrate preference, acid tolerance and bile tolerance in five strains of Bifidobacterium |
| 135 | Aachary | 2011 | Xylooligosaccharides (XOS) as an Emerging Prebiotic: Microbial Synthesis, Utilization, Structural Characterization, Bioactive Properties, and Applications |
| 136 | Barrett | 2012 | gamma-Aminobutyric acid production by culturable bacteria from the human intestine |
| 137 | Crociani | 1994 | Degradation of complex carbohydrates by Bifidobacterium spp. |
| 138 | Hojo | 2007 | Reduction of vitamin K concentration by salivary Bifidobacterium strains and their possible nutritional competition with Porphyromonas gingivalis |
| 139 | Kaplan | 2000 | Fermentation of Fructooligosaccharides by Lactic Acid Bacteria and Bifidobacteria |
| 140 | Wilson | 2004 | Microbial inhabitants of humans (Table 9.9) |
| 141 | Corfield | 1992 | Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria |
| 142 | Hoskins | 1981 | Mucin degradation in human colon ecosystems |
| 143 | Katayama | 2005 | Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins |
| 144 | Peterson | 1945 | Relation of bacteria to vitamins and other growth factors |
| 145 | Ruas-Madiedo | 2008 | Mucin degradation by bifidobacterium strains isolated from the human intestinal microbiota |
| 146 | Menard | 2004 | Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport |
| 147 | Pokusaeva | 2010 | Ribose utilization by the human commensal Bifidobacterium breve UCC2003 |
| 148 | Scott | 2011 | Substrate-driven gene expression in Roseburia inulinivorans: Importance of inducible enzymes in the utilization of inulin |
| 149 | Degnan | 1995 | Arabinogalactan utilization in continuous cultures of bifidobacterium longum: Effect of co-culture with bacteroides thetaiotamicrobon |
| 150 | Kamra | 2005 | Rumen microbial ecosystem |
| 151 | Sela | 2008 | The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome |
| 152 | Vitali | 2010 | Impact of a synbiotic food on the gut microbial ecology and metabolic profiles |
| 153 | Wang | 2008 | Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine |
| 154 | Barcenilla | 2000 | Phylogenetic Relationships of Butyrate-Producing Bacteria from the Human Gut |
| 155 | Li | 2008 | Symbiotic gut microbes modulate human metabolic phenotypes |
| 156 | David | 2013 | Diet rapidly and reproducibly alters the human gut microbiome |
| 157 | Devkota | 2012 | Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice |
| 158 | Laue | 1997 | Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU |
| 159 | Nava | 2012 | Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human |
| 160 | Silva | 2008 | Hydrogen as an energy source for the human pathogen Bilophila wadsworthia |
| 161 | Liu | 2008 | Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces |
| 162 | Nakamura | 2010 | Mechanisms of microbial hydrogen disposal in the human colon and implications for health and disease |
| 163 | Bernalier | 1996 | Ruminococcus hydrogenotrophicus sp. nov., a new H2/CO2-utilizing acetogenic bacterium isolated from human feces |
| 164 | Chassard | 2006 | H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut |
| 165 | Jorda | 1982 | Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a Genus of Slow-Growing, Root Nodule Bacteria from Leguminous Plants |
| 166 | Douglas | 1998 | Nutritional Interactions in Insect-Microbial Symbioses: Aphids and Their Symbiotic Bacteria Buchnera |
| 167 | Perez-Brocal | 2006 | A small microbial genome: the end of a long symbiotic relationship? |
| 168 | Park | 2007 | Characterization of an Extracellular Lipase in Burkholderia sp. HY-10 Isolated from a Longicorn Beetle |
| 169 | Jaeger | 1994 | Bacterial lipases |
| 170 | Chistoserdova | 2009 | The expanding world of methylotrophic metabolism |
| 171 | Moon | 2008 | Reclassification of Clostridium proteoclasticum as Butyrivibrio proteoclasticus comb. nov., a butyrate-producing ruminal bacterium |
| 172 | Russell | 1985 | Fermentation of Cellodextrins by Cellulolytic and Noncellulolytic Rumen Bacteria |
| 173 | Wallace | 1997 | Metabolism of nitrogen-contraining compounds |
| 174 | Wallace | 1985 | The Role of Different Species of Bacteria in the Hydrolysis of Protein in the Rumen |
| 175 | Wallace | 1985 | Synergism between different species of proteolytic rumen bacteria |
| 176 | Cotta | 1986 | Proteolytic Activity of the Ruminal Bacterium Butyrivibrio fibrisolvens |
| 177 | Dehority | 1991 | Effects of microbial synergism on fibre digestion in the rumen |
| 178 | Duncan | 2002 | Acetate utilization and butyryl coenzyme A(CoA):acetate-CoA transferase in butyrate-producing bacteria from human large intestine |
| 179 | Harfoot | 1997 | Lipid metabolism in the rumen |
| 180 | Martin | 1994 | Nutrient transport by ruminal bacteria |
| 181 | Kelly | 2010 | The Glycobiome of the Rumen Bacterium Butyrivibrio proteoclasticus B316T Highlights Adaptation to a Polysaccharide-Rich Environment |
| 182 | Brunecky | 2013 | Revealing natures cellulase diversity: the digestion mechanism of caldicellulosirutpor bescii |
| 183 | Iino | 2008 | Calditerrivibrio nitroreducens gen. nov., sp. nov., a thermophilic, nitrate-reducing bacterium isolated from a terrestrial hot spring in Japan |
| 184 | Voordeckers | 2005 | Caminibacter mediatlanticus sp. nov., a thermophilic, chemolithoautotrophic, nitrate-ammonifying bacterium isolated from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge |
| 185 | Wexler | 1996 | Sutterella wadsworthensis gen. nov., sp. nov., Bile-Resistant Microaerophilic CampyZobacter gracilis-Like Clinical Isolates |
| 186 | Hanfrey | 2011 | Alternative spermidine biosynthetic route is critical for growth of Campylobacter jejuni and is the dominant polyamine pathway in human gut microbiota |
| 187 | McCutcheon | 2007 | Parallel genomic evolution and metabolic interdependence in an ancient symbiosis |
| 188 | Wu | 2006 | Metabolic Complementarity and Genomics of the Dual Bacterial Symbiosis of Sharpshooters |
| 189 | Kageyama | 2000 | Catenibacterium mitsuokai gen. nov., sp. nov., a Gram-positive anaerobic bacterium isolated from human faeces |
| 190 | Miller | 2011 | Complete genome sequence of the cellulose-degrading bacterium cellulosilyticum lentocellum |
| 191 | Garrity | 2005 | Bergey's Manual of Systematic Bacteriology, Volume 2 : The Proteobacteria |
| 192 | Mead | 1971 | The Amino Acid-fermenting Clostridia |
| 193 | Milne | 2011 | Metabolic network reconstruction and genome-scale model of butanol-producing strain clostridium beijerinckii ncimb 8052 |
| 194 | Salimi | 2010 | Genome-scale metabolic modeling of a clostridial co-culture for consolidated bioprocessing |
| 195 | Lee | 2005 | Evidence for the presence of an alternative glucose transport system in clostridium beijerinckii ncimb 8052 and the solvent-hyperproducing mutant ba101 |
| 196 | Chen | 1999 | Effect of acetate on molecular and physiological aspects of clostridium beijerinckii ncimb 8052 solvent production and strain degeneration |
| 197 | McSweeney | 1999 | Isolation and Characterization of Proteolytic Ruminal Bacteria from Sheep and Goats Fed the Tannin-Containing Shrub Legume Calliandra calothyrsus |
| 198 | Elsden | 1979 | Amino acid utilization patterns in clostridial taxonomy |
| 199 | Nakanishi | 2003 | Effects of high amylose maize starch and Clostridium butyricum on metabolism in colonic microbiota and formation of azoxymethane-induced aberrant crypt foci in the rat colon |
| 200 | Seedorf | 2008 | The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features |
| 201 | Aklujkar | 2012 | The genome of Pelobacter carbinolicus reveals surprising metabolic capabilities and physiological features |
| 202 | Payot | 1998 | Metabolism of cellobiose by Clostridium cellulolyticum growing in continuous culture: evidence for decreased nadh reoxidation as a factor limiting growth |
| 203 | McGarr | 2005 | Diet, anaerobic bacterial metabolism, and colon cancer |
| 204 | Stams | 1994 | Metabolic interactions between anaerobic bacteria in methanogenic environments |
| 205 | Stams | 2003 | Metabolic interactions between methanogenic consortia and anaerobic respiring bacteria |
| 206 | Holmstrom | 2004 | Subdoligranulum variabile gen. nov., sp. nov. from human feces |
| 207 | Eudes | 2008 | Identification of genes encoding the folate and thiamine binding membrane proteins in firmicutes |
| 208 | Macfarlane | 1988 | Contribution of the microflora to proteolysis in the human large intestine |
| 209 | Smith | 1997 | Dissimilatory amino acid metabolism in human colonic bacteria |
| 210 | Starr | 2006 | Role of hyaluronidase in subcutaneous spread and growth of group A streptococcus |
| 211 | Vince | 1980 | Ammonia production by intestinal bacteria: The effect of lactose, lactulose and glucose |
| 212 | Zukaite | 2000 | Acceleration of hyaluronidase production in the course of batch cultivation of Clostridium perfringens can be achieved with bacteriolytic enzymes |
| 213 | van B. Robertson | 1940 | Mucinase: A bacterial enzyme which hydrolyzes synovial fluid mucin and other mucins |
| 214 | Attwood | 1998 | Ammonia-Hyperproducing Bacteria from New Zealand Ruminants |
| 215 | Johnson | 2009 | Interspecies Signaling between Veillonella atypica and Streptococcus gordonii Requires the Transcription Factor CcpA |
| 216 | Taras | 2002 | Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces |
| 217 | Kageyama | 2000 | Emendation of genus Collinsella and proposal of Collinsella stercoris sp. nov. and Collinsella intestinalis sp. nov. |
| 218 | Newsholme | 1994 | Quantitative aspects of glucose and glutamine metabolism by intestinal cells |
| 219 | Roediger | 1997 | Human colonocyte detoxification |
| 220 | Thiele | 2013 | A community-driven global reconstruction of human metabolism |
| 221 | Turnberg | 1970 | Electrolyte absorption from the colon |
| 222 | Yu | 2010 | Bestrophin-2 mediates bicarbonate transport by goblet cells in mouse colon |
| 223 | Bergen | 2009 | Intestinal Nitrogen Recycling and Utilization in Health and Disease |
| 224 | Chen | 2010 | Microbial and Bioconversion Production of D-xylitol and Its Detection and Application |
| 225 | Conly | 1994 | The contribution of vitamin K2(menaquinones) produced by the intestinal microflora to human nutritional requirements for vitamin K |
| 226 | Ganong | 1991 | Review of medical physiology. Section V. 25. Digestion & absorption |
| 227 | Hughes | 2011 | Protein Degradation in the Large Intestine: Relevance to Colorectal Cancer |
| 228 | Suen | 2011 | The Complete Genome Sequence of Fibrobacter succinogenes S85 Reveals a Cellulolytic and Metabolic Specialist |
| 229 | Russell | 1981 | Degradation of Protein by Mixed Cultures of Rumen Bacteria: Identification of Streptococcus Bovis as an Actively Proteolytic rumen bacterium |
| 230 | Holland | 2006 | Development of a defined medium supporting rapid growth for Deinococcus radiodurans and analysis of metabolic capacities |
| 231 | Myhr | 2000 | Denitrovibrio acetiphilus, a novel genus and species of dissimilatory nitrate-reducing bacterium isolated from an oil reservoir model column |
| 232 | Schmidt | 1995 | Interspecies Electron Transfer during Propionate and Butyrate Degradation in Mesophilic, Granular Sludge |
| 233 | Imachi | 2002 | Pelotomaculum thermopropionicum gen. nov., sp. nov., an anaerobic, thermophilic, syntrophic propionate-oxidizing bacterium |
| 234 | Cord-Ruwisch | 1998 | Growth of Geobacter sulfurreducens with Acetate in Syntrophic Cooperation with Hydrogen-Oxidizing Anaerobic Partners |
| 235 | Klitgord | 2010 | Environments that induce synthetic microbial ecosystems |
| 236 | Stolyar | 2007 | Metabolic modeling of a mutualistic microbial community |
| 237 | Kosaka | 2008 | The genome of Pelotomaculum thermopropionicum reveals niche-associated evolution in anaerobic microbiota |
| 238 | Lovley | 1993 | Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals |
| 239 | Downes | 2003 | Dialister invisus sp. nov., isolated from the human oral cavity |
| 240 | Pircher | 2007 | Formation of cadaverine, histamine, putrescine and tyramine by bacteria isolated from meat, fermented sausages and cheeses |
| 241 | Schleifer | 1984 | Transfer of Streptococcus faecalis and Streptococcus faecium to the Genus Enterococcus norn. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. |
| 242 | Vos | 2009 | Bergey's Manual of Systematic Bacteriology, Volume 3: The Firmicutes |
| 243 | Deibel | 1964 | Pyruvate fermentation by Streptococcus faecalis |
| 244 | Delk | 1975 | Biosynthesis of Ribosylthymine in the TransferRNA of Streptococcus faecalis: A Folate-Dependent Methylation Not Involving S-Adenosylmethionine |
| 245 | Bos | 2013 | Volatile Metabolites of Pathogens: A Systematic Review |
| 246 | Mulligan | 1977 | Transport and metabolism of vitamin B6 in lactic acid bacteria |
| 247 | Ryu | 2001 | Characteristics and Glycerol Metabolism of Fumarate-Reducing Enterococcus faecalis RKY1 |
| 248 | Wang | 2009 | Bio-hydrogen production from cellulose by sequential co-culture of cellulosic hydrogen bacteria of Enterococcus gallinarum G1 and Ethanoigenens harbinense B49 |
| 249 | Ryan | 2004 | Sherris Medical Microbiology (4th ed.) |
| 250 | Tang | 2005 | Xanthomonas campestris pv. campestris possesses a single gluconeogenic pathway that is required for virulence |
| 251 | Sawers | 2005 | Formate and its role in hydrogen production in Escherichia coli |
| 252 | Clark | 1989 | The fermentation pathways of Escherichia coli. |
| 253 | Peekhaus | 1998 | What’s for Dinner?: Entner-Doudoroff Metabolism in Escherichia coli |
| 254 | Jones | 2011 | Anaerobic Respiration of Escherichia coli in the Mouse Intestine |
| 255 | Van der Werf | 1997 | Environmental and physiological factors affecting the succinate product ratio during carbohydrate fermentation by Actinobacillus sp. 130Z |
| 256 | Sirko | 1995 | Sulfate and Thiosulfate Transport in Escherichia coli K-12: Evidence for a Functional Overlapping of Sulfate-and Thiosulfate-Binding Proteins |
| 257 | Walker | 2011 | Dominant and diet-responsive groups of bacteria within the human colonic microbiota |
| 258 | Duncan | 2002 | Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces |
| 259 | Duncan | 2003 | Effects of Alternative Dietary Substrates on Competition between Human Colonic Bacteria in an Anaerobic Fermentor System |
| 260 | Chen | 1989 | More Monensin-Sensitive, Ammonia-Producing Bacteria from the Rumen |
| 261 | Venkataraman | 2014 | Metabolite transfer with the fermentation product 2,3-butanediol enhances virulence by Pseudomonas aeruginosa |
| 262 | McBride | 2009 | Novel Features of the Polysaccharide-Digesting Gliding Bacterium Flavobacterium johnsoniae as Revealed by Genome Sequence Analysis |
| 263 | Chen | 1988 | Fermentation of peptides and amino acids by monensin-sensitive ruminal peptostreptococcus |
| 264 | Loesche | 1968 | Amino acid fermentation by Fusobacterium nucleatum |
| 265 | Yang | 2013 | Isolation and Characterization of Novel Denitrifying Bacterium Geobacillus sp. SG-01 Strain from Wood Chips Composted with Swine Manure |
| 266 | Heider | 1999 | Anaerobic bacterial metabolism of hydrocarbons |
| 267 | Nagarajan | 2013 | Characterization and modeling of interspecies electron transfer mechanisms and microbial community dynamics of a syntrophic association |
| 268 | Schleinitz | 2009 | Phenol Degradation in the Strictly Anaerobic Iron-Reducing Bacterium Geobacter metallireducens GS-15 |
| 269 | Summers | 2010 | Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobe bacteria |
| 270 | Manzoni | 2001 | Biotransformation of D-galactitol to tagatose by acetic acid bacteria |
| 271 | Sato | 2011 | Novel metabolic pathways in archaea |
| 272 | Severina | 1991 | Glucose transport into the extremely halophilic archaebacteria |
| 273 | Albers | 2004 | Insights into ABC transport in archaea |
| 274 | Deplancke | 2001 | Microbial modulation of innate defense: goblet cells and the intestinal mucus layer |
| 275 | Gibson | 1996 | Fermentation of non-digestible oligosaccharides by human colonic bacteria |
| 276 | Marshall | 1990 | Urea protects Helicobacter (Campylobacter) pylori from the bactericidal effect of acid |
| 277 | McNulty | 2013 | Mechanisms of molecular transport through the urea channel of Helicobacter pylori |
| 278 | Wilems | 1997 | Phenotypic and Phylogenetic Characterization of some Eubacterium-like isolates containing a novel type B wall murein from human feces: description of Holdemania filiformis gen. nov., sp. nov. |
| 279 | Urakami | 1995 | Characterization and Description of Hyphomicrobium denitrificans sp. nov. |
| 280 | Wrede | 2012 | Archaea in symbioses |
| 281 | Oppenberg | 1990 | Anaerobic degradation of 1,3-propanediol by sulfate-reducing and by fermenting bacteria |
| 282 | Snell | 1976 | Transfer of Some Saccharolytic Moraxella Species to Kingella Henriksen and Bovre 1976, with Descriptions of Kingella indologenes sp. nov. and Kingella denitrificans sp. nov. |
| 283 | Ahrens | 1997 | Kinetic, Dynamic, and Pathway Studies of Glycerol Metabolism by Klebsiella pneumoniae in Anaerobic Continuous Culture: III. Enzymes and Fluxes of Glycerol Dissimilation and 1,3-Propanediol Formation |
| 284 | Hung | 2011 | Facultative methylotrophs from the human oral cavity and methylotrophy in strains of Gordonia, Leifsonia, and Microbacterium |
| 285 | Keuth | 1994 | Vitamin B12 production by Citrobacter freundii or Klebsiella pneumoniae during tempeh fermentation and proof of enterotoxin absence by PCR |
| 286 | Liao | 2012 | An Experimentally Validated Genome-Scale Metabolic Reconstruction of Klebsiella pneumoniae MGH 78578, iYL1228 |
| 287 | Begley | 2005 | The interaction between bacteria and bile |
| 288 | Rao | 1984 | Biosynthesis and Utilization of Folic Acid and Vitamin B12 by Lactic Cultures in Skim Milk |
| 289 | Rooj | 2010 | Metabolites produced by probiotic lactobacilli rapidly increase glucose uptake by caco-2 cells |
| 290 | Almståhl | 2013 | Fermentation of sugars and sugar alcohols by plaque Lactobacillus strains |
| 291 | Veiga | 1992 | Sugar-Glycerol Cofermentations in Lactobacilli: the Fate of Lactate |
| 292 | González-Pajuelo | 2006 | Microbial Conversion of Glycerol to 1,3-Propanediol: Physiological Comparison of a Natural Producer, Clostridium butyricum VPI 3266, and an Engineered Strain, Clostridium acetobutylicum DG1(pSPD5) |
| 293 | Hugenschmidt | 2010 | Screening of a natural biodiversity of lactic and propionic acid bacteria for folate and vitamin B12 production in supplemented whey permeate |
| 294 | Oude Elferink | 2001 | Anaerobic Conversion of Lactic Acid to Acetic Acid and 1,2-Propanediol by Lactobacillus buchneri |
| 295 | Cselovsky | 1992 | Production of formate, acetate, and succinate by anaerobic fermentation of Lactobacillus pentosus in the presence of citrate |
| 296 | Henderson | 1979 | Mechanism of Folate Transport in Lactobacillus casei: Evidence for a Component Shared with the Thiamine and Biotin Transport Systems |
| 297 | Thompson | 1987 | Regulation of sugar transport and metabolism in lactic acid bacteria |
| 298 | Bertelsen | 2001 | Fermentation of D-Tagatose by Human Intestinal Bacteria and Dairy Lactic Acid Bacteria |
| 299 | Beshkova | 1998 | Production of Amino Acids by Yogurt Bacteria |
| 300 | Piveteau | 1995 | Interactions between lactic and propionic acid bacteria |
| 301 | Taranto | 2003 | Lactobacillus reuteri CRL1098 Produces Cobalamin |
| 302 | Robbins | 1940 | Fermentation of Sugar Acids by Bacteria |
| 303 | Wegkamp | 2004 | Transformation of Folate-Consuming Lactobacillus gasseri into a Folate Producer |
| 304 | Klein | 1998 | Taxonomy and physiology of probiotic lactic acid bacteria |
| 305 | Fujisawa | 1992 | Taxonomic Study of the Lactobacillus acidophilus Group, with Recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and Synonymy of Lactobacillus acidophilus Group A3 (Johnson et l. 1980) with the Type Strain of Lactobacillus amylovorus (Nakamura 1981) |
| 306 | Collins | 1989 | Deoxyribonucleic Acid Homology Studies of Lactobacillus casei, Lactobacillus paracasei sp. nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb. nov. |
| 307 | Hedberg | 2008 | Sugar fermentation in probiotic bacteria – an in vitro study |
| 308 | Hugenschmidt | 2011 | Concurrent high production of natural folate and vitamin B12 using a co-culture process with lactobacillus plant arum SM39 and propionibacterium freudenreichii DF13 |
| 309 | Lindgren | 1990 | Anaerobic L-lactate degradation by lactobacillus plantarum |
| 310 | Santos | 2008 | High-Level Folate Production in Fermented Foods by the B12 Producer Lactobacillus reuteri JCM1112 |
| 311 | Talarico | 1990 | Utilization of Glycerol as a Hydrogen Acceptor by Lactobacillus reuteri: Purification of 1,3-Propanediol:NAD+ Oxidoreductaset |
| 312 | Zelante | 2013 | Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22 |
| 313 | Rodríguez | 2012 | Mannitol production by heterofermentative Lactobacillus reuteri CRL 1101 and Lactobacillus fermentum CRL 573 in free and controlled pH batch fermentations. |
| 314 | Mao | 2013 | Bacteroides fragilis polysaccharide A is necessary and sufficient for acute activation of intestinal sensory neurons |
| 315 | Eribe | 2004 | Genetic diversity of Leptotrichia and description of Leptotrichia goodfellowii sp. nov., Leptotrichia hofstadii sp. nov., Leptotrichia shahii sp. nov. and Leptotrichia wadei sp. nov |
| 316 | Kihal | 2007 | Carbon dioxide production by Leuconostoc mesenteroides grown in single and mixed culture with Lactococcus lactis in skimmed milk |
| 317 | Erten | 1998 | Metabolism of fructose as an electron acceptor by Leuconostoc mesenteroides |
| 318 | Begley | 2006 | Bile Salt Hydrolase Activity in Probiotics |
| 319 | Wolin | 2003 | Formate-Dependent Growth and Homoacetogenic Fermentation by a Bacterium from Human Feces: Description of Bryantella formatexigens gen. nov., sp. nov |
| 320 | Sakon | 2008 | Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces |
| 321 | Latham | 1977 | Fermentation of cellulose by Ruminococcus flavefaciens in the presence and absence of Methanobacterium ruminantium |
| 322 | Le Chatelier | 2013 | Richness of human gut microbiome correlates with metabolic markers |
| 323 | Woese | 2012 | The Bacteria. A Treatise on Structure and Function. Vol. VIII Archaebacteria |
| 324 | Liu | 2008 | Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea |
| 325 | DHaeze | 2002 | A hyperthermophilic methanogen sequenced |
| 326 | Fischer | 2005 | Structures and reaction mechanisms of riboflavin synthases of eubacterial and archaeal origin |
| 327 | Blaut | 1994 | Metabolism of methanogens |
| 328 | Benedict | 2011 | Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon methanosarcina acetivorans c2a |
| 329 | Gonnerman | 2013 | Genomically and biochemically accurate metabolic reconstruction of methanosarcina barkeri fusaro, iMG746 |
| 330 | Cadillo-Quiroz | 2009 | Methanosphaerula palustris gen. nov., sp. nov., a hydrogenotrophic methanogen isolated from a minerotrophic fen peatland |
| 331 | Struchtemeyer | 2011 | Evidence for syntrophic butyratemetabolismunder sulfatereducing conditions ina hydrocarbon-contaminated aquifer |
| 332 | Chistoserdova | 2011 | Modularity of methylotrophy, revisited |
| 333 | Jourand | 2004 | Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria |
| 334 | Ward | 2004 | Genomic Insights into Methanotrophy: The Complete Genome Sequence of Methylococcus capsulatus |
| 335 | Sakai | 2007 | Degradation of Glyoxylate and Glycolate with ATP Synthesis by a Thermophilic Anaerobic Bacterium, Moorella sp. Strain HUC22-1 |
| 336 | Enright | 1997 | Moraxella (Branhamella) catarrhalis - clinical and molecular aspects of a rediscovered pathogen |
| 337 | Grant | 1981 | Denitrification by Strains of Neisseria, Kingella, and Chromobacterium |
| 338 | Goker | 2011 | Complete genome sequence of Odoribacter splanchnicus type strain (1651/6T) |
| 339 | Kuhner | 1996 | Generation of a proton motive force by the anaerobic oxalate-degrading bacterium Oxalobacter formigene |
| 340 | Sakamoto | 2006 | Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. |
| 341 | Sugawara | 1991 | Digestion and Fermentation by Human Intestinal Bacteria of Corn Fiber and Its Hemicellulose in Vitro |
| 342 | Sakamoto | 2007 | Parabacteroides johnsonii sp. nov., isolated from human faeces |
| 343 | Tanasupawat | 1993 | Characterization of Pediococcus pentosaceus and Pediococcus acidilactici Strains and Replacement of the Type Strain of P. acidilactici with the Proposed Neotype DSM 20284 |
| 344 | Papagianni | 2009 | Pediocins: The bacteriocins of Pediococci. Sources, production, properties and applications |
| 345 | Schink | 1984 | Fermentation of 2,3-butanediol by Pelobacter carbinolicus sp. nov. and Pelobacter propionicus sp. nov., and evidence for propionate formation from C2 compounds |
| 346 | Muller | 2010 | Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanisms |
| 347 | Ng | 2013 | Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens |
| 348 | Theriot | 2014 | Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection |
| 349 | Holmes | 2011 | Understanding the role of gut microbiome-host metabolic signal disruption in health and disease |
| 350 | Accetto | 2007 | Studies on Prevotella nuclease using a system for the controlled expression of cloned genes in P. bryantii TC1-1 |
| 351 | Hayashi | 2007 | Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces |
| 352 | Chassard | 2005 | Interaction between H2-producing and non-H2-producing cellulolytic bacteria from the human colon |
| 353 | Herbeck | 1974 | Nutritional Features of the Intestinal Anaerobe Ruminococcus bromii |
| 354 | Kabel | 2011 | Biochemical Characterization and Relative Expression Levels of Multiple Carbohydrate Esterases of the Xylanolytic Rumen Bacterium Prevotella ruminicola 23 Grown on an Ester-Enriched Substrate |
| 355 | Pittman | 1964 | Peptides and other nitrogen sources for growth of bacteroides ruminicola |
| 356 | Russell | 1988 | Enrichment and Isolation of a Ruminal Bacterium with a Very High Specific Activity of Ammonia Production |
| 357 | Strobel | 1992 | Vitamin B12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23 |
| 358 | Varel | 1974 | Nutritional Features of Bacteroides fragilis subsp. fragilis |
| 359 | Yanagisawa | 2006 | Proteinase Activity of Prevotella Species Associated with Oral Purulent Infection |
| 360 | Ingram | 1983 | Studies of the extracellular proteolytic activity produced by Propionibacterium acnes |
| 361 | Jeter | 1984 | Salmonella typhimurium Synthesizes Cobalamin (Vitamin B12) De Novo Under Anaerobic Growth Conditions |
| 362 | Kosmider | 2010 | Propionic Acid Production by Propionibacterium freudenreichiissp. shermanii Using Crude Glycerol and Whey Lactose Industrial Wastes |
| 363 | Roth | 1996 | COBALAMIN (COENZYME B12): Synthesis and Biological Significance |
| 364 | Vince | 1973 | Ammonia production by intestinal bacteria |
| 365 | Carlier | 2009 | Proposal to unify Clostridium orbiscindens Winter et al. 1991 and Eubacterium plautii (Sguin 1928) Hofstad and Aasjord 1982, with description of Flavonifractor plautii gen. nov., comb. nov., and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. |
| 366 | Chou | 2008 | Transcriptome analysis of agmatine and putrescine catabolism in Pseudomonas aeruginosa PAO1 |
| 367 | Stuer | 1986 | Purification of extracellular lipase from Pseudomonas aeruginosa |
| 368 | Valentini | 2011 | Identification of C4-Dicarboxylate Transport Systems in Pseudomonas aeruginosa PAO1 |
| 369 | Sias | 1979 | Isolation and analysis of mutants of Pseudomonas aeruginosa unable to assimilate nitrate. |
| 370 | Samuelsson | 1988 | Heat Production by the Denitrifying Bacterium Pseudomonas fluorescens and the Dissimilatory Ammonium-Producing Bacterium Pseudomonas putrefaciens during Anaerobic Growth with Nitrate as the Electron Acceptor |
| 371 | Sharma | 2001 | Production, purification, characterization, and applications of lipases |
| 372 | Daum | 1998 | Physiological and Molecular Biological Characterization of Ammonia Oxidation of the Heterotrophic Nitrifier Pseudomonas putida |
| 373 | Downes | 2009 | Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum Synergistetesisolated from the human oral cavity |
| 374 | Völkl | 1993 | Pyrobaculum aerophilum sp. nov., a Novel Nitrate-Reducing Hyperthermophilic Archaeum |
| 375 | Amo | 2002 | Pyrobaculum calidifontis sp. nov., a novel hyperthermophilic archaeon that grows in atmospheric air |
| 376 | Koning | 2001 | Cellobiose Uptake in the Hyperthermophilic Archaeon Pyrococcus furiosus Is Mediated by an Inducible, High-Afnity ABC Transporter |
| 377 | Hemachander | 2001 | Whole cell immobilization of Ralstonia pickettii for lipase production |
| 378 | Reichardt | 2014 | Phylogenetic distribution of three pathways for propionate production within the human gut microbiota |
| 379 | Doi | 1991 | Enhancement of Denitrifying Activity in Cells of Roseobacter denitrificans Grown Aerobically in the Light |
| 380 | Gold | 2007 | Global view of the clostridium thermocellum cellulosome revealed by quantitative proteomic analysis |
| 381 | Ng | 1982 | Differential metabolism of cellobiose and glucose by clostridum thermocellum and clostridium thermohydrosulfuricum |
| 382 | Rincon | 2005 | Unconventional Mode of Attachment of the Ruminococcus flavefaciens Cellulosome to the Cell Surface |
| 383 | Sparling | 2006 | Formate synthesis by clostridium thermocellum |
| 384 | Bryant | 1960 | Studies on the Nitrogen Requirements of Some Ruminal Cellulolytic Bacteria |
| 385 | Louis | 2007 | Understanding the effects of diet on bacterial metabolism in the large intestine |
| 386 | Shi | 1997 | Formation of formate and hydrogen, and flux of reducing equivalents and carbon in Ruminococcus flavefaciens FD-1 |
| 387 | Allison | 1962 | Metabolic function of branched-chain volatile fatty acids, growth factors for ruminococci. II. Biosynthesis of higher branched-chain fatty acids and aldehydes |
| 388 | Brettar | 2002 | Shewanella denitrificans sp. nov., a vigorously denitrifying bacterium isolated from the oxic–anoxic interface of the Gotland Deep in the central Baltic Sea |
| 389 | Sadovski | 1969 | Extracellular Nuclease Activity of Fish Spoilage Bacteria, Fish Pathogens, and Related Species |
| 390 | Woodward | 2005 | Identification and characterization of Shigella boydii 20 serovar nov., a new and emerging Shigella serotype |
| 391 | Snyder | 2010 | Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts |
| 392 | Arnone | 1969 | The extracellular nuclease of staphylococcus aureus: Structures of the native enzyme and an enzyme-inhibitor complex at 4A resolution |
| 393 | Beenken | 2012 | Impact of Extracellular Nuclease Production on the Biofilm Phenotype of Staphylococcus aureus under In Vitro and In Vivo Conditions |
| 394 | Willcox | 2001 | Streptococcus australis sp. nov., a novel oral streptococcus |
| 395 | Luppens | 2008 | Effect of Veillonella parvula on the antimicrobial resistance and gene expression of Streptococcus mutans grown in a dual-species biofilm |
| 396 | Loscalzo | 2011 | Lipid metabolism by gut microbes and atherosclerosis |
| 397 | Whiley | 1990 | Streptococcus parasanguis sp. nov., an atypical viridans Streptococcus from human clinical specimens |
| 398 | Benedik | 1998 | Serratia marcescens and its extracellular nuclease |
| 399 | Whiley | 1988 | Streptococcus vestibularis sp. nov. from the Human Oral Cavity |
| 400 | Nisole | 2006 | Extracellular production of Streptomyces lividans acetyl xylan esterase A in Escherichia coli for rapid detection of activity |
| 401 | Albers | 1999 | Glucose Transport in the Extremely Thermoacidophilic Sulfolobus solfataricus Involves a High-Affinity Membrane-Integrated Binding Protein |
| 402 | Mukhopadhya | 2011 | A Comprehensive Evaluation of Colonic Mucosal Isolates of Sutterella wadsworthensis from Inflammatory Bowel Disease |
| 403 | Boone | 1989 | Diffusion of the Interspecies Electron Carriers H2 and Formate in Methanogenic Ecosystems and Its Implications in the Measurement of Km for H2 or Formate Uptake |
| 404 | Sousa | 2007 | Syntrophomonas zehnderi sp. nov., an anaerobe that degrades long-chain fatty acids in co-culture with methanobacterium formicicum |
| 405 | Stieb | 1985 | Anaerobic oxidation of fatty acids by clostridium byrantii sp. nov., a sporeforming, obligately syntrophic bacterium |
| 406 | Wu | 1994 | Anaerobic degradation of normal- and branched-chain fatty acids with four or more carbons to methane by a syntrophic methanogenic triculture |
| 407 | Jackson | 2002 | Anaerobic microbial metabolism can proceed close to thermodynamic limits |
| 408 | Fulcinos | 2005 | Identification of extracellular lipases/esterases produced by Thermus thermophilus HB27: partial purification and preliminary biochemical characterisation |
| 409 | Kelly | 2000 | Confirmation of Thiobacillus denitrificans as a species of the genus Thiobacillus, in the β- subclass of the Proteobacteria, with strain NCIMB 9548 as the type strain |
| 410 | Baalsrud | 1954 | Studies on Thiobacillus denitrificans |
| 411 | Paralonov | 2012 | Comparative genome analysis of 19 Ureaplasma urealyticum and Ureaplasma parvum strains |
| 412 | Ng | 1971 | Lactate metabolism by Veillonella parvula |
| 413 | Seper | 2011 | Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation |
| 414 | Blokesch | 2008 | The Extracellular Nuclease Dns and Its Role in Natural Transformation of Vibrio cholerae |
| 415 | Balch | 1997 | Acetobacterium, a New Genus of Hydrogen-Oxidizing, Carbon Dioxide-Reducing,Anaerobic Bacteria |
| 416 | Buschhorn | 1989 | Production and Utilization of Ethanol by the Homoacetogen Acetobacterium woodii |
| 417 | Peters | 1998 | Efficiency of hydrogen utilization during unitrophic and mixotrophic growth of Acetobacterium woodii on hydrogen and lactate in the chemostat |
| 418 | Heise | 1989 | Sodium Dependence of Acetate Formation by the Acetogenic Bacterium Acetobacterium woodii |
| 419 | Zitomersky | 2013 | Characterization of Adherent Bacteroidales from Intestinal Biopsies of Children and Young Adults with Inflammatory Bowel Disease |
| 420 | Mishra | 2012 | Genome sequence and description of Alistipes senegalensis sp. nov. |
| 421 | Downes | 2013 | Description of Alloprevotella rava gen. nov., sp. nov., isolated from the human oral cavity, and reclassification of Prevotella tannerae Moore et al. 1994 as Alloprevotella tannerae gen. nov., comb. nov. |
| 422 | Baena | 1999 | Aminomonas paucivorans gen. nov., sp. nov., a mesophilic, anaerobic, amino-acid-utiIizing bacterium |
| 423 | Allen-Vercoe | 2012 | Anaerostipes hadrus comb. nov., a dominant species within the human colonic microbiota; reclassification of Eubacterium hadrum Moore et al. 1976 |
| 424 | Moore | 1976 | Emendation of Bacteroidaceae and Butyrivibrio and Descriptions of Desulfornonas gen. nov. and Ten New Species in the Genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus |
| 425 | Whitehead | 2005 | Bacteroides coprosuis sp. nov., isolated from swine-manure storage pits |
| 426 | Fenner | 2005 | Bacteroides massiliensis sp. nov., isolated from blood culture of a newborn |
| 427 | Song | 2004 | “Bacteroides nordii” sp. nov. and “Bacteroides salyersae” sp. nov. Isolated from Clinical Specimens of Human Intestinal Origin |
| 428 | Love | 1986 | Bacteroides tectum sp. nov. and Characteristics of Other Nonpigmented Bactevoides Isolates from Soft-Tissue Infections from Cats and Dogs |
| 429 | Benno | 1983 | Bacteroides pyogenes sp. nov., Bacteroides suis sp. nov., and Bacteroides helcogenes sp. nov., New Species from Abscesses and Feces of Pigs |
| 430 | Ezaki | 1994 | 16s Ribosomal DNA Sequences of Anaerobic Cocci and Proposal of Ruminococcus hansenii comb. nov. and Ruminococcus productus comb. nov. |
| 431 | Kinyon | 1979 | Treponema innocens, a New Species of Intestinal Bacteria, and Emended Description of the Type Strain of Treponema hyodysenteriae Harris et al. |
| 432 | Stanton | 1997 | Recognition of Two New Species of Intestinal Spirochetes: Serpulina intermedia sp. nov. and Serpulina murdochii sp. nov. |
| 433 | Fardeau | 2000 | Thermoanaerobacter subterraneus sp. nov., a novel thermophile isolated from oilfield water |
| 434 | Mori | 2009 | Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. nov., Caldisericales ord. nov. and Caldisericia classis nov. |
| 435 | Itoh | 2003 | Caldisphaera lagunensis gen. nov., sp. nov., a novel thermoacidophilic crenarchaeote isolated from a hot spring at Mt Maquiling, Philippines |
| 436 | Miroshnichenko | 2003 | Caldithrix abyssi gen. nov., sp. nov., a nitrate- reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage |
| 437 | Ogg | 2011 | Caloramator mitchellensis sp. nov., a thermoanaerobe isolated from the geothermal waters of the Great Artesian Basin of Australia, and emended description of the genus Caloramator |
| 438 | Ogg | 2009 | Caloramator australicus sp. nov., a thermophilic, anaerobic bacterium from the Great Artesian Basin of Australia |
| 439 | Vandamme | 2010 | Reclassification of Bacteroides ureolyticus as Campylobacter ureolyticus comb. nov., and emended description of the genus Campylobacter |
| 440 | Askew | 2009 | Transcriptional Regulation of Carbohydrate Metabolism in the human pathogen candida albicans |
| 441 | Williamson | 1986 | Biotypes of Candida albicans using the API 20C system |
| 442 | Sullivan | 1995 | Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals |
| 443 | Wong | 1993 | D-Arabitol Metabolism in Candida albicans: Studies of the Biosynthetic Pathway and the Gene That Encodes NAD-Dependent D-Arabitol Dehydrogenase |
| 444 | Granstrom | 2002 | Metabolic flux analysis of candida tropicalis growing on xylose in an Oxygen-limited chemostat |
| 445 | Rehman | 2010 | Cadmium biosorption by yeats, Candida tropical is CBL-1, isolated from industrial wastewater |
| 446 | Sulman | 2013 | Isolation and Characterization of Cellulose Degrading Candida tropicalis W2 from Environmental Samples |
| 447 | West | 2009 | Xylitol production by Candida species grown on a grass hydrolysate |
| 448 | Lohmeier-Vogel | 1989 | 31P Nuclear Magnetic Resonance Study of the Effect of Azide on Xylose Fermentation by Candida tropicalis |
| 449 | Jiang | 2007 | Biodegradation of phenol and 4-chlorophenol by the yeast Candida tropicalis |
| 450 | Sudha | 2010 | Comparative study for the production, characterisation and antimicrobial studies of Sophorolipids using Candida tropicalis |
| 451 | Nakamura | 1968 | Transglucosyl-Amylase of Candida tropicalis |
| 452 | Brenner | 1989 | Capnocytophaga canimorsussp.nov.(FormerlyCDC GroupDF-2), a Cause of Septicemia following Dog Bite, and C. cynodegmi sp. nov., a Cause of Localized Wound Infection following Dog Bite |
| 453 | Lawson | 2006 | Catellicoccus marimammalium gen. nov., sp. nov., a novel Gram-positive, catalase-negative, coccus- shaped bacterium from porpoise and grey seal |
| 454 | Finegold | 2003 | Cetobacterium somerae sp. nov. from Human Feces and Emended Description of the Genus Cetobacterium |
| 455 | Jung | 2010 | Clostridium arbusti sp. nov., an anaerobic bacterium isolated from pear orchard soil |
| 456 | Abrini | 1994 | Clostridium autoethanogenum, sp. nov., an anaerobic bacterium that produces ethanol from carbon monoxide |
| 457 | Chamkha | 2001 | Isolation of Clostridium bifermentans from Oil Mill Wastewaters Converting Cinnamic Acid to 3-phenylpropionic Acid and Emendation of the Species |
| 458 | Dai | 2011 | Amino acid metabolism in intestinal bacteria: links between gut ecology and host health |
| 459 | Hauschild | 1974 | Clostridium celatum sp.nov., Isolated from Normal Human Feces |
| 460 | Warren | 2006 | Clostridium aldenense sp. nov. and Clostridium citroniae sp. nov. Isolated from Human Clinical Infections |
| 461 | Kaneuchi | 1976 | Taxonomic Study of Bacteroides dostridiiformis subsp. clostridiiformis (Burri and Ankersmit) Holdeman and Moore and of Related Organisms: Proposal of Clostridium clostridiiformis (Burri and Ankersmit) comb. nov. and Clostridium symbiosum (Stevens) comb. nov. |
| 462 | Greetham | 2003 | Clostridium colicanis sp. nov., from canine faeces |
| 463 | Smith | 1962 | Clostridium innocuum, sp. n., a spore-forming anaerobe isolated from human infections |
| 464 | Dabrock | 1992 | Parameters Affecting Solvent Production by Clostridium pasteurianum |
| 465 | Keis | 2001 | Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov. and Clostridium saccharobutylicum sp. nov. |
| 466 | Partansky | 1935 | Anaerobic Bacteria Capable of the Fermentation of Sulfite Waste Liquor |
| 467 | Madden | 1983 | Isolation and Characterization of Clostridium stercorarium sp. nov., Cellulolytic Thermophile |
| 468 | Fardeau | 2001 | Transfer of Thermobacteroides leptospartum and Clostridium thermolacticum as Clostridium stercorarium subsp. leptospartum subsp. nov., comb. nov. and C. stercorarium subsp. thermolacticum subsp. nov., comb. nov. |
| 469 | Hethener | 1992 | Clostridium termitidis sp. nov., a Cellulolytic Bacterium from the Gut of the Wood-feeding Termite, Nasutitermes lujae |
| 470 | Jonsson | 1990 | Enumeration and Confirmation of Clostridium tyrobutyricum in Silages Using Neutral Red, D-Cycloserine, and Lactate Dehydrogenase Activity |
| 471 | Kunzelmann | 2002 | Electrolyte Transport in the Mammalian Colon: Mechanisms and Implications for Disease |
| 472 | Rabus | 1993 | Complete Oxidation of Toluene under Strictly Anoxic ConditionsbyaNew Sulfate-ReducingBacterium |
| 473 | Trinkerl | 1990 | Desulfovibrio termitidis sp. nov., a Carbohydrate-Degrading Sulfate-Reducing Bacterium from the Hindgut of a Termite |
| 474 | Sorokin | 2008 | Dethiobacter alkaliphilus gen. nov. sp. nov., and Desulfurivibrio alkaliphilus gen. nov. sp. nov.: two novel representatives of reductive sulfur cycle from soda lakes. |
| 475 | L'Haridon | 1998 | Desulfurobacterium thermolithotrophum gen. nov., sp. nov., a novel autotrophic, sulphur-reducing bacterium isolated from a deep-sea hydrothermal vent |
| 476 | Hofstad | 2000 | Dysgonomonas gen. nov. to accommodate Dysgonomonas gadei sp. nov., an organism isolated from a human gall bladder, and Dysgonomonas capnocytophagoides (formerly CDC group DF-3) |
| 477 | Lawson | 2002 | Dysgonomonas mossii sp. nov., from Human Sources* |
| 478 | Doran | 1978 | Eimeria tenella: vitamin requirements for development in primary cultures of chicken kidney cells. |
| 479 | Smith | 1986 | Monosaccharide Transport by Eimeria tenella Sporozoites |
| 480 | Kampfer | 2011 | Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae |
| 481 | Kim | 2005 | Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. |
| 482 | Holmes | 1982 | Flavobacteriurn breve sp. nov., norn. rev. |
| 483 | Saha | 2006 | Emticicia oligotrophica gen. nov., sp. nov., a new member of the family ‘Flexibacteraceae’, phylum Bacteroidetes |
| 484 | Mda | 2006 | Enterococcus caccae sp. nov., isolated from human stools |
| 485 | Svec | 2001 | Enterococcus haemoperoxidus sp. nov. and Enterococcus moraviensis sp. nov., isolated from water |
| 486 | Law-Brown | 2003 | Enterococcus phoeniculicola sp. nov., a novel member of the enterococci isolated from the uropygial gland of the Red-billed Woodhoopoe, Phoeniculus purpureus |
| 487 | Vancanneyt | 2001 | Enterococcus villorum sp. nov., an enteroadherent bacterium associated with diarrhoea in piglets |
| 488 | Holdeman | 1971 | Clostridium ramosum (Vuillemin)comb.nov.:Emended Description and Proposed-NeotypeStrain |
| 489 | Holdeman | 1980 | Descriptions of Eubacterium timidum sp. nov., Eubacterium brachy sp. nov., and Eubacterium nodatum sp. nov. Isolated from Human Periodontitis |
| 490 | Margaret | 1986 | Eubacterium yurii subsp. yurii sp. nov. and Eubacterium yurii subsp. margaretiae subsp. nov.: Test Tube.Brush Bacteria from Subgingival Dental Plaque |
| 491 | Cato | 1985 | Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the Human Gingival Sulcus |
| 492 | Siqueira | 2003 | Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases |
| 493 | Wakabayashi | 1989 | Flavobacterium branchiophila sp. nov. a Causative Agent of Bacterial Gill Disease of Freshwater Fishes |
| 494 | Bernardet | 1986 | Cutting a Gordian Knot: Emended Classification and Description of the Genus Flavobacterium, Emended Description of the Family Flavobacteriaceae, and Proposal of Flavobacterium hydatis norn. nov. (Basonym, Cytophaga aquatilis Strohl and Tait 1978) |
| 495 | Lewin | 1969 | A Classification of Flexibacteria |
| 496 | Hosoya | 2007 | Reclassification of Flexibacter aggregans (Lewin 1969) Leadbetter 1974 as a later heterotypic synonym of Flexithrix dorotheae Lewin 1970 |
| 497 | Shinjo | 1981 | Proposal of Two Subspecies of Fusobacterium necrophorum (Flugge) Moore and Holdeman: Fusobacterium necrophorum subsp. necrophorum subsp. nov., nom. rev. (ex Flugge 1886), and Fusobacterium necrophorum subsp. funduliforme subsp. nov., nom. rev. (ex Hall6 1898) |
| 498 | Collins | 1998 | Gemella bergeriae sp. nov., Isolated from Human Clinical Specimens |
| 499 | Kilpper-Balz | 1988 | Transfer of Streptococcus morbillorum to the Genus Gemella as Gemella morbillorum comb. nov. |
| 500 | Collins | 1998 | Description of Gemella sanguinis sp. nov., Isolated from Human Clinical Specimens |
| 501 | Oren | 1984 | Halobacteroides halobius gen. nov., sp. nov., a Moderately Halophilic Anaerobic Bacterium from the Bottom Sediments ofthe Dead Sea |
| 502 | Peel | 1997 | Helcococcus kunzii as Sole Isolate from an Infected Sebaceous Cyst |
| 503 | Harper | 2002 | Helicobacter cetorum sp. nov., a Urease-Positive Helicobacter Species Isolated from Dolphins and Whales |
| 504 | Fox | 2007 | Isolation and Characterization of a Novel Helicobacter Species, “Helicobacter macacae,” from Rhesus Monkeys with and without Chronic Idiopathic Colitis” |
| 505 | Flores | 2012 | Hippea jasoniae sp. nov. and Hippea alviniae sp. nov., thermoacidophilic members of the class Deltaproteobacteria isolated from deep-sea hydrothermal vent deposits |
| 506 | Eggerth | 1935 | The Gram-positive Non-spore-bearing Anaerobic Bacilli of Human Feces |
| 507 | Iino | 2010 | Ignavibacterium album gen. nov., sp. nov., a moderately thermophilic anaerobic bacterium isolated from microbial mats at a terrestrial hot spring and proposal of Ignavibacteria classis nov., for a novel lineage at the periphery of green sulfur bacteria |
| 508 | Moore | 1994 | Oribaculum catoniae gen. nov., sp. nov.; Catonella morbi gen. nov., sp. nov.; Hallella seregens gen. nov., sp. nov.; Johnsonella ignava gen. nov., sp. nov.; and Dialister pneumosintes gen. nov., comb. nov., nom. rev., Anaerobic Gram-Negative Bacilli from the Human Gingival Crevice |
| 509 | Whitford | 2001 | Lachnobacterium bovis gen. nov., sp. nov., a novel bacterium isolated from the rumen and faeces of cattle |
| 510 | Morita | 2010 | Lactobacillus equicursoris sp. nov., isolated from the faeces of a thoroughbred racehorse |
| 511 | Endo | 2010 | Lactobacillus florum sp. nov., a fructophilic species isolated from flowers |
| 512 | Cousin | 2012 | Lactobacillus gigeriorum sp. nov., isolated from chicken crop |
| 513 | Cousin | 2013 | Lactobacillus pasteurii sp. nov. and Lactobacillus hominis sp. nov. |
| 514 | Chen | 201 | Lactobacillus pobuzihii sp. nov., isolated from pobuzihi (fermented cummingcordia) |
| 515 | Zou | 2013 | Lactobacillus shenzhenensis sp. nov., isolated from a fermented dairy beverage |
| 516 | Rodas | 2006 | Lactobacillus vini sp. nov., a wine lactic acid bacterium homofermentative for pentoses |
| 517 | Hellemond | 1997 | Leishmania infantum promastigotes have a poor capacity for anaerobic functioning and depend mainly on respiration for their energy generation |
| 518 | Vieira | 1995 | Amino acid uptake and intracellular accumulation in Leishmannia major promastigotes are largely determined by an H+-pump generated membrane potential |
| 519 | Ellenberger | 1987 | Biochemistry and Regulation of Folate and Methotrexate transport in Leishmania major |
| 520 | Naderer | 2010 | Evidence that Intracellular stages of Leishmania major utilize amino sugars as a major carbon source |
| 521 | Darling | 1989 | Carbon dioxide abolishes the reverse Pasteur effect in Leishmania major promastigotes |
| 522 | Chevrot | 2008 | Megamonas rupellensis sp. nov., an anaerobe isolated from the caecum of a duck |
| 523 | Podosokorskaya | 2013 | Characterization of Melioribacter roseus gen. nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae |
| 524 | Bellack | 2011 | Methanocaldococcus villosus sp. nov., a heavily flagellated archaeon that adheres to surfaces and forms cell–cell contacts |
| 525 | Takai | 2004 | Methanotorris formicicus sp. nov., a novel extremely thermophilic, methane-producing archaeon isolated from a black smoker chimney in the Central Indian Ridge |
| 526 | Burrgraf | 1990 | Methanococcus igneus sp. nov., a Novel Hyperthermophilic Methanogen from a Shallow Submarine Hydrothermal System |
| 527 | Leach | 1973 | Further Studies on Classification of Bovine Strains of Mycoplasmatales, with Proposals for New Species, Acholeplasma modicum and Mycoplasma alkalescens |
| 528 | Tully | 1972 | Synonymy of Mycoplasma arginini and Mycoplasma leonis |
| 529 | DaMassa | 1994 | Mycoplasma auris sp. nov., Mycoplasma cottewii sp. nov., and Mycoplasma yeatsii sp. nov., New Sterol-Requiring Mollicutes from the External Ear Canals of Goats |
| 530 | Jordan | 1981 | Isolation and Characterization of Mycoplasma columbium and Mycoplasma columborale, Two New Species from Pigeons |
| 531 | Rosendal | 1973 | Mycoplasma cynos, a New Canine Mycoplasma Species |
| 532 | Jordan | 1982 | Characterization and Taxonomic Description of Five Mycoplasma Serovars (Serotypes) of Avian Origin and Their Elevation to Species Rank and Further Evaluation of the Taxonomic Status of Mycoplasrna synoviae |
| 533 | Madden | 1974 | Mycoplasma moatsii, a New Species Isolated from Recently Imported Grivit Monkeys (Cercopithecus aethiops) |
| 534 | Tully | 1974 | Characterization of Some Caprine Mycoplasmas, with Proposals for New Species, Mycoplasma capricolum and M ycoplasma putvefaciens |
| 535 | Paek | 2015 | Myroides injenensis sp. nov., a new member isolated from human urine |
| 536 | Vancanneyt | 1996 | Reclassification of Flavobacterium odoraturn (Stutzer 1929) Strains to a New Genus, Myroides, as Myroides odoratus comb. nov. and Myroides odoratimimus sp. nov. |
| 537 | Kurtzman | 2011 | The Yeasts: A Taxonomic Study |
| 538 | Brew | 1990 | The rapid nitrosation of 2,3‐diaminonaphthalene by gastric isolates of Neisseria subflava |
| 539 | Nagai | 2010 | Alistipes indistinctus sp. nov. and Odoribacter laneus sp. nov., common members of the human intestinal microbiota isolated from faeces |
| 540 | Iino | 2007 | Oscillibacter valericigenes gen. nov., sp. nov., a valerate-producing anaerobic bacterium isolated from the alimentary canal of a Japanese corbicula clam |
| 541 | Takayuki | 1983 | Transfer of Peptococcus indolicus, Peptococcus asaccharolyticus, Peptococcus prevotii, and Peptococcus magnus to the Genus Peptostreptococcus and Proposal of Peptostreptococcus tetradius sp. nov. |
| 542 | Jung | 2014 | Peptoniphilus rhinitidis sp. nov., isolated from specimens of chronic rhinosinusitis |
| 543 | Downie | 2006 | Transport of nucleosides across the Plasmodium falciparum parasite plasma membrane has characteristics of PfENT1 |
| 544 | Sherman | 1979 | Biochemistry of Plasmodium (Malarial Parasites) |
| 545 | Vander-Jagt | 1990 | D-Lactate production in erythrocytes infected with Plasmodium falciparum |
| 546 | Lwoff | 1951 | Biochemistry and Physiology of Protozoa |
| 547 | Lewis-Hughes | 1984 | In Vitro culture of plasmodium yoelii blood stages |
| 548 | Gosink | 1998 | Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the C'ophaga- Flavobacterium-Bacteroides group and reclassificationof 'Flectobacillusglomeratus as Polaribacter glomeratus comb. nov. |
| 549 | Chen | 2005 | Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater |
| 550 | Dobson | 1993 | Direct Sequencing of the Polymerase Chain Reaction- Amplified 16s rRNA Gene of Flavobacten'um gondwanense sp. nov. and Flavobacten'um salegens sp. nov., Two New Species from a Hypersaline Antarctic Lake |
| 551 | Duncan | 2006 | Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces |
| 552 | Chandel | 2011 | Bioconversion of pentose sugars into ethanol: A review and future directions |
| 553 | Senac | 1990 | Intermediary Metabolite Concentrations in Xylulose- and Glucose- Fermenting Saccharomyces cerevisiae Cells |
| 554 | Kradolfer | 1982 | Tryptophan degradation in Saccharomyces cerevisiae: Characterization of two aromatic aminotransferases |
| 555 | Brayant | 1956 | The characteristics of strains of Selenomonas isolated from bovine rumen contents |
| 556 | Whitcomb | 1997 | Spiroplasma chrysopicola sp. nov., Spiroplasma gladiatoris sp. nov., Spiroplasma helicoides sp. nov., and Spiroplasma tabanidicola sp. nov., from Tabanid (Diptera: Tabanidae) Flies |
| 557 | Williamson | 1996 | Spiroplasma diminutum sp. nov., from Culex annulus Mosquitoes Collected in Taiwan |
| 558 | Clark | 1985 | Spiroplasrna melliferum, a New Species from the Honeybee (Apis mellifera) |
| 559 | Whitcomb | 1996 | Spiroplasma syrphidicola sp. nov., from a Syrphid Fly (Diptera: Syrphidae) |
| 560 | Abalain-Colloc | 1988 | Spiroplasma taiwanense sp. nov. from Culex tritaeniorhynchus Mosquitoes Collected in Taiwan |
| 561 | Chesneau | 1993 | Staphylococcus pasteuri sp. nov., Isolated from Human, Animal, and Food Specimens |
| 562 | Whiley | 1991 | Emended Descriptions and Recognition of Streptococcus constellatus, Streptococcus intermedius, and Streptococcus anginosus as Distinct Species |
| 563 | Poyart | 2002 | Taxonomic dissection of the Streptococcus bovis group by analysis of manganese- dependent superoxide dismutase gene (sodA) sequences: reclassification of ‘Streptococcus infantarius subsp. coli’ as Streptococcus lutetiensis sp. nov. and of Streptococcus bovis biotype II.2 as Streptococcus pasteurianus sp. nov. |
| 564 | Schlegel | 2003 | Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. |
| 565 | Zbinden | 2012 | Streptococcus tigurinus sp. nov., isolated from blood of patients with endocarditis, meningitis and spondylodiscitis |
| 566 | Janssen | 1999 | Succinispira mobilis gen. nov., sp. nov., a succinate-decarboxylatinganaerobic bacterium |
| 567 | Chamkha | 2001 | Isolation of a cinnamic acid-metabolizing Clostridium glycolicum strain from oil mill wastewaters and emendation of the species description |
| 568 | Chen | 2013 | Tetrapisispora taiwanensis sp. nov. and Tetrapisispora pingtungensis sp. nov., two ascosporogenous yeast species isolated from soil |
| 569 | Ueda-Nishimura | 1999 | A new yeast genus, Tetrapisisporagen. nov.: Tetrapisisporairiornotensis sp. nov., Tetrapisisporananseiensis sp. nov. and Tetrapisisporaarboricolasp. nov., frornthe Nansei Islands, and reclassification of Kluyveromyces phaffii (van der Walt) van der Walt as Tetrapisispora phaffii comb. nov. |
| 570 | Van der Walt | 1963 | Fabospora phaffii sp.n. |
| 571 | Lee | 1993 | Taxonomic Distinction of Saccharolytic Thermophilic Anaerobes: Description of Thermoanaerobacteriumxylanolyticumgen. nov.,sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp~. nov.;Reclassificationof Thermoanaerobiumbrockii,Clostridium thermosulfurogenes, and Clostridium thermohydrosulfiricum ElO0-69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacteriumthermosulfurigenescomb. nov.,and Thermoanaerobacter thermohydrosulfuricus comb. nov., Respectively; and Transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus |
| 572 | Moussard | 2004 | Thermodesulfatator indicus gen. nov., sp. nov., a novel thermophilic chemolithoautotrophic sulfate-reducing bacterium isolated from the Central Indian Ridge |
| 573 | Hamilton-Brehm | 2013 | Thermodesulfobacterium geofontis sp. nov., a hyperthermophilic, sulfate-reducing bacterium isolated from Obsidian Pool, Yellowstone National Park |
| 574 | Mori | 2003 | A novel lineage of sulfate-reducing microorganisms: Themodesulfobiaceae farm. no., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring |
| 575 | Chung | 2000 | Thermus igniterrae sp. nov. and Thermus antranikianii sp. nov., two new species from Iceland |
| 576 | Bjornsdottir | 2009 | Thermus islandicus sp. nov., a mixotrophic sulfur-oxidizing bacterium isolated from the Torfajokull geothermal area |
| 577 | Williams | 1996 | Thermus oshimai sp. nov., Isolated from Hot Springs in Portugal, Iceland, and the Azores, and Comment on the Concept of a Limited Geographical Distribution of Themus Species |
| 578 | Stanton | 1980 | Treponema bryantii sp. nov., a Rumen Spirochete that Interacts with Cellulolytic Bacteria |
| 579 | Graber | 2004 | Description of Treponema azotonutricium sp. nov. and Treponema primitia sp. nov., the First Spirochetes Isolated from Termite Guts |
| 580 | Cwyk | 1979 | Treponema succinifaciens sp. nov., an Anaerobic spirochete from the swine intestine |
| 581 | Heyworth | 1984 | Pyrimidine metabolism in Trichomonas vaginalis |
| 582 | Beach | 1990 | Fatty acid and sterol metabolism of cultured Trichomonas vaginalis and Tritichomonas foetus |
| 583 | Petrin | 1998 | Clinical and Microbiological Aspects of Trichomonas vaginalis |
| 584 | Mack | 1980 | End products of carbohydrate metabolism in Trichomonas vaginalis |
| 585 | Muller | 2012 | Biochemistry and Evolution of Anaerobic Energy Metabolism in Eukaryotes |
| 586 | Linstead | 1983 | The pathway of arginine catabolism in the parasitic flagellate Trichomonas vaginalis |
| 587 | Tsukahara | 1961 | Respiratory metabolism of trichomonad vaginalis |
| 588 | Heyworth | 1982 | Purine metabolism in Trichomonas vaginalis |
| 589 | Yarlett | 1988 | Polyamine biosynthesis and inhibition in Trichomonas vaginalis |
| 590 | Kleydman | 2004 | Production of ammonia by Tritrichomonas foetus and Trichomonas vaginalis |
| 591 | Chapman | 1999 | Hydrogen peroxide is a product of oxygen consumption by Trichomonas vaginalis |
| 592 | Ninomiya | 1952 | The metabolism of Trichomonas vaginalis, with comparative aspects of Trichomonads |
| 593 | Lehker | 1999 | Trichomonad invasion of the mucous layer requires adhesions, mucinases, and motility |
| 594 | Ginger | 2007 | Comparative genomics of trypanosome metabolism |
| 595 | Rogerson | 1980 | Catabolic metabolic in Trypanosoma cruzi |
| 596 | Bringaud | 2006 | Energy metabolism of typanosomatids: Adaptation to available carbon sources |
| 597 | Silber | 2002 | Active Transport of L-Proline in Trypanosoma cruzi |
| 598 | Taylor | 2008 | Validation of spermidine synthase as a drug target in African trypanosomes |
| 599 | Oliveira | 2000 | Inositol metabolism in Trypanosoma cruzi: Potential target for chemotherapy against chagas disease |
| 600 | Silber | 2005 | Amino Acid Metabolic Routes in Trypanosoma cruzi: Possible Therapeutic Targets Against Chagas’ Disease |
| 601 | Bosshard | 2002 | Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Gram-positive bacterium |
| 602 | Chung | 2009 | Varibaculum cambriense Infections in Hong Kong, China, 2006 |
| 603 | Denariaz | 1989 | A Halophilic Denitrifier, Bacillus halodenitrificans sp. nov. |
| 604 | Yoon | 2004 | Transfer of Bacillus halodenitrificans Denariaz et al. 1989 to the genus Virgibacillus as Virgibacillus halodenitrificans comb. nov. |
| 605 | Sharpe | 1972 | Some Slime-Forming Heterofermentative Species of the Genus Lactobacillus |
| 606 | Choi | 2002 | Weissella kimchii sp. nov., a novel lactic acid bacterium from kimchi |
| 607 | Tohno | 2014 | Aerococcus vaginalis sp. nov., isolated from the vaginal mucosa of a beef cow, and emended descriptions of Aerococcus suis, Aerococcus viridans, Aerococcus urinaeequi, Aerococcus urinaehominis, Aerococcus urinae, Aerococcus christensenii and Aerococcus sanguinicola |
| 608 | Deibel | 1960 | Comparative study of Gaffkya homari, Aerococcus viridans, tetrad-forming cocci from meat curing brines, and the genus Pediococcus |
| 609 | Sakamoto | 2009 | Butyricimonas synergistica gen. nov., sp. nov. and Butyricimonas virosa sp. nov., butyric acid-producing bacteria in the family ‘Porphyromonadaceae’ isolated from rat faeces |
| 610 | Sakamoto | 2014 | Butyricimonas faecihominis sp. nov. and Butyricimonas paravirosa sp. nov., isolated from human faeces, and emended description of the genus Butyricimonas |
| 611 | Cooke | 1927 | A type of urea-splitting bacterium found in the human intestinal tract |
| 612 | Collins | 1987 | Transfer of Brevibacterium ammoniagenes (Cooke and Keith) to the Genus Corynebacterium as Corynebacterium ammoniagenes comb. nov. |
| 613 | Wilkins | 1974 | Eubacterium plexicaudatum sp. nov., an Anaerobic Bacterium with a sub polar tuft of flagella, isolated from a mouse cecum |
| 614 | Dent | 1982 | Lactobacillus animalis sp. nov., a New Species ofLactobacillus from the Alimentary Canal of Animals |
| 615 | Wood | 1992 | Genera of Lactic Acid Bacteria |
| 616 | Schleifer | 1983 | Elevation of Staphylococcus sciuri subsp. lentus (Kloos et al.) to Species Status: Staphylococcus lentus (Kloos et al.) comb. nov. |
| 617 | Cook | 1994 | Emendation of the Description of Acidaminococcus fermentans, a trans-Aconitate- and Citrate-Oxidizing Bacterium |
| 618 | Miller | 1970 | Nutritional Requirements for Growth of Aerococcus viridans |
| 619 | Bergaust | 2008 | Transcription and activities of NOx reductases in Agrobacterium tumefaciens: the influence of nitrate, nitrite and oxygen availability; Agrobacterium tumefaciens C58 Uses ActR and FnrN To Control nirK and nor Expression |
| 620 | Weelink | 2008 | Alicycliphilus denitrificans gen. nov., sp. nov., a cyclohexanol-degrading, nitrate-reducing b-proteobacterium; Isolation and Characterization of Alicycliphilus denitrificans Strain BC, Which Grows on Benzene with Chlorate as the Electron Acceptor |
| 621 | Schwiertz | 2002 | Anaerostipes caccae gen. nov., sp. nov., a New Saccharolytic, Acetate-utilising, Butyrate-producing Bacterium from Human Faeces |
| 622 | Karlsson | 2011 | Prospects for systems biology and modeling of the gut microbiome |
| 623 | Sirotek | 2004 | Fermentation of pectin and glucose, and activity of pectin-degrading enzymes in the rabbit caecal bacterium Bacteroides caccae |
| 624 | Christophersen | 2013 | Xylo-Oligosaccharides and Inulin Affect Genotoxicity and Bacterial Populations Differently in a Human Colonic Simulator Challenged with Soy Protein |
| 625 | Gibson | 1993 | Sulphate reducing bacteria and hydrogen metabolism in the human large intestine |
| 626 | Tannock | 1977 | Characteristics of Bacteroides Isolates from the Cecum of Conventional Mice |
| 627 | Reilly | 1980 | The carbon dioxide requirements of anaerobic bacteria |
| 628 | Balamurugan | 2008 | Real-time polymerase chain reaction quantification ofspecific butyrate-producing bacteria,Desulfovibrio andEnterococcus faecalis in the feces of patients withcolorectal cancer |
| 629 | Vandamme | 1994 | New Perspectives in the Classification of the Flavobacteria: Description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter norn. rev. |
| 630 | Holmes | 1986 | Weeksella zoohelcum sp. nov. (Formerly Group IIj), from Human Clinical Specimens |
| 631 | Goodfellow | 2012 | Bergey's Manual of Systematic Bacteriology, Volume 5: The Actinobacteria |
| 632 | Stanton | 1991 | Reclassification of Treponema hyodysenteriae and Treponema innocens in a New Genus, Serpula gen. nov., as Serpula hyodysenteriae comb. nov. and Serpula innocens comb. nov. |
| 633 | Donohoe | 2011 | The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon |
| 634 | Fardeau | 2004 | Isolation from oil reservoirs of novel thermophilic anaerobes phylogenetically related to Thermoanaerobacter subterraneus: reassignment of T. subterraneus, Thermoanaerobacter yonseiensis, Thermoanaerobacter tengcongensis and Carboxydibrachium pacificum to Caldanaerobacter subterraneus gen. nov., sp. nov., comb. nov. as four novel subspecies |
| 635 | Colina | 1996 | Evidence for Degradation of Gastrointestinal Mucin by Candida albicans Secretory Aspartyl Proteinase |
| 636 | Leadbetter | 1979 | Capnocytophaga:New Genus of Gram-Negative Gliding Bacteria I. General Characteristics, Taxonomic Considerations and Significance |
| 637 | Pfennig | 1968 | Chlorobium phaeobacteroides nov. spec. und C. phaeovibrioides nov. spec. |
| 638 | Bergstein | 1979 | Uptake and metabolism of organic compounds by Chlorobium phaeobacteroides isolated from Lake Kinneret |
| 639 | Hofman | 1985 | Ecological significance of acetate assimilation by Chlorobium phaeobacteroides |
| 640 | Weber | 2006 | Anaerobic Nitrate-Dependent Iron(II) Bio-Oxidation by a Novel Lithoautotrophic Betaproteobacterium, Strain 2002 |
| 641 | Srivastava | 2003 | Understanding Bacteria (Table 6.2 Type of fermentative pathway in different bacteria) |
| 642 | Fuchs | 1957 | The Nutritional Requirements of Clostridium perfringens |
| 643 | Steer | 2001 | Clostridium hathewayi sp. nov., from Human Faeces |
| 644 | Hall | 1927 | Bacillus sordellii, a cause of malignant edema in man |
| 645 | Ramachandran | 2008 | Hydrogen production and end-product synthesis patterns by Clostridium termitidis strain CT1112 in batch fermentation cultures with cellobiose or a-cellulose |
| 646 | Salminen | 2004 | Lactic Acid Bacteria: Microbiological and Functional Aspects, Third Edition |
| 647 | Mackie | 1991 | Lipid metabolism in anaerobic ecosystems |
| 648 | Widdel | 1977 | A New Anaerobic, Sporing, Acetate-Oxidizing, Sulfate-Reducing Bacterium, Desulfotomaculum (emend.)acetoxidans |
| 649 | Junier | 2010 | The genome of the Gram-positive metal- and sulfate-reducing bacterium Desulfotomaculum reducens strain MI-1 |
| 650 | Gibson | 1988 | Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut |
| 651 | Sakaguchi | 2002 | Desulfovibrio magneticus sp. nov., a novel sulfate-reducing bacterium that produces intracellular single-domain-sized magnetite particles |
| 652 | Zellner | 1989 | Desulfovibrio simplex spec. nov., a new sulfate-reducing bacterium from a sour whey digester |
| 653 | Wolfe | 2005 | The acetate switch |
| 654 | Mackie | 1979 | Changes in Lactate-Producing and Lactate-Utilizing Bacteria in Relation to pH in the Rumen of Sheep During Stepwise Adaptation to a High-Concentrate Diet |
| 655 | Duncan | 2002 | Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. |
| 656 | Scheifinger | 1973 | Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium |
| 657 | Collins | 1994 | The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. |
| 658 | Van Trappen | 2004 | Flavobacterium degerlachei sp. nov., Flavobacterium frigoris sp. nov. and Flavobacterium micromati sp. nov., novel psychrophilic bacteria isolated from microbial mats in Antarctic lakes |
| 659 | Zhilina | 1992 | Ecology, Physiology and Taxonomy Studies on a New Taxon of Haloanaerobiaceae, Haloincola saccharolytica gen. nov., sp. nov. |
| 660 | Collins | 1993 | Phylogenetic Analysis of Some Aerococcus-Like Organisms from Clinical Sources: Description of Helcococcus kunzii gen. nov., sp. nov. |
| 661 | Vandamme | 1991 | Revision of Campylobacter, Helicobacter, and Wolinella Taxonomy: Emendation of Generic Descriptions and Proposal of Arcobacter gen. nov. |
| 662 | De Maesschalck | 2014 | Faecalicoccus acidiformans gen. nov., sp. nov., isolated from the chicken caecum, and reclassification of Streptococcus pleomorphus (Barnes et al. 1977), Eubacterium biforme (Eggerth 1935) and Eubacterium cylindroides (Cato et al. 1974) as Faecalicoccus pleomorphus comb. nov., Holdemanella biformis gen. nov., comb. nov. and Faecalitalea cylindroides gen. nov., comb. nov., respectively, within the family Erysipelotrichaceae |
| 663 | Hedberg | 2012 | Lachnoanaerobaculum gen. nov., a new genus in the Lachnospiraceae: characterization of Lachnoanaerobaculum umeaense gen. nov., sp. nov., isolated from the human small intestine, and Lachnoanaerobaculum orale sp. nov., isolated from saliva, and reclassification of Eubacterium saburreum (Prevot 1966) Holdeman and Moore 1970 as Lachnoanaerobaculum saburreum comb. nov. |
| 664 | Gummalla | 1999 | Tryptophan Catabolism by Lactobacillus casei and Lactobacillus helveticus Cheese Flavor Adjuncts |
| 665 | Ishii | 1999 | Identification of Compounds Causing Symbiotic Growth of Lactobacillus paracasei subsp. tolerans and Kluyveromyces marxianus var. lactis in Chigo, Inner Mongolia, China |
| 666 | Teusink | 2005 | In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: comparing predictions of nutrient requirements with those from growth experiments. |
| 667 | Golbach | 2007 | Adaptation of Lactobacillus rhamnosus riboflavin assay to microtiter plates |
| 668 | Rogosa | 1953 | Species differentiation of oral lactobacilli from man including description of Lactobacillus salivarius nov spec and lactobacillus Cellobiosus nov spec. |
| 669 | Embley | 1989 | Lactobacillus vaginalis sp. nov. from the Human Vagina |
| 670 | Flahaut | 2013 | Genome-scale metabolic model for Lactococcus lactis MG1363 and its application to the analysis of flavor formation |
| 671 | Liu | 2011 | Molecular Detection of Human Bacterial Pathogens |
| 672 | Haroun | 1982 | Reclassification of Bacteroides hypermegas (Harrison and Hansen) in a New Genus Megamonas, as Megamonas hypermegas comb. nov. |
| 673 | Bailey | 1982 | Reclassification of ‘Streptococcus pluton’ (White)in a new genus Melissococcus, as Melissococcus pluton nom. rev.; comb. nov. |
| 674 | Garrity | 2001 | Bergey's Manual of Systematic Bacteriology: Volume One : The Archaea and the Deeply Branching and Phototrophic Bacteria |
| 675 | Bordbar | 2010 | Insight into human alveolar macrophage and m. tuberculosis interactions via metabolic reconstructions |
| 676 | Pastink | 2009 | Genome-scale model of Streptococcus thermophilus LMG18311 for metabolic comparison of lactic acid bacteria. |
| 677 | Lee | 2013 | Oscillibacter ruminantium sp. nov., isolated from the rumen of Korean native cattle |
| 678 | Larocque | 2014 | A curated C. difficile strain 630 metabolic network: prediction of essential targets and inhibitors |
| 679 | Stanton | 1983 | Roseburia cecicola gen. nov., sp. nov., a Motile, Obligately Anaerobic Bacterium from a Mouse Cecum |
| 680 | Gradel | 1972 | Fermentation of isolated pectin and pectin from intact forages by pure cultures of rumen bacteria. |
| 681 | Yoon | 2013 | Shewanella spp. Use Acetate as an Electron Donor for Denitrification but Not Ferric Iron or Fumarate Reduction |
| 682 | Ahmed | 1988 | Nutritional requirements of shigellae for growth in a minimal medium. |
| 683 | Slobodkin | 1999 | Thermoanaerobater siderophilus sp. nov., a novel dissimilarity Fe(III)-reducing, anaerobic, thermophilic bacterium |
| 684 | Alain | 2010 | Thermodesulfatator atlanticus sp. nov., a thermophilic, chemolithoautotrophic, sulfate- reducing bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent |
| 685 | Jeanthon | 2002 | Thermodesulfobacterium hydrogeniphilum sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a deep-sea hydrothermal vent at Guaymas Basin, and emendation of the genus Thermodesulfobacterium |
| 686 | Nobre | 1996 | Transfer of Thermus ruber (Loginova et al. 1984), Themus silvans (Tenreiro et al. 1999, and Themus chliarophilus (Tenreiro et al. 1995) to Meiothemzus gen. nov. as Meiothermus ruber comb. nov., Meiothermus silvanus comb. nov., and Meiothermus chliarophilus comb. nov., Respectively, and Emendation of the Genus Thermus |
| 687 | Kane | 1978 | Early Detection and Identification of Trichophyton verrucosum |
| 688 | Hall | 2003 | Characterization of some actinomyces-like isolates from human clinical sources: description of Varibaculum cambriensis gen nov, sp nov. |
| 689 | Rogosa | 1964 | THE GENUS VEILLONELLA II. Nutritional Studies |
| 690 | Boyer | 2013 | Bile Formation and Secretion |
| 691 | Alnouti | 2008 | Quantitative-profiling of bile acids and their conjugates in mouse liver, bile, plasma, and urine using LC-MS/MS. |
| 692 | Birchenough | 2015 | New developments in goblet cell mucus secretion and function |
| 693 | Nakano | 1997 | Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. |
| 694 | Bezkorovainy | 1989 | Biochemistry and Physiology of Bifidobacteria |
| 695 | Duerden | 1989 | A comparison of Bacteroides ureolyticus isolates ftom different clinical sources |
| 696 | Sarantinopoulos | 2001 | Citrate metabolism by Enterococcus faecalis FAIR-E 229 |
| 697 | Kim | 2005 | Dissimilatory Fe(III) reduction by an electrochemically active lactic acid bacterium phylogenetically related to Enterococcus gallina um isolated from submerged soil |
| 698 | Jacobson | 1997 | Cellulase activity of Leishmania major in the sandfly vector and in culture. |
| 699 | Scherer | 1981 | Effect of trace elements and vitamins on the growth of Methanosarcina barkeri |
| 700 | Boden | 2012 | Emended description of the genus Methylophaga Janvier et al. 1985 |
| 701 | Bowman | 1993 | Revised Taxonomy of the Methanotrophs: Description of Methylobacter gen. nov., Emendation of Methylococcus, Validation of Methylosinus and Methylocystis Species, and a Proposal that the Family Methylococcaceae Includes Only the Group I Methanotrophs |
| 702 | Cato | 1983 | Synonymy of Strains of “Lactobacillus acidophilus” Group A2 (Johnson et 81. 1980) with the Type Strain of Lactobacillus crispatus (Brygoo and Aladame 1953) Moore and Holdeman 1970 |
| 703 | Chervaux | 2000 | Physiological study of Lactobacillus delbrueckii subsp. bulgaricus strains in a novel chemically defined medium. |
| 704 | Helanto | 2006 | Characterization of genes involved in fructose utilization by Lactobacillus fermentum |
| 705 | Francl | 2010 | The PTS transporters of Lactobacillus gasseri ATCC 33323 |
| 706 | Su | 2011 | Physiological and fermentation properties of Bacillus coagulans and a mutant lacking fermentative lactate dehydrogenase activity |
| 707 | De Clerck | 2004 | Polyphasic characterization of Bacillus coagulans strains, illustrating heterogeneity within this species, and emended description of the species. |
| 708 | Creczynski-Pasa | 2004 | Energetic metabolism of Chromobacterium violaceum |
| 709 | Takai | 2006 | Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas |
| 710 | Kopke | 2013 | Clostridium difficile is an autotrophic bacterial pathogen. |
| 711 | Christiansen | 1996 | Desulfitobacterium hafniense sp. nov., an Anaerobic, Reductively Dechlorinating Bacterium |
| 712 | Pfennig | 1976 | Desulfuromonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. |
| 713 | Atherly | 2014 | Genotypic and Phenotypic comparison of Bacteroides ovatus, B. thetaiotaomicron, and B. xylanisolvens isolates obtained from cow, goat, human, and pig feces |
| 714 | Akagi | 1967 | Electron carries for the phosphoroclastic reaction of Desulfovibrio desulfuricans. |
| 715 | Lewis | 2012 | Host sialoglycans and bacterial sialidases: a mucosal perspective |
| 716 | Reddy | 1983 | Wheat straw hemicelluloses: Composition and fermentation by human colon bacteroides |
| 717 | Postgate | 1966 | Classification of Desulfovibrio species, the nonsporulating sulfate-reducing bacteria. |
| 718 | Warren | 2005 | Biochemical differentiation and comparison of Desulfovibrio species and other phenotypically similar genera. |
| 719 | Halpern | 1961 | Utilization of L-glutamic and 2-oxoglutaric acid as sole sources of carbon by Escherichia coli. |
| 720 | Hebert | 2000 | Nutritional requirements and nitrogen-dependent regulation of proteinase activity of Lactobacillus helveticus CRL 1062. |
| 721 | Horn | 2005 | Growth of Lactobacillus plantarum in media containing hydrolysates of fish viscera |
| 722 | Kalyuzhnaya | 2006 | Methylotenera mobilis gen. nov., sp. nov., an obligately methylamine-utilizing bacterium within the family Methylophilaceae |
| 723 | Helaszek | 1991 | Cellobiose uptake and metabolism by Ruminococcus flavefaciens. |
| 724 | Tong | 2003 | Streptococcus oligofermentans sp. nov., a novel oral isolate from caries-free humans |
| 725 | Feio | 2004 | Desulfovibrio alaskensis sp. nov., a sulphate- reducing bacterium from a soured oil reservoir |
| 726 | Niamsup | 2003 | Lactobacillus thermotolerans sp. nov., a novel thermotolerant species isolated from chicken faeces |
| 727 | Le Gall | 1963 | A New Species of Desulfovibrio |
| 728 | Collins | 2000 | An unusual Streptococcus from human urine, Streptococcus urinalis sp. nov. |
| 729 | Giebel | 1990 | Isolation of Mycoplasma moatsii from the Intestine of Wild Norway Rats (Rattus norvegicus) |
| 730 | Baele | 2003 | Lactobacillus ingluviei sp. nov., isolated from the intestinal tract of pigeons |
| 731 | Chander | 2012 | Phenotypic and molecular characterization of a novel strongly hemolytic Brachyspira species, provisionally designated “Brachyspira hampsonii ” |
| 732 | Schlegel | 2000 | Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov. and Streptococcus infantarius subsp. coli subsp. nov., isolated from humans and food |
| 733 | Kloos | 1976 | Characterization of Staphylococcus sciuri sp.nov. and its subspecies |
| 734 | Robert | 2007 | Bacteroides cellulosilyticus sp. nov., a cellulolytic bacterium from the human gut microbial community |
| 735 | Chassard | 2007 | Characterization of the xylan-degrading microbial community from human faeces |
| 736 | Salyers | 1979 | Energy sources of major intestinal fermentative anaerobes |
| 737 | Attwood | 1996 | Clostridium proteoclasticum sp. nov., a Novel Proteolytic Bacterium from the Bovine Rumen |
| 738 | Gylswyk | 1986 | Description and Designation of a Neotype Strain of Eubacterium cellulosolvens (Cillobacterium cellulosolvens Bryant, Small, Bouma and Robinson) Holdeman and Moore |
| 739 | Lopez-Siles | 2011 | Cultured Representatives of Two Major Phylogroups of Human Colonic Faecalibacterium prausnitzii Can Utilize Pectin, Uronic Acids, and Host-Derived Substrates for Growth |
| 740 | Wallnofer | 1967 | Pathway of Propionate Formation in Bacteroides ruminicola |
| 741 | Zanoni | 1987 | Lactobacillus pentosus (Fred, Peterson, and Anderson) sp. nov., nom, rev. |
| 742 | Paterek | 1988 | Methanohalophilus mahii gen. nov. sp. nov. a Methylotrophic Halophilic Methanogen |
| 743 | Dedysh | 2005 | Methylocella Species Are Facultatively Methanotrophic |
| 744 | Cho | 2003 | Parvularcula bermudensis gen. nov., sp. nov., a marine bacterium that forms a deep branch in the a-Proteobacteria |
| 745 | Nicholson | 2012 | Host-Gut Microbiota Metabolic Interactions |
| 746 | Seetharam | 1982 | Absorption and Transport of Cobalamin (Vitamin B12) |
| 747 | Mekhjian | 1979 | Colonic Absorption of Unconjugated Bile Acids |
| 748 | Davila | 2013 | Re-print of “Intestinal luminal nitrogen metabolism: Role of the gut microbiota and consequences for the host” |
| 749 | Pal | 2005 | Hexachlorocyclohexane-degrading bacterial strains Sphingomonas paucimobilis B90A, UT26 and Sp+, having similar lin genes, represent three distinct species, Sphingobium indicum sp. nov., Sphingobium japonicum sp. nov. and Sphingobium francense sp. nov., and reclassification of [Sphingomonas] chungbukensis as Sphingobium chungbukense comb. nov. |
| 750 | Houghton | 2015 | Thermorudis pharmacophila sp. nov., a novel member of the class Thermomicrobia isolated from geothermal soil, and emended descriptions of Thermomicrobium roseum, Thermomicrobium carboxidum, Thermorudis peleae and Sphaerobacter thermophilus |
| 751 | Shiratori | 2009 | Clostridium clariflavum sp. nov. and Clostridium caenicola sp. nov., moderately thermophilic, cellulose-/cellobiose-digesting bacteria isolated from methanogenic sludge |
| 752 | Madden | 1982 | Isolation and Characterization of an Anaerobic, Cellulolytic Bacterium, Clostridium papyrosolvens sp. nov. |
| 753 | Warnick | 2002 | Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil |
| 754 | Motamedi | 1998 | Desulfovibrio aespoeensis sp. nov., a mesophilic sulfate-reducing bacterium from deep groundwater at Aspo hard rock laboratory, Sweden |
| 755 | Khelaifia | 2011 | Desulfovibrio piezophilus sp. nov., a piezophilic, sulfate-reducing bacterium isolated from wood falls in the Mediterranean Sea |
| 756 | Youn | 2009 | Characterization of the Dicarboxylate Transporter DctA in Corynebacterium glutamicum |
| 757 | Pokusaeva | 2011 | Carbohydrate metabolism in Bifidobacteria |
| 758 | Shah | 1988 | Proposal for Reclassification of Bacteroides asaccharolyticus, Bacteroides gingivalis, and Bacteroides endodontalis in a New Genus, Porphyromonas |
| 759 | Marshall | 1967 | Growth of Bacillus coagulans in Chemically Defined Media |
| 760 | Holländer | 1975 | Energy metabolism of some representatives of the Haemophilus group |
| 761 | Jensen | 1986 | Bacteroides pectinophilus sp. nov. and Bacteroides galacturonicus sp. nov.: Two Pectinolytic Bacteria from the Human Intestinal Tract |
| 762 | Kindberg | 1987 | Menaquinone production and utilization in germ-free rats after inoculation with specific organisms |
| 763 | Song | 2004 | Clostridium bartlettii sp. nov., isolatedfrom human faeces |
| 764 | Marquet | 2009 | Lactate has the potential to promote hydrogen sulphide formation in the human colon |
| 765 | Loubinoux | 2002 | Reclassification of the only species of the genus Desulfomonas, Desulfomonas pigra, as Desulfovibrio piger comb. nov. |
| 766 | Scott | 2013 | Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro |
| 767 | Shetty | 2018 | Reclassification of Eubacterium hallii as Anaerobutyricum hallii gen. nov., comb. nov., and description of Anaerobutyricum soehngenii sp. nov., a butyrate and propionate-producing bacterium from infant faeces |
| 768 | Cambell | 1966 | Desulfovibrio africanus sp. n., a New Dissimilatory Sulfate-reducing Bacterium |
| 769 | Abildgaard | 2006 | Desulfovibrio alkalitolerans sp. nov., a novel alkalitolerant, sulphate-reducing bacterium isolated from district heating water |
Collection and integration of metabolic information for NJC19 construction
Using the repertoire of the aforementioned microbial species, metabolic information primarily collected for NJC19 construction was direct experimental evidence of the import and export of small-molecule metabolites (e.g., sugars, vitamins, organic acids, and gases) and the degradation of macromolecules (e.g., starch, cellulose, hemicellulose, and mucin), reported in literature. For the small-molecule metabolites, we mostly considered primary metabolites, i.e., nutrients and metabolic byproducts associated with microbial growth or reproduction. In addition, literature sources that report the mRNA or protein expression for metabolite-specific enzymes or transporters were considered. When encountering the information of which chemical compounds are not able to be transported or degraded by a given organism, we recorded this negative information as well, as part of our collected data. Despite technically not being a part of the gut microbiota, some host cells directly affect or are affected by microbial metabolism, and thus were considered to be a functional extension of the microbial community. In a similar fashion to our previous network NJS16 for the human case, the specific mouse tissue cells that we considered were the intestinal absorptive cell, the mucin-secreting goblet cell, and the bile acid-secreting hepatocyte. Although the hepatocyte is not part of intestinal tissue, its secreted bile acids are utilized by microbes in the gut. In the case of the mouse intestinal absorptive cell, the information from a manually-curated, genome-scale metabolic model (iSS1393) was adopted33. All annotated metabolite transport or macromolecule degradation processes for different strains of the same species were consolidated for that species as its collective feature. Because degradation of a given macromolecule is often performed by multiple species in the gut, we considered the corresponding degradation products to be indirect export products of all species participating in that macromolecule degradation. In this work, we differentiated two macromolecules, xylan and mannan, from a “hemicellulose” macromolecule in NJS16, for more specific representation of their degradation products.
In parallel, we carefully re-examined the existing components of NJS1610,34, and removed the incorrectly-placed components and added new links found from literature, according to more specific and accurate information. The revised NJS16 was finally connected to the above mouse gut microbiota interaction network through the common chemical compounds shared by the both networks, to form the mouse and human gut microbiota interaction network, NJC19 (Fig. 1 and Table 1). NJC19 is provided in both human- and machine-readable forms, through Online-only Tables 1–5(XLSX files) and JavaScript Object Notation (JSON) files deposited in the Dryad Digital Repository35, respectively. In addition, the Cytoscape Session (cys) file of NJC19 is provided for interactive network visualization35.
Table 1.
Datasets used for the construction and validation of NJC19.
| Source | Processing | Data |
|---|---|---|
| Mouse fecal and cecal microbiome data (Online-only Table 1). | Application of taxonomic analysis tools to the microbiome data (Methods). | Output flies of the taxonomic analysis tools, which include the lists of identified microbial taxa and their relative abundances35. |
| Lists of identified microbial taxa and their relative abundances from mouse fecal and cecal microbiome data35. | Selection of microbial species in metagenome samples based on the frequency of the species occurrence across the samples (Methods). | List of selected microbial species in the metagenome samples35. |
| Literature (Online-only Table 2) and NJS1634. | Manual collection of metabolic information from literature, and revision of NJS16 (Methods). | Interaction network of microbial species/host cells mediated by metabolic compounds, NJC19 (Online-only Tables 3–5 and the corresponding JSON files35). |
| Mouse microbiome and metabolome data4,37,38. | Extraction of taxonomic compositions and metabolite levels (see Technical Validation and Fig. 3 legend). | Taxonomic compositions35, and microbial producer and metabolite levels for NJC19 validation (Fig. 3). |
Online-only Table 4.
List of small-molecule metabolites and macromolecules in NJC19.
| Compound name | KEGG compound identifier | |
|---|---|---|
| D-Glucose (Glucose) | C00031 | |
| Sucrose | C00089 | |
| D-Fructose (Fructose) | C00095 | |
| Glycerol | C00116 | |
| D-Ribose (Ribose) | C00121 | |
| D-Galactose | C00124 | |
| N-Acetyl-D-Glucosamine (N-Acetylglucosamine) | C00140 | |
| D-Mannose (Mannose) | C00159 | |
| D-Xylose (Xylose) | C00181 | |
| Cellobiose | C00185 | |
| D-Glucuronic acid (D-Glucuronate) | C00191 | |
| Maltose | C00208 | |
| D-Arabinose (L-Arabinose, Arabinose, L-Arabinopyranose, L-Arabinofuranose) | C00216, C00259 | |
| Lactose | C00243 | |
| L-Sorbose (Sorbose) | C00247 | |
| Isomaltose | C00252 | |
| D-Gluconate (D-Gluconic acid, Gluconate) | C00257 | |
| D-Glycerate (Glycerate, [R]-Glycerate, Glyceric acid) | C00258 | |
| N-Acetylneuraminic acid (N-acetylneuraminate, Neu5Ac, Sialic acid) | C00270 | |
| D-Xylulose (Xylulose, L-Xylulose) | C00310, C00312 | |
| D-Glucosamine (Glucosamine) | C00329 | |
| D-Galacturonate | C00333 | |
| Xylitol | C00379 | |
| D-Mannitol (Mannitol) | C00392 | |
| Ribitol (Adonitol) | C00474 | |
| D-Lyxose (L-Lyxose) | C00476, C01508 | |
| Raffinose | C00492 | |
| Erythritol | C00503 | |
| L-Rhamnose (Rhamnose, D-Rhamnose) | C00507, C01684 | |
| L-Arabitol (L-Arabinitol, D-Arabitol) | C00532, C01904 | |
| D-Tagaturonate | C00558 | |
| N-Acetyl-D-mannosamine | C00645 | |
| Dextrin (Maltotriose, Maltodextrin, Maltohexaose, Maltotetraose, Maltooligosaccharide) | C00721, C01835, C01935, C01936, C02052 | |
| L-Idonate | C00770 | |
| D-Sorbitol (D-Glucitol, Sorbitol, Glucitol, L-Sorbitol, Sorbitol) | C00794, C01722 | |
| D-Tagatose | C00795 | |
| D-Fructuronate | C00905 | |
| L-Fucose | C01019 | |
| N-Acetylgalactosamine (N-Acetyl-D-galactosamine) | C01074 | |
| Trehalose | C01083 | |
| Stachyose | C01613 | |
| Galactitol (Dulcitol) | C01697 | |
| Palatinose (Isomaltulose) | C01742 | |
| Cellotetraose (Cellohexaose, Cellopentaose, Cellotriose) | C02013, C06217, C06218, C06219 | |
| D-Galactosamine (Galactosamine) | C02262 | |
| Melibiose | C05402 | |
| D-Psicose | C06468 | |
| L-iduronate | C06472 | |
| D-Turanose | C19636 | |
| FOS (Fructooligosaccharide) | - | |
| XOS (Xylooligosaccharide) | - | |
| L-Glutamate (L-Glutamic acid, Glutamate, D-Glutamate) | C00025, C00217 | |
| L-Glycine (Glycine) | C00037 | |
| L-Alanine (D-Alanine, Alanine) | C00041, C00133 | |
| L-Lysine (Lysine, D-Lysine) | C00047, C00739 | |
| L-Aspartate (Aspartate, D-Aspartate) | C00049, C00402 | |
| L-Arginine (Arginine) | C00062 | |
| L-Glutamine (D-Glutamine, Glutamine) | C00064, C00819 | |
| L-Serine (Serine, D-Serine) | C00065, C00740 | |
| L-Methionine (D-Methionine) | C00073, C00855 | |
| L-Ornithine (Ornithine) | C00077 | |
| L-Tryptophan (Tryptophan) | C00078 | |
| L-Phenylalanine (Phenylalanine, D-Phenylalanine) | C00079, C02265 | |
| L-Tyrosine (Tyrosine) | C00082 | |
| L-Cysteine (Cysteine, D-Cysteine) | C00097, C00793 | |
| beta-alanine (3-Aminopropionate) | C00099 | |
| L-Leucine (Leucine) | C00123 | |
| L-Histidine (Histidine) | C00135 | |
| L-Proline (Proline, D-Proline) | C00148, C00763 | |
| L-Asparagine (Asparagine) | C00152 | |
| L-Valine (Valine, D-Valine) | C00183, C06417 | |
| L-Threonine (Threonine) | C00188 | |
| L-Homoserine | C00263 | |
| L-Carnitine (D-Carnitine) | C00318, C15025 | |
| L-Isoleucine (Isoleucine) | C00407 | |
| L-Selenocysteine | C05688 | |
| Uracil | C00106 | |
| Adenine | C00147 | |
| Thymine | C00178 | |
| Adenosine | C00212 | |
| Thymidine | C00214 | |
| Guanine | C00242 | |
| Inosine | C00294 | |
| Uridine | C00299 | |
| Cytosine | C00380 | |
| Xanthine | C00385 | |
| Guanosine | C00387 | |
| Cytidine | C00475 | |
| Ascorbic acid (Vitamin C, L-Ascorbate, Ascorbate) | C00072 | |
| Choline | C00114 | |
| Biotin (Vitamin B7) | C00120 | |
| Adenosylcobalamin (Vitamin B12, Cobamide coenzyme, Cob[II]alamin, Vitamin B12r, Cob[I]alamin, Vitamin B12s, Aquacobalamin, Cobalamin [III]) | C00194 | |
| Pyridoxal (Vitamin B6, Pyridoxine, Pyridoxamine, Vitamin B6) | C00250, C00314, C00534 | |
| Niacin (Vitamin B3, Nicotinic acid, Nicotinate, Nicotinamide, Vitamin B3) | C00153, C00253 | |
| Riboflavin (Vitamin B2) | C00255 | |
| Thiamine (Vitamin B1, Thiamin) | C00378 | |
| Retinol (Vitamin A) | C00473 | |
| Folic acid (Folate, Vitamin B9) | C00504 | |
| Retinoate | C00777 | |
| Menaquinone (Vitamin K2) | C00828 | |
| Pantothenic acid (Vitamin B5, Pantothenate) | C00864 | |
| Phylloquinone (Vitamin K1) | C02059 | |
| alpha-Tocopherol (Vitamin E, alpha-Tocotrienol) | C02477, C14155, C14153 | |
| Arachidonate | C00219 | |
| Cholecalciferol (Vitamin D3, Vitamin D2) | C05443, C05441 | |
| Palmitate (Palmitic acid, Hexadecanoic acid, Hexadecanoate) | C00249 | |
| 1,2-diacylglycerol (1-acylglycerol, Monoacylglycerol, Monoglyceride, Monoacylglycerol, Diacylglycerol) | C00641, C01885, C00165 | |
| Oleic acid ([9Z]-Octadecenoic acid) | C00712 | |
| Lipoic acid (Lipoate) | C00725 | |
| Stearic acid (Stearate, Octadecanoate) | C01530 | |
| Decanoic acid (Decanoate) | C01571 | |
| Caproate (Hexanoate) | C01585 | |
| Linoleic acid (Omega-6 fatty acid) | C01595 | |
| Pelargonic acid (Pelargonate, Nonanoic acid) | C01601 | |
| Octanoate (Caprylic acid) | C06423 | |
| Myristic acid (Tetradecanoic acid, Tetradecanoate) | C06424 | |
| Arachidate (Arachidic acid) | C06425 | |
| alpha-Linolenic acid (Omega-3 fatty acid, eicosapentaenoic acid) | C06427, C12083 | |
| Acetate | C00033 | |
| Propanoate (Propionate) | C00163 | |
| Butyrate | C00246 | |
| Valerate (Pentanoic acid, Pentanoate) | C00803 | |
| Isobutyrate (2-Methylpropanoic acid) | C02632 | |
| Isovalerate (3-Methylbutanoic acid) | C08262 | |
| Methanol | C00132 | |
| Inositol (myo-Inositol, Inositol 1-phosphate, Myo-inositol phosphate) | C00137, C01177 | |
| Phenol | C00146 | |
| Acetoin (3-hydroxybutanone, acetyl methyl carbinol, [R]-Acetoin) | C00466, C00810 | |
| Ethanol | C00469 | |
| Benzyl alcohol | C00556 | |
| 1,2-propanediol (Propene diol, Propylene glycol, [R]-1,2-propanediol, [R]-propane-1,2-diol, [S]-1,2-propanediol, [S]-propane-1,2-diol) | C00583, C02912, C02917 | |
| 1,2-Ethanediol (Ethylene glycol) | C01380 | |
| Isopropanol (2-Propanol) | C01845 | |
| 1,3-Propanediol | C02457 | |
| [R,R]-2,3-Butanediol ([R,R]-Butanediol, [S,S]-2,3-Butanediol, [S,S]-Butanediol) | C03044, C03046 | |
| Propanol (n-propanol, 1-Propanol) | C05979 | |
| Isobutyl alcohol (isobutanol, 2-Methyl-1-propanol) | C14710 | |
| Butanol | C06142 | |
| Pentanol | C16834 | |
| Formaldehyde | C00067 | |
| Acetaldehyde | C00084 | |
| Benzaldehyde | C00261 | |
| Propanal | C00479 | |
| Putrescine | C00134 | |
| Ethanolamine (Phosphatidylethanolamine) | C00189, C00350 | |
| Methylamine (Monomethylamine) | C00218 | |
| Spermidine | C00315 | |
| Histamine | C00388 | |
| Tryptamine | C00398 | |
| Tyramine | C00483 | |
| Dimethylamine | C00543 | |
| Trimethylamine | C00565 | |
| Spermine | C00750 | |
| Trimethylamine N-oxide (Trimethylamine-N-oxide) | C01104 | |
| Cadaverine | C01672 | |
| Phenylethylamine | C02455 | |
| Butylamine | C18706 | |
| Urea | C00086 | |
| Acetone | C00207 | |
| Indole | C00463 | |
| Toluene | C01455 | |
| p-Cresol (4-methylphenol, 4-cresol) | C01468 | |
| Anthranilate (o-amino-benzoic acid) | C00108 | |
| Benzoate | C00180 | |
| 4-Aminobenzoate (para-aminobenzoic acid, PABA, p-Aminobenzoate) | C00568 | |
| Indole-3-acetate (Indoleacetate) | C00954 | |
| Ferulate | C01494 | |
| Hippurate | C01586 | |
| 3-phenylpropionic acid (Phenylpropanoate, 3-phenylpropionate) | C05629 | |
| Vanillate | C06672 | |
| Phenylacetate | C07086 | |
| Pyruvate | C00022 | |
| alpha-ketoglutarate (2-oxoglutarate) | C00026 | |
| Oxaloacetate | C00036 | |
| Succinate | C00042 | |
| Glyoxylate (Glyoxylic acid, Glyoxalate) | C00048 | |
| Formate | C00058 | |
| 2-Oxobutyrate (Alpha-ketobutyrate, 2-Oxobutanoate) | C00109 | |
| L-Malate ([S]-Malate, L-Malic acid, Malate, D-Malate, [R]-Malate) | C00149, C00497 | |
| Fumarate | C00122 | |
| Citrate | C00158 | |
| Glycolate | C00160 | |
| L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate) | C00186, C00256 | |
| Oxalate | C00209 | |
| Taurine | C00245 | |
| Orotic acid (Orotate) | C00295 | |
| 4-Aminobutyrate (GABA) | C00334 | |
| Malonate | C00383 | |
| cis-Aconitate (trans-Aconitate, trans-Aconitic acid) | C00417, C02341 | |
| 5-Aminovalerate | C00431 | |
| Glutarate | C00489 | |
| Itaconate | C00490 | |
| Shikimate | C00493 | |
| meso-Tartrate (meso-Tartaric acid, Tartrate, L-tartrate, L-Tartaric acid) | C00552, C00898 | |
| Cholic acid (Cholate) | C00695 | |
| Betaine (Glycine betaine) | C00719 | |
| Crotonate (2-Butenoate, 2-Butenoic acid, Crotonic acid, 3-Methylacrylic acid) | C01771 | |
| Glycocholate | C01921 | |
| 2-Aminobutyric acid (2-Aminobutyrate) | C02261, C02356 | |
| Chenodeoxycholic acid (Chenodeoxycholate) | C02528 | |
| Taurolithocholate | C02592 | |
| Pimelate | C02656 | |
| Lithocholic acid | C03990 | |
| Deoxycholic acid | C04483 | |
| Taurocholate | C05122 | |
| Isethionate | C05123 | |
| Taurodeoxycholate | C05463 | |
| Glycodeoxycholate | C05464 | |
| Taurochenodeoxycholate | C05465 | |
| Glycochenodeoxycholate | C05466 | |
| Adipate | C06104 | |
| Glycolithocholate | C15557 | |
| 2-methylbutyrate (2-methylbutanoic acid) | C18319 | |
| H2O2 | C00027 | |
| Zn2+ (Zinc) | C00038 | |
| Sulfate (Sulfuric acid, Sulfite) | C00059, C00094 | |
| Cu2+ (Copper) | C00070 | |
| Ca2+ (Calcium) | C00076 | |
| K+ (Potassium) | C00238 | |
| Nitrate (NO3-) | C00244 | |
| Bicarbonate (HCO3-, H2CO3, Carbonic acid, Carbonate) | C00288 | |
| Mg2+ (Magnesium) | C00305 | |
| Thiosulfate | C00320 | |
| Dimethyl sulfide (Methyl sulfide) | C00580 | |
| Cl− (Chloride) | C00698 | |
| I− (Iodide) | C00708 | |
| Na+ (Sodium) | C01330 | |
| Selenite | C05684 | |
| Fe3+ (Ferric ion, Fe2+, Ferrous ion) | C14819, C14818 | |
| Sulfur (Elemental sulfur) | C00087 | |
| CO2 | C00011 | |
| NH3 (Ammonia, NH4+, Ammonium) | C00014 | |
| Carbon monoxide (CO) | C00237 | |
| H2 (Hydrogen) | C00282 | |
| H2S (HS-) | C00283 | |
| Nitric Oxide (NO) | C00533 | |
| Nitrite (NO2-) | C00088 | |
| N2 | C00697 | |
| Methane (CH4) | C01438 | |
| Dichloromethane (Methylene chloride) | C02271 | |
| DNA RNA Polynucleotide | C00039, C00046, C00419 | |
| Starch (Amylopectin, Amylose, 1,4-alpha-D-Glucan, Pullulan, Resistant starch, Glycogen) | C00317, C00369, C00480, C00718, C00182 | |
| Polypeptide | C00403 | |
| Triglyceride | C00422 | |
| Chitin | C00461 | |
| Mannan | C00464 | |
| Hyaluronan (Hyaluronate, Hyaluronic acid) | C00518 | |
| Arabinogalactan | C00569 | |
| Chondroitin 4-sulfate (Chondroitin 6-sulfate) | C00634, C00635 | |
| Pectin | C00714 | |
| Cellulose (Beta-D-glucan) | C00760 | |
| Fructan (Inulin, Levan) | C01355, C03323, C06215 | |
| Galactan | C05796 | |
| Mucin (Mucus Glycoprotein) | C02705 | |
| * | Hemicellulose, not specified | — |
| Xylan | C00707 | |
| Arabinan | C02474 | |
| Hyodeoxycholic acid | — | |
| alpha-Muricholic acid | C17647 | |
| beta-Muricholic acid | C17726 | |
| omega-Muricholic acid | C17727 | |
| Tauro-b-muricholic acid | — | |
| Tauroursodeoxycholic acid | C16868 | |
| Ursodeoxycholic acid | C07880 | |
| Indole-3-lactic acid | — | |
| Indole-3-carboxaldehyde | C08493 | |
| 7-Oxodeoxycholic acid | — | |
| 7-Oxolithocholic acid | — | |
| Carnosine | C00386 | |
| Glycylsarcosine | — | |
| Prolylglycine | — | |
| Glycyl-Phenylalanine | — | |
| Levomefolic acid | — | |
| H+ | C00080 | |
| Leucyl-leucine | C11332 | |
| Alanylalanine | — | |
| Glycylproline | — | |
| Glycyl-leucine | C02155 | |
| Leucylglycine | — | |
| L-Dopa | C00355 | |
| Lysophosphatidylcholine (1-Lysophosphatidylcholine, 2-Lysophosphatidylcholine) | C04230, C04233 | |
| Cholesterol | C00187 | |
| Glycylglycine | C02037 | |
*Denotes hemicellulose that is not yet specified as xylan, mannan, or others in NJC19.
Data Records
Our network NJC19 offers the reference map of the mammalian gut microbiota and chemical compound relationships (from 769 literature sources), which can be adapted for each context of mouse, human, and humanized mouse microbiomes. In NJC19, one set of nodes corresponds to organisms (i.e., microbial species and host cells), while the other set corresponds to chemical compounds (i.e., small-molecule metabolites or macromolecules). An organism and a chemical compound are connected if the organism imports, exports, or degrades the chemical compound. NJC19 comprises 838 microbial species (766 bacteria, 53 archaea, and 19 eukaryotes) in the mouse and human gut, 6 mouse and human cell types metabolically interacting with those microbes, and 283 chemical compounds (266 small molecules and 17 macromolecules)—all interconnected by 8,224 small-molecule transport or macromolecule degradation events. In addition, NJC19 provides information on small molecules and macromolecules that are reportedly not transportable or degradable by certain organisms—described through 912 negative metabolic associations. These negative associations can be particularly useful for the curation of automatically-generated metabolic models, which may include false-positive transport reactions derived from inaccurate genome annotations.
Figure 2a shows the overall phylogenetic composition of microbial species included in NJC19. To overview the network topology of NJC19, we counted the number of metabolites imported or exported by each microbial species. Each species in the network imports 5.8 and exports 3.5 metabolites on average, and the probability that a given species imports (or exports) k metabolites follows an exponential distribution P(k) ∝ e−rk (r ≈ 0.2 and 0.3 for the import and export cases, respectively; see Fig. 2b,c). Bacteroides thetaiotaomicron is one of the most promiscuous species, importing 33 and exporting 29 metabolites. Conversely, for each metabolite, we counted the number of species importing or exporting that metabolite. The probability that a given metabolite is imported (or exported) by k species follows a power-law distribution P(k) ∝ k−γ (γ ≈ 1.4 for both import and export cases; Fig. 2d,e), which is much broader than the above exponential distributions. Among metabolites, glucose and acetate are the most frequent substrate and product, respectively, and are imported by 303 species (36.2% of the total species) and exported by 461 species (55.0% of the total species). In contrast, an average metabolite is imported by 21.8 species and exported by 13.0 species. Collectively, metabolites are highly uneven in terms of the ranges of their transporting species.
Fig. 2.
Microbial taxonomic composition and network structural properties of NJC19. (a) Fraction of microbial species in the network, which belong to each domain (left) or phylum (right). The right panel shows several phyla with the largest fractions in each domain. Both left and right panels show bacteria in blue, archaea in tan, and eukaryotes in green. (b,c) The vertical axis represents the distribution of the probability P(k) that a given microbial species imports (b) or exports (c) k metabolites on the horizontal axis. (d,e) The vertical axis represents the distribution of the probability P(k) that a given metabolite is imported (d) or exported (e) by k species on the horizontal axis.
As noted above, the full details of NJC19 are available in both human- and machine-readable forms, through Online-only Table 1 and JSON files in the Dryad Digital Repository35, respectively. As noted above, the cys file of NJC19 is available for network visualization35, and can be accessed by Cytoscape v3.7.236.
Online-only Table 1 shows the detailed sources of mouse metagenome and 16S rRNA gene sequence data that were used for microbial species identification when we constructed NJC19. Online-only Table 2 shows the literature sources of metabolic information used for NJC19 construction. Online-only Table 3 shows the list of microbial species and host cell types in NJC19. The name of each microbial species is presented with the NCBI taxonomy ID. Online-only Table 4 includes the list of small-molecule metabolites and macromolecules in NJC19. The name of each compound is presented with the KEGG compound ID. Supplementary Table 1 provides all the metabolic associations between chemical compounds and microbial species/host cells in NJC19, along with their literature sources. These metabolic associations include both positive and negative associations (see above). Online-only Table 5 shows the degradation products of macromolecules in NJC19.
Online-only Table 3.
List of microbial species and host cell types in NJC19.
| NCBI ID | Species name | Synonym | |
|---|---|---|---|
| * | 31971 | Absiella dolichum | Eubacterium dolichum |
| 1511 | Acetoanaerobium sticklandii | Clostridium sticklandii | |
| 438 | Acetobacter pasteurianus | ||
| * | 33952 | Acetobacterium woodii | |
| 28187 | Acetohalobium arabaticum | ||
| 2148 | Acholeplasma laidlawii | ||
| 72556 | Achromobacter piechaudii | ||
| 85698 | Achromobacter xylosoxidans | ||
| 905 | Acidaminococcus fermentans | ||
| 53635 | Acidimicrobium ferrooxidans | ||
| 524 | Acidiphilium cryptum | ||
| 33059 | Acidithiobacillus caldus | ||
| 920 | Acidithiobacillus ferrooxidans | ||
| 28049 | Acidothermus cellulolyticus | ||
| 80867 | Acidovorax avenae | ||
| 47920 | Acidovorax delafieldii | ||
| 471 | Acinetobacter calcoaceticus | ||
| 40216 | Acinetobacter radioresistens | ||
| 67854 | Actinobacillus succinogenes | ||
| 103618 | Actinomyces coleocanis | ||
| 544580 | Actinomyces oris | ||
| 103621 | Actinomyces urogenitalis | ||
| 1656 | Actinomyces viscosus | ||
| 40567 | Actinosynnema mirum | ||
| * | 1377 | Aerococcus viridans | |
| 219314 | Aeromicrobium marinum | ||
| 644 | Aeromonas hydrophila | ||
| 645 | Aeromonas salmonicida | ||
| 56636 | Aeropyrum pernix | ||
| 714 | Aggregatibacter actinomycetemcomitans | ||
| 358 | Agrobacterium tumefaciens | ||
| 239935 | Akkermansia muciniphila | ||
| 179636 | Alicycliphilus denitrificans | ||
| * | 214856 | Alistipes finegoldii | |
| * | 626932 | Alistipes indistinctus | |
| * | 1118061 | Alistipes obesi | Bacteroidales bacterium ph8 |
| * | 328813 | Alistipes onderdonkii | |
| 28117 | Alistipes putredinis | ||
| * | 1288121 | Alistipes senegalensis | |
| 328814 | Alistipes shahii | ||
| * | 908612 | Alistipes sp. HGB5 | |
| * | 671218 | Alloprevotella rava | Prevotella sp. oral taxon 302 |
| 76122 | Alloprevotella tannerae | ||
| 81468 | Aminobacterium colombiense | ||
| * | 81412 | Aminomonas paucivorans | |
| 39488 | Anaerobutyricum hallii | Eubacterium hallii | |
| 33029 | Anaerococcus hydrogenalis | ||
| 33032 | Anaerococcus lactolyticus | ||
| * | 1287640 | Anaerococcus obesiensis | |
| 33034 | Anaerococcus prevotii | ||
| 33036 | Anaerococcus tetradius | ||
| 33037 | Anaerococcus vaginalis | ||
| 105841 | Anaerostipes caccae | ||
| * | 649756 | Anaerostipes hadrus | Eubacterium hadrum |
| 169435 | Anaerotruncus colihominis | ||
| 2234 | Archaeoglobus fulgidus | ||
| 1382 | Atopobium parvulum | ||
| 1383 | Atopobium rimae | ||
| 82135 | Atopobium vaginae | ||
| 354 | Azotobacter vinelandii | ||
| 1390 | Bacillus amyloliquefaciens | ||
| 1392 | Bacillus anthracis | ||
| 1452 | Bacillus atrophaeus | ||
| 1413 | Bacillus cellulosilyticus | ||
| 1396 | Bacillus cereus | ||
| 79880 | Bacillus clausii | ||
| 1398 | Bacillus coagulans | ||
| 408580 | Bacillus coahuilensis | ||
| 86665 | Bacillus halodurans | ||
| 1402 | Bacillus licheniformis | ||
| 1404 | Bacillus megaterium | ||
| 1405 | Bacillus mycoides | Bacillus weihenstephanensis | |
| 79885 | Bacillus pseudofirmus | ||
| 64104 | Bacillus pseudomycoides | ||
| 1408 | Bacillus pumilus | ||
| 85683 | Bacillus selenitireducens | ||
| 1423 | Bacillus subtilis | ||
| 1428 | Bacillus thuringiensis | ||
| * | 376804 | Bacteroides barnesiae | |
| 47678 | Bacteroides caccae | ||
| 246787 | Bacteroides cellulosilyticus | ||
| * | 626929 | Bacteroides clarus | |
| 310298 | Bacteroides coprocola | ||
| 387090 | Bacteroides coprophilus | ||
| * | 151276 | Bacteroides coprosuis | |
| 357276 | Bacteroides dorei | ||
| 28111 | Bacteroides eggerthii | ||
| * | 674529 | Bacteroides faecis | |
| 338188 | Bacteroides finegoldii | ||
| * | 626930 | Bacteroides fluxus | |
| 817 | Bacteroides fragilis | ||
| * | 376806 | Bacteroides gallinarum | |
| 290053 | Bacteroides helcogenes | ||
| 329854 | Bacteroides intestinalis | ||
| * | 204516 | Bacteroides massiliensis | |
| * | 291645 | Bacteroides nordii | |
| * | 626931 | Bacteroides oleiciplenus | |
| 28116 | Bacteroides ovatus | ||
| 384638 | Bacteroides pectinophilus | ||
| 310297 | Bacteroides plebeius | ||
| * | 392838 | Bacteroides propionicifaciens | |
| * | 310300 | Bacteroides pyogenes | |
| 376805 | Bacteroides salanitronis | ||
| * | 291644 | Bacteroides salyersiae | |
| * | 469589 | Bacteroides sp. 2_1_33B | |
| 46506 | Bacteroides stercoris | ||
| 818 | Bacteroides thetaiotaomicron | ||
| 820 | Bacteroides uniformis | ||
| 821 | Bacteroides vulgatus | ||
| 371601 | Bacteroides xylanisolvens | ||
| * | 1015 | Bergeyella zoohelcum | |
| * | 999 | Bernardetia litoralis | Flexibacter litoralis |
| 1680 | Bifidobacterium adolescentis | ||
| 1683 | Bifidobacterium angulatum | ||
| 28025 | Bifidobacterium animalis | ||
| 1681 | Bifidobacterium bifidum | ||
| 1685 | Bifidobacterium breve | ||
| 1686 | Bifidobacterium catenulatum | ||
| 1689 | Bifidobacterium dentium | ||
| 78342 | Bifidobacterium gallicum | ||
| 216816 | Bifidobacterium longum | ||
| 28026 | Bifidobacterium pseudocatenulatum | ||
| * | 1694 | Bifidobacterium pseudolongum | |
| 35833 | Bilophila wadsworthia | ||
| 1322 | Blautia hansenii | ||
| 53443 | Blautia hydrogenotrophica | ||
| 40520 | Blautia obeum | Ruminococcus obeum | |
| * | 33035 | Blautia producta | Ruminococcus productus |
| * | 1287055 | Brachyspira hampsonii | |
| * | 159 | Brachyspira hyodysenteriae | |
| * | 13264 | Brachyspira innocens | |
| * | 84377 | Brachyspira intermedia | |
| * | 84378 | Brachyspira murdochii | |
| * | 52584 | Brachyspira pilosicoli | |
| 375 | Bradyrhizobium japonicum | ||
| 1393 | Brevibacillus brevis | ||
| 235 | Brucella abortus | ||
| 29459 | Brucella melitensis | ||
| 29461 | Brucella suis | ||
| 9 | Buchnera aphidicola | ||
| 152480 | Burkholderia ambifaria | ||
| 95486 | Burkholderia cenocepacia | ||
| 292 | Burkholderia cepacia | ||
| 337 | Burkholderia glumae | ||
| 87883 | Burkholderia multivorans | ||
| 60552 | Burkholderia vietnamiensis | ||
| * | 544644 | Butyricimonas synergistica | |
| 45851 | Butyrivibrio crossotus | ||
| 831 | Butyrivibrio fibrisolvens | ||
| 43305 | Butyrivibrio proteoclasticus | ||
| * | 911092 | Caldanaerobacter subterraneus | |
| 31899 | Caldicellulosiruptor bescii | ||
| 44001 | Caldicellulosiruptor saccharolyticus | ||
| * | 693075 | Caldisericum exile | |
| * | 200415 | Caldisphaera lagunensis | |
| 477976 | Calditerrivibrio nitroreducens | ||
| * | 187145 | Caldithrix abyssi | |
| * | 515264 | Caloramator australicus | |
| 291048 | Caminibacter mediatlanticus | ||
| * | 195 | Campylobacter coli | |
| * | 199 | Campylobacter concisus | |
| * | 200 | Campylobacter curvus | |
| * | 196 | Campylobacter fetus | |
| 824 | Campylobacter gracilis | ||
| * | 76517 | Campylobacter hominis | |
| 197 | Campylobacter jejuni | ||
| * | 201 | Campylobacter lari | |
| * | 203 | Campylobacter rectus | |
| * | 204 | Campylobacter showae | |
| * | 28080 | Campylobacter upsaliensis | |
| * | 827 | Campylobacter ureolyticus | |
| * | 5476 | Candida albicans | |
| * | 42374 | Candida dubliniensis | |
| * | 5482 | Candida tropicalis | |
| 186490 | Candidatus Baumannia cicadellinicola | ||
| * | 28188 | Capnocytophaga canimorsus | |
| * | 28189 | Capnocytophaga cynodegmi | |
| * | 1017 | Capnocytophaga gingivalis | |
| * | 45242 | Capnocytophaga granulosa | |
| * | 1018 | Capnocytophaga ochracea | |
| * | 1019 | Capnocytophaga sputigena | |
| * | 300419 | Catellicoccus marimammalium | |
| 100886 | Catenibacterium mitsuokai | ||
| 1711 | Cellulomonas flavigena | ||
| 29360 | Cellulosilyticum lentocellum | Clostridium lentocellum | |
| * | 188913 | Cetobacterium somerae | |
| 1096 | Chlorobium phaeobacteroides | ||
| 1108 | Chloroflexus aurantiacus | ||
| 536 | Chromobacterium violaceum | ||
| 545 | Citrobacter koseri | ||
| 67825 | Citrobacter rodentium | ||
| 133448 | Citrobacter youngae | ||
| 1496 | Clostridioides difficile | Clostridium difficile, Peptoclostridium difficile | |
| 1488 | Clostridium acetobutylicum | ||
| * | 1137848 | Clostridium arbusti | |
| 333367 | Clostridium asparagiforme | ||
| * | 84023 | Clostridium autoethanogenum | |
| 1520 | Clostridium beijerinckii | ||
| 208479 | Clostridium bolteae | ||
| 1491 | Clostridium botulinum | ||
| 1492 | Clostridium butyricum | ||
| 217159 | Clostridium carboxidivorans | ||
| * | 36834 | Clostridium celatum | |
| 1493 | Clostridium cellulovorans | ||
| * | 358743 | Clostridium citroniae | |
| * | 1531 | Clostridium clostridioforme | |
| * | 179628 | Clostridium colicanis | |
| 89152 | Clostridium hiranonis | ||
| 89153 | Clostridium hylemonae | ||
| * | 1522 | Clostridium innocuum | |
| 1534 | Clostridium kluyveri | ||
| 1535 | Clostridium leptum | ||
| 1538 | Clostridium ljungdahlii | ||
| 84026 | Clostridium methylpentosum | ||
| 1542 | Clostridium novyi | Clostridium oedematiens | |
| * | 1501 | Clostridium pasteurianum | |
| 1502 | Clostridium perfringens | ||
| * | 169679 | Clostridium saccharobutylicum | |
| 84030 | Clostridium saccharolyticum | ||
| * | 36745 | Clostridium saccharoperbutylacetonicum | |
| * | 84031 | Clostridium sartagoforme | Clostridium sartagoformum |
| 29347 | Clostridium scindens | ||
| * | 97138 | Clostridium sp. ASF356 | |
| * | 97139 | Clostridium sp. ASF502 | |
| 1509 | Clostridium sporogenes | ||
| 1512 | Clostridium symbiosum | ||
| 1513 | Clostridium tetani | ||
| * | 219748 | Clostridium tunisiense | |
| * | 1519 | Clostridium tyrobutyricum | |
| * | 45497 | Clostridium ultunense | |
| 74426 | Collinsella aerofaciens | ||
| 147207 | Collinsella intestinalis | ||
| 147206 | Collinsella stercoris | ||
| 285 | Comamonas testosteroni | ||
| 116085 | Coprococcus catus | ||
| 410072 | Coprococcus comes | ||
| 33043 | Coprococcus eutactus | ||
| * | 1697 | Corynebacterium ammoniagenes | |
| 1717 | Corynebacterium diphtheriae | ||
| 1718 | Corynebacterium glutamicum | ||
| 1747 | Cutibacterium acnes | Propionibacterium acnes | |
| 985 | Cytophaga hutchinsonii | ||
| 1299 | Deinococcus radiodurans | ||
| 118000 | Denitrovibrio acetiphilus | ||
| 453230 | Desulfarculus baarsii | ||
| 259354 | Desulfatibacillum alkenivorans | ||
| * | 49338 | Desulfitobacterium hafniense | |
| 2296 | Desulfobacterium autotrophicum | ||
| * | 28223 | Desulfobacula toluolica | |
| 894 | Desulfobulbus propionicus | ||
| * | 873 | Desulfocurvibacter africanus | Desulfovibrio africanus |
| 58138 | Desulfofarcimen acetoxidans | ||
| * | 293256 | Desulfohalovibrio alkalitolerans | Desulfovibrio alkalitolerans |
| 1565 | Desulfotomaculum nigrificans | Desulfotomaculum carboxydivorans | |
| 59610 | Desulfotomaculum reducens | ||
| * | 58180 | Desulfovibrio alaskensis | |
| 876 | Desulfovibrio desulfuricans | ||
| 878 | Desulfovibrio fructosivorans | ||
| * | 879 | Desulfovibrio gigas | |
| * | 191026 | Desulfovibrio hydrothermalis | |
| * | 889 | Desulfovibrio longus | |
| 184917 | Desulfovibrio magneticus | ||
| * | 63560 | Desulfovibrio oxyclinae | |
| 901 | Desulfovibrio piger | ||
| 880 | Desulfovibrio salexigens | ||
| * | 42252 | Desulfovibrio termitidis | |
| 881 | Desulfovibrio vulgaris | ||
| * | 427923 | Desulfurivibrio alkaliphilus | |
| * | 64160 | Desulfurobacterium thermolithotrophum | |
| 891 | Desulfuromonas acetoxidans | ||
| * | 427926 | Dethiobacter alkaliphilus | |
| 218538 | Dialister invisus | ||
| 39486 | Dorea formicigenerans | Eubacterium formicigenerans | |
| 88431 | Dorea longicatena | ||
| * | 156974 | Dysgonomonas gadei | |
| * | 163665 | Dysgonomonas mossii | |
| 84112 | Eggerthella lenta | ||
| * | 5802 | Eimeria tenella | |
| * | 1117645 | Elizabethkingia anophelis | |
| * | 238 | Elizabethkingia meningoseptica | |
| * | 247 | Empedobacter brevis | |
| * | 312279 | Emticicia oligotrophica | |
| 69218 | Enterobacter cancerogenus | Enterobacter taylorae | |
| 550 | Enterobacter cloacae | ||
| * | 57732 | Enterococcus asini | |
| * | 33945 | Enterococcus avium | |
| * | 317735 | Enterococcus caccae | |
| 37734 | Enterococcus casseliflavus | ||
| * | 44008 | Enterococcus cecorum | |
| * | 1355 | Enterococcus columbae | |
| * | 44009 | Enterococcus dispar | |
| * | 53345 | Enterococcus durans | |
| 1351 | Enterococcus faecalis | ||
| 1352 | Enterococcus faecium | ||
| 1353 | Enterococcus gallinarum | ||
| * | 160453 | Enterococcus gilvus | |
| * | 155618 | Enterococcus haemoperoxidus | |
| * | 1354 | Enterococcus hirae | |
| 246144 | Enterococcus italicus | ||
| * | 71451 | Enterococcus malodoratus | |
| * | 155617 | Enterococcus moraviensis | |
| * | 53346 | Enterococcus mundtii | |
| * | 160454 | Enterococcus pallens | |
| * | 154621 | Enterococcus phoeniculicola | |
| * | 71452 | Enterococcus raffinosus | |
| * | 41997 | Enterococcus saccharolyticus | |
| * | 1356 | Enterococcus sulfureus | |
| * | 112904 | Enterococcus villorum | Enterococcus porcinus |
| * | 1547 | Erysipelatoclostridium ramosum | Clostridium ramosum |
| 1648 | Erysipelothrix rhusiopathiae | ||
| 208962 | Escherichia albertii | Escherichia/Shigella albertii | |
| 562 | Escherichia coli | ||
| 564 | Escherichia fergusonii | Escherichia/Shigella fergusonii | |
| 253239 | Ethanoligenens harbinense | ||
| * | 35517 | Eubacterium brachy | |
| 29322 | Eubacterium cellulosolvens | ||
| 39485 | Eubacterium eligens | ||
| 1736 | Eubacterium limosum | ||
| * | 97253 | Eubacterium plexicaudatum | |
| * | 39490 | Eubacterium ramulus | |
| 39491 | Eubacterium rectale | ||
| 51123 | Eubacterium saphenum | ||
| 39492 | Eubacterium siraeum | ||
| 39496 | Eubacterium ventriosum | ||
| * | 39498 | Eubacterium yurii | |
| 853 | Faecalibacterium prausnitzii | ||
| 833 | Fibrobacter succinogenes | ||
| * | 143361 | Filifactor alocis | |
| 1260 | Finegoldia magna | Peptostreptococcus magnus | |
| * | 271155 | Flavobacterium antarcticum | |
| * | 55197 | Flavobacterium branchiophilum | |
| * | 510946 | Flavobacterium cauense | |
| * | 996 | Flavobacterium columnare | |
| * | 1341165 | Flavobacterium enshiense | |
| * | 229204 | Flavobacterium frigoris | |
| 986 | Flavobacterium johnsoniae | ||
| * | 1401027 | Flavobacterium limnosediminis | |
| * | 96345 | Flavobacterium psychrophilum | |
| * | 498301 | Flavobacterium rivuli | |
| * | 329186 | Flavobacterium saliperosum | |
| * | 70993 | Flexithrix dorotheae | |
| 849 | Fusobacterium gonidiaformans | Fusobacterium gonidiiformans | |
| 850 | Fusobacterium mortiferum | Clostridium rectum | |
| * | 859 | Fusobacterium necrophorum | |
| 851 | Fusobacterium nucleatum | ||
| 860 | Fusobacterium periodonticum | ||
| * | 854 | Fusobacterium russii | |
| 861 | Fusobacterium ulcerans | ||
| 856 | Fusobacterium varium | ||
| * | 84136 | Gemella bergeri | |
| * | 29391 | Gemella morbillorum | |
| * | 84135 | Gemella sanguinis | |
| 33940 | Geobacillus thermodenitrificans | Bacillus thermodenitrificans | |
| 225194 | Geobacter bemidjiensis | ||
| 313985 | Geobacter lovleyi | ||
| 28232 | Geobacter metallireducens | ||
| 35554 | Geobacter sulfurreducens | ||
| 351604 | Geobacter uraniireducens | ||
| 442 | Gluconobacter oxydans | Gluconobacter uchimurae | |
| 364410 | Granulibacter bethesdensis | ||
| 727 | Haemophilus influenzae | ||
| 729 | Haemophilus parainfluenzae | ||
| * | 656519 | Halanaerobium hydrogeniformans | |
| * | 2331 | Halanaerobium praevalens | |
| * | 43595 | Halanaerobium saccharolyticum | |
| 2238 | Haloarcula marismortui | ||
| 2242 | Halobacterium salinarum | ||
| * | 42422 | Halobacteroides halobius | |
| 2246 | Haloferax volcanii | ||
| * | 40091 | Helcococcus kunzii | |
| * | 212 | Helicobacter acinonychis | |
| * | 37372 | Helicobacter bilis | |
| * | 56877 | Helicobacter bizzozeronii | |
| * | 123841 | Helicobacter canadensis | |
| * | 29419 | Helicobacter canis | |
| * | 138563 | Helicobacter cetorum | |
| * | 213 | Helicobacter cinaedi | |
| * | 214 | Helicobacter felis | |
| * | 215 | Helicobacter fennelliae | |
| * | 32025 | Helicobacter hepaticus | |
| * | 398626 | Helicobacter macacae | |
| * | 217 | Helicobacter mustelae | |
| * | 35818 | Helicobacter pullorum | |
| 210 | Helicobacter pylori | ||
| * | 104628 | Helicobacter suis | |
| * | 157268 | Helicobacter winghamensis | |
| * | 1279027 | Hippea alviniae | |
| * | 84405 | Hippea maritima | |
| * | 1735 | Holdemanella biformis | Eubacterium biforme |
| 61171 | Holdemania filiformis | ||
| 35830 | Hungateiclostridium cellulolyticum | Acetivibrio cellulolyticus | |
| * | 288965 | Hungateiclostridium clariflavum | Clostridium clariflavum |
| 1515 | Hungateiclostridium thermocellum | Clostridium thermocellum, Ruminiclostridium thermocellum | |
| 154046 | Hungatella hathewayi | Clostridium hathewayi | |
| 53399 | Hyphomicrobium denitrificans | ||
| * | 591197 | Ignavibacterium album | |
| 160233 | Ignicoccus hospitalis | ||
| 167642 | Ilyobacter polytropus | ||
| 261299 | Intestinibacter bartlettii | ||
| * | 43995 | Johnsonella ignava | |
| 92945 | Ketogulonicigenium vulgare | ||
| 502 | Kingella denitrificans | ||
| 573 | Klebsiella pneumoniae | ||
| 244366 | Klebsiella variicola | ||
| 467210 | Lachnoanaerobaculum saburreum | Eubacterium saburreum | |
| * | 140626 | Lachnobacterium bovis | |
| 66219 | Lachnoclostridium phytofermentans | Clostridium phytofermentans | |
| * | 89059 | Lactobacillus acidipiscis | |
| 1579 | Lactobacillus acidophilus | ||
| 83683 | Lactobacillus amylolyticus | ||
| 1604 | Lactobacillus amylovorus | Lactobacillus sobrius | |
| * | 1605 | Lactobacillus animalis | |
| 227943 | Lactobacillus antri | ||
| 1580 | Lactobacillus brevis | ||
| 1581 | Lactobacillus buchneri | ||
| 1582 | Lactobacillus casei | ||
| 181675 | Lactobacillus coleohominis | ||
| * | 1610 | Lactobacillus coryniformis | |
| 47770 | Lactobacillus crispatus | ||
| * | 28038 | Lactobacillus curvatus | |
| 1584 | Lactobacillus delbrueckii | ||
| * | 137357 | Lactobacillus equi | |
| * | 420645 | Lactobacillus equicursoris | |
| * | 1612 | Lactobacillus farciminis | |
| 1613 | Lactobacillus fermentum | ||
| * | 640331 | Lactobacillus florum | |
| * | 1614 | Lactobacillus fructivorans | |
| 1596 | Lactobacillus gasseri | ||
| * | 227942 | Lactobacillus gastricus | |
| * | 1203069 | Lactobacillus gigeriorum | |
| 1587 | Lactobacillus helveticus | ||
| 1588 | Lactobacillus hilgardii | ||
| * | 1203033 | Lactobacillus hominis | |
| 147802 | Lactobacillus iners | Lactobacillus sp. 7_1_47FAA | |
| * | 148604 | Lactobacillus ingluviei | Lactobacillus thermotolerans |
| 109790 | Lactobacillus jensenii | ||
| 33959 | Lactobacillus johnsonii | ||
| * | 267818 | Lactobacillus kefiranofaciens | |
| * | 176292 | Lactobacillus malefermentans | |
| * | 1618 | Lactobacillus mali | |
| * | 97478 | Lactobacillus mucosae | |
| * | 1622 | Lactobacillus murinus | |
| 1632 | Lactobacillus oris | ||
| 1597 | Lactobacillus paracasei | ||
| * | 872327 | Lactobacillus pasteurii | |
| * | 1589 | Lactobacillus pentosus | |
| 1590 | Lactobacillus plantarum | Lactobacillus arizonensis | |
| * | 449659 | Lactobacillus pobuzihii | |
| 1598 | Lactobacillus reuteri | ||
| 47715 | Lactobacillus rhamnosus | ||
| * | 231049 | Lactobacillus rossiae | |
| 1623 | Lactobacillus ruminis | ||
| * | 228229 | Lactobacillus saerimneri | |
| 1599 | Lactobacillus sakei | ||
| 1624 | Lactobacillus salivarius | ||
| * | 1625 | Lactobacillus sanfranciscensis | |
| * | 1231337 | Lactobacillus shenzhenensis | |
| * | 97137 | Lactobacillus sp. ASF360 | |
| * | 152335 | Lactobacillus suebicus | |
| 227945 | Lactobacillus ultunensis | ||
| 1633 | Lactobacillus vaginalis | ||
| * | 194326 | Lactobacillus versmoldensis | |
| * | 238015 | Lactobacillus vini | |
| 1358 | Lactococcus lactis | ||
| * | 5671 | Leishmania infantum | |
| * | 5664 | Leishmania major | |
| 40542 | Leptotrichia buccalis | ||
| 157692 | Leptotrichia goodfellowii | ||
| 157688 | Leptotrichia hofstadii | ||
| * | 157691 | Leptotrichia shahii | |
| * | 157687 | Leptotrichia wadei | |
| 33964 | Leuconostoc citreum | ||
| * | 1244 | Leuconostoc gelidum | |
| 136609 | Leuconostoc kimchii | ||
| 1245 | Leuconostoc mesenteroides | ||
| 1642 | Listeria innocua | ||
| 1639 | Listeria monocytogenes | ||
| 168384 | Marvinbryantia formatexigens | ||
| * | 437897 | Megamonas funiformis | |
| 158847 | Megamonas hypermegale | ||
| * | 491921 | Megamonas rupellensis | |
| 187326 | Megasphaera micronuciformis | ||
| * | 1134405 | Melioribacter roseus | |
| * | 33970 | Melissococcus plutonius | |
| 381 | Mesorhizobium loti | ||
| 43687 | Metallosphaera sedula | ||
| 83816 | Methanobrevibacter ruminantium | ||
| 2173 | Methanobrevibacter smithii | ||
| 83171 | Methanocaldococcus fervens | ||
| 67760 | Methanocaldococcus infernus | ||
| 2190 | Methanocaldococcus jannaschii | ||
| * | 667126 | Methanocaldococcus villosus | |
| 73913 | Methanocaldococcus vulcanius | ||
| 29291 | Methanococcoides burtonii | ||
| 42879 | Methanococcus aeolicus | ||
| 39152 | Methanococcus maripaludis | ||
| 2187 | Methanococcus vannielii | ||
| 2188 | Methanococcus voltae | ||
| 83984 | Methanocorpusculum labreanum | ||
| 2198 | Methanoculleus marisnigri | ||
| 2322 | Methanohalobium evestigatum | ||
| 2176 | Methanohalophilus mahii | ||
| 54120 | Methanolacinia petrolearia | ||
| 2320 | Methanopyrus kandleri | ||
| 2214 | Methanosarcina acetivorans | ||
| 2208 | Methanosarcina barkeri | ||
| 2209 | Methanosarcina mazei | ||
| 2317 | Methanosphaera stadtmanae | ||
| 475088 | Methanosphaerula palustris | ||
| 2203 | Methanospirillum hungatei | ||
| 145263 | Methanothermobacter marburgensis | ||
| 145262 | Methanothermobacter thermautotrophicus | Methanobacterium thermautotrophicum | |
| 155863 | Methanothermococcus okinawensis | ||
| 2180 | Methanothermus fervidus | ||
| 2224 | Methanothrix thermoacetophila | ||
| * | 213185 | Methanotorris formicicus | |
| * | 2189 | Methanotorris igneus | |
| 511746 | Methylacidiphilum infernorum | ||
| 105560 | Methylibium petroleiphilum | ||
| 405 | Methylobacillus flagellatus | ||
| 114616 | Methylobacterium nodulans | ||
| 199596 | Methylocella silvestris | ||
| 414 | Methylococcus capsulatus | ||
| 392484 | Methylophaga thiooxydans | Methylophaga thiooxidans | |
| 408 | Methylorubrum extorquens | Methylobacterium dichloromethanicum, Methylobacterium chloromethanicum | |
| 426 | Methylosinus trichosporium | ||
| 359408 | Methylotenera mobilis | ||
| 1270 | Micrococcus luteus | ||
| 52226 | Mitsuokella multacida | ||
| 1525 | Moorella thermoacetica | ||
| 480 | Moraxella catarrhalis | ||
| 1764 | Mycobacterium avium | ||
| 1769 | Mycobacterium leprae | ||
| 1773 | Mycobacterium tuberculosis | ||
| 1772 | Mycolicibacterium smegmatis | ||
| 2110 | Mycoplasma agalactiae | ||
| * | 45363 | Mycoplasma alkalescens | |
| 47687 | Mycoplasma alligatoris | ||
| * | 2094 | Mycoplasma arginini | |
| 2111 | Mycoplasma arthritidis | ||
| * | 51363 | Mycoplasma auris | |
| 28903 | Mycoplasma bovis | ||
| 2095 | Mycoplasma capricolum | ||
| * | 114881 | Mycoplasma columbinum | |
| 45361 | Mycoplasma conjunctivae | ||
| 50052 | Mycoplasma crocodyli | ||
| * | 171284 | Mycoplasma cynos | |
| 2115 | Mycoplasma fermentans | ||
| 2096 | Mycoplasma gallisepticum | ||
| 2097 | Mycoplasma genitalium | ||
| 29501 | Mycoplasma haemofelis | ||
| 2098 | Mycoplasma hominis | ||
| 2099 | Mycoplasma hyopneumoniae | ||
| 2100 | Mycoplasma hyorhinis | ||
| * | 2116 | Mycoplasma iowae | |
| 2105 | Mycoplasma leachii | ||
| * | 171287 | Mycoplasma moatsii | |
| 2118 | Mycoplasma mobile | ||
| 2102 | Mycoplasma mycoides | ||
| 28227 | Mycoplasma penetrans | ||
| 2104 | Mycoplasma pneumoniae | ||
| 2107 | Mycoplasma pulmonis | ||
| * | 2123 | Mycoplasma putrefaciens | |
| 2109 | Mycoplasma synoviae | ||
| * | 51365 | Mycoplasma yeatsii | |
| * | 1183151 | Myroides injenensis | |
| * | 76832 | Myroides odoratimimus | |
| * | 256 | Myroides odoratus | |
| * | 27289 | Naumovozyma dairenensis | |
| 484 | Neisseria flavescens | ||
| 485 | Neisseria gonorrhoeae | ||
| 488 | Neisseria mucosa | ||
| * | 490 | Neisseria sicca | |
| * | 28449 | Neisseria subflava | |
| * | 626933 | Odoribacter laneus | |
| 28118 | Odoribacter splanchnicus | Bacteroides splanchnicus | |
| * | 351091 | Oscillibacter valericigenes | |
| 847 | Oxalobacter formigenes | ||
| 1464 | Paenibacillus larvae | ||
| 1406 | Paenibacillus polymyxa | ||
| * | 1505 | Paeniclostridium sordellii | Clostridium sordellii |
| 823 | Parabacteroides distasonis | Bacteroides distasonis | |
| * | 328812 | Parabacteroides goldsteinii | Bacteroides goldsteinii |
| 387661 | Parabacteroides johnsonii | ||
| 46503 | Parabacteroides merdae | Bacteroides merdae | |
| 148447 | Paraburkholderia phymatum | Burkholderia phymatum | |
| * | 1490 | Paraclostridium bifermentans | Clostridium bifermentans |
| 266 | Paracoccus denitrificans | ||
| 208216 | Parvularcula bermudensis | ||
| 1254 | Pediococcus acidilactici | ||
| 19 | Pelobacter carbinolicus | ||
| 29543 | Pelobacter propionicus | ||
| 110500 | Pelotomaculum thermopropionicum | ||
| * | 507750 | Peptoniphilus duerdenii | |
| 54005 | Peptoniphilus harei | Peptostreptococcus harei | |
| * | 33030 | Peptoniphilus indolicus | |
| 33031 | Peptoniphilus lacrimalis | Peptostreptococcus lacrimalis | |
| * | 1175452 | Peptoniphilus rhinitidis | |
| * | 1111134 | Peptoniphilus sp. BV3C26 | Clostridiales bacterium BV3C26 |
| * | 671216 | Peptoniphilus sp. oral taxon 836 | |
| * | 1268254 | Peptoniphilus timonensis | |
| 1261 | Peptostreptococcus anaerobius | ||
| 341694 | Peptostreptococcus stomatis | ||
| 82076 | Picrophilus torridus | ||
| * | 5821 | Plasmodium berghei | |
| * | 5833 | Plasmodium falciparum | |
| * | 5850 | Plasmodium knowlesi | |
| * | 5855 | Plasmodium vivax | |
| * | 5861 | Plasmodium yoelii | |
| * | 37453 | Polaribacter franzmannii | |
| * | 531 | Polaribacter irgensii | |
| 28123 | Porphyromonas asaccharolytica | ||
| 837 | Porphyromonas gingivalis | ||
| 419005 | Prevotella amnii | ||
| 242750 | Prevotella bergensis | ||
| 28125 | Prevotella bivia | ||
| 77095 | Prevotella bryantii | ||
| 28126 | Prevotella buccae | ||
| 28127 | Prevotella buccalis | ||
| 165179 | Prevotella copri | ||
| 28130 | Prevotella disiens | ||
| 189722 | Prevotella marshii | ||
| 28132 | Prevotella melaninogenica | ||
| 282402 | Prevotella multiformis | ||
| 28134 | Prevotella oralis | ||
| 28135 | Prevotella oris | ||
| 839 | Prevotella ruminicola | ||
| 228604 | Prevotella salivae | ||
| 386414 | Prevotella timonensis | ||
| 28137 | Prevotella veroralis | ||
| 1744 | Propionibacterium freudenreichii | ||
| * | 294710 | Proteiniphilum acetatigenes | |
| 584 | Proteus mirabilis | ||
| 102862 | Proteus penneri | ||
| 182210 | Pseudodesulfovibrio aespoeensis | Desulfovibrio aespoeensis | |
| * | 879567 | Pseudodesulfovibrio piezophilus | Desulfovibrio piezophilus |
| 106588 | Pseudoflavonifractor capillosus | Bacteroides capillosus | |
| 287 | Pseudomonas aeruginosa | ||
| 312306 | Pseudomonas entomophila | ||
| 294 | Pseudomonas fluorescens | ||
| 300 | Pseudomonas mendocina | ||
| 303 | Pseudomonas putida | ||
| 29438 | Pseudomonas savastanoi | ||
| 316 | Pseudomonas stutzeri | ||
| 317 | Pseudomonas syringae | ||
| * | 251 | Psychroflexus gondwanensis | |
| 638849 | Pyramidobacter piscolens | ||
| 13773 | Pyrobaculum aerophilum | ||
| 181486 | Pyrobaculum calidifontis | ||
| 29292 | Pyrococcus abyssi | ||
| 2261 | Pyrococcus furiosus | ||
| 53953 | Pyrococcus horikoshii | ||
| 329 | Ralstonia pickettii | ||
| 384 | Rhizobium leguminosarum | ||
| 1061 | Rhodobacter capsulatus | ||
| 1076 | Rhodopseudomonas palustris | ||
| 1085 | Rhodospirillum rubrum | ||
| 29549 | Rhodothermus marinus | Rhodothermus obamensis | |
| * | 301301 | Roseburia hominis | |
| 166486 | Roseburia intestinalis | ||
| 360807 | Roseburia inulinivorans | ||
| 2434 | Roseobacter denitrificans | ||
| * | 29355 | Ruminiclostridium cellobioparum | Clostridium termitidis, Ruminiclostridium cellobioparum subsp. termitidis |
| 1521 | Ruminiclostridium cellulolyticum | Clostridium cellulolyticum | |
| 29362 | Ruminiclostridium papyrosolvens | Clostridium papyrosolvens | |
| 1264 | Ruminococcus albus | ||
| 40518 | Ruminococcus bromii | ||
| 1265 | Ruminococcus flavefaciens | ||
| 33038 | Ruminococcus gnavus | ||
| 46228 | Ruminococcus lactaris | ||
| 33039 | Ruminococcus torques | ||
| 2287 | Saccharolobus solfataricus | Sulfolobus solfataricus | |
| * | 4932 | Saccharomyces cerevisiae | |
| 28901 | Salmonella enterica | ||
| 1660 | Schaalia odontolytica | Actinomyces odontolyticus | |
| 671224 | Selenomonas artemidis | ||
| 135080 | Selenomonas flueggei | ||
| 135083 | Selenomonas noxia | ||
| * | 971 | Selenomonas ruminantium | |
| 69823 | Selenomonas sputigena | ||
| 192073 | Shewanella denitrificans | ||
| 24 | Shewanella putrefaciens | ||
| 621 | Shigella boydii | ||
| 622 | Shigella dysenteriae | ||
| 623 | Shigella flexneri | Escherichia/Shigella flexneri | |
| 624 | Shigella sonnei | ||
| 382 | Sinorhizobium meliloti | ||
| 63612 | Sodalis glossinidius | ||
| 2057 | Sphaerobacter thermophilus | ||
| 332056 | Sphingobium japonicum | ||
| 154 | Spirochaeta thermophila | ||
| * | 216933 | Spiroplasma chrysopicola | |
| * | 216936 | Spiroplasma diminutum | |
| * | 2134 | Spiroplasma melliferum | |
| * | 216945 | Spiroplasma syrphidicola | |
| * | 2145 | Spiroplasma taiwanense | |
| 1280 | Staphylococcus aureus | ||
| 29388 | Staphylococcus capitis | ||
| 1281 | Staphylococcus carnosus | ||
| 1282 | Staphylococcus epidermidis | ||
| 1283 | Staphylococcus haemolyticus | ||
| 1290 | Staphylococcus hominis | ||
| * | 42858 | Staphylococcus lentus | |
| 28035 | Staphylococcus lugdunensis | ||
| * | 45972 | Staphylococcus pasteuri | |
| 283734 | Staphylococcus pseudintermedius | ||
| 29385 | Staphylococcus saprophyticus | ||
| 1292 | Staphylococcus warneri | ||
| 40324 | Stenotrophomonas maltophilia | ||
| 1311 | Streptococcus agalactiae | ||
| 1328 | Streptococcus anginosus | ||
| 113107 | Streptococcus australis | ||
| * | 439220 | Streptococcus caballi | |
| * | 1329 | Streptococcus canis | |
| * | 76860 | Streptococcus constellatus | |
| * | 1333 | Streptococcus criceti | |
| 45634 | Streptococcus cristatus | Streptococcus oligofermentans | |
| * | 102886 | Streptococcus didelphis | |
| 1317 | Streptococcus downei | ||
| 1334 | Streptococcus dysgalactiae | ||
| * | 155680 | Streptococcus entericus | |
| 1336 | Streptococcus equi | ||
| 1335 | Streptococcus equinus | Streptococcus bovis | |
| * | 1345 | Streptococcus ferus | |
| 315405 | Streptococcus gallolyticus | ||
| 1302 | Streptococcus gordonii | ||
| * | 439219 | Streptococcus henryi | |
| * | 380397 | Streptococcus ictaluri | |
| 102684 | Streptococcus infantarius | ||
| 68892 | Streptococcus infantis | ||
| * | 1346 | Streptococcus iniae | |
| * | 1338 | Streptococcus intermedius | |
| * | 150055 | Streptococcus lutetiensis | Streptococcus infantarius subsp. coli |
| * | 1339 | Streptococcus macacae | |
| * | 59310 | Streptococcus macedonicus | Streptococcus gallolyticus subsp. macedonicus |
| * | 269666 | Streptococcus marimammalium | |
| * | 313439 | Streptococcus massiliensis | |
| * | 400065 | Streptococcus merionis | |
| * | 229549 | Streptococcus minor | |
| 28037 | Streptococcus mitis | ||
| 1309 | Streptococcus mutans | ||
| 1303 | Streptococcus oralis | Streptococcus tigurinus, Streptococcus oralis subsp. tigurinus | |
| * | 114652 | Streptococcus orisratti | |
| * | 82806 | Streptococcus ovis | |
| 1318 | Streptococcus parasanguinis | Streptococcus gallolyticus subsp. pasteurianus | |
| * | 1348 | Streptococcus parauberis | |
| * | 197614 | Streptococcus pasteurianus | |
| 68891 | Streptococcus peroris | ||
| 1313 | Streptococcus pneumoniae | ||
| * | 1340 | Streptococcus porcinus | |
| * | 257758 | Streptococcus pseudopneumoniae | |
| 361101 | Streptococcus pseudoporcinus | ||
| 1314 | Streptococcus pyogenes | ||
| * | 1341 | Streptococcus ratti | |
| 1304 | Streptococcus salivarius | ||
| 1305 | Streptococcus sanguinis | ||
| * | 1310 | Streptococcus sobrinus | |
| * | 1169673 | Streptococcus sp. GMD4S | |
| 1307 | Streptococcus suis | ||
| 1308 | Streptococcus thermophilus | ||
| * | 55085 | Streptococcus thoraltensis | |
| 1349 | Streptococcus uberis | ||
| * | 149016 | Streptococcus urinalis | |
| 1343 | Streptococcus vestibularis | ||
| 1911 | Streptomyces griseus | ||
| 1916 | Streptomyces lividans | ||
| 1938 | Streptomyces viridochromogenes | ||
| 214851 | Subdoligranulum variabile | ||
| * | 78120 | Succinispira mobilis | |
| 2285 | Sulfolobus acidocaldarius | ||
| 39766 | Sulfurimonas denitrificans | ||
| 40545 | Sutterella wadsworthensis | ||
| 119484 | Syntrophobacter fumaroxidans | ||
| 51197 | Syntrophobotulus glycolicus | ||
| 863 | Syntrophomonas wolfei | ||
| 86170 | Syntrophothermus lipocalidus | ||
| 316277 | Syntrophus aciditrophicus | ||
| * | 36841 | Terrisporobacter glycolicus | Clostridium glycolicum |
| * | 1071379 | Tetrapisispora blattae | |
| * | 113608 | Tetrapisispora phaffii | |
| * | 29323 | Thermoanaerobacter brockii | |
| * | 1757 | Thermoanaerobacter ethanolicus | |
| * | 1125974 | Thermoanaerobacter indiensis | |
| * | 108150 | Thermoanaerobacter italicus | |
| * | 583357 | Thermoanaerobacter mathranii | |
| * | 496866 | Thermoanaerobacter pseudethanolicus | |
| * | 106578 | Thermoanaerobacter siderophilus | |
| * | 1516 | Thermoanaerobacter thermohydrosulfuricus | Clostridium thermohydrosulfuricum |
| * | 46354 | Thermoanaerobacter wiegelii | |
| * | 28896 | Thermoanaerobacterium saccharolyticum | |
| * | 1517 | Thermoanaerobacterium thermosaccharolyticum | |
| * | 29329 | Thermoanaerobacterium xylanolyticum | Thermoanaerobacter xylanolyticum |
| 2021 | Thermobifida fusca | ||
| * | 1510 | Thermoclostridium stercorarium | Clostridium stercorarium |
| 55802 | Thermococcus barophilus | ||
| 187878 | Thermococcus gammatolerans | ||
| 311400 | Thermococcus kodakarensis | ||
| 342948 | Thermococcus onnurineus | ||
| 172049 | Thermococcus sibiricus | ||
| * | 501497 | Thermodesulfatator atlanticus | |
| * | 171695 | Thermodesulfatator indicus | |
| * | 1295609 | Thermodesulfobacterium geofontis | |
| * | 886 | Thermodesulfobacterium thermophilum | |
| * | 184064 | Thermodesulfobium narugense | |
| 2303 | Thermoplasma acidophilum | ||
| 2336 | Thermotoga maritima | ||
| 2337 | Thermotoga neapolitana | ||
| * | 271 | Thermus aquaticus | |
| * | 88189 | Thermus igniterrae | |
| * | 540988 | Thermus islandicus | |
| * | 56957 | Thermus oshimai | |
| * | 37636 | Thermus scotoductus | |
| 274 | Thermus thermophilus | ||
| 36861 | Thiobacillus denitrificans | ||
| * | 163 | Treponema bryantii | |
| * | 88058 | Treponema primitia | |
| * | 167 | Treponema succinifaciens | |
| * | 5722 | Trichomonas vaginalis | |
| * | 63417 | Trichophyton verrucosum | |
| * | 5691 | Trypanosoma brucei | |
| * | 5693 | Trypanosoma cruzi | |
| * | 154288 | Turicibacter sanguinis | |
| 29361 | Tyzzerella nexilis | Clostridium nexile | |
| 134821 | Ureaplasma parvum | ||
| 2130 | Ureaplasma urealyticum | ||
| * | 184870 | Varibaculum cambriense | |
| 39777 | Veillonella atypica | ||
| 39778 | Veillonella dispar | ||
| 29466 | Veillonella parvula | ||
| 666 | Vibrio cholerae | ||
| 672 | Vibrio vulnificus | ||
| * | 1482 | Virgibacillus halodenitrificans | |
| * | 1583 | Weissella confusa | |
| 51229 | Wigglesworthia glossinidia | ||
| 844 | Wolinella succinogenes | ||
| 339 | Xanthomonas campestris | ||
| 2371 | Xylella fastidiosa | ||
| 542 | Zymomonas mobilis | ||
| human colonocyte | |||
| human goblet cell | |||
| human hepatocyte | |||
| * | mouse goblet cell | ||
| * | mouse hepatocyte | ||
| * | mouse intestinal cell | ||
*Denotes a species/host cell type that was not included in the predecessor, NJS16 (Sung et al., Nat. Commun. 8, 15393 (2017)).
On the other hand, our JSON files35 include “NJC19_network.json”, “NJC19_organism.json”, “NJC19_compound.json”, and “NJC19_reference.json”. Among them, “NJC19_network.json” is equivalent to in Supplementary Table 1, in terms of its contents. This file consists of a total of 9,136 items. Each object in the file is exactly matched with one association in Supplementary Table 1. Each object has its own identification number that starts with “NJC19_” followed by a five-digit number. The object includes four key-value pairs. The keys are “Species”, “Small-molecule metabolite or macromolecule”, “Metabolic activity”, and “Ref. #”, reminiscent of the column names in Supplementary Table 1. The other files “NJC19_organism.json”, “NJC19_compound.json”, and “NJC19_reference.json” include detailed information on the values of the keys “Species”, “Small-molecule metabolite or macromolecule”, and “Ref. #” in the file “NJC19_network.json”, respectively. In a similar fashion to Online-only Tables 3 and 4, each microbial species in “NJC19_organism.json” is annotated with the NCBI taxonomy ID, and each compound in “NJC19_compound.json” is annotated with the KEGG compound ID. Furthermore, the specific sample sources of these microbial species are also present in “NJC19_organism.json”. Full metadata of these JSON files are provided in another file “README_NJC19.txt”, which is available in the Dryad Digital Repository together with the JSON files35.
As described in Methods, our NJC19 construction was started with taxonomic identification of mouse gut microbiome samples. The comprehensive repertoire of those microbial taxa, identified before the collection of their metabolic information, is provided in the Dryad Digital Repository35 (Table 1). In the case of the metagenome samples, it also provides the list of the selected species based on the frequency of their occurrence across the samples (Methods).
Technical Validation
Metabolic information collected in this study was primarily experimental evidence of small molecule transport and macromolecule degradation events, reported in the literature. Given the information dispersed across research papers, review articles, and textbooks (Online-only Table 2), a careful read of these sources was done to distinguish experimentally-verified information from the predictions solely based on automated bioinformatics algorithms. To check the accuracy of our network, the entire individual links in the compiled network were thoroughly re-examined by the independent authors who had not participated in the initial construction of the network. If potential errors were identified from the examined links (e.g., errors from the possible misinterpretations of the literature), these errors were carefully corrected based on the discussion of multiple authors.
To further assess the validity of our network, we examined the correlations between microbe-metabolite links in the network and measured metabolite levels in the mouse gut and portal vein plasma. Specifically, we examined whether the abundance increase/decrease of microbes associated with a particular metabolite in our network is consistent with the shift of the metabolite level across different experimental conditions. Regarding this analysis, three published mouse studies were found to provide the information of both microbial and metabolite levels in their collected samples: one study is for antibiotics (cefoperazone) treatment and recovery37, another is for fecal microbiota transplantation from twins discordant for obesity4, and the other for gnotobiotic mice with multiple diets38. From these studies, we considered only the cases with clear variations in the microbial and metabolite levels, which span at least 1.5-fold changes across different mouse groups for the metabolites and their microbial producers/consumers in NJC19. We further excluded host- and diet-derived metabolites, which may confound our analysis focusing on the effects of microbial metabolism. For all the resulting metabolites, Fig. 3 presents the levels of their microbial producers in NJC19 and those metabolite levels across the mouse groups with varying experimental conditions (Table 1). In Fig. 3, we did not consider microbial consumers because they were relatively deficient in their abundance, less than a half of the producers in each case. Here, microbial producers of each metabolite from the gnotobiotic mouse study38 in Fig. 3 are defined as the microbial species that produce this metabolite in NJC19. However, for the other two studies in Fig. 3, the finest taxonomic information is available at the genus level35, from the 16S rRNA gene sequence data processed by the Ribosomal Database Project Classifier in this analysis (RDP Naive Bayesian rRNA Classifier Version 2.11 with 16S rRNA training set 16)39. Therefore, for these two studies, microbial producers of a given metabolite are defined as the microbial genera, with each having the species whose majority (>50%) can produce that metabolite in NJC19. We also defined microbial consumers in a similar way, although they were excluded from Fig. 3 as discussed above.
Fig. 3.
Comparison of mouse microbiome and metabolome data based on NJC19. (a,b) In the left panels, the abundances of the propionate (a) and acetate (b) producers in NJC19 were obtained from cecal 16S rRNA gene sequence data35 in ref. 37 Cecal metabolite concentrations in the right panels were obtained from Fig. 3c of the same study. Mouse group I consists of mice six weeks after 10-day cefoperazone treatment on mouse group II, cefoperazone-naive mice. (c) In the left panel, the abundances of the butyrate producers in NJC19 were obtained from fecal 16S rRNA gene sequence data35 in ref. 4 Cecal butyrate concentrations in the right panel were obtained from Fig. 2c of the same study. Although we used the fecal 16S rRNA gene sequence data for the left panel due to the limited data availability, fecal and cecal microbial compositions were found to strongly correlate in other samples from that study, allowing us to use the fecal sequence data as a proxy for the cecal ones. All mice were initially germ-free in the study. Mouse groups I to V comprise mice transplanted with an obese twin’s microbiota (Ob; group I), mice co-housed with Ob and Ln mice (group II; see next for the definition of Ln mice), mice transplanted with the lean co-twin’s microbiota (Ln; group III), Ob mice co-housed with Ln and germ-free mice (group IV), and Ln mice co-housed with Ob and germ-free mice (group V). (d–f) In the left panels, the abundances of the succinate (d,e) and isovalerate (f) producers in NJC19 were obtained from cecal 16S rRNA gene copy levels in Fig. 2a of ref. 38 We re-scaled succinate producers in (d) to the total 16S rRNA gene copy levels, because the corresponding succinate concentrations (d) were available per cecal dry weight (i.e., per cecal microbial load). Cecal and portal vein metabolite concentrations in the right panels were obtained from Fig. 2b and Supplementary Table 4 of the same study, respectively. Mouse groups ‘HF/HS’, ‘ZF/HS’, and ‘Chow’ represent gnotobiotic mice on a high-fat/high-sucrose diet, a zero-fat/high-sucrose diet, and a chow diet, respectively. In (a–d), each error bar represents standard deviation across replicates. Error bars are missing for (e,f), as well as for the left panel in (d), and units are missing for the right panels in (e,f), because of the information unavailability from the data sources.
Figure 3 indeed demonstrates that alternations in the producer and metabolite levels tend to agree with each other, with the overall 71.4% matches of their increasing or decreasing tendencies across the mouse groups. For example, the propionate producers in NJC19 decreased by 90.0% in the cecum after cefoperazone treatment and recovery, consistent with an 88.5% decrease in the cecal propionate concentration (Fig. 3a). Likewise, the acetate producers in NJC19 decreased by 64.5% at the same time, consistent with a 59.3% decrease in the cecal acetate concentration (Fig. 3b). In these examples, the total microbial loads remained similar during the experiments37, and thus the metabolite concentration changes here are not likely to be a mere consequence of the microbial load changes. To test the statistical significance of these correlations, we introduce quantities fij, gij, and fij’ for each pair of mouse groups i and j: fij (gij) denotes a fold change from group i to group j in the measured producer (metabolite) abundance averaged over the replicates in the group i or j. fij’ denotes a group-i-to-j fold change in the average abundance of randomly-assigned producers, while the number of those randomly-assigned producers from all the data sources in Fig. 3 is maintained as the same as the number of the total observed producers. To assess the significance of the observed producer and metabolite correlations against a scenario that the producer information in NJC19 may not be more correct than expected by chance, we computed the P value as the probability of satisfying fij’ ≥ fij (fij’ ≤ fij) for all (i, j)-pairs that have fij and gij with both ≥ 1 (≤1). Accordingly, our P value calculation reveals that the producers in NJC19 and the detected metabolic compounds are significantly correlated in Fig. 3, thereby supporting the validity of the microbe-compound associations in NJC19 (P = 0.02 for Fig. 3a,c–e, P < 10–4 for Fig. 3f, and P = 0.06 for Fig. 3b).
Supplementary information
Acknowledgements
We thank John F. Baines and Philipp Rausch for kindly providing wild-caught mouse metagenome data, and Jonathan W. Johnson for preparing most icons in Fig. 1. This work was supported by Hong Kong Baptist University, Faculty Research Grant Category II (FRG2/17-18/097) and Start-up Grant Tier 2 (RC-SGT2/18-19/SCI/001). This work was conducted using the resources of the High Performance Cluster Computing Centre, Hong Kong Baptist University, which receives funding from Research Grant Council, University Grant Committee of the HKSAR and Hong Kong Baptist University.
Online-only Table
Online-only Table 5.
Degradation products of macromolecules in NJC19.
| Macromolecule | Degradation product(s) | Remark |
|---|---|---|
| Arabinogalactan | D-Galactose, D-Arabinose (L-Arabinose, Arabinose, L-Arabinopyranose, L-Arabinofuranose) | |
| Cellulose (Beta-D-glucan) | D-Glucose (Glucose), Cellobiose, Cellotetraose (Cellohexaose, Cellopentaose, Cellotriose) | |
| Chitin | N-Acetyl-D-Glucosamine (N-Acetylglucosamine) | |
| Chondroitin 4-sulfate (Chondroitin 6-sulfate) | D-Glucuronic acid (D-Glucuronate), N-Acetylgalactosamine (N-Acetyl-D-galactosamine), Sulfate (Sulfuric acid, Sulfite) | |
| Fructan (Inulin, Levan) | D-Fructose (Fructose), FOS (Fructooligosaccharide) | |
| Galactan | D-Galactose | |
| Hemicellulose, not specified | D-Glucose (Glucose), D-Ribose (Ribose), D-Galactose, D-Mannose (Mannose), D-Xylose (Xylose), D-Glucuronic acid (D-Glucuronate), D-Arabinose (L-Arabinose, Arabinose, L-Arabinopyranose, L-Arabinofuranose), D-Galacturonate, L-Rhamnose (Rhamnose, D-Rhamnose), XOS (Xylooligosaccharide) | |
| Hyaluronan (Hyaluronate, Hyaluronic acid) | N-Acetyl-D-Glucosamine (N-Acetylglucosamine), D-Glucuronic acid (D-Glucuronate) | |
| Mucin (Mucus Glycoprotein) | D-Galactose, N-Acetyl-D-Glucosamine (N-Acetylglucosamine), D-Glucuronic acid (D-Glucuronate), N-Acetylneuraminic acid (N-acetylneuraminate, Neu5Ac, Sialic acid), D-Galacturonate, L-Fucose, N-Acetylgalactosamine (N-Acetyl-D-galactosamine), L-Serine (Serine, D-Serine), L-Cysteine (Cysteine, D-Cysteine), L-Proline (Proline, D-Proline), L-Threonine (Threonine), Sulfate (Sulfuric acid, Sulfite) | |
| Pectin | D-Galacturonate | |
| Starch (Amylopectin, Amylose, 1,4-alpha-D-Glucan, Pullulan, Resistant starch, Glycogen) | D-Glucose (Glucose), Maltose, Dextrin (Maltotriose, Maltodextrin, Maltohexaose, Maltotetraose, Maltooligosaccharide) | |
| Triglyceride | 1,2-diacylglycerol (1-acylglycerol, Monoacylglycerol, Monoglyceride, Monoacylglycerol, Diacylglycerol), Glycerol | |
| Xylan | D-Xylose (Xylose), XOS (Xylooligosaccharide) | |
| Polypeptide | L-Alanine (D-Alanine, Alanine), L-Arginine (Arginine), L-Asparagine (Asparagine), L-Aspartate (Aspartate, D-Aspartate), L-Cysteine (Cysteine, D-Cysteine), L-Glutamine (D-Glutamine, Glutamine), L-Leucine (Leucine), L-Histidine (Histidine), L-Isoleucine (Isoleucine), L-Threonine (Threonine), L-Lysine (Lysine, D-Lysine), L-Methionine (D-Methionine), L-Phenylalanine (Phenylalanine, D-Phenylalanine), L-Proline (Proline, D-Proline), L-Serine (Serine, D-Serine), L-Tryptophan (Tryptophan), L-Tyrosine (Tyrosine), L-Valine (Valine, D-Valine), L-Glycine (Glycine), L-Glutamate (L-Glutamic acid, Glutamate, D-Glutamate), Prolylglycine, Glycyl-Phenylalanine, Leucyl-leucine, Alanylalanine, Glycylproline, Glycyl-leucine, Leucylglycine, Glycylglycine | Polypeptides can be broken down into amino acids and small peptides. Among them, only the compounds which have associations with the microbial species or the host cells in NJC19 are listed as the degradation products in Column B. |
| Arabinan | D-Arabinose (L-Arabinose, Arabinose, L-Arabinopyranose, L-Arabinofuranose) | |
| Mannan | D-Mannose (Mannose) | |
| DNA RNA Polynucleotide | Adenine, Cytosine, Guanine, Thymine, Uracil |
Author contributions
P.-J.K. designed the research. R.L., J.J.T.C., T.L.P.M., H.K., S.K., J.S. and P.-J.K. performed the research. R.L., J.J.T.C., T.L.P.M., H.K., C.-M.G. and P.-J.K. analyzed the data. R.L., J.J.T.C., J.S., C.-M.G. and P.-J.K. wrote the manuscript.
Code availability
Our Python code that converts the JSON format of NJC19 network data (NJC19_network.json35) to the format of Supplementary Table 1 can be downloaded from the Dryad Digital Repository35. For the taxonomic profiling of microbiome samples for the NJC19 construction, we used MetaPhlAn v2.0 with the “sensitive-local” mapping option and QIIME v1.8.0 with Greengenes v13_8_pp reference files30,31, as described above. The aforementioned cys file of NJC19 for network visualization was produced by Cytoscape v3.7.236.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41597-020-0516-5.
References
- 1.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mackie, R. I., White, B. A. & Isaacson, R. E. Gastrointestinal Microbiology (Chapman & Hall, New York, 1997).
- 3.Wang J, et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc. Natl. Acad. Sci. USA. 2014;111:E2703–E2710. doi: 10.1073/pnas.1402342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 6.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 7.Zhao L, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 8.Blaut M. Ecology and physiology of the intestinal tract. Curr. Top. Microbiol. Immunol. 2013;358:247–272. doi: 10.1007/82_2011_192. [DOI] [PubMed] [Google Scholar]
- 9.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 10.Sung J, et al. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat. Commun. 2017;8:15393. doi: 10.1038/ncomms15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. Plos Comput. Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc. Natl. Acad. Sci. USA. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy R, Borenstein E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl. Acad. Sci. USA. 2013;110:12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zelezniak A, et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. USA. 2015;112:6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinken A, Thiele I. Systematic prediction of health-relevant human-microbial co-metabolism through a computational framework. Gut Microbes. 2015;6:120–130. doi: 10.1080/19490976.2015.1023494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kettle H, Louis P, Holtrop G, Duncan SH, Flint HJ. Modelling the emergent dynamics and major metabolites of the human colonic microbiota. Environ. Microbiol. 2015;17:1615–1630. doi: 10.1111/1462-2920.12599. [DOI] [PubMed] [Google Scholar]
- 17.Magnúsdóttir S, et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 2017;35:81–89. doi: 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- 18.Tramontano M, et al. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat. Microbiol. 2018;3:514–522. doi: 10.1038/s41564-018-0123-9. [DOI] [PubMed] [Google Scholar]
- 19.Babaei P, Shoaie S, Ji B, Nielsen J. Challenges in modeling the human gut microbiome. Nat. Biotechnol. 2018;36:682–686. doi: 10.1038/nbt.4213. [DOI] [PubMed] [Google Scholar]
- 20.Magnúsdóttir S, Heinken A, Fleming RMT, Thiele I. Reply to “Challenges in modeling the human gut microbiome”. Nat. Biotechnol. 2018;36:686–691. doi: 10.1038/nbt.4212. [DOI] [PubMed] [Google Scholar]
- 21.Bleich A, Fox J. The mammalian microbiome and its importance in laboratory animal research. ILAR J. 2015;56:153–158. doi: 10.1093/ilar/ilv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson KE. An update on the status of current research on the mammalian microbiome. ILAR J. 2015;56:163–168. doi: 10.1093/ilar/ilv033. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen TLA, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis. Model Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickard JM, et al. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullender TC, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langille MG, et al. Microbial shifts in the aging mouse gut. Microbiome. 2014;2:50. doi: 10.1186/s40168-014-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rooks MG, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linnenbrink M, Wang J, Hardouin EA, Künzel S, Metzler D, Baines JF. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol. Ecol. 2013;22:1904–1916. doi: 10.1111/mec.12206. [DOI] [PubMed] [Google Scholar]
- 30.Truong DT, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 31.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouladoux N, Harrison OJ, Belkaid Y. The mouse model of infection with Citrobacter rodentium. Curr. Protoc. Immunol. 2017;119:19.15.1–19.15.25. doi: 10.1002/cpim.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinken A, Sahoo S, Fleming RMT, Thiele I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes. 2013;4:28–40. doi: 10.4161/gmic.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung J, 2017. Data from: Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Dryad Digital Repository. [DOI] [PMC free article] [PubMed]
- 35.Lim R, 2020. Data from: Large-scale metabolic interaction network of the mouse and human gut microbiota. Dryad Digital Repository. [DOI] [PMC free article] [PubMed]
- 36.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theriot CM, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovatcheva-Datchary P, et al. Simplified intestinal microbiota to study microbe-diet-host interactions in a mouse model. Cell Rep. 2019;26:3772–3783. doi: 10.1016/j.celrep.2019.02.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Sung J, 2017. Data from: Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Dryad Digital Repository. [DOI] [PMC free article] [PubMed]
- Lim R, 2020. Data from: Large-scale metabolic interaction network of the mouse and human gut microbiota. Dryad Digital Repository. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Our Python code that converts the JSON format of NJC19 network data (NJC19_network.json35) to the format of Supplementary Table 1 can be downloaded from the Dryad Digital Repository35. For the taxonomic profiling of microbiome samples for the NJC19 construction, we used MetaPhlAn v2.0 with the “sensitive-local” mapping option and QIIME v1.8.0 with Greengenes v13_8_pp reference files30,31, as described above. The aforementioned cys file of NJC19 for network visualization was produced by Cytoscape v3.7.236.