Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a major nosocomial pathogen worldwide. Malta is one of the countries with the highest MRSA prevalence in Europe, as identified from hospital blood cultures [1]. However, community prevalence of MRSA has never previously been investigated. This study aimed at establishing the prevalence of community MRSA nasal colonization in Maltese individuals and identifying the clonal characteristics of the detected isolates. Nasal swabs were collected from 329 healthy individuals who were also asked to complete a brief questionnaire about risk factors commonly associated with MRSA carriage and infection. The swabs were transported and enriched in a nutrient broth supplemented with NaCl. The presence of MRSA was then determined by culturing on MRSA Select chromogenic agar and then confirming by several assays, including catalase, coagulase and PBP2a agglutination tests. The isolates were assayed for antibiotic susceptibilities and typed by microarray analysis to determine the clonal characteristics of each strain. The prevalence of MRSA nasal colonization in the healthy Maltese population was found to be 8.81% (95% confidence interval [CI], 5.75–11.87%), much higher than that found in other studies carried out in several countries. No statistical association was found between MRSA carriage and demographics or risk factors; however, this was hindered by the small sample size. Almost all the isolates were fusidic-acid resistant. The majority were found to belong to a local endemic clone (CC5) which seems to be replacing the previously prevalent European clone UK-EMRSA-15 in the country. A new clone (CC50-MRSA-V) was also characterized. The presence of such a significant community reservoir of MRSA increases the burdens already faced by the local healthcare system to control the MRSA epidemic. Colonization of MRSA in otherwise healthy individuals may represent a risk for endogenous infection and transmission to hospitalized patients after admission to a healthcare facility, leading to longer hospital stays and, consequently, increased healthcare costs.
Keywords: MRSA, Antibiotics, Surveillance, Colonization
1. Introduction
Methicillin-resistant S. aureus (MRSA) is a major nosocomial pathogen worldwide [2]. Its acquisition of multiple antibiotic resistance mechanisms has led to increasingly challenging infections [3]. Multi-resistant strains, only sensitive to glycopeptide antibiotics, have been increasingly reported from hospitals in several countries [4].
In the past, MRSA was mainly associated with hospital settings. Risk factors for infection and colonization included recent or prolonged hospitalization, nursing home admission, recent antimicrobial therapy, chronic disease and contact with a colonized individual [5]. Reports of MRSA outbreaks among individuals without any healthcare-associated risk factors [6,7] have led to an increased awareness of community-associated MRSA (CA-MRSA). This “changing epidemiology” of MRSA has several important clinical implications, since increasing prevalence will increase antibiotic consumption leading to greater risk of multidrug-resistant MRSA within the community [8].
Apart from the absence of healthcare-associated risk factors, CA-MRSA can often be distinguished from healthcare-associated strains by different microbiological, epidemiological and molecular characteristics. Unlike healthcare-associated MRSA (HA-MRSA), which is often associated with bloodstream, respiratory tract and urinary infections, CA-MRSA is more implicated in skin and soft tissue infections, tends to be more prevalent in younger patients and also generally shows a greater susceptibility to non-β-lactam antibiotics [9]. CA-MRSA is often characterized by the presence of an SCC mec type IV or V allele in the mec element [10,11] and genes coding for specific endotoxins, especially the Panton–Valentine leukocidin (PVL) associated with an increased risk for skin and soft tissue infections [9,12]. All these factors indicate that new MRSA strains can arise de novo from the community. Although the United States Centers for Disease Control and Prevention (CDC) has established a set of guidelines to distinguish CA-MRSA from HA-MRSA, these are mostly based on the absence of healthcare-associated risk factors in the individual rather than microbiological or molecular criteria [13].
HA-MRSA has been documented to be a major challenge to the Maltese healthcare. Malta currently has one of the highest proportions of MRSA in Europe [1]. However, no other studies on the prevalence of MRSA within the general community have been carried out. This epidemiological study thus aimed to determine the prevalence of MRSA nasal carriage in the general population and to find out any relationship between colonization and published risk factors.
2. Materials and methods
The study, which took place between August 2010 and March 2011, was approved by the University of Malta Research Ethics Committee. 329 volunteers were recruited by random sampling as they attended eight primary healthcare centers around Malta. These recruits were not actual patients, but their accompanying adult relatives or friends, all of whom were asked to participate in the study. Individuals who had been hospitalized within the previous twelve months were excluded from the study according to the established CDC guidelines [13]. Upon accepting to take part in the study, the volunteers were admitted to a private room where a consent form with all the relevant details was signed. A questionnaire was also administered, aimed at determining whether the respondent had any of the healthcare and/or community-associated risk factors for MRSA colonization reported in previous studies.
Swabbing of both nostrils was carried out by a single investigator (J.S.) using a sterile cotton swab moistened in deionized sterile water. A standard technique of swabbing both anterior nares was employed using the same swab and rubbing it against the interior skin of the nose up to a depth of about 2.5 cm while gently rotating the swab for five times to sample the entire area. The swab was immediately snapped into a 5 ml bijoux bottle in an enrichment medium containing 4 ml of sterile nutrient broth enriched with 2.5% sodium chloride and incubated overnight. Each sample was then cultured on MRSA Select chromogenic agar (Bio-Rad Laboratories, Marnes-la-Coquette, France). Presumptive MRSA strains, which grew as pink colonies on the chromogenic medium, were confirmed by Gram stain, catalase, slide coagulase by the Prolex™ Staph Latex kit (Pro-Lab Diagnostics, Toronto, Canada), and PBP 2′ testing using the Mastalex™ MRSA kit (Mast Diagnostic, Bootle, Merseyside, UK). Antimicrobial susceptibility testing was also carried out using the VITEK® 2 system (bioMérieux, Marcy-l’Etoile, France), by AST-P580 Gram positive susceptibility cards. Antibiotics tested included a cefoxitin screen (used to confirm the presence of MRSA and detect low level methicillin resistance), benzylpenicillin, oxacillin, fusidic acid and mupirocin, clindamycin and inducible clindamycin resistance, tetracycline, as well as aminoglycosides (gentamicin, tobramycin), quinolones (levofloxacin, moxifloxacin), and glycopeptides (teicoplanin, vancomycin).
Microarray-based characterization was carried out on all the isolated and confirmed strains at the Dresden University of Technology (refer to [14] and [15] for detailed description of the technique). The simultaneous detection of several molecular targets allowed the assignation of the isolates to clonal complexes, as well as genotype-based assessment of antibiotic resistance and virulence factors. Antibiotic resistance genes included those for penicillinase (blaZ), mecA defining methicillin-resistance, and genes encoding resistance to aminoglycosides, chloramphenicol, fusidic acid, lincosamides, macrolides, tetracyclines, trimethoprim and vancomycin. Virulence factors included various enterotoxins, toxic shock syndrome toxin (tst1), exfoliative toxins and various haemolysins and leukocidins, including Panton-Valentine leukocidin. Probes allowing the identification of different SCCmec types were also included.
Pearson’s chi-square test (SPSS Statistics for Windows, Version 17.0, Chicago: SPSS Inc.) was used to determine whether there was any statistically significant correlation between the nasal carriage of MRSA and the demographics and risk factors addressed in the questionnaire. A p-value of 0.05 or less was taken to determine if differences were statistically significant.
The z-ratio for the significance of the difference between two independent proportions (http://www.mccallum-layton.co.uk/stats/ZTestTwoTailSampleValues.aspx) was also used to test whether prevalent clones varied significantly in terms of age group and risk factors at the 95% CI.
3. Results
The prevalence of MRSA nasal carriage in the Maltese general population was 8.81% (95% CI, 5.75–11.87%). Prevalence was higher in females (9.5%) than in males (7.6%) as well as in lower age groups; however, these differences were both not statistically significant (Table 1).
Table 1.
Results for chi–square test for association between MRSA carriage and different demographics.
| Demographics | MRSA carriage | Degrees of freedom | p-value | ||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Gender | Male | 9 (7.6%) | 109 (92.4%) | 1 | 0.570 |
| Female | 20 (9.5%) | 191 (90.5%) | |||
| Age Group | 16–40 | 11 (14.7%) | 64 (85.3%) | 3 | 0.177 |
| 41–55 | 6 (8.0%) | 69 (92.0%) | |||
| 56–65 | 8 (8.3%) | 88 (91.7%) | |||
| > 65 | 4 (4.8%) | 79 (95.2%) | |||
No statistically significant association was found between MRSA carriage and each community- or healthcare-related risk factors using the chi-square test (data not shown). In order to obtain some more statistical strength owing to the small sample size, risk factors were grouped according to category; however, again no statistically significant association was found (Table 2).
Table 2.
Organization of risk factors in the questionnaire and results for chi–square test for association between MRSA carriage and different risk factor categories. For the association between carriage and having any risk factor, the result of the Fisher’s exact test is shown in brackets as one of the cells had an expected count of less than 5.
| Risk Factors | Categories | MRSA carriage | X2 value | D.F. | p-value | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||
| Gym membership Attendance at a day care center Sharing of personal items |
Having a community associated risk factor | Yes | 13 (11.3%) | 102 (88.7%) | 1.364 | 1 | 0.243 |
| No | 16 (7.5%) | 198 (92.5%) | |||||
| Healthcare workers Elderly relatives living in nursing homes Chronic relatives making frequent hospital visits |
Having relatives with healthcare contacts | Yes | 16 (10.5%) | 137 (89.5%) | 0.960 | 1 | 0.327 |
| No | 13 (7.4%) | 163 (92.6%) | |||||
| Chronic disease History of skin infection with boils/abscesses Chronic skin disease Nasal disease |
Having a medical history associated with MRSA | Yes | 12 (7.1%) | 158 (92.9%) | 1.349 | 1 | 0.245 |
| No | 17 (10.7%) | 142 (89.3%) | |||||
| Antibiotic consumption Surgery Proximity to hospitalized patients |
Exposure to risk factors in past year | Yes | 20 (7.6%) | 242 (92.4%) | 2.233 | 1 | 0.135 |
| No | 9 (13.4%) | 58 (86.6%) | |||||
| All of the above | Having any risk factor | Yes | 26 (8.3%) | 288 (91.7%) | 2.446 | 1 | 0.118 (0.136) |
| No | 3 (20.0%) | 12 (80.0%) | |||||
The antibiotic resistance patterns and the genotypic characteristics of the MRSA clones as determined by Vitek 2™ and microarray analysis, respectively, are summarized in Table 3. All the isolates were found to be resistant to at least one non-β-lactam antibiotic. Resistance to fusidic acid and streptogramins was the most prominent; all strains but one were resistant to one, or the other, or both. Resistance to other antibiotics included levofloxacin and moxifloxacin (5 isolates, later confirmed to be UK-EMRSA-15), erythromycin and clindamycin (1 isolate), and tetracycline (1 isolate).
Table 3.
Results of antibiotic resistance results obtained by the VITEK 2 system and strain characterization by microarray. mecA is the methicillin resistance gene; blaZ, blaI, blaR are â-lactamase resistance genes; fosB is a resistance gene for phosphomycin and bleomycin; Q6GD50 is a putative gene for fusidic acid resistance; ermC is an erythromycin and clindamycin resistance gene; sat is a streptothricin resistance gene; far1 is a fusidic acid resistance gene; tetK is a tetracycline resistance gene. MLSB = inducible clindamycin resistance; PVL = Panton–Valentine leukocidin; tsst-1 = toxic shock syndrome toxin 1; agr = accessory gene regulator; EDIN-B = epidermal cell differentiation inhibitor B.
| Strain Type | No. of isolates (% of total; n = 29) | VITEK® 2 | Microarray | ||||
|---|---|---|---|---|---|---|---|
| Antibiotic Resistance | Antibiotic Resistance genes | PVL | TSST-1 | Entero-toxins | Remarks | ||
| CC5-MRSA-IV (Maltese Clone); agr group II | 16 (55.2%) | Oxacillin, Fusidic Acid, Streptogramins | mecA, blaZ, blaI, blaR, fosB, Q6GD50 | Neg | Pos | A, C, L, egc-cluster | |
| 1 (3.4%) | Fusidic Acid, Streptogramins | mecA, fosB, Q6GD50 | Neg | Neg | A, egc-cluster | ||
| 2 (6.8%) | Fusidic Acid, Streptogramins | mecA, blaZ, blaI, blaR, fosB, Q6GD50 | Neg | Pos | A, C, L, egc-cluster | ||
| 1 (3.4%) | N/A | mecA, blaZ, blaI, blaR, fosB, Q6GD50 | Neg | Pos | A, C, L, egc-cluster | ||
| 1 (3.4%) | N/A | mecA, blaZ, blaI, blaR, fosB, Q6GD50 | Neg | Neg | A, ecg-cluster | ||
| CC22-MRSA-IV (Barnim/UK-EMRSA-15); agr group I | 1 (3.4%) | Oxacillin, Levofloxacin, Moxifloxacin, Fusidic Acid, Streptogramins, Vancomycin-intermediate | mecA, blaZ, blaI, blaR, Q6GD50, ermC | Neg | Neg | C, L, egc-cluster | |
| 1 (3.4%) | Oxacillin, Levofloxacin, Moxifloxacin, Streptogramins | mecA, blaZ, blaI, blaR, | Neg | Neg | C, L, egc-cluster | ||
| 1 (3.4%) | Oxacillin, Levofloxacin, Moxifloxacin, Streptogramins | mecA, blaZ, blaI, blaR, | Neg | Neg | B, C, L, egc-cluster | ||
| 2 (6.8%) | Oxacillin, Levofloxacin, Moxifloxacin-intermediate, Streptogramins | mecA, blaZ, blaI, blaR, | Neg | Neg | B, C, L, egc-cluster | ||
| ST6-MRSA-IV (WA-MRSA-51); agr group I | 1 (3.4%) | Oxacillin, Erythromycin, Clindamycin, MLSB inducible | mecA, blaZ, blaI, blaR, ermC; sat, fosB | Neg | Neg | A | |
| CC80-MRSA-IV; agr group III | 1 (3.4%) | Oxacillin, Tetracycline, Fusidic Acid, Streptogramins | mecA, blaZ, blaI, blaR, aphA, sat, far1, tetK | Neg | Neg | Neg | Other toxins: exfoliative toxin D, EDIN-B |
| CC50 (New Strain); agr group IV | 1 (3.4%) | Oxacillin, Fusidic Acid, Streptogramins | mecA, blaZ, blaI, blaR, Q6GD50 | Neg | Neg | ecg-cluster | |
Upon microarray-based typing, the majority of the strains (72.4%, 21 isolates) were assigned to the clonal complex (CC) 5, termed the “Maltese clone” because it appears to be, so far, endemic to Malta [25]. This clone harbored the SCCmec IV element. All isolates belonging to this clone carried the putative fusidic acid resistance element, Q6GD50 (fusC), and were genotypically resistant only to fosfomycin and bleomycin. Furthermore, all carried the gene for enterotoxin A (sea) and the egc enterotoxin locus, and all but two also carried genes for enterotoxins C (sec) and L (sel) and toxic shock syndrome toxin (tst1). One of the two isolates negative for sec, sel and tst1 was also negative for the beta-lactamase resistance genes blaZ, blaI and blaR.
Five isolates (17.2%) belonged to ST22-MRSA-IV (UK-EMRSA-15 or Barnim Epidemic MRSA Strain). All of these isolates carried enterotoxins C and L. Only one isolate carried a gene for macrolide/lincosamide resistance (ermC), as well as Q6GD50 (fusC); no additional antibiotic resistance genes were found in the other strains.
A single isolate belonged to the CC80-MRSA-IV European CA-MRSA clone. While this strain is normally described as PVL-positive, however, the actual isolate resulted as PVL-negative on the microarray. The isolate was positive for neomycin, streptothricin, fusidic acid and tetracycline resistance genes (aphA3, sat, far1 and tetK, respectively), and exfoliative toxin D (etd); this confirmed results of antibiotic susceptibility testing (Table 3).
One isolate belonged to the ST6-MRSA-IV strain. This is a rare strain which is usually found in Western Australia (where it is designated WA MRSA-51). It carried the gene for resistance to erythromycin and clindamycin (ermC), which confirmed phenotypic resistance to these antibiotics (Table 3), as well as enterotoxin A.
Finally, a single isolate of a CC50-MRSA-V clone was also characterized, which has never been described as yet. This clone had a SCCmec V element, and was positive for Q6GD50 (fusC) indicating fusidic acid resistance. MLST showed it to be a double locus variant of ST50, with the profile 16-16-12-2-39-13-2. It carried the egc enterotoxin gene cluster but was negative for the toxic shock syndrome toxin gene and other enterotoxin genes, as well as other antibiotic resistance gene loci. It did not carry PVL genes.
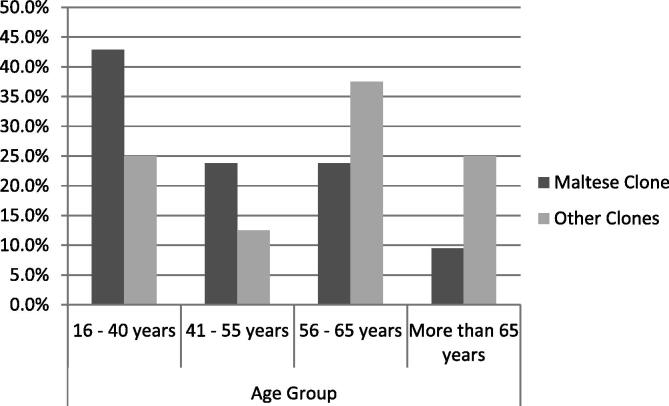
When MRSA carriers were grouped according to strain type, no significant difference was observed between genders. The Maltese clone was predominant in the younger age groups (16–40 years; Fig. 1) and in individuals with more than three risk factors, while the other clones were more prevalent in older individuals (>56 years) and in those with two risk factors for carriage. The z-ratio for the significance of the difference between two independent proportions showed a statistically significant difference in the proportion of colonization with the Maltese clone between the age groups of 16–40 and >56 years.
Figure 1.
Distribution of the different MRSA clones across age groups. The z-ratio calculation showed a statistically significant difference in colonization with the Maltese clone between the smallest and largest age groups.
4. Discussion
The prevalence of MRSA nasal carriage found in this study is much higher than that reported in similar studies from other locations, including Birmingham, the United Kingdom (1.5%) [16], Greece (0.94%) [17], the United States (1.5%) [18], and Taiwan (3.5%) [19]. This discrepancy could be owing to several reasons. First of all, mentioned studies predominantly employed direct inoculation of the nasal swabs onto nutrient agar with horse blood serum [16], oxacillin resistance screening agar (ORSA) [17], or mannitol salt agar (MSA) [18,19]. Meanwhile, the sensitivity of this technique was increased by incubation of the swabs in an enrichment broth before inoculation onto chromogenic medium [20], and this could have contributed at a certain degree to an increased prevalence. Secondly, although “healthy” individuals were sampled, they were nevertheless accompanying relatives to primary healthcare centers; these relatives could be regular attendees to the healthcare centers and could thus have been themselves colonized and transmitted the MRSA to the participants.
No significant association was found between MRSA colonization and gender or age (p > 0.05). This contrasts with other studies reporting a correlation with ages of 60 years or more [18,21] and gender [18,22]. Older individuals are more likely to suffer from chronic illnesses and to have frequent hospital contact, and would thus be expected to be more at risk for colonization. However, in this study, MRSA prevalence was greater in younger ages and would support a truly community-acquired etiology [9].
The majority of carriers (26/29, 89.7%) had at least one risk factor for MRSA carriage; 25 individuals (86.2%) had at least one healthcare-related risk factor, namely relatives with healthcare contacts, risk factors related to medical history and/or healthcare-associated risk factor exposure within the previous year. This was consistent with the conclusion arrived at by Salgado et al. [5] that many MRSA carriers in the community have one or more healthcare-associated risk factors and that more adequate criteria for distinguishing between CA- and HA-MRSA should be set-up. Most studies have been carried out in populations attending hospital outpatient centers and other healthcare settings [22,23]; this study, although itself conducted in a healthcare setting, was based on participants who were not themselves patients and who, consequently, were supposed to be less at risk for MRSA carriage.
Several outbreaks of community-acquired MRSA have been reported in specific settings, such as athletic centers, and community-associated risk factors have been associated with these outbreaks, including sharing of towels [24]. Out of the 32 isolates found in this study, only one came from an individual who had been exposed to a community-associated risk factor only, namely the sharing of personal items, such as towels or razors, with other individuals in the household. The others also had healthcare-related risk factors. Furthermore, no community-associated risk factors were linked with MRSA carriage in our subjects. Therefore, these isolates are better termed community-associated rather than community-acquired, since their acquisition could not be traced to a particular community origin.
The rate of fusidic acid resistance in the isolated MRSA strains was rather remarkable. In fact, 82.8% of the strains (24/29, including two strains which were not tested by VITEK® 2, but which carried the genes for resistance) were found to be resistant. Fusidic acid-resistant MRSA is not common in other parts of Europe. In the study by Karapsias et al. [17], only two out of nine isolates (22.2%) were fusidic acid-resistant. The increased carriage of the fusidic acid resistance gene may be related to a higher selective pressure, resulting from the widespread use of this antibiotic in the country. In fact, there is evidence of abuse of topical formulations of fusidic acid in Malta [25]. Apart from fusidic acid and streptogramins, the majority of strains were susceptible to other non-β-lactam antibiotics. This correlates well with the view that community-associated strains generally show a greater susceptibility to non-β-lactam antibiotics and are less likely than healthcare-associated strains to be multidrug-resistant [9]. A single UK-EMRSA-15 strain resulted as vancomycin-intermediate upon antibiotic susceptibility testing; this was confirmed with Etest (MIC = 4 μg/ml), however, the strain did not result in having any resistance genes by microarray analysis. Such a finding was also reported in a case of an S. aureus bloodstream infection in Greece where the strain was also teicoplanin-resistant [26].
The majority of strains isolated belonged to the endemic Maltese clone, a CC5-MRSA-IV variant. This clone has already been described in a previous Maltese study carried out on hospital strains, and likewise carried the putative fusidic acid resistance element, Q6GD50 (fusC), usually corresponding to phenotypic resistance to fusidic acid [25] and previous work indicated a possible linkage to SCCmec IV, or the presence of a composite SCC element [15]. This study also described the presence of the enterotoxin A gene (sea) and the egc enterotoxin locus in all isolates, and sec, sel and tst1 in nine out of ten isolates, which correlates well with the findings as described in the results section and also in Table 3. Fortunately, the strains isolated in this study were susceptible to most antibiotics tested.
The Maltese clone seems to be replacing UK EMRSA-15, the dominant strain across Western Europe and previously the predominant clone in Maltese hospital isolates [25]. The Malta clone was more prevalent in younger individuals with no history of hospitalization (Fig. 1), suggesting community acquisition [8]. It is possible that this clone emerged from the community and has disseminated to the hospital; in fact, data from tertiary care suggest that the Maltese clone has become predominant even in this setting (personal communication).
A strain belonging to a new clone (CC50-V) was also encountered. The presence of the novel SCCmec V element and the absence of antibiotic resistance genes on characterization of this strain indicated community acquisition [11]; however, toxic shock syndrome toxin gene and many other enterotoxin genes were also absent. Unfortunately, no travel history or other data which might have been useful in determining the source of such a strain was present.
Microarray typing confirmed all the isolates but one to be of SCCmec type IV, which is associated with, but not exclusive to, community strains. This finding was rather consistent with the small degree of non-β-lactam antibiotic resistance found in the majority of the isolates, since SCCmec IV strains often lack several antibiotic resistance genes [10]. The same applies to SCCmec V found in CC50 [11].
No strain was found to carry the Panton-Valentine leukocidin toxin, including the single isolate belonging to the CC80 clone, which is usually PVL-positive. Such a strain has been reported only once in the Maltese literature, where it was isolated from a hospital employee with no recorded patient contact or travel history in continental Europe [25]. Once more, no travel history was available for this case.
This study faced several limitations. Like all cross-sectional prevalence studies, it provided a snapshot of the prevalence of MRSA nasal colonization only at a given time period. Consequently, intermittently colonized individuals might not have been detected at the time. Risk factor analysis depended on volunteer recall at the time of questioning and thus could not be fully accurate. Most importantly, the sample size was relatively small, even for a country of less than half a million inhabitants, resulting in a wide confidence interval. The sample size did not allow the accurate determination of any association between MRSA carriage and reported risk factors. The site where this study was conducted constituted yet another limitation, since primary health centers are still healthcare settings, and although healthy volunteers who had not been recently admitted to hospital were selected, their relatives who were attending the clinics could have been colonized themselves through frequent visits to the clinics.
5. Conclusions
Even with these limitations, the prevalence of MRSA carriage in the Maltese community seems to be very high, especially when compared with that found in other countries. The presence of such a significant community reservoir of MRSA increases the burdens already faced by the local healthcare system to control the MRSA epidemic. The presence of MRSA strains in up to 10% of admissions increases the risk for cross transmission to hospitalized patients and poses a major challenge to control initiatives. Secondly, the local endemic MRSA clone seems to be rather established in the community and has replaced UK-EMRSA-15 as the prevailing clone. Finally, the absence of PVL-positive strains is encouraging, considering the problems associated with community skin and soft tissue infections caused by these strains.
Acknowledgements
We would like to thank Dr. Liberato Camilleri and Ms. Elizabeth Scicluna for assistance in statistical analysis and the staff of the Microbiology Laboratory of Mater Dei Hospital as well as Antje Ruppelt (Dresden University of Technology) and Elke Müller (Alere, Jena) for their assistance and support.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jegh.2013.05.003.
6. Conflict of interest
None of the authors have any conflict of interest to declare.
References
- [1].European Antimicrobial Resistance Surveillance System(EARSS) ECDC; 2009. EARSS annual report 2008: On-going surveillance of S. pneumoniae, S. aureus, E. coli, E. faecium, E. faecalis, K. pneumoniae, P. aeruginosa. http://www.ecdc.europa.eu/en/activities/surveillance/EARS-Net/publications/Pages/documents.aspx, accessed 30 April 2011. [Google Scholar]
- [2].Emori TG, Gaynes RP. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 2009;6:428–42. doi: 10.1128/CMR.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rybak MJ, LaPlante KL. Community associated methicillin-resistant Staphylococcus aureus: a review. Pharmacotherapy. 2005;25:74–85. doi: 10.1592/phco.25.1.74.55620. [DOI] [PubMed] [Google Scholar]
- [4].Sievert DM, Rudrik JT, Patel JB, Clifford McDonald L, Wilkins MJ, Hageman JC. Vancomycin-resistant Staphylococcus aureus in the United States, 2002–2006. Clin Infect Dis. 2008;46:668–74. doi: 10.1086/527392. [DOI] [PubMed] [Google Scholar]
- [5].Salgado CD, Farr BM, Calfee DP. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin Infect Dis. 2003;36:131–9. doi: 10.1086/345436. [DOI] [PubMed] [Google Scholar]
- [6].Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279:593–8. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- [7].Centers for Disease Control and Prevention (CDC) Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus – Minnesota and North Dakota, 1997–1999. Morb Mortal Wkly Rep. 1999;48:707–10. [PubMed] [Google Scholar]
- [8].Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001;7:178–82. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, et al. Comparison of community- and healthcare-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290:2976–84. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- [10].Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002;46:1147–52. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 2004;48:2637–51. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–84. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].CDC . Community-associated MRSA information for clinicians. CDC; 2005. http://www.cdc.gov/ncidod/dhqp/ar_mrsa_ca_clinicians.html#7, accessed 28 April 2011. [Google Scholar]
- [14].Monecke S, Slickers P, Ehricht R. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol Med Microbiol. 2008;53:237–51. doi: 10.1111/j.1574-695X.2008.00426.x. [DOI] [PubMed] [Google Scholar]
- [15].Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg MA, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistantStaphylococcus aureus. PLoS One. 2001;6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Abudu L, Blair I, Fraise A, Cheng KK. Methicillin-resistant Staphylococcus aureus (MRSA): a community-based prevalence survey. Epidemiol Infect. 2001;126:351–6. doi: 10.1017/s0950268801005416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karapsias S, Piperaki ET, Spiliopoulou I, Katsanis G, Tseleni-Kotsovili A. Methicillin-resistant Staphylococcus aureus nasal carriage among healthy employees of the Hellenic Air Force. Euro Surveill. 2008;13 doi: 10.2807/ese.13.40.18999-en. pii: 18999. [DOI] [PubMed] [Google Scholar]
- [18].Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, Fosheim G, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J Infect Dis. 2006;193:172–9. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- [19].Lu PL, Chin LC, Peng CF, Chiang YH, Chen TP, Ma L, Siu LK. Risk factors and molecular analysis of community methicillin-resistant Staphylococcus aureus carriage. J Clin Microbiol. 2005;43:132–9. doi: 10.1128/JCM.43.1.132-139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee S, Park YJ, Yoo JH, Kahng J, Jeong IH, Kwon YM, et al. Comparison of culture screening protocols for methicillin-resistant Staphylococcus aureus (MRSA) using a chromogenic agar (MRSA-Select) Ann Clin Lab Sci. 2008;38:254–7. [PubMed] [Google Scholar]
- [21].Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis. 2008;197:1226–34. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- [22].Kenner J, O’Connor T, Piantanida N, Fishbain J, Eberly B, Viscount H, et al. Rates of carriage of methicillin-resistant and methicillin-susceptible Staphylococcus aureus in an outpatient population. Infect Control Hosp Epidemiol. 2003;24:439–44. doi: 10.1086/502229. [DOI] [PubMed] [Google Scholar]
- [23].Jernigan JA, Pullen AL, Flowers L, Bell M, Jarvis WR. Prevalence of and risk factors for colonization with methicillin-resistant Staphylococcus aureus at the time of hospital admission. Infect Control Hosp Epidemiol. 2003;24:409–14. doi: 10.1086/502230. [DOI] [PubMed] [Google Scholar]
- [24].CDC Methicillin-resistant Staphylococcus aureus infections among competitive sports participants - Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000–2003. Morb Mortal Wkly Rep. 2003;52:793–5. [PubMed] [Google Scholar]
- [25].Scicluna EA, Shore AC, Thürmer A, Ehricht R, Slickers P, Borg MA, et al. Characterisation of MRSA from Malta and the description of a Maltese epidemic MRSA strain. Eur J Clin Microbiol Infect Dis. 2010;29:163–70. doi: 10.1007/s10096-009-0834-1. [DOI] [PubMed] [Google Scholar]
- [26].Tsakris A, Papadimitriou E, Douboyas J, Stylianopoulou F, Manolis E. Emergence of vancomycin-intermediate Staphylococcus aureus and S. Sciuri, Greece. Emerg Infect Dis. 2002;8:536–7. doi: 10.3201/eid0805.010387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jegh.2013.05.003.