Abstract
Background
Order Chaetophorales currently includes six families, namely Schizomeridaceae, Aphanochaetaceae, Barrancaceae, Uronemataceae, Fritschiellaceae, and Chaetophoraceae. The phylogenetic relationships of Chaetophorales have been inferred primarily based on short and less informative rDNA sequences. This study aimed to phylogenetically reconstruct order Chaetophorales and determine the taxonomic scheme, and to further understand the evolution of order Chaetophorales.
Results
In the present study, seven complete and five fragmentary chloroplast genomes were harvested. Phylogenomic and comparative genomic analysis were performed to determine the taxonomic scheme within Chaetophorales. Consequently, Oedogoniales was found to be a sister to a clade linking Chaetophorales and Chaetopeltidales. Schizomeriaceae, and Aphanochaetaceae clustered into a well-resolved basal clade in Chaetophorales, inconsistent with the results of phylogenetic analysis based on rDNA sequences. Comparative genomic analyses revealed that the chloroplast genomes of Schizomeriaceae and Aphanochaetaceae were highly conserved and homologous, highlighting the closest relationship in this order. Germination types of zoospores precisely correlated with the phylogenetic relationships.
Conclusions
chloroplast genome structure analyses, synteny analyses, and zoospore germination analyses were concurrent with phylogenetic analyses based on the chloroplast genome, and all of them robustly determined the unique taxonomic scheme of Chaetophorales and the relationships of Oedogoniales, Chaetophorales, and Chaetopeltidales.
Keywords: Chlorophyta, Classification, Gene order rearrangement, Green algae, Nuclear genes
Background
Class Chlorophyceae comprises two primary lineages based on molecular phylogeny, one comprising orders Sphaeropleales and Volvocales (SV clade) and another comprising orders Oedogoniales, Chaetophorales, and Chaetopeltidales (OCC clade) [1–6]. Order Chaetophorales, a lesser known member of Chlorophyceae (Chlorophyta) first circumscribed by Wille [7], containing nine families, as reported by Printz [8] and six families, as reported by Bourrelly [9]. Based on ultrastructural studies (mitosis-cytokinesis, motile cell) and molecular phylogenetic analyses, six families (Schizomeridaceae, Aphanochaetaceae, Barrancaceae Uronemataceae, Chaetophoraceae, and Fritschiellaceae) have been reported in this order and numerous traditional families were transferred to other green algal orders or classes [10–23]. Although Chaetophorales has exhibited uncontested monophyly upon improvement in gene and taxon sampling [20, 21], certain key relationships within this order and the OCC clade are less prominent and warrant further investigation. Previous molecular phylogenetic analyses focusing on taxonomic schemes in this order simply included few species or single molecular marker, which failed to reveal relationships within Chaetophorales [1, 2, 5, 24]. The preliminary taxonomic scheme was not presented until Caisová et al. [20] reported certain additional representative species and 18S rDNA sequences in Chaetophorales. Thereafter, family Barrancaceae, as a new member was included in Chaetophorales [21] and the broadly defined family Chaetophoraceae was revised with an additional family, i.e., Fritschiellaceae [23]. The three most common and well-known genera of Chaetophorales, i.e., Chaetophora, Stigeoclonium, and Aphanochaete are polyphyletic [20, 21]. Relationships among families remain unclear, indicating the need to better understand the taxonomic scheme of this order. Most of the aforementioned phylogenetic studies are based on one or a few rRNA genes and were performed with partial Chaetophoralean taxa, and few studies have focused on chloroplast genes and the chloroplast genome.
Thus far, only two organelle genomes have been published in Chaetophorales [25, 26], thus restricting our understanding of the taxonomic status and evolution of this group. Taxon sampling, especially the lack of important species, e.g., type species in each genus, is still the most prominent obstacle for phylogenetic analysis of Chaetophorales. Chloroplast phylogenomics has recently been considered a useful approach to elucidate enigmatic evolutionary relationships among different plant taxa [27–32]. The plastome has been increasingly applied for phylogenetic and evolutionary studies on green algae with its unique advantages. The acquisition of high-density plastid genomic data, coupled with the establishment of various complex analytical methods, could greatly help understand the evolution of green plants [31, 33–38]. This study attempted to obtain 12 chloroplast genomes in Chaetophorales. This study aimed to phylogenetically reconstruct order Chaetophorales and determine the taxonomic scheme and to further the current understanding of the evolution of the order Chaetophorales.
Results
General features of cpDNA
This study contains data from 14 chloroplast genomes representing the existing major branches of Chaetophorales. Seven of twelve newly added chloroplast genomes were with complete genomic maps (Additional file 1).
All complete chloroplast genomes of Chaetophorales (Table 1) consistently contained 67 protein-coding genes and 3 rRNA genes without inverted repeats (IR). Protein-coding genes primarily included 5 psa, 15 psb, 11 rps, 8 rpl, 6 atp, 5 rpo, 4 pet, 3 chl and 4 ycf genes. Furthermore, some genes appeared only once, such as the rbc, cem, fts, clp, tuf, and ccs. Significant differences were observed in genome size, GC content, total number of genes, number of tRNAs, number of introns, and number of protein-coding genes distributed on the positive and negative strands of the genome respectively. The chloroplast genome size ranged 150,157–223,902 bp. Aphanochaete elegans (HB201732) had the smallest chloroplast genome, and Stigeoclonium helveticum (UTEX 441) had the largest chloroplast genome. The GC content ranged 23.88–31.70%, of which Aphanochaete elegans (HB201732) had the lowest GC content, and Chaetophoropsis polyrhium (HB201646) had the highest GC content. The number of tRNAs ranged 25–30, which was markedly different. Introns varied between 2 and 33. Aphanochaete elegans (HB201732) only contained two introns, displaying the most compact genome, while Schizomeris leibleinii (UTEX LB 1228) contained 33 introns. Furthermore, the distribution of genes on the coding strand was skewed and varied among species. The protein-coding genes were distributed among both strands, and the number of genes at the plus or minus strand varied among different species. The distribution of protein-coding genes of Aphanochaete elegans (HB201732) was the most uneven (+/−, 51/16). The total length of the coding region accounted for 45.15–65.79%, and Aphanochaete elegans (HB201732) accounted for the highest proportion, while Stigeoclonium sp. (bmA10) accounted for the lowest proportion.
Table 1.
The complete chloroplast genome features of the Chaetophorales
| Taxa | Size (bp) | GC content (%) | Number of gene | CDS percent | CDS (plus/minus) | tRNA | rRNA | Intron | |
|---|---|---|---|---|---|---|---|---|---|
| + | – | ||||||||
| Uronema confervicolum | 182,093 | 27.21 | 96 | 58.42 | 37 | 30 | 26 | 3 | 27 |
| Aphanochaete confervicola | 157,920 | 27.04 | 99 | 64.29 | 48 | 19 | 29 | 3 | 9 |
| Aphanochaete elegans | 150,157 | 23.88 | 99 | 65.79 | 51 | 16 | 29 | 3 | 2 |
| Chaetophora sp. | 208,126 | 30.27 | 98 | 49.87 | 47 | 20 | 28 | 3 | 14 |
| Stigeoclonium sp. | 193,940 | 27.92 | 98 | 45.15 | 23 | 44 | 28 | 3 | 28 |
| Chaetophoropsis polyrhizum | 214,786 | 31.70 | 95 | 45.68 | 27 | 40 | 25 | 3 | 26 |
| Draparnaldia mulabilis | 202,965 | 30.55 | 98 | 49.15 | 21 | 46 | 28 | 3 | 24 |
| Schizomeris leibleinii | 182,759 | 27.20 | 98 | 51.60 | 49 | 18 | 30 | 3 | 33 |
| Stigeoclonium helveticum | 223,902 | 28.90 | 97 | 48.60 | 24 | 43 | 28 | 3 | 21 |
Furthermore, five fragmentary chloroplast genomes were obtained. Despite different degrees of deletions in the chloroplast genome, partial genome sequences we generated, including complete sequences of all 58 protein-coding genes shared among the completely sequenced cpDNAs; therefore, protein-coding genes were maximally extracted for phylogenetic analyses (Table 1).
Phylogenetic analyses based on the four nuclear concatenated markers (18S + 5.8S + ITS2 + partial 28S rDNA)
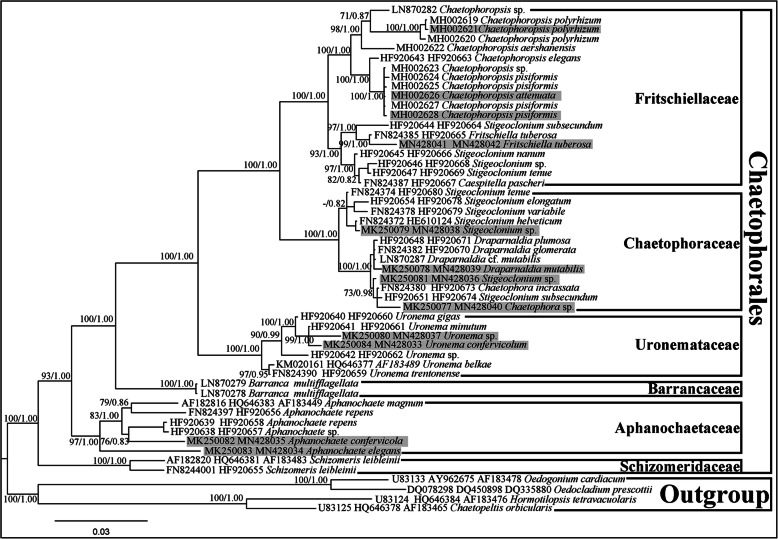
The 53-taxa alignment comprised 3032 bp. In total, 664 sites among these nucleotides were variable, of which 496 sites were parsimoniously informative and 168 sites were singleton sites. The average content of A, T, C, and G was 24.17, 25.67, 21.46, and 28.70%, respectively, of which the G + C content (50.16%) was greater than that of the A + T content (49.84%). The transition/transversion ratio was 1.77. Chloroplast genomes from 12 strains represented four families herein and are shaded in grey. The phylogenetic trees generated using the Bayesian and ML methods displayed similar topologies to those reported previously [21, 39, 40]. Phylogenetic analyses of both alignments resolved six currently recognized monophyletic families in Chaetophorales [23]. Family Schizomeridaceae, as a sister family of those in Chaetophorales, was the basal clade of Chaetophorales with robust support (100/1.00) and was markedly separated from Aphanochaetaceae (Fig. 1).
Fig. 1.
ML and Bayesian phylogenetic tree of the Chaetophorales constructed by using a concatenated data set of four markers (18S + 5.8S + ITS2 + partial 28S rDNA). The best-fit model was GTR + I + G. The numbers on the nodes represent the posterior probabilities (PP)/bootstrap support values (BP) above 50/0.50. The tree was rooted with two species of Oedogoniales and Chaetopeltidales respectively. Strains for chloroplast genomes investigated in this study are shaded in grey
Phylogenetic analyses based on the chloroplast protein-coding genes
Both data sets were assembled from the following 58 protein-coding genes: atpA, atpB, atpE, atpF, atpH, atpI, ccsA, cemA, chlB, chlN, clpP, petB, petD, petG, petL, psaA, psaB, psaC, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ, rbcL, rpl2, rpl5, rpl14, rpl16, rpl20, rpl23, rpl36, rpoA, rpoC2, rps3, rps4, rps7, rps8, rps9, rps11, rps12, rps14, rps18, rps19, tufA, ycf12, ycf3, and ycf4.
These aforementioned genes formed a concatenated nucleotide (nt) dataset comprising 32,019 and 21,346 base pairs (without 3rd codon positions). In total, 18,578 sites and 10,706 in these nucleotides were variable, of which 16,752 and 9429 sites were parsimoniously informative and 1826 and 1277 sites were singleton sites. The average content of A, T, C, and G was 31.44, 33.89, 15.25, and 19.42% for the complete data set, and 29.71, 32.57, 16.03, and 21.69% for the dataset without 3rd codon positions, wherein the A + T content was markedly greater than that of G + C. The 58 protein-coding genes concatenated amino acid (aa) dataset comprised 10,673 characters.
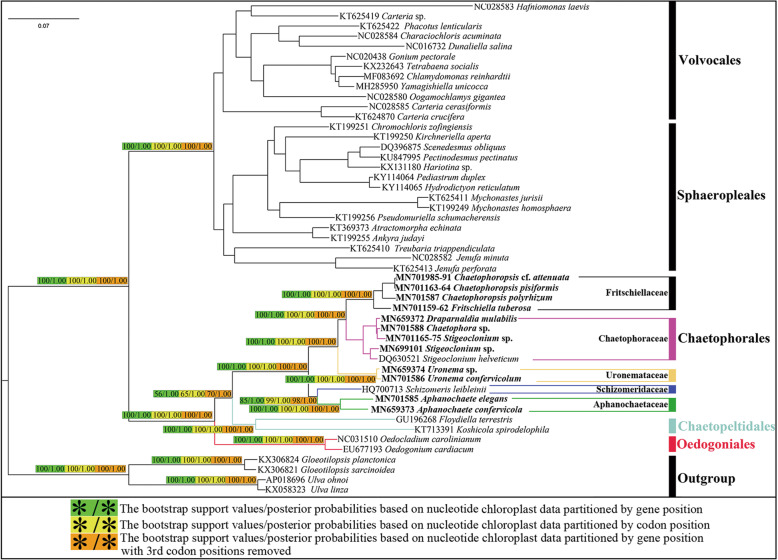
Maximum likelihood (ML) phylogenetic trees generated with the concatenated nucleotide (nt) data set treated with three methods (partitioned by gene position, codon position, and gene position without 3rd codon positions) had low support values at the node of the clade (orders Chaetophorales and Chaetopeltidales) (56/65/70) (Fig. 2).
Fig. 2.
ML and Bayesian phylogenetic tree of the Chlorophyceae constructed by using 58 protein-coding genes of the chloroplast genomes. The concatenated nucleotide (nt) data set treated by three methods (partitioned by gene position, codon position and gene position without 3rd codon positions). The posterior probabilities (PP)/bootstrap support values (BP) above 50/0.50 are only shown on the key nodes. The tree was rooted with four species of the Ulvophyceae. Strains for chloroplast genomes investigated in this study are in bold
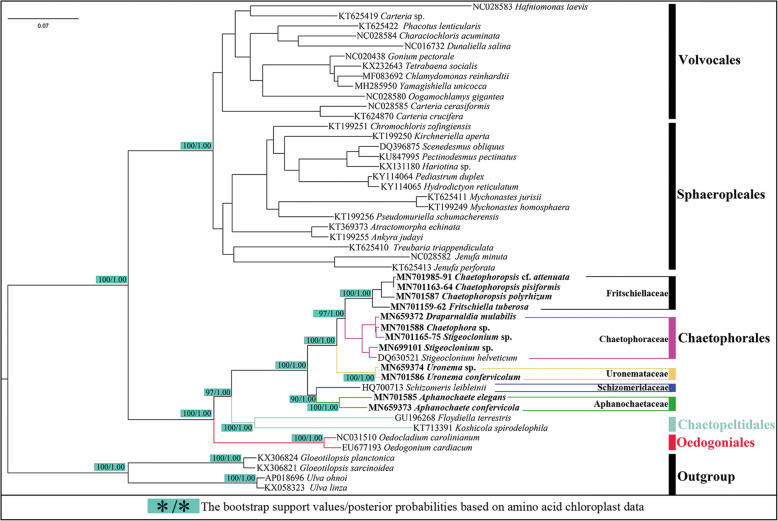
Nonetheless, the topologies of phylogenetic trees generated with concatenated datasets (nt and aa) were almost identical to each other and the support values in the amino acid (aa) data set were high at almost all nodes (Fig. 3), in contrast with previous studies with rDNA datasets [20, 21, 39]; this can be visualized on the basis of two aspects: the topologies and the support value, especially in the OCC clade. The support values in concatenated datasets of the chloroplast were markedly higher than those on rDNA datasets. Chlorophyceae diverged into two well-supported clades: VS and OCC clades. In the OCC clade, Oedogoniales was located at the base of the branch, and Chaetophorales and Chaetopeltidales were most closely related. Regarding the marked differences in the inner branching in Chaetophorales, Chaetophorales diverged into four well-supported clades, including five currently approved families except for Barrancaceae: Schizomeriaceae, Aphanochaetaceae, Uronemataceae, Fritschiellaceae, and Chaetophoraceae. Schizomeriaceae and Aphanochaetaceae could not be adequately separated, as rDNA datasets instead clustered into one branch at the base of order Chaetophorales. Chaetophoraceae sensu lato was located at the top branch of the Chaetophorales, displaying a basal split into the two well-supported clades, representing Fritschiellaceae and Chaetophoraceae sensu stricto, respectively. Family Uronemataceae as the sister was most closely related to Chaetophoraceae sensu lato.
Fig. 3.
ML and Bayesian phylogenetic tree of the Chlorophyceae constructed by using concatenated 58 amino acid (aa) data set of the chloroplast genomes. The posterior probabilities (PP)/bootstrap support values (BP) above 50/0.50 are only shown on the key nodes. The tree was rooted with four species of the Ulvophyceae. Strains for chloroplast genomes investigated in this study are in bold
Synteny analysis
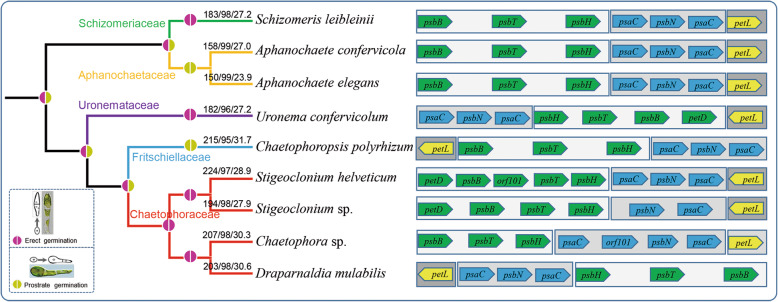
ProgressiveMauve was used to analyze synteny in the chloroplast genome in Chaetophorales and set Schizomeris leibleinii as the reference genome [26]. Synteny analysis is illustrated in Fig. 4. Nine genomes from five families were used, including seven genera, and more than 27 locally collinear blocks (LCBs) were identified. The LCB connecting lines were confounding among chloroplast genomes and considerable rearrangements and inversions were noted, especially in Fritschiellaceae and Chaetophoraceae. The largest LCB was more than 40 kb (Fig. 4a). Synteny was highly homogenous among Schizomeris leibleinii (Schizomeridaceae), Aphanochaete confervicola, and Aphanochaete elegans (Aphanochaetaceae) (Fig. 4b). Three conserved LCBs comprising common genes (psbB, psbT, and psbH), (psaC and psbN), and (petL), respectively, were somewhat modified within most members of Chaetophorales. For example, compared to Schizomeris leibleinii, LCB (psbB, psbT, psbH) included another gene petD and orf101, and gene petL was inverted in Stigeoclonium helveticum. Similar patterns were observed in other species. Moreover, gene psbN was proximal to psaC; however, it did not split and transsplice psaC in Stigeoclonium sp. Nonetheless, the aforementioned three LCBs between Schizomeridaceae and Aphanochaetaceae were highly conserved (Fig. 5). Furthermore, the guide tree inferred from chloroplast genomes, using progressiveMauve, clearly indicated that Schizomeridaceae and Aphanochaetaceae clustered into one clade at the base of Chaetophorales (Fig. 5).
Fig. 4.
Synteny comparison of the Chaetophorales chloroplast genomes using progressiveMauve. a, Synteny comparison of nine chloroplast genomes representing five family; b, Synteny comparison of the family Schizomeridaceae (Schizomeris leibleinii HQ700713) and Aphanochaetaceae (Aphanochaete confervicola MN659373; Aphanochaete elegans MN701585). The coloured syntenic blocks are local collinear blocks; blocks above the centre line indicate they are on the same strand, and blocks below the centre line indicate they are on the opposite strand
Fig. 5.
Chloroplast genomic feature and evolutionary relationship based on the zoospore germination of the Chaetophorales. The guide tree inferred from the chloroplast genomes by using progressiveMauve. Three conserved Locally Collinear Blocks (LCB) and genes of Chaetophorales are shaded in different colour. The numbers on the node represent the size/number of gene/GC content of the chloroplast respectively
Evolution of the Chaetophorales based on the germination type of zoospores
Morphological and life history observations clearly revealed that in the order Chaetophorales, zoospores of Schizomeriaceae contained zoospores for erect germination; Aphanochaetaceae, prostrate germination. Uronemataceae only contained zoospores for erect germination. In Chaetophoraceae sensu lato, zoospores of the family Chaetophoraceae sensu stricto and family Fritschiellaceae were present for erect germination and prostrate germination, respectively [23].
Based on the germination type of zoospores, the evolutionary hypothesis of Chaetophorales was proposed: the clade including Schizomeriaceae and Aphanochaetaceae including zoospores for erect and prostrate germination, respectively, was most closely related to the original ancestors of Chaetophorales, wherein the aforementioned two families were clustered together and located at the base of Chaetophorales; Uronemataceae displayed a loss of traits [20], only retaining zoospores for erect germination. In Chaetophoraceae sensu lato, the Stigeoclonium-like ancestors evolved independently in two directions. Some of them evolved into a group only with zoospores for prostrate germination and the highly differentiated prostrate in genera Fritschiella and Chaetophoropsis (Fritschiellaceae). The other part evolved into a group containing only zoospores for erect germination and the highly differentiated erect part in genus Draparnaldia (Chaetophoraceae) [41, 42], which were located at the top branch of Chaetophorales representing the most evolved taxa (Fig. 5).
Disscussion
Unlike most green algae, the chloroplast genome of Chaetophorales does not have a typical quadripartite structure (a large single-copy region, a small single-copy region, and two inverted repeats separated by the single-copy region), and the inverted repeat region (IR) is obliterated. This phenomenon is not unique to species in Chaetophorales, some green algal groups have also presented a loss of this structure [6, 25, 43–49]. Inverted repeats have been lost numerous times during evolution in green algae, even in the same group [45]. Within Chlorophycean green algae, IR loss may be a synapomorphy marking the common ancestry of Chaetophorales and Chaetopeltidales [49], because the IR is obliterated in the plastomes of Floydiella (Chaetopeltidales), Stigeoclonium, and Schizomeris (Chaetophorales); however, it is present in Oedogoniales and other remaining investigated Chlorophyceae [5, 25, 26, 47]. The mechanisms leading to IR loss are still largely unknown [50].
In general, the size of the chloroplast genome of Chaetophorales tends to increase among families from Schizomeriaceae to Chaetophoraceae. The smallest chloroplast genome belongs to Aphanochaete elegans (family Aphanochaetaceae) and the largest one belongs to Fritschiella tuberosa (family Fritschiellaceae), despite its fragmentary chloroplast genomes. This difference in chloroplast genome size results primarily from differences in non-coding regions. Aerial or subaerial algae may have the larger chloroplast genomes than freshwater algae, e.g., Fritschiella tuberosa in this study, Floydiella terrestris in Chaetopeltidales [47] and Trentepohlia odorata in Trentepohliales [51]. Subaerial genera Fritschiella and Floydiella include only one species thus far [52]. Large genome constraints during speciation influence the species distribution and abundance, and plant physiology [53]. However, further studies are required to determine whether this phenomenon occurs in this order. Plant evolution in Chaetophorales has become more complex, consistent with that in the chloroplast genome, which tends to expand from the base to the top.
Evolution of the chloroplast genome in Chaetophorales tends to cause AT enrichment, consistent with other green algal groups [6]. In contrast with Chaetophorales plastomes, contiguous genes in the Floydiella chloroplast genome markedly tend to be clustered on the same strand [47]. The distribution of protein-coding genes in two chains of the chloroplast genome vary among different species; this distribution is most balanced in family Uronemataceae but gravely imbalanced in family Aphanochaetaceae, which can be explained by gene inversions and rearrangements [54]. Synteny analyses have accounted for numerous complex rearrangements and inversions among the chloroplast genomes of Chaetophorales; however, families Schizomeriaceae and Aphanochaetaceae displayed another trend. The plastome structures and conserved gene blocks in both Schizomeriaceae and Aphanochaetaceae were more similar to each other than to those of other families according to synteny comparison performed herein, as evident from their close phylogenetic relationship.
Phylogenetic analyses based on nuclear rDNA were incongruent with chloroplast genes, especially on the relative position of families Schizomeriaceae and Aphanochaetaceae, resulting from taxon sampling and characteristics of genes themselves. In contrast with numerous nuclear genes with limited resolving power and multi-copy in nature, thus potentially confounding phylogenetic reconstruction, organellar genes are typically single-copy by nature and do not present these issues [6, 55]. Furthermore, because of their relatively high and condensed gene content, chloroplast genomes are particularly useful for phylogenetic reconstruction [6]. Although support values of phylogenetic trees based on nt datasets were lower than those based on aa datasets at certain nodes, all of them supported the same unique topologies of the OCC clade and Chaetophorales.
Attempts to resolve relationships among the OCC clade met with limited success owing to the lack of samples and few genes [1, 2, 5, 20, 26, 47, 56–59]. By increasing the sizes of the chloroplast genomes of Chaetophorales, the present results confirmed that Chaetophorales and Chaetopeltidales constituted a clade, in contrast with basal Oedogoniales in the OCC group.
Conclusions
In conclusion, chloroplast genome structure analyses, synteny analyses, and the zoospore germination analyses were concurrent with phylogenetic analyses based on the chloroplast genome, and all of them robustly determined the unique taxonomic scheme of Chaetophorales. Further studies are required to carry out phylogenetic analysis with a large number of samples to yield more convincing genomic data regarding phylogenetic relationships within Chaetophorales, thus furthering the current understanding of chloroplast genome evolution.
Methods
Taxon sampling and culture conditions
All strains described herein were sampled from different districts of China and voucher species was stored at the Freshwater Algal Herbarium (HBI), Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China. Detailed information was given in Table 2.
Table 2.
Detail information on the species with Chloroplast genome data of the order Chaetophorales. Taxa newly of this study are in bold
| Family | Taxa | Isolator, isolation data | Voucher specimen | GenBank accession number | ||
|---|---|---|---|---|---|---|
| 18S rDNA | 5.8S-ITS2-partial 28S 28S |
cpDNA | ||||
| Schizomeridaceae | Schizomeris leibleinii | unknown | UTEX LB 1228 | unkown | unkown | HQ700713 [26] |
| Aphanochaetaceae | Aphanochaete confervicola | B. W. Liu, 2017,. Hubei province, China. An epiphyte on the Oedogonium, freshwater. | HB201725 | MK250082 | MN428035 | MN659373 |
| Aphanochaete elegans | B. W. Liu, 2017,. Hubei province, China, on the water grass, freshwater. | HB201732 | MK250083 | MN428034 | MN701585 | |
| Uronemataceae | Uronemasp. | B. W. Liu, 2017,. Guizhou province, China, on the Lotus leaf in a pool, freshwater. | GZ201706 | MK250080 | MN428037 | MN659374 |
| Uronema confervicolum | B. W. Liu, 2017,. Henan province, China, on water grasses in a river, freshwater. | LY201701 | MK250084 | MN428033 | MN701586 | |
| Chaetophoraceae | Stigeocloniumsp. | B. W. Liu, 2016, Tibet, China, on the stones in a stream, freshwater. | bmA10 | MK250079 | MN428038 | MN699101 |
| Draparnaldia mulabilis | B. W. Liu, 2017, Aershan, Hinggan, Inner mongolia province, China, on the stones in Halaha river, freshwater. | AES201713 | MK250078 | MN428039 | MN659372 | |
| Chaetophorasp. | B. W. Liu, 2017, Aershan, Hinggan, Inner mongolia province, China, on the stones in Halaha river, freshwater. | AES201704 | MK250077 | MN428040 | MN701588 | |
| Stigeoclonium helveticum | unknown | UTEX 441 | EU123941 | unkown | DQ630521 [25] | |
| Stigeocloniumsp. | B. W. Liu, 2016,. Hubei province, China. Wuhan Botanical Garden, Chinese Academy of Sciences, on the stick in a pool, freshwater. | HB201635 | MK250081 | MN428036 | MN701165–75 | |
| Fritschiellaceae | Fritschiella tuberosa | B. W. Liu, 2018,. Hubei province, China. Sanbar of Yangtze River, on the moist soil. | HB201823 | MN428041 | MN428042 | MN701159–62 |
| Chaetophoropsis polyrhizum | B. W. Liu, 2016,. Hubei province, China. Wuhan Botanical Garden, Chinese Academy of Sciences, on water grasses in a pool, freshwater. | HB201646 | MF497328 | MH002621 | MN701587 | |
| Chaetophoropsis pisiformis | B. W. Liu, 2017, Guizhou province, China. Shuangrufeng, on the Lotus leaf in a pool, freshwater. | XR201704 | MH002618 | MH002628 | MN701163–64 | |
| Chaetophoropsiscf.attenuata | B. W. Liu, 2016, Hubei province, China. Wuhan Botanical Garden, Chinese Academy of Sciences, on the stick in a pool, freshwater. | FHB201644 | MH002616 | MH002626 | MN701985–91 | |
Each sample was preserved in 4% formalin for the morphological study. Natural samples were isolated using an Olympus SZX7 microscope (Olympus Corp., Tokyo, Japan) and rinsed with double-distilled H2O. The algae were first grown in culture dishes on sterilized BBM medium [60] solidified with 1.2% agar under the photon fluence rate of 15–35 μmol m− 2 s− 1 in a 14:10 h light:dark cycle at 20 °C and transferred into the fresh medium every week untill to be the unialgal strains. The unialgal strains were cultivated in liquid BBM medium [60] at 20 °C, under the photon fluence rate of 45–60 μmol m− 2 s− 1, in a 14/10 h light/dark cycle. The algae were grown at 20 °C in the dark for approximately 48 h to induce the liberation of zoospores. Microphotographs were taken with an Olympus BX53 light microscope (Olympus Corp., Tokyo, Japan) using the differential interference contrast method. The photographs were taken under an oil immersion objective lens.
Nuclear DNA extraction, polymerase chain reaction amplification and phylogenetic analyses
Nuclear DNA extraction and phylogenetic analyses were conducted according to Liu et al. [40]. The polymerase chain reaction (PCR) of the 18S rDNA was amplified according to Medlin et al. [61]. Amplifications of internal transcribed spacer of nuclear ribosomal DNA (ITS) and of partial 28S rDNA were performed using (1) EAF3 forw + ITS055 rev or (2) 1380 forw + 1495 rev [39]. ContigExpress Project (Invitrogen, Grand Island, New York USA) was used to edit low-quality regions and assemble the partial sequences.
Chloroplast DNA sequencing, assembly, and annotation
Twelve species were used to isolate the chloroplast DNA. Chloroplast DNA (cpDNA) was isolated using an improved extraction method [62]. Purified DNA was fragmented and used to construct short-insert libraries according to the manufacturer’s instructions (Illumina), and sequenced on the Illumina Hiseq 4000 [63]. The data were trimmed using SOAPnuke 1.3.0 [64] and assembled with SPAdes 3.13.0 [65].
The chloroplast genes were annotated using an online DOGMA tool [66]. Protein-coding and ribosomal RNA genes were further polished using Blast with genes from the available the Chaetophorales Chloroplast DNA [25, 26]. The tRNA genes were redetected using tRNAScan-SE 1.21 [67]. The circular chloroplast genome maps were drawn using OrganellarGenomeDRAW 1.2 [68]. Intron boundaries were determined by modeling intron secondary structures and by comparing intron-containing genes with intronless homologs [69, 70]. The annotated chloroplast genomes were submitted to GenBank under the accession numbers given in Table 2.
Phylogenetic analyses based on chloroplast genome
Total 49 Chlorophyceae (Chlorophyta) taxa were used to generate the analysed nucleotide and amino acid data sets. The large, hypervariable genes ftsH, ycf1, rpoC1, and rps2 were not included in analyses, and only genes present in all ingroup and outgroup taxa were used for phylogenetic analyses [31].
Each gene was aligned using mafft 7.0 [71]. The ambiguously aligned regions were further manually edited and adjusted by eye and translated into amino acid using MEGA6 [72]. All genes were then concatenated using Phyutility [73]. The evolutionary models and partitioning of each data set were determined by PartitionFinder2 [74].
The concatenated data set of nucleotide were partitioned by gene position, codon position and gene position without 3rd codon positions respectively. The concatenated data set of amino acid was partitioned by gene position. Phylogenies were inferred from both data sets using maximum likelihood (ML) and Bayesian (BI) methods. Phylogenetic trees were conducted using MrBayes 3.2.6 [75] and RAxML 8.2.10 [76]. MrBayes was ran for 5,000,000 generations, sampling and printing every 500 and bootstrap analyses with 1000 replicates of the ML dataset were performed to estimate the statistical reliability. The synteny comparison was visualized using progressiveMauve [77].
Supplementary information
Additional file 1 Figs. S1-S7. Gene map of seven complete chloroplast genomes of the Chaetophorales. Arrows show the direction of transcription. The same colour block shows the functional gene group (legend at bottom left). Transfer RNAs are represented by their one-letter amino acid code. The grey circle on the inside shows a graph of the GC content. Fig. S1.Aphanochaete repens (HB201725). Fig. S2.Aphanochaete elegans (HB201732). Fig. S3.Uronema repens (LY201701).Fig. S4.Chaetophoropsis polyrhiza (HB201646).Fig. S5.Stigeoclonium sp. (bmA10).Fig. S6.Draparnaldia mutabilis (AES201713).Fig. S7.Chaetophora sp. (AES201704).
Acknowledgements
Special thanks go to two anonymous reviewers for their insightful and helpful comments and editorial corrections to improve our manuscript.
Abbreviations
- SV clade
Sphaeropleales and volvocales
- OCC clade
Oedogoniales, chaetophorales, and chaetopeltidales
- IR
Inverted repeats
- nt
Nucleotide
- aa
Amino acid
- ML
Maximum likelihood
- HBI
Freshwater algal herbarium
- PCR
Polymerase chain reaction
- ITS
Internal transcribed spacer of nuclear ribosomal DNA
- cpDNA
Chloroplast DNA
Authors’ contributions
BL, ZH, GL and HZ conceived, designed the study. BL, YH and HZ analyzed the data. BL wrote the manuscript. All authors have read and approved the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 31770220), the China Postdoctoral Science Foundation (Grant No. 2019 M662748), and the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (Grant No. 2019QZKK05010102). The funding agency had no role in the design of the study, collection, analysis, or interpretation of data, or in writing the manuscript.
Availability of data and materials
All data generated and analysed during this study are included in this published article and its supplementary information files. Raw sequencing data of all species are available from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). Accession numbers of cpDNA: MN659373, MN701585, MN659374, MN701586, MN699101, MN659372, MN701588, MN701165-MN701175, MN701159-MN701162, MN701587, MN701163-MN701164 and MN701985-MN701991.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-06845-y.
References
- 1.Booton GC, Floyd GL, Fuerst PA. Origins and affinities of the filamentous green algal orders Chaetophorales and Oedogoniales based on 18SrRNA gene sequences. J Phycol. 1998;34(2):312–318. [Google Scholar]
- 2.Buchheim MA, Michalopulos EA, Buchheim JA. Phylogeny of the Chlorophyceae with special reference to the Sphaeropleales: a study of 18S and 26Sr RNA data. J Phycol. 2001;37(5):819–835. [Google Scholar]
- 3.Wolf M, Buchheim M, Hegewald E, Krienitz L, Hepperle D. Phylogenetic position of the Sphaeropleaceae (Chlorophyta) Plant Syst Evol. 2002;230(1):161–171. [Google Scholar]
- 4.Wolf M, Hepperle D, Krienitz L. On the phylogeny of Radiococcus, Planktosphaeria and Schizochlamydella (Radiococcaceae, Chlorophyta) Biologia. 2003;58(4):759–765. [Google Scholar]
- 5.Turmel M, Brouard J-S, Gagnon C, Otis C, Lemieux C. Deep division in the Chlorophyceae (Chlorophyta) revealed by chloroplast phylogenomic analyses. J Phycol. 2008;44(3):739–750. doi: 10.1111/j.1529-8817.2008.00510.x. [DOI] [PubMed] [Google Scholar]
- 6.Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, Clerck OD. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31(1):1–46. [Google Scholar]
- 7.Wille N. Algologische Notizen VII, VIII. Nyt Magazin Naturvidenskapens. 1901;39:1–24. [Google Scholar]
- 8.Printz H. Die Chaetophoralen der Binnengewässer. Hydrobiologia. 1964;24(1–2):1–376. [Google Scholar]
- 9.Bourrelly P. Les Algues d’ eau Douce I. les Algues Vertes. Paris: Boubée; 1966. [Google Scholar]
- 10.Manton I. Observations on fine structure of zoospore and young germling of Stigeoclonium. J Exp Bot. 1964;15(2):399–411. [Google Scholar]
- 11.Stewart KD, Mattox KR, Floyd GL. Mitosis, cytokinesis, distribution of Plasmodesmata, and other cytological characteristics in Ulotrichales, Ulvales, and Chaetophorales: phylogenetic and taxonomic considerations. J Phycol. 1973;9(2):128–141. [Google Scholar]
- 12.Melkonian M. Fine structure of zoospores of Fritschiella tuberosa Iyeng (Chaetophorineae, Chlorophyceae) with special reference to flagellar apparatus. Protoplasma. 1975;86(4):391–404. [Google Scholar]
- 13.Silva PC. Chlorophycota. In: Parker S, editor. Synopsis and classification of living organisms. New York: Mc Graw-Hill; 1982. pp. 133–161. [Google Scholar]
- 14.Mattox KR, Stewart KD. Classification of the green algae: a concept based on comparative cytology. In: Irvine DEG, John DM, editors. Systematics of the green algae. London: Academic Press; 1984. pp. 29–72. [Google Scholar]
- 15.O’Kelly CJ, Floyd GL. Flagellar apparatus absolute orientations and the phylogeny of the green algae. Biosystems. 1984;16(3–4):227–251. doi: 10.1016/0303-2647(83)90007-2. [DOI] [PubMed] [Google Scholar]
- 16.Melkonian M. Chlorophyte orders of uncertain affinities: order Pedinomonadales. In: Margulis L, Corliss JO, Melkonian M, Chapman DJ, editors. Handbook of Protoctista. The Structure, Cultivation, Habitats and Life Histories of the Eukaryotic Microorganisms and Their Descendants Exclusive of Animals, Plants and Fungi. Boston: Jones and Bartlett; 1990. pp. 649–651. [Google Scholar]
- 17.Friedl T, Zeltner C. Assessing the relationships of some coccoid green lichen algae and the Microthamniales (Chlorophyta) with 18S ribosomal-RNA gene sequence comparisons. J Phycol. 1994;30(3):500–506. [Google Scholar]
- 18.O’Kelly CJ, Watanabe S, Floyd GL. Ultrastructure and phylogenetic relationships of Chaetopeltidales Ord. Nov. (Chlorophyta, Chlorophyceae) J Phycol. 1994;30(1):118–128. [Google Scholar]
- 19.Lewis LA, McCourt RM. Green algae and the origin of land plants. Am J Bot. 2004;91(10):1535–1556. doi: 10.3732/ajb.91.10.1535. [DOI] [PubMed] [Google Scholar]
- 20.Caisová L, Marin B, Sausen N, Pröschold T, Melkonian M. Polyphyly of Chaetophora and Stigeoclonium within the Chaetophorales (Chlorophyceae), revealed by sequence comparisons of nuclear-encoded SSU rRNA genes. J Phycol. 2011;47(1):164–177. doi: 10.1111/j.1529-8817.2010.00949.x. [DOI] [PubMed] [Google Scholar]
- 21.Caisová L, Pérez Reyes C, Álamo VC, Quintana AM, Surek B, Melkonian M. Barrancaceae: a new green algal lineage with structural and behavioral adaptations to a fluctuating environment. Am J Bot. 2015;102(9):1482–1492. doi: 10.3732/ajb.1500199. [DOI] [PubMed] [Google Scholar]
- 22.Caisová L. Dicranochaete – an enigmatic green alga with surprising adaptive capabilities. Phycologia. 2016;55(2):219–229. [Google Scholar]
- 23.Caisová L, Melkonian M. The Chaetophorales (Chlorophyceae) – a taxonomic revision at family level. Eur J Phycol. 2018;53(3):1–12. [Google Scholar]
- 24.Brouard JS, Otis C, Lemieux C, Turmel M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics. 2008;9:290–316. doi: 10.1186/1471-2164-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bélanger AS, Brouard JS, Charlebois P, Otis C, Lemieux C, Turmel M. Distinctive architecture of the chloroplast genome in the chlorophycean green alga Stigeoclonium helveticum. Mol Genet Genomics. 2006;276(5):464–477. doi: 10.1007/s00438-006-0156-2. [DOI] [PubMed] [Google Scholar]
- 26.Brouard JS, Otis C, Lemieux C, Turmel M. The chloroplast genome of the green alga Schizomeris leibleinii (Chlorophyceae) provides evidence for bidirectional DNA replication from a single origin in the Chaetophorales. Genome Biol Evol. 2011;3:505–515. doi: 10.1093/gbe/evr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen RK, Cai ZQ, Raubeson LA, Daniell H, de Pamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, Peery R, JR MN, Kuehl JV, Boore JL. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. P Natl Acad Sci USA. 2007;104(49):19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. P Natl Acad Sci USA. 2010;107(10):4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 2011;3:1284–1295. doi: 10.1093/gbe/evr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong BJ, Deusch O, Goremykin VV, Penny D, Biggs PJ, Atherton RA, Nikiforova SV, Lockhart PJ. Systematic error in seed plant phylogenomics. Genome Biol Evol. 2011;3:1340–1348. doi: 10.1093/gbe/evr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fučíková K, Lewis PO, Lewis LA. Chloroplast phylogenomic data from the green algal order Sphaeropleales (Chlorophyceae, Chlorophyta) reveal complex patterns of sequence evolution. Mol Phylogenet Evol. 2016;98:176–183. doi: 10.1016/j.ympev.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 32.Sun LH, Fang L, Zhang ZH, Chang X, Penny D, Zhong BJ. Chloroplast phylogenomic inference of green algae relationships. Sci Rep. 2016;6:20528. doi: 10.1038/srep20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turmel M, Otis C, Lemieux C. Tracing the evolution of streptophyte algae and their mitochondrial genome. Genome Biol Evol. 2013;5(10):1817–1835. doi: 10.1093/gbe/evt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turmel M, Otis C, Lemieux C. Dynamic evolution of the chloroplast genome in the green algal classes Pedinophyceae and Trebouxiophyceae. Genome Biol Evol. 2015;7(7):2062–2082. doi: 10.1093/gbe/evv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fučíková K, Leliaert F, Cooper ED, Škaloud P, D'Hondt S, Clerck OD, Gurgel CFD, Lewis LA, Lewis PO, Lopez-Bautista JM, Delwiche CF, Verbruggen H. New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front Ecol Evol. 2014;2:63. [Google Scholar]
- 36.Lemieux C, Otis C, Turmel M. Chloroplast phylogenomic analysis resolves deep-level relationships within the green algal class Trebouxiophyceae. BMC Evol Biol. 2014;14:211. doi: 10.1186/s12862-014-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 2014;14:23. doi: 10.1186/1471-2148-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nie Y, Foster CSP, Zhu T, Yao R, Duchêne DA, Ho SYW, Zhong BJ. Accounting for uncertainty in the evolutionary timescale of green plants through clock-partitioning and fossil calibration strategies. Syst Biol. 2020;69:1–16. doi: 10.1093/sysbio/syz032. [DOI] [PubMed] [Google Scholar]
- 39.Caisová L, Marin B, Melkonian MA. Consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist. 2013;164(4):482–496. doi: 10.1016/j.protis.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Liu BW, Xiong Q, Liu XD, Liu GX, Hu ZY. Molecular phylogeny and taxonomy of the genus Chaetophora (Chlorophyceae, Chlorophyta), including descriptions of Chaetophoropsis aershanensis gen. Et sp. nov. J Phycol. 2019;55(1):74–83. doi: 10.1111/jpy.12803. [DOI] [PubMed] [Google Scholar]
- 41.Forest HS. A study of the genera Draparnaldia Bory and Draparnaldiopsis smith and klyver. Castanea. 1956;21(1):1–27. [Google Scholar]
- 42.Islam AKMN. A revision of the genus Stigeoclonium. Nova Hedwigia. 1963;10:1–164. [Google Scholar]
- 43.Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales. BMC Biol. 2005;3:22. doi: 10.1186/1741-7007-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Koning AP, Keeling PJ. Nucleus-encoded genes for plastidtargeted proteins in Helicosporidium: functional diversity of a cryptic plastid in a parasitic alga. Eukaryot Cell. 2004;3:1198–1205. doi: 10.1128/EC.3.5.1198-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Cambiaire JC, Otis C, Turmel M, Lemieux C. The chloroplast genome sequence of the green alga Leptosira terrestris: multiple losses of the inverted repeat and extensive genome rearrangements within the Trebouxiophyceae. BMC Genomics. 2007;8:213. doi: 10.1186/1471-2164-8-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turmel M, Otis C, Lemieux C. The chloroplast genomes of the green algae Pedinomonas minor, Parachlorella kessleri, and Oocystis solitatia reveal a shared ancestry between the Pedinomonadales and Chlorellales. Mol Biol Evol. 2009;26(10):2317–2331. doi: 10.1093/molbev/msp138. [DOI] [PubMed] [Google Scholar]
- 47.Brouard J-S, Otis C, Lemieux C, Turmel M. The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae. Genome Biol Evol. 2010;2:240–256. doi: 10.1093/gbe/evq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lü F, Xü W, Tian C, Wang GC, Niu JF, Pan GH, Hu SN. The Bryopsis hypnoides plastid genome: multimeric forms and complete nucleotide sequence. PLoS One. 2011;6:e14663. doi: 10.1371/journal.pone.0014663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe S, Fučíková K, Lewis LA, Lewis PO. Hiding in plain sight: Koshicola spirodelophila gen. Et sp. nov. (Chaetopeltidales, Chlorophyceae), a novel green alga associated with the aquatic angiosperm Spirodela polyrhiza. Am J Bot. 2016;103(5):865–875. doi: 10.3732/ajb.1500481. [DOI] [PubMed] [Google Scholar]
- 50.Turmel M, Lemieux C. Evolution of the plastid genome in green algae. Adv Bot Res. 2017;85:157–193. [Google Scholar]
- 51.Zhu H, Hu Y, Liu F, Hu Z, Liu G. Characterization of the Chloroplast Genome of Trentepohlia odorata (Trentepohliales, Chlorophyta), and Discussion of its Taxonomy. Int J Mol Sci. 2019;20:1774. [DOI] [PMC free article] [PubMed]
- 52.Guiry MD, Guiry GM. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org. Accessed 12 Jan 2020.
- 53.Knight CA, Molinari NA, Petrov DA. The large genome constraint hypothesis: evolution, ecology and phenotype. Ann Bot. 2005;95(1):177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui LY, Leebens-Mack J, Wang LS, Tang JJ, Rymarquis L, Stern DB, de Pamphilis CW. Adaptive evolution of chloroplast genome structure inferred using a parametric bootstrap approach. BMC Evol Biol. 2006;6:13. doi: 10.1186/1471-2148-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fučíková K, Lewis PO, Neupane S, Karol KG, Lewis LA. Order, please! Uncertainty in the ordinal-level classification of Chlorophyceae. PeerJ. 2019;7:e6899. doi: 10.7717/peerj.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shoup S, Lewis LA. Polyphyletic origin of parallel basal bodies in swimming cells of chlorophycean green algae (Chlorophyta) J Phycol. 2003;39(4):789–796. [Google Scholar]
- 57.Sanchez-Puerta MV, Leonardi PI, O’Kelly CJ, Cáceres EJ. Pseudulvella americana belongs to the order Chaetopeltidales (class Chlorophyceae), evidence from ultrastructure and SSU rDNA sequence. J Phycol. 2006;42(4):943–950. [Google Scholar]
- 58.Buchheim MA, Sutherland DM, Schleicher T, Förster F, Wolf M. Phylogeny of Oedogoniales, Chaetophorales and Chaetopeltidales (Chlorophyceae): inferences from sequence-structure analysis of ITS2. Ann Bot. 2012;109(1):109–116. doi: 10.1093/aob/mcr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tippery NP, Fučíková K, Lewis PO, Lewis LA. Probing the monophyly of the Sphaeropleales using data from five genes. J Phycol. 2012;48(6):1482–1493. doi: 10.1111/jpy.12003. [DOI] [PubMed] [Google Scholar]
- 60.Bischoff HW, Bold HC. Phycological studies IV. Some soil algae from enchanted rock and related algal species. Texas: University of Texas Publication; 1963. [Google Scholar]
- 61.Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA coding regions. Gene. 1988;71(2):491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 62.McPherson H, van der Merwe M, Delaney SK, Edwards MA, Henry RJ, McIntosh E, Rymer PD, Milner ML, Siow J, Rossetto M. Capturing chloroplast variation for molecular ecology studies: a simple next generation sequencing approach applied to a rainforest tree. BMC Ecol. 2013;13:8. doi: 10.1186/1472-6785-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borgstrom E, Lundin S, Lundeberg J. Large scale library generation for high throughput sequencing. PLoS One. 2011;6:e19119. doi: 10.1371/journal.pone.0019119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen YX, Chen YS, Shi CM, Huang ZB, Zhang Y, Li SK, Li Y, Ye J, Yu C, Li Z, Zhang XQ, Wang J, Yang HM, Fang L, Chen Q. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7(1):1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 67.Lowe TM, Chan PP. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44(W1):W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 69.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82(1):5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 70.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 71.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith SA, Dunn CW. Phyutility: a phyloinformatics tool for trees, alignments and moleculardata. Bioinformatics. 2008;24(5):715–716. doi: 10.1093/bioinformatics/btm619. [DOI] [PubMed] [Google Scholar]
- 74.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2017;34(3):772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 75.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 76.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS one. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Figs. S1-S7. Gene map of seven complete chloroplast genomes of the Chaetophorales. Arrows show the direction of transcription. The same colour block shows the functional gene group (legend at bottom left). Transfer RNAs are represented by their one-letter amino acid code. The grey circle on the inside shows a graph of the GC content. Fig. S1.Aphanochaete repens (HB201725). Fig. S2.Aphanochaete elegans (HB201732). Fig. S3.Uronema repens (LY201701).Fig. S4.Chaetophoropsis polyrhiza (HB201646).Fig. S5.Stigeoclonium sp. (bmA10).Fig. S6.Draparnaldia mutabilis (AES201713).Fig. S7.Chaetophora sp. (AES201704).
Data Availability Statement
All data generated and analysed during this study are included in this published article and its supplementary information files. Raw sequencing data of all species are available from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). Accession numbers of cpDNA: MN659373, MN701585, MN659374, MN701586, MN699101, MN659372, MN701588, MN701165-MN701175, MN701159-MN701162, MN701587, MN701163-MN701164 and MN701985-MN701991.