Abstract
Summary
We report Spark-based INFERence of the molecular mechanisms of NOn-coding genetic variants (SparkINFERNO), a scalable bioinformatics pipeline characterizing non-coding genome-wide association study (GWAS) association findings. SparkINFERNO prioritizes causal variants underlying GWAS association signals and reports relevant regulatory elements, tissue contexts and plausible target genes they affect. To achieve this, the SparkINFERNO algorithm integrates GWAS summary statistics with large-scale collection of functional genomics datasets spanning enhancer activity, transcription factor binding, expression quantitative trait loci and other functional datasets across more than 400 tissues and cell types. Scalability is achieved by an underlying API implemented using Apache Spark and Giggle-based genomic indexing. We evaluated SparkINFERNO on large GWASs and show that SparkINFERNO is more than 60 times efficient and scales with data size and amount of computational resources.
Availability and implementation
SparkINFERNO runs on clusters or a single server with Apache Spark environment, and is available at https://bitbucket.org/wanglab-upenn/SparkINFERNO or https://hub.docker.com/r/wanglab/spark-inferno.
Contact
lswang@pennmedicine.upenn.edu
Supplementary information
Supplementary data are available at Bioinformatics online
1 Introduction
Genome-wide association studies (GWASs) have successfully identified over 70 000 genetic variants associated with more than 3000 human diseases and phenotypes (Buniello et al., 2019). Interpretation of these associations remain difficult (Amlie-Wolf et al., 2018; Watanabe et al., 2017) as most GWAS hits are in the non-coding genome. Resolution of GWAS is limited as neighboring variants have similar associations due to linkage disequilibrium (LD) (Amlie-Wolf et al., 2018). Our recently developed INFERNO method (Amlie-Wolf et al., 2018) focuses on identifying potentially causal variants underlying observed GWAS associations by integrating with hundreds of functional genomics datasets. The current INFERNO implementation is not optimized for big data, and a scalable framework for annotating genetic variants and genomic regions generated by various human genetic studies in a high-throughput manner is in need for systematic large-scale genomic and genetic analyses.
The scale and heterogeneity of functional genomics datasets and annotations necessitate systematic, integrative analysis and interpretation of GWAS association findings. For example, while INFERNO uses relatively small set of functional genomics datasets, projects, such as GTEx (Aguet et al., 2017), FANTOM5 (Andersson et al., 2014), ENCODE (Bernstein et al., 2012) and Roadmap Epigenomics (Kundaje et al., 2015), produce >60 000 experimental datasets across >1100 tissues, cell types, biological conditions, each with millions to billions of records across the genome. In order to pair these functional annotations with modern population-level studies, such as UK Biobank (500 000 individuals with >2500 phenotypes), we need a scalable, high-throughput, robust and easy to use software that can systematically interpret hundreds of millions of genotypes across millions of participants.
We implemented Spark-based INFERence of the molecular mechanisms of NOn-coding genetic variants (SparkINFERNO) as a scalable, high-throughput automated workflow that integrates a large-scale functional genomics data repository and processes GWAS results by performing LD analysis, functional evidence evaluation and aggregation, Bayesian colocalization analysis of GWAS and expression quantitative trait loci (eQTL) signals, characterize the downstream regulatory effects including the tissue contexts, regulatory elements and target genes that they affect. We applied SparkINFERNO on inflammatory bowel disease (IBD) (Liu et al., 2015) and the International Genomics of Alzheimer’s disease (AD) GWAS datasets (Lambert et al., 2013) and show that this scalable framework is at least 60 times more efficient and able to identify the molecular mechanisms underlying non-coding GWAS signals.
2 Materials and methods
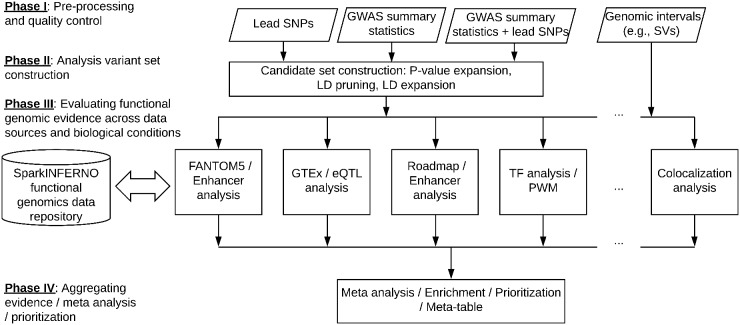
We chose Apache Spark (Zaharia et al., 2016) and Python for a scalable implementation of INFERNO (Amlie-Wolf et al., 2018) (see Supplementary Table S1 and ‘Comparison with original INFERNO implementation’ section in Supplementary Methods). The new SparkINFERNO is highly scalable, modular and coupled with an integrated functional genomics data repository (Fig. 1 and Supplementary Fig. S1 and Tables S1 and S2). Analysis modules perform various types of genomic data integration to produce functional evidence including tissue-specific regulatory elements (enhancers), transcription factor (TF) activity, chromatin states and genetic regulation (eQTL) information. SparkINFERNO implements scalable genomic querying (Supplementary Figs S2 and S3) using Spark parallel transformations and Giggle-based genomic indexing (Layer et al., 2018). SparkINFERNO can be extended with additional annotation data and/or customized evaluation modules. Results are reported by individual evaluation modules and as combined summaries (Supplementary Methods).
Fig. 1.
Overview of SparkINFERNO
SparkINFERNO accepts complete GWAS summary statistics or top GWAS association variants as the input and generates a list of potentially causal variants, affected tissue-specific enhancers and target gene(s) as the output. The entire workflow consists of four phases (Fig. 1): (i) Pre-processing and QC of GWAS input; (ii) Generating candidate set of potentially causal variants; (iii) Evaluating functional genomic evidence across genomic datasets in a tissue-specific manner including regulatory elements (enhancers), eQTL colocalization, transcriptional factor binding sites (TFBSs) and others for each GWAS locus/signal; and (iv) Aggregating evidence to infer prioritization of causal variants, including information on affected tissues/cell types, regulatory elements, TFs and target genes. See Supplementary Methods for technical details.
The pre-processing phase takes raw GWAS summary statistics in a tab-separated values format as input, resolves reference and alternative alleles, checks allele frequencies in the reference population (e.g. 1000 Genomes Project) and produces quality control flags. Quality control steps mark GWAS variants with inconsistent alleles that could not be matched with reference genotype data (Supplementary Methods and Fig. S4).
The candidate set construction phase expands genome-wide significant associations into a putative causal variant set by pruning significant variants into a smaller set of independent variants using publicly available LD data (e.g. 1000 Genome), and then expanding these signals into putative causal sets consisting of nearby variants in LD. The user can specify the reference population in LD pruning/expansion to match the population underlying the input GWAS study. Supplementary Methods and Figure S4 provide details of the workflow for generating putative variant sets.
The evaluation phase executes Spark-based annotation jobs in parallel (Fig. 1). SparkINFERNO uses an integrated repository of annotations for genomic elements (promoters, exons, introns, etc.), non-coding RNAs, regulatory elements, such as enhancers, TFBSs and others (integrated data and data repository implementation in Supplementary Table S2 and Fig. S1). The current SparkINFERNO implementation contains 3.5 billion genomic intervals from 2342 tracks for 32 tissue categories.
In the final aggregation phase, SparkINFERNO combines functional evidence from individual genomic analyses and produces a list of candidate variants, enhancer elements and their target genes as supported by FANTOM5, Roadmap, GTEx, TF binding and other functional evidence. SparkINFERNO performs colocalization analysis (Supplementary Fig. S5) of the GWAS and eQTL signals across genome-wide significant loci using COLOC (Giambartolomei et al., 2014).
To install SparkINFERNO, users can either install the package (https://bitbucket.org/wanglab-upenn/SparkINFERNO) on their own Spark cluster, or use a pre-created Docker image (wanglab/spark-inferno). To run SparkINFERNO, the user first edits the configuration file and provides input GWAS specifications. A complete run of SparkINFERNO produces candidate potentially causal variants, target genes, tissue contexts, regulatory elements and detailed BED files documenting overlaps with functional genomics and annotation datasets.
3 Results
We evaluated SparkINFERNO on our AWS Spark cluster using IGAP AD and IBD GWAS datasets containing 8 080 502 and 11 555 676 variants, respectively. For the IGAP GWAS dataset, SparkINFERNO took 993 s on a 16-core Linux server to complete the analysis, whereas the original INFERNO took 60 973 s. SparkINFERNO is 61 times faster (Supplementary Fig. S2). SparkINFERNO scales well with the amount of computational resources both in local and cluster modes (Supplementary Figs S3A and S3B), including parallel Giggle-based genomic querying (Supplementary Fig. S8). SparkINFERNO identified 1418 and 15 343 candidate causal variants and 97 and 317 colocalized target gene–tissue combinations for IGAP and IBD, respectively (Supplementary Table S3). As can be seen from distribution of identified overlaps across functional genomics datasets and tissue types (Supplementary Figs S6 and S7) SparkINFERNO identifies genes and tissues that are likely important for the disease etiology.
Funding
This work was supported by the National Institute on Aging [U24-AG041689, U54-AG052427, U01-AG032984 and T32-AG00255]; Biomarkers Across Neurodegenerative Diseases (BAND 3) (award number 18062), co-funded by Michael J Fox Foundation, Alzheimer's Association, Alzheimer's Research UK and the Weston Brain institute.
Conflict of Interest: none declared.
Supplementary Material
References
- Aguet F. et al. (2017) Genetic effects on gene expression across human tissues. Nature, 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amlie-Wolf A. et al. (2018) INFERNO: inferring the molecular mechanisms of noncoding genetic variants. Nucleic Acids Res., 46, 8740–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R. et al. ; The FANTOM Consortium. (2014) An atlas of active enhancers across human cell types and tissues. Nature, 507, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E. et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A. et al. (2019) The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res., 47, D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C. et al. (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet., 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A. et al. ; Roadmap Epigenomics Consortium. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.C. et al. ; European Alzheimer's Disease Initiative (EADI). (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet., 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer R.M. et al. (2018) GIGGLE: a search engine for large-scale integrated genome analysis. Nat. Methods, 15, 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.Z. et al. (2015) Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet., 47, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K. et al. (2017) Functional mapping and annotation of genetic associations with FUMA. Nat. Commun., 8, 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharia M. et al. (2016) Apache Spark: a unified engine for big data processing. Commun. ACM, 59, 56–65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.