Abstract
Motivation
Protein domains are subunits that can fold and function independently. Correct domain boundary assignment is thus a critical step toward accurate protein structure and function analyses. There is, however, no efficient algorithm available for accurate domain prediction from sequence. The problem is particularly challenging for proteins with discontinuous domains, which consist of domain segments that are separated along the sequence.
Results
We developed a new algorithm, FUpred, which predicts protein domain boundaries utilizing contact maps created by deep residual neural networks coupled with coevolutionary precision matrices. The core idea of the algorithm is to retrieve domain boundary locations by maximizing the number of intra-domain contacts, while minimizing the number of inter-domain contacts from the contact maps. FUpred was tested on a large-scale dataset consisting of 2549 proteins and generated correct single- and multi-domain classifications with a Matthew’s correlation coefficient of 0.799, which was 19.1% (or 5.3%) higher than the best machine learning (or threading)-based method. For proteins with discontinuous domains, the domain boundary detection and normalized domain overlapping scores of FUpred were 0.788 and 0.521, respectively, which were 17.3% and 23.8% higher than the best control method. The results demonstrate a new avenue to accurately detect domain composition from sequence alone, especially for discontinuous, multi-domain proteins.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Protein domains are the basic building blocks of protein structures, which fold and function independently. Therefore, correct detection of the domain boundaries of proteins is an essential step for determining their structural folds, understanding their biological functions and/or annotating their evolutionary mechanisms.
Due to the importance of the problem, many methods have been proposed to determine the domain boundaries of proteins. One class of methods delineates and defines domains directly from the experimental structures of proteins; these methods include PDP (Alexandrov and Shindyalov, 2003), DomainParser (Guo, 2003), DDOMAIN (Zhou et al., 2007) and SWORD (Postic et al., 2017). Another important class of methods predicts domain boundaries from the amino acid sequences. These domain prediction methods can typically be categorized into three general groups. The first group is mainly based on machine learning, with representative examples including DOMPro (Cheng et al., 2006), DoBo (Eickholt et al., 2011), ConDo (Hong et al., 2018) and DNN-dom (Shi et al., 2019). Here, DOMpro trains recursive neural networks for domain models with training features including sequence profiles, predicted secondary structure and solvent accessibility, while DoBo, which was developed by the same lab, detects domain boundaries using similar features but is based on support vector machines. ConDo utilizes neural networks that are trained on long-range, coevolutionary features in addition to conventional local window features. DNN-dom adopts a hybrid deep-learning method to predict protein domain boundaries based on features such as protein position-specific matrices, secondary structure and solvent accessibility. Additionally, balanced random forests are used to solve the classification problem. The second group of methods is based on structural templates detected from the PDB, typically by threading (Söding, 2004; Wu and Zhang, 2008). For example, ThreaDom (Xue et al., 2013) deduces domain boundary locations based on multiple threading alignments (Wu and Zhang, 2007), where the profile distribution of a domain conservation score, which combines the template domain structure and terminal/internal alignment gaps, is used to assign the domain boundary locations. Because ThreaDom cannot directly detect discontinuous domains beyond the templates, an extended version, ThreaDomEx (Wang et al., 2017), was further developed to assign discontinuous domains by domain-segment assembly. Here, a discontinuous domain is defined as a domain that contains two or more segments from separate regions of the query sequence. Finally, the third group is based on 3D structure prediction (Cheng, 2007; George and Heringa, 2002; Kim et al., 2005; Wu et al., 2009), which first models the full-length 3D structures by ab initio folding, where the domain boundaries are then deduced from the 3D structure models.
Despite their successes, each of these methods have their own limitations. The methods, which deduce domains from experimental protein structures, e.g. generally have higher accuracies than the methods that start from sequences but can only be applied to a small portion of proteins that have known experimental structures. Furthermore, the methods that predict domain boundaries from sequences are in principle more generally applicable but have their own restrictions depending on the approach. For machine learning-based methods, e.g. the accuracy of prediction is often low, although they have the advantage of being able to generate de novo predictions from sequence alone. The threading-based methods generally have higher accuracy when close templates are identified, but the accuracy decreases sharply for targets lacking homologous templates. Finally, 3D model-based methods rely on the quality of the ab initio 3D models, which can only be applied to proteins with short lengths because of the limited ability of ab initio structure prediction. Furthermore, most approaches cannot deal with the prediction of discontinuous multi-domains, except for ThreaDomEx.
In this work, we propose a new method, named FUpred (Folding Unit predictor), to detect domain boundaries from protein sequences based on contact map prediction, partly motivated by the quick progress recently achieved in the field of contact prediction (Li et al., 2019). Following the intuition of domain definition, the major procedure of FUpred is to derive an FUscore (Folding Unit score) that maximizes the number of intra-domain contacts, while minimizing the number of inter-domain contacts. Although there are some methods that utilize contact information as a machine learning feature to help predict domains, FUpred is the first method to deduce domain boundary structure directly from contact map predictions. The large-scale benchmark results presented in this study demonstrate the significant advantages associated with employing contact map-based domain prediction for domain classification and domain boundary detection, especially for discontinuous domain proteins, compared to other approaches. In particular, the case study demonstrates that FUpred can accurately detect domain boundaries for proteins with complex domain structures.
2 Materials and methods
2.1 Dataset
To train and test FUpred, we collected a set of non-redundant proteins with known domain structures from the SCOPe2.07-stable database (Chandonia et al., 2014, 2017, 2019), using a pair-wise sequence identity cutoff <30% and a sequence length cutoff >30 residues. This dataset contained 3400 single-domain and 1698 multi-domain proteins. For the multi-domain proteins, we further classified them into a continuous domain subset (1494 entries), for which every domain of each protein was a continuous segment along the query sequence, and a discontinuous domain subset (204 entries), for which at least one domain contained discontinuous segments from separate regions along the query sequence.
Table 1 lists a summary of the 1698 multi-domain proteins, where the continuous domain proteins are split into seven subsets ranging from 2- to 10-domain entries and the discontinuous domain proteins are split into nine subsets ranging from 2- to 15-domain entries. Note that the number of proteins with >3 domains is relatively small, and there are neither continuous 8-, 9- nor more than 10-domain proteins, nor discontinuous 9- to 13-domain proteins in the dataset due to the low statistics for high-order domain proteins. These proteins were randomly split into 849 training and 849 test proteins. Similarly, the 3400 single-domain proteins were also randomly split and used as the negative control dataset. The split datasets can be downloaded at https://zhanglab.ccmb.med.umich.edu/FUpred.
Table 1.
Breakdown of the 1698 multi-domain entries split into each category
| Continuous multi-domain |
Discontinuous multi-domain |
||
|---|---|---|---|
| #Domain | #Target | #Domain | #Target |
| 2 | 1175 | 2 | 129 |
| 3 | 234 | 3 | 49 |
| 4 | 63 | 4 | 11 |
| 5 | 13 | 5 | 7 |
| 6 | 7 | 6 | 3 |
| 7 | 1 | 7 | 1 |
| 10 | 1 | 8 | 2 |
| 14 | 1 | ||
| 15 | 1 | ||
| Total | 1494 | Total | 204 |
2.2 Multiple sequence alignment construction and contact map prediction
Starting from an input protein sequence, FUpred generates a multiple sequence alignment (MSA) using the DeepMSA program (Zhang et al., 2019), which searches the query sequence against multiple whole-genome and metagenomic sequence databases utilizing an iterative process (see the explanations in Supplementary Fig. S1 and Text S1). Then, using the MSA as an input, the contact map for the query sequence (with Cβ−Cβ distance <8 Å) is predicted using ResPRE (Li, 2019) by coupling evolutionary precision matrices with deep residual neural networks. Here, a precision matrix is generated by the inverse covariance matrix from an MSA, which is represented by an L × L × 21 × 21 array of evolutionary couplings between L pairs of residues in a query protein. For each residue pair, the 21 × 21 coupling matrix is fed directly into the deep residual convolutional networks composed of a set of 22 residual blocks, each adding the output of the feedforward neural networks to an identity map of the input. ResPRE was trained using the Adam method (Kingma and Ba, 2014) under the supervision of binary cross entropy loss.
2.3 FUscore for continuous two-domain proteins
The FUscore for a continuous two-domain protein, which is the simplest example of a multi-domain protein, is defined as
| (1) |
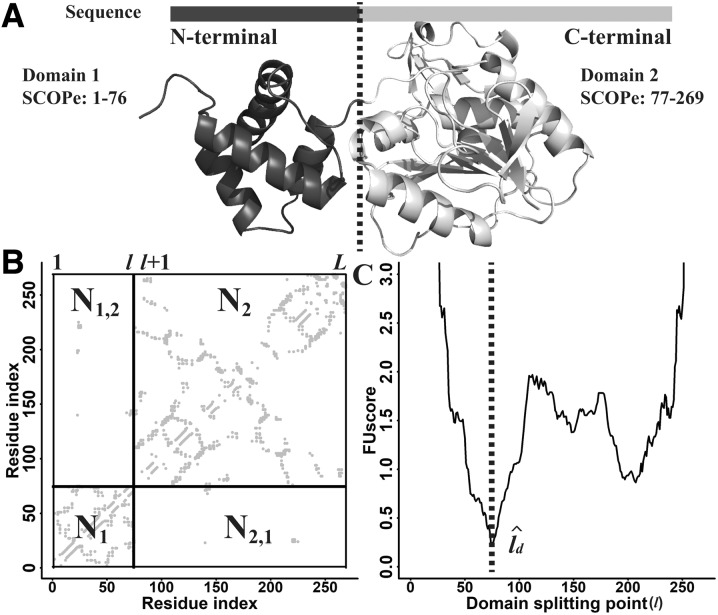
where l is the domain splitting point of a protein, N1(l) and N2(l) represent the number of contacts within the first and second domains, respectively and N1,2(l) = N2,1(l) indicates the number of contacts between the first and second domains (Fig. 1).
Fig. 1.
Illustration of FUscore calculation for a continuous two-domain protein from Chemotaxis protein methyltransferase (PDB ID: 1BC5A). (A) Experimental structure of the protein. (B) Predicted contact map for the protein. (C) FUscore versus domain splitting point l. The splitting line in panel (B) corresponds to the domain boundary in panel (C) where the lowest FUscore is located
The domain boundary, , for a continuous two-domain protein is predicted to be the position where the lowest FUscore2c is obtained (Fig. 1C).
| (2) |
where L is the protein length. By taking the lowest FUscore2c, we are maximizing the number of intra-domain contacts, N1() and N2(), while minimizing the number of inter-domain contacts, N1,2(). Furthermore, the secondary structure of the query is predicted by PSSpred (Yan et al., 2013). For cases when is located within a helix or a strand, it will be shifted to the coil residue that is closest to the estimated based on Equation (2), since domain boundaries occur more often at loop regions than on regular secondary structures.
2.4 FUscore for discontinuous two-domain proteins
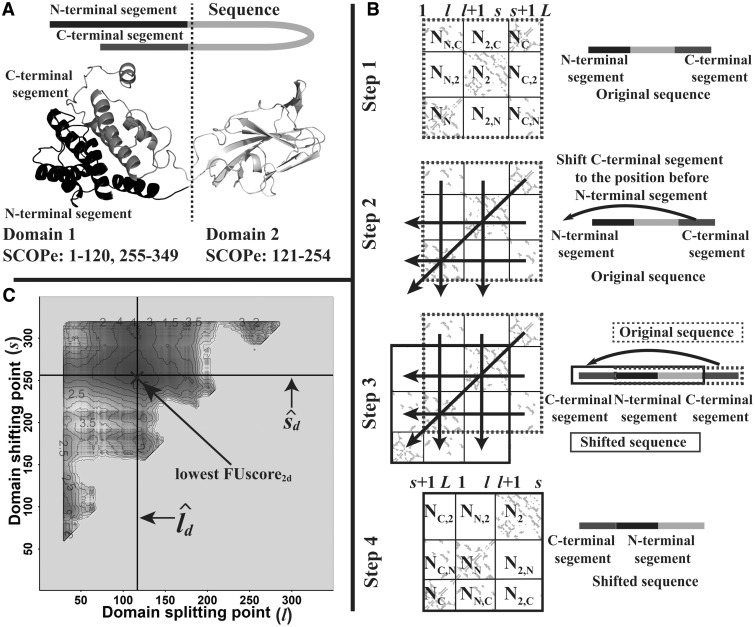
A 2D contact map for a two-domain protein with one continuous and one discontinuous domain is illustrated in Figure 2. Since there is no clear splitting point in the map, this type of domain boundary cannot be directly calculated by Equation (1). However, there is still some pattern similarity between the two cases since they both have many intra-domain contacts but few inter-domain contacts (as shown in Fig. 2A). Inspired by the design principle behind FUscore2c, we derived FUscore2d for discontinuous two-domain proteins by shifting the C-terminal contact map to the N-terminal and converting the discontinuous case into a continuous two-domain case (Fig. 2B), i.e.
| (3) |
where l and s are the domain splitting point and shifting point for a protein (l < s), i.e. ([1,l],[s + 1,L]) represents the two regions of the first discontinuous domain and [l + 1,s] the region of the second continuous domain, where L is the length of the protein. NN(l,s), N2(l,s) and NC(l,s) represent the number of contacts within the segments [1,l], [l + 1,s] and [s + 1,L], respectively. NN,C(l,s) [or NC,N(l,s)], NN,2(l,s) [or N2,N(l,s)] and N2,C(l,s) [or NC,2(l,s)] indicate the number of contacts between the segment pair [1,l] and [s + 1,L], the segment pair [1,l] and [l + 1,s] and the segment pair [s + 1,L] and [l + 1,s], respectively.
Fig. 2.
Illustration of FUscore calculation for a discontinuous two-domain protein from bluetongue virus coat protein VP7 (PDB ID: 1BVP1). (A) Experimental structure of the protein. (B) The contact map shifting procedure for transforming a discontinuous two-domain protein into a continuous two-domain protein. Nx in each sub-block of the panel represents the number of contacts in the sub-block. (C) The 2D FUscore heatmap. The splitting lines in panel (B) correspond to the domain boundaries and in panel (C) where the lowest FUscore is located
Similar to continuous two-domain proteins, the estimated domain boundaries , for discontinuous two-domain proteins are predicted to be located at the location where the lowest FUscore2d is obtained (as shown in Fig. 2C), i.e.
| (4) |
The complexity of using FUscore2d to calculate domain boundaries is O(L4), which is time-consuming since we need to repeatedly count the number of contacts in each block (such as NN,C). In order to improve the efficiency of our algorithm, we implemented a dynamic programming algorithm to speed up the procedure, which uses the recurrence relationship of contact numbers to calculate the increment of contacts in each block and bypasses the need to repeatedly count the number of contacts. As shown in Supplementary Text S2 and Figure S2, the time complexity of FUscore2d using this strategy was reduced to O(L3).
2.5 Pipeline of FUpred
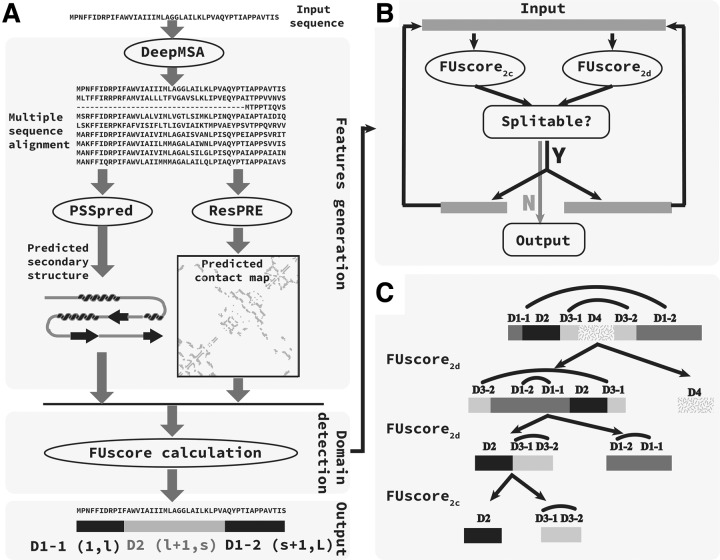
The pipeline of the FUpred algorithm is shown in Figure 3. Starting from the input protein sequence, a deep MSA is generated by iterative sequence homology searches against multiple sequence databases. Then, using the deep MSA as input, the secondary structure of a query sequence is predicted by PSSpred (Yan et al., 2013), and the contact map (with Cβ-Cβ distance <8 Å) is predicted by the ResPRE method. Both the contact map and secondary structure information are used to calculate the FUscore, where the domain pattern and boundary locations are determined by the recursion procedure outlined in Figure 3B.
Fig. 3.
Pipeline of the FUpred algorithm. (A) Overall pipeline of the FUpred algorithm. (B) Recursion strategy for domain boundary detection. (C) Case illustration. The curves on the tops of the domain bars indicate separate regions of a discontinuous domain
As shown in Figure 3B, the recursion strategy is built on the difference between the distributions of multiple and single domains. In detail, using the contact map and secondary structure information for the query sequence as input, both FUscore2c and FUscore2d are calculated for the input sequence. Due to the subtle differences between the FUscore distributions for multi- and single-domain proteins in the training set (see Supplementary Fig. S3), FUpred uses two cutoff parameters, Cutoff2c and Cutoff2d, to distinguish between continuous multi- and single-domain proteins, as well as discontinuous multi- and single-domain proteins, respectively. If the FUscore2c/FUscore2d of the input protein is smaller than Cutoff2c/Cutoff2d, the input protein is predicted to be a continuous/discontinuous two-domain protein, where the domain boundaries are generated according to the FUscore; otherwise, the input protein is predicted to be a single-domain protein. If both the FUscore2c and FUscore2d of the input protein are lower than Cutoff2c and Cutoff2d, respectively, we compare the difference between the value of Cutoff2c minus FUscore2c and the value of Cutoff2d minus FUscore2d, and adopt the domain boundaries predicted by the method with the larger difference. Then, the two possible domains recursively perform the same procedure as described above. The recursion procedure is stopped when none of the domains can be further split.
Figure 3C presents an example to illustrate the applicability of the FUpred algorithm. The input protein (‘D1-1, D2, D3-1, D4, D3-2, D1-2’ representing the first part of Domain 1, Domain 2, the first part of Domain 3, Domain 4, the second part of Domain 3, and the second part of Domain 1, respectively) had two discontinuous domains, i.e. ‘D1-1, D1-2’ and ‘D3-1, D3-2’ and two continuous domains, i.e. ‘D2’ and ‘D4’. First, the input protein was split into two parts, ‘D3-2, D1-2, D1-1, D2, D3-1’ and ‘D4’, based on the FUscore2d. In detail, ‘D3-2, D1-2’ in the C-terminal was first shifted to the N-terminal to get ‘D3-2, D1-2, D1-1, D2, D3-1, D4’, and then ‘D3-2, D1-2, D1-1, D2, D3-1, D4’ was split into ‘D3-2, D1-2, D1-1, D2, D3-1’ and ‘D4’. The ‘D4’ domain could not be further split, while the ‘D3-2, D1-2, D1-1, D2, D3-1’ segment was further divided into two parts, ‘D2, D3-1, D3-2’ and ‘D1-2, D1-1’, based on the FUscore2d. The ‘D1-2, D1-1’ segment could not be further split, while the ‘D2, D3-1, D3-2’ could be further divided into two parts, ‘D2’ and ‘D3-1, D3-2’, based on the FUscore2c. Both ‘D2’ and ‘D3-1, D3-2’ could not be split any further. Finally, the input protein was predicted to be a four-domain protein, with two continuous domains, ‘D2’ and ‘D4’, as well as two discontinuous domains, ‘D3-1, D3-2’ and ‘D1-1, D1-2’. Unlike methods that predict domains by homologous protein searching and alignment [e.g. PfamScan (Mistry et al., 2007)], which assign residues with high confidence scores to a domain, while the residues with low confidence scores or in ‘linker’ regions are not assigned to any domain, FUpred is able to split the entire protein into different domains. This means each residue (even those in ‘linker’ regions) is assigned to a domain by FUpred.
A multi-domain protein may have more than one distinct splitting order when generating the final domain boundaries. Taking a three continuous domain protein, ‘D1, D2, D3’, as an example, the FUpred algorithm may first split ‘D1’ from ‘D2, D3’, and then further split ‘D2’ and ‘D3’. On the other hand, it can also first split ‘D3’ from ‘D1, D2’, and then further split ‘D1’ and ‘D2’. Nevertheless, different types of splitting orders do not influence the final domain boundary results. To illustrate this, we assembled a subset of 38 three-domain proteins from our dataset, where the total length of the two adjacent domains was less than the length of the third one. We also modified the original FUpred algorithm into FUpreds (FUpredb), which forced the algorithm to search for the domain splitting point between the two adjacent small domains (or between the adjacent small domain and large domain) in the first iteration round, where the splitting point was located where the local minimum FUscore was obtained around the SCOPe2.07 domain boundary definition (±20 residues). The comparison of FUpred and FUpreds (FUpredb) is shown in Supplementary Table S1. We found that there was no significant difference between FUpred and FUpreds (FUpredb), indicating that the performance of the FUpred algorithm is robust and does not depend on the order in which domains are split.
Since FUpred is built on the calculation of two-domain FUscores, it is important to examine the applicability of the FUpred method, especially the iterative recursion strategy, to modeling proteins of various domain structure patterns. While it is straightforward to use FUscore2c to calculate domain boundaries of continuous multi-domain proteins, the situation is more complex for discontinuous multi-domain proteins. Supplementary Table S2 lists all of the discontinuous multi-domain patterns in the SCOPe2.07 database. There are in total 26 distinct patterns for discontinuous multi-domain proteins, where FUpred was able to reproduce the domain patterns for 24 of them. Here, we give an illustration of how FUpred can deal with the complex domain pattern ‘D1-1, D2, D1-2, D3, D1-3’, where ‘D1-1, D1-2, D1-3’ is a single discontinuous domain with three parts. As shown in Supplementary Figure S4A, ‘D1-1, D2, D1-2, D3, D1-3’ is a discontinuous three-domain protein. FUpred first split the protein into ‘D1-3, D1-1, D2, D1-2’ and ‘D3’ using FUscore2d. In detail, ‘D1-3’ in the C-terminal was first shifted to the N-terminal to get ‘D1-3, D1-1, D2, D1-2, D3’, and then ‘D1-3, D1-1, D2, D1-2, D3’ was split into ‘D1-3, D1-1, D2, D1-2’ and ‘D3’. Then, by repeatedly using FUscore2d, ‘D1-3, D1-1, D2, D1-2’ was further divided into ‘D2’ and ‘D1-2, D1-3, D1-1’.
However, there were two patterns, ‘D1-1, D2-1, D1-2, D2-2’ and ‘D1, D2-1, D3-1, D2-2, D3-2’, that the FUpred procedure could not solve. We took the domain pattern ‘D1-1, D2-1, D1-2, D2-2’ here as an example to illustrate why this pattern could not be solved by FUpred. As shown in Supplementary Figure S4B, ‘D1-1, D2-1, D1-2, D2-2’ is a discontinuous two-domain protein, where the two domains in the protein are both discontinuous, so this pattern cannot be directly dealt with by FUscore2c, which is applicable to continuous multi-domain proteins. When considering FUscore2d, whether we shifted ‘D2-2’ to the N-terminal to make the protein domain pattern ‘D2-2, D1-1, D2-1, D1-2’ or shifted ‘D1-2, D2-2’ to the N-terminal to get ‘D1-2, D2-2, D1-1, D2-1’, the new pattern would still remain the same. Thus, FUscore2d cannot deal with the pattern ‘D1-1, D2-1, D1-2, D2-2’. Similarly, the pattern ‘D1, D2-1, D3-1, D2-2, D3-2’ cannot be dealt with by the current strategy of FUpred either. Nevertheless, since these patterns only make up a tiny portion (0.0235%) of the SCOPe2.07 database (Supplementary Table S2), the issue does not impact the overall performance of the FUpred pipeline.
2.6 Assessment metrics
The performance of the proposed algorithm was evaluated in terms of its protein classification and domain boundary prediction ability. The following criteria were used to assess the ability of FUpred to classify whether proteins are composed of single or multiple domains:
| (5) |
where TM/TS represent the number of cases that were correctly predicted to be multi-domain/single-domain proteins, and FM/FS signify the number of cases that were incorrectly predicted to be multi-domain/single-domain proteins. Pre(multi)/Pre(single) represent the precision of multi-domain/single-domain classification and Rec(multi)/Rec(single) symbolize the recall of multi-domain/single-domain classification. The ACC and MCC are the accuracy of protein classification and the Matthew’s correlation coefficient, respectively.
Moreover, the normalized domain overlap (NDO) score (Tai et al., 2005) and the domain boundary distance (DBD) score (Tress et al., 2007), which were used to assess domain splitting in the CASP experiments, were utilized to assess the domain boundary prediction. The NDO score calculates the overlap between the predicted domain regions and true domain regions, while the DBD score is defined as the distance of the predicted domain boundaries from the true domain boundaries, where all linker regions of the domains are considered as the true boundaries.
2.7 Parameter training
There were three parameters that had to be trained in FUpred, which were optimized based on the protein training set. First, the top αL contact pairs ranked by predicted confidence scores were used to form the final contact map for an input sequence, where L refers to the length of a query protein. Based on the training dataset, we varied the parameter α from 0.5 to 5 and obtained the optimal parameter α = 2.6 based on the balance of the MCC, ACC, NDO and DBD scores (see Supplementary Fig. S5). The other two parameters were Cutoff2c and Cutoff2d, which we varied from 0.3 to 1.5, and finally assigned Cutoff2c = 0.85 and Cutoff2d = 0.66 (see Supplementary Fig. S6).
3 Results
In this section, we tested the performance of FUpred on the benchmark dataset, where its performance was compared to the threading-based method ThreaDomEx (Wang et al., 2017), and three machine learning-based methods, including ConDo (Hong et al., 2018), DOMpro (Cheng et al., 2006) and DoBo (Eickholt et al., 2011). Note that ConDo also utilizes contact map information as an input feature for neural network training, while DoBo and DOMpro predict domain boundaries utilizing sequence and sequence profile information as the input features.
3.1 Classification of single- and multi-domain proteins
First, we analyzed the domain classification ability of the aforementioned methods, where Table 2 shows the overall comparison of the domain classification performance of FUpred and the four control methods. In our test set of 849 multi-domain proteins and 1700 single-domain proteins, FUpred correctly assigned 91% of the proteins as multi- or single-domain proteins, which was 3% higher than the second-best method, ThreaDomEx. Among all five predictors, FUpred produced the highest MCC (0.799), followed by the threading-based method ThreaDomEx (0.759), and machine learning-based methods, ConDo (0.671), DOMpro (0.408) and DoBo (0.371).
Table 2.
Single- and multi-domain classification results on 2549 test proteins
| Methods | Multi |
Single |
All |
|||
|---|---|---|---|---|---|---|
| Pre | Rec | Pre | Rec | ACC | MCC | |
| FUpred | 0.860 | 0.873 | 0.936 | 0.929 | 0.910 | 0.799 |
| ThreaDomEx | 0.767 | 0.933 | 0.962 | 0.858 | 0.883 | 0.759 |
| ConDo | 0.803 | 0.751 | 0.880 | 0.908 | 0.856 | 0.671 |
| DOMpro | 0.629 | 0.561 | 0.792 | 0.835 | 0.743 | 0.408 |
| DoBo | 0.436 | 0.973 | 0.965 | 0.371 | 0.571 | 0.371 |
Note: ‘Pre’, ‘Rec’, ‘ACC’ and ‘MCC’ are the precision, recall, accuracy and Matthew’s correlation coefficient, respectively, as defined by Equation (5). Bold values indicate the best performer in each category.
Considering the individual metrics, DoBo achieved the highest recall (0.973) for multi-domain proteins, and the highest precision (0.965) for single-domain proteins, but had the lowest MCC; these data imply that DoBo tends to classify most proteins as multi-domain. In fact, 3% (=23/849) of the multi-domain proteins in the test set were predicted to be single domains by DoBo, resulting in the extremely high multi-domain recall and high single-domain precision. Nevertheless, 63% (=1070/1700) of the single-domain proteins in the test set were predicted to be composed of multiple domains by DoBo, which means that DoBo is biased toward over-predicting the number of multi-domain proteins. On the other hand, 44% (=373/849) of the multi-domain proteins were predicted to be single-domain by DOMpro, which was much higher than ConDo (25%), FUpred (13%), ThreaDomEx (7%) or DoBo (3%), indicating that DOMpro is biased toward under-predicting the number of multi-domain proteins.
Due to the inclusion of contact map information, ConDo did a better job balancing single- and multi-domain protein recognition, achieving the highest MCC score among the machine learning-based approaches. However, due to the lower recall for multi-domain assignment, the overall MCC and accuracy of ConDo was lower than that of ThreaDomEx and FUpred.
3.2 Prediction of domain boundary locations
To examine the ability of various methods to predict the location of domain boundaries, we present in Table 3 a summary of the NDO and DBD scores for FUpred in comparison to the other four methods (Cheng et al., 2006; Eickholt et al., 2011; Hong et al., 2018; Wang et al., 2017).
Table 3.
Summary of domain boundary prediction for the 849 multi-domain proteins
| Methods | NDO | DBD |
|---|---|---|
| FUpred | 0.791 | 0.498 |
| ThreaDomEx | 0.760 (2.31E04) | 0.471 (3.17E02) |
| ConDo | 0.742 (1.75E08) | 0.376 (9.43E14) |
| DOMpro | 0.584 (3.49E87) | 0.087 (3.20E115) |
| DoBo | 0.568 (9.15E−88) | 0.205 (8.08E67) |
Note: The values in parentheses are P-values between the FUpred results and the other control methods results calculated using one-sided Student’s t-tests. Bold values indicate the best performer in each category.
Both the NDO and DBD scores for FUpred were significantly higher than those for the other four methods with P-values <0.05 as determined by paired one-sided Student’s t-tests. For the 849 multi-domain proteins in the test set, ConDo had comparable performance to ThreaDomEx in terms of the NDO scores, indicating that for both methods at least 74.2% of the residues in the predicted domains overlapped with the correct domains. The DBD score of ConDo was at least 10% worse than that by FUpred or ThreaDomEx, indicating that the predicted domain boundaries assigned by ConDo were much worse than those assigned by FUpred or ThreaDomEx. DoBo had the second worst performance in domain boundary detection, where 45% (=382/849) of the domain boundaries in the test set were incorrectly predicted, while DOMpro had the worst performance, since it predicted 44% of the multi-domain proteins as single-domain proteins.
In our test dataset construction, the homologous entries with sequence identities >30% to the training proteins were filtered out. However, sequences with >30% sequence identity to the proteins in the ResPRE training set, whose contact predictions are used by FUpred, were not excluded from our test datasets. This is partly because ResPRE and FUpred are different methods and trained on independent protein sets. Furthermore, the ResPRE training set is large, including about 5600 high-resolution protein structures to facilitate effective deep-learning training, and the filtering of homologous proteins from this training set would result in an insufficient number of proteins in the benchmark dataset. Nevertheless, to examine the impact of the ResPRE training set on the comparison between FUpred and the other control predictors, we constructed a new test dataset by removing proteins with a 30% sequence identity to not only the ResPRE training set but also the training sets of two relatively accurate methods, ThreaDomEx and ConDo, resulting in there being only 136 multi-domain proteins and 355 single-domain proteins left in our benchmark dataset. Supplementary Tables S3 and S4 show the results for domain classification and domain boundary prediction on this reduced test dataset, respectively. The accuracy of FUpred models in this reduced dataset is slightly lower than that of the entire benchmark set (compared to Tables 2 and 3), which is probably due to the fact that this sub-dataset is more difficult for domain prediction as the average accuracy is reduced for all the control methods (including those whose training proteins were not included in the homologous filtering). Nevertheless, FUpred still significantly outperformed all the control methods on this reduced dataset as shown in Supplementary Tables S3 and S4.
3.3 Prediction results for discontinuous domain proteins
Due to the difficulty of modeling them, here, we separately discuss the prediction of discontinuous domains, which consist of more than one non-consecutive segment. Out of the 133 discontinuous multi-domain proteins in the test dataset, FUpred correctly classified 94 of them, resulting in a recall rate of 70.7%. ThreaDomEx (Wang et al., 2017) is another method designed for discontinuous domain prediction but it only detected 39.1% (=52/133) of the targets containing discontinuous domains. Meanwhile, we found that ThreaDomEx had a tendency to over-predict the number of discontinuous multi-domains, since it predicted that the average number of discontinuous multi-domains was 3.36 in the test set, which was higher than the actual number (2.81), while the FUpred predictions were closer to the real value (2.95). On the other hand, none of the three-machine learning-based methods, ConDo, DOMpro and DoBo, could detect any proteins containing discontinuous domains.
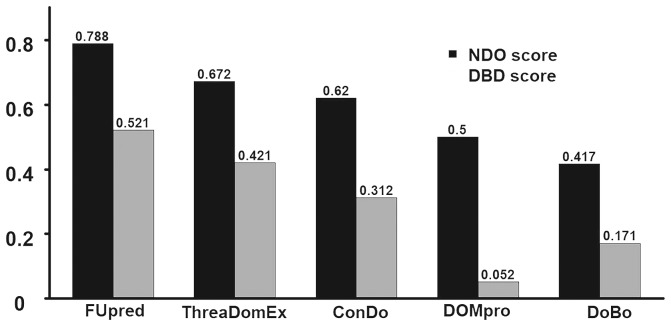
Figure 4 lists a summary of the domain boundary prediction results for the 133 discontinuous domain proteins. The results show that FUpred achieved average NDO and DBD scores of 0.788 and 0.521, respectively, which were 17.3% and 23.8% higher than the second-best method, ThreaDomEx, with P-values <0.05 as calculated using one-side Student’s t-tests (Supplementary Table S5).
Fig. 4.
Summary of the domain boundary prediction results by different methods for the 133 discontinuous multi-domain proteins. Data are taken from Supplementary Table S5
We note that the domain boundary prediction performance of FUpred was very close for continuous and discontinuous domain proteins as listed in Supplementary Table S5. More specifically, the NDO/DBD scores were 0.791/0.494 and 0.788/0.521 for continuous and discontinuous domain proteins, respectively, the difference of which corresponds to P-values of 0.88/0.44 as calculated by two-sided Student’s t-tests. These results suggest that FUpred’s performance does not obviously depend on the domain type for multi-domain proteins, thus highlighting the effectiveness of FUpred’s iterative recursion procedure for recognizing complex domain structures.
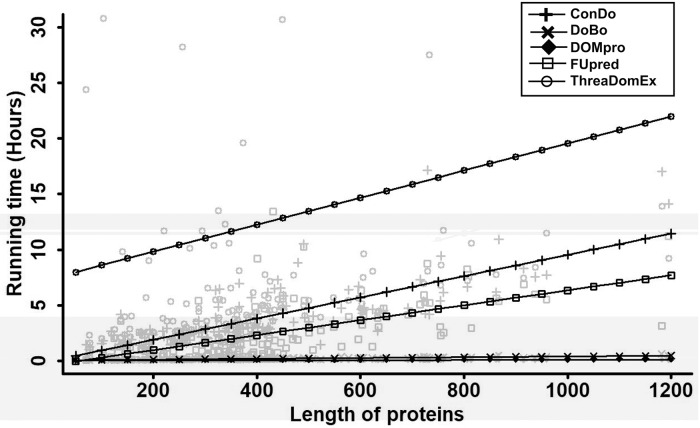
3.4 Analysis of time complexity
We also compared the time complexity for FUpred and the other four methods on the 136 multi-domain proteins with lengths ranging from 70 to 1200 amino acids, which were non-redundant to the training datasets for ResPRE, ThreaDomEx and ConDo. All five methods were run as standalone packages with only one CPU and the pure running times for the jobs were counted. The results are shown in Figure 5. ThreaDomEx was the most time-consuming method, followed by ConDo. Among the top three most accurate methods (FUpred, ThreaDomEx and ConDo), FUpred required the least amount of time. Although the DOMpro and DoBo methods ran faster than the other three methods, as previously discussed, these two methods were not as accurate as the others. Overall, FUpred had good performance in terms of both accuracy and running time. Note that ThreaDomEx and FUpred also provide online servers. Thus, the user waiting time includes both the pending time and actual running time for the jobs.
Fig. 5.
The time complexity comparison between FUpred and the other four methods. All five methods, FUpred, ThreaDomEx, DOMpro, DoBo and ConDo, were run as standalone packages using only one CPU and the pure running times for the jobs were counted. Linear regression was used to fit the correlation relationship between running time and protein length for different methods
3.5 A case study for predicting complex multi-domain segments and boundaries
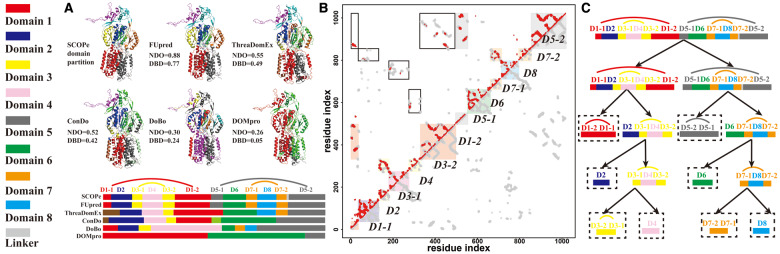
Although most of the multi-domain proteins in the SCOPe database were two-domain or three-domain proteins, there were some relatively complex multi-domain proteins, which are particularly challenging for nearly all domain prediction algorithms. In Figure 6, we present an example from the AcrB bacterial multi-drug efflux transporter (PDB ID: 2dhhA) in our test set to illustrate how FUpred recognizes complex domain structures.
Fig. 6.
Case study of domain boundary prediction for the AcrB bacterial multi-drug efflux transporter (PDB ID: 2dhhA). (A) NDO and DBD scores for domain boundary prediction by different methods. (B) Native (gray) and ResPRE-predicted (red) contact maps for the target protein, where colored solid squares indicate the domain boundaries of the native structure, and the square frames mark the inter-domain contacts that are key to the domain boundary prediction in FUpred. (C) Iterative recursion procedure of domain boundary detection in FUpred. Different domains are marked by distinct colors as indicated at the bottom of the panel
2dhhA has 1022 residues and contains eight domains (‘D1-1, D2, D3-1, D4, D3-2, D1-2, D5-1, D6, D7-1, D8, D7-2, D5-2’) where four are continuous domains (‘D2’, ‘D4’, ‘D6’ and ‘D8’) and the other four are discontinuous domains (‘D1-1, D1-2’, ‘D3-1, D3-2’, ‘D5-1, D5-2’ and ‘D7-1, D7-2’), as indicated by the SCOPe database. Figure 6A shows the predicted domain boundaries by five different methods, in comparison with the assignment based on the experimental structure. FUpred achieved NDO and DBD scores of 0.88 and 0.77, respectively, which were significantly higher than all of the control methods, each of which had both NDO and DBD scores below 0.55.
For most cases, the higher quality of the FUpred models could be mainly attributed to the effective iterative recursion procedure and the relatively high accuracy of the contact map predictions generated by ResPRE (as shown in Fig. 6B). While there is no doubt that high accuracy contact map prediction can help FUpred correctly predict the domain boundaries, for this case, the inter- and intra-domain contact prediction was not highly accurate. The precision of intra-domain contact prediction for 2dhhA by ResPRE was 0.36, which was much lower than the average precision for all of the discontinuous domain proteins (0.54). Additionally, 90% of the inter-domain contacts were not predicted by ResPRE. Despite the low accuracy of inter- and intra-domain contact prediction, FUpred very accurately split the whole sequence into eight domains recursively based on the FUscore2d and FUscore2c (as shown in Fig. 6C) guided by the contact map information. In the first step, FUpred detected the continuous domain boundary between ‘D1-2’ (the second part of the discontinuous domain 2) and ‘D5’ (the continuous domain 5), so ‘D1-1, D2, D3-1, D4, D3-2, D1-2’ and ‘D5-1, D6, D7-1, D8, D7-2, D5-2’ were approximately split into two domains (Fig. 6C). Then, ‘D1-1, D2, D3-1, D4, D3-2, D1-2’ and ‘D5-1, D6, D7-1, D8, D7-2, D5-2’ had similar domain patterns, which FUpred separately predicted with the same iterative procedure. Taking ‘D1-1, D2, D3-1, D4, D3-2, D1-2’ as an example, this part was further split into ‘D1-2, D1-1’ and ‘D2, D3-1, D4, D3-2’ by discontinuous domain detection, since continuous domain detection could not split it. After that, ‘D2, D3-1, D4, D3-2’ was split by continuous domain detection into ‘D2’ and ‘D3-1, D4, D3-2’, where ‘D3-1, D4, D3-2’ followed a very typical discontinuous two-domain protein pattern. Finally, ‘D3-1, D4, D3-2’ was correctly split into ‘D3-2, D3-1’ and ‘D4’.
We have also provided four more representative cases with complex domain patterns in Supplementary Fig. S7. The four multi-domain proteins are 1we3F (‘D1-1, D2-1, D3, D2-2, D1-2’), 1dq3A (‘D1-1, D2, D3, D4, D1-2’), 3ac0A (‘D1, D2-1, D3, D2-2, D4’) and 1miuA (‘D1, D2, D3-1, D4, D3-2, D5’). FUpred was able to almost perfectly predict the domain boundaries for each of these four proteins with NDO scores (DBD score) of 0.954, 0.888, 1.000 and 0.951 (0.844, 0.750, 1.000 and 0.800) for 1we3F, 1dq3A, 3ac0A and 1miuA, respectively.
3.6 Complementarity between threading and contact map-based domain prediction
The benchmark results demonstrated that both contact map-based methods (FUpred) and threading-based methods (ThreaDomEx) have considerable advantages over machine learning approaches. Here, we further examine the complementarity between the threading-based methods and contact map-based methods.
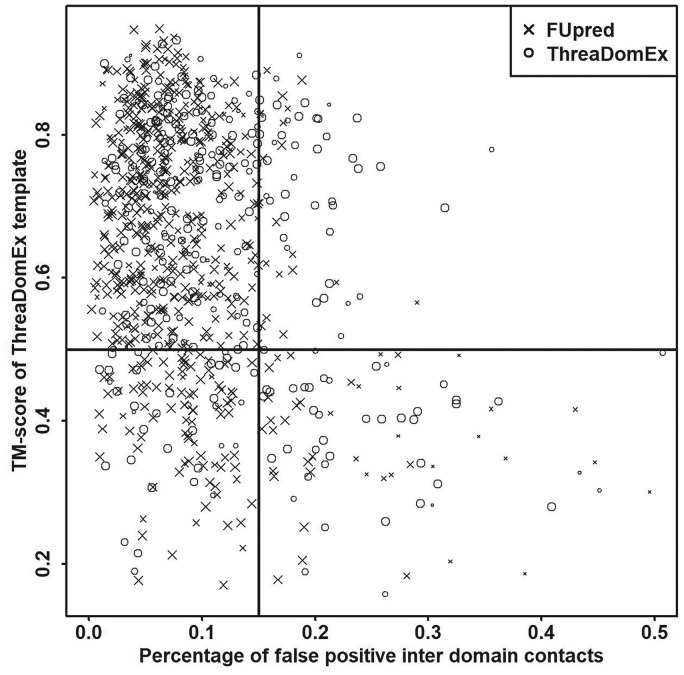
As a threading-based method, ThreaDomEx can accurately detect domain boundaries when the correct templates are identified. In Supplementary Table S6, we split the protein samples into two groups based on NDO score using a cutoff of 0.4. We show that the average TM-score of the best threading templates (0.642) for the high-performance cases (NDO ≥ 0.4) was much higher than that (TM-score = 0.584) for the low-performance cases (NDO < 0.4). Although homology templates can be found for most proteins by threading methods, there are still many targets where homology templates cannot be easily identified by current threading approaches. Thus, an alternative method is needed. Similarly, contact map-guided methods, such as FUpred, can accurately predict domain boundaries when the contact maps are accurately predicted. To assess the quality of the predicted contact maps, we calculated four types of metrics: precision of intra/inter-domain (PREintra/PREinter) contact prediction and percentage of false positive predicted intra/inter-domain contacts (PFPintra/PFPinter) based on the top 2.6L predicted contacts. As shown in Supplementary Table S7, the PFPinter is more relevant to the performance of FUpred, i.e. the high-performance cases (with NDO ≥ 0.4) had significantly lower PFPinter values (=0.096) than the values (=0.139) for the low-performance cases (NDO < 0.4). This is easy to understand since the FUscore will be high when too many false positive contacts appear in inter-domain regions.
Figure 7 shows the relationship between a threading-based method (ThreaDomEx) and a contact map-based method (FUpred). For each multi-domain protein in the test set, we only show one point representing the method (ThreaDomEx or FUpred) with the higher NDO score in the figure. In the top left region, which represents targets that have both good templates and accurate contact maps, there are many circular points (173) and cross points (404) and the sizes of the points are generally large, indicating that both threading-based and contact map-based methods performed excellently on targets in this region. However, the sizes of the points in the bottom-right region are much smaller, indicating that neither threading-based nor contact map-based methods generated accurate predictions since the targets in this region did not have good templates or accurate contact maps.
Fig. 7.
Performance relationship between threading-based and contact map-based methods. The point size corresponds to the value of the NDO score, where a larger point indicates a higher score
The complementarity of the two methods can be found in the two remaining regions. In the bottom-left region, e.g. the targets had accurate contact maps but failed to detect good templates. Accordingly, there are 90 cross points with reasonable predictions from FUpred and only 35 circular points from ThreaDomEx predictions. On the other hand, in the upper-right region, there are more accurately predicted cases from ThreaDomEx than from FUpred (41 circles versus 21 crosses). In the previous test, the average NDO and DBD scores for FUpred were 0.791 and 0.498, respectively (see Table 3). However, if we combine the FUpred and ThreaDomEx results together by taking the higher performing model for each target, the NDO and DBD scores increased to 0.869 and 0.660, respectively, which again demonstrates the complementarity of the two approaches, despite the fact that FUpred significantly outperformed ThreaDomEx on its own.
4 Conclusion
We have developed a new pipeline, FUpred, which utilizes contact map prediction, in conjunction with secondary structure information, to detect domain boundary locations for protein sequences. Given a 2D contact map, the optimal domain splitting can be obtained in principle by maximizing the number of intra-domain contacts, while minimizing the number of inter-domain contacts. Quantitatively, this was implemented by optimizing the FUscore, which balances the number of inter- and intra-domain contacts, while an iterative recursion strategy was developed for further domain splitting and refinement in order to detect higher-order, more complex domain structures, including multiple continuous and discontinuous domains.
FUpred was tested in large-scale benchmark experiments and showed significant advantages over a state-of-the-art threading-based method (ThreaDomEx) and leading machine learning-based methods (ConDo, DoBo and DOMpro). In particular, the FUpred algorithm demonstrated excellent performance for modeling discontinuous domains, with an accuracy comparable to that for continuous domains. In fact, FUpred is the first computational domain prediction method to achieve this for the challenging problem of discontinuous domain modeling. Furthermore, FUpred had the fastest running time among the most accurate methods, including ThreaDomEx and ConDo.
Nevertheless, the performance of FUpred was still unsatisfactory for several targets. These occurred in particular when the effective number of homologous sequences detected by DeepMSA was low and the contact map prediction was poor. Given the complementarity between contact map prediction and threading template identification, one way to alleviate this issue is through the combination of the threading alignments and contact map information for composite domain prediction. Work along this line is in progress.
Supplementary Material
Acknowledgements
The authors thank Chengxin Zhang for insightful discussions. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation (ACI-1548562).
Funding
This work is supported in part by the National Institute of General Medical Sciences [GM083107, GM116960 and GM136422], the National Institute of Allergy and Infectious Diseases [AI134678] and the National Science Foundation [DBI1564756 and IIS1901191].
Conflict of Interest: none declared.
References
- Alexandrov N., Shindyalov I. (2003) PDP: protein domain parser. Bioinformatics, 19, 429–430. [DOI] [PubMed] [Google Scholar]
- Chandonia J.-M. et al. (2014) SCOPe: structural classification of proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res., 42, D304–D309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandonia J.-M. et al. (2017) SCOPe: manual curation and artifact removal in the structural classification of proteins-extended database. J. Mol. Biol., 429, 348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandonia J.-M. et al. (2019) SCOPe: classification of large macromolecular structures in the structural classification of proteins-extended database. Nucleic Acids Res., 47, D475–D481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. (2007) DOMAC: an accurate, hybrid protein domain prediction server. Nucleic Acids Res., 35, W354–W356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. et al. (2006) DOMpro: protein domain prediction using profiles, secondary structure, relative solvent accessibility, and recursive neural networks. Data Min. Knowl. Disc., 13, 1–10. [Google Scholar]
- Eickholt J. et al. (2011) DoBo: protein domain boundary prediction by integrating evolutionary signals and machine learning. BMC Bioinform., 12, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R.A., Heringa J. (2002) SnapDRAGON: a method to delineate protein structural domains from sequence data. J. Mol. Biol., 316, 839–851. [DOI] [PubMed] [Google Scholar]
- Guo J. (2003) Improving the performance of DomainParser for structural domain partition using neural network. Nucleic Acids Res., 31, 944–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.H. et al. (2018) ConDo: protein domain boundary prediction using coevolutionary information. Bioinformatics, 14, 2411–2417. [DOI] [PubMed] [Google Scholar]
- Kim D.E. et al. (2005) Automated prediction of domain boundaries in CASP6 targets using Ginzu and RosettaDOM. Proteins, 61, 193–200. [DOI] [PubMed] [Google Scholar]
- Kingma D.P., Ba J. (2014) Adam: a method for stochastic optimization. arXiv:1412.6980.
- Li Y. et al. (2019) Ensembling multiple raw coevolutionary features with deep residual neural networks for contact-map prediction in CASP13. Proteins, 87, 1082–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J. et al. (2007) Predicting active site residue annotations in the Pfam database. BMC Bioinform., 8, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic G. et al. (2017) An ambiguity principle for assigning protein structural domains. Sci. Adv., 3, e1600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q. et al. (2019) DNN-Dom: predicting protein domain boundary from sequence alone by deep neural network. Bioinformatics, 35, 5128–5136. [DOI] [PubMed] [Google Scholar]
- Söding J. (2004) Protein homology detection by HMM–HMM comparison. Bioinformatics, 21, 951–960. [DOI] [PubMed] [Google Scholar]
- Tai C.-H. et al. (2005) Evaluation of domain prediction in CASP6. Proteins, 61, 183–192. [DOI] [PubMed] [Google Scholar]
- Tress M. et al. (2007) Assessment of predictions submitted for the CASP7 domain prediction category. Proteins, 69, 137–151. [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. (2017) ThreaDomEx: a unified platform for predicting continuous and discontinuous protein domains by multiple-threading and segment assembly. Nucleic Acids Res., 45, W400–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhang Y. (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res., 35, 3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhang Y. (2008) MUSTER: improving protein sequence profile–profile alignments by using multiple sources of structure information. Proteins, 72, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. et al. (2009) OPUS-Dom: applying the folding-based method VECFOLD to determine protein domain boundaries. J. Mol. Biol., 385, 1314–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z. et al. (2013) ThreaDom: extracting protein domain boundary information from multiple threading alignments. Bioinformatics, 29, i247–i256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R. et al. (2013) A comparative assessment and analysis of 20 representative sequence alignment methods for protein structure prediction. Sci. Rep., 3, 2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Li J.H. et al. (2019) ResPRE: high-accuracy protein contact prediction by coupling precision matrix with deep residual neural networks. Bioinformatics, 35, 4647–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. et al. (2020) DeepMSA: constructing deep multiple sequence alignment to improve contact prediction and fold-recognition for distant-homology proteins. Bioinformatics, btz863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. et al. (2007) DDOMAIN: dividing structures into domains using a normalized domain–domain interaction profile. Prot. Sci., 16, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.