Abstract
Summary
QuartataWeb is a user-friendly server developed for polypharmacological and chemogenomics analyses. Users can easily obtain information on experimentally verified (known) and computationally predicted (new) interactions between 5494 drugs and 2807 human proteins in DrugBank, and between 315 514 chemicals and 9457 human proteins in the STITCH database. In addition, QuartataWeb links targets to KEGG pathways and GO annotations, completing the bridge from drugs/chemicals to function via protein targets and cellular pathways. It allows users to query a series of chemicals, drug combinations or multiple targets, to enable multi-drug, multi-target, multi-pathway analyses, toward facilitating the design of polypharmacological treatments for complex diseases.
Availability and implementation
QuartataWeb is freely accessible at http://quartata.csb.pitt.edu.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
It is now widely accepted that many complex diseases are associated with multiple targets, which in turn affect multiple pathways, requiring the adoption of quantitative systems pharmacology (QSP) approaches for assessing the mechanisms of disease etiology, progression and treatment (Stern et al., 2016). The possibility of exploiting the promiscuity of drugs via drug repurposing and polypharmacological treatments also emerged in recent years as a means of reducing risk and cost in drug development (Ashburn and Thor, 2004; Pantziarka et al., 2018; Sachs et al., 2017). In parallel, chemogenomics studies assist in improving our understanding of disease mechanisms and developing therapeutic strategies by providing phenotypic information on ensembles of active compounds screened against families of targets. Such studies underscore the importance of developing computational tools that would harness the rapidly accumulating data to predict new chemical–target interactions (CTIs) over a broad space of chemicals and enable their mapping to pathways and function.
Several resources have been developed in the last decade to address different aspects of the emerging needs but an integrated server designed to automate the association of multiple CTIs with enriched pathways and functions remains to be developed. For example, the servers SEA (Keiser et al., 2007), SwissTargetPrediction (Daina et al., 2019) and SuperPred (Nickel et al., 2014) predict new CTIs, but not corresponding pathways. DINIES (Yamanishi et al., 2014) and DT-Web (Alaimo et al., 2015) incorporate pathway information, but not large-scale CTIs, their respective data being limited to Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2017) and DrugBank (Wishart et al., 2018). Furthermore, existing interfaces are not designed to use as input multiple drugs/targets for polypharmacological strategies and/or for complementing chemogenomics efforts.
We developed the QuartataWeb server to address those needs. QuartataWeb uses known (experimentally verified) CTIs from DrugBank and STITCH (Szklarczyk et al., 2016; Supplementary Table S1 and Fig. S1) in a probabilistic matrix factorization algorithm (Cobanoglu et al., 2013) to predict new CTIs in the extended space of more than 300 000 chemicals and 9000 human proteins. The engine parameters have been optimized (see Supplementary Table S2) to ensure high CTI prediction accuracy (see Supplementary Table S3 and Fig. S2). The outputs are linked to KEGG pathways and Gene Ontology (GO) annotations (GOAs) (Huntley et al., 2015) to predict the most probable pathways, functions and processes affected by one or more chemicals and to efficiently assist in interpreting and/or guiding chemogenomics and polypharmacological studies.
2 QuartataWeb implementation and pipeline
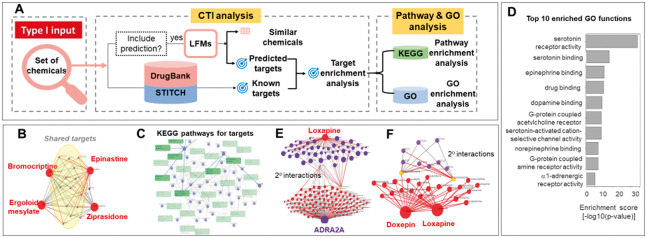
The QuartataWeb server pipeline is schematically depicted in Figure 1A and Supplementary Figure S4. The server can be flexibly queried with three types of input: (I) a list of chemicals (or targets) for chemogenomics-like screening in silico (Fig. 1A); (II) one or more pairs of chemicals to be administered in combination for polypharmacological purposes (Supplementary Fig. S4); and (III) a single chemical and/or a single target to be characterized (Supplementary Fig. S4). In response to a list of chemicals entered in Type I query, QuartataWeb releases newly predicted CTIs and chemical–chemical similarities based on pre-computed latent factor models learned from DrugBank or STITCH data, in addition to retrieving known CTIs from these datasets, as schematically described in Figure 1A. The outputted targets are then subjected to target enrichment, which also lead to enrichment scores for associated pathways and GOAs (P-values; see Supplementary Theory and Methods]. The same sequence of tasks can be carried out for a list of targets entered as input. In the case of Type II input, the same tasks are carried out for pairs of chemicals to obtain shared targets, and their enrichment, along with enriched pathways and GOAs (Supplementary Fig. S4). Type III input is the simplest query where users enter one chemical, one target or a chemical–target pair to identify associated CTIs, similar chemicals or targets, and enriched pathways and GOAs. Furthermore, the secondary interactions (2°, beyond the immediate neighbors) in the bipartite network of chemical/targets can be visualized. In all cases, outputs are presented as tables with several specifications (e.g. drug/chemical identifiers, Gene IDs and names, PDB IDs, confidence scores and enrichment P-values), in addition to visuals such as network representations, bar plots or heatmaps. The force-directed layout in JavaScript D3 package has been customized and designed to interactively display the CTIs and pathways networks.
Fig. 1.
QuartataWeb workflow for Type I input, example outputs on CTIs, pathways and GOAs. (A) In Type I input, users enter either a list of chemicals or a list of targets of interest. Here, the workflow for a list of chemicals is illustrated. See details in the text. (B) Identification of targets (dark violet dots, in yellow ellipse) shared by four drugs (Input Type I) indicated by red nodes. (C) KEGG pathways (green boxes) corresponding to targets in (B). (D) Top 10 enriched GO molecular functions for targets in (B). Bar plot shows enrichment P-values. (E) Illustration of ligand–target interactions obtained by Type III input. Second generation of nodes with degrees < 3 are hidden by applying ‘Trim 2nd generation nodes’ button. (F) Chemical–chemical similarities. The option ‘Display secondary interactions’ displays targets shared by selected drugs (yellow)
3 Applications
Chemogenomics analysis for a list of chemicals. In many cases, a set of chemicals exhibiting comparable phenotypes are analyzed in phenotypic screening. Suppose, we are interested in finding out the common mechanism of action of this set of chemicals (Type I input). Figure 1B illustrates such a case where four drugs are inputted having same phenotype. QuartataWeb permits to identify the common targets along with the interaction confidence scores and enrichment scores of the targets. One may further learn about the pathways associated with the shared targets (Fig. 1C) and corresponding GOAs (Fig. 1D). The interface also provides tables with detailed information on the pathways and GOAs, including their P-values (Supplementary Fig. S5), which could help assessing the dominant pathways and processes that underlie the shared phenotype. Our recent QSP analysis of 50 drugs of abuse serves as an example of the utility of this type of integrated studies (Pei et al., 2019).
Polypharmacological evaluation of drug pairs. Similarly, Supplementary Figure S6 illustrates the output from QuartataWeb for identifying common targets given pairs of chemicals (Type II input) that trigger comparable responses, e.g. aripiprazole/olanzapine, clozapine/trimipramine, methotrimeprazine/epinastine and cabergoline/mianserin. The corresponding CTIs are listed in tables, and also displayed in a network viewer with an interactive control panel. Links to pathways and GOAs result pages are indicated. In this example, 15 among 192 known and 80 predicted (confidence scores > 0.9) targets were identified as common targets. Pathways shared by each chemical pair are also listed in the pathway enrichment table. This type of analysis applied to drug combinations used in a Huntington’s disease model helped elucidate the origin (shared pathways) of observed synergistic effects (Pei et al., 2017).
Drug repurposing or identification of side-effects (Type III). Consider loxapine and its target, α2A adrenergic receptor (gene name: ADRA2A) as an example. CTIs corresponding to both drug (red sphere) and target (blue sphere) can be viewed in peacock representation (Fig. 1E), where known and predicted CTIs being distinguished by the gray and red edges, respectively. Users can interactively select nodes to view primary and 2° interactions. Loxapine is an antipsychotic agent approved for treating schizophrenia, whose primary targets are dopamine and serotonin receptors. The figure displays the 2° interactions of a serotonin receptor (HTR3C, node colored cyan) which turns out to be a target of many drugs associated ADRA2A, some of which are repurposable. Finally, Type III input also permits to identify similar drugs and shared targets as illustrated for the pair doxepin and loxapine (Fig. 1F).
4 Conclusion
We presented QuartataWeb, an integrated server that offers multiple capabilities for QSP analyses using both known associations and machine-learning predictions. We showed that the interface can help identify repurposable drugs, side-effects, enriched pathways, as well as shared functions, cellular processes and environment for different types of queries. QuartataWeb is expected to serve as a first filter toward designing more effective phenotypic screens and polypharmacological strategies.
Supplementary Material
Acknowledgements
The authors thank Dr Bing Liu for useful feedback.
Funding
Support by the National Institutes of Health awards [P41 GM103712 and P30 DA035778] is gratefully acknowledged by I.B.
Conflict of Interest: none declared.
References
- Alaimo S. et al. (2015) DT-Web: a web-based application for drug-target interaction and drug combination prediction through domain-tuned network-based inference. BMC Syst. Biol., 9, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn T.T., Thor K.B. (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov., 3, 673–683. [DOI] [PubMed] [Google Scholar]
- Cobanoglu M.C. et al. (2013) Predicting drug-target interactions using probabilistic matrix factorization. J. Chem. Inf. Model, 53, 3399–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A. et al. (2019) SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res., 47, W357–W364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley R.P. et al. (2015) The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res., 43, D1057–D1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res., 45, D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser M.J. et al. (2007) Relating protein pharmacology by ligand chemistry. Nat. Biotechnol., 25, 197–206. [DOI] [PubMed] [Google Scholar]
- Nickel J. et al. (2014) SuperPred: update on drug classification and target prediction. Nucleic Acids Res., 42, W26–W31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantziarka P. et al. (2018) ReDO_DB: the repurposing drugs in oncology database. Ecancermedicalscience, 12, 886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei F. et al. (2017) Connecting neuronal cell protective pathways and drug combinations in a Huntington’s disease model through the application of quantitative systems pharmacology. Sci. Rep., 7, 17803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei F. et al. (2019) Quantitative systems pharmacological analysis of drugs of abuse reveals the pleiotropy of their targets and the effector role of mTORC1. Front. Pharmacol., 10, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs R.E. et al. (2017) Encouraging new uses for old drugs. JAMA, 318, 2421–2422. [DOI] [PubMed] [Google Scholar]
- Stern A.M. et al. (2016) A perspective on implementing a quantitative systems pharmacology platform for drug discovery and the advancement of personalized medicine. J. Biomol. Screen., 21, 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D. et al. (2016) STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res., 44, D380–D384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S. et al. (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res., 46, D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi Y. et al. (2014) DINIES: drug-target interaction network inference engine based on supervised analysis. Nucleic Acids Res., 42, W39–W45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.