Abstract
Protein composition is restricted by the genetic code to a relatively small number of natural amino acids. Similarly, the known three-dimensional structures adopt a limited number of protein folds. However, proteins exert a large variety of functions and show a remarkable ability for regulation and immediate response to intracellular and extracellular stimuli. To some degree, the wide variability of protein function can be attributed to the post-translational modifications. Post-translational modifications have been observed in all kingdoms of life and give to proteins a significant degree of chemical and consequently functional and structural diversity. Their importance is partly reflected in the large number of genes dedicated to their regulation. So far, hundreds of post-translational modifications have been observed while it is believed that many more are to be discovered along with the technological advances in sequencing, proteomics, mass spectrometry and structural biology. Indeed, the number of studies which report novel post translational modifications is getting larger supporting the notion that their space is still largely unexplored. In this review we explore the impact of post-translational modifications on protein structure and function with emphasis on catalytic activity regulation. We present examples of proteins and protein families whose catalytic activity is substantially affected by the presence of post translational modifications and we describe the molecular basis which underlies the regulation of the protein function through these modifications. When available, we also summarize the current state of knowledge on the mechanisms which introduce these modifications to protein sites.
Keywords: Backbone proline hydroxylation, CE4 deacetylases, Asparagine deamidation, Norovirus, AMPylation, Lysine lactoylation, SelO pseudokinase, BiP chaperone, Stress response modifications, Multiple modified active sites, DJ-1, Pathogenicity, Neurodegenerative disease
1. Introduction
Post-translational modifications (PTMs) are the chemical changes occurring at the side chains or the main chain of a protein after the completion of its translation in the ribosome. These modifications involve different types of chemical reactions and their final products may include addition of new chemical moieties, double bond formation and formation of cross-linked products, L-to d-amino acid conversion, heteroatom substitution and proteolytic cleavages (Bischoff and Schlüter, 2012, Lin, Du, & Jiang, 2008, Müller, 2018, Okeley and Van Der Donk, 2000). Addition of small or larger chemical moieties to the side chains are by far the most common PTMs in proteins. Hydroxylation, methylation, acetylation, phosphorylation and glycosylation are some of the commonest and best studied PTMs as far as the source of the prosthetic group, the type of reaction and the biological impact is concerned (Lin, Du, & Jiang, 2008). Ubiquitylation is another major modification which involves the addition of the whole ubiquitin protein at specific sites of another protein to target it for degradation. Similar to the addition of chemical groups to the side chains, small groups usually hydroxyl or methyl moieties can also be added to the protein backbone (Müller, 2018). Cross linking of spatially adjacent side chains is generally a rare modification. However, the disulphide bond which is one of the commonest and more frequent PTM is formed by the cross linking of side chains. Likewise, a side chain could react with an adjacent backbone atom and form a ring product or a new residue. Deamidation of asparagine provides a characteristic example which proceeds through a cyclic succinimide intermediate. Dependent on the local environment succinimide may be the final product (Kumar et al., 2016), even though it is usually unstable and opens to isoAsp which has a shortened side chain compared to Asp and an one-carbon atom longer backbone compared to usual residues (Bischoff and Schlüter, 2012, Ravikiran and Mahalakshmi, 2014, Müller, 2018). Heteroatom substitution, as for example thioamide formation and Cα-Cβ double bond formation are two unusual PTMs with impact on backbone chemistry and flexibility. On the other hand, the backbone hydrolytic cleavage is a well-established and common modification.
PTMs can have multiple effects on a protein such as, for example, they can change the physicochemical properties, increase the steric hindrances or change the chemical reactivity of a specific site. Thus, a prosthetic group might substantially change hydrophobicity/hydrophilicity, charge and hydrogen bonding potential of the site where it is introduced. For instance, histone acetylation activates transcription, in part, by relaxation of chromatin structure through masking the lysine’s positive charge and thus, attenuating interactions with the negatively charged DNA. Likewise, histone methylation influences the histone-DNA interactions by the addition of a hydrophobic group. The prosthetic group might impose steric hindrances and restrict accessibility to a biological important site i.e. an active, binding or recognition site. For example, such a role is usually assigned to AMPylations, which restrict access to significant sites of the modified protein. Moreover, a prosthetic group may induce structural changes with impact on the local or overall structure. Typically, backbone modifications influence structural and conformational stability and flexibility with a local or long-range effect (Craveur et al., 2019, Müller, 2018). In addition, a PTM may provide a novel functional group which complement the chemistry of the site where it is introduced and potentially generate novel cofactors (Okeley & Van Der Donk, 2000).
PTMs may be reversible or irreversible, enzymatically installed or spontaneous, strictly regulated by cell mechanisms or not under any cell control. They may be dependent on the concentration of metabolism intermediate products and environmental factors such as oxidation stress and adaptation pressure. Thus, PTMs have been associated with feedback mechanisms of metabolism, regulation of biochemical pathways and often reflect a cellular ability for rapid and local response to a sharp stimulus. Sometimes PTMs are the result of chemical or radiation damage due to exposure to stressful conditions or due to the natural accumulation of deleterious factors during the cell’s lifespan. In these cases, they are associated with aging and age-related disorders and can be utilized as biomarkers or as tools to guide drug design procedures.
Current literature affords many excellent reviews which thoroughly and systematically cover specific and general topics related to PTMs. Here, our aim is not to provide an exhaustively detailed overview of the field. Instead, we anthologize and briefly present a few examples from the very recent literature which either describe a novel modification or provide new findings which expand our knowledge and understanding of how PTMs regulate protein function and especially how they modify enzymatic activity and consequently control biological pathways. These examples show the chemical and structural complexity involved in such processes and underline the still-to-be-discovered biological potential of PTMs.
2. Post-translational modifications at protein backbones
2.1. A backbone proline hydroxylation promotes active-site maturation
Proline hydroxylation, primarily at Cγ and secondarily at Cβ atoms (Bischoff & Schlüter, 2012), is a common enzymatic post-translational modification mostly catalyzed by prolyl hydroxylases. For example, Cγ-hydroxylated proline is a recurrent feature of collagen with a profound structural role at the stabilization of the triple helix (Myllyharju & Kivirikko, 2001). Hydroxyproline is also present at the plant cell wall and in proteins related to hypoxia response (Lamport et al., 2011, Palmer and Clegg, 2014). On the other hand, proline hydroxylation is largely unexplored in bacteria. Although a putative bacterial peptidyl-propyl hydroxylase has been detected, its specific substrate and biochemical pathway remain unknown (Culpepper, Scott, & Limburg, 2010).
In this context, it was of great surprise the discovery of a modified proline hydroxylated at its Cα atom in a bacterial enzyme named Bc0361 (Fadouloglou et al., 2013). Bc0361, is a Bacillus cereus polysaccharide deacetylase from CE4 family (Fig. 1 A) and the modified residue is a conserved proline lying in the active site (Balomenou & Arnaouteli, 2015). The modification was first detected when during refinement of the crystal structure extra electron density attached to that proline was best modeled as Cα-hydroxylated-l-proline (Fadouloglou et al., 2017). The modification was then confirmed by tandem mass spectrometry. In addition, it was detected in many other CE4 family polysaccharide deacetylases from the gram positive bacteria B. cereus and B . anthracis (Fadouloglou et al., 2017). Indeed, mass spectrometric (MS/MS) characterization of the hydroxylation levels at the CE4 family of deacetylases showed significant percent occurrence at the conserved proline position for many members of the family i.e. Ba1961, Ba0330, Ba3942, Bc1960, produced by different expression systems and under different conditions (Fadouloglou et al., 2017). In addition to that, Cα-hydroxylation of the conserved proline has been observed in the crystal structures of B. anthracis Ba0330 (Arnaouteli et al., 2015), B. cereus Bc1960 (Fadouloglou et al., 2017) and Bdellovibrio bacteriovorus Bd3279 (Lambert et al., 2016) polysaccharide deacetylases (Fig. 1, A–C).
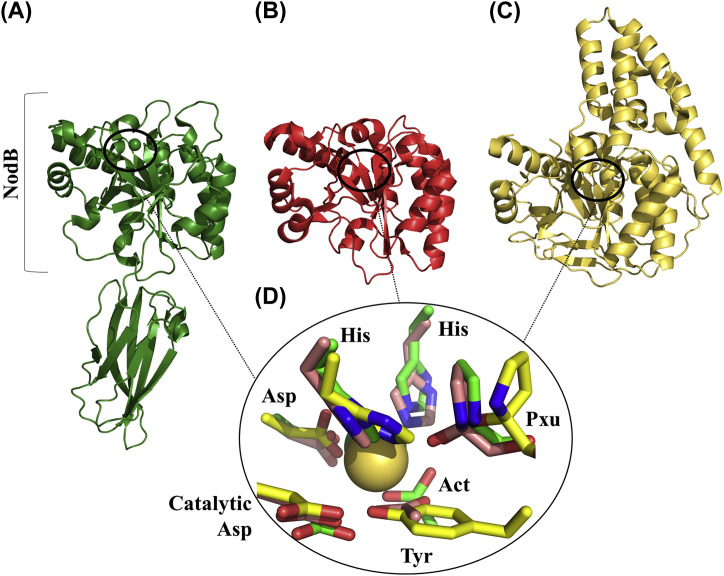
Fig. 1.
Crystal structures of three CE4 deacetylases whose active site proline is hydroxylated at the Cα (Pxu). For comparison the molecules are shown at the same orientation. Ribbon representations of the (A) Bacilluscereus Bc0361 (PDB 4HD5), (B) B. cereus Bc1960 (PDB 4L1G) and (C) Bdellovibriobacteriovorus Bd3279 (PDB 5JP6). (D) Close-up view of the superimposed active sites. The following residues are shown (coloring as in panels (A)–(C)): Cα hydroxylated Pro (Pxu); His-His-Asp metal coordination triad; catalytic Asp; the fourth ion-ligand is an acetate molecule (Act) for Bc0361 and Bc1960 and a Tyr residue for Bd3279. The ion is shown as sphere (it is only present in the structures of Bc0361 and Bd3279).
In the CE4 family of polysaccharide deacetylases, catalysis is performed by a conserved NodB domain, which folds as a distorted TIM-like barrel (Fig. 1, A–C; Balomenou & Arnaouteli, 2015). The active site contains a divalent ion coordinated by a His-His-Asp triad and it is complemented by a catalytic Asp and a His/Asp electron relay (Fig. 1D; Fadouloglou et al., 2013). The modified proline is located at the one side of the active site and the prosthetic hydroxyl group points toward the metal ion and the substrate binding site (Fig. 1D). Although the mechanism of hydroxylation is still elusive, recent findings suggest a non-enzymatic hydroxylation implicating at least one of the deacetylation reaction catalytic residues i.e. the catalytic Asp. Moreover, the origin of the hydroxyl moiety appears to be molecular oxygen rather than water (Fadouloglou et al., 2017).
Measurement of the deacetylase activities of modified and unmodified Bc1960 shows that the modified enzyme is more active by a factor of 10. Inspection of the structure and a recent computational analysis (Prejanò, Romeo, Sgrizzi, Russo, & Marino, 2019) underline that the proline hydroxylation substantially extends the hydrogen-bonding network in the active site and contributes to the stability of the intermediate state. Prejanò et al. performed a quantum mechanistics (QM) cluster model study on the reaction mechanism of Bc1960 considering the canonical and Cα-hydroxylated active site proline and showed that the hydroxyl moiety on the modified Cα establishes an extended interaction network with proline’s closest neighbors. The result is a substantial stabilization of the transition state and a decrease in the activation energy. Thus, the addition of such a small chemical moiety in the active site can cause significant improvements in enzymatic activity and contributes to active site maturation.
Collectively, Cα-hydroxylation at a strictly conserved, active-site proline residue contributes to the active site maturation by enhancing the hydrogen-bonding network and thus stabilizing the transition state.
2.2. Asparagine deamidation to isoAspartate impairs binding interactions with ligand
Asparagine deamidation is a commonly occurring spontaneous modification which starts with the nucleophilic attack of a backbone amide nitrogen to the side chain amide of an adjacent Asn to form a cyclic succinimide intermediate (Bischoff & Schlüter, 2012). Succinimide is usually unstable and is hydrolyzed to either Asp or isoAsp. Asn deamidation is usually accumulated in proteins with time, therefore it has been strongly correlated with protein aging such as, for example, calmodulin, crystallin, β-amyloid and human growth hormone aging as well as age-related disorders such as cataract and neurodegenerative diseases (Fujii et al., 2016, Johnson et al., 1989, Moro et al., 2018, Müller, 2018, Potter et al., 1993, Reissner and Aswad, 2003). However, their involvement to other biological processes is increasingly reported (Eschenburg & Schönbrunn, 2000) revealing a functional potential which extends from protein thermostability (Kumar et al., 2016) to viral pathogenicity (Mallagaray et al., 2019).
Asn deamidation to isoAsp was recently detected on the glycan-binding site of norovirus capsid protein by triple-resonance 3D NMR experiments and verified by crystallography and hydrogen/deuterium exchange (HDX) MS (Mallagaray et al., 2019). The dense layer of glycans which cover the surface of red cell membrane and consist the major part of histo-blood group antigens (HBGAs) can be exploited by microorganisms and viruses to be attached to the host-cell membrane (Heggelund et al., 2017, Shanker et al., 2017). Such a virus is the human norovirus, the causative agent of acute gastroenteritis, which recognizes and binds HBGAs to achieve infection (Taube, Mallagaray, & Peters, 2018). The norovirus capsid protein comprises a shell domain and a protruding (P) domain which exists as a dimer (Fig. 2 A) and is responsible for host-cell interactions and strain diversity (Singh, Leuthold, & Hansman, 2015).
Fig. 2.
Crystal structure of norovirus P domain dimer. (A) Superimposed ribbon diagrams of P domain dimers containing the unmodified Asn 373 (yellow, PDB 4X06) and the modified isoAsp 373 (red, PDB 6H9V). Bound glycan is shown with space filling model. The modification site is shown with sticks and highlighted in the insertion. (B) Comparison of the conformations adopted by the peptides which contain unmodified Asn 373 and modified isoAsp 373. (C) Close-up view of the P domain-glycan complex structure. The fucose moiety of the glycan is held by two direct hydrogen bonds from the side chain of Asp 374 when 373 is unmodified Asn. Instead, only one hydrogen bond remains when 373 is converted to isoAsp.
The virus-glycan interaction implicates a HBGA binding pocket on the surface of the P domain dimer. Mutations highlighting the evolution of P protein among the viral variants are mainly located on the protein surface except for the binding pocket beneath the HBGA and the surrounding regions which show little variation. In particular, on the rim of the binding pocket is found a common set of conserved residues which form numerous interactions with divergent HBGA types (Singh et al., 2015). One of them is Asp374 immediately after Asn373, the residue which is irreversible modified to isoAsp373 (Fig. 2; Mallagaray et al., 2019).
The formation of isoAsp dramatically changes the binding potential of P protein for HBGAs and causes a loss of affinity of approximately one order of magnitude (Mallagaray et al., 2019). It has been shown that isoAsp can affect protein in two ways which could explain the observed decrease in HBGA binding affinity. Firstly, there is a local effect on recognition and binding of glycans. It has been shown that Asp374 forms two direct hydrogen bonds with a fucose residue of the glycan ligand. Inspection of the structure reveals that in the deamidated P-protein the side chain of Asp374 adopts a different rotamer (Fig. 2B)-probably induced by the backbone changes occurring on the adjacent residue 373- which results in loss of one hydrogen bond (Fig. 2C).
Secondly, isoAsp formation has a long-range effect on protein dynamics, which is known to have significant impact on the ability of P-dimers to efficiently recognize and bind divergent HBGAs (Singh et al., 2015). NMR and HDX MS data agree that in the deamidated protein certain regions of P dimer display higher flexibility compared to the native protein independently on the presence of glycans. Interestingly, the elevated flexibility is not detected in the peptide which contains the isoAsp.
The deamidation of Asn373 is an irreversible modification and depends on the temperature (Mallagaray et al., 2019). It was shown that at room temperature or lower the modification occurs slowly displaying a half-life of several days or weeks. Conversely, in body temperature the half-time of modification is just 1–2 days. Moreover, it is shown that the Asn373-Asp374 motif (Fig. 2) as well as the conformation of the loop which incorporates the motif are well conserved. Thus, the Asn deamidation may represent a conserved viral mechanism for modulating recognition and binding of glycans in the course of infection.
Collectively, Asn deamidation to isoAsp occurs on an area which is critically involved in glycan binding. In addition, the modification induces changes in overall protein dynamics and protein’s binding potential. Accordingly, the modification causes a significant loss of binding affinity for glycans.
3. Metabolism regulation by post-translational modifications
3.1. Multiple post-translational modifications in a single active site
Methyl coenzyme M reductase (MCR) is universally used by methanogenic archaea to catalyze both the final step of methane-forming reaction and the first step of the anaerobic oxidation of methane (Wongnate & Ragsdale, 2015). Protein homologs share a common overall structure and a highly conserved active site. They are assembled in hexamers of three different subunits (αβγ). The subunits are organized in a dimer of trimers forming two equivalent active sites (Fig. 3 A; Ermler et al., 1997, Grabarse et al., 2001). The active site pocket incorporates a redox-sensitive nickel tetrahydrocorphinoid (coenzyme F430) and a coenzyme B. The enzyme needs both these factors to catalyze the reduction of methyl coenzyme M to produce methane (Fig. 3A; Grabarse et al., 2001). Interestingly, the α subunit of MCR features several, mostly unusual but well conserved post-translational modifications located within the active site (Fig. 3A). The modifications, which have been confirmed by numerous crystal structures and mass spectrometric data (Ermler et al., 1997, Grabarse et al., 2000, Grabarse et al., 2001) include a group of methylated residues i.e. methyl-histidine, methyl-arginine, methyl-cysteine and methyl-glutamine, a thio-glycine, a hydroxyl-tryptophan and a didehydro-aspartic acid (Fig. 3B).
Fig. 3.
Crystal structure of Methyl coenzyme M reductase (MCR). (A) Ribbon representation of the Methanothermococcus thermolithotrophicus MCR III (PDB 5N1Q). The different colors represent each of the α, β and γ subunits. The insert is a close-up view of the active site which shows the arrangement of the modified residues. The tetrahydrocorphinoid (coenzyme F430) is represented as a space-filling model. CoB-SH and CoM-SH are also shown. (B) Stick models of modified residues commonly found in the MCR active site.
Besides a heavily modified active site, MCR incorporates quite uncommon modifications. Arginine is modified at C-5 instead of the guanidino nitrogen atom where arginine methylation usually occurs (Fig. 3B). Histidine is methylated at Nπ which is a common modification site for this residue. However, histidine methylation is an unusual modification. Methyl-His and thio-Gly are strictly conserved in all MCRs examined to date (Mahanta et al., 2018, Nayak et al., 2017). Thio-Gly is a highly unusual protein modification, where the peptide amide bond is converted to thioamide i.e. the carbonyl oxygen has been replaced by sulfur. As a result, a higher multiple bond character is observed along the C—N bond and consequently a larger rotational barrier. Unlike the other methylated residues methyl-Gln is methylated at the Cα atom instead of the side chain. The didehydro-Asp contains a double bond between Cα and Cβ atoms (Siodłak, 2015, Wagner et al., 2016) and it is present only in a subgroup of MCR isoenzymes (Wagner, Wegner, Kahnt, Ermler, & Shima, 2017). Likewise, the 6-hydroxyl-Trp, which is a novel modification is not highly conserved among MCRs (Wagner et al., 2017).
Although post-translational modifications in the MCR have been known for many years, the elucidation of their exact impact on catalytic mechanism and the identification of their source remain largely unresolved. The newly discovered Trp hydroxylation to C-6 introduces an additional polar group and therefore increases the hydrogen bonding network in the active site (Wagner et al., 2017). Indeed, inspection of the crystal structures demonstrates that the hydroxy-group is in hydrogen-bonding distance with main chain and side chain atoms. On the other hand, the double bond between Cα and Cβ in the didehydro aspartic acid causes a local backbone distortion which also affects the interaction distances of the polar side chain group with adjacent residues. Recent findings show that the modification is reversible since it is easily reduced to aspartate (Wagner et al., 2016). However, no evidence exists for the biochemical pathway and the mechanism which generates this modification.
The didehydro-Asp is adjacent to the thio-Gly and both residues are found on a bend structure. Indeed, the functional role which has been proposed for thio-Gly is stabilization of the protein secondary structure in the active site (Nayak et al., 2017). The formation of thio-Gly is enzymatic and at least two enzymes i.e. YcaO and TfuA have been identified to be involved in its biosynthesis (Mahanta et al., 2018). Thio-Gly is not essential for MCR activity but it does affect methanotrophic archaea growth under specific conditions of nutrient and temperature (Nayak et al., 2017). Furthermore, thio-Gly does not influence the MCR global stability since the thermal stability of the protein is not significantly affected by the presence of this modification. Accordingly, it has been proposed that the role of the thioamide is to maintain the architecture of the active site and the conformations of the surrounding amino acids.
Methylation of methyl-modified residues in MCR has long been presumed to be catalyzed by S-adenosylmethionine-dependent methyltransferases (Selmer et al., 2000). However, only recently a study was able to correlate the MCR methyl-Arg to MmpO protein, which is a S-adenosylmethionine methylase (Lyu et al., 2020, Radle et al., 2019). The study not only demonstrates that MmpO and methyl-Arg are correlated in a highly specific manner but also shows that arginine methylation has an impact in methanogenesis and cell growth (Lyu et al., 2020).
MCR serves a chemically challenging and complex catalytic mechanism. PTMs complement the active site with important chemical features and largely contribute to the necessary fine-tuning (Nayak et al., 2020). Thus, PTMs optimize the catalysis in such a way that they are indispensable.
3.2. Lysine acylations: novel discoveries of PTMs of a commonly modified residue
Lysine is commonly modified at the side-chain nitrogen atom by alkylation and acylation reactions. Mono-, di- and tri-methylations as well as acetylation, formylation and ubiquitinylation belong to the most well-known lysine modifications (Zee & Garcia, 2012) with significant impact on biological systems. Particularly, lysine methylation and acetylation have attracted much attention mainly because of their relation to gene transcription regulation through histone modifications (Karve and Cheema, 2011, Lin, Du, & Jiang, 2008). Histone acetylation is primarily correlated with transcription activation through a relaxation of chromatin packing on the nucleosome. In addition, acetylated Lys is able to recruit other proteins such as bromodomain containing proteins which help with gene transcription activation. On the other hand, histone methylation is associated with both transcription activation and inactivation. Histone acetylases and methylases catalyze these modifications during reactions which chiefly use acetyl-CoA and S-adenylosyl-methionine (SAM) as acetylating and methylating agents, respectively. Likewise, removal of these modifications is enzymatically catalyzed by histone deacetylases and demethylases. At least two types of histone deacetylases are known, the zinc-dependent hydrolases and the NAD-dependent sirtuins. It is interesting that sirtuins can couple NAD-degradation to Lys-deacetylation by sensing the cell metabolic state i.e. sirtuins sense the NAD concentration in the cell and regulate Lys deacetylation accordingly (Imai et al., 2000, Sauve et al., 2006).
In the last decade a growing number of new, non-enzymatic Lys-acylations has been reported and their significance as metabolism regulators has been demonstrated (Lin et al., 2012, Harmel and Fiedler, 2018). The acylation group typically originates from a cellular metabolite which is converted to acyl-CoA that is the most common acylating agent. This kind of spontaneous modifications are usually enzymatically reversible, occur to metabolic enzymes and regulate their function -activate or inactivate them (Lin et al., 2012). For instance, it was shown that lysine acetylation, apart from its well established role in histone regulation, is also present in enzymes that catalyze metabolic processes including glycolysis, gluconeogenesis, glycogen and fatty acid metabolism, urea and tricarboxylic acid cycle (Zhao et al., 2010). The acetylation state of the enzymes is greatly dependent on the concentration of metabolites such as glucose, amino acids and fatty acids. Moreover, it was shown that Lys-acetylation can cause different effects i.e. activation, deactivation or destabilization on different enzymes.
Another example is the selective modification of lysine residues in the active site of glycolytic enzymes by the glycolytic intermediate 1,3-bisphosphoglycerate. The formation of 3-phosphoglyceryl-lysines results in enzymatic inhibition which is believed that is a feedback mechanism for redirection of glycolytic intermediates to other biosynthetic processes (Moellering & Cravatt, 2013).
In 2014, Tan and workers detected for the first time a lysine glutarylation in metabolic enzymes and mitochondrial proteins (Tan et al., 2014). In particular, it was shown that the carbamoyl phosphate synthase 1, an enzyme important for ammonia detoxification in the urea cycle, is Lys-glutarylated and that the effect of this modification is enzyme inhibition. Moreover, it was demonstrated that the modification is reversible and that lysine deglutarylation is performed by sirtuin 5, an enzyme previously annotated as deacetylase (Tan et al., 2014).
The latest addition to the list of acylated lysines is the lactoylysine which was reported recently by two different groups. Gaffney and co-workers (Gaffney et al., 2020) showed that the new lysine modification is derived from methylglyoxal, a glycolytic by-product. According to the study, methylglyoxal is rapidly conjugated to glutathione to produce lactoylglutathione. Enzymatic hydrolysis of lactoylglutathione produces glutathione and lactate. Lactate can then modify lysine residues mainly of glycolytic enzymes to generate the lactoylysine modification which result in a decreased glycolytic output. Therefore, it was proposed that lysine lactoylation serves as a regulatory feedback mechanism for glycolysis. The lactoylysine levels are directly dependent on the levels of lactoylglutathione and its hydrolytic enzyme. Moreover, findings support a non-enzymatic acyl transfer of the lactoyl-group similar to the non-enzymatic acyl-transfer mechanism of lysine acetylation from acetyl-CoA and acetyl-glutathione.
The existence and biological relevance of lysine lactoylation was further verified by another group which reported a histone lactoylysine modification (Zhang et al., 2019), derived by a lactoyl-CoA intermediate. Zhang and co-workers showed that lactate production and histone lysine lactoylation levels were induced by glucose and regulated by glycolysis. Use of a non-metabolizable glucose analog resulted in a decrease of both lactate production and histone lysine lactoylation. Moreover, histone lysine lactoylation by lactate was proposed to serve as an epigenetic mechanism which directly stimulates gene transcription.
Collectively, a growing number of new Lys-acylations has been discovered the last decade. The source of these modifications are metabolic intermediate products and their targets are primarily enzymes involved in metabolism. Glucose is the major source of intracellular lactate and lactate is used to modify Lys to lactoylated-Lys which is the newest reported Lys-acylation and plays a role as regulator of glucose metabolism and stimulator of gene transcription.
4. AMPylation: new insights to an emerging modification
AMPylation is the transfer of AMP to a protein substrate during an enzymatically catalyzed reaction called adenylylation (Casey & Orth, 2018). AMPylases (or AMPylators) have the same substrate as kinases i.e. an ATP molecule and primarily modify Ser, Thr and Tyr residues. Two classes of AMPylases have been identified so far, those containing an adenyl transferase (ATase) domain and those containing a Fic domain. AMPylation is an emerging modification which influences the enzymatic activity of the modified proteins or alters the pattern of interactions with molecular partners. In this way, for instance, it is used for the rapid regulation of a protein folding chaperon in response to its substrate fluctuations or it is exploited by bacterial pathogens to manipulate the host cellular machinery.
4.1. AMPylation regulates protein folding
BiP is an Hsp70 protein folding chaperone located at the endoplasmatic reticulum (ER). The protein comprises a nucleotide binding domain (NBD) which is responsible for ATP binding and hydrolysis, a substrate binding domain (SBD) which binds unfolded protein and a linker between them (Fig. 4 ). It is known that in the course of BiP-mediated protein folding, the chaperon binds the unfolded client-protein in a process that is coupled to ATP hydrolysis and also involves major conformational changes. When BiP is in the ADP state (and in the apo form) adopts an extended, domain-undocked, conformation (Fig. 4B) and binds with high affinity the exposed hydrophobic residues of unfolded or misfolded proteins (Fig. 4C; Bertelsena, Chang, Gestwicki, & Zuiderweg, 2009). ATP binding, on the other hand, induce a conformational change to a more compact, domain-docked, structure (Fig. 4A), which results in release of the client protein (Yan, Rato, Rohland, Preissler, & Ron, 2019).
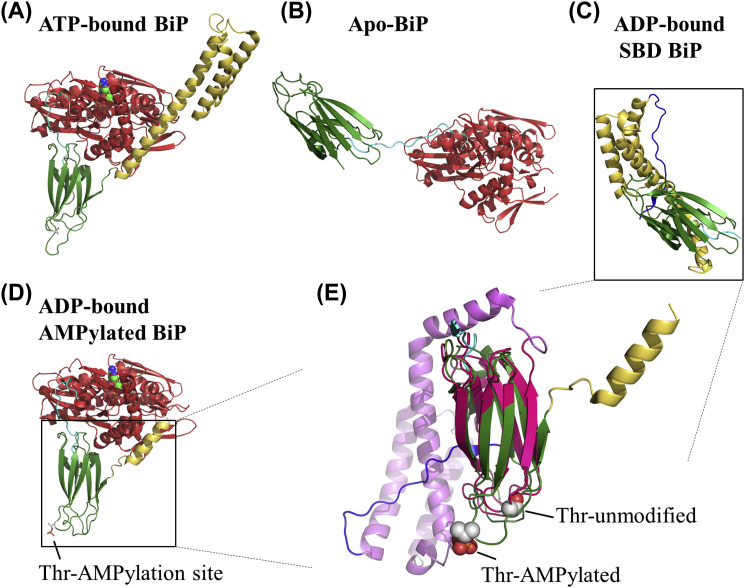
Fig. 4.
Crystal structures of BiP protein in different conformations. In panels (A)–(D) the color scheme is as follows: the nucleotide binding domain (NBD) is shown in red, the substrate binding domain (SBD) is shown in green, the C-terminal helical extension of SBD is shown in yellow. The structures are superimposed on their NBDs. (A) Ribbon representation of BiP in complex with ATP (space filling model). (B) Apo BiP which is structurally equivalent to the ADP-bound conformation. (C) SDB and its C-terminal helical extension. A peptide from the C-terminus occupies the peptide binding pocket and it is shown in blue. (D) Ribbon representation of AMPylated BiP in complex with ADP (space filling model). (E) Comparison of the SDBs and their C-terminal helical extensions from panels (C) and (D). Here, the structure from (C) panel is shown in magenta for clarity.
Because the load of unfolded proteins is continually fluctuating, the levels of active BiP must be accordingly regulated by a rapid and highly effective mechanism. It has long been speculated that such a mechanism should be regulated at the post-translational level. Indeed, recent evidence supports a BiP regulation mechanism which is mediated by BiP AMPylation through the FICD enzyme of the ER. The mechanism is regulated as follows: when the load of unfolded proteins is low, FICD transfers an AMP moiety to a specific Thr residue of BiP and AMPylates it (Fig. 4D and E). This modification inactivates BiP since the modified BiP binds less firmly peptide-substrates than the unmodified or it has an impaired response in particular stimuli. Notably, when there is a high load of unfolded proteins, FICD activates a reverse mechanism and deAMPylates Thr (Perera et al., 2019).
Crystal structure determination of the AMPylated BiP complexed with ADP revealed that modification favors a domain-docked BiP conformation (Fig. 4D) similar to that adopted by the ATP-bound chaperone (Fig. 4A) instead of the one adopted by the ADP-bound unmodified BiP (Fig. 4B; Preissler et al., 2017). This conformation corresponds to the low affinity binding of BiP for client-proteins (Fig. 4E). Moreover, it was shown that apart from the consequences that AMPylation has on the substrate binding, it also affects the interactions of BiP on J-domain co-chaperones. J-domain are non-client bound proteins which assist BiP by stimulating the ATPase activity.
4.2. AMPylation mediates bacterial pathogenicity and host-defense mechanisms
Legionella pneumophila is a gram-negative bacterium, the causative agent of Legionnaires’ disease. During infection, L. pneumophila is internalized into the host cells and forms a protective membrane-vacuole where replicates. To achieve replication, the bacterium secretes to the host an arsenal of effector proteins which modulate host-cell functions. DrrA and SdeA are two L. pneumophila effector proteins whose activity is related to AMPylation and they will be discussed in detail below.
L. pneumophila DrrA protein AMPylates the eukaryotic host’s Rab1b GTPase therefore provides an example of AMPylation performed by a bacterial protein to a eukaryotic one. Crystal structure determination of DrrA revealed structural similarities with nucleotidyl transferases. In particular, the N-terminal domain of DrrA folds similarly to the C-terminal domain of glutamine synthetase adenylyl transferase (GS-ATPase) and contains a motif similar to the GS-ATPase catalytic motif. Initially, these observations raised speculations for the AMPylase activity of DrrA and led to the experiments which confirmed this activity. Subsequently, it was shown that the high cytotoxicity of DrrA for mammalian cells is associated with its activity to AMPylate the Rab1b GTPase (Marcucci et al., 2010).
In particular, tandem MS and site-directed mutagenesis demonstrated that the AMPylation site is a surface Tyr residue on the switch II region of Rab1b GTPase (Fig. 5Α; Marcucci et al., 2010). Switch II region is known to be involved in binding to effector and GTPase-activating proteins (GAPs). Structural comparison of unmodified and modified Rab1b GTPases showed that no significant structural changes are induced from the AMPylation consistent with the location of AMP group to the exterior of the structure (Fig. 5Α). On the other hand, AMPylation significantly inhibits GAPs stimulating GTP hydrolysis and prolongs Rab1b GTP state presumably by restricting the access of GAPs to switch II region. Likewise, AMPylation selectively affects interactions with effector proteins. For instance, it was shown that AMPylation abolish binding to the mammalian effector MICAL-3 but not to the bacterial LidA (Marcucci et al., 2010).
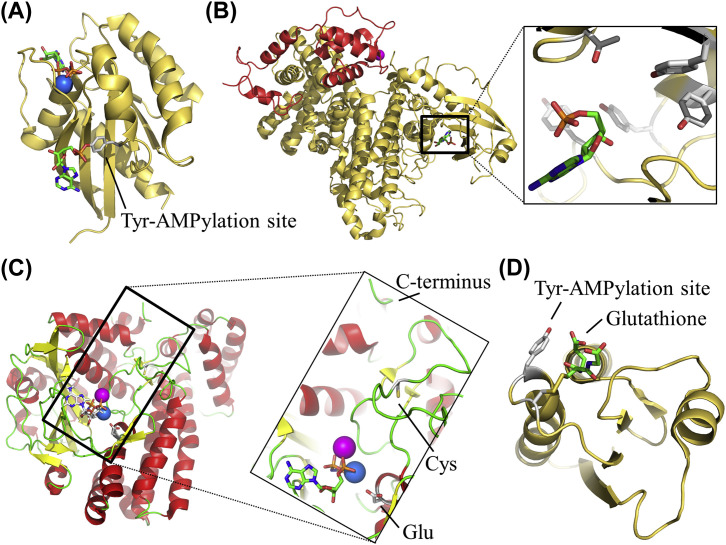
Fig. 5.
Crystal structures of proteins associated with AMPylation. Nucleotides and analogs are shown as green sticks, side chains as gray sticks and metal ions as spheres (blue for Mg, magenta for Ca). (A) Ribbon representation of the human Rab 1b covalently modified with AMP at Tyr 77 (PDB 3NKV). It is shown (i) a GppNHp and a magnesium ion (blue sphere) bound in the GTP-binding site (ii) the AMPylated Tyr on the switch II area of the protein. (B) Ribbon representation of Legionella pneumophila SidJ (yellow) in complex with CaM (red) and AMP (stick model) (PDB 6K4R) (PDB 6K4R). The insertion is a close-up view of the AMP-binding pocket. Several residues within the pocket could be possible AMPylation sites (sticks in gray). (C) Ribbon representation of the Pseudomonas syringae SelO in complex with AMP-PNP (PDB 6EAC). Insertion is a close-up view of the nucleotide binding site. It is shown a Glu residue with a possible catalytical role and the Cys in the enzyme’s activating loop which forms a disulfide bridge with the C-terminal Sec/Cys. (D) Ribbon representation of the E.coli Glutaredoxin (PDB 1GRX). The AMPylation site (Tyr) and the Cys which interacts with glutathione are shown in stick representation.
L. pneumophila SdeA is a mono-ADP ribosyltransferase involved in ubiquitination of host proteins associated with the ER. Bacterial dependent ubiquitination of host proteins starts with the ADP-ribosylation of specific ubiquitin sites, an activity catalyzed by SdeA. The modified ubiquitin impairs the function of eukaryotic cells by inhibiting canonical ubiquitination. Thus, SdeA is an important bacterial virulence factor. The deleterious function of SdeA for the host is inhibited by the post-translational modification of a catalytically important residue. Activation of this residue for modification is achieved through an AMPylated intermediate (Gan et al., 2019). Especially, it was shown that SdeA toxicity can be suppressed by another bacterial protein named SidJ. SidJ inhibits the mono-ADP ribosyltransferase activity of SdeA through glutamylation of a catalytically important Glu residue. SidJ exerts the glutamylase activity and modifies Glu by adding one or more glutamate moieties (Gan et al., 2019).
Interestingly, the inhibitory effects of SidJ to SdeA were only evident when the proteins were expressed in eukaryotic cells supporting the hypothesis that the SidJ/SdeA interaction is mediated by a eukaryotic factor. Indeed, it was found that SdeA can only be modified in the presence of calmodulin (CaM) and that CaM/SidJ/l-glutamate complex suppressed SdeA-mediated ubiquitination (Fig. 5B; Gan et al., 2019).
Further examination of the SidJ catalyzed reaction revealed that glutamylation mechanism involves ATP hydrolysis to AMP (Fig. 5B). Moreover, when the reaction cannot be productive i.e. in the absence of substrate (SdeA) or glutamate, SidJ undergoes self-AMPylation. Taking together these findings it was proposed that SdeA-Glu glutamylation from SidJ proceeds through an AMPylated-Glu intermediate as follows: First, SidJ activates its target i.e. SdeA-Glu by its adenylation (AMPylation). Then, the glutamate amino group attacks the activated carbonyl group of the unstable intermediate Glu-AMP to form a Glu-glutamate final modification (Gan et al., 2019).
Collectively, bacterial SidJ is a CaM-dependent glutamylase that catalyzes the ligation of glutamates to a catalytically important Glu of bacterial SdeA. As a result, SdeA is inactivated and its eukaryotic toxicity is abolished. The mechanism of inhibition includes an intermediate state where a transient Glu-AMP is formed. In addition, when SidJ cannot catalyze a productive reaction undergoes self-AMPylation.
4.3. The SelO pseudokinase is actually an AMPylase
Perhaps the most exciting example of AMPylation comes from an evolutionary conserved seleno pseudokinase (SelO). Members of SelO family have been characterized as pseudokinases because they share sequence similarity with kinases and they lack amino acids which are important for catalysis. Interestingly, SelO homologs from higher eukaryotes incorporate the uncommon 21st genetically encoded amino acid namely Sec. Sec is structurally equivalent to Cys except for the sulfur atom which is replaced by a selenium. Despite their similarities, Sec has a lower pKa than Cys and it is deprotonated at physiological pH; therefore, it is a stronger nucleophile with higher oxidoreductase efficiency. There is a single Sec at the C-terminus of the Sec-containing SelO homologs, while in lower eukaryotes and prokaryotes the equivalent position is invariably occupied by a Cys residue (Sreelatha et al., 2018).
Crystal structure determination of the Pseudomonas syringae SelO homolog in complex with ATP (Fig. 5C) confirmed previous predictions that the protein adopts a protein kinase fold and revealed an unexpected flipped binding orientation for ATP (Sreelatha et al., 2018). This accidental observation raised questions about a possible previously overlooked protein functionality and triggered a series of experiments. It was demonstrated that SelO can transfer AMP from ATP to protein substrates, therefore it is an AMPylase (Sreelatha et al., 2018). SelO activity depends on the presence of divalent cations in the active site (Fig. 5C), similarly to kinases. Interestingly, SelO activity also depends on the formation of a disulfide bond between the conserved Cys/Sec at the C-terminus and a Cys in the enzyme’s activation loop (Fig. 5C). Furthermore, the structural inspection of SelO suggests that the originally considered as impaired active site, because of the lack of a catalytically important Asp, may not be actually impaired. The apparently absent catalytic Asp could be afforded by a different position which better suits the new, flipped substrate orientation.
Two proteins have been identified as direct protein targets for SelO namely Glutaredoxin (GlxA) and the SucA component of the α-ketoglutarate dehydrogenase. Both, SucA and GlxA are involved in redox processes i.e. the first is a redox sensor and the latter removes glutathione from cysteines in an oxidative stress defense mechanism conserved in eukaryotes and prokaryotes. It was shown that SelO AMPylates a Tyr residue of the highly conserved redox active site of GlxA (Fig. 5D). AMPylation inactivates GlxA and protect yeast cells from oxidative stress. It was proposed that Tyr-AMPylation imposes steric hindrance which renders the active site inaccessible to glutathione and inhibits GlxA activity.
In conclusion, SelO-mediated GlxA AMPylation regulates protein glutathionylation and protects against oxidative stress.
5. Stress responsive post-translational modifications: cysteine oxidation and S-nitrosylation
Human DJ-1 is a small (21 kDa) ubiquitously expressed redox protein associated with many neurodegenerative diseases and the cellular oxidative stress response (Fig. 6 A; Antipova & Bandopadhyay, 2017). DJ-1 has been associated with neurodegenerative disorders because deletion of the DJ-1 gene or loss-of-function mutations causes early-onset Parkinson’s disease. Other studies implicate DJ-1 to oxidative stress response as, for instance, that activates SOD1 (superoxide dismutase) by acting as a metallochaperone which transfers copper to SOD1 (Girotto et al., 2014). DJ-1 shows analogies with thiol peroxidases and sulfoxide reductases. There are studies which attribute to DJ-1 a translational/transcriptional regulation role or even a function outside the cell (Tashiro, Caaveiro, Wu, Hoang, & Tsumoto, 2014). Although the precise biochemical function remains still elusive, accumulating evidence correlates DJ-1 function with the ability of a specific Cys to undergo post-transcriptional modifications.
Fig. 6.
Crystal structure of DJ-1 protein. (A) Ribbon diagram of the human DJ-1 (PDB 1P5F). (B)–(C) Close-up views of the modified Cys in three different cases: (B) Sulfinic-Cys (Csd; PDB 1SOA), (C) S-nitrosylated-Cys (Snc; PDB 4RKY), (D) Carboxymethylated- Cys (Ccs; PDB 6E5Z).
An acidic isoform of DJ-1 is formed in vivo in response to oxidative stress suggesting the presence of a sensor on the protein for reactive oxygen species. Indeed, a conserved Cys residue, extremely sensitive to radiation damage during DJ-1 crystal structure determination (Wilson, Collins, Hod, Ringe, & Petsko, 2003), was identified as the preferred target for post-translational modifications by reactive oxygen species generated by oxidative stress. This Cys (Cys 106), which is surrounded by a cluster of conserved acidic residues, is a catalytic residue in homologs and systematically adopts an unfavorable backbone conformation in the Ramachandran plot. The sulfinic modification of Cys 106 was directly observed by X-ray crystallography (Fig. 6B; Canet-Avilés et al., 2004), was also confirmed by mass spectrometry and it is consistent with the formation of an acidic DJ-1 isoform whose accumulation is observed under oxidative stress. Moreover, it was shown that oxidation of Cys 106 is reversible and signals the DJ-1 re-localization to mitochondria where exerts a protective activity against mitochondrial damage. For example, toxicity induced by the MPP + toxin was eliminated by overexpression of wt DJ-1 but not by Cys106Ala variant (Blackinton et al., 2009, Canet-Avilés et al., 2004). Apart from the mitochondrial localization, the sulfinic Cys106 modification is implicated in neuroprotection by at least two more mechanisms i.e. regulating a redox-regulated chaperone activity and directly scavenging reactive oxygen species (Andres-Mateos et al., 2007, Girotto et al., 2014). Furthermore, the reduced structure of DJ-1 -stabilized by a Glu-Leu motif of a peptide bound to a site close to Cys106-was captured and showed no structural differences compared to the oxidized protein (Premkumar, Dobaczewska, & Riedl, 2011).
An unusually low pKa (5.4) has been recorded spectrophotometrically for Cys106 and has been attributed to the formation of a thiolate anion which is substantially stabilized by the local protein environment (Witt et al., 2008). Structural inspection reveals a Cys106-Glu18 interaction which is conserved in every structurally characterized member of the DJ-1 superfamily (Fig. 6). The significance of this interaction was demonstrated by site-directed mutagenesis and shows that even minor substitutions significantly influence the pKa value of C106. A bond length analysis, after unrestrained bond length refinement of a high-resolution structure (1.2 Å) of DJ-1 shows that Glu18 is protonated (and so uncharged) and able to donate a hydrogen bond to Cys106 and thus to stabilize the thiolate anion.
Similar to oxidative stress, nitrosative stress, namely overproduction of reactive NO species, causes neuron death and it is associated with many neurodegenerative disorders. On the other hand, small concentrations of NO play multiple regulatory roles in cell pathways. S-nitrosylation is a post-translational modification often involved in processes that regulate the biological functions of NO. It is known that the phosphatase activity of PTEN (phosphatase and tensin homolog) causes neuronal death and that DJ-1 is a negative regulator of PTEN. Choi and co-workers showed that under mild-NO stress DJ-1 is post-translationally S-nitrosylated at Cys106 (Fig. 6C) and this modification has a neuroprotective effect through regulation of the PTEN pathway (Choi et al., 2014). In particular, S-nitrosylated DJ-1 transfers the NO group to PTEN (transnitrosylation) and thus inhibits the phosphatase activity of PTEN and suppresses the PTEN-dependent cell death.
Recent findings imply that Cys106 may undertake even more modifications such as a transient carboxymethylation (Fig. 6D) mediating an, as yet, uncharacterized metabolic pathway (Mussakhmetov et al., 2018). Collectively, oxidative/nitrosative stress contribute to the pathogenesis of neurodegenerative diseases, whereas detoxification of the reactive species is neuroprotective. DJ-1 is a conserved multifunctional protein which is implicated in a number of cellular pathways. The ability of DJ-1 to respond to oxidative/nitrosative stress mitigates cellular toxicity and results in neuroprotection. Cys106 is a redox-active site of oxidation and S-nitrosylation which trigger different processes of neuroprotection.
6. Conclusions
Post-translational modifications are used by all kingdoms of life to generate molecular diversity and afford biological macromolecules a huge potential for functional and structural flexibility and adaptation. Based on chemistry rather than genetics, this mechanism expands, modifies and regulates the physiological roles of proteins so that they are able for quick and selective responses to numerous stimuli. Thus, PTMs can generate permanent or transient alterations, in the whole or in part of the population of a protein by using either enzymatic or spontaneous processes.
Introducing PTMs to enzymes is a highly versatile tool for controlling catalytic activity. In this way, it is possible to accurately and promptly regulate a wide range of processes such as protein folding, cellular metabolism, stress response, defense against pathogens and pathogenicity itself. It has long been known that pathogens can use PTMs as a virulence mechanism to manipulate the host’s cellular machinery (Ribet & Cossart, 2010). Numerous bacterial pathogens including Streptococcus, Staphylococcus, Yersinia, Pseudomonas, Salmonella and Legionella species use, in part, PTMs to achieve survival and replication within the host. In these cases, the virulence factors namely the virulence proteins produced by pathogens and mediating the host-pathogen interactions can use PTMs to target host’s signaling pathways and evade immune responses (Chiang and Gack, 2017, Ravikumar et al., 2015, Salomon and Orth, 2013). Likewise, viral pathogens can utilize PTMs in order to overcome the host defense mechanisms and hijack its cellular machinery for its own replication and propagation (Loboda, Soond, Piacentini, & Barlev, 2019). It is well established, for example, that structural and non-structural proteins from the coronavirus family of RNA viruses are extensively post-translationally modified by a variety of modifications. These modifications facilitate utilization of the host’s protein synthesis machinery for viral replication and promote pathogenesis (Fung & Liu, 2018).
Besides the infectious diseases, aging and age-related diseases have often been related to PTMs (Santos & Lindner, 2017). Such PTMs are usually those which are no-under cellular control, for example those which are accumulated in proteins during the cell lifespan or those which are generated as the result of oxidative stress (Banks and Andersen, 2019, Svistunova et al., 2019). Moreover, disturbances in the pattern of PTMs affect the cellular control pathways and causes abnormalities. Neurodegenerative and autoimmune diseases as well as heart failure and cancer have been associated with PTMs. Thus, PTMs are often used as biomarkers, indicators of abnormalities and attractive targets for vaccines (Brentville et al., 2020, Carubbi et al., 2019, Didonna and Benetti, 2016, Toya et al., 2020).
Although PTMs have long been considered as potential drug targets (Parekh & Rohlff, 1997), it is the progress of the last decade that has given new impetus to the discovery of medicine based on them. Our better understanding of the mechanisms that control PTMs and the mechanisms through which PTMs regulate interaction networks into the cell makes possible the identification of relevant molecular targets and the design of specific drugs. Thus, PTMs especially those which affect catalytic activity emerge as a highly promising field for investigation and development of new therapeutics for a wide range of infectious and non-infectious diseases.
References
- Andres-Mateos E., Perier C., Zhang L., Blanchard-Fillion B., Greco T.M., Thomas B., et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(37):14807–14812. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antipova D., Bandopadhyay R. Expression of DJ-1 in neurodegenerative disorders. Advances in Experimental Medicine & Biology. 2017;1037:25–43. doi: 10.1007/978-981-10-6583-5_3. [DOI] [PubMed] [Google Scholar]
- Arnaouteli S., Giastas P., Andreou A., Tzanodaskalaki M., Aldridge C., Tzartos S.J., et al. Two putative polysaccharide deacetylases are required for osmotic stability and cell shape maintenance in Bacillus anthracis. Journal of Biological Chemistry. 2015;290(21):13465–13478. doi: 10.1074/jbc.M115.640029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balomenou S., Arnaouteli S. Polysaccharide deacetylases: New antibacterial drug targets. Frontiers in Anti-Infective Drug Discovery. 2015:68–130. doi: 10.2174/9781681080826115040005. [DOI] [Google Scholar]
- Banks C.J., Andersen J.L. Mechanisms of SOD1 regulation by post-translational modifications. Redox Biology. 2019;26(May):101270. doi: 10.1016/j.redox.2019.101270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsena E.B., Chang L., Gestwicki J.E., Zuiderweg E.R.P. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(21):8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff R., Schlüter H. Amino acids: Chemistry, functionality and selected non-enzymatic post-translational modifications. Journal of Proteomics. 2012;75(8):2275–2296. doi: 10.1016/j.jprot.2012.01.041. [DOI] [PubMed] [Google Scholar]
- Blackinton J., Lakshminarasimhan M., Thomas K.J., Ahmad R., Greggio E., Raza A.S., et al. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. Journal of Biological Chemistry. 2009;284(10):6476–6485. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentville V.A., Vankemmelbeke M., Metheringham R.L., Durrant L.G. Post-translational modifications such as citrullination are excellent targets for cancer therapy. Seminars in Immunology. 2020;47(January):101393. doi: 10.1016/j.smim.2020.101393. [DOI] [PubMed] [Google Scholar]
- Canet-Avilés R.M., Wilson M.A., Miller D.W., Ahmad R., McLendon C., Bandyopadhyay S., et al. The Parkinson’s disease DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(24):9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carubbi F., Alunno A., Gerli R., Giacomelli R. Post-translational modifications of proteins: Novel insights in the autoimmune response in rheumatoid arthritis. Cells. 2019;8(7):657. doi: 10.3390/cells8070657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey A.K., Orth K. Enzymes involved in AMPylation and deAMPylation. Chemical Reviews. 2018;118(3):1199–1215. doi: 10.1021/acs.chemrev.7b00145. American Chemical Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C., Gack M.U. Post-translational control of intracellular pathogen sensing pathways. Trends in Immunology. 2017;38(1):39–52. doi: 10.1016/j.it.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M.S., Nakamura T., Cho S.J., Han X., Holland E.A., Qu J., et al. Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in Parkinson’s disease models. Journal of Neuroscience. 2014;34(45):15123–15131. doi: 10.1523/JNEUROSCI.4751-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craveur P., Narwani T.J., Rebehmed J., de Brevern A.G. Investigation of the impact of PTMs on the protein backbone conformation. Amino Acids. 2019;51(7):1065–1079. doi: 10.1007/s00726-019-02747-w. [DOI] [PubMed] [Google Scholar]
- Culpepper M.A., Scott E.E., Limburg J. Crystal structure of prolyl 4-hydroxylase from Bacillus anthracis. Biochemistry. 2010;49(1):124–133. doi: 10.1021/bi901771z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didonna A., Benetti F. Post-translational modifications in neurodegeneration. AIMS Biophysics. 2016;3(1):27–49. doi: 10.3934/biophy.2016.1.27. [DOI] [Google Scholar]
- Ermler U., Grabarse W., Shima S., Goubeaud M., Thauer R.K. Crystal structure of methyl-coenzyme M reductase: The key enzyme of biological methane formation. Science. 1997;278(5342):1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- Eschenburg S., Schönbrunn E. Comparative X-ray analysis of the un-liganded fosfomycin-target MurA. Proteins: Structure, Function, and Genetics. 2000;40(2):290–298. doi: 10.1002/(SICI)1097-0134(20000801)40:2<290::AID-PROT90>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Fadouloglou V.E., Balomenou S., Aivaliotis M., Kotsifaki D., Arnaouteli S., Tomatsidou A., et al. Unusual α-carbon hydroxylation of proline promotes active-site maturation. Journal of the American Chemical Society. 2017;139(15):5330–5337. doi: 10.1021/jacs.6b12209. [DOI] [PubMed] [Google Scholar]
- Fadouloglou V.E., Kapanidou M., Agiomirgianaki A., Arnaouteli S., Bouriotis V., Glykos N.M., et al. Structure determination through homology modelling and torsion-angle simulated annealing: Application to a polysaccharide deacetylase from Bacillus cereus. Acta Crystallographica Section D Biological Crystallography. 2013;69(2):276–283. doi: 10.1107/S0907444912045829. [DOI] [PubMed] [Google Scholar]
- Fujii N., Takata T., Fujii N., Aki K. Isomerization of aspartyl residues in crystallins and its influence upon cataract. Biochimica et Biophysica Acta (BBA) - General Subjects. 2016;1860(1):183–191. doi: 10.1016/j.bbagen.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: Roles and function. Future Virology. 2018;13(6):405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney D.O., Jennings E.Q., Anderson C.C., Marentette J.O., Shi T., Schou Oxvig A.M., et al. Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chemical Biology. 2020;27(2):206–213.e6. doi: 10.1016/j.chembiol.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan N., Zhen X., Liu Y., Xu X., He C., Qiu J., et al. Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. Nature. 2019;572(7769):387–391. doi: 10.1038/s41586-019-1439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotto S., Cendron L., Bisaglia M., Tessari I., Mammi S., Zanotti G., et al. DJ-1 Is a copper chaperone acting on SOD1 activation. Journal of Biological Chemistry. 2014;289(15):10887–10899. doi: 10.1074/jbc.M113.535112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarse W., Mahlert F., Duin E.C., Goubeaud M., Shima S., Thauer R.K., et al. On the mechanism of biological methane formation: Structural evidence for conformational changes in methyl-coenzyme M reductase upon substrate binding. Journal of Molecular Biology. 2001;309(1):315–330. doi: 10.1006/jmbi.2001.4647. [DOI] [PubMed] [Google Scholar]
- Grabarse W., Mahlert F., Shima S., Thauer R.K., Ermler U. Comparison of three methyl-coenzyme M reductases from phylogenetically distant organisms: Unusual amino acid modification, conservation and adaptation. Journal of Molecular Biology. 2000;303(2):329–344. doi: 10.1006/jmbi.2000.4136. [DOI] [PubMed] [Google Scholar]
- Harmel R., Fiedler D. Features and regulation of non-enzymatic post-translational modifications. Nature Chemical Biology. 2018;14(3):244–252. doi: 10.1038/nchembio.2575. [DOI] [PubMed] [Google Scholar]
- Heggelund J.E., Varrot A., Imberty A., Krengel U. Histo-blood group antigens as mediators of infections. Current Opinion in Structural Biology. 2017;44:190–200. doi: 10.1016/j.sbi.2017.04.001. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- Imai S.I., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Johnson B.A., Shirokawa J.M., Hancock W.S., Spellman M.W., Basa L.J., Aswad D.W. Formation of isoaspartate at two distinct sites during in vitro aging of human growth hormone. Journal of Biological Chemistry. 1989;264(24):14262–14271. http://www.ncbi.nlm.nih.gov/pubmed/2760065 [PubMed] [Google Scholar]
- Karve T.M., Cheema A.K. Small changes huge impact: The role of protein posttranslational modifications in cellular homeostasis and disease. Journal of Amino Acids. 2011;2011:1–13. doi: 10.4061/2011/207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Prakash S., Gupta K., Dongre A., Balaram P., Balaram H. Unexpected functional implication of a stable succinimide in the structural stability of Methanocaldococcus jannaschii glutaminase. Nature Communications. 2016;7 doi: 10.1038/ncomms12798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C., Lerner T.R., Bui N.K., Somers H., Aizawa S.I., Liddell S., et al. Interrupting peptidoglycan deacetylation during Bdellovibrio predator-prey interaction prevents ultimate destruction of prey wall, liberating bacterial-ghosts. Scientific Reports. 2016;6(May):1–19. doi: 10.1038/srep26010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D.T.A., Kieliszewski M.J., Chen Y., Cannon M.C. Role of the extensin superfamily in primary cell wall architecture. Plant Physiology. 2011;156(1):11–19. doi: 10.1104/pp.110.169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Du J., Jiang H. Post-translational modifications to regulate protein function. Wiley Encyclopedia of Chemical Biology. 2008 doi: 10.1002/9780470048672.wecb467. [DOI] [Google Scholar]
- Lin H., Su X., He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chemical Biology. 2012;7(6):947–960. doi: 10.1021/cb3001793. American Chemical Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A.P., Soond S.M., Piacentini M., Barlev N.A. Lysine-specific post-translational modifications of proteins in the life cycle of viruses. Cell Cycle. 2019;18(17):1995–2005. doi: 10.1080/15384101.2019.1639305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu Z., Shao N., Chou C.W., Shi H., Patel R., Duin E.C., et al. Posttranslational methylation of arginine in methyl coenzyme M reductase has a profound impact on both methanogenesis and growth of Methanococcus maripaludis. Journal of Bacteriology. 2020;202(3):1–18. doi: 10.1128/JB.00654-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanta N., Liu A., Dong S., Nair S.K., Mitchell D.A. Enzymatic reconstitution of ribosomal peptide backbone thioamidation. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(12):3030–3035. doi: 10.1073/pnas.1722324115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallagaray A., Creutznacher R., Dülfer J., Mayer P.H.O., Grimm L.L., Orduña J.M., et al. A post-translational modification of human Norovirus capsid protein attenuates glycan binding. Nature Communications. 2019;10(1) doi: 10.1038/s41467-019-09251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Bloomfield C.D., Caligiuri M.A., Tachibana K.E., Laskey R.A., Coleman N., et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab 1b. Science. 2010;329(August):946–950. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- Moellering R.E., Cravatt B.F. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science. 2013;341(6145):549–553. doi: 10.1126/science.1238327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro M.L., Phillips A.S., Gaimster K., Paul C., Mudher A., Nicoll J.A.R., et al. Pyroglutamate and Isoaspartate modified Amyloid-Beta in ageing and Alzheimer’s disease. Acta Neuropathologica Communications. 2018;6(1):3. doi: 10.1186/s40478-017-0505-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.M. Post-translational modifications of protein backbones: Unique functions, mechanisms, and challenges. Biochemistry. 2018;57(2):177–185. doi: 10.1021/acs.biochem.7b00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussakhmetov A., Shumilin I.A., Nugmanova R., Shabalin I.G., Baizhumanov T., Toibazar D., et al. A transient post-translational modification of active site cysteine alters binding properties of the parkinsonism protein DJ-1. Biochemical and Biophysical Research Communications. 2018;504(1):328–333. doi: 10.1016/j.bbrc.2018.08.190. [DOI] [PubMed] [Google Scholar]
- Myllyharju J., Kivirikko K.I. Collagens and collagen-related diseases. Annals of Medicine. 2001;33(1):7–21. doi: 10.3109/07853890109002055. Royal Society of Medicine Press Ltd. [DOI] [PubMed] [Google Scholar]
- Nayak D.D., Liu A., Agrawal N., Rodriguez-Carerro R., Dong S.H., Mitchell D.A., et al. Functional interactions between posttranslationally modified amino acids of methyl-coenzyme M reductase in Methanosarcina acetivorans. PLoS Biology. 2020;18(2) doi: 10.1371/journal.pbio.3000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak D.D., Mahanta N., Mitchell D.A., Metcalf W.W. Post-translational thioamidation of methyl-coenzyme M reductase, a key enzyme in methanogenic and methanotrophic archaea. ELife. 2017;6(I):1–18. doi: 10.7554/eLife.29218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeley N.M., Van Der Donk W.A. Novel cofactors via post-translational mocifications of enzyme active sites. Chemistry & Biology. 2000;7(7):159–171. doi: 10.1016/S1074-5521(00)00140-X. [DOI] [PubMed] [Google Scholar]
- Palmer B.F., Clegg D.J. Oxygen sensing and metabolic homeostasis. Molecular and Cellular Endocrinology. 2014;397(1–2):51–58. doi: 10.1016/j.mce.2014.08.001. Elsevier Ireland Ltd. [DOI] [PubMed] [Google Scholar]
- Parekh R.B., Rohlff C. Post-translational modification of proteins and the discovery of new medicine. Current Opinion in Biotechnology. 1997;8(6):718–723. doi: 10.1016/S0958-1669(97)80126-7. [DOI] [PubMed] [Google Scholar]
- Perera L.A., Rato C., Yan Y., Neidhardt L., McLaughlin S.H., Read R.J., et al. An oligomeric state-dependent switch in the ER enzyme FICD regulates AMP ylation and de AMP ylation of BiP. The EMBO Journal. 2019;38(21) doi: 10.15252/embj.2019102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S.M., Henzel W.J., Aswad D.W. In vitro aging of calmodulin generates isoaspartate at multiple Asn–Gly and Asp–Gly sites in calcium-binding domains II, III, and IV. Protein Science. 1993;2(10):1648–1663. doi: 10.1002/pro.5560021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S., Rohland L., Yan Y., Chen R., Read R.J., Ron D. AMPylation targets the rate-limiting step of BiP’s ATPase cycle for its functional inactivation. ELife. 2017;6 doi: 10.7554/eLife.29428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prejanò M., Romeo I., Sgrizzi L., Russo N., Marino T. Why hydroxy-proline improves the catalytic power of the peptidoglycan: N -deacetylase enzyme: Insight from theory. Physical Chemistry Chemical Physics. 2019;21(42):23338–23345. doi: 10.1039/c9cp03804c. [DOI] [PubMed] [Google Scholar]
- Premkumar L., Dobaczewska M.K., Riedl S.J. Identification of an artificial peptide motif that binds and stabilizes reduced human DJ-1. Journal of Structural Biology. 2011;176(3):414–418. doi: 10.1016/j.jsb.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radle M.I., Miller D.V., Laremore T.N., Booker S.J. Methanogenesis marker protein 10 (Mmp10) from methanosarcina acetivorans is a radical S-adenosylmethionine methylase that unexpectedly requires cobalamin. Journal of Biological Chemistry. 2019;294(31) doi: 10.1074/jbc.RA119.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikiran B., Mahalakshmi R. Unusual post-translational protein modifications: The benefits of sophistication. RSC Advances. 2014;4(64):33958–33974. doi: 10.1039/c4ra04694c. Royal Society of Chemistry. [DOI] [Google Scholar]
- Ravikumar V., Jers C., Mijakovic I. Elucidating host-pathogen interactions based on post-translational modifications using proteomics approaches. Frontiers in Microbiology. 2015;6(NOV):1–7. doi: 10.3389/fmicb.2015.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissner K.J., Aswad D.W. Deamidation and isoaspartate formation in proteins: Unwanted alterations or surreptitious signals? Cellular and Molecular Life Sciences. 2003;60(7):1281–1295. doi: 10.1007/s00018-003-2287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D., Cossart P. Pathogen-mediated posttranslational modifications: A re-emerging field. Cell. 2010;143(5):694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D., Orth K. What pathogens have taught us about posttranslational modifications. Cell Host & Microbe. 2013;14(3):269–279. doi: 10.1016/j.chom.2013.07.008. Cell Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A.L., Lindner A.B. Protein posttranslational modifications: Roles in aging and age-related disease. Oxidative Medicine and Cellular Longevity. 2017;2017 doi: 10.1155/2017/5716409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve A.A., Wolberger C., Schramm V.L., Boeke J.D. The biochemistry of sirtuins. Annual Review of Biochemistry. 2006;75(1):435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Selmer T., Kahnt J., Goubeaud M., Shima S., Grabarse W., Ermler U., et al. The biosynthesis of methylated amino acids in the active site region of methyl-coenzyme M reductase. Journal of Biological Chemistry. 2000;275(6) doi: 10.1074/jbc.275.6.3755. [DOI] [PubMed] [Google Scholar]
- Shanker S., Hu L., Ramani S., Atmar R.L., Estes M.K., Venkataram Prasad B.V. Structural features of glycan recognition among viral pathogens. Current Opinion in Structural Biology. 2017;44:211–218. doi: 10.1016/j.sbi.2017.05.007. Elsevier Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.K., Leuthold M.M., Hansman G.S. Human noroviruses’ fondness for histo-blood group Antigens. Journal of Virology. 2015;89(4):2024–2040. doi: 10.1128/jvi.02968-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siodłak D. α,β-Dehydroamino acids in naturally occurring peptides. Amino Acids. 2015;47(1):1–17. doi: 10.1007/s00726-014-1846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreelatha A., Yee S.S., Lopez V.A., Park B.C., Kinch L.N., Pilch S., et al. Protein AMPylation by an evolutionarily conserved pseudokinase. Cell. 2018;175(3):809–821. doi: 10.1016/j.cell.2018.08.046. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistunova D.M., Simon J.N., Rembeza E., Crabtree M., Yue W.W., Oliver P.L., et al. Oxidation resistance 1 regulates post-translational modifications of peroxiredoxin 2 in the cerebellum. Free Radical Biology and Medicine. 2019;130(July 2018):151–162. doi: 10.1016/j.freeradbiomed.2018.10.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Peng C., Anderson K.A., Chhoy P., Xie Z., Dai L., et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metabolism. 2014;19(4):605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S., Caaveiro J.M.M., Wu C.X., Hoang Q.Q., Tsumoto K. Thermodynamic and structural characterization of the specific binding of Zn(II) to human protein DJ-1. Biochemistry. 2014;53(14):2218–2220. doi: 10.1021/bi500294h. [DOI] [PubMed] [Google Scholar]
- Taube S., Mallagaray A., Peters T. Norovirus, glycans and attachment. Current Opinion in Virology. 2018;31:33–42. doi: 10.1016/j.coviro.2018.04.007. Elsevier B.V. [DOI] [PubMed] [Google Scholar]
- Toya T., Ito K., Kagami K., Osaki A., Sato A., Kimura T., et al. Impact of oxidative posttranslational modifications of SERCA2 on heart failure exacerbation in young patients with non-ischemic cardiomyopathy: A pilot study. IJC Heart and Vasculature. 2020;26:100437. doi: 10.1016/j.ijcha.2019.100437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Kahnt J., Ermler U., Shima S. Didehydroaspartate modification in methyl-coenzyme M reductase catalyzing methane formation. Angewandte Chemie International Edition. 2016;55(36):10630–10633. doi: 10.1002/anie.201603882. [DOI] [PubMed] [Google Scholar]
- Wagner T., Wegner C.E., Kahnt J., Ermler U., Shima S. Phylogenetic and structural comparisons of the three types of methyl coenzyme M reductase from Methanococcales. Journal of Bacteriology. 2017;199(16):1–15. doi: 10.1128/JB.00197-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.A., Collins J.L., Hod Y., Ringe D., Petsko G.A. The 1.1-Å resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt A.C., Lakshminarasimhan M., Remington B.C., Hasim S., Pozharski E., Wilson M.A. Cysteine pKa depression by a protonated glutamic acid in human DJ-1. Biochemistry. 2008;47(28):7430–7440. doi: 10.1021/bi800282d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongnate T., Ragsdale S.W. The reaction mechanism of methyl-coenzyme M reductase. Journal of Biological Chemistry. 2015;290(15):9322–9334. doi: 10.1074/jbc.M115.636761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Rato C., Rohland L., Preissler S., Ron D. MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP. Nature Communications. 2019;10(1):1–15. doi: 10.1038/s41467-019-08450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee B.M., Garcia B.A. Discovery of lysine post-translational modifications through mass spectrometric detection. Essays in Biochemistry. 2012;52(1):147–163. doi: 10.1042/BSE0520147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]