Abstract
Human placental development during early pregnancy is poorly understood. Many conceptuses are lost at this stage. It is thought that preeclampsia, intrauterine growth restriction and other placental syndromes that manifest later in pregnancy may originate early in placentation. Thus, there is a need for models of early human placental development. Treating human embryonic stem cells (hESCs) with BMP4 (bone morphogenic protein 4) plus A83-01 (ACTIVIN/NODAL signaling inhibitor) and PD173074 (fibroblast growth factor 2 or FGF2 signaling inhibitor) (BAP conditions) induces differentiation to the trophoblast lineage (hESCBAP), but it is not clear which stage of trophoblast differentiation these cells resemble. Here, comparison of the hESCBAP transcriptome to those of trophoblasts from human blastocysts, trophoblast stem cells and placentas collected in the first–third trimester of pregnancy by principal component analysis suggests that hESC after 8 days BAP treatment most resemble first trimester syncytiotrophoblasts. To further test this hypothesis, transcripts were identified that are expressed in hESCBAP but not in cultures of trophoblasts isolated from term placentas. Proteins encoded by four genes, GABRP (gamma-aminobutyric acid type A receptor subunit Pi), WFDC2 (WAP four-disulfide core domain 2), VTCN1 (V-set domain containing T-cell activation inhibitor 1) and ACTC1 (actin alpha cardiac muscle 1), immunolocalized to placentas at 4–9 weeks gestation, and their expression declined with gestational age (R2 = 0.61–0.83). None are present at term. Expression was largely localized to syncytiotrophoblast of both hESCBAP cells and placental material from early pregnancy. WFDC2, VTCN1 and ACTC1 have not previously been described in placenta. These results support the hypothesis that hESCBAP represent human trophoblast analogous to that of early first trimester and are a tool for discovery of factors important to this stage of placentation.
Keywords: embryonic stem cells, trophoblast differentiation, placental development, primitive syncytium, human blastocyst implantation
Introduction
The villous human placenta is readily obtained after delivery and easily separated from maternal decidual tissue, allowing its structure, its main physiological features and gene expression profiles to be extensively characterized at term and to a lesser extent from the mid-first trimester onwards (Burton and Fowden, 2015; Jedrusik et al., 2015). It comprises several types of trophoblast, fetal endothelial cells and mesenchyme. The villi themselves contain a layer of cytotrophoblast (CTB) that is self-renewing, and that gives rise to the syncytiotrophoblast (STB) and extravillous trophoblast cells (EVTs) (Harris, 2010; Fowden et al., 2015; Michelsen et al., 2017). These cell types emerge at Weeks 5–6 since last menstrual period or LMP, but the developmental period from implantation, at about 3 weeks LMP, or 7 days post-fertilization, through their formation (Fig. 1) is for practical purposes inaccessible to investigators, who have had to rely primarily on archived material such as the Boyd Collection at the University of Cambridge and the Carnegie Collection at the Human Developmental Anatomy Center, Washington DC (Hertig et al., 1956; Burton et al., 1999) or on a few studies in primates (Enders, 1989; Enders et al., 1997; Fazleabas et al., 1997). Barriers to understanding this stage of pregnancy involve practical and ethical constraints. In particular, blastocyst implantation and pre-villous placental development occur just prior to the time of missed menses and before the time that a pregnancy can be detected clinically.
Figure 1.
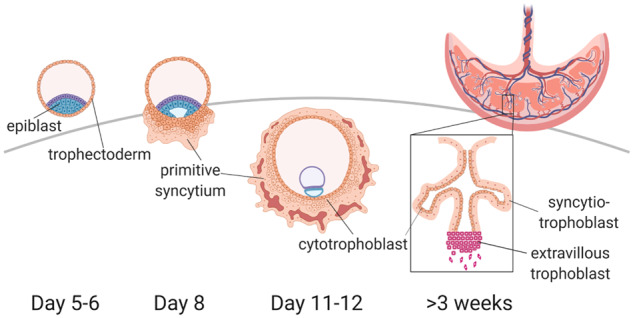
Human placental trophoblasts emerge from the trophectoderm layer of the blastocyst (left). During embryo implantation, these cells differentiate into proliferative cytotrophoblasts, and an outer syncytiotrophoblast mass that is invasive, called the primitive syncytium. By Day 12 post-fertilization, primary villi, consisting of cytotrophoblast, begin to penetrate the primitive syncytium layer. These become filled with fetal mesenchyme to form secondary villi and are then vascularized by fetal endothelium to form the tertiary villi that are characteristic of the mature placenta (right). By the third week after conception, it is extravillous trophoblast cells, emerging from the tips of anchoring villi that are invasive, and the syncytiotrophoblast, surrounded by maternal blood, functions primarily in transport (Boyd and Hamilton, 1970). Human embryonic stem cells (hESC) are derived from blastocyst epiblast cells. Trophoblast stem cells (TSC) are derived from either blastocyst trophectoderm or first trimester villous cytotrophoblast. Transcriptomic data, invasive behavior and syncytial morphology collectively suggest that BAP-treated human embryonic stem cells (hESCBAP) cells may resemble the primitive syncytium.
The lack of a model system has hindered characterization of trophoblast gene and protein expression during this period, which likely has features distinct from later stages of pregnancy, although it is part of a continuum of placental developmental and functional change lasting until term. This can be inferred from microarray studies performed over the first and second trimesters of pregnancy in which it was observed that the transcriptomes of Weeks 6–7, 9–10, 10–11 and 14–16 week placentae clustered separately from each other and dramatically from placental tissue obtained at 4–5 weeks LMP (Soncin et al., 2018). Importantly, trophoblast dysfunction that originates during this timeframe could result in complications later in pregnancy, including intrauterine growth restriction, preeclampsia and preterm birth (Villar et al., 2012; Fisher, 2015; Nardozza et al., 2017). Alternatively, failure of pre-villous placental development may lead to implantation failure or idiopathic early pregnancy loss (Norwitz et al., 2001; Home et al., 2017). This highlights the pressing need to model peri-implantation placental development so as to enable detailed study at the transcript, protein and functional levels.
We hypothesize that human embryonic stem cells (hESCs) treated with bone morphogenic protein 4 (BMP4, B) and the inhibitors A83-01 (Activin A/Nodal signaling inhibitor, A) and PD173074 (fibroblast growth factor 2 or FGF2 receptor inhibitor, P) BAP treated embryonic stem cells most closely model the early first trimester period of trophoblast differentiation. Although the trophoblastic identity of this cell culture model has been extensively investigated, the stage of pregnancy that it represents has not yet been determined. These BAP-differentiated hESC (hESCBAP) syncytialize (Das et al., 2007; Schulz et al., 2008; Amita et al., 2013; Yabe et al., 2016; Roberts et al., 2018), are invasive (Telugu et al., 2013), and express genes encoding a range of proteins involved in trophoblast invasion and migration (Horii et al., 2016; Yabe et al., 2016; Jain et al., 2017) and placental hormone production, (e.g. chorionic gonadotropin subunits b and a—CGB, CGA) at much higher levels than term trophoblast (Amita et al., 2013; Yabe et al., 2016). They express more than 63 individual gene makers characteristic of trophoblast and a global transcriptome profile that exclusively resembles placental trophoblast (Telugu et al., 2013; Roberts et al., 2014; Yabe et al., 2016; Jain et al., 2017; Roberts et al., 2018). Nevertheless, CTB and STB isolated from BAP-differentiated colonies of hESC, express some genes not found in CTB and STB cultured from primary human trophoblast at term (Yabe et al., 2016; Jain et al., 2017). Based on the hypothesis that hESCBAP most resemble the placenta of very early pregnancy, we predict that these genes are expressed in the human placenta in the first trimester.
To test this, here the global transcriptomic profiles of hESCBAP were compared not only to our cultured term primary human trophoblast datasets but to publicly available transcriptomes of trophoblast from the blastocyst stage through term. Secondly, genes differentially expressed between hESCBAP and cultures of primary term human trophoblasts were identified as putative markers of trophoblast from early in the first trimester of human pregnancy. Four of the putative markers, GABRP (gamma-aminobutyric acid type A receptor subunit Pi), WFDC2 (WAP Four-Disulfide Core Domain 2), VTCN1 (V-set domain containing T-cell activation inhibitor 1) and ACTC1 (actin alpha cardiac muscle 1), were selected for further study because they are largely or entirely unstudied in the placenta, and because their functions in cancers and other tissues suggest potential roles in trophoblast invasion, which is unique to the first trimester placenta (Ferretti et al., 2007). GABRP, a GABAergic receptor subunit, is highly expressed within the uterus and placenta (Hedblom and Kirkness, 1997), and its overexpression in HTR8-SV/neo cells inhibits invasion through Matrigel (Lu et al., 2016). WFDC2 has not previously been identified in trophoblast, but in ovarian cancer cells it promotes expression of MMP2 (matrix metalloproteinase 2) and other genes required for cell migration (Chen et al., 2017). VTCN1 has previously been identified in immune cells during pregnancy and recently was found in the transcriptome of trophoblast derived from hESCs via treatment with BMP4 alone (Krendl et al., 2017). VTCN1 is expressed also in several cancers, with heightened expression coinciding with larger tumor size, increased malignancy score, decreased survival and decreased number of tumor-infiltrating T-cells (Podojil and Miller, 2017). Finally, ACTC1 is an alpha-actin protein not previously studied in the placenta, but whose identity as a cytoskeletal protein makes a role in invasion possible (Zhou et al., 2014). Here, we examine for the first time when and where these four proteins are expressed in the placenta across gestation, also thereby testing whether hESCBAP may be a tool for the discovery of novel markers of the first trimester placenta.
Materials and methods
Principal component analysis
Principal component analysis (PCA) was used to compare trophoblast derived by BAP-treatment of hESC to other sources of trophoblast. RNAseq data for hESCBAP was obtained from a previous study (Yabe et al., 2016) (NCBI Gene Expression Omnibus, GSE73017). For that study (Yabe et al., 2016), H1 hESC were BAP-treated for eight days. Then, colonies were dispersed non-enzymatically, and the cell suspension sequentially passed through 70 µm and 40 µm cell strainers. The fraction larger than 70 µm (hESCBAP > 70) comprised syncytialized patches, or STB. Cells smaller than 40 µm were largely CTB, with some small STB aggregates. CTB were also isolated from term placentae at Magee-Women’s Hospital of the University of Pittsburgh Medical Center and cultured. RNA was collected from the undifferentiated CTB after 9 h of in vitro cell culture, or after 48 h when the majority of cells had spontaneously syncytialized (Kliman et al., 1986; Nelson et al., 1999; Yabe et al., 2016).
Four RNAseq datasets were selected to collectively profile gene expression across gestation in individual trophoblast subtypes. Petropoulos et al. (2016a) accessed via ArrayExpress: E-MTAB-3929, contains single-cell RNAseq data from preimplantation human blastocysts, including trophectoderm cells. Liu et al. (2018) NCBI Gene Expression Omnibus (GEO) GSE89497, contains single-cell RNAseq data from CTB, STB and EVT at 8 weeks gestation and CTB and EVT at 24 weeks gestation. Pavlicev et al. (2017) NCBI GEO GSE87692, contains single cell RNA-seq of enzymatically digested, percoll-isolated trophoblast from villous tissue and bulk-sequenced, laser-captured STB, all from term placenta. Okae et al. (2018) contains bulk RNAseq data from trophoblast stem cells (TSCs), CTB, STB and EVT derived from those TSCs, as well as CTB and EVT purified from first trimester placentas, and STB shed from first trimester placentas. Accession numbers from Okae et al include JGA00000000074, JGA00000000117 and JGA00000000122. Gene identifiers from each data set were cross-referenced from the supplied identifier from the Sequence Read Archive (National Center for Biotechnology Information) to the Ensembl gene ID by using annotation data from the BioConductor package org.Hs.eg.db (Carlson, 2018), and the union of all identifiers was used for subsequent processing. To capture the most biologically relevant subset of this combined dataset, only the top 25% most variant genes were used for PCA. The dataset was first mean centered and scaled to unit variance, while the Eigen values were calculated by using singular value decomposition (R Core Team, 2018).
Human embryonic stem cell culture
The hESC line H1 (WA01) obtained from WiCell Research Institute in Madison, WI (Thomson et al., 1998) was maintained in mTeSR1 medium (Stem Cell Technologies) in Matrigel (Corning)-coated dishes. Cells were passaged every 5–6 days by using Dispase (Stem Cell Technologies) and the Stempro EZpassage tool (Invitrogen) dispersion method to obtain uniformly-sized square sheets ∼100 cells. The differentiation protocol employed was that of Amita et al. (2013). Accordingly, 50 000 hESC were passaged onto a Matrigel-coated coverslip in each well of a 6-well plate. The cells were first cultured on mTeSR1 medium (Stem Cell Technologies) for 24 h and then for a further 24 h on basal DMEM/F12 medium supplemented with 20% knock-out serum replacement, nonessential amino acids, 2 mM L-glutamine and 4 ng/ml FGF2. At this stage, the FGF2-containing medium was replaced with one lacking FGF2 but containing BMP4 (10 ng/ml), A83-01 (1 µM) and PD173074 (0.1 µM), with daily medium changes for either 6 or 8 days.
Immunocytochemistry
H1 hESCs were plated onto Matrigel-coated coverslips and differentiated for 6 days with BAP before fixation in 4% paraformaldehyde. Coverslips were placed in 5% bovine serum albumin + 2.5% v/v donkey serum for 1 h to block non-specific staining and then incubated overnight at 4°C with primary antibody (Table I). After incubation with secondary antibody for 1 h at room temperature, coverslips were mounted with Vectashield + DAPI (4,6-diamidino-2-phenylindole). Images were captured either by a Zeiss Axiovert 200M fluorescent microscope equipped with a monochrome ORCA-ER camera or a Leica GSD 3D super-resolution microscope, equipped with 405, 488, 560 and 642 nm lasers for multicolor detection and with a resolution 20 nm in lateral plane and 50 nm in the axial plane, and a Andor iXon Ultra 897 EMCCD camera. Both are located in the University of Missouri Molecular Cytology Core. Coverslips incubated with only secondary antibodies were used as negative controls to determine image acquisition settings for each experiment (Supplementary Fig. S1).
Table I.
Antibodies used for immunostaining.
| Antigen | Source | Catalog number | Host species | Dilution |
|---|---|---|---|---|
| ACTC1 | Millipore | MABT823 | Mouse | 1:100 |
| CGA | R&D | MAB4169 | Mouse | 1:200 |
| CGB | abcam | ab53087 | Rabbit | 1:200 |
| GABRP | Aviva Systems | AVARP13034_P050 | Rabbit | 1:70 |
| HLAG | Santa Cruz | Sc-21799 | Mouse | 1:100 |
| KRT7 | Santa Cruz | sc-17116 | Goat | 1:100 |
| VTCN1 | Cell Signaling (IHC); abcam (IF) | D1M8I (IHC); ab209242 (IF) | Rabbit | 1:100 |
| WFDC2 | abcam | ab109298 | Rabbit | 1:70 |
| Rabbit, mouse or goat IgG | Invitrogen Life Technologies | A11055, A21206, A31570, A31573, A21447 | Donkey | 1:300 |
ACTC1, Actin Alpha Cardiac Muscle 1; CGA, Chorionic Gonadotrophin Subunit Alpha; CGB, Chorionic Gonadotropin Subunit Beta; GABRP, Gamma-Aminobutyric Acid Type A Receptor Subunit Pi; HLAG, Major Histocompatibility Complex, Class I, G; KRT7, Cytokeratin 7; VTCN1, V-Set Domain Containing T Cell Activation Inhibitor 1; WFDC2, WAP Four-Disulfide Core Domain 2.
Human placental tissue samples
Paraffin-embedded placental tissue sections were obtained from the University of California, San Diego Perinatal Biorepository. Blastocyst sections consist of maternal decidual tissue, CTB shell, primitive syncytium and secondary villi from developmental Day 16. First and second trimester samples consist of embedded villous fragments received post-surgically. For each third trimester placenta, two samples were taken from the pericentral area, approximately half way between the cord insertion site and the margin.
Immunohistochemistry
Placental sections were rehydrated through a xylene substitute and graded alcohol series. Antigen retrieval was performed by boiling slides in 10 mM citrate buffer pH 6.0 in a pressure cooker on a hot plate set to maximum temperature (153°C) and pressure for 3 min (Norton et al., 1994). Immunofluorescent staining was then performed as described above. Immunoperoxidase staining of primary placental sections was performed for VTCN1 by using the Vectastain Elite ABC HRP kit (Vector Laboratories) for rabbit primary antibodies. Sections were counterstained in Mayer’s hematoxylin (DAKO #S3309).
Image analysis
Immunofluorescence was quantified in ImageJ (v2.0, National Institute of Health) by calculating the Corrected Total Cell Fluorescence (CTCF = integrated density − (area of selected region × mean background fluorescence)) (McCloy et al., 2014). For each section, two to four fields of view were photographed with a 20× objective lens. All CTB and STB regions of each villus within each field of view were manually outlined in Image J to calculate the average CTCF per villus. To analyze changes in immunofluorescence across gestational age, CTCF values at each age were fit to a second order polynomial equation (x + x2) by using the nonlinear regression function in Prism 5.0 software (GraphPad Inc.).
Ethical approvals
Human placental tissues had been collected under a protocol approved by the Human Research Protections Program Committee of the University of California San Diego Institutional Review Board and transferred to the University of Missouri with approval from the University of Missouri Institutional Review Board. All patients gave informed consent for collection and use of these tissues and samples were de-identified.
Results
Comparison of the hESCBAP transcriptome to those of other human trophoblast
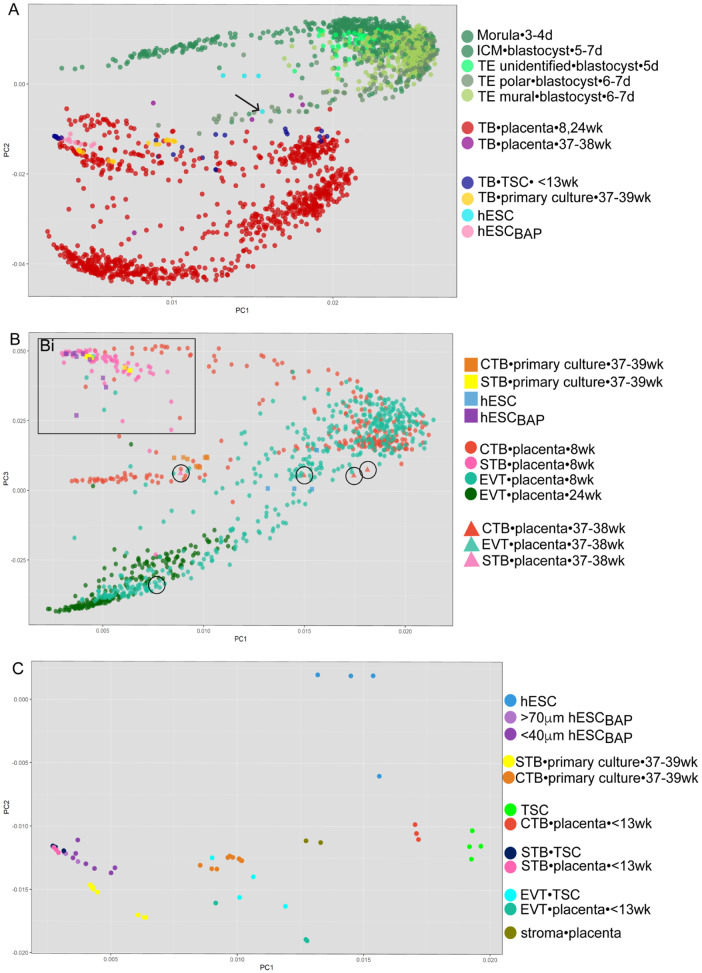
To test whether the hESCBAP model most closely represents trophoblast of early pregnancy, we compared the hESCBAP RNA sequencing data with simultaneously sequenced CTB cultured from term placentas and STB derived from them, along with four other publicly deposited RNAseq datasets spanning various gestational ages and trophoblast cell types. The earliest timepoint we employed is a single cell RNAseq (scRNAseq) dataset from human embryos at the morula and blastocyst stage (Petropoulos et al., 2016a). PCA shows that hESCBAP, as well as the cultured primary term human trophoblasts, cluster distinctly from all lineages of human morula and blastocyst cells (Fig. 2A, Supplementary Fig. S3), although they are closer to these cells than are late pregnancy trophoblast. Undifferentiated embryonic stem cells, hESC, do cluster close to cells of the blastocyst lineage (Fig. 2A), although one hESC sample, which had shown morphological signs of spontaneous differentiation, clusters further away from the other hESC, and closer to the hESCBAP, as denoted by the arrow in Fig. 2A. A series of polar trophectoderm (TE) datapoints leads toward the early STB points, in proximity to the spontaneously differentiated hESC sample (Fig. 2A, darker olive green). Cells of the morula and inner cell mass cluster closely together, near but separate from hESC (Supplementary Fig. S3). The PCA was also computed with an scRNAseq dataset containing 8 week CTB, STB and EVT and 24 week EVT (Liu et al., 2018) and a dataset containing CTB, STB, and EVT lineages from term placenta (Pavlicev et al., 2017). Figure 2B shows that hESCBAP cluster closely with 8 week CTB and STB, more distantly from 24 week EVT and all trophoblast lineages from term placenta (Fig. 2B, term placental samples circled) and cluster most closely with 8 week STB (Fig. 2Bi). Finally, the hESCBAP model was compared with TSCs and first trimester trophoblast sequenced by Okae et. al. (Okae et al., 2018). As shown in Fig. 2C, hESCBAP most closely cluster with primary first trimester STBs and STB derived from TSCs. Thus, hESCBAP transcriptionally most resemble STB from the first trimester and human TSCs differentiated toward the STB lineage.
Figure 2.
Principal component analysis comparing five human trophoblast RNAseq datasets from the following sources: morula-blastocyst stage human embryos (Petropoulos et al., 2016a), trophoblast stem cells (Okae et al., 2018), placentas (Pavlicev et al., 2017; Liu et al., 2018), and embryonic stem cells and primary trophoblast cultures (Yabe et al., 2016). For each datapoint, cell type•cell source•gestational age are indicated in the legend on the right. (A) Principal component (PC) 1 and 2 separated by cell origin. hESCBAP cluster separately from cells of the blastocyst and from term trophoblast. Arrow indicates an hESC sample with morphological signs of spontaneous differentiation. (B) hESCBAP and trophoblast isolated from first trimester placentas do not cluster with primary trophoblast lineages from term trophoblast isolated from term placenta (circles). The hESCBAP cluster closer to 8 weeks CTB and STB than to 8 weeks or 24 weeks EVT. (C) STB derived from TSCs, hESCBAP, and first trimester STB cluster together. First trimester CTB and TSCs cluster close to one another yet distinctly from hESCBAP. The hESCBAP cluster closely with STB from the first trimester across multiple datasets. CTB, cytotrophoblast; D, days since conception; EPI, epiblast; EVT, extravillous trophoblast; hESC, human embryonic stem cell; embryonic stem cell; hESCBAP, hESC differentiated to TB with BAP; ICM, inner cell mass; PE, primitive endoderm; STB, syncytiotrophoblast; TB, trophoblast; TE, trophectoderm; TSC, trophoblast stem cell; Wk, weeks since last menstrual period.
Identification of transcripts that distinguish trophoblast derived from BAP-differentiated hESC and term trophoblast
Next, transcripts were identified that are highly expressed by CTB and STB isolated from differentiated colonies of ESC (hESCBAP < 40 µm and hESCBAP > 70 µm), but are not well expressed in CTB and STB cultured from human placentae at term to find potential markers of the first trimester placenta. Nineteen transcripts were identified for which there was differential expression (P < 0.0001) between hESCBAP < 40 µm and primary cultures of term human CTB fractions, high expression (FPKM > 100) in the hESCBAP < 40 µm fraction, extremely low expression (Fragments Per Kilobase of transcript per Million mapped reads or FPKM < 1) in the primary cultured term CTB fraction, and relatively weak expression in undifferentiated hESC (Table II). Four of these genes (GABRP, WFDC2, ACTC1 and VTCN1) and their protein products were selected for further investigation, beginning with the RNAseq data sets used in the PCA (Supplementary Fig. S4). None of the four genes were expressed in any of the trophoblast lineages in the term trophoblast dataset, consistent with results from the term trophoblast cultures. ACTC1, VTCN1 and WFDC2 mRNA were all present in trophectoderm from preimplantation embryos, though VTCN1 and WFDC2 mRNAs were also found in morula. Later in the first trimester, ACTC1 and VTCN1 mRNA were detected in primary EVT and CTB, while WFDC2 was expressed exclusively in EVT. ACTC1 and WFDC2 were also expressed by TSCs. Strikingly, GABRP mRNA, which is very highly expressed in hESCBAP, was not detected in any of the other trophoblast datasets.
Table II.
Genes highly expressed in trophoblast derived from embryonic stem cells but not in cultured term trophoblast.
| Gene | hESC | hESCBAP <40 µm | hESCBAP >70 µm | primary human CTB | primary human STB |
|---|---|---|---|---|---|
| *GABRP | 1.3 | 1994.8 | 1074.2 | <0.1 | <0.1 |
| DIO3 | 5.8 | 248.5 | 1076.7 | 0.0 | 0.0 |
| NTRK2 | 0.1 | 142.0 | 99.1 | <0.1 | <0.1 |
| NDNF | 0.8 | 151.4 | 99.7 | <0.1 | 0 |
| CPE | 7.8 | 327.25 | 227.1 | .25 | 0.1 |
| *VTCN1 | 0.4 | 443.5 | 259.0 | 0.6 | 3.8 |
| MATN2 | 3.4 | 288.0 | 208.9 | 0.4 | 0.2 |
| *WFDC2 | 37.7 | 224.5 | 181.3 | 0.3 | 0.2 |
| IGFBPL1 | 21.3 | 123.1 | 111.5 | 0.25 | 0 |
| *ACTC1 | 56.2 | 388.2 | 273.9 | 0.8 | 0.2 |
| IGFBP5 | 1.4 | 275.1 | 195.6 | 0.7 | 0.2 |
| COL4A5 | 26.1 | 242.0 | 85.2 | 0.7 | 1.3 |
| NID2 | 8.2 | 237.6 | 136.3 | 0.7 | 0.6 |
| CTHRC1 | 2.7 | 286.5 | 180.7 | 0.8 | 0.6 |
| TNC | 9.0 | 299.5 | 180.3 | 0.9 | 1.2 |
| AADAT | 8.1 | 158.8 | 88.8 | 0.85 | 7.6 |
| PXDN | 57.8 | 100.2 | 99.6 | 0.6 | 0.3 |
| SESN3 | 28.3 | 134.1 | 77.1 | 0.9 | 1.2 |
| CGB8 | 0.2 | 102.1 | 1008.3 | 0.8 | 41.4 |
hESC, human embryonic stem cell; hESCBAP, hESC differentiated to TB with BAP.
Localization of proteins encoded by transcripts unique to hESCBAP within BAP-treated hESC colonies
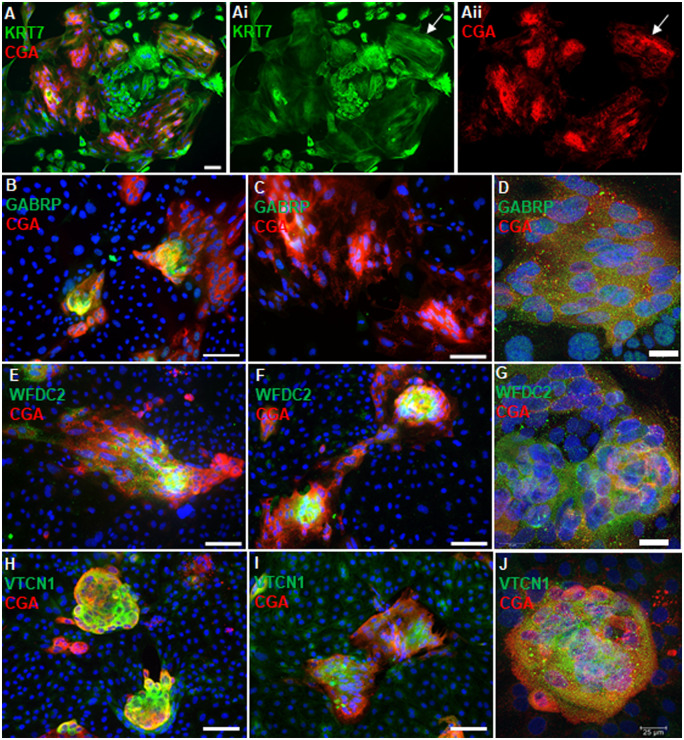
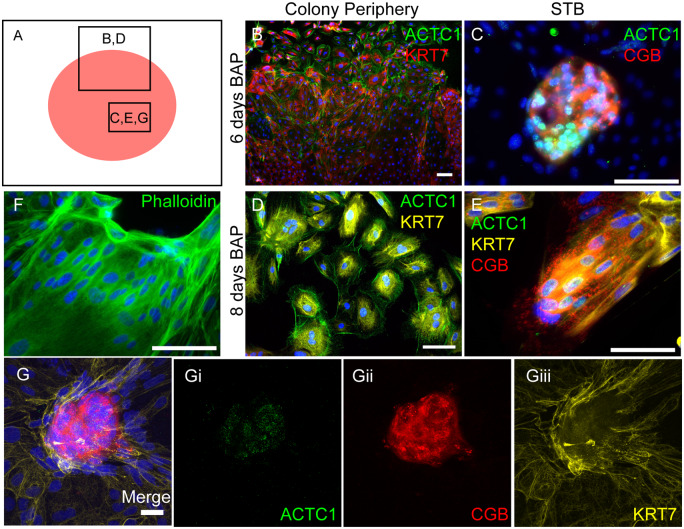
GABRP, WFDC2, VTCN1 and ACTC1 were localized by immunofluorescence within hESCBAP cultures to determine which trophoblast subtypes express these proteins. Following 6–8 days of BAP treatment, cytokeratin 7 (KRT7), a pan-trophoblast marker, was expressed throughout the cell colonies, indicating complete differentiation to trophoblast (Fig. 3A). Areas stained less intensely for KRT7 were cell clusters positive for CGA, a marker of emerging STB (Fig. 3Ai, ii, arrows). Staining for GABRP, WFDC2, and VTCN1 strongly overlapped with these CGA+ syncytialized areas (Fig. 3B–J). In contrast, ACTC1 staining was present in both CGB+ and CGB− cells (Fig. 4). ACTC1 presented in a filamentous fashion within CGB− cells near the periphery of the colony, consistent with a cytoskeletal localization (Fig. 4A, B, E). However, ACTC1 was most consistently found in cells expressing CGB (Fig. 4C, F, G). Within these CGB+ cells, ACTC1 immunoreactivity appeared to be in the nucleus where staining was punctate (Fig. 4C, F, G, Gi). Confocal imaging confirmed that ACTC1 localizes to the nucleus of these cells, whereas phalloidin staining, which marks filamentous actin, was not present within the nucleus (Fig. 4D, G), suggesting that the nuclear ACTC1 is in a globular form.
Figure 3.
Gamma-aminobutyric acid type A receptor subunit Pi (GABRP), WAP four-disulfide core domain 2 (WFDC2) and V-set domain containing T-cell activation inhibitor 1 (VTCN1) are expressed within specific cell types in BAP-treated hESCs (hESCBAP). (A) At 6–8 days of BMP4+A83-01+PD173074 (BAP) differentiation, all cells present as cytokeratin 7 (KRT7) positive with a mixture of single-nucleated CTB and multi-nucleated STB made apparent by a continuous cytoplasm and long stretches of KRT7 strands across multiple nuclei (arrows). (B–D) GABRP is expressed by the patches of chorionic gonadotropin-a positive (CGA+) STB in the cytoplasm and nucleus on days 6 (B, D), though decreasing at Day 8 of BAP treatment (C). (E–G) WFDC2 expression is within CGA+ STB areas at Days 6 (E, G) and 8 (F) of BAP treatment. (G) Confocal microscopy shows that WFDC2 is expressed in the cytoplasm. (H–J) VTCN1 expression also overlaps strongly with CGA+ STB regions at Days 6 (H, J) and 8 (I) of BAP treatment. (J) Confocal microscopy shows that VTCN1 is also contained within the cytoplasm. Panels A, B, C, E, F, H, I scale bar = 100 µm. Panels D, G, J scale = 25 µm. All blue fluorescence represents DAPI nuclear staining.
Figure 4.
Actin alpha cardiac muscle 1 (ACTC1) is expressed in 6- and 8-day treated hESCBAP. (A–E) ACTC1 is expressed in CGB+ STB in two distinct patterns. (A) The red circle represents a colony of hESCBAP and boxes indicate the approximate location of images shown in panels B–E, G. (B, D) Single cells found in the periphery of the colonies showed a filamentous, cytoskeletal pattern of expression. (C, E) Within CGB+ syncytialized cells, ACTC1 presented in a nuclear pattern of expression. (F) Phalloidin staining for filamentous actin. (Gi–iii) Maximum projections of z stacks created from confocal images on 6-day BAP treated ESCs. Punctate staining for ACTC1 is visible within the nucleus. (B, C, D, E, F Scale = 100 µm; G scale = 25µm). DAPI nuclear staining is shown in blue.
Undifferentiated hESC did not express either GABRP or WFDC2, and had only weak, scattered signal for ACTC1 (Supplementary Fig. S1A–C). There was an immunofluorescent signal for VTCN1 in undifferentiated hESC, but it was not detected in undifferentiated hESC by western blot, suggesting that the immunofluorescent signal in hESC is non-specific (Supplementary Fig. S1D and E). However, hESCBAP with siRNA knockdown of VTCN1 showed no signal, indicating that the VTCN1 immunofluorescence in hESCBAP is specific (Supplementary Fig. S1F).
Immunofluorescent localization of GABRP, WFDC2 and ACTC1 in human placental sections across gestation
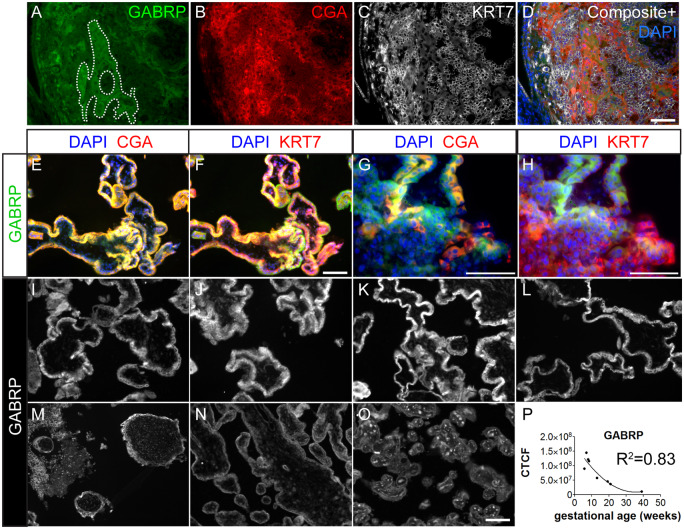
In primary sections at approximately 4 weeks LMP (16 days post-fertilization), GABRP is expressed most strongly by primitive syncytium (dotted outline, Fig. 5A) and STB in emerging secondary villi (SV). The areas strongest in CGA staining (Fig. 5B) show an extended expression pattern of KRT7, encircling multiple nuclei (Fig. 5C) and overlap with areas positive for GABRP (Fig. 5D), further confirming GABRP’s association with syncytium at this stage. GABRP localization in the first trimester villous placenta was similar to its immunolocalization in the hESCBAP (Figs 3B–D and 5E–H). Specifically, GABRP was expressed throughout the STB, and colocalized with CGA as shown by yellow fluorescent signal (Fig. 5E, G). GABRP was also localized in floating villi to CTB (Fig. 5F) and in anchoring villi to both CTB and EVT (Fig. 5G and H), which stain intensely for KRT7. EVT localization was confirmed by co-staining for HLAG (Supplementary Fig. S5).
Figure 5.
GABRP expression in human placenta samples over gestation. (A–D) Human blastocyst developmental age 16 days. (A) GABRP expression is most prominent in primitive syncytium highlighted in dashes. GABRP is also expressed by emerging STB from the secondary villus (SV). (B) CGA is strongly expressed in areas of primitive syncytium while KRT7 (B) is expressed at a low level in these areas. (E–H) Six weeks 6 days human placenta was immunostained for STB markers CGA (E, G) and CTB/EVT marker KRT7 (F, H) in floating (E, F) and anchoring (G, H) villi. G) Solid arrow = STB; arrowhead = CTB; line arrow = EVT. I–O are placental villi from the first trimester at gestational ages (I) 5 weeks 5 days, (J) 6 weeks 6 days, (K) 7 weeks 6 days, (L) 8 weeks 2 days, (M) 13 weeks, (N) 20 weeks 6 days, (O) Term. Signal detected within the mesenchymal villous core at term is erythrocyte autofluorescence. (P) Corrected total cell fluorescence (CTCF) quantification of positive CTB/STB regions. First trimester samples showed higher fluorescence intensity values than second trimester and term. Best fit line is y = x + x2. R2 = 0.83. Scale bars = 100 µm.
GABRP expression was highest in villous samples at Weeks 5–7 LMP, declining gradually over the course of gestation, and it was still detectable early in the second trimester, though it was absent at term (Fig. 5I–P). Quantification of the average fluorescent intensity across gestation fit closely to a quadratic curve (Fig. 5P).
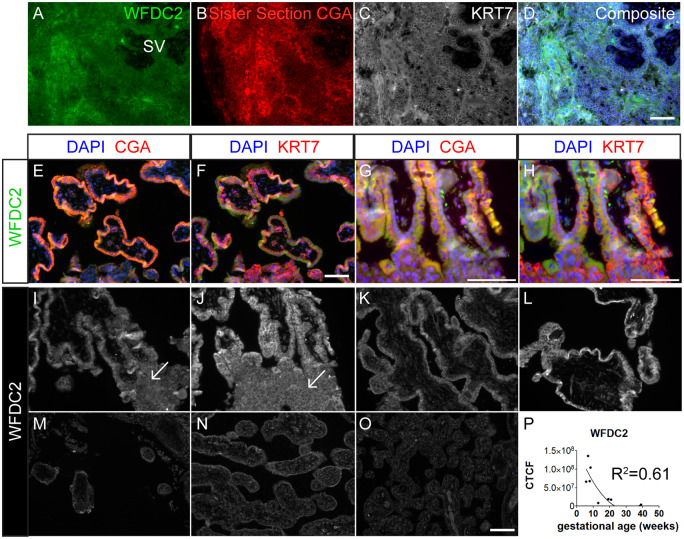
WFDC2 was ubiquitously expressed in the 4-week LMP placenta but was most strongly expressed in CGA+ areas, and these areas also showed a KRT7 localization pattern characteristic of syncytium (Fig. 6A–D). In a 6 week, 6 day sample, WFDC2 followed a similar pattern of expression to GABRP, as it was mostly seen within the STB in both floating and anchoring villi (Fig. 6E, G), just as it localized to CGA+ cells in the hESC-derived trophoblast cultures. In addition, WFDC2 appeared within EVT regions (Fig. 6G–J, arrows). This was confirmed by overlapping expression of HLAG and WFDC2 in the more distal regions of the AV, similar to what was observed with GABRP (Supplementary Fig. S6). WFDC2 expression was somewhat variable among the four first trimester samples, and then declined dramatically (Fig. 6I–P); it was essentially undetectable from 13 weeks onward (Fig. 6M–P).
Figure 6.
WFDC2 expression in human placenta samples over gestation. (A–D) Human blastocyst developmental age 16 days. WFDC2 most strongly associates with primitive syncytium high in CGA and low in KRT7 staining. (E and F) Six weeks 6 days human placenta was immunostained for STB marker CGA (E, G) and CTB/EVT marker KRT7 (F, H) in floating (E, F) and anchoring (G, H) villi. (I–O) are placental villi from the first trimester at gestational ages (I) 5 weeks 5 days, (J) 6 weeks 6 days, (K) 7 weeks 6 days, (L) 8 weeks 2 days, (M) 13 weeks, (N) 20 weeks 6 days and (O) Term. Line arrows indicate WFDC2+ anchoring villi. (P) CTCF quantification of positive CTB/STB regions. First trimester samples showed higher fluorescence intensity values than second trimester and term. Best fit line is y = x + x2. R2 = 0.61. Scale bars = 100 µm.
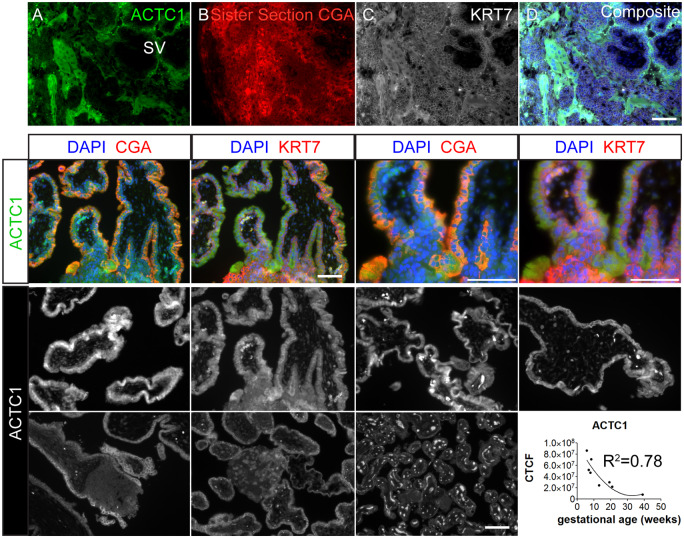
ACTC1 presented strongly in the syncytium and emerging secondary villous (SV) STB of the week four conceptus (Fig. 7A–D). Later in the first trimester, as with GABRP and WFDC2, ACTC1 showed strong staining in the STB layer, with weaker staining in the CTB and EVT (Fig. 7E–H). Although there was some overlap with HLAG, ACTC1 was largely absent from HLAG-positive areas (Supplementary Fig. S7). In some syncytial areas within the first trimester samples, ACTC1 stain was not limited to the cytoplasm alone and also appears to be present in nuclei (Supplementary Fig. S8), as was observed in the hESCBAP. In the second trimester, ACTC1 was present primarily within the cytoplasmic compartment of primary villous sections (Supplementary Fig. S8). As with GABRP and WFDC2, ACTC1 immunofluorescence was brightest before 9 weeks LMP, with declining intensity through the remainder of gestation, fitting a quadratic curve (Fig. 7I–P).
Figure 7.
ACTC1 expression in human placenta samples over gestation. (A–D) ACTC1 strongly associates with primitive syncytium and emerging STB on the secondary villus. (E–H) Six weeks 6 days human placenta was immunostained for STB markers CGA (E, G) and CTB/EVT marker KRT7 (F, H) at lower (E, F) and higher (G, H) magnifications. (I–O) are placental villi from the first trimester at gestational ages (I) 5 weeks 5 days, (J) 6 weeks 6 days, (K) 7 weeks 6 days, (L) 8 weeks 2 days, (M) 13 weeks, (N) 20 weeks 6 days and (O) term. Signals detected within the mesenchymal villous core in O, and intervillous space in K are erythrocyte autofluoresence. (P) CTCF quantification of positive CTB/STB regions. First trimester samples showed higher fluorescence intensity values than second trimester and term. Best fit line is y = x + x2. R2 = 0.61. (M–P) Scale bars = 100 µm.
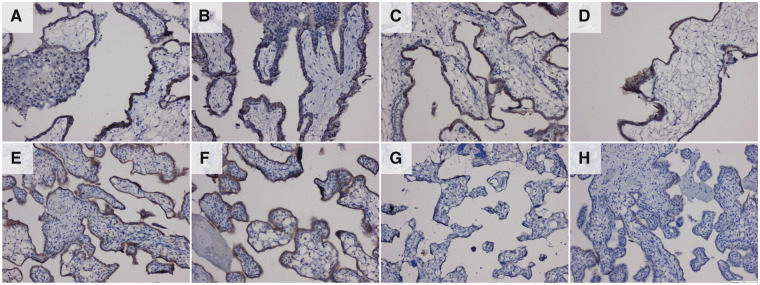
The VTCN1 antibody used for immunofluorescence detection in hESCBAP was incompatible with immunofluorescence in paraffin-embedded samples, and so immunoperoxidase detection was used to localize VTCN1 in the primary placental samples (Fig. 8). VTCN1 was present in both first and second trimester samples but was absent at term (Fig. 8A–G). As in the hESCBAP (Fig. 3H–J), staining was most intense in STB areas of the placental sections (Fig. 8A–F), although staining was also visible in CTB, and weakly in EVT in anchoring villi (Fig. 8B).
Figure 8.
Immunohistochemistry localization of VTCN1 protein (brown). VTCN1 is present in CTB/STB cells throughout the first and second trimesters of pregnancy but is absent at term. Samples include first trimester placenta (A–D): (A) 5weeks 5 days, (B) 6 weeks 6 days, (C) 7 weeks 6 days and (D) 8 weeks 2 days. Second trimester samples: (E) 19 weeks 1 day, (F) 20 weeks 6 days. Negative control using secondary antibody only (H) was performed on a 19 weeks, 1 day sample.
Discussion
The goal of this study was to test the suitability of the hESCBAP model for the discovery of genes and proteins uniquely expressed by trophoblasts in early pregnancy. We predicted that transcripts expressed by trophoblast derived from hESC—but not term trophoblast—would likely be markers of early first trimester trophoblast. PCA shows that the transcriptome of hESCBAP most closely resembles that of STB obtained from first trimester placentas or from TSCs. The four proteins examined here in detail, ACTC1, GABRP, VTCN1 and WFCD2, are all highly expressed in hESC-derived trophoblast and in placental samples from early in gestation. Expression of ACTC1, GABRP and WFDC2 decreased over the course of the first trimester to almost undetectable levels by the end of the second trimester. Although VTCN1 persists longer than the other three antigens, all four proteins are virtually absent from placental samples collected at term. These findings support the broader hypothesis that trophoblast arising from BAP-treated hESC represent an early stage of trophoblast differentiation in vivo. Furthermore, it shows the utility of this model for understanding the biology of the human placenta at this stage. ACTC1, VTCN1 and WFDC2, for example, were not previously known to be expressed in trophoblast at any stage of pregnancy.
Each of the four genes was expressed predominately in placental STB, perhaps reflecting the fact that hESCBAP most closely represent first trimester STB, based on the PCA of the hESCBAP transcriptome with transcriptomes from early, late, and in vitro-produced trophoblast cell types. Trophectoderm cells from blastocyst (Days 5–7), despite expressing typical trophoblast markers (C-C Motif Chemokine Receptor 7—CCR7, Cytochrome P450 Family 19 Subfamily A Member 1—CYP19A1, Distal-Less Homeobox 5—DLX5, Endogenous Retrovirus Group FRD Member 1, Envelope—ERVFRD-1, Glial Cells Missing Transcription Factor 1–GCM1, Gremlin 2—GREM2, Mucin 15—MUC15 and Ovo Like Transcriptional Repressor 1—OVOL1), cluster distinctly away from the later trophoblast cell types, and cluster more closely to undifferentiated hESC than to hESCBAP (Fig. 2A). When day 8 hESCBAP were compared to placental trophoblast from 6 to 9 weeks (Okae et al., 2018), 8 and 24 weeks (Liu et al., 2018), and term (Pavlicev et al., 2017), they most closely clustered with STB from 8 weeks than with any trophoblast type later in gestation.
Although the hESCBAP fraction >70 µM most closely clustered to STB, as expected, the hESCBAP <40 fraction was also similar to STB. This is consistent with our previous findings that, although markers characteristic of CTB and EVT are present on Days 2–6 of BAP treatment, by Day 8, when the RNAseq was performed, and certainly by day 10, most of the cells are STB (Amita et al., 2013; Yabe et al., 2016) and the PCA results suggest that even cells that have not yet fused are already differentiating toward STB. Thus, in order to determine whether CTB and EVT-like cells derived from hESC also closely resemble their first trimester counterparts, it would be necessary to either collect hESCBAP a few days earlier, or use a protocol to maximize EVT differentiation (Horii et al., 2019). Unfortunately, transcriptomes of these cells have only been assessed by microarray, and could not be accurately included in the present PCA (Telugu et al., 2013; Horii et al., 2019).
Seemingly inconsistent with an early stage identity of hESCBAP, was the relatively close clustering of hESCBAP with primary cultures of term human STB, whereas previously we had emphasized their distinctiveness (Yabe et al., 2016; Jain et al., 2017). This apparent closeness is probably because of the inclusion of other, more distantly related cell types, such as those from preimplantation blastocysts and whole villous tissues from term placentas in the newer PCA. Also surprising was the observation that transcriptomes of primary cultures of term human STB differed from those of STB directly isolated from the placenta. The most likely explanation is that culture alters STB gene expression. Human trophoblasts change their gene expression within 3 h of culture with the peak of differentially expressed genes occurring at 15 h post-seeding (Robinson et al., 2017). Furthermore, STB freshly generated by culturing term CTB likely represent an earlier developmental stage than villous STB that had existed in vivo on the villous surface for days or even weeks prior to tissue collection. Consistent with this hypothesis, the primary cultured term STB samples cluster quite closely to STB isolated from first trimester placentas and hESCBAP (Fig. 2B and C).
One concern with the PCA method generally is that transcriptomes vary in part based on laboratory conditions and sequencing methods used by different laboratories and in different experiments. Some of the cell types that were analyzed in the PCA, including the cultured term STB and the freshly isolated term STB, were only present in a dataset from one laboratory, making it impossible to fully separate cell type differences from technical variation. Had the technical variation strongly influenced the results, cells from each laboratory would have clustered together, independent of cell type or gestational age, but this was not the case. Instead, principal components PC1 and PC2 were associated with cell age, and PC3 with cell type. In Fig. 2B and C, STB from different laboratories and derived from different cell types all cluster with one another and each STB sample is separate from other cell types sequenced by the same labs.
The expression of the four genes of interest—GABRP, WFDC2, VTCN1 and ACTC1 among the various datasets demonstrates that all but GABRP are expressed in early, first trimester trophoblast and even blastocysts but are absent at term (Supplementary Fig. S4). A curious finding is that while the protein products of all four genes immunolocalize to first trimester STB, their mRNAs were low or absent in analyses of STB obtained from first trimester placentas. RNA isolation from STB is a challenge, especially for single cell RNA sequencing. As a large, multinucleated syncytium, STB cannot be separated by automated single-cell sorting procedures; even defining a single STB cell is a challenge. Thus, Okae et al. (2018) collected STB fragments shed from villi in culture, while Liu et al. (2018) used a mouth pipet to select STB fragments from villous digests. These methodological variations may contribute to some differences in gene expression due to selective transcript degradation. Overall, however, the expression of ACTC1, WFDC2 and VTCN1 across multiple datasets, despite being low, was consistent with our hypothesis that hESCBAP is a model of early, first trimester trophoblast. In contrast, the GABRP transcript appears to be present only in hESCBAP. This is puzzling given the robust expression of GABRP protein in the archived placental specimens and the previously published findings of GABRP in a transformed EVT cell line, HTR-8, as well as in mouse placentas (Luo et al., 2013; Lu et al., 2016). The relative absence of GABRP immunostaining in undifferentiated hESC colonies and second and third trimester placental specimens argues against non-specific antibody binding. It is possible that the protein is more stable than its RNA. Alternatively, hESCBAP culture conditions may enhance GABRP expression.
The human placenta changes dramatically over the course of gestation, particularly during the first trimester. Trophectoderm first emerges around Days 5–6 post-conception, often considered to be the first developmental differentiation event in embryos, although there is recent evidence that hypoblast arises at about the same time (Petropoulos et al., 2016b). When implantation begins around Day 7 (3 weeks LMP), polar trophectoderm adjoining the uterine epithelium forms a syncytium that pushes through the epithelial barrier and forms an invasive front as the conceptus embeds itself in the decidua and forms the first surface for exchange between mother and fetus (Hertig et al., 1956; Enders, 1976, 1989; Herzog, 1909). Between the second and third week after conception (4–5 weeks LMP), columns of CTB protrude through the primitive syncytium to begin formation of chorionic villi, so that over the next few weeks, the general morphology of the mature villous placenta is established (Boyd and Hamilton, 1970). Although term placental samples are widely available, first trimester samples are more difficult to obtain, while it is extremely rare to obtain samples before the end of the fourth week of gestation. This underscores the need for a cell type that recapitulates early pregnancy trophoblast development in vitro. To date, all characteristics of hESCBAP are consistent with early stage trophoblast.
There is a paradox, however, in that the very need for a model of early trophoblast development makes it difficult to compare hESCBAP to a placenta at 3–4 weeks gestation. We have examined primitive syncytium and emerging STB in just three sister sections of a single 16 day developmental age (4 weeks LMP) human blastocyst. As our data are derived from single samples at fixed stages of pregnancy, and because some technical and biological variation is anticipated, precise quantitative differences in expression levels with stage across the first trimester cannot be established for any of the four antigens we examined. For example, GABRP appears to persist longer than WFDC2 and ACTC1, but many more samples would be required to prove this contention. Information is less precise for the fourth antigen, VTCN1, where the immunoperoxidase staining protocol was not optimized to reveal quantitative differences occurring over time. Nonetheless, it is clear that GABRP, WFDC2 and ACTC1 levels are highest during 4–9 weeks gestation, and the best-fit models of fluorescent intensity for each indicates a quadratic decline, with highest expression in the samples recovered between 5 weeks and 8 weeks of pregnancy. While all three antigens were highly expressed in the primitive syncytium still visible in the 4-week sample, none proved to be uniquely expressed at this stage of gestation. If markers unique to the placenta at 3–5 weeks of pregnancy are to be found, additional candidate antigens must be sought.
Additional studies, including knockdown or overexpression models, are also needed to determine the function of ACTC1, GABRP, VTCN1 and WFCD2 in trophoblast cells from early in pregnancy. Each was predominately expressed in STB, both in the hESCBAP and in the human placental samples. This observation supports the idea that their expression and function in the hESCBAP model likely reflects their roles in trophoblast development and function in vivo.
GABRP is a subunit of the GABA class A of GABAergic receptors. These receptors are ligand-binding Cl- channels that act to reduce activity in neurons by hyperpolarization of the neuronal membrane (Hedblom and Kirkness, 1997). Its expression is associated with an aggressive, metastatic phenotype of certain cancers (Takehara et al., 2007; Sizemore et al., 2014; Sung et al., 2017), leading us to speculate that its role in trophoblast is linked to invasive potential. Alternatively, GABRP may alter steroid hormone signaling, as it has been shown to alter the interaction of GABA receptors with pregnenolone in other tissues (Hedblom and Kirkness, 1997).
Neither WFDC2 nor VTCN1 has previously been studied in trophoblast, but their functions in other tissues, as well as their localization to both STB and EVT, suggest potential functions in placental invasion and immunity. WFDC2 was originally identified in human epididymal cells and named human epididymis 4 (HE4) but was more recently found in several non-reproductive tissues that included trachea, nasal epithelium, salivary gland, kidney and lung (Bingle et al., 2002). It is secreted in the oral cavity and lungs, where it serves as part of the innate immune response. WFDC2 works in concert with the antiproteinases SLPI (Secretory Leukocyte Peptidase Inhibitor) and PI3 (Peptidase Inhibitor 3) to protect against proteolytic enzymes released by inflammatory cells (Bingle et al., 2006). WFDC2 also has been extensively studied in cancers of the ovary, uterus and colon (Chen et al., 2017; Kemal et al., 2017; Abbink et al., 2018) and it is a predictor of ovarian cancer severity (Goff et al., 2017; Scaletta et al., 2017). In ovarian cancer cells, it promotes expression of MMP2 and other genes required for metastasis and cell migration (Chen et al., 2017).
VTCN1 is expressed on either the cell surface as a transmembrane protein or in a soluble form that functions to suppress the immune system (Sica et al., 2003). In CD303+ dendritic cells, VTCN1 is highest during the first and second trimesters, falling at term, and rising again during and immediately following parturition (Darmochwal-Kolarz et al., 2013; Mach et al., 2015). Like GABRP and WFDC2, VTCN1 is expressed in several cancers including endometrial, breast, renal, melanoma, lung, gastric, colorectal, pancreatic and prostatic cancers, with heightened expression of VTCN1 predicting poor outcomes (Podojil and Miller, 2017).
ACTC1 is an alpha-actin protein found in cardiac muscle, developing skeletal muscle and vascular endothelium (Abdelwahid et al., 2004). Although ACTC1 is not normally expressed in adult skeletal muscle, it is induced in satellite cells and skeletal myocytes during regeneration (Franke et al., 1996; Dennis et al., 2008) and when satellite cells syncytialize with muscle fibers, raising the possibility that ACTC1 plays a role in trophoblast syncytialization. Syncytialization of BeWo choriocarcinoma cells and primary term trophoblast is dependent on actin remodeling and actin binding proteins, and is accompanied by an increase in the relative ratio of globular to filamentous actin. Curiously in syncytialized areas of hESCBAP, confocal images showed ACTC1 to be globular, nuclear and perinuclear. ACTC1 also appeared to be nuclear in some of the first and second trimester trophoblast cells. The role of nuclear actin in trophoblast is unknown, but it is found in a variety of cell types including cardiomyocytes (Asumda and Chase, 2012), keratinocytes (Sharili et al., 2016), oocytes (Misu et al., 2017) and mesenchymal stem cells (Asumda and Chase, 2012), in which it regulates transcription and chromatin remodeling (de Lanerolle, 2012; Sen et al., 2015; Misu et al., 2017).
In summary, our hypothesis that trophoblast derived from hESCs after BAP exposure, i.e. hESCBAP, is transcriptionally more similar to primary trophoblast from the early first trimester of pregnancy following implantation than trophoblast isolated at term appears to be correct. In particular, four proteins, ACTC1, GABRP, VTCN1 and WFCD2, identified through the high expression of their genes in hESCBAP and by an absence of such expression in cells from term placentae, are also associated with villous trophoblast development and particularly STB during the first trimester of human pregnancies. Expression of all four antigens has also been correlated with a metastatic phenotype for several common cancers. The finding that these pro-metastatic proteins are expressed by the STB associated with hESCBAP and early villous trophoblast suggests that early syncytium may be invasive. Their loss as pregnancy progresses may correlate with a loss of invasiveness. We conclude that the hESCBAP model has the potential to be used to investigate trophoblast characteristics unique to early pregnancy and particularly to implantation and the establishment of a functional placenta.
Supplementary Material
Acknowledgements
The University of Missouri Cytology Core provided much help for the visualization of the immunostaining. We particularly thank Dr Frank Baker and Dr Alexander Jurkevich. Diane McConnell from the University of Missouri OneHealth Biorepository aided with immunostaining. Paraffinized primary trophoblast sections were provided by the University of California-San Diego Perinatal Biorepository.
Authors’ roles
R.M.K. designed and carried out experiments, analyzed data, interpreted results and drafted the manuscript. S.M. designed and carried out the principal component analysis. J.Z. carried out experiments pertaining to VTCN1 antibody specification. T.E. participated in design of experiments, interpretation of results and manuscript revision. D.J.S. participated in design of experiments, interpretation of results and manuscript revision. R.M.R. participated in design of experiments, analysis of RNA sequencing data, interpretation of results and manuscript revision. L.C.S. participated in design of experiments, analysis of data, interpretation of results and manuscript drafting and revision. All authors have approved the final version of the manuscript.
Funding
This work was supported by the National Institutes of Health (HD R01067759, HD R01077108, HD R01HD094937) and Missouri Mission Enhancement.
Conflict of interest
The authors have no conflicts of interest to declare.
Data availability
Data underlying this article are available in the article and its online supplementary material. Transcriptome data are available at NCBI Gene Expression Omnibus (GEO), accession number GSE73017. Additional data were accessed from ArrayExpress: E-MTAB-3929, NCBI GEO GSE89497, NCBI GEO GSE87692 and the Japanese Genotype-phenotype Archive JGA00000000074, JGA00000000117 and JGA00000000122 as described in materials and methods.
References
- Abbink K, Zusterzeel PL, Geurts-Moespot AJ, Herwaarden AEV, Pijnenborg JM, Sweep FC, Massuger LF.. HE4 is superior to CA125 in the detection of recurrent disease in high-risk endometrial cancer patients. Tumour Biol 2018;40:1010428318757103. [DOI] [PubMed] [Google Scholar]
- Abdelwahid E, Pelliniemi LJ, Szucsik JC, Lessard JL, Jokinen E.. Cellular disorganization and extensive apoptosis in the developing heart of mice that lack cardiac muscle alpha-actin: apparent cause of perinatal death. Pediatr Res 2004;55:197–204. [DOI] [PubMed] [Google Scholar]
- Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM, Ezashi T.. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA 2013;110:E1212–E1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asumda FZ, Chase PB.. Nuclear cardiac troponin and tropomyosin are expressed early in cardiac differentiation of rat mesenchymal stem cells. Differentiation 2012;83:106–115. [DOI] [PubMed] [Google Scholar]
- Bingle L, Cross SS, High AS, Wallace WA, Rassl D, Yuan G, Hellstrom I, Campos MA, Bingle CD.. WFDC2 (HE4): a potential role in the innate immunity of the oral cavity and respiratory tract and the development of adenocarcinomas of the lung. Respir Res 2006;7:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle L, Singleton V, Bingle CD.. The putative ovarian tumour marker gene HE4 (WFDC2), is expressed in normal tissues and undergoes complex alternative splicing to yield multiple protein isoforms. Oncogene 2002;21:2768–2773. [DOI] [PubMed] [Google Scholar]
- Boyd J, Hamilton W.. The Human Placenta. Cambridge: W Heffer & Sons Ltd, 1970. [Google Scholar]
- Burton GJ, Fowden AL.. The placenta: a multifaceted, transient organ. Philos Trans R Soc Lond B Biol Sci 2015;370:20140066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton GJ, Jauniaux E, Watson AL.. Maternal arterial connections to the placental intervillous space during the first trimester of human pregnancy: the Boyd collection revisited. Am J Obstet Gynecol 1999;181:718–724. [DOI] [PubMed] [Google Scholar]
- Carlson M. org.Hs.eg.db: Genome wide annotation for Human. R package version 3.8, 2018.
- Chen Y, Huang L, Wang S, Liu T, Wu Y, Li JL, Li M.. WAP four-disulfide core domain protein 2 promotes metastasis of human ovarian cancer by regulation of metastasis-associated genes. J Ovarian Res 2017;10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmochwal-Kolarz D, Kludka-Sternik M, Kolarz B, Chmielewski T, Tabarkiewicz J, Rolinski J, Leszczynska-Gorzelak B, Oleszczuk J.. The expression of B7-H1 and B7-H4 co-stimulatory molecules on myeloid and plasmacytoid dendritic cells in pre-eclampsia and normal pregnancy. J Reprod Immunol 2013;99:33–38. [DOI] [PubMed] [Google Scholar]
- Das P, Ezashi T, Schulz LC, Westfall SD, Livingston KA, Roberts RM.. Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res 2007;1:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lanerolle P. Nuclear actin and myosins at a glance. J Cell Sci 2012;125:4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis RA, Przybyla B, Gurley C, Kortebein PM, Simpson P, Sullivan DH, Peterson CA.. Aging alters gene expression of growth and remodeling factors in human skeletal muscle both at rest and in response to acute resistance exercise. Physiol Genomics 2008;32:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders AC. Cytology of human early implantation. Res Reprod 1976;8:1–2. [PubMed] [Google Scholar]
- Enders AC. Trophoblast differentiation during the transition from trophoblastic plate to lacunar stage of implantation in the rhesus monkey and human. Am J Anat 1989;186:85–98. [DOI] [PubMed] [Google Scholar]
- Enders AC, Lantz KC, Peterson PE, Hendrickx AG.. From blastocyst to placenta: the morphology of implantation in the baboon. Hum Reprod Update 1997;3:561–573. [DOI] [PubMed] [Google Scholar]
- Fazleabas AT, Bell SC, Fleming S, Sun J, Lessey BA.. Distribution of integrins and the extracellular matrix proteins in the baboon endometrium during the menstrual cycle and early pregnancy. Biol Reprod 1997;56:348–356. [DOI] [PubMed] [Google Scholar]
- Ferretti C, Bruni L, Dangles-Marie V, Pecking AP, Bellet D.. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update 2007;13:121–141. [DOI] [PubMed] [Google Scholar]
- Fisher SJ. Why is placentation abnormal in preeclampsia? Am J Obstet Gynecol 2015;213:S115–S122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowden AL, Forhead AJ, Sferruzzi-Perri AN, Burton GJ, Vaughan OR.. Review: endocrine regulation of placental phenotype. Placenta 2015;36(Suppl 1):S50–S59. [DOI] [PubMed] [Google Scholar]
- Franke WW, Stehr S, Stumpp S, Kuhn C, Heid H,, Rackwitz HR, Schnolzer M, Baumann R, Holzhausen HJ, Moll R.. Specific immunohistochemical detection of cardiac/fetal alpha-actin in human cardiomyocytes and regenerating skeletal muscle cells. Differentiation 1996;60:245–250. [DOI] [PubMed] [Google Scholar]
- Goff BA, Agnew K, Neradilek MB, Gray HJ, Liao JB, Urban RR.. Combining a symptom index, CA125 and HE4 (triple screen) to detect ovarian cancer in women with a pelvic mass. Gynecol Oncol 2017;147:291–295. [DOI] [PubMed] [Google Scholar]
- Harris LK. Review: trophoblast-vascular cell interactions in early pregnancy: how to remodel a vessel. Placenta 2010;31(Suppl):S93–S98. [DOI] [PubMed] [Google Scholar]
- Hedblom E, Kirkness EF.. A novel class of GABAA receptor subunit in tissues of the reproductive system. J Biol Chem 1997;272:15346–15350. [DOI] [PubMed] [Google Scholar]
- Hertig AT, Rock J, Adams EC.. A description of 34 human ova within the first 17 days of development. Am J Anat 1956;98:435–493. [DOI] [PubMed] [Google Scholar]
- Herzog M. A contribution to our knowledge of the earliest known stages of placentation and embryonic development in man. Dev Dynam 1909;9:361–400. [Google Scholar]
- Home P, Kumar RP, Ganguly A, Saha B, Milano-Foster J, Bhattacharya B, Ray S, Gunewardena S, Paul A, Camper SA. et al. Genetic redundancy of GATA factors in the extraembryonic trophoblast lineage ensures the progression of preimplantation and postimplantation mammalian development. Development 2017;144:876–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii M, Bui T, Touma O, Cho HY, Parast MM.. An improved two-step protocol for trophoblast differentiation of human pluripotent stem cells. Curr Protoc Stem Cell Biol 2019;50:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii M, Li Y, Wakeland AK, Pizzo DP, Nelson KK, Sabatini K, Laurent LC, Liu Y, Parast MM.. Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc Natl Acad Sci USA 2016;113:E3882–E3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Ezashi T, Roberts RM, Tuteja G.. Deciphering transcriptional regulation in human embryonic stem cells specified towards a trophoblast fate. Sci Rep 2017;7:17257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A, Cox A, Wicher KB, Glover DM, Zernicka-Goetz M.. Maternal-zygotic knockout reveals a critical role of Cdx2 in the morula to blastocyst transition. Dev Biol 2015;398:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal YN, Demirag GN, Bedir AM, Tomak L, Derebey M, Erdem DL, Gor U, Yucel I.. Serum human epididymis protein 4 levels in colorectal cancer patients. Mol Clin Oncol 2017;7:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman HJ, Nestler JE, Sermasi E, Sanger JM, Strauss JF 3rd. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology 1986;118:1567–1582. [DOI] [PubMed] [Google Scholar]
- Krendl C, Shaposhnikov D, Rishko V, Ori C, Ziegenhain C, Sass S, Simon L, Muller NS, Straub T, Brooks KE. et al. GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proc Natl Acad Sci USA 2017;114:E9579–E9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fan X, Wang R, Lu X, Dang YL, Wang H, Lin HY, Zhu C, Ge H, Cross JC. et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res 2018;28:819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhang Q, Tan D, Luo W, Zhao H, Ma J, Liang H, Tan Y.. GABA A receptor pi subunit promotes apoptosis of HTR-8/SVneo trophoblastic cells: Implications in preeclampsia. Int J Mol Med 2016;38:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Liu Z, Tan D, Zhang Q, Peng H, Wang Y, Tan Y.. Gamma-amino butyric acid and the A-type receptor suppress decidualization of mouse uterine stromal cells by down-regulating cyclin D3. Mol Reprod Dev 2013;80:59–69. [DOI] [PubMed] [Google Scholar]
- Mach P, Gellhaus A, Wicherek L, Schmidt B, Kimmig R, Kasimir-Bauer S, Koninger A.. Changes in the blood serum levels of the costimulatory soluble B7-H4 molecule in pregnant women during the peripartal phase. Am J Reprod Immunol 2015;74:209–215. [DOI] [PubMed] [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A.. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 2014;13:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen TM, Holme AM, Henriksen T.. Transplacental nutrient transfer in the human in vivo determined by 4 vessel sampling. Placenta 2017;59(Suppl 1):S26–S31. [DOI] [PubMed] [Google Scholar]
- Misu S, Takebayashi M, Miyamoto K.. Nuclear actin in development and transcriptional reprogramming. Front Genet 2017;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardozza LM, Caetano AC, Zamarian AC, Mazzola JB, Silva CP, Marcal VM, Lobo TF, Peixoto AB, Araujo Junior E.. Fetal growth restriction: current knowledge. Arch Gynecol Obstet 2017;295:1061–1077. [DOI] [PubMed] [Google Scholar]
- Nelson DM, Johnson RD, Smith SD, Anteby EY, Sadovsky Y.. Hypoxia limits differentiation and up-regulates expression and activity of prostaglandin H synthase 2 in cultured trophoblast from term human placenta. Am J Obstet Gynecol 1999;180:896–902. [DOI] [PubMed] [Google Scholar]
- Norton AJ, Jordan S, Yeomans P.. Brief, high-temperature heat denaturation (pressure cooking): a simple and effective method of antigen retrieval for routinely processed tissues. J Pathol 1994;173:371–379. [DOI] [PubMed] [Google Scholar]
- Norwitz ER, Schust DJ, Fisher SJ.. Implantation and the survival of early pregnancy. N Engl J Med 2001;345:1400–1408. [DOI] [PubMed] [Google Scholar]
- Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T.. Derivation of human trophoblast stem cells. Cell Stem Cell 2018;22:50–63.e56. [DOI] [PubMed] [Google Scholar]
- Pavlicev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, Jones H.. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res 2017;27:349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S, Edsgard D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F.. Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 2016. a;165:1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S, Edsgard D, Reinius B, Deng Q, Panula SP, Codeluppi S, Reyes AP, Linnarsson S, Sandberg R, Lanner F.. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 2016. b;167:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podojil JR, Miller SD.. Potential targeting of B7-H4 for the treatment of cancer. Immunol Rev 2017;276:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RM, Ezashi T, Sheridan MA, Yang Y.. Specification of trophoblast from embryonic stem cells exposed to BMP4. Biol Reprod 2018;99:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RM, Ezashi T, Sheridan M, Yang Y.. Specification of trophoblast from embryonic stem cells exposed to BMP4. Biol Reprod 2018;doi:10.1093/biolre/ioy070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RM, Loh KM, Amita M, Bernardo AS, Adachi K, Alexenko AP, Schust DJ, Schulz LC, Telugu BP, Ezashi T. et al. Differentiation of trophoblast cells from human embryonic stem cells: to be or not to be? Reproduction 2014;147:D1–D12. [DOI] [PubMed] [Google Scholar]
- Robinson JF, Kapidzic M, Gormley M, Ona K, Dent T, Seifikar H, Hamilton EG, Fisher SJ.. Transcriptional dynamics of cultured human villous cytotrophoblasts. Endocrinology 2017;158:1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaletta G, Plotti F, Luvero D, Capriglione S, Montera R, Miranda A, Lopez S, Terranova C, De Cicco Nardone C, Angioli R.. The role of novel biomarker HE4 in the diagnosis, prognosis and follow-up of ovarian cancer: a systematic review. Expert Rev Anticancer Ther 2017;17:827–839. [DOI] [PubMed] [Google Scholar]
- Schulz LC, Ezashi T, Das P, Westfall SD, Livingston KA, Roberts RM.. Human embryonic stem cells as models for trophoblast differentiation. Placenta 2008;29(Suppl A):S10–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen B, Xie Z, Uzer G, Thompson WR, Styner M, Wu X, Rubin J.. Intranuclear actin regulates osteogenesis. Stem Cells 2015;33:3065–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharili AS, Kenny FN, Vartiainen MK, Connelly JT.. Nuclear actin modulates cell motility via transcriptional regulation of adhesive and cytoskeletal genes. Sci Rep 2016;6:33893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica GL, Choi IH, Zhu G, Tamada K, Wang SD, Tamura H, Chapoval AI, Flies DB, Bajorath J, Chen L.. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity 2003;18:849–861. [DOI] [PubMed] [Google Scholar]
- Sizemore GM, Sizemore ST, Seachrist DD, Keri RA.. GABA(A) receptor pi (GABRP) stimulates basal-like breast cancer cell migration through activation of extracellular-regulated kinase 1/2 (ERK1/2). J Biol Chem 2014;289:24102–24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncin F, Khater M, To C, Pizzo D, Farah O, Wakeland A, Arul Nambi Rajan K, Nelson KK, Chang CW, Moretto-Zita M. et al. Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development 2018;145:dev156273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HY, Yang SD, Ju W, Ahn JH.. Aberrant epigenetic regulation of GABRP associates with aggressive phenotype of ovarian cancer. Exp Mol Med 2017;49:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehara A, Hosokawa M, Eguchi H, Ohigashi H, Ishikawa O, Nakamura Y, Nakagawa H.. Gamma-aminobutyric acid (GABA) stimulates pancreatic cancer growth through overexpressing GABAA receptor pi subunit. Cancer Res 2007;67:9704–9712. [DOI] [PubMed] [Google Scholar]
- Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2018. [Google Scholar]
- Telugu BP, Adachi K, Schlitt JM, Ezashi T, Schust DJ, Roberts RM, Schulz LC.. Comparison of extravillous trophoblast cells derived from human embryonic stem cells and from first trimester human placentas. Placenta 2013;34:536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ,, Marshall VS, Jones JM.. Embryonic stem cell lines derived from human blastocysts. Science 1998;282:1145–1147. [DOI] [PubMed] [Google Scholar]
- Villar J, Papageorghiou AT, Knight HE, Gravett MG, Iams J, Waller SA, Kramer M, Culhane JF, Barros FC, Conde-Agudelo A. et al. The preterm birth syndrome: a prototype phenotypic classification. Am J Obstet Gynecol 2012;206:119–123. [DOI] [PubMed] [Google Scholar]
- Yabe S, Alexenko AP, Amita M, Yang Y, Schust DJ, Sadovsky Y, Ezashi T, Roberts RM.. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci USA 2016;113:E2598–E2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yuge A, Rajah AM, Unek G, Rinaudo PF, Maltepe E.. LIMK1 regulates human trophoblast invasion/differentiation and is down-regulated in preeclampsia. Am J Pathol 2014;184:3321–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data underlying this article are available in the article and its online supplementary material. Transcriptome data are available at NCBI Gene Expression Omnibus (GEO), accession number GSE73017. Additional data were accessed from ArrayExpress: E-MTAB-3929, NCBI GEO GSE89497, NCBI GEO GSE87692 and the Japanese Genotype-phenotype Archive JGA00000000074, JGA00000000117 and JGA00000000122 as described in materials and methods.