Abstract
Background
In our previous study, serine protease inhibitor Kazal-type 5 (SPINK5), which encodes the product of serine protease inhibitor lymphoepithelial Kazal-type-related inhibitor (LEKTI) was found to be down-regulated in head and neck squamous cell carcinoma (HNSCC) using oligonucleotide microarrays. However, the function and clinical implications of SPINK5/LEKTI remain obscure in HNSCC.
Methods
The endogenous expression level of SPINK5/LEKTI was further verified in 9 HNSCC cell lines and HNSCCs by means of reverse transcription-polymerase chain reaction, real-time PCR, Western blotting and immunohistochemistry. The biological function of SPINK5/LEKTI was investigated in vitro and in vivo experiments. Kaplan–Meier survival analysis and Cox proportional hazards regression model were used to determine the correlation between SPINK5/LEKTI expression and clinical outcome.
Results
Down-regulation expression of SPINK5/LEKTI was found in six out of nine HNSCC cell lines and in 85.7% HNSCC specimens (P<0.0001). Upon silencing of SPINK5/LEKTI, the cell proliferation, plate colony formation and cell invasion of WU-HN6 cells were significantly increased, while exogenous overexpression of SPINK5/LEKTI, the proliferation, plate colony and invasion of WU-HN13 and HN30 cells were remarkably inhibited with the arrest of G1 cell cycle (P=0.0001, P=0.003, respectively). HNSCC patients with lower LEKTI levels had significantly inferior overall survival compared to those patients with higher LEKTI (P=0.0017) by Kaplan–Meier survival analysis. Univariate and multivariate Cox proportional hazards regression model analysis revealed that LEKTI expression was an independent prognostic predictor for HNSCC patients (HR=0.114, 95% CI:0.044–0.292, P<0.001).
Conclusion
Our results demonstrate that SPINK5/LEKTI might be a tumor suppressor in HNSCCs and serve as an independent prognostic predictor for HNSCC patients.
Keywords: SPINK5/LEKTI, biological function, head and neck squamous cell carcinoma, prognostic predictor
Introduction
Squamous cell carcinoma is a common type of malignancies (account for over 80%) in the head and neck. According to the report,1 the incidence of HNSCC accounts for 8% of the whole malignant tumors. In recent years, the incidence of HNSCC keeps increasing year by year and more than 600,000 new cases occur in the world each year.2,3 Due to the corresponding treatment including surgery, radiotherapy and chemotherapy, HNSCC often causes serious impairment of the appearance and vital function of patients with a five-year overall survival of only about 60%.4 In previous study, we found that genes dysregulation plays an irreplaceable role in the initiation and the progression of HNSCC. These genes not only are closely related to the prognosis of patients but also are sensitive to the radiotherapy and chemotherapy.5 Therefore, these genes can be used as the biomarkers in clinic to predict the prognosis of the patients. More excitingly, they also act as the important targets for gene targeting therapy to improve the recovery rate. In our previous study, serine protease inhibitor Kazal-type 5 (SPINK5) was found down-regulation with 9.7-fold in 22 HNSCC tissues compared to the paired adjacent normal tissues using oligonucleotide microarrays analysis-the efficient way to select the different express genes.6 However, the functional role of SPINK5 in the carcinogenesis of HNSCC and its clinical significance were rarely reported.
SPINK5 gene which contained 33 exons is located on chromosome 5q32.7 LEKTI, the protein product of SPINK5, is a representative serine protease inhibitor which contains a 15-domain Kazal-type-related serine protease inhibitor.8 It expressed in the most differentiated layers of stratified epithelial tissues including the skin.9,10 In the epidermis, LEKTI protein is confined to the granular layer and stratum corneum where it co-localized with serine proteases of the kallikrein family such as the stratum corneum chymotryptic enzyme (SCCE) and the stratum corneum tryptic enzyme (SCTE) that are encoded by the kallikrein 5 and kallikrein 7 (KLK 5, 7) respectively.11–13
Several domains of LEKTI have shown to inhibit KLK5 and KLK7 that are involved in the degradation of corneodesmosomes such as desmoglein 1 (Dsg1), desmocollin 1 (Dsc 1) and corneodesmosin.14 Normally, the balance between the inhibitory pressure of LEKTI and the proteolytic activity of KLK5 and KLK7 is well kept leading to an intact epithelial barrier.15 Mutations of SPINK5 result in reduction or loss of LEKTI, which subsequently a reduced inhibition of serine proteases and finally lead to premature degradation of corneodesmosomal cadherins, over-desquamation and stratum corneum thinning caused by the increased activity of KLK5, KLK7 and reduction of Dsg1, Dsc1.16,17
Based on the biological properties and the localization of SPINK5, many studies have focused on searching for associations between the genetic variants of SPINK5 and skin barrier defeats disease.18 For example, mutation of the SPINK5 gene has been identified as disease-causing in Nethetonsydrome (NS) - a rare autosomal-recessive skin disorder clinically characterized by congenital erythroderma, trichorrhexis invatinata and atopic diathesis.19,20 Liu found that the G allele at SNP −206G>A in the SPINK5 was associated with asthma susceptibility in a case-control Chinese Han population study including 669 asthma patients and 711 healthy controls.21 Another study found that the lower SPINK5 expression might be a contributing factor leading to chronic rhinosinusitis in patients who suffered from aspirin tolerance and allergies.22 Recently, it has been found that LEKTI was able to inhibit the cysteine protease caspase 14 which implicated in both epidermal differentiation and hydration in vitro.23 These indicated that LEKTI was a multifunction protease inhibitor and played critical role in the epidermal barrier function by regulating protease activity.
There are still some unconformities in diverse cancer types, suggesting the complex function of SPINK5/LEKTI. For example, the expression level of SPINK5 was reported to be obviously up-regulated in bladder cancer tissues compared to the normal bladder tissues.24 However, the expression level of SPINK5 was detected to be remarkably down-regulated in esophageal cancer and the decreased expression of SPINK5 was significantly associated with the poor survival of patients with OSCC.25,26 In our previous study, down-regulation of SPINK5 has been found in HNSCCs as compared to the adjacent normal tissues by using oligonucleotide microarrays.6 In this study, we further validated the expression level of SPINK5/LEKTI expression in HNSCC cell lines and HNSCCs and investigated its biological roles and clinical implications in HNSCC.
Materials and Methods
Cell Culture
HNSCC cell lines include WSU-HN4, HN6, HN12, HN13, HN30 (provided by the University of Maryland Dental School, USA and approved by the Institutional Ethics Committee of the Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine) and CAL-27 (American Type Culture Collection, ATCC, USA) were cultured in DMEM (GIBCO-BRL, USA) added with 10% fetal bovine serum (FBS) (GIBCO-BRL, USA). SCC-4, SCC-9, SCC-25 cell lines (American Type Culture Collection, ATCC, USA) were cultured in DMEM/F12 medium (GIBCO-BRL) supplemented with 10% FBS. Normal primary gingival epithelial cells taken from the patient with impacted tooth served as normal control and were cultured in keratinocyte serum-free medium (GIBCO-BRL) with 0.2% ng/mL recombinant epidermal growth factor (rEGF) (Invitrogen, USA). All cells were cultured at 37°C in a humidified 5% CO2 atmosphere.27
Patients and Specimens
Samples were obtained from patients who were first diagnosed with primary HNSCC and treated with surgical treatment in Department of Oral and Maxillofacial-Head and Neck in Ninth People’s Hospital between June 2008 and December 2010. In this study, 35 paired clinical samples including cancerous tissues and their apparently normal surgical margins and a cohort of samples including 142 cancerous tissues and 92 apparently normal surgical margins were used. The required samples were quickly frozen in liquid nitrogen then stored at −80°C. Before using these samples must be accomplished pathological diagnosis according to the design and principle of the study. The tissue microarray used in this study consisted of paraffin-embedded tissues from 101 patients who were pathological diagnosed with primary HNSCC and accomplished surgical treatment between 1989 and 1993. The exclusion criteria of this study included preoperative radiotherapy, preoperative chemotherapy, distant metastasis, recurrent tumors and incomplete medical materials. In addition to two patients had no follow-up data, all patients had perfect information. The summarized materials of patients are shown in Table 1. In this study, samples were stained with hematoxylin and eosin to determine the pathological differentiation, and clinical stages were made according to the TNM classification system of the International Union against Cancer (1988) prior to further analysis. All of the patients signed written informed consent in accordance with the institutional guidelines and the study was approved by the Institutional Ethics Committee of the Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine.
Table 1.
Demographic Characteristics of the Patient Population by LEKTI Levels
| Characteristic | N (%) | LEKTI Expression (M±SD) | P |
|---|---|---|---|
| Age | |||
| <60y | 53 (60.2%) | 2.233±0.604 | 0.7665a |
| ≥60y | 35 (39.8%) | 2.276±0.732 | |
| Gender | |||
| Male | 48 (54.5%) | 2.197±0.740 | 0.4770a |
| Female | 40 (45.5%) | 2.297±0.536 | |
| Smoking history | |||
| No | 59 (67%) | 2.281±0.581 | 0.5247a |
| Yes | 29 (33%) | 2.186±0.790 | |
| Alcohol history | |||
| No | 68 (77.3%) | 2.293±0.609 | 0.2622a |
| Yes | 20 (22.7%) | 2.105±0.788 | |
| TNM stage | |||
| I/II | 43 (48.9%) | 2.459±0.559 | 0.0029a |
| III/IV | 45 (51.1%) | 2.05±0.681 | |
| Disease site | |||
| Tongue | 68 (77.3%) | 2.294±0.585 | 0.0733a |
| Others (cheek, gingiva) | 20 (22.7%) | 2.10±0.850 | |
| Lymph node metastasis | |||
| pN0 | 64 (72.7%) | 2.309±0.643 | 0.1717a |
| pN1-pN2 | 24 (27.3%) | 2.094±0.672 | |
| Pathological differentiation | |||
| I | 49 (55.7%) | 2.345±0.483 | 0.0408b |
| II | 29 (32.9%) | 2.29±0.610 | |
| III | 10 (11.4%) | 1.76±0.533 |
Note: aThe P value was analyzed by two-tailed Student’s t-test. bThe P value was analyzed by one-way ANOVA analysis.
Immunohistochemical Analysis
Formalin-fixed, paraffin-embedded tissues were cut into 4 μm tissue sections. Tissue sections were deparaffinized in xylene, hydrated in graded ethanol, treated with heated citrate buffer for epitope retrieval then cooled to room temperature naturally. Tissue sections were blocked with hydrogen peroxide, incubated with anti-LEKTI antibody (1:150) (Bioworld, USA) overnight at 4°C followed by biotinylated secondary antibody then diaminobenzidine reagent (Dako, Denmark). Eleven patients were excluded due to the tissues shedding in the process of immunohistochemistry. The LEKTI labeling index was defined as the intensity of staining (0, 1, 2 and 3) multiplied by the percentage of positive epithelial thickness (25%, 50%, 75% and 100%). In this study, the scores including LEKTI staining index and positive epithelial cell index were analyzed by two experienced pathologists. The median score of 2.4 was set as the cut-off value to determine high or low LEKTI expression. A value <2.4 was considered low expression and a value ≥2.4 was considered high expression.28
RNA Extraction and Reverse Transcriptase-Polymerase Chain Reaction Analysis
Total RNA was extracted from tissue by using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instruction. Approximately 1 µg of total RNA derived from each sample was converted to cDNA in a total volume of 20 μL using SuperScript reverse transcriptase reagent kit (Takara, Japan). Conventional reverse transcriptase-polymerase chain reaction was performed using E×Tap DNA polymerase (Takara, Japan) in a total volume of 20 μL according to the manufacturer’s instruction. The primer sequences were as follow: for SPINK5, the forward primer was 5ʹ-TGCTTTACGTGGCAAGGAG-3ʹ, the reverse primer was 5ʹ- GTAAGAACCTTTGTGATTACATGGG-3ʹ; for β-actin, the forward primer was 5ʹ-CCTGGCACCCAGCACAAT-3ʹ, the reverse primer was 5ʹ-GGGCCGGACTCGTCATACT-3ʹ. An initial denaturation step was performed for 5 min at 95°C, total 35 cycles were performed with the following program that denatured at 95°C for 20 s, annealed at 60°C for 30 s and elongated at 72°C for 30 s. Finally, the program was finished with a final extension step at 72°C for 5 min and kept at 4°C.29
Real-Time Polymerase Chain Reaction Analysis
In this study, all real-time PCR reactions were performed using the SYBR Premix E×Tap DNA polymerase kit (Takara, Japan) and an ABI 7300 real-time PCR system (ABI, USA). The primer sequences for SPINK5 and β-actin were the same as described above. Real-time PCR amplification was performed according to the kit instruction in a volume of 20 μL with 2 μL of cDNA, 10 μL SYBR Premix E×Tap, 0.4 μL ROX Reference Dye, 0.8 μL of primer for the target gene, 6.8 μL distilled water and β-actin as internal control. The procedure was completed at 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 60°C for 31 s. Ct values that represent the cycle number at which the fluorescent signal of the samples passes a given threshold above the baseline indicate the measured gene content in each sample. ∆Ct was the difference between the Ct value of the target gene and the Ct value of the β-actin of the same sample. Then, the relative expression level of SPINK5 in a tumor tissue compared to the adjacent normal tissue was calculated by 2 −∆∆Ct, where ∆∆Ct=∆Ct of HNSCC samples - ∆Ct of adjacent normal sample. Therefore, 2 −∆∆Ct indicate the fold change in the HNSCC samples relative to the adjacent normal samples.30
Western Blot Analysis
Cells were harvested in sodium dodecyl sulfate (SDS) lysis buffer (Beyotime, China) which added with complete protease inhibitors (Roche, USA). Total protein was separated in sodium dodecyl sulfate-polyacrylamide gel electrophoresis then transferred the separated protein to a polyvinylidene fluoride (PVDF) membrane. Then, the membranes were blocked with 1% nonfat milk for 1 h at room temperature, incubated with primary antibodies overnight at 4°C, followed by secondary antibody labeled with IRDyeTM 800 and signals were observed to detect specific immunoreactivity using an Odyssey Infrared Imaging System (Rockland, USA).31 The antibodies against LEKTI (1:1000, Epitomics, USA), Cyclin D1 (CCND1) (1:5000, Epitomics, USA), CDK4, p21, MMP2 and MMP9 (1:1000, Proteintech, USA) were used in this study. The mouse monoclonal β-actin (1:5000, Sigma-Aldrich, USA) served as an internal control.
Small Interfering RNA (siRNA) Transfection
Chemically synthesized siRNA special target to SPINK5 was purchased from Shanghai Genepharma Co., Ltd. In this study, the siRNA was named si-1771 and its sequences were 5ʹ-GCAGUGAAUAUCGUCAUUATTUAAUGACGAUAUUCACUGCTT-3ʹ. The FAM-labeled Scrambled siRNA was also provided from Shanghai Genepharma Co., Ltd. Lipofectamine 2000 (Invitrogen, USA) was used to transient transfection and accomplished according to the manufacturer’s protocol.32
SPINK5 Overexpression Vector Transfection
Human SPINK5 cDNA was cloned into the eukaryotic expression vector pcDNA3.0 to construct the pcDNA3.0-SPINK5 expression vector (Thermo). PCR was used to amplify SPINK5 gene from the plasmid pcDNA-SPINK5 and then cloned into the plasmid pLenti-CMV-EGFP-3FLAG-PGK-Puro to construct the lentiviral expression plasmid pLenti-CMV-SPINK5-3FLAG-PGK-Puro. In this study, the forward primer used for the amplification of SPINK5 was 5ʹ-CGAGCTCAAGCTTCGAATTCGCCACCATGAAGATAGCCACAGTGTCAG-3ʹ, containing an EcoR I enzyme site; the reverse primer was 5ʹ-TCATCCTTGTAGTCGGATCCTTCGTCAGACGGGGGCAT-3ʹ, containing a BamH I enzyme site. The plasmid pLenti-CMV-SPINK5-3FLAG-PGK-Puro, the packaging plasmid Δ8.91 and the envelope plasmid pVSVG was co-transfected into 293T cells to generate the recombinant lentivirus of pLenti-CMV-SPINK5. The recombinant lentivirus pLenti-CMV-SPINK5 (SPINK5 over-expression) was used to transfect HNSCC cell lines. PLenti-CMV- EGFP was used as control.
Cell Proliferation Assay
Cell proliferation assays were used to determine the SPINK5 gene on cell growth. Cell proliferation assays were performed to analyze the proliferation ability of WSU-HN6 cells transiently transfected with si-1771 or Scrambled siRNA and cells of WSU-HN13 and HN30 transfected with the lentivirus pLenti-CMV-SPINK5 or the lentivirus pLenti-CMV-EGFP. The cells were cultured in a 96-well plate at a density of 1×103 cells/well in triplicate, then cell viability was assayed using Cell-Counting Kit-8 (CCK-8) (Dojindo, Japan) at 24-h interval. 10 μL of CCK-8 reagent was added to each well then incubated at 37°C for 2 h. The viable cells in each well were measured at 450 nm by reading the optical density of absorbance.33
Plate Colony Formation Assay
Twenty-four hours after WSU-HN6 cells transiently transfected with si-1771 or Scrambled siRNA and 2 weeks after WSU-HN13, HN30 cells transfected with the lentivirus pLenti-CMV-SPINK5 or the lentivirus pLenti-CMV-EGFP, ihe cells were cultured in 10 cm dish at a density of 1×103 cells/well for 2 weeks. When the cell number of colonies more than 50 cells under a microscope, the colonies were fixed with formalin for 30 min, stained with crystal violet solution for 2 h and washed with double-distilled water. The number was counted according to the size and density of the colonies. All experiments were independently repeated 3 times.34
Cell Cycle Analysis
WSU-HN6 cells were harvested after transfection with siRNA for 72 h; WSU-HN13 and HN30 cells transfected with the lentivirus pLenti-CMV-SPINK5 or pLenti-CMV-EGFP were harvested after transfection for 2 weeks. The cells were washed with phosphate-buffer saline (PBS) and fixed in 70% cold ethanol overnight at 4°C. Then, the fixed cells were washed with cold PBS and stained with PI/RNase Staining Buffer (BD Pharmingen) on ice in the dark for 30 min. Cell cycle analysis was performed using flow cytometry (FCM).35
Cell Invasion Array in vitro
24-well Millicell Hanging Cell Culture Inserts (Millipore Corporation, USA) and Matrigel (BD Biosciences) were used. Matrigel and serum-free DMEM mixed in a ratio of 1:8 then paved 35 μL the mixture into Millicell Hanging Cell Culture Inserts. After rehydration of the chambers, 1×105 transfected cells in 150 μL DMEM added with 1% fetal bovine serum were cultured in the upper chambers and 500 μL DMEM supplemented with 10% fetal bovine serum was placed in the lower chambers. The cells were fixed in 4% formaldehyde after cultured of 24 h and stained with 0.5% crystal violet. Cells in the upper chamber were removed using cotton swab.36 Invading cells lie on the lower surface of the membrane were photographed and counted at least 5 random microscopic fields (100×). All experiments were repeated 3 times.
Tumorigenesis in Xenografted Mice
To further determine SPINK5 gene effects on cancerous cell in vivo, xenograft model in BALB/C nude mice (4 weeks old, female) was generated by injecting 1.5˟106 HN13 cells transfected with the lentivirus pLenti-CMV-SPINK5 or pLenti-CMV-EGFP into the right and left backside of the nude mice, respectively. The xenografted tumors were regularly observed and the tumors volume was recorded. Four weeks after the cell injection, the BALB/C nude mice were dislocated put to death. The tumor weights were measured to analyze SPINK5 effect on cancerous cells in vivo. All animal studies were performed in accordance with the NIH Guide for Care and Use Laboratory Animals, and have been approved by the Shanghai Ninth People’s Hospital Institute Animal Care and Use Committee.
Statistical Analysis
Student’s t - test was used in comparison between two groups such as the results of the real-time PCR, plate colony formation, and cell cycle. More groups’ comparison was analyzed by using one-way ANOVA. The Log-rank test was used to analyze the overall survival of patients with high or low LEKTI expression. Univariate and multivariate Cox proportional hazards regression model analyses were used to analyze various parameters associated with overall survival of 88 patients with HNSCC. In this study, all analyses were conducted using SPSS 13.0 software, and all analyses were two-tailed test, P<0.05 was considered statistically significant.
Results
Decreased SPINK5/LEKTI Expression in HNSCC Cell Lines and Specimens
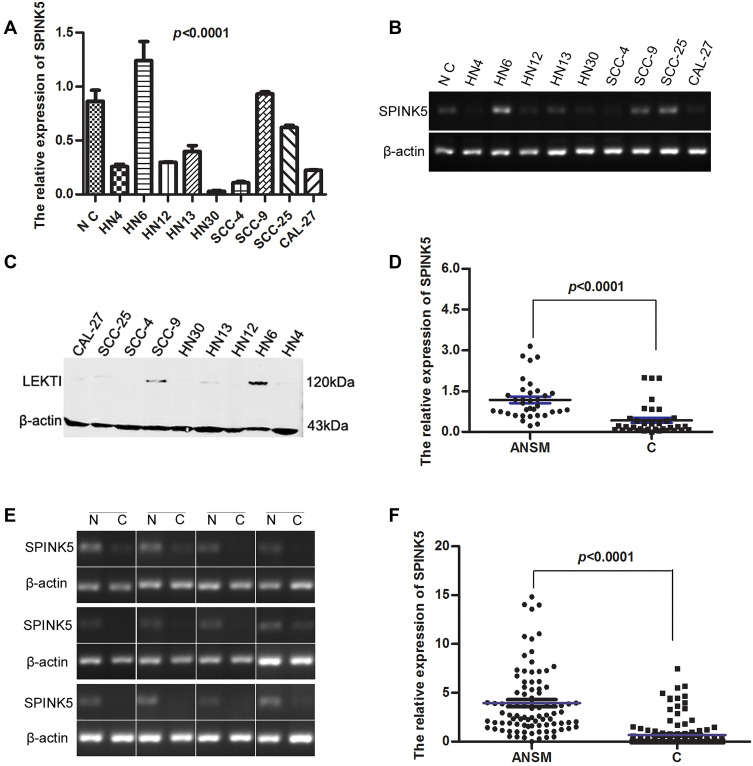
To determine SPINK5/LEKTI expression in HNSCC, SPINK5 mRNA and LEKTI protein levels were detected in representative 9 HNSCC cell lines as well as normal gingival epithelial cells and in HNSCC specimens including cancerous tissue and their apparently normal surgical margins. The expression of SPINK5 mRNA and LEKTI protein in HNSCC cells were significantly lower than normal epithelial cells in addition to WSU-HN6, SCC-9 and SCC-25 cells in representative 9 HNSCC cell lines (P<0.0001, One-way ANOVA) (Figure 1A–C). Then, we further to determine the SPINK5 expression in clinical HNSCC specimens. The result expectedly showed that SPINK5 mRNA level in 85.7% (30/35) paired HNSCC specimens was remarkably lower than that of their apparently normal surgical margins (P<0.0001, Student’s t-test, paired) (Figure 1D and E). Moreover, other cohort of HNSCC samples including 142 cancerous tissues and 92 apparently normal surgical margins were used and confirmed that SPINK5 mRNA level were significantly reduced in the cancerous tissues (P<0.0001, Student’s t test, unpaired) (Figure 1F). We analyzed Gene Expression profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/index.html) and found that SPINK5 was remarkably down-regulated in HNSCC (Supplemental Figure 1A).
Figure 1.
SPINK5/LEKTI expressions were showed in HNSCC cell lines and specimens. (A–C) SPINK5 mRNA and LEKTI protein levels were determined in 9 representative HNSCC cell lines and primary normal epithelial cells using real-time PCR, semi-quantitative RT-PCR and Western blotting; (D) SPINK5 mRNA expression was analyzed in 35 paired HNSCC specimens by using real-time PCR (ANSM: apparently normal surgical margins; C cancerous tissues); (E) It was shown that the result of SPINK5 mRNA in 12 representative HNSCC specimens by means of RT-PCR (N, adjacent normal tissue; C, cancerous tissue); (F) It was illustrated that the results of real-time PCR analysis about unpaired HNSCC specimens including 142 cancerous tissues and 92 apparently normal surgical margins (ANSM: apparently normal surgical margins; C cancerous tissues).
SPINK5/LEKTI Expression Represses the Proliferation, Colony-Formation and Induce the G1-Arrest of HNSCC cells
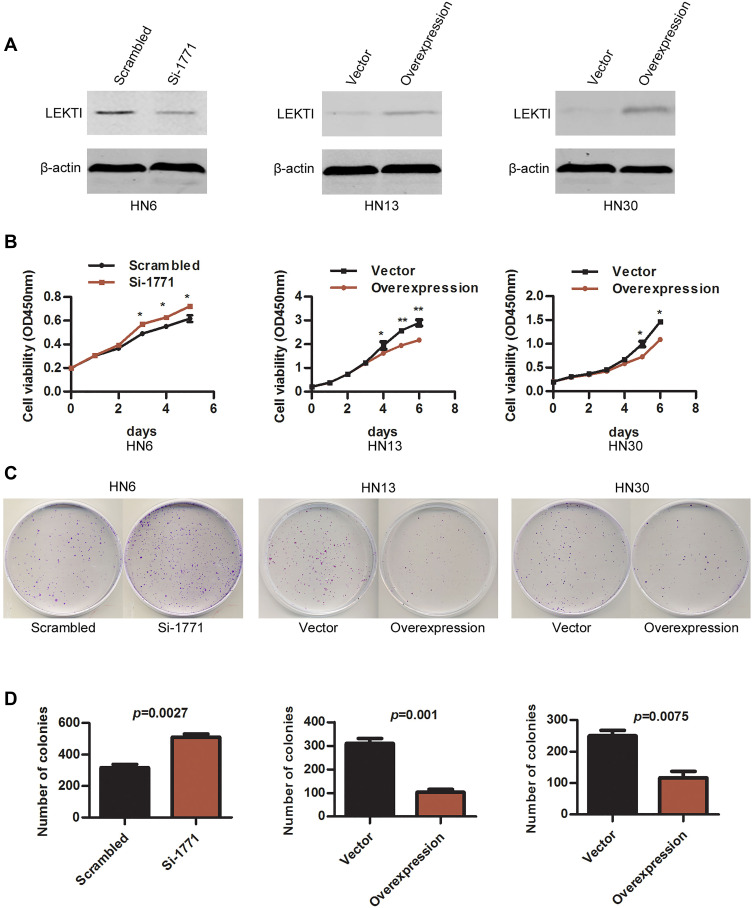
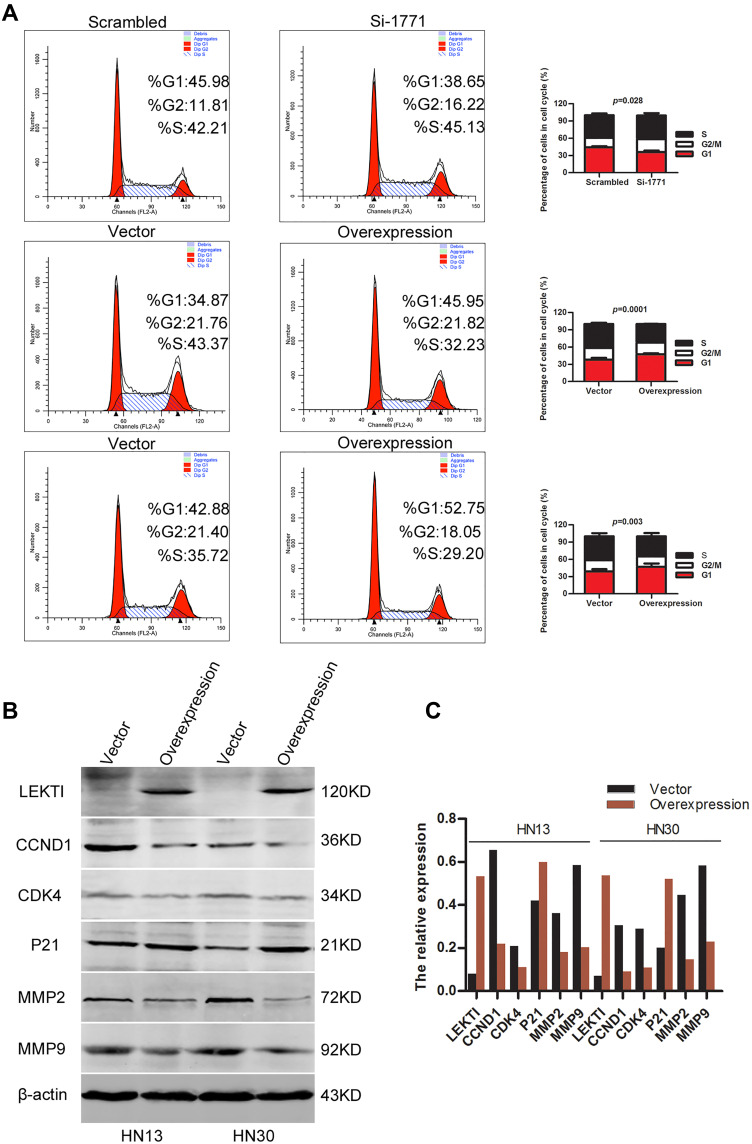
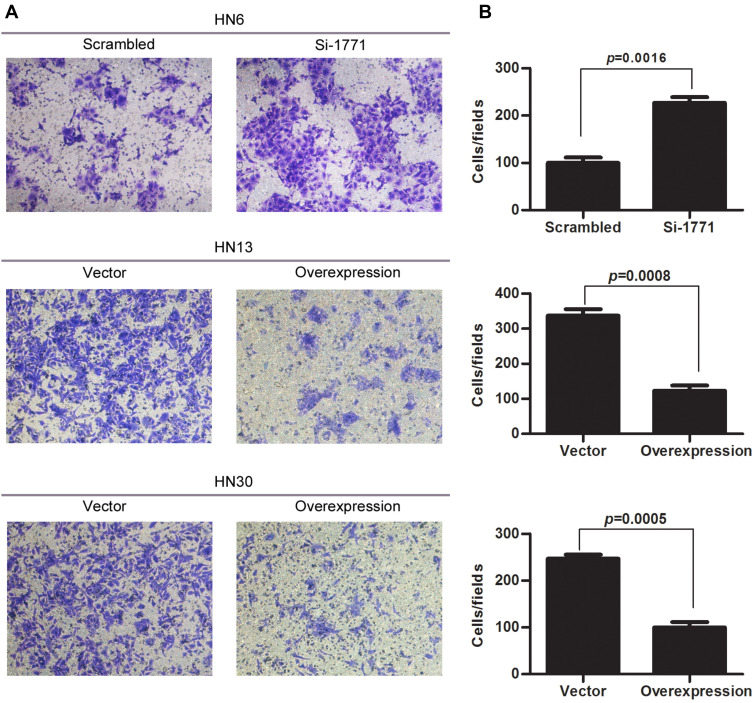
SPINK5/LEKTI was down-regulated in HNSCC cell lines and specimens, so far it has not been reported how it affects the cancerous cells. In this study, WSU-HN6 cells that expressed high levels of SPINK5/LEKTI and HN13, HN30 cells that had lower or few SPINK5/LEKTI expressions were selected for investigation of the impact of SPINK5/LEKTI on cancerous cells. In WSU-HN6 cells, si-1771, the best sequence of interference in our study, was used to silence SPINK5/LKETI expression. The result showed that transfection with si-1771 reduced LEKTI protein level by 70% (Figure 2A). Cell proliferation and colony formation assays showed that reducing SPINK5/LEKTI expression could significantly increase the proliferation and colony formation capability of WSU-HN6 (Figure 2B–D). On the contrary, the cell proliferation and colony formation were remarkably decreased in WSU-HN13 and HN30 cells transfected with the lentivirus pLenti-CMV-SPINK5 to increase exogenous SPINK5/LEKTI expression (Figure 2A–D). To determine the mechanism of SPINK5/LEKTI affect the cell proliferation and colony formation. Cell cycle and apoptosis were analyzed in these transfected HNSCC cells by FCM. The results showed that reducing SPINK5/LEKTI in WSU-HN6 cells leads to the arrest of the cells in G1 phase, while increasing SPINK5/LEKTI in WSU-HN13 and HN30 cells lead to the accumulation of the cells in G1 phase (Figure 3A). However, the apoptosis of these transfected cells were not shown changed (data not shown). These indicated that SPINK5/LEKTI affecting cell proliferation and colony formation were implemented by influencing the cell cycle of HNSCC cells. In addition, we found that reducing SPINK5/LEKTI expression in WSU-HN6 cells showed a significant increase in migration and invasion ability. However, the ability of WSU-HN13 and HN30 cells transfected with the lentivirus pLenti-CMV-SPINK5 to migrate and invade were remarkably reduced by transwell migration and invasion assays (Figure 4A and B).
Figure 2.
The effect of SPINK5/LEKTI on cell proliferation and colony formation. (A) LEKTI levels were confirmed in WSU-HN6, HN13 and HN30 cell lines using Western blotting after transfection; (B) Cell proliferation of WSU-HN6, HN13 and HN30 cell lines after transfection was analyzed using the CCK-8 kit, each point showed the mean values of triplicate wells (mean ± SD), * P<0.05, ** P<0.01; (C) Plate colony formation of WSU-HN6, HN13 and HN30 cell lines after transfection was analyzed; (D) The count of colonies was analyzed using statistical software. Each experiment repeated at least three times.
Figure 3.
The impact of SPINK5/LEKTI on the cell cycle. (A) Flow cytometry was used to analyze the cell cycle of WSU-HN6, HN13 and HN30 cells with reducing or increasing SPINK5/LEKTI expression; (B) Cell cycle proteins (CCND1, CDK4, p21) and matrix metalloproteinase (MMP2, MMP9) were determined in WSU-13 and HN30 cells transfected with the pLenti-CMV-SPINK5 or pLenti-CMV-EGFP; (C) The grey value analysis was made about the proteins in Figure 3B.
Figure 4.
The effect of SPINK5/LEKTI on invasion of HNSCC cells. (A) The result of cell invasion and migration assay showed that reducing SPINK5/LEKTI expression in WSU-HN6 cells could significantly increase the cell invasion capability. On the contrary, increasing SPINK5/LEKTI expression leads to reduced cell invasion capability of WSU-HN13 and HN30 cells; (B) The invaded cells were counted and made a comparison using SPSS 13.0 software. Each experiment repeated at least three times.
Effect of Increasing SPINK5/LEKTI Expression on Cell Cycle and Metastatic Regulators
To identify the molecular mechanisms of SPINK5/LEKTI affecting G1 phase of cell cycle in HNSCC cells, we measured a series of key proteins involved in G1 phase regulation. The results showed that CCND1 and CDK4 protein levels were significantly reduced while on the other hand, p21 was increased in WSU-HN13 and HN30 cells transfected with exogenous SPINK5. Considering SPINK5/LEKTI was involved in migration and invasion in HNSCC cells. The two important matrix metalloproteinases 2 and 9 (MMP2 and MMP9) were measured in WSU-HN13 and HN30 cells transfected with the lentivirus pLenti-CMV-SPINK5. The results showed that the protein levels of MMP2 and MMP9 were decreased in these transfected cells, which were in accordance with the results about phenotype research (Figure 3B), and the changes were directly showed in these proteins after the quantification of these proteins (Figure 3C). However, the proteins related apoptosis were not changed. These results further confirm the above phenotypic alterations and may partly explain the molecular mechanism underlying these phenotypes.
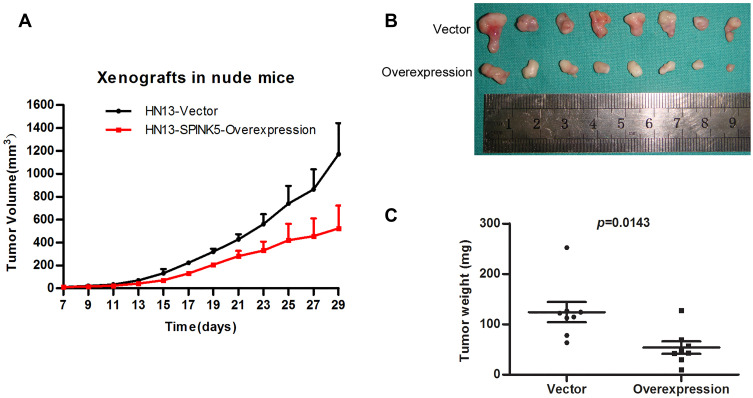
Exogenous Overexpression of SPINK5/LEKTI Suppresses Tumor Growth of HNSCC in vivo
To evaluate whether exogenous SPINK5/LEKTI suppresses the growth of cancerous cells in vivo, WSU-HN13 cells transfected with the lentivirus pLenti-CMV-SPINK5 or pLenti-CMV-EGFP were used to inject into the subcutaneous tissues of BAL/C nude mice then xenografted tumors were generated. The xenografted tumors were formed after subcutaneously injection and they grew gradually after 1 week. The tumor volume was recorded every other day after the tumors were formed. We found that the growth of xenografted tumors formed by HN13 cells transfected with the lentivirus pLenti-CMV-SPINK5 was slower than that of control group (Figure 5A). The used BALB/C nude mice were put to death 4 weeks after injection. The tumor sizes and weights of xenografted tumors derived from HN13 cells exogenously expressed SPINK5/LEKTI were significantly reduced in comparison to the control group (Figure 5B and C).
Figure 5.
The effect of SPINK5/LEKTI on tumorigenicities in xenografted mice. (A) The time course was from growth of HN13 xenograft tumors to the end of 1 month after the cell inoculation; (B) Over-expression of SPINK5/LEKTI inhibited the growth of xenografted tumor formed by WSU-HN13 cells transfected with the lentivirus pLenti-CMV-SPINK5; (C) The weights of xenografted tumor made a comparison.
Decreased SPINK5/LEKTI Expression Was Significantly Correlated with Poor Clinical Outcome of Patients with HNSCC
To determine the association between LEKTI protein level and the prognosis as well as clinical parameters in patients with HNSCCs, tissue microarray containing cancerous specimens from 101 patients who had a confirmed diagnosis of HNSCCs and a complete clinical & follow-up data was performed and measured by immunohistochemistry. Then, the correlation of LEKTI protein levels with clinical parameters and outcomes was analyzed to estimate whether SPINK5/LEKTI can be used as an efficient biomarker in predicting the prognosis of patients with HNSCCs. Except for 11 specimens were taken the tissues off in the process of immunohistochemistry experiment and two patients had no follow-up data, we observed that LEKTI protein was expressed as cytoplasmic staining in the rest cases. Among these patients, the age ranged from 28 to 74 years old, 40 of the patients were female and 48 patients were male. Twenty-nine of patients had a history of smoking and 20 patients had a history of alcohol. The early stage patients (stage I and stage II) account for 48.9% of all patients, the follow-up time ranged from 1 to 122 months, and with a median follow-up time was 75 months. The patients were divided into two parts by LEKTI expression median of 2.4, the patients of high LEKTI expression account for 54.5%, and the remaining of 45.5% patients had low LEKTI expression. The detailed documents of the patients are summarized in Table 1.
To determine whether LEKTI protein level affects the overall survival of these patients, Kaplan–Meier survival analysis was used. The results showed that the patients with low LEKTI protein levels had significantly inferior clinical outcomes compared with those with high LEKTI protein levels (P=0.0017, Figure 6B). In GEPIA database, we found that the HNSCC patients with higher expression levels of SPINK5 mRNA had prognosis (Supplemental Figure 1B). In this study, we found that there was positive association between LEKTI protein level and the clinical TNM stage. LEKTI level in patients with early stage was higher than that in patients with advanced stage (P=0.0029) (Figure 6A and C). We also observed that LKETI expression was more reduced in patients with poor pathological differentiation (P=0.0408) (Table 1), and their expressions gradually decreased with tumor differentiation (Figure 6D). Moreover, there was no significant correlation between LEKTI level and age, gender, lymph node metastasis, smoking history, alcohol history and anatomic sites. In the univariate Cox proportional analyses, in addition to pathological differentiation, lymph node metastasis, TNM stage and LEKTI expression were found to be strongly associated with overall survival of patients with HNSCCs (hazard ratio: 0.123; 95% CI: 0.050–0.304; P<0.001). In multivariate Cox proportional analyses, LEKTI levels, together with pathological differentiation and TNM stage, were strongly associated with overall survival of patients with HNSCC (hazard ratio: 0.114; 95% CI: 0.044–0.292; P<0.001). LEKTI could serve as an independent prognostic predictor of patients with HNSCC (Table 2).
Figure 6.
The correlation of LEKTI levels with clinical outcome and clinicopathological parameters in 88 patients with HNSCCs. (A) LEKTI expression was detected in clinical stage I, stage II, stage III and stage IV as shown by Immunohistochemistry; (B) Kaplan–Meier survival curves showed that the overall survival of patients with low LEKTI expressions was worse than that of patients with high LEKTI expression; (C) Compared to early HNSCCs, the LEKTI expression in advanced HNSCC was significantly reduced; (D) Representative images show immunohistochemical staining for LEKTI expression in well-differentiated HNSCC, moderately differentiated HNSCC, and poorly differentiated HNSCC from the left to the right side.
Table 2.
Univariate and Multivariate Cox Regression Models for Estimating the Overall Survival
| Characteristics | HR | 95% CI | P |
|---|---|---|---|
| Univariate analysis | |||
| Overall survival | |||
| Age (<60 y vs 60 y) | 0.612 | 0.279 to 1.340 | 0.219 |
| Gender (male vs female) | 1.840 | 0.840 to 4.030 | 0.127 |
| Smoking history (smoker vs nonsmoker) | 1.325 | 0.636 to 2.761 | 0.452 |
| Alcohol history (drinker vs nondrinker) | 1.273 | 0.566 to 2.863 | 0.560 |
| Pathological differentiation | 2.037 | 1.143 to 3.631 | 0.016 |
| Lymph node metastasis (pN0 vs pN1 to pN2) |
2.297 | 1.115 to 4.374 | 0.024 |
| TNM stage | 2.049 | 1.379 to 3.046 | <0.001 |
| LEKTI expression (high vs low) | 0.123 | 0.050 to 0.304 | <0.001 |
| Disease site | 1.015 | 0.608 to 1.697 | 0.954 |
| Multivariate analysis | |||
| Overall survival | |||
| Pathological differentiation | 2.835 | 1.504 to 5.345 | 0.001 |
| Lymph node metastasis (pN0 vs pN1 to pN2) |
0.790 | 0.363 to 1.719 | 0.552 |
| TNM stage | 2.082 | 1.333 to 3.250 | 0.001 |
| LEKTI expression (high vs low) | 0.114 | 0.044 to 0.292 | <0.001 |
Abbreviations: HR, hazard ration; CI, confidence interval; N, lymph node; TNM, tumor-lymph node-metastasis classification.
Discussion
HNSCC is a life-threatening disease with severe dysfunction, pain and cachexia. Even the great progress was made in the treatment including surgery, radiotherapy, chemotherapy and targeted therapy, the 5-year survival rate of patients with HNSCC remains about 55%. It is generally believed that HNSCC is a multi-factorial disease that probably arises as a result of environmental factors such as the irritant of cigarettes, alcohol and genetic diversity.37,38 In recent years, with the development of high-throughput sequencing, disease-causing genes and genes polymorphism was effectively found to be closely related to some tumor types.39–41 Further functional analysis of cancer-related genes not only uncovers the molecular mechanisms underlying in cancers, but also plays a role in adjuvant predicting the clinical implications of patients with cancer.
SPINK5 could keep intact skin barrier by inhibiting KLK family. It was shown that mutation of SPINK5 leads to dermatoses such as NS and atopic disease. In our prior study, the expression level of SPINK5 was found to be notably reduced in HNSCC tissues by using oligonucleotide microarrays. Here our results indicated that SPINK5 mRNA and LEKTI protein levels were remarkably decreased in HNSCCs compared to adjacent normal tissues, suggesting SPINK5 may be a potential tumor suppressor in HNSCC. Furthermore, silencing of SPINK5/LEKTI prominently increased the proliferation, colony formation and invasion abilities of WSU-HN6 cells Whereas SPINK5/LEKTI overexpression remarkably suppressed the proliferation, colony formation and invasion abilities of WSU-HN13 and HN30 cells in vitro. Definitively, SPINK5/LEKTI knock down led to the tumor inhibitory rate of xenograft tumors in BALB/C nude mice was more than 50% as compared with control groups in vivo. Our finding demonstrated that SPINK5 is a tumor suppressor in HNSCC which was not consistent with the findings described in bladder transitional cell carcinoma.23 The inconsistency is likely due to the diversity of tumor genetic background, also can be caused by different pathogenesis in different cancers. Based on our results, SPINK5 is a multifunctional protease inhibitor more than serine protease inhibitor. It not only inhibits the endogenous protease such as kallikreins, caspase and cathepsin, but also inhibit the exogenous proteases derived from microorganisms. Down-regulation of SPINK5, on the one hand, results in the enhanced activity of KLKs which leads to increase of epidermal thickness, hyperkeratosis and dermal inflammation; on the other hand, give rise to the accumulation of exogenous protease from bacteria, fungi and virus result in the integrity and protective barrier function of the skin were damaged severely.
In the study, we found that silencing of SPINK5/LEKTI induced the arrest of G1 phase in WSU-HN6 cells, whereas overexpression of SPINK5/LEKTI led to accelerating the transition of G1-S phase in WSU-HN13 and HN30 cells. And the above changes were accompanied by the reduced expression levels of CCND1 and CDK4, indicating overexpression of SPINK5/LEKTI might reduce the activities of CDK4 leading to the inhibitory role on cell proliferation and colony formation abilities. Additionally, the expression level of caspase-3 and PARP was found to be unchanged in WSU-HN13 and HN30 cells with SPINK5/LEKTI overexpression (data not shown). Our data suggest that down-regulation of SPINK5/LEKTI contributes to the proliferation of HNSCC cells by regulating the cell cycle rather than cell apoptosis.
In addition, we also found that SPINK5/LEKTI dysregulation was associated with other aggressive behaviors of HNSCC cells such as the abilities of migration and invasion. The study showed that the abilities of migration and invasion were significantly reduced in WSU-HN13 and HN30 cells transfected with exogenous SPINK5, respectively. The result is consistent with the result described in esophageal cancer.25 Moreover, MMP2 and MMP9, two key proteins related to metastasis, were decreased remarkably in our resulting. According to the literature,42,43 down-regulation of SPINK5 and up-regulation of matrix metalloproteinases was found in other squamous cell carcinoma tissues. This suggests direct or indirect interactions between the over-expression of MMP2, MMP9 and down-regulation of SPINK5/LEKTI.
To determine the clinical significance of SPINK5/LEKTI expression in HNSCC, we measured the expression levels of LEKTI protein in 101 patients with HNSCCs by using IHC and analyzed the correlation between LEKTI expression and clinical outcome. Our data indicated that downregulated LEKTI was closely associated with advanced clinical TNM stage in HNSCC. Moreover, lower LEKTI expression was also found to be reduced in poorer pathological differentiation. Kaplan–Meier curve analysis showed that patients with low LEKTI expression had poorer survival compared to that of the patients with high LEKTI expression. In univariate and multivariate analyses, our resulting indicates LEKTI was an independent prognostic predictor for patients with HNSCC. To a certain extent, the expression level of SPINK5/LEKTI serves as an adjuvant measure to guide the treatment of patients with HNSCC.
There were some limitations in the current study. Firstly, the down-regulation mechanism of SPINK5/LEKTI remains unclear in HNSCC. In the study, 5-aza-2ʹ-deoxycytidine (5-Aza-dC), a kind of methylation inhibitors, could not improve the expression level of SPINK5. Furthermore, methylations of primers were designed to analyze 5 paired bisulfate-treated DNA from clinical samples including cancerous tissues and adjacent normal tissues, no changes were found between their productions. So, the down-regulation mechanism of SPINK5/LEKTI in HNSCC was needed to be investigated in further study. Secondly, the HNSCC sample size was relatively small and collected in single institution. A multicenter and large sample was used to analyze the correlation of SPINK5/LEKTI with clinical pathological parameters.
Conclusions
SPINK5/LEKTI is down-regulated in HNSCCs and the downregulated SPINK5/LEKTI contributes to malignant phenotypes including the proliferation, colony formation, invasion and tumorigenesis abilities of HNSCC cells partly due to the dysregulation of cell cycle. Our findings also demonstrated that LEKTI expression is significantly associated with the clinical TNM stage and tumor differentiation, and SPINK5/LEKTI is an independent predictor for overall survival of patients with HNSCC.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81672745 and 81972589) and the Natural Science Foundation of Jiangsu Province (BK20160225).
Data Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent Statement
This study was approved by the independent Ethics Committee of Shanghai Ninth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine.
Informed Consent
Written informed consents were obtained from all participants.
Disclosure
The authors have declared no competing interests exist.
References
- 1.Gaykalova DA, Mambo E, Choudhary A, et al. Novel insight into mutational landscape of head and neck squamous cell carcinoma. PLoS One. 2014;9(3):e93102. doi: 10.1371/journal.pone.0093102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 4.Zheng M, Li L, Tang Y, Liang XH. How to improve the survival rate of implants after radiotherapy for head and neck cancer? J Periodontal Implant Sci. 2014;44:2. doi: 10.5051/jpis.2014.44.1.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyss AB, Weissler MC, Avery CL, et al. Single nucleotide polymorphisms in nucleotide excision repair genes, cancer treatment, and head and neck cancer survival. Cancer Causes Control. 2014;25(4):437–450. doi: 10.1007/s10552-014-0346-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuriakose MA, Chen WT, He ZM, et al. Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci. 2004;61(11):1372–1383. doi: 10.1007/s00018-004-4069-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birben E, Sackesen C, Turgutoğlu N, Kalayci O. The role of SPINK5 in asthma related physiological events in the airway epithelium. Respir Med. 2012;106(3):349–355. doi: 10.1016/j.rmed.2011.11.007 [DOI] [PubMed] [Google Scholar]
- 8.Roedl D, Oji V, Buters JT, Behrendt H, Braun-Falco M. rAAV2-mediated restoration of LEKTI in LEKTI-deficient cells from Netherton patients. J Dermatol Sci. 2011;61(3):194–198. doi: 10.1016/j.jdermsci.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 9.Elias PM, Wakefield JS. Therapeutic implications of a barrier-based pathogenesis of atopic dermatitis. Clin Rev Allergy Immunol. 2010;41(3):282–295. doi: 10.1007/s12016-010-8231-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer-Hoffert U. Reddish, scaly, and itchy: how proteases and their inhibitors contribute to inflammatory skin diseases. Arch Immunol Ther Exp (Warsz). 2009;57:345–354. doi: 10.1007/s00005-009-0045-6 [DOI] [PubMed] [Google Scholar]
- 11.Nin M, Katon N, Kokura S, Handa O, Yoshikawa T, Kishimoto S. Dichotomous effect of ultraviolet B on the expression of corneodesmosomal enzymes in human epidermal keratinocytes. J Dermatol Sci. 2009;54:17–24. doi: 10.1016/j.jdermsci.2008.11.004 [DOI] [PubMed] [Google Scholar]
- 12.Wood SH, Clements DN, Ollier WE, Nuttall T, McEwan NA, Carter SD. Gene expression in canine atopic dermatitis and correlation with clinical severity scores. J Dermatol Sci. 2009;55:27–33. doi: 10.1016/j.jdermsci.2009.03.005 [DOI] [PubMed] [Google Scholar]
- 13.Bennett K, Heywood W, Di WL, et al. The identification of a new role for LEKTI in the skin: the use of protein ‘bait’ arrays to detect defective trafficking of dermcidin in the skin of patients with Netherton syndrome. J Proteomics. 2012;75:3925–3937. doi: 10.1016/j.jprot.2012.04.045 [DOI] [PubMed] [Google Scholar]
- 14.Ishida-Yamamoto A, Kishibe M. Involvement of corneodesmosome degradation and lamellar granule transportation in the desquamation process. Med Mol Morphol. 2011;44(1):1–6. doi: 10.1007/s00795-010-0513-4 [DOI] [PubMed] [Google Scholar]
- 15.Parate PN, Wang DY, Chew FT. Linkage disequilibrium pattern in asthma candidate genes from 5q31-q33 in the Singapore Chinese Population. Ann Hum Genet. 2010;74:137–145. doi: 10.1111/j.1469-1809.2009.00558.x [DOI] [PubMed] [Google Scholar]
- 16.Bingol B, Tasdemir S, Gunenc Z, et al. Prenatal diagnosis of Comel-Netherton syndrome with PGD, case report and review article. J Assist Reprod Genet. 2011;28(7):615–620. doi: 10.1007/s10815-011-9568-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortugno P, Furio L, Teson M, et al. The 420K LEKTI variant alters LEKTI proteolytic activation and results in protease deregulation: implications for atopic dermatitis. Hum Mol Genet. 2012;21(19):4187–4200. doi: 10.1093/hmg/dds243 [DOI] [PubMed] [Google Scholar]
- 18.Roelandt T, Thys B, Heughebaet C, et al. LEKTI-1 in sickness and in health. Int J Cosmet Sci. 2009;31(4):247–254. doi: 10.1111/j.1468-2494.2009.00516.x [DOI] [PubMed] [Google Scholar]
- 19.Cork MJ, Danby SG, Vasilopoulos Y, et al. Epidermal barrier dysfunction in atopic dermatitis. J Invest Dermatol. 2009;129(8):1892–1908. doi: 10.1038/jid.2009.133 [DOI] [PubMed] [Google Scholar]
- 20.Namkung JH, Lee JE, Kim E, et al. Hint for association of single nucleotide polymorphisms and haplotype in SPINK5 gene with atopic dermatitis in Koreans. Exp Dermatol. 2010;19:1048–1053. doi: 10.1111/j.1600-0625.2010.01142.x [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Xia Y, Zhang W, et al. A functional polymorphism in the SPINK5 gene is associated with asthma in a Chinese Han Population. BMC Med Genet. 2009;10(1):59. doi: 10.1186/1471-2350-10-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fruth K, Goebel G, Koutsimpelas D, et al. Low SPINK5 expression in chronic rhinosinusitis. The Laryngoscope. 2012;122(6):1198–1204. doi: 10.1002/lary.23300 [DOI] [PubMed] [Google Scholar]
- 23.Bennett K, Callard R, Heywood W, et al. New role for LEKTI in skin barrier formation: label-free quantitative proteomic identification of caspase 14 as a novel target for the protease inhibitor LEKTI. J Proteome Res. 2010;9:4289–4294. doi: 10.1021/pr1003467 [DOI] [PubMed] [Google Scholar]
- 24.Tyan YC, Yang MH, Chen SC, et al. Urinary protein profiling by liquid chromatography/tandem mass spectrometry: ADAM28 is overexpressed in bladder transitional cell carcinoma. Rapid Commun Mass Spectrom. 2011;25:2851–2862. doi: 10.1002/rcm.5169 [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Lv Q, Bian H, et al. A novel tumor suppressor SPINK5 targets Wnt/β-catenin signaling pathway in esophageal cancer. Cancer Med. 2019;8:2360–2371. doi: 10.1002/cam4.2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao C, Zou H, Zhang J, Wang J, Liu H. An integrated methylation and gene expression microarray analysis reveals significant prognostic biomarkers in oral squamous cell carcinoma. Oncol Rep. 2018;40:2637–2647. doi: 10.3892/or.2018.6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao W, Younis RH, Li J, et al. EZH2 promotes malignant phenotypes and is a predictor of oral cancer development in patients with oral leukoplakia. Cancer Prev Res (Phila). 2011;4:1816–1824. doi: 10.1158/1940-6207.CAPR-11-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Q, Liu X, Chen W, Zhang Z. Inhibiting adenoid cystic carcinoma cells growth and metastasis by blocking the expression of ADAM 10 using RNA interference. J Transl Med. 2010;8(1):136. doi: 10.1186/1479-5876-8-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Z, Guo W, Zhang C, et al. CCND1 as a predictive biomarker of neoadjuvant chemotherapy in patients with locally advanced head and neck squamous cell carcinoma. PLoS One. 2011;6:e26399. doi: 10.1371/journal.pone.0026399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Q, Sun Q, Zhang J, Yu J, Chen W, Zhang Z. Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis. 2013;34:539–549. doi: 10.1093/carcin/bgs374 [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Guo H, Qian G, et al. MiR-145, a new regulator of the DNA Fragmentation Factor-45 (DFF45)-mediated apoptotic network. Mol Cancer. 2010;9(1):211. doi: 10.1186/1476-4598-9-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui Z, Cwo W, Li J, Song X, Mao L, Chen W. TRIM24 overexpression is common in locally advanced head and neck squamous cell carcinoma and correlates with aggressive malignant phenotypes. PLoS One. 2013;8(5):e63887. doi: 10.1371/journal.pone.0063887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Cao W, Wang X, et al. TGM3, a candidate tumor suppressor gene, contributes to human head and neck cancer. Mol Cancer. 2013;12(1):151. doi: 10.1186/1476-4598-12-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao W, Zhang ZY, Xu Q, et al. Epigenetic silencing of MAL, a putative tumor suppressor gene, can contribute to human epithelium cell carcinoma. Mol Cancer. 2010;9(1):296. doi: 10.1186/1476-4598-9-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan M, Xu Q, Zhang P, Zhou XJ, Zhang ZY, Chen WT. Correlation of NF-kappaB signal pathway with tumor metastasis of human head and neck squamous cell carcinoma. BMC Cancer. 2010;10:437. doi: 10.1186/1471-2407-10-437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J, He Y, Yan M, et al. Dose dependent activation of retinoic acid-inducible gene-I promotes both proliferation and apoptosis signals in human head and neck squamous cell carcinoma. PLoS One. 2013;8(3):e58273. doi: 10.1371/journal.pone.0058273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen D, Cabay RJ, Jin Y, et al. MicroRNA deregulations in head and neck squamous cell carcinomas. J Oral and Maxillofac Res. 2013;4(1):e2. doi: 10.5037/jomr.2013.4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seven D, Yavuz E, Kilic E, et al. DLEC1 is not silenced solely by promoter methylation in head and neck squamous cell carcinoma. Gene. 2015;563(1):83–86. doi: 10.1016/j.gene.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 39.Kim TM, Lee SH, Chung YJ. Clinical applications of next-generation sequencing in colorectal cancers. World J Gastroenterol. 2013;19:6784. doi: 10.3748/wjg.v19.i40.6784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei JS, Johansson P, Chen L, et al. Massively parallel sequencing reveals an accumulation of de novo mutations and an activating mutation of LPAR1 in a patient with metastatic neuroblastoma. PLoS One. 2013;8:e77731. doi: 10.1371/journal.pone.0077731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu MD, Wu H, Wang S, et al. MiR-1275 promotes cell migration, invasion and proliferation in squamous cell carcinoma of head and neck via up-regulating IGF-1R and CCR7. Gene. 2018;646:1–7. doi: 10.1016/j.gene.2017.12.049 [DOI] [PubMed] [Google Scholar]
- 42.Nair J, Jain P, Chandola U, et al. Gene and miRNA expression changes in squamous cell carcinoma of larynx and hypopharynx. Genes Cancer. 2015;6(7–8):328–340. doi: 10.18632/genesandcancer.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye H, Yu T, Teman S, et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;6(9):69. doi: 10.1186/1471-2164-9-69 [DOI] [PMC free article] [PubMed] [Google Scholar]