Abstract
Purpose
To evaluate the susceptibility rates of carbapenem-resistant (CR)-Enterobacterales strains from Chinese intra-abdominal infections (IAI), respiratory tract infections (RTI) and urinary tract infections (UTI) between 2015 and 2017 to colistin.
Methods
In total, 7138 Enterobacterales including 1074 CR-Enterobacterales strains were isolated from IAI+UTI+RTI samples and collected in 21 hospitals across 7 regions of China. Antimicrobial susceptibilities were determined at a central laboratory using CLSI broth microdilution and interpretive standards.
Results
From 2015 to 2017, E. coli (51.4%) and K. pneumoniae (30.0%) accounted for the majority of Enterobacterales isolated from IAIs, UTIs and RTIs. The percentage of CR strains within the species was highest for S. marcescens (27.9%), followed by K. pneumoniae (24.8%), P. mirabilis (22.6), K. oxytoca (19.5%), E. cloacae (17.7%), C. freundii (12.5%), K. aerogenes (11.0%) and lowest for E. coli (6.9%). Colistin susceptibilities were generally higher in CS than in CR isolates and were 83.5% for CR-E. coli, 88.6% for CR-K. pneumoniae, 79.2% for CR-E. cloacae and 87.5% for CR-K. aerogenes. For IAI and UTI isolates in particular, CR-E. coli and CR-K. pneumoniae showed a trend of decreasing susceptibility, which was especially noted for CR-E. coli in UTI isolates, and for both organisms in IAI isolates susceptibility dropped markedly in 2017.
Conclusion
Colistin was a last resort antibiotics for empirical CR-Enterobacterales treatments, since especially the percentage of CR-K. pneumoniae was 30.0% of all IAI, UTI and RTI isolates, with an incidence of 24.8% CR strains, of which 88.6% were susceptible to colistin. Also other analyzed CR-Enterobacterales showed colistin susceptibilities of ≥80.0%. However, resistance rates of IAI derived CR-K. pneumoniae and CR-E. coli, and CR-K. pneumoniae UTI isolates to colistin increased between 2015 and 2017, which should further be closely monitored.
Keywords: Enterobacterales, carbapenem, colistin, intra-abdominal infection, respiratory tract infection, urinary tract infection
Introduction
Multi-drug resistance (MDR) has become a significant problem worldwide1 with carbapenems being commonly used for the treatment of MDR bacterial infections. However, carbapenem-resistant (CR)-Enterobacterales induce infections that have high mortality rates and limited treatment options.2 The mechanisms of carbapenem resistance in Enterobacterales include AmpC expression in combination with the loss of porin due to alterations of Omp genes, and the production of acquired carbapenemases.3,4 In China, the incidence of CR-Enterobacterales infections has been steadily increasing,5 with resistance rates to E. coli and K. pneumoniae of 0% and 0.7% in 20046 raised to 1.0% and 13.4% in 2014, respectively.7 Therefore, we investigated the susceptibility rates of major Enterobacterales strains isolated from hospitalized patients with intra-abdominal infections (IAIs), urinary tract infections (UTIs) and respiratory tract infections (RTI) between 2015 and 2017. Isolates were collected for the Study for Monitoring Antimicrobial Resistance Trends (SMART) global surveillance program, which was established in China in 2002 for IAIs and in 2012 for UTIs to monitor in vitro antimicrobial susceptibility profiles of clinical isolates collected from Chinese patients with IAIs and UTIs.8
Polymyxins are one of the last-line antibiotic choices for such infections. The World Health Organization (WHO) has designated polymyxin as the “highest priority critically important antimicrobial for human medicine”9 as it is often one of the only therapies available for treating serious bacterial infections in critically ill patients. However, polymyxin resistance rates of 36.1% and 15.9% have been reported in some parts of the world and were found to be independently associated with mortality.10,11 Emergence of resistance towards antibiotics of last-resort has also been reported in China,12 but while colistin has been in clinical use for about five years, the sensitivities of CR-Enterobacterales to this drug remain unclear. In the present study, we analyzed the prevalence of CR-Enterobacterales isolates from Chinese IAIs, RTIs and UTIs between 2015 and 2017 focusing on the incidence of colistin resistance.
Methods
Isolates from IAI, UTI and RTI Infections
The Human Research Ethics Committee of our hospital approved the study protocols (Et. Number: S-K238). Since some Enterobacterales members have an acquired resistance against colistin but others including Proteus spp., Providencia spp., Serratia spp., and Morganella spp. have an intrinsic resistance,13 we focused on Enterobacterales isolates, which are known not to be naturally resistant to colistin.
In total, 7113 Enterobacterales (2015, n = 2203; 2016, n = 2375; 2017, n = 2535) and 1070 CR-Enterobacterales (2015, n = 259; 2016, n = 371; 2017, n = 440) isolates were detected in IAI+UTI+RTI samples from 2015 to 2017 obtained from 21 centers in 7 regions of China (northeast, north, central south, southwest, east Jiangzhe and east non-Jiangzhe areas), with a range of 77 to 250 samples per year per hospital, which were consecutively collected. The majority of the IAI specimens were taken from the appendix, gall bladder, small intestine, colon, rectum, pancreas, stomach, liver and peritoneal fluid or from abscesses. The majority of the UTI specimens were sampled from urine and the urethra. The majority of the RTI specimens were taken from bronchial brushings, bronchoalveolar lavage, sputum and thoracentesis. Isolates were identified using local hospital procedures and then sent for analysis to the clinical microbiology laboratory of Peking Union Medical College Hospital for re-identification using MALDI-TOF MS (Vitek MS, BioMérieux, France). All duplicate isolates (the same genus and species from the same patient) were excluded.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed at the Peking Union Medical College Hospital center laboratory using a customized MicroScan broth microdilution techniques in 2014, with the aid of a Trek Diagnostic System (Thermo Fisher Scientific, Cleveland, US) between 2015 and 2017, with susceptibility interpretations based on the European Committee on Antimicrobial Susceptibility Testing clinical breakpoints.14 The antibiotics colistin, imipenem, ertapenem, amikacin, ceftazidime and cefoxitin were tested following the recommendations of the Surgical Infection Society and the Infectious Diseases Society of America in their guidelines for the diagnosis and management of complicated IAIs, UTIs and RTIs.15 For each MIC test batch, reference strains of Escherichia coli (E. coli) American Type Culture Collection (ATCC) ATCC 27853 and Klebsiella pneumoniae (K. pneumoniae) (ATCC 700603) were used as quality control strains. The results were analyzed only when the quality control test results fell within acceptable ranges. In the present study, a CR-strain was defined as an organism resistant to any drug in the carbapenem class. A carbapenem sensitive (CS) strain was defined as an organism susceptible to all of the tested carbapenems.
Statistical Analysis
The susceptibility rates of all the Enterobacterales, CR-Enterobacterales and CS-Enterobacterales isolates were determined using appropriate breakpoints for each species. The trend of each year was analyzed with a chi-squared trend test for distribution and susceptibility rates. A P-value <0.05 was considered to be a statistically significant difference.
Results
General Distribution of Enterobacterales and CR-Enterobacterales from 2015 to 2017
A total of 7113 strains of Enterobacterales were collected from 2015 to 2017, and the yearly number of strains showed an increasing trend. E. coli (3657, 51.4%) accounted for more than half of the strains in the three years of study, followed by K. pneumoniae (2150, 30.2%). Other strains with more than 1% occurrence accounted for <10% of bacteria, being Enterobacter cloacae (E. cloacae) (407, 5.7%), Proteus mirabilis (P. mirabilis) (186, 2.6%), Klebsiella aerogenes (K. aerogenes) (146, 2.1%), Serratia marcescens (S. marcescens) (140, 2.0%), Klebsiella oxytoca (K. oxytoca) (123, 1.7%) and Citrobacter freundii (C. freundii) (96, 1.3%). The complete list of all Enterobacterales is provided in Supplementary Table 1. A total of 1070 CR-Enterobacterales strains were collected from 2015 to 2017, and the number of strains increased year by year. The percentage of CR strains within species was highest for S. marcescens (27.9%), followed by K. pneumoniae (24.8%), P. mirabilis (22.6%), K. oxytoca (19.5%), E. cloacae (17.7%), C. freundii (12.5%), K. aerogenes (11.0%) and lowest for E. coli (6.9%) (Table 1).
Table 1.
Distribution of Enterobacterales, CR-Enterobacterales and CS-Enterobacterales from 2015 to 2017 in IAIs, UTIs and RTIs (N, %)
| 2015 | 2016 | 2017 | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IAI | UTI | RTI | Subtotal | IAI | UTI | RTI | Subtotal | IAI | UTI | RTI | Subtotal | ||
| Enterobacterales | 1216 | 742 | 245 | 2203 | 1136 | 741 | 498 | 2375 | 899 | 1000 | 636 | 2535 | 7113 |
| CR-Enterobacterales | 148 (12.2) | 59 (8.0) | 52 (21.2) | 259 (11.8) | 159 (14.0) | 76 (10.3) | 136 (27.3) | 371 (15.6) | 155 (17.2) | 113 (11.3) | 172 (27.0) | 440 (17.4) | 1070 (15.0) |
| CS-Enterobacterales | 970 (79.8) | 613 (82.6) | 176 (71.8) | 1759 (79.8) | 899 (79.1) | 608 (82.1) | 325 (65.3) | 1832 (77.1) | 679 (75.5) | 830 (83.0) | 424 (66.7) | 1933 (76.3) | 5524 (77.7) |
| CI-Enterobacterales | 98 (8.1) | 70 (9.4) | 17 (6.9) | 185 (8.4) | 78 (6.9) | 57 (7.7) | 37 (7.4) | 172 (7.2) | 65 (7.2) | 57 (5.7) | 40 (6.3) | 162 (6.4) | 519 (7.3) |
| Escherichia coli | 648 | 531 | 38 | 1217 | 618 | 519 | 72 | 1209 | 456 | 692 | 83 | 1231 | 3657 |
| CR-Escherichia coli | 47 (7.3) | 18 (3.4) | 5 (13.2) | 70 (5.8) | 52 (8.4) | 35 (6.7) | 10 (13.9) | 97 (8.0) | 39 (8.6) | 36 (5.2) | 12 (14.5) | 87 (7.1) | 254 (6.9) |
| CS-Escherichia coli | 573 (88.4) | 486 (91.5) | 33 (86.8) | 1092 (89.7) | 543 (87.9) | 463 (89.2) | 61 (84.7) | 1067 (88.3) | 408 (89.5) | 637 (92.1) | 69 (83.1) | 1114 (90.5) | 3273 (89.5) |
| CI-Escherichia coli | 28 (4.3) | 27 (5.1) | 0 (0.0) | 55 (4.5) | 23 (3.7) | 21 (4.0) | 1 (1.4) | 45 (3.7) | 9 (2.0) | 19 (2.7) | 2 (2.4) | 30 (2.4) | 130 (3.6) |
| Klebsiella pneumoniae | 331 | 114 | 150 | 595 | 330 | 137 | 289 | 756 | 223 | 179 | 397 | 799 | 2150 |
| CR-Klebsiella pneumoniae | 55 (16.6) | 22 (19.3) | 36 (24.0) | 113 (19.0) | 69 (20.9) | 28 (20.4) | 87 (30.1) | 184 (24.3) | 69 (30.9) | 45 (25.1) | 123 (31.0) | 237 (29.7) | 534 (24.8) |
| CS-Klebsiella pneumoniae | 264 (79.8) | 81 (71.1) | 112 (74.7) | 457 (76.8) | 249 (75.5) | 97 (70.8) | 187 (64.7) | 533 (70.5) | 144 (64.6) | 129 (72.1) | 266 (67.0) | 539 (67.5) | 1529 (71.1) |
| CI-Klebsiella pneumoniae | 12 (3.6) | 11 (9.6) | 2 (1.3) | 25 (4.2) | 12 (3.6) | 12 (8.8) | 15 (5.2) | 39 (5.2) | 10 (4.5) | 5 (2.8) | 8 (2.0) | 23 (2.9) | 87 (4.0) |
| Enterobacter cloacae | 100 | 16 | 19 | 135 | 60 | 22 | 39 | 121 | 83 | 31 | 37 | 151 | 407 |
| CR-Enterobacter cloacae | 15 (15.0) | 5 (31.3) | 3 (15.8) | 23 (17.0) | 11 (18.3) | 3 (13.6) | 9 (23.1) | 23 (19.0) | 11 (13.3) | 9 (29.0) | 6 (16.2) | 26 (17.2) | 72 (17.7) |
| CS-Enterobacter cloacae | 62 (62.0) | 8 (50.0) | 13 (68.4) | 83 (61.5) | 39 (65.0) | 12 (54.5) | 26 (66.7) | 77 (63.6) | 61 (73.5) | 20 (64.5) | 27 (73.0) | 108 (71.5) | 268 (65.8) |
| CI-Enterobacter cloacae | 23 (23.0) | 3 (18.8) | 3 (15.8) | 29 (21.5) | 10 (16.7) | 7 (31.8) | 4 (10.3) | 21 (17.4) | 11 (13.3) | 2 (6.5) | 4 (10.8) | 17 (11.3) | 67 (16.5) |
| Proteus mirabilis | 29 | 26 | 7 | 62 | 20 | 18 | 11 | 49 | 24 | 38 | 13 | 75 | 186 |
| CR-Proteus mirabilis | 9 (31.0) | 5 (19.2) | 1 (14.3) | 15 (24.2) | 2 (10.0) | 4 (22.2) | 3 (27.3) | 9 (18.4) | 7 (29.2) | 9 (23.7) | 2 (15.4) | 18 (24.0) | 42 (22.6) |
| CS-Proteus mirabilis | 8 (27.6) | 5 (19.2) | 0 (0.0) | 13 (21.0) | 8 (40.0) | 6 (33.3) | 6 (54.5) | 20 (40.8) | 2 (8.3) | 9 (23.7) | 2 (15.4) | 13 (17.3) | 46 (24.7) |
| CI-Proteus mirabilis | 12 (41.4) | 16 (61.5) | 6 (85.7) | 34 (54.8) | 10 (50.0) | 8 (44.4) | 2 (18.2) | 20 (40.8) | 15 (62.5) | 20 (52.6) | 9 (69.2) | 44 (58.7) | 98 (52.7) |
| Klebsiella aerogenes | 27 | 9 | 2 | 38 | 23 | 9 | 17 | 49 | 30 | 12 | 17 | 59 | 146 |
| CR-Klebsiella aerogenes | 4 (14.8) | 0 (0.0) | 1 (50.0) | 5 (13.2) | 5 (21.7) | 1 (11.1) | 1 (5.9) | 7 (14.3) | 2 (6.7) | 1 (8.3) | 1 (5.9) | 4 (6.8) | 16 (11.0) |
| CS-Klebsiella aerogenes | 13 (48.1) | 7 (77.8) | 1 (50.0) | 21 (55.3) | 8 (34.8) | 4 (44.4) | 10 (58.8) | 22 (44.9) | 22 (73.3) | 7 (58.3) | 14 (82.4) | 43 (72.9) | 86 (58.9) |
| CI-Klebsiella aerogenes | 10 (37.0) | 2 (22.2) | 0 (0.0) | 12 (31.6) | 10 (43.5) | 4 (44.4) | 6 (35.3) | 20 (40.8) | 6 (20.0) | 4 (33.3) | 2 (11.8) | 12 (20.3) | 44 (30.1) |
| Serratia marcescens | 9 | 4 | 10 | 23 | 10 | 15 | 35 | 60 | 15 | 5 | 37 | 57 | 140 |
| CR-Serratia marcescens | 2 (22.2) | 1 (25.0) | 0 (0.0) | 3 (13.0) | 3 (30.0) | 1 (6.7) | 12 (34.3) | 16 (26.7) | 9 (60.0) | 0 (0.0) | 11 (29.7) | 20 (35.1) | 39 (27.9) |
| CS-Serratia marcescens | 5 (55.6) | 1 (25.0) | 6 (60.0) | 12 (52.2) | 6 (60.0) | 11 (73.3) | 16 (45.7) | 33 (55.0) | 2 (13.3) | 2 (40.0) | 16 (43.2) | 20 (35.1) | 65 (46.4) |
| CI-Serratia marcescens | 2 (22.2) | 2 (50.0) | 4 (40.0) | 8 (34.8) | 1 (10.0) | 3 (20.0) | 7 (20.0) | 11 (18.3) | 4 (26.7) | 3 (60.0) | 10 (27.0) | 17 (29.8) | 36 (25.7) |
| Klebsiella oxytoca | 17 | 5 | 3 | 25 | 21 | 7 | 22 | 50 | 16 | 5 | 27 | 48 | 123 |
| CR-Klebsiella oxytoca | 1 (5.9) | 1 (20.0) | 1 (33.3) | 3 (12.0) | 3 (14.3) | 0 (0.0) | 7 (31.8) | 10 (20.0) | 3 (18.8) | 1 (20.0) | 7 (25.9) | 11 (22.9) | 24 (19.5) |
| CS-Klebsiella oxytoca | 15 (88.2) | 4 (80.0) | 2 (66.7) | 21 (84.0) | 16 (76.2) | 7 (100.0) | 13 (59.1) | 36 (72.0) | 13 (81.3) | 4 (80.0) | 18 (66.7) | 35 (72.9) | 92 (74.8) |
| CI-Klebsiella oxytoca | 1 (5.9) | 0 (0.0) | 0 (0.0) | 1 (4.0) | 2 (9.5) | 0 (0.0) | 2 (9.1) | 4 (8.0) | 0 (0.0) | 0 (0.0) | 2 (7.4) | 2 (4.2) | 7 (5.7) |
| Citrobacter freundii | 19 | 18 | 5 | 42 | 17 | 5 | 2 | 24 | 15 | 12 | 3 | 30 | 96 |
| CR-Citrobacter freundii | 1 (5.3) | 1 (5.6) | 0 (0.0) | 2 (4.8) | 2 (11.8) | 0 (0.0) | 1 (50.0) | 3 (12.5) | 3 (20.0) | 3 (25.0) | 1 (33.3) | 7 (23.3) | 12 (12.5) |
| CS-Citrobacter freundii | 15 (78.9) | 13 (72.2) | 5 (100.0) | 33 (78.6) | 12 (70.6) | 5 (100.0) | 1 (50.0) | 18 (75.0) | 11 (73.3) | 9 (75.0) | 1 (33.3) | 21 (70.0) | 72 (75.0) |
| CI-Citrobacter freundii | 3 (15.8) | 4 (22.2) | 0 (0.0) | 7 (16.7) | 3 (17.6) | 0 (0.0) | 0 (0.0) | 3 (12.5) | 1 (6.7) | 0 (0.0) | 1 (33.3) | 2 (6.7) | 12 (12.5) |
| Others | 36 (3.0%) | 19 (2.6%) | 12 (4.9%) | 66 (3.0%) | 37 (3.3%) | 9 (1.2%) | 11 (2.2%) | 57 (2.4%) | 37 (4.1%) | 26 (2.6%) | 22 (3.5%) | 85 (3.4%) | 208 (2.9%) |
Note: Other refers to isolates <1% of the total.
Abbreviations: IAI, intra-abdominal infection, UTI, urinary tract infection RTI respiratory tract infection; CS, carbapenem sensitive; CR, carbapenem-resistant; CI, carbapenem-intermediate.
Susceptibility Analysis of Colistin to All Enterobacterales from 2015 to 2017
We found that colistin was less effective against CR-Enterobacterales (76.3%) than against CS-Enterobacterales (92.9%) (Table 2). This pattern was similar for E. coli (CR, 83.5%; CS, 95.9%), K. pneumoniae (CR, 88.6%; CS, 95.8%) and K. aerogenes (CR, 87.5%; CS, 91.9%). (Table 2). The complete list of MIC 90 value for all Enterobacterales is provided in Supplementary Table 1. These findings indicated that CR-Enterobacterales had a lower colistin susceptibility compared with CS-Enterobacterales.
Table 2.
Ranges of MIC50, MIC90 and Susceptible (%), Intermediate (%) and Resistant (%) Rates of Enterobacterales to Carbapenems and Colistin Between 2015 and 2017
| N | Range | MIC50 (mg/L) | MIC90 (mg/L) | S% | I% | R% | |
|---|---|---|---|---|---|---|---|
| All Enterobacterales | |||||||
| Colistin | 7113 | ≤ 1 to > 8 | ≤ 1 | > 4 | 87.7 | 0.0 | 12.3 |
| Ertapenem | 7113 | ≤ 0.03 to > 4 | ≤ 0.06 | > 4 | 83.6 | 3.0 | 13.4 |
| Imipenem | 7113 | ≤ 0.5 to > 32 | ≤ 0.5 | 8 | 83.2 | 5.1 | 11.7 |
| Meropenem | 4910 | ≤ 0.12 to > 16 | ≤ 0.12 | 16 | 88.0 | 0.4 | 11.6 |
| CR-Enterobacterales | |||||||
| Colistin | 1070 | ≤ 1 to > 8 | ≤ 1 | > 4 | 76.3 | 0.0 | 23.7 |
| Ertapenem | 1070 | ≤ 0.03 to > 4 | > 4 | > 4 | 10.7 | 0.5 | 88.9 |
| Imipenem | 1070 | ≤ 0.5 to > 32 | 32 | > 32 | 18.4 | 3.6 | 78.0 |
| Meropenem | 811 | ≤ 0.12 to > 16 | > 16 | > 16 | 27.9 | 2.2 | 69.9 |
| CS-Enterobacterales | |||||||
| Colistin | 5524 | ≤ 1 to > 8 | ≤ 1 | ≤ 1 | 92.9 | 0.0 | 7.1 |
| Ertapenem | 5524 | ≤ 0.03 to 0.5 | ≤ 0.06 | 0.25 | 100.0 | 0.0 | 0.0 |
| Imipenem | 5524 | ≤ 0.5 to 1 | ≤ 0.5 | 1 | 100.0 | 0.0 | 0.0 |
| Meropenem | 3765 | ≤ 0.12 to 1 | ≤ 0.12 | ≤ 0.12 | 100.0 | 0.0 | 0.0 |
| Escherichia coli | |||||||
| Colistin | 3657 | ≤ 1 to > 8 | ≤ 1 | ≤ 1 | 94.6 | 0.0 | 5.4 |
| Ertapenem | 3657 | ≤ 0.03 to > 4 | ≤ 0.06 | 0.5 | 90.6 | 2.7 | 6.7 |
| Imipenem | 3657 | ≤ 0.5 to > 32 | ≤ 0.5 | 1 | 94.8 | 1.4 | 3.9 |
| Meropenem | 2440 | ≤ 0.12 to >16 | ≤ 0.12 | ≤ 0.12 | 95.8 | 0.3 | 3.9 |
| CR-Escherichia coli | |||||||
| Colistin | 254 | ≤ 1 to > 8 | ≤ 1 | > 4 | 83.5 | 0.0 | 16.5 |
| Ertapenem | 254 | ≤ 0.03 to > 4 | > 4 | > 4 | 3.5 | 0.4 | 96.1 |
| Imipenem | 254 | ≤ 0.5 to > 32 | 4 | > 32 | 38.6 | 5.9 | 55.5 |
| Meropenem | 184 | ≤ 0.12 to > 16 | 4 | > 16 | 44.6 | 3.3 | 52.2 |
| CS-Escherichia coli | |||||||
| Colistin | 3273 | ≤ 1 to > 8 | ≤ 1 | ≤ 1 | 95.9 | 0.0 | 4.1 |
| Ertapenem | 3273 | ≤ 0.03 to 0.5 | ≤ 0.06 | 0.25 | 100.0 | 0.0 | 0.0 |
| Imipenem | 3273 | ≤ 0.5to 1 | ≤ 0.5 | ≤ 0.5 | 100.0 | 0.0 | 0.0 |
| Meropenem | 2181 | ≤ 0.12to 0.5 | ≤ 0.12 | ≤ 0.12 | 100.0 | 0.0 | 0.0 |
| Klebsiella pneumoniae | |||||||
| Colistin | 2150 | ≤ 1 to > 8 | ≤ 1 | 2 | 93.4 | 0.0 | 6.6 |
| Ertapenem | 2150 | ≤ 0.03 to > 4 | ≤ 0.06 | > 4 | 73.4 | 2.3 | 24.3 |
| Imipenem | 2150 | ≤ 0.5 to > 32 | ≤ 0.5 | > 32 | 75.6 | 2.7 | 21.7 |
| Meropenem | 1555 | ≤ 0.12 to >16 | ≤ 0.12 | > 16 | 75.9 | 0.6 | 23.5 |
| CR-Klebsiella pneumoniae | |||||||
| Colistin | 534 | ≤ 1 to > 8 | ≤ 1 | 4 | 88.6 | 0.0 | 11.4 |
| Ertapenem | 534 | ≤ 0.06 to > 4 | > 4 | > 4 | 1.9 | 0.4 | 97.8 |
| Imipenem | 534 | ≤ 0.5 to > 32 | > 32 | > 32 | 9.9 | 2.6 | 87.5 |
| Meropenem | 421 | ≤ 0.12 to > 16 | > 16 | > 16 | 11.2 | 2.1 | 86.7 |
| CS-Klebsiella pneumoniae | |||||||
| Colistin | 1529 | ≤ 1 to > 8 | ≤ 1 | ≤ 1 | 95.8 | 0.0 | 4.3 |
| Ertapenem | 1529 | ≤ 0.03to 0.5 | ≤ 0.06 | 0.25 | 100.0 | 0.0 | 0.0 |
| Imipenem | 1529 | ≤ 0.5 to 1 | ≤ 0.5 | 1 | 100.0 | 0.0 | 0.0 |
| Meropenem | 1072 | ≤ 0.12 to 1 | ≤ 0.12 | ≤ 0.12 | 100.0 | 0.0 | 0.0 |
| Enterobacter cloacae | |||||||
| Colistin | 407 | ≤ 1 to > 8 | ≤ 1 | > 4 | 73.5 | 0.0 | 26.5 |
| Ertapenem | 407 | ≤ 0.03 to > 4 | 0.12 | > 4 | 71.5 | 12.0 | 16.5 |
| Imipenem | 407 | ≤ 0.5 to > 32 | ≤ 0.5 | 8 | 82.6 | 6.6 | 10.8 |
| Meropenem | 272 | ≤ 0.12 to > 16 | ≤ 0.12 | 16 | 88.2 | 0.0 | 11.8 |
| CR-Enterobacter cloacae | |||||||
| Colistin | 72 | ≤ 1 to > 8 | ≤ 1 | > 4 | 79.2 | 0.0 | 20.8 |
| Ertapenem | 72 | ≤ 0.06 to > 4 | > 4 | > 4 | 6.9 | 0.0 | 93.1 |
| Imipenem | 72 | ≤ 0.5 to > 32 | 16 | > 32 | 31.9 | 6.9 | 61.1 |
| Meropenem | 49 | ≤ 0.12 to > 16 | 16 | > 16 | 34.7 | 0.0 | 65.3 |
| CS-Enterobacter cloacae | |||||||
| Colistin | 268 | ≤ 1 to > 8 | ≤ 1 | > 4 | 74.6 | 0.0 | 25.4 |
| Ertapenem | 268 | ≤ 0.03 to 0.5 | ≤ 0.06 | 0.5 | 100.0 | 0.0 | 0.0 |
| Imipenem | 268 | ≤ 0.5 to 1 | ≤ 0.5 | 1 | 100.0 | 0.0 | 0.0 |
| Meropenem | 185 | ≤ 0.12 to 0.25 | ≤ 0.12 | ≤ 0.12 | 100.0 | 0.0 | 0.0 |
| Klebsiella aerogenes | |||||||
| Colistin | 146 | ≤ 1 to > 8 | ≤ 1 | ≤ 1 | 92.5 | 0.0 | 7.5 |
| Ertapenem | 146 | ≤ 0.03 to > 4 | 0.25 | 2 | 87.0 | 2.1 | 11.0 |
| Imipenem | 146 | ≤ 0.5 to > 32 | 1 | 2 | 65.1 | 28.1 | 6.9 |
| Meropenem | 108 | ≤ 0.12 to > 16 | ≤ 0.12 | 0.25 | 94.4 | 0.0 | 5.6 |
| CR-Klebsiella aerogenes | |||||||
| Colistin | 16 | ≤ 1 to > 8 | ≤ 1 | > 8 | 87.5 | 0.0 | 12.5 |
| Ertapenem | 16 | 2 to > 4 | > 4 | > 4 | 0.0 | 0.0 | 100.0 |
| Imipenem | 16 | ≤ 0.5 to > 32 | 4 | > 32 | 37.5 | 0.0 | 62.5 |
| Meropenem | 11 | ≤ 0.12 to > 16 | 4 | > 16 | 45.5 | 0.0 | 54.6 |
| CS-Klebsiella aerogenes | |||||||
| Colistin | 86 | ≤ 1 to > 8 | ≤ 1 | 2 | 91.9 | 0.0 | 8.1 |
| Ertapenem | 86 | ≤ 0.03 to 0.5 | 0.25 | 0.5 | 100.0 | 0.0 | 0.0 |
| Imipenem | 86 | ≤ 0.5 to 1 | 1 | 1 | 100.0 | 0.0 | 0.0 |
| Meropenem | 65 | ≤ 0.12 to 0.25 | ≤ 0.12 | 0.25 | 100.0 | 0.0 | 0.0 |
Abbreviations: CS, carbapenem sensitive; CR, carbapenem-resistant; MIC, minimum inhibitory concentration; S, susceptibility; I, intermediate; R, resistance.
MIC Distribution of Colistin to Enterobacterales in IAI, UTI and RTI
For Enterobacterales, E. coli, and K. pneumoniae there was a general trend that isolates from IAIs, UTIs and RTIs which were CS included a higher percentage with MIC values ≤1 and a lower percentage with MIC values >4 compared to CR isolates. However, this was valid for E. cloacae only in IAIs, whereas the percentages of CR- E. cloacae in the MIC values ≤1 groups were higher in UTIs and the same in RTIs compared to CS-E. cloacae (Table 3).
Table 3.
MIC Distribution of Colistin Between 2015 and 2017 in IAI, RTI and UTI Isolates
| Isolates (n, %) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IAI | UTI | RTI | ||||||||||
| Total | MIC (mg/L) | Total | MIC (mg/L) | Total | MIC (mg/L) | |||||||
| ≤1* | 2–4* | >4* | ≤1* | 2–4* | >4* | ≤1* | 2–4* | >4* | ||||
| All Enterobacterales | 3251 (100.0) | 2795 (86.0) | 121 (3.7) | 335 (10.3) | 2483 (100.0) | 2154 (86.7) | 100 (4.0) | 229 (9.2) | 1379 (100.0) | 1108 (80.3) | 85 (6.2) | 186 (13.5) |
| CR-Enterobacterales | 462 (100.0) | 320 (69.3) | 28 (6.1) | 114 (24.7) | 248 (100.0) | 163 (65.7) | 20 (8.1) | 65 (26.2) | 360 (100.0) | 286 (79.4) | 20 (5.6) | 54 (15.0) |
| CS-Enterobacterales | 2548 (100.0) | 2335 (91.6) | 83 (3.3) | 130 (5.1) | 2051 (100.0) | 1893 (92.3) | 74 (3.6) | 84 (4.1) | 925 (100.0) | 783 (84.6) | 61 (6.6) | 81 (8.8) |
| Escherichia coli | 1722 (100.0) | 1605 (93.2) | 59 (3.4) | 58 (3.4) | 1742 (100.0) | 1619 (92.9) | 64 (3.7) | 59 (3.4) | 193 (100.0) | 171 (88.6) | 19 (9.8) | 3 (1.6) |
| CR-Escherichia coli | 138 (100.0) | 112 (81.2) | 6 (4.3) | 20 (14.5) | 89 (100.0) | 64 (71.9) | 8 (9.0) | 17 (19.1) | 27 (100.0) | 24 (88.9) | 2 (7.4) | 1 (3.7) |
| CS-Escherichia coli | 1524 (100.0) | 1442 (94.6) | 49 (3.2) | 33 (2.2) | 1586 (100.0) | 1502 (94.7) | 53 (3.3) | 31 (2.0) | 163 (100.0) | 144 (88.3) | 17 (10.4) | 2 (1.2) |
| Klebsiella pneumoniae | 884 (100.0) | 802 (90.7) | 39 (4.4) | 43 (4.9) | 430 (100.0) | 374 (87.0) | 28 (6.5) | 28 (6.5) | 836 (100.0) | 746 (89.2) | 48 (5.7) | 42 (5.0) |
| CR-Klebsiella pneumoniae | 193 (100.0) | 160 (82.9) | 13 (6.7) | 20 (10.4) | 95 (100.0) | 73 (76.8) | 9 (9.5) | 13 (13.7) | 246 (100.0) | 218 (88.6) | 11 (4.5) | 17 (6.9) |
| CS-Klebsiella pneumoniae | 657 (100.0) | 617 (93.9) | 22 (3.3) | 18 (2.7) | 307 (100.0) | 276 (89.9) | 17 (5.5) | 14 (4.6) | 565 (100.0) | 512 (90.6) | 35 (6.2) | 18 (3.2) |
| Enterobacter cloacae | 243 (100.0) | 165 (67.9) | 10 (4.1) | 68 (28.0) | 69 (100.0) | 55 (79.7) | 5 (7.2) | 9 (13.0) | 95 (100.0) | 68 (71.6) | 8 (8.4) | 19 (20.0) |
| CR-Enterobacter cloacae | 37 (100.0) | 22 (59.5) | 4 (10.8) | 11 (29.7) | 17 (100.0) | 16 (94.1) | 1 (5.9) | 0 (0.0) | 18 (100.0) | 13 (72.2) | 4 (22.2) | 1 (5.6) |
| CS-Enterobacter cloacae | 162 (100.0) | 117 (72.2) | 4 (2.5) | 41 (25.3) | 40 (100.0) | 33 (82.5) | 3 (7.5) | 4 (10.0) | 66 (100.0) | 48 (72.7) | 3 (4.5) | 15 (22.7) |
| Klebsiella aerogenes | 80 (100.0) | 74 (92.5) | 1 (1.3) | 5 (6.3) | 30 (100.0) | 26 (86.7) | 1 (3.3) | 3 (10.0) | 36 (100.0) | 33 (91.7) | 3 (8.3) | 0 (0.0) |
| CR-Klebsiella aerogenes | 11 (100.0) | 9 (81.8) | 0 (0.0) | 2 (18.2) | 2 (100.0) | 2 (100.0) | 0 (0.0) | 0 (0.0) | 3 (100.0) | 3 (100.0) | 0 (0.0) | 0 (0.0) |
| CS-Klebsiella aerogenes | 43 (100.0) | 39 (90.7) | 1 (2.3) | 3 (7.0) | 18 (100.0) | 16 (88.9) | 1 (5.6) | 1 (5.6) | 25 (100.0) | 22 (88.0) | 3 (12.0) | 0 (0.0) |
Notes: The data are expressed as n (%). *MIC breakpoint (mg/L). MIC breakpoint (mg/L) of colistin to Enterobacterales for sensitivity is ≤2; resistance is >2 according to European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters Version 7.1, valid from 2017-03-10.
Abbreviations: MIC, minimum inhibitory concentration; IAI, intra-abdominal infection; UTI, urinary tract infection; RTI respiratory tract infection; CS, carbapenem sensitive; CR, carbapenem-resistant.
Susceptibility Changes of Enterobacterales to Colistin in IAI, UTI and RTI Isolates from 2015 to 2017
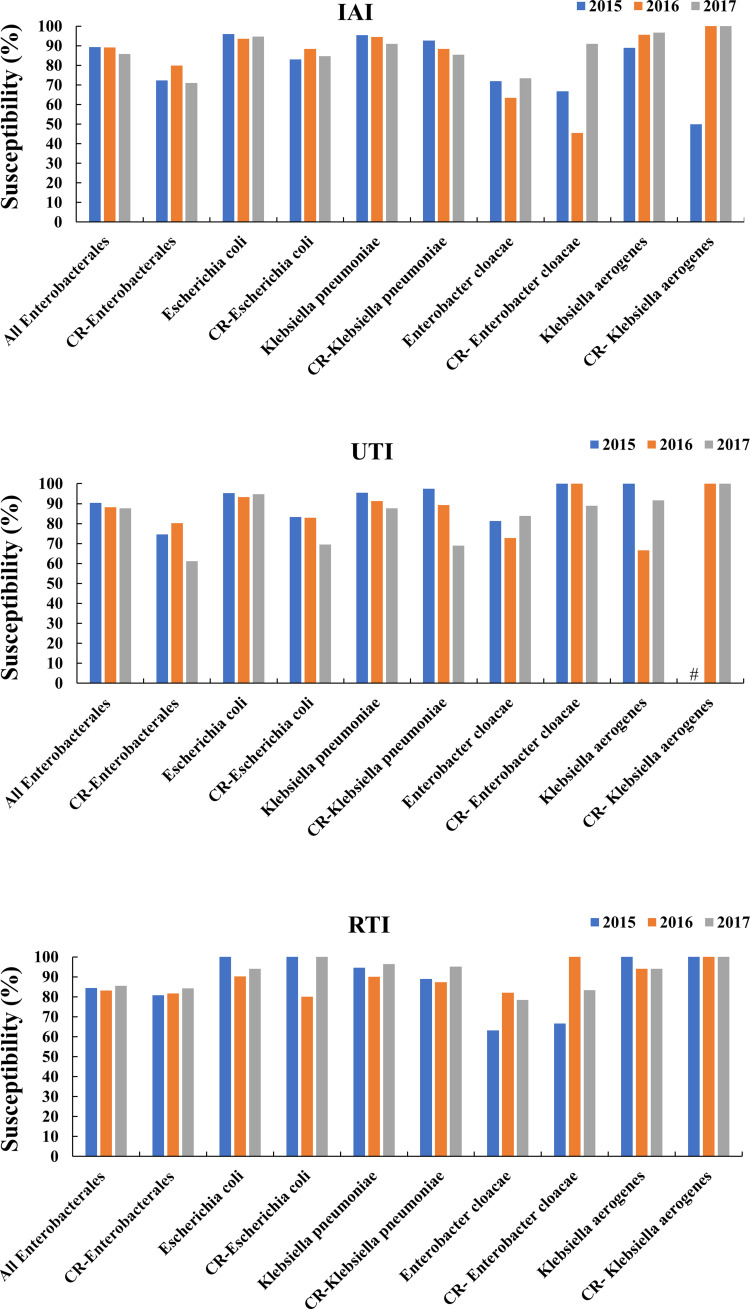
In IAI and UTI isolates, all Enterobacterales, and particularly CR-E. coli and CR-K. pneumoniae, showed a trend of decreasing susceptibility. Especially for CR-K. pneumoniae in IAIs and for both in UTIs, the susceptibility decreased markedly in 2017. In RTI isolates, unlike UTI and IAI isolates, all Enterobacterales and CR-Enterobacterales showed trends of increasing or fairly constant susceptibilities. However, the vast majority of isolates from RTIs exhibited sensitivities of ≥80.0% (Figure 1).
Figure 1.
Changes in the of indicated Enterobacterales sensitivity to colistin over time for the indicated infection types.
Note: #means that the susceptibility of CR-K. aerogenes in 2015 was not available.
Abbreviations: IAI, intra-abdominal infections; UTI, urinary tract infections; RTI, respiratory tract infections; CR, carbapenem-resistant.
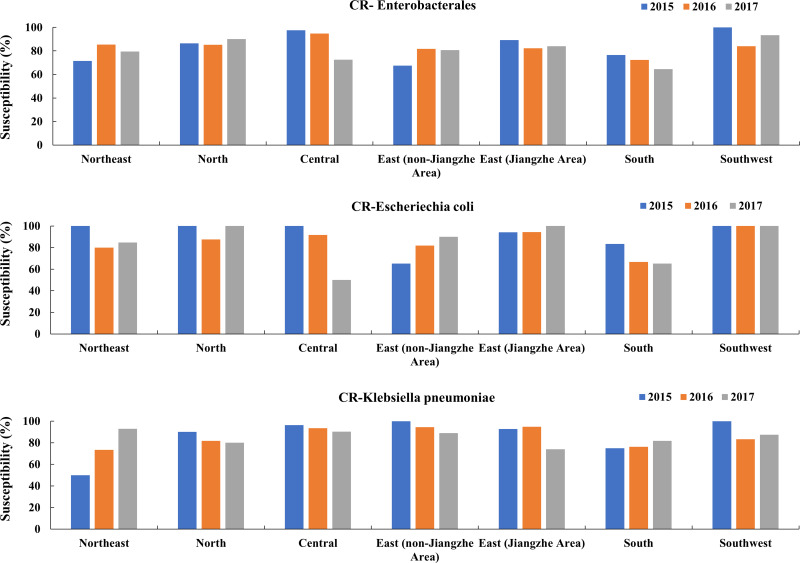
Susceptibility Changes of CR-Enterobacterales to Colistin in Different Regions of China in IAI, UTI and RTI Isolates from 2015 to 2017
In south and central areas, the susceptibility rates of CR-Enterobacterales to colistin decreased yearly becoming <80.0% (64.5% and 72.5%, respectively). The susceptibility of CR-Enterobacterales to colistin increased from 2016 to 2017 in the north, east (Jiangzhe) and southwest areas. In IAI isolates, the susceptibility rates to colistin in the northeast and east (non-Jiangzhe) areas increased year by year. In UTI isolates, the susceptibility rates to colistin increased in the northeast, north, central, east (non-Jiangzhe) and east (Jiangzhe) areas from 2016 to 2017. In RTI, the susceptibility rates to colistin in the northeast and central areas decreased year by year (Figure 2). The susceptibility of CR-E. coli to colistin increased in the east (Jiangzhe) and east (non-Jiangzhe) areas, and decreased in the central and south areas. In 2017 the susceptibility rate to colistin in the central area was only 50.0%. The susceptibility of CR-K. pneumoniae to colistin increased in the northeast, south and southwest areas, and exhibited a downward trend in the north, central, east (non-Jiangzhe) and east (Jiangzhe) regions from 2016 to 2017 (Figure 2). MIC 90 values and the susceptibilities to colistin of CR-Enterobacterales, CR-E. coli and CR-K. pneumoniae isolated from IAIs, UTIs and RTIs in different Chinese regions between 2015 and 2017 are shown in Supplementary Figure 1 and in Supplementary Table 2.
Figure 2.
Changes in the susceptibility of CR-Enterobacterales, CR-E. coli and CR-K. pneumoniae to colistin over time in different regions of China.
Abbreviation: CR, carbapenem-resistant.
Discussion
According to the Centre for Disease Dynamics, Economics and Policy (CDDEP), 80% of Indian K. pneumoniae isolates were up to 60% resistant to carbapenems. In Italy the prevalence of CR-K. pneumoniae isolates, which did not exist in 2008, had risen to 60% by 2013 (CDDEP: https://cddep.org/). This rapid rise in carbapenem resistance reflects a worrisome trend. In the present study, we observed that CR-Enterobacterales comprised 15% of all Enterobacterales collected in IAI, UTI and RTI isolates from 2015 to 2017. In particular, the proportion of CR-K. pneumoniae in all K. pneumoniae IAI, UTI and RTI isolates increased from 19% in 2015 to 24.3% and 24.8%, respectively in 2017.
Polymyxins, introduced in the 1950s to treat infections caused by Gram-negative bacteria, exert bactericidal activity by displacing membrane-stabilizing calcium and magnesium ions and targets the polyanionic lipopolysaccharide components.16,17 Despite their toxicity, they are considered to be one of the treatments of last resort for CR-Enterobacterales infections.18
Several studies investigated resistance to colistin between 2005 and 2010 and reported resistance rates of 6.8% for K. pneumoniae in South Korea,19 0.09% for E. coli, 1.2% for K. pneumoniae and 1.5% for E. cloacae in Tunisia.20 A global survey reported colistin susceptibility rates of 97–99% for K. pneumoniae and E. coli isolates, respectively in North America, Europe, Latin America and Asia pacific.21 Available data from China collected in 2009 are 7.5% resistance of CR-Enterobacterales to colistin.22
Later studies, however, revealed a rise of colistin resistant rates particularly for CR-K. pneumoniae strains in countries such as Greece (20.8% in 2010),23 Spain (31.70% in 2012)24 and Italy (36.1% in 2011 and 43% in 2013–2014).10,25 The overall susceptibility rates of E. coli and K. pneumoniae to colistin were ≥80% throughout the years 2015 to 2017 in the present study, but particularly for CR-K. pneumoniae strains from IAI isolates, and CR-K. pneumoniae and CR-E. coli isolates from UTIs, the colistin susceptibilities decreased in 2017. This decrease might be attributed to the increased treatment of infections caused by CR-Enterobacterales with colistin, which has been used in humans in China since 2015. In addition, the local distribution of CR-E. coli and CR-K. pneumoniae in 7 regions of China showed that in the east Jiangzhe area, susceptibility of CR-E. coli to colistin was >80.0% throughout the study years, but it is noteworthy that CR-K. pneumoniae susceptibility decreased in 2017 compared to previous years to <80%, which might indicate enhanced colistin usage in this region where the 2013 incidence of CR-K. pneumoniae was the highest in China.26 On the other hand, it should be considered that other studies have suggested that the cause of colistin resistance is excessive veterinary usage.22,27–29 In addition, previous studies about PK/PD parameters of colistin indicated that sub-optimal doses of colistin might have increased the resistance rates to colistin with persistent levels being organ specific. There were high levels in muscle, kidney and liver, but poor distribution to the cerebrospinal fluid, bones, lung parenchyma and pleural cavity, which might lead to organ specific susceptibility differences,30 though in the present study RTI isolates showed consistent susceptibility rates of ≥80.0% throughout the years.
Limitations of the present study are that the paper only provides details about phenotypical testing and completely omits strain typing, such as MLST, detection of resistance genes and others.
Conclusions
Colistin is a last resort antibiotic for empirical CR-Enterobacterales therapy. E. coli (51.4%) and K. pneumoniae (30.0%) with percentages of CR-K. pneumoniae and CR-E. coli strains of 24.8% and 6.9%, accounted for the majority of Enterobacterales isolated from IAIs, UTIs and RTIs in 2015–2017. Colistin susceptibilities were generally higher in CS than in CR isolates and were 83.5% for CR-E. coli and 88.6% for CR-K. pneumoniae, respectively. However, the resistance rates of IAI derived CR-K. pneumoniae as well as CR-E. coli and CR-K. pneumoniae UTI isolates to colistin increased between 2015 and 2017, which should further be closely monitored.
Acknowledgments
Medical writing and editorial assistance was provided by Shanghai BIOMED Science Technology (Shanghai, China) through funding provided by MSD China. The authors were solely responsible for the conception and implementation of this study and for writing the manuscript.
Funding Statement
This study was supported by funding from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA. Further support was provided by the National Key Research and Development Program of China (2018YFC1200100, 2018YFC1200105), the Chinese Academy of Medical Sciences (CAMS) Initiative for Innovative Medicine (Grant No. 2016-I2M-3-014), the CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant No. 2016-I2M-1-014) and the Outstanding Talents Training Funding Project of Dongcheng District, Beijing (2017).
Data Sharing Statement
The SMART database is not public and is only accessible for SMART investigators, but the data that support the findings of this study are directly available from MSD China or from the authors upon reasonable request and with permission of MSD China.
Ethics Approval and Consent to Participate
The protocol has been reviewed by the human research ethics committee of the Institutional Review Board (IRB) of the Peking Union Medical College Hospital and since the project falls under the category observational study and all bacterial strains were from residual samples used in clinical diagnosis or were strains from their subcultures, it has been determined they meet the criteria for exemption. This project does not involve any patient information nor does it affect the normal diagnosis and treatment of patients, and after consultation with the IRB, formal ethical approval was reviewed and waived and written patient consent was not required (Ethics Approval Number: S-K238).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
Disclosure
Yue Kang is an employee of MSD China. The authors report no other conflicts of interest in this work.
References
- 1.WHO. Antimicrobial resistance: global report on surveillance; 2014. Available from: http://www.who.int/drugresistance/documents/surveillancereport/en. Accessed June5, 2020.
- 2.Schwaber MJ, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: a potential threat. JAMA. 2008;300(24):2911–2913. [DOI] [PubMed] [Google Scholar]
- 3.Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi: 10.1093/infdis/jiw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wise MG, Horvath E, Young K, Sahm DF, Kazmierczak KM. Global survey of Klebsiella pneumoniae major porins from ertapenem non-susceptible isolates lacking carbapenemases. J Med Microbiol. 2018;67(3):289–295. doi: 10.1099/jmm.0.000691 [DOI] [PubMed] [Google Scholar]
- 5.Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao YH, Wang J, Li Y. Bacterial resistance surveillance in China: a report from Mohnarin 2004–2005. Eur J Clin Microbiol Infect Dis. 2008;27(8):697–708. doi: 10.1007/s10096-008-0494-6 [DOI] [PubMed] [Google Scholar]
- 7.Hu FP, Guo Y, Zhu DM, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect. 2016;22(Suppl 1):S9–S14. doi: 10.1016/j.cmi.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 8.Biedenbach D, Bouchillon S, Hackel M, et al. Dissemination of NDM metallo-β-lactamase genes among clinical isolates of Enterobacteriaceae collected during the SMART global surveillance study from 2008 to 2012. Antimicrob Agents Chemother. 2015;59(2):826–830. doi: 10.1128/AAC.03938-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. Critically important antimicrobials for human medicine, 5th revision; 2017. Available from: https://www.who.int/foodsafety/publications/antimicrobials-fifth/en/. Accessed June5, 2020.
- 10.Capone A, Giannella M, Fortini D, et al. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect. 2013;19(1):E23–E30. doi: 10.1111/1469-0691.12070 [DOI] [PubMed] [Google Scholar]
- 11.Qamar S, Shaheen N, Shakoor S, Farooqi J, Jabeen K, Hasan R. Frequency of colistin and fosfomycin resistance in carbapenem-resistant Enterobacteriaceae from a tertiary care hospital in Karachi. Infect Drug Resist. 2017;10:231–236. doi: 10.2147/IDR.S136777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 13.Aghapour Z, Gholizadeh P, Ganbarov K, et al. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist. 2019;12:965–975. doi: 10.2147/IDR.S199844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Testing TECoAS. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1, 2017; 2017. Available from: http://www.eucast.org. Accessed June5, 2020.
- 15.Solomkin JS, Mazuski JE, Bradley JS, et al. Diagnosis and management of complicated intra-abdominal infection in adults and children: guidelines by the Surgical Infection Society and the Infectious Diseases Society of America. Clin Infect Dis. 2010;50(2):133–164. doi: 10.1086/649554 [DOI] [PubMed] [Google Scholar]
- 16.Newton BA. The properties and mode of action of the polymyxins. Bacteriol Rev. 1956;20(1):14–27. doi: 10.1128/MMBR.20.1.14-27.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schindler M, Osborn MJ. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry. 1979;18(20):4425–4430. doi: 10.1021/bi00587a024 [DOI] [PubMed] [Google Scholar]
- 18.Sheu -C-C, Chang Y-T, Lin S-Y, Chen Y-H, Hsueh P-R. Infections caused by carbapenem-resistant enterobacteriaceae: an update on therapeutic options. Front Microbiol. 2019;10:80. doi: 10.3389/fmicb.2019.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suh JY, Son JS, Chung DR, Peck KR, Ko KS, Song JH. Nonclonal emergence of colistin-resistant Klebsiella pneumoniae isolates from blood samples in South Korea. Antimicrob Agents Chemother. 2010;54(1):560–562. doi: 10.1128/AAC.00762-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mezghani Maalej S, Rekik Meziou M, Mahjoubi F, Hammami A. Epidemiological study of Enterobacteriaceae resistance to colistin in Sfax (Tunisia). Med Mal Infect. 2012;42(6):256–263. doi: 10.1016/j.medmal.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 21.Gales AC, Jones RN, Sader HS. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY antimicrobial surveillance program (2006-09). J Antimicrob Chemother. 2011;66(9):2070–2074. doi: 10.1093/jac/dkr239 [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Hu F, Zhang X, et al. Independent emergence of colistin-resistant Enterobacteriaceae clinical isolates without colistin treatment. J Clin Microbiol. 2011;49(11):4022–4023. doi: 10.1128/JCM.01233-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ah Y-M, Kim A-J, Lee J-Y. Colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents. 2014;44(1):8–15. doi: 10.1016/j.ijantimicag.2014.02.016 [DOI] [PubMed] [Google Scholar]
- 24.Pena I, Picazo JJ, Rodríguez-Avial C, Rodríguez-Avial I. Carbapenemase-producing Enterobacteriaceae in a tertiary hospital in Madrid, Spain: high percentage of colistin resistance among VIM-1-producing Klebsiella pneumoniae ST11 isolates. Int J Antimicrob Agents. 2014;43(5):460–464. doi: 10.1016/j.ijantimicag.2014.01.021 [DOI] [PubMed] [Google Scholar]
- 25.Monaco M, Giani T, Raffone M, et al. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Euro Surveill. 2014;19(42). doi: 10.2807/1560-7917.ES2014.19.42.20939. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Yang Q, Liao K, et al. Antimicrobial susceptibilities of aerobic and facultative gram-negative bacilli from intra-abdominal infections in patients from seven regions in China in 2012 and 2013. Antimicrob Agents Chemother. 2016;60(1):245–251. doi: 10.1128/AAC.00956-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016;21(9):30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155 [DOI] [PubMed] [Google Scholar]
- 28.Yang YQ, Li YX, Song T, et al. Colistin resistance gene mcr-1 and its variant in escherichia coli isolates from chickens in China. Antimicrob Agents Chemother. 2017;61(5). doi: 10.1128/AAC.01204-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Wang Y, Wang Y, Zhang S, Shen Z, Wang S. Emergence of the colistin resistance gene mcr-1 and its variant in several uncommon species of Enterobacteriaceae from commercial poultry farm surrounding environments. Vet Microbiol. 2018;219:161–164. doi: 10.1016/j.vetmic.2018.04.002 [DOI] [PubMed] [Google Scholar]
- 30.Gurjar M. Colistin for lung infection: an update. J Intensive Care. 2015;3(1):3. doi: 10.1186/s40560-015-0072-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- WHO. Antimicrobial resistance: global report on surveillance; 2014. Available from: http://www.who.int/drugresistance/documents/surveillancereport/en. Accessed June5, 2020.
- WHO. Critically important antimicrobials for human medicine, 5th revision; 2017. Available from: https://www.who.int/foodsafety/publications/antimicrobials-fifth/en/. Accessed June5, 2020.
- Testing TECoAS. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1, 2017; 2017. Available from: http://www.eucast.org. Accessed June5, 2020.