Abstract
Glutathione-S transferase (GST) is a most ancient protein superfamily of multipurpose roles and evolved principally from gene duplication of an ancestral GSH binding protein. They have implemented in diverse plant functions such as detoxification of xenobiotic, secondary metabolism, growth and development, and majorly against biotic and abiotic stresses. The vital structural features of GSTs like highly divergent functional topographies, conserved integrated architecture with separate binding pockets for substrates and ligand, the stringent structural fidelity with high Tm values (50º–60º), and stress-responsive cis-regulatory elements in the promoter region offer this protein as most flexible plant protein for plant breeding approaches, biotechnological applications, etc. This review article summarizes the recent information of GST evolution, and their distribution and structural features with emphasis on the assorted roles of Ser and Cys GSTs with the signature motifs in their active sites, alongside their recent biotechnological application in the area of agriculture, environment, and nanotechnology have been highlighted.
Keywords: Active site residues, Detoxification, Evolution, Glutathionylation, Ser, cys GST
Introduction
Plant GSTs and their relevance
Plant glutathione-S transferases (GSTs; EC 2.5.1.1.8) are key phase II detoxification enzymes that work downstream of Cyt P450s in cellular metabolism. These enzymes are universally reported in eukaryotes and prokaryotes. Earlier, GSTs were reported to be found specifically in the cytosol (Frova 2003), but later, with the concurrent research, two GSTs, Nt ParA in tobacco, and GTSU12 in Arabidopsis thaliana, were first found to be present in the nucleus (Zettl et al. 1994; Dixon et al. 2009). Currently, GSTs are also reported in chloroplasts, mitochondria, and also in the ER, etc. (Lallement et al. 2014a, b). In kingdom planta, GSTs were first documented in the 1970s in maize that was found to be involved in conjugation reaction and hence detoxify the chloro-s-triazine photosystem II inhibitor atrazine in maize (Frova 2003). In plants, several GSTs have been acknowledged for their functional involvement in biotic and abiotic stress management; plant growth and development, phytohormone signaling, cell signaling and regulation of redox homeostasis, biosynthesis, and transport of secondary metabolites such as anthocyanin as well as controlled cell death (apoptosis) (Nianiou-Obeidat et al. 2017; Chronopoulou et al. 2017a; Chen et al. 2007; Singhal et al. 2015; Singhal et al. 2015; Dixon et al. 2010 and Hu et al. 2016). The nucleophilic attack of the thiol group of the tripeptide glutathione GSH (γ-Glu-Cys-Gly) to various electrophilic molecules and hydrophobic toxic molecules is mainly catalyzed by GSTs. Glutathionylation reaction is important in herbicide selectivity because both cereal and broadleaf crops holding high GST activities are tolerant to herbicides (Chronopoulou et al. 2017b). According to various biochemical studies, in contrast to the glutathionylation activity, several GSTs despite catalyzing glutathionylation activity perform deglutathionylation. This is due to the differential active site residue replacement from Ser to Cys. It is also revealed that GSTs bind hormones such as auxin and cytokinin and can be induced by a wide variety of phytohormones, including ethylene, auxin, MeJ, SA, and abscisic acid (Smith et al. 2003; Shi et al. 2014). Certain GSTs like tau, phi, theta, and zeta exhibit peroxidase activity (Zheng et al. 2008). The involvement of GSTs in endogenous metabolism is less understood. However, some secondary metabolites such as steroids, leukotriene, sulfur-containing volatiles, and glucosinolates are synthesized by some isoenzymes (Nianiou-Obeidat et al. 2017; Hu et al. 2016).
Plant GSTs classification and distribution
GSTs are a huge gene family of multidisciplinary functions. The GST protein family consists of three members: cytosolic GSTs, mitochondrial GSTs, and microsomal GSTs. Among them, the cytosolic GSTs are great in number compared to the other two families. From plants, animals, fungi, and bacteria, a total of 36 GST classes have been recognized. Based on the phylogenetic study of photosynthetic organisms a refined classification was proposed. Cd00570 Sequence Cluster of the “conserved domains” tool in NCBI was used as a source, which includes the GST classes that contain the typical N-terminal thioredoxin (Trx) domain with a β1α1β2α2β3β4α3 topology and a C-terminal all-helical domain that together form a typical GST fold. By taking the sequences of all GSTs from a model organisms such as Pinus tabulaeformis (gymnosperm), A. thaliana, Populus trichocarpa, Oryza sativa, Solanum lycopersicum, and Hordeum vulgare (angiosperms), Selaginella moellendorffii (lycophyte), and P. patens (bryophyte) for phylogenetic analysis it is concluded that eukaryote photosynthetic organisms can be classified into 14 classes. Among them, Tau, Phi, Zeta, Theta, and TCHQ classes contain GSTs with a serine as active site residue. The nature of the catalytic residue in the EF1Bγ and Ure2p classes is not yet well known. The other seven classes Iota (GSTIs), Hemerythrin (GSTHs), DHARs, Lambda (GSTLs), GHRs, mPGES-2 s, and metaxins belong to Cys- GSTs as they contain members that possess a very conserved cysteine in the active site motif (Lallement et al. 2014a, b). The cysteine of the CPxC signature is conserved to plants only. Initially, mPGES-2 s were not considered as GSTs because they showed a low similarity with GSTs identified at that time, and also GSH was not required as their substrate for a functional activity like in the isomerization of ProstaGlandinH2 (PGH2) (Tanikawa et al. 2002). However, by extensive analysis it was proved that mPGES-2 s do belong to the GST family based on (i) its typical GST structure, (ii) the credentials of another activity requiring GSH, and (iii) the identification of additional more closely related Cys-GSTs (Yamada et al. 2005; Takusagawa et al. 2013). Moreover, particular proteins that are listed as putative GST members under the name “2-GST_N” possess two repeated N-terminal Trx domains and a rather conserved CPFC motif, which lack the C-terminal domain (Lallement et al. 2014a, b).
In vascular plants, maize was the first which was reported with GSTs. After then, GSTs have been identified in several crops. To date, soybean is having the highest number of GSTs 101, with a total of eight classes. Additionally, Arabidopsis, rice, barley, wheat, poplar, mungbean, and Medicago contain a total 55, 82, 84, 52, 81, 44, and 73 GSTs, respectively. The tau and phi GSTs are the most numerous among all the plants. Exceptionally, wheat contains lesser tau GSTs represented by eight GST genes. Zeta and theta GSTs are present in both animals and plants. The number of genes for DHARs is ranging from two to four. Both DHAR and lambda class GSTs function as thiol-transferases and are central to terrestrial plants. Only one TCHQD GST has been identified in the genome of Arabidopsis, rice, poplar, barley, tomato, and broccoli while a maximum of three is known in soybean. It is localized in the plasma membrane. The functional characterization of this membrane-bound protein, TCHQD is so far not clear. This GST family member showed (25% identity) resemblance to the TCHQD enzyme recognized in Sphingobium chlorophenolicum, which catalyzes the reductive dehalogenation of TCHQ and trichloro hydroquinone, key steps in the degradation of the pesticide pentachlorophenol (Warner et al. 2008). Microsomal class of GST belongs to MAPEG superfamily and due to their membrane-bound localization, they perform the differential function from cytosolic GSTs (Basantani et al. 2011). A new class of GST, Omega GSTs, was reported in V. radiata. but authors have detected error in Vaish et al. (2018) because it contain CPFA motif that it is the key feature of glutathionyl hydroquinone reductases (GHR) family and hence the reported V. radiata omega GST belongs to the well characterized glutathionyl hydroquinone reductases (GHR) and not to the Omega class.
Earlier there was no report on GSTs gene family or their molecular characterization in nonvascular plants. But from the past severals years GST gene family has been identified and characterized (Table 1) in bryophyte and gymnosperms. The presence of GSTs has also been reported in lower organisms like fungi algae, etc.
Table 1.
Distribution of GSTs in different plant species
| Plant GSTs | Types of GSTs | References | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tau | Phi | Theta | Zeta | Lambda | DHAR | TCHQD | EF1Bγ | GHR | Iota | Hemerythrin | Other | Total | ||
| Arabidopsis | 28 | 13 | 2 | 2 | 3 | 4 | 1 | 2 | 4 | – | – | 1 | 60 | Wagner et al. (2002), Lallement et al. (2014a, b) |
| (A. thaliana isoforms) | 7 | 4 | – | – | – | – | – | – | – | – | – | – | 11 | Sylvestre-Gonon et al. (2019) |
| Rice | 52 | 17 | 1 | 4 | 3 | 2 | 1 | 2 | – | – | – | – | 82 | Jain et al. 2010 |
| Soybean | 63 | 14 | 3 | 3 | 7 | 4 | 3 | 4 | – | – | – | – | 101 | Liu et al. (2015) |
| Maize | 28 | 12 | – | 2 | – | – | – | – | – | – | – | – | 42 | McGonigle et al. (2000) |
| (Z. mays isoforms) | 2 | 1 | – | – | – | – | – | – | – | – | – | 4 | 7 | Sylvestre-Gonon et al. (2019) |
| Wheat | 8 | 25 | 9 | 10 | - | - | – | – | – | – | – | – | 52 | Gallé et al. (2005) |
| Poplar | 58 | 9 | 2 | 2 | 3 | 3 | 1 | 3 | – | – | – | – | 81 | Lan et al. (2009) |
| Tomato | 56 | 5 | 4 | 2 | 5 | 6 | 1 | 1 | – | – | – | 1 | 81 | Csiszar et al. (2014) |
| Broccoli | 28 | 14 | 2 | 2 | 3 | 4 | 1 | 3 | 5 | – | – | 3 | 65 | Vijayakumar et al. (2016) |
| Barley | 50 | 21 | 1 | 5 | 2 | 2 | 1 | 2 | – | – | – | – | 84 | Rezaei et al. (2013) |
| Vigna | 12 | 4 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | – | – | 4 | 31 | Vaish et al. (2018) |
| Moss | 0 | 10 | 3 | 1 | 1 | 3 | 5 | 4 | – | 1 | 8 | 1 | 37 | Liu et al. (2013) |
| Larix | 11 | 7 | 1 | 2 | 3 | 1 | 1 | 1 | – | – | – | – | 27 | Yang et al. (2014) |
| Dracaena | 9 | 5 | 1 | – | 1 | 2 | 1 | – | 1 | – | – | – | 20 | Zhu et al. (2016) |
| Sweet orange | 29 | 20 | 2 | 3 | 6 | – | – | – | – | – | – | 1 | 61 | Licciardello et al. (2014) |
| Capsicum | 59 | 4 | 2 | 4 | 2 | 1 | 2 | 3 | – | – | 4 | 2 | 85 | Islam et al. (2019) |
| Sweet potato | 27 | 2 | 2 | 3 | 3 | 2 | – | 3 | – | – | – | – | 42 | Ding et al. (2017) |
| Chinese cabbage | 40 | 22 | 5 | 3 | 4 | 4 | 1 | – | – | – | – | – | 79 | Du et al. (2018) |
The different gene numbers of their respective classes of the plant organisms. There are no data depicted in context to pseudogenes. The isoforms of Arabidopsis and maize have also been shown
GSTs in lower organisms
Whole work on GST gene family which is reported focused mainly on vascular plants, especially on agricultural plants. Providentially, the data of the sequenced genome of the nonvascular plant P. patens (moss) allowed to molecularly characterize the whole GST gene family. It is the only bryophyte whose whole genome has been sequenced till now. In P. patens 37 GST genes were identified; out of these, 37 GSTs and 28 GSTs were clustered into eight GST classes: the phi, DHAR, theta, zeta, lambda, EF1Bγ, Ure2p, and TCHQD classes, and the rest nine P. patens GSTs were grouped into two separate clades called iota and hemerythrin, the two novel GSTs classes. In P. patens GST classes, phi, and hemerythrin GSTs were the most abundant, with ten and eight members, respectively. The TCHQD and EF1Bγ classes had five and four members, respectively; both the DHAR and theta classes had three members; and the zeta, lambda, Ure2p, and iota classes were represented by one member each (Liu et al. 2013). Gymnosperms are such a large group of plants with a great evolutionary past. However, due to the lack of genome information, natural enzyme family evolution in conifers is limited. P. tabuliformis (pine) is the first gymnosperm in which the GST gene family was characterized. Based on the NCBI conserved domain analysis, the 44 genes predicted to be GSTs were confirmed divided into eight classes: Tau, Phi, Theta, Zeta, DHAR, Lambda, TCHQD, and EF1B. The Tau GSTs were highly abundant (26) followed by seven Phi and three Lambda classes, then three members each of Zeta, DHAR, and EF1B classes, and finally one member each of Theta and TCHQD classes (Lan et al. 2013). Larix kaempferi is the second gymnosperm in which a total of 27 GST genes were cloned and classified into eight classes. Tau GSTs are the most numerous among all other classes (Yang et al. 2014).
Distribution of GSTs was reported in Fucus species and its varieties (Fucus ceranoides, Fucus spiralis var. platycarpus, Fucus spiralis var. spiralis and Fucus vesiculosus var. vesiculosus) and the potential of GST activity of Fucus as a biomarker of environmental contamination in coastal zones and estuaries (Cairrao et al. 2004). The first examination of GST in freshwater algae (Chlamydomonas sp., Chlorella sp., Pediastrum sp., and Scenedesmus quadricauda,) and diatoms including Cyclotella gamma, Cyelotella meneghiniana, Synedra acus, and Synedra radians were proposed. The enzyme was relatively stable at high temperatures and exhibited higher activity under alkaline conditions (Tang et al. 1998).
GSTs in diverse plant functions
Plant growth and development
GSTs are known to be induced by phytohormones such as auxins, cytokinin, SA, methyl jasmonate, ethylene, and other hormones (Gong et al. 2005; Moons 2005) implies that plant GSTs may play dynamic roles in plant growth and development. A systematic analysis was done by Jain et al. (2010) in response to different phytohormones by taking 74GST genes of rice and it was found that all these genes were responsive to those hormones. Gong et al. (2005), demonstrated the role of glutathione S-transferase in plant growth and development and shoot morphogenesis in vivo and in vitro, respectively. Kumar et al. (2013), developed the Arabidopsis transgenic variety expressing rice lambda GST gene OsGSTL2 which was found to be involved in early plant growth and development, especially in seed germination. During their study, it was found that though transgenic lines did not show much phenotypic difference when grown in soil rite, whenseeds were germinated on half-strength MS media, then during the growth of the seedling, increased root length was observed for the transgenic lines as compared to the wild type. The rate of germination of seedlings was about twofold for the transgenic lines compared to the wild type. The same results were observed in cotyledon development. These results suggest the role of OsGSTL2 in early plant development, exclusively in seed germination.
Jiang et al. (2010), reported a tau class GST from Arabidopsis AtGSTU17 which is involved in seedling development, hypocotyl elongation, anthocyanin accumulation, and far-red light-mediated inhibition of greening. It is revealed by RT-PCR that the expression of AtGSTU17 is controlled by various photoreceptors, especially phytochrome A (phyA) under all light conditions and different phytohormones, including auxin and ABA. Shi et al. (2014) depicted the role of the two Pear (Pyrus pyrifolia) GST genes,PpGST1 and PpGST2, in fruit ripening and senescence of pear. During fruit development, the expression activity of PpGST2 gene was higher than in PpGST1. Moreover, PpGST2 transcripts were detected at a relatively high level in 140 days after full bloom fruit, suggesting that PpGST2 gene might be involved in pear fruit ripening. In this study, it is also reported that when the pear fruit was treated with glucose, salicylic acid (SA), and indole-3-acetic acid (IAA), the expression level of PpGST1 and PpGST2 gets induced. The same result was reported with the diseased fruit. Conclusively, it can be suggested that these two genes (PpGST1 and PpGST2) play a role in response to sugar, SA, and IAA signaling during fruit development.
Plant secondary metabolism
Interestingly, GSTs are also involved in secondary metabolism and signaling. The GST protein of Arabidopsis is not only involved in conjugating natural products but also signaling and transport of secondary metabolites. For instance, AtGSTF12 is involved in anthocyanin and proanthocyanidin accumulation (Kitamura et al. 2004), whereas AtGSTU20 modulates responses to light reception (Chen et al. 2007). Different Phi GSTs of the same organism A. thaliana perform different roles. For example, AtGSTF2 make a tight association with indole-derived phytoalexin, camalexin and may be involved in its transport Arabidopsis GSTF2 selectively bind the flavonol quercetin-3- O-rhamnoside, suggesting a role in regulating the binding and transport of defense-related compounds in plants (Dixon et al. 2011), whereas a defense compound camalexin is synthesized by catalyzing the conjugation of glutathione onto indole-3-acetonitrile (Su et al. 2011) by the AtGSTF6. On the other hand, AtGSTF8 catalyzes glutathione conjugation to two stress signaling molecules, prostaglandin12-oxophytodienoic acids, and A1-phytoprostanes (Mueller et al. 2008).
Conn et al. (2008), reported that the ligandin activity of specific glutathione S-transferases (GSTs) is necessary for the transport of anthocyanins from the cytosol to the plant vacuole. The same is observed in anthocyanin-transporting GSTs in Vitis vinifera. VvGST1 and VvGST4 are involved in anthocyanin transport. In V. vinifera, the GST activity and anthocyanin accumulation were enhanced by treatment with sucrose, jasmonic acid, and light (Conn et al. 2008). Kitamura et al. (2012), performed degenerate PCR using total RNA from immature young petals of cyclamen, to identify the anthocyanin-related GSTs in cyclamen, and results indicate the functional participation of CkmGST3 in anthocyanin accumulation in cyclamen (Kitamura et al. 2012).
Biotic stress
It is reported in many studies that during biotic stresses, pathogen infections, phytohormone signaling, etc., plant GSTs exhibit variant expressions. Tau GSTs are defense-specific GST enzymes. A tau class GST from G. max, GmGSTU10-10, has been characterized by their structure and function. It shows differential over-expression in response to the infection of soybean mosaic virus (SMV). Upon infection by the SMV, the infected soybean leaf tissues show significant upregulation of GmGSTU10-10, suggesting that GmGSTU10-10 represents a defense-specific GST enzyme (Skopelitou et al. 2015). Specifically, it is the only GST among the 25 different GST isoenzymes in soybean that are induced upon infection by the SMV (Babu et al. 2008). G. max tau GST, GmGSTU10-10 catalyzes several other different reactions and shows extensive substrate specificity and high antioxidant catalytic function, and acts as hydro-peroxidase. Additionally, its substrate affinity (km) for GSH is considerably lower, suggesting that GmGSTU10-10 can perform efficient catalysis under reduced GSHconcentration (e.g., oxidative stress) (Skopelitou et al. 2015). The biological role of GmGSTU10-10 in biotic stress is also supported by the recent finding that this enzyme is differentially expressed in soybean in response to Phakopsora pachyrhizi infections (Morales et al. 2013). Reports also have cleared that enzyme is down-regulated by abiotic stress (salt stress, NaCl) as established using proteomic analysis (Xu et al. 2011). It is found that the GmGSTU10-10 gene was found to be constitutively expressed in soybean, suggesting that the enzyme has housekeeping roles and is involved in various developmental processes of soybean (Ali et al. 2012).
A tau GST from Phaseolus vulgaris, PvGSTU3-3, which shows high substrate specificity, got induced by infection of a pathogen Uromyces appendiculatus and thus detoxifies the toxic chemicals produced and protects the plants again biotic stress. The enzyme exhibits high antioxidant catalytic function and also functions as hydroperoxides, thioltransferase, and dehydroascorbate reductase (Chronopoulou et al. 2012). It has been reported by Chen et al. (2013) that when Nicotiana benthamiana was infected with Bamboo mosaic virus (BaMV), the expression levels of tau GSTs, NbGSTU1, and NbGSTU3 were found to be upregulated post-infection, while those of NbGSTU2 and NbGSTF1 were unaffected. On the other hand, NbGSTU4 makes binding with viral RNA and deliver GSH to the replication complex to create an unfavorable condition for BaMV minus-strand RNA synthesis. With this study of a plant GST protein involving the replication of a plant, a virus could help to gain insights into the relationship between viral replicating proteins, its process, and the GST mediated metabolic pathway.
Abiotic stress
Abiotic stresses such as high and low temperature, salinity, drought, heavy metals, UV radiation, etc. extensively affect plant growth and development and, therefore, plants are developed with refined defense systems to overcome them. GSTs are one among the naturally inbuilt defense mechanisms in plants to fight against such stress and lead a healthy life. In this area of resistance against abiotic stress confer by GSTs many studies have been done. A GST gene from tomato (Lycopersicon esculentum) LeGSTU2, when cloned in Arabidopsis, is highly expressed in flowers and roots. The developed transgenic lines show an increased rate of germination and root length and exhibit enhanced resistance to salt and osmotic stress induced by NaCl and mannitol. It was also reported that LeGSTU2 may act as a stress regulator through increasing the activity of antioxidant enzymes (SOD, POD) to strengthen the ROS scavenging ability or maintain ROS homeostasis (Xu et al. 2014). Further, in addition to the role of GSTs in abiotic stress management, a transgenic approach was performed by Li Cicero et al. (2015) in which they developed transgenic tobacco plants overexpressing tau GSTs of Citrus sinensis CsGSTU1 and CsGSTU2 and found that the transformed tobacco lines demonstrate tolerance against a herbicide fluorodifen. The mechanism behind this protective role against fluorodifen is mainly due to the GSH conjugating activity of the GSTs and there is no GSH-peroxidase activity involved in scavenging of the oxidative stress by-products. Developed transgenic plants also displayed resistance against drought and salt stress when grown on mannitol (8%) and NaCl (150 and 200 mM) containing substrates.
SbGSTU gene from Salicornia brachiata plays a vital role in abiotic stress tolerance. Tiwari et al. (2016) have studied the long promoter region of GSTU from S. brachiata, a halophyte that grows in salt marshes. They observed that the abiotic stress reactive cis-elements like ABRE, MYB, GATA, GT1, etc. phytohormones, pathogen, and wound responsive motifs from S. brachiata. MYB and MYC are regulatory genes that code for a transcription factor. Binding sites for ABRE and MYB transcription factors on the SbGSTU promoter region are indicative of the SbGSTU regulation by the ABA-mediatedd signaling pathway under abiotic stress. GATA and GT1 are themselves a class of transcription factors involved in promoting or repressing gene expression under altered circumstances. These are up-regulated in the salinity, drought, and cold stress and also by ABA treatment thus revealing that these promoters are involved in conferring stress tolerance against drought and salinity and can be used in developing genetically engineered crops. Salt, drought, and cold stress generate ROS in plants, and the induction of the SbGSTU gene then reduces secondary noxious by-products generated during oxidative stress. Over-expression of tau GST gene SbGSTU in transgenic tobacco is central to enhance seed germination and grow under salt stress (Tiwari et al. 2016). In Pumpkin, GSTs play a major role in combatting against cold stress (Kayum et al. 2018).
Sharma et al. (2014) categorized the role of a tau class GST gene from rice, OsGSTU4. They developed and analyzed Arabidopsis transgenic plants over-expressing OsGSTU4. The developed transgenic plants showed increased tolerance to salinity and oxidative stresses at various developmental stages and also proposed that OsGSTU4 might be involved in auxin- and ABA-dependent processes that provide stress tolerance to transgenic plants. OsGSTU4 may represent a component mediating cross-talk between auxin and redox signaling to regulate abiotic stress responses. The transgenic Arabidopsis plants expressing OsGSTU4 also showed better seed germination, root and plant growth, and increased capacity to retain chlorophyll content in plant leaves as compared to WT plants under salinity and oxidative stress conditions. The Arabidopsis transgenic lines showed lower accumulation of ROS and increased GST activity. The increased salinity and oxidative stress tolerance might be related to the higher level of GST activity in transgenic lines due to the overexpression of OsGSTU4. Roxas et al. (1997) have shown in their studies that transgenic tobacco seedlings overexpressing Nt107, a GST encoding cDNA, with twofold high GST/GPOX activity than wild-type plants, were more tolerant to chilling and oxidative stress and reveals increased seed germination and growth under extreme chilling and oxidative stress. AmGSTF1, a phi class GST possesses GPOX activity and is highly active in the herbicide-resistant form of the weed black-grass (Alopecurus myosaroides) but is only detected in low amounts in the herbicide sensitive form (Cummins et al. 2013). Phi class GST AtGSTF2 was functionally characterized in Arabidopsis against phenol treatment. Transgenic plants exhibit increased tolerance to oxidative stress when treated with phenol. It is also reported that the activity of SOD and POD is higher in transgenic plants. This work suggested the application of GSTs in bioremediation under phenol contaminated areas (Xu et al. 2017).
The involvement of GSTs in apoptosis has been shown by a research group. They transformed yeast cells expressing Bax, a mammalian pro-apoptotic Bax protein which induces apoptosis, with a tomato cDNA library and found a tau class tomato GST which inhibits the Bax directly or indirectly. This “Bax-inhibitor GST” (BI-GST, SlGSTU24) possesses GST/weak GPOX activity when expressed in E. coli. Its expression in yeast decreases the intracellular level of total GSH and total cellular phospholipids and changes the mitochondrial membrane potential. The Co-expression of the BI-GST/GPX expressively enhanced resistance to H2O2-induced stress re-established the mitochondrial membrane potential and brought the total glutathione back to normal. With this approach, it would be possible to practice a yeast-based genetic strategy for the isolation of novel antioxidant and antiapoptotic genes (Kampranis et al. 2000).
Juglans regia is a nutritional nut tree (Abdallah et al. 2015). It possesses a tau GST (JrGSTTau1) which is responsible for chilling tolerance of J. regia. To confirm this specific activity of this plant, GST transgenic tobacco lines were developed by transforming the 35S::JrGSTTau1 into tobacco and it was found that JrGSTTau1is highly inducible under low temperature in plant's root stems and leaves and provides chilling tolerance to the plant. The transgenic lines also exhibited much higher GST, GPX, SOD, and POD activities, and lower H2O2 content under low-temperature stress. Transgenic tobacco lines were also increased in their biomass accumulation and lower cell damage. The promoters of J. regia JrGSTTau1 could be a possible candidate in developing chilling resistant transgenic crops (Yang et al. 2016).
Major involvement of GSTs in combatting against herbicidal stress
GSTs are centrally known for their role against herbicide tolerance in plants. They work by forming a GST/GSH conjugation complex that is involved in detoxifying several classes of herbicides like alachlor, fluorodifen, glyphosate, atrazine, metolachlor, thiocarbamates, etc. Glyphosate is a broad-spectrum herbicide and widely used for weed control in many crop plants. According to a report by Basantani et al. (2011) the activity of tau class GST of V. radiate gets elevated in response to glyphosate treatment. It is involved in glyphosate detoxification and aids plant survival against stress conditions. A novel sigma class GST from a freshwater planaria (flatworm) Dugesia japonica was cloned and found to be involved in defending against oxidative stress induced by glyphosate (Zhang et al. 2020). Multiple herbicide resistance (MHR) has become a very major and common agricultural issue in many countries. Keeping this in mind Georgakis et al. (2020) identified a specific GST that belongs to the Phi class and responsible for multiple herbicide resistance named MHR-GSTFs. They have a high hydroperoxidase activity and function by diminishing the toxic hydroperoxides. Alopecurus myosuroides and Lolium rigidum are the two grass weeds possesing MHR-GSTFS, and H. vulgare and T. aestivum are the two crop plants in which MHR-GSTFs have been reported. The catalytic properties and substrate specificities are different in grass weeds and crops. These MHR-GSTFs can be of great interest to the plant breeders for developing herbicide-tolerant crops. In a recent review on herbicide resistance, it is mentioned that GSTs are the natural inbuilt defense system in plants against oxidative stress caused by herbicide treatment (Nakka et al. 2019). The upregulation of GST activity in response to glyphosate is also reported in goosegrass (Eleusine indica (L.) Gaertn.). The study was done taking two populations, resistant (R) and susceptible (S). On the treatment of glyphosate, the GST activity of both the plants R and S increases but S plants exhibit low activity than R plants. The enhanced GST activity against glyphosate indicates its role in herbicide resistance in goosegrass (Chen et al. 2017). A comparative study was done in maize shoots and roots about the effect of the herbicide metolachlor and it was correlated with GST activity and expression in these plant parts. In conclusion it was found that the expression of GST gene and GST activity is much higher in roots than shoots which leads to enhanced tolerance to metolachlor in maize roots (Li et al. 2017). From the past few years, many herbicide-tolerant transgenic crops have been developed with recombinant DNA technology. A transgenic tobacco plant was developed by Agrobacterium-mediated transformation of the GmGSTU4-4 gene from G. max, which is a diphenyl ether herbicide, fluorodifen inducible GST gene. The resulting transformants exhibit enhanced tolerance towards fluorodifen and alachlor (Benekos et al. 2010). A transgenic approach was performed by Li Cicero et al. (2015) in which they developed transgenic tobacco plants overexpressing tau GSTs of Citrus sinensis CsGSTU1 and CsGSTU2 and found that the transformed tobacco lines unveil tolerance against a herbicide fluorodifen. The mechanism behind this protective role against fluorodifen is mainly the GSH conjugating activity of the GSTs and there is no GSH-peroxidase activity involved in scavenging of the oxidative stress by-products. Developed transgenic plants also displayed resistance against drought and salt stress when grown on mannitol (8%) and NaCl (150 and 200 mM) containing substrates.
GSTs in tetrapyrrole metabolism
There are several reports related to binding of GSTs with tetrapyrolic compounds in animals but the first report of the interaction of tetrapyrroles with oat GSTs (photosynthetic organism) was reported by (Singhal et al. 2015). It was found that the four tetrapyrroles (bilirubin, biliverdin, chlorophyllin, and hemin) have an inhibitory effect on the activity of oat glutathione-S-transferases. This, in turn, can enhance the plant senescence because of the inhibition of GSTs as GSTs play an important role in the plant detoxification system. GSTs also play an important role in tetrapyrrole metabolism because GSTs can bind different porphyrins such as protoporphyrin ix, mesoporphyrin, coproporphyrin, Mg protoporphyrin, and uroporphyrin. They also have a preventive role. A GST from Zea mays Zm GST III-III, which binds protoporphyrin with high affinity, prevents autoxidation of protoporphyrinogen (Lederer and Böger 2003). It is also reported that ZmGSTU1, ZmGSTU2 reduces the heme levels and accumulates porphyrin precursors such as harderoporphyrin in the chloroplast (Dixon et al. 2008). In current research by Sylvestre-Gonon et al. (2020), a tau class GST from A. thailiana ATGSTU8 binds protoporphyrin IX (PPIX) and haem moieties in their substrate binding sites suggesting their role in binding and transport. The GSTU members are involved in chaperon function more significantly in the cytosolic trafficking of haem as GSTs can also interact with ABC transporters. The involvement of GSTs has also been reported in plastid to nucleus retrograde signaling to regulate the expression of photosynthesis associated nuclear genes.
The mystery of the evolution of GSTs
GSTs are evolutionarily primordial protein. In animals, these enzymes were first discovered in the 1960s as a result of their importance in the metabolism and detoxification of drugs. In plants, GSTs were discovered in the 1970s in maize. The structures of GSTs from prokaryotes and eukaryotes so far characterized through crystallography and X-ray diffraction exhibit a remarkable level of structural conservation (Dixon and Edwards 2009). Initially, plant GSTs have progressed from a common ancestral GST into four distinct classes, namely Tau, Phi, Theta, and Zeta, based on sequence assessment and gene organization. Due to high sequence similarity, Theta and Zeta share common ancestral evolution. The first model of evolution, Theta class GSTs was present in bacteria. The mode of evolution of soluble GSTs is duplications followed by divergent evolution from this ancestral gene that has been reported in the early 1990s in plants, animals, and fungi (Sylvestre-Gonon et al. 2019). Reports suggested Theta, Zeta, and Omega GSTs as the most ancestral classes (da Fonseca et al. 2010; Frova 2003). The superfamily of mitochondrial and microsomal GSTs (MAPEG) is evolutionarily distinct compared to cytosolic GSTs having a restricted ability to conjugate xenobiotics with GSH. Three evolutionary mechanisms play a pivotal role in gene family expansion such as tandem duplication, segmental duplication, and transposition due to the presence of high repeating units in plant chromosomes. Besides this unequal crossing over, alternative splicing (around C-terminal domain) (Armstrong 1998; Wongsantichon and Ketterman 2005), swapping and mutagenesis (around N-terminal domain), domain combinations are also key factors to gene distribution and functional heterogeneity of GSTs. In Capsicum annuum, the members of the GST gene family arise largely due to several rounds of tandem and segmental gene duplications (Islam et al. 2019). The same factor is mentioned in sweet potato and Medicago truncatula (Kou et al. 2019; Han et al. 2018 and Du et al. 2018). The selective pressure is very strong in GSTs subfamilies, it has been proved by their potential to respond to various xenobiotics in cell metabolism, and also in expressing functional divergence that facilitates evolutionary divergence and attainment of novel activities (Lan et al. 2013). The evolution of GTSs has also been reported with GSH in aerobic organisms such as bacteria, fungi, higher plants, and parasites. Phylogenetic analysis reveals the rise of soluble GSTs from an ancient progenitor gene through convergent and divergent pathways. The evolution of GSTs has also been reported from a thioredoxin-like ancestor in response to oxidative stress (Koonin et al. 1994; Martin 1995), and glutaredoxins are the proposed ancestors of the GST_N domain. Several GST_N domains, for instance, in the sorghum locus Sb02g003090, possess very high sequence similarity to the glutaredoxin domain, supporting the putative origin of GSTs from glutaredoxins (Oakley 2005). Among several identified GSTs, both GST_N and GST_C domains are usually encoded in a single gene to support the co-evolution of both GST_N and GST_C domains with unlike evolutionary rates. Interestingly, few GST genes have been identified that encode only GST_N or GST_C domain indicating the loss of GST_N or GST_C domain during long evolution (Apic et al. 2001; Chothia et al. 2003) or suggesting the limited domain combination in this family. In a study on plant sorghum, the Ka/Ks ratios of these two domains suggested that both domains were under differential selective pressures and the GST_C domain might have been exposed to more relaxed functional constraints (Chi et al. 2011).
In monocots and dicots, GSTs have been evolved differentially and exhibit a considerable difference in their evolutionary history. They have been reported to be involved in the lineage-specific expansion and species diversification. Most of the dicot members fall into their subclasses, separating from monocot plants. These results suggested the rapid gene expansion for monocots occurred after the divergence of monocots from dicot plants. The large-scale expansion of Tau or Phi members for dicot plants occurred during their divergence from their most recent common ancestors thus signifying the differential tau and phi subfamily expansion history of both monocots and dicots plants. The expansion rate of tau or phi in monocot plants was earlier than in dicot plants. Furthermore, GST_N and GST_C domain evolved faster in dicot plants than that in monocot plants as proved in an analysis when the frequency distribution of the Ka/Ks ratios was tested. In a study, genome-wide phylogenetic analysis of Tau GST revealed two major clades: A and B. Clade A contained Tau GSTs of eight seed plant species but not lycophyte, indicating that this group of GSTs might have been lost in lycophyte. The clade B can be divided into two major groups, B1 and B2. B1 contained GSTs from all species, whereas B2 contained only lycophyte and gymnosperm GSTs. This phylogeny suggests that the B2 GSTs might have been lost in angiosperms (Chi et al. 2011) (Table 2).
Table2.
Important events in the plant GSTs research (1970–2020)
| Year | Major event(s) | References |
|---|---|---|
| 2020 | Role of GSTs in tertrapyrrole metabolism | Sylvestre-Gonon et al. (2020) |
| Role of Phi class GST in multiple-herbicide resistance of grass weeds and crops | Georgakis et al. (2020) | |
| 82 GST genes identified in radish | Gao et al. (2020) | |
| 2019 | GST gene family identification and characterization in capsicum | Islam et al. (2019) |
| A review on Plant GSTs and light | Gallé et al. (2019) | |
| 2018 | First report of 73 GSTs in Medicago truncatula | Han et al. (2018) |
| First report of 44GSTs in Vigna radiata | Vaish et al. (2018) | |
| First report of 88 GST genes in Oak (Quercus robur) | Plomion et al. (2018) | |
| First report of GST’s specific activity, kinetic behavior, substrate specificity, and protein expression levels in the leaves, fruits, stems, and roots of olive tree cv.Picual | Peragón and Amores-Escobar (2018) | |
| 2017 | Review on plant GSTs: Functions and biotechnological applications | Nianiou-Obeidat et al. (2017) |
| First report of 42 GST genes in sweet potato | Ding et al. (2017) | |
| 2016 | Identification of the role of Tau GST gene promoter SBGSTU of Salicornia brachiata under salinity and osmotic Stress | Tiwari et al. (2016) |
| 2015 | Very first characterized all GST classes of Glycine max | Liu et al. (2015) |
| First report of essential role of GSTs in pathogenicity of Alternaria brassicicola | Cicero et al. (2015) | |
| Review on plant GSTs | Labrou et al. (2015) | |
| 2014 | Report of 27 GST genes in gymnosperm Larix kaempferi | Yang et al. (2014) |
| Characterization of GST gene family in Citrus Sinensis | Licciardello et al. (2014) | |
| 2013 | First report of total 37 GSTs in Physcomitrella patens, a nonvascular plant (bryophyte) with two novel classes’ iota and hemerythrin and no tau GST | Liu et al. (2013) |
| 2012 | First report of maximum number of GSTs that possess α- helix followed by turns and β- sheets and also that they undergo post translational reversible phosphorylation activation | Puglisi et al. (2013) |
| 2011 | Expansion mechanisms and functional divergence of the Glutathione S-Transferase family in sorghum and other higher plants | Chi et al. (2011) |
| 2010 | Total 79 GSTs in rice | Jain et al. (2010) |
| 2009 | Physiological role of GSTs in root nodule of Glycine max | Dalton et al. (2009) |
| 2008 | Review on plant GSTs | Öztetik (2008) |
| Report on presence of GSTs in red and brown algae | Herv’E et al. (2008) | |
| 2007 | Review published on plant GSTs | Basantani and Srivastava (2007) |
| 2005 | Molecular characterization of GSTs for the first time in gymnosperm Pinus tabulaeformis (Pinaceae) | Zeng et al. (2005) |
| 2004 | First characterization of GST gene family in rice | Soranzo et al. (2004) |
| 2003 | GSTs undergo glycosylation | Moons (2003) |
| 2002 | Identification of two novel GSTs DHAR and Lambda in Arabidopsis thaliana | Dixon et al. (2002) |
| 2001 | Structure of Zeta class GST from A. thaliana was proposed with its putative role in Tyrosine catabolism | Thom et al. (2001) |
| 2000 | Review of plant GSTs with new modified nomenclature | Dixon and Edwards (2009) |
| Development of transgenic tobacco overexpressing GST with high stress tolerance | Roxas et al. (1997) | |
| 1999 | A phi class GST was reported in black grass Alopecurus myosuroides | Cummins et al.(1999) |
| 1998 | First classification of GSTs into Type I,II and III | Dixon et al. (1998) |
| Identification of GSTs in fresh water algae | Tang et al. (1998) | |
| 1997 | Identification of additional zeta class GSTs | Board et al. (1997) |
| Purification of multiple glutathione transferases involved in herbicide detoxification from wheat (Triticum aestivum L.) treated with the safener fenchlorazole-ethyl | Cummins et al. (1998) | |
| 1996 | Review on plant GSTs | Marrs (1996) |
| 1994 | Cloning and characterization of maize herbicide safener-induced cDNAs encoding subunits of glutathione S-transferase isoforms I, II, and IV | Jepson et al. (1994) |
| 1980 | Very first report on presence of GSTs in microorganism bacteria, fungi, protozoa, etc | Edward et al. (1980) |
| 1970 | First report of plant GSTs in maize | Frear and Swanson (1970) |
Genomics of plant GSTs
The genomic organization of plant GSTs is very well known and their chromosomal locations are well established in many plant species such as Arabidopsis, rice, soybean, wheat, maize, etc. Recently, a review reported a typical gene structure, i.e., one-intron/two-exon in tau class GSTs and two-intron/three-exon in a phi class GSTs in higher plants (Chronopoulou et al. 2017a). The number of introns and exons differ from the type of GSTs. With the growing studies on GSTs, these patterns of GST gene are shown to be present in many plant species from moss to angiosperms including gymnosperms, etc. In P. patens, number of intron/exon among classes and within-class varied greatly. Due to the diverse gene structure of P. patens phi, theta, EF1Bγ, TCHQD, and DHAR classes, a high rate of intron gain/loss has been documented. Intron number in phi GST genes varied from 2 to 3, whereas three theta GSTs possess four, five, and six introns. Like angiosperms, four PpEF1Bγ contained both a GST and an EF1Bγ domain with conserved intron positions and numbers. Intron number in five PpTCHQD GSTs ranges from one to three with conserved numbers and positions in the GST domain regions. Highly variable gene structures were observed in three PpDHAR genes with four, five, and seven introns, respectively (Liu et al. 2013). Recently, in mung bean and pepper same gene architecture of tau and phi has been reported with few variations (Vaish et al. 2018; Islam et al. 2019).
Protein architecture of plant GSTs
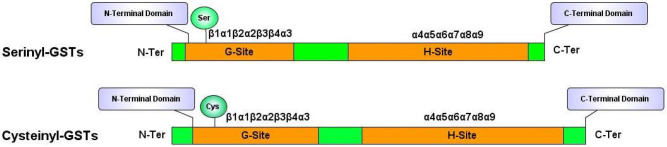
The comparative molecular masses of plant GSTs are nearby 50 kDa. It is a homodimer or heterodimer composed of two similarly sized (~ 25 kDa) subunits with an isoelectric point in the pH range of 4–5. Both the subunits are linked via an interaction between the N-terminal domain of one subunit and the C-terminal domain of the other. Plant GSTs have a propensity to attain α-helical structure followed by a random coil and then by β-sheet (Martin 1995). Thus, the loop α2-β3, the strand β4, and the helix α3 of one subunit interact with the helices α4 and α5 of the other subunit. Theses subunits are along a structural C2 axis roughly parallel to the helix bundle axis (binary axial symmetry) (Lallement et al. 2014a, b). In Theta, the interaction surface is rather hydrophilic, whereas in Tau, Phi, and Zeta GSTs, the surface is more hydrophobic (Frova 2003). Most of the GSTs attain an established dimeric quaternary structure. A conserved N-terminal domain with G-site is hydrophilic for binding of GSH adopts the typical TRX-fold (with β1α1β2α2β3β4α3 topology); and relatively less conserved C-terminal domain, the H-site, with which hydrophobic toxic molecules interact (Fig. 1). The G-site of each subunit in plant GSTs is strictly restrained and autonomous that facilitates thiol binding without requiring the firm co-alignment of subunits (Prade et al. 1997). In GSTFs, the H-site residues are chiefly located around the active site serine (N-terminal end of helix α1), in the loop β2-α2, and the C-terminal end of helix α4. The C-terminal domain is a bundle of minimum five helices (α4–α8). Tau GSTs have supplementary α9 helix that is oriented toward the active site without blocking it (Sylvestre-Gonon et al. 2019). In Cys-GSTs, two distinct motifs, an N-terminal motif (β1α1β2) and a C-terminal motif (β3β4α3) of a common N-terminal thioredoxin domain, were explained which are connected together with α2 helix and form a four β-sheet in the order 2134 with β3 anti-parallel to the others. In GSTUs both subunits are connected with conserved salt bridges. whereas, in GSTFs, the nature and the number of polar interactions varied considerably with one isoform to another. In AtGSTF2, a single H2 bond connects two subunits whereas in PtGSTF1 nine H2 bonds are present (Pégeot et al. 2014). α6–α7 linkage analytically embraces a small helix (α60) in GSTFs (Pégeot et al. 2014, 2017).
Fig. 1.
Schematic representation of the protein architecture of plant Ser and Cys GSTs. The N-terminal and C-terminal domain are represented in orange containing G and H site, respectively. The position of the active site residue, i.e., Ser and Cys are shown in green and blue circles, respectively, at the G site. Secondary structures are shown as α-helices and β-strands. The stretch of the boxes is directly correlated with the length of the amino acids
The crystal structure of zeta class GST was characterized in A. thaliana (AtGSTZ1). The N-terminal 2° structure of AtGSTZ1, specifically the loop structure between β2 and β3, was found identical to theta class. AtGSTZ1 contains four proline residues out of eight in its linker region that promotes this region to adopt the extended structure, not observed in any other classes previously. The helices α4 and α5 of AtGSTZ1 aligned differentially with α4. The core of the C-terminal domain of AtGSTZ1 carries more aliphatic than aromatic amino acid residues. The superimposition of other GST classes with AtGSTZ1 reveals the dominant hydrophobic core of the subunit interface (Thom et al. 2001). GSTs are diverse proteins and also associated with various non- catalytic activities such as secondary metabolism. For their ligandin role, GSTs possess an L-site other than the active site to which a wide range of compounds binds in a non-catalytic manner. The crystallographic structure of AtGSTF2 was revealed in complex with an organic ligand indole-3-aldehyde. The 2° structure analysis explored the presence of 12 α helices with two 310 helices and four β strands. Two ligands binding site L1 and L2 were observed. L1 site was located in a hydrophobic binding pocket formed between helices α4 and L2 site was present at the base of the dimer interface, including helices α3 of one subunit and α4 of its neighboring subunit (Ahmad et al. 2017).
Among cysteinyl-GSTs, DHAR and Lambda are monomeric, whereas GHRs and mPGES2s are dimeric. Only a few plant Cys-GSTs’ protein structures are solved such as the first 3D structures of lambda GST was solved in P. trichocarpa (Pt-GSTL1 and Pt-GSTL3) and it was established that Pt-GSTL1 and Pt-GSTL3 are monomeric. These two monomeric enzymes elucidate the standard GST fold which consists of an N-terminal domain adopting a thioredoxin-fold (β1α1β2α2β3β4α3) and α-helical C-terminal domain (α4α5α6α7α8α9 in PtGSTL3). Glutathione is covalently bound to the catalytic cysteine residue, i.e., Cys36 in Pt-GSTL1 and Cys41 in Pt-GSTL (Lallement et al. 2014a, b). DHAR GST from a gymnosperm Pinus bungeana has been molecularly characterized, as it has been found that the PbDHAR gene encodes a protein of 215 amino acid residues with a molecular mass of 24.26 kDa. The predicted 3‐D structure of PbDHAR showed a typical glutathione S‐transferase fold (Yang et al. 2009). In comparison to other GSTs classes, GSTL members have an extended N-terminal analogous to that of GSTOs (Lallement et al. 2014a, b). Structure of GHR1 of Phanerochaete chrysosporium which is a crust fungi possesses two monomers to interact via their C-terminal domain and are related to each other by a twofold symmetry axis. The structure of mPGES-2 s GST isoform is solved in only Macaca fascicularis which is a crab-eating macaque (Yamada et al. 2005).
Differential functioning of GSTs, containing Ser and Cys active site residue
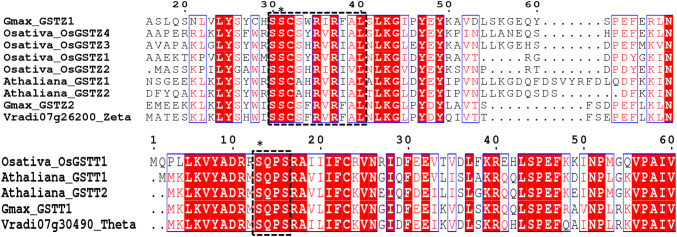
Structural studies of plant GSTs revealed that plant-specific tau, phi, theta, and zeta GSTs contain Ser as active site residue involved in GSH binding and activation (Axarli et al. 2009), whereas DHAR, Lambda, GHR, mPGES2, Hemerythrin, Iota, and metaxin classes hold in their active site Cys residue that facilitates deglutathionylation reaction. EF1Bγ beenpredicted to hold Ser or Tyr residue (Koonin et al. 1994). The nature of the residue promoting GSH activation in class Ure2p is yet not known. Traditionally, GSTUs are mainly associated with plant xenobiotic detoxification, specifically in herbicide detoxification. Later on, the functional diversity of GSTU extended from xenobiotic detoxification to other chemical pollutants such as 2,4,6-trinitrotoluene (TNT) by GSH-conjugation reaction (Gunning et al. 2014). When A. thaliana plants were treated with TNT, the overexpression of tau GST, AtGSTU24, and AtGSTU25 endure and detoxify TNT. Additionally, GSTUs are also associated in response to light signals. GSTU of A. thaliana GSTU5 and GSTU14 is also involved in response to light stress (Lv et al. 2015). Recently, the Arabidopsis Phi class GST (AtGSTF2) has been suggested to have a role in regulating the binding and transport of defense-related compounds in plants (Dixon et al. 2011). Mutants of Atgstf9 have elevated levels of AsA and GSH levels with increased GST activity. It is also reported that Atgstf9 plays a significant role in salt stress (Horváth et al. 2015). The Zeta-class GSTs from a range of species contain a characteristic motif [SSCX (W/H) RVIAL] in the N terminal region and expected to carry active site Ser residue (Board et al. 1997, 2003; Oztetik et al. 2015) (Fig. 2). It functions as maleylacetone isomerase that catalyzes the isomerization of maleylacetoacetate to fumarylacetoacetate. Thus, it is found to be involved in tyrosine degradation. Moreover, AtGSTZ1 is found to be involved in glutathione-dependent dehalogenation of dichloroacetic acid to glyoxylic acid in plants, suggesting another additional possible function in planta (Sylvestre-Gonon et al. 2019). Theta GSTs are generally about 250 amino acids long. The conserved active site residue is found around position 10 of the signature motif SQPS. This signature motif is conserved among mammals (SQPC) and insects [S(/A)PC] with few exceptions (Fig. 2). In vitro analysis reveals weak GSH-conjugation reaction but high GSH associated peroxidase activity towards fatty acids (Dixon et al. 2009). AtGSTT2 functions as an interacting factor of FLD (FLOWERING LOCUS D), which is required for systemic acquired resistance (SAR), SAR‐associated epigenetic modifications, and SAR activation (Banday and Nandi 2018).
Fig. 2.
Alignment of Ser-GST sequences of Zeta and Theta class is done, taking the amino acid sequences of A. thaliana, O. sativa, G. max, and V. radiata through Clustal omega alignment tool, and the protein alignments are rendered in ESPript 3.0. Conserved residues are marked with red background in white characters. The catalytic Ser residue is marked with ( ∗). The conserved motifs of Zeta GST [SSCX (W/H) RVIAL] and Theta GSTs (SQPS) are marked with black dashed box (Fig adopted from
Vaish et al. 2018)
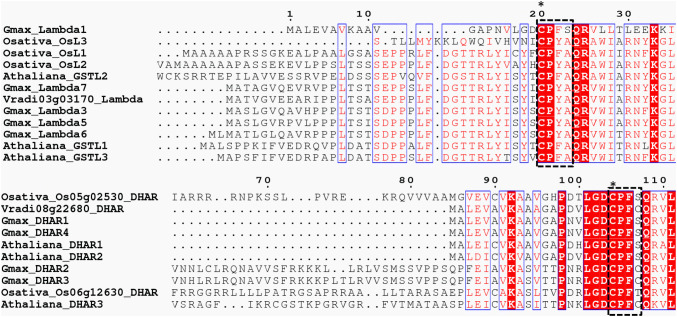
The biochemical and structural characterization of Cys-GSTs reveals that it follows a ping pong reaction so the regeneration of these glutathionylated GSTs requires a GSH molecule to produce GSSG as another end product and the reduced Cys-GSTs are ready for another catalytic cycle. Most Cys-GSTs require a single cysteine in the active site motif to follow this reaction mechanism. Conversely, a few isoforms have an additional cysteine in the active site, for instance, some DHAR isoforms which have CPFC or CPFS active sites motifs (Fig. 3). In a report, it is mentioned that not all but most of the GSTLs, GHRs, GSTOs, and DHARs that have been characterized unveil thiol-transferase and DHAR activities but no transferase, peroxidase or isomerase activities have been reported. GHRs are involved in the catabolism of chlorinated quinones, probably preventing noxiousness of quinones, either found naturally or present as environmental pollutants and in lignin degradation through the deglutathionylation of metabolic intermediates (Lallement et al. 2014a, b). Among GSTLs, the active site motif, which is CP(F/Y)A, (Fig. 3) is found around position 40 and is similar to GSTOs might endorse a common origin. Plant GSTLs may be involved in the metabolism or trafficking of flavonoids and combatting against Arsenic (As) stress (Kumar and Trivedi 2018) (Table 3).
Fig. 3.
Alignment of Cys-GST amino acid sequences of Lambda and DHAR GST classes. The GST amino acid sequences of A. thaliana, O. sativa, G. max, and V. radiata are aligned on Clustal omega alignment tool and the protein alignments are rendered in ESPript 3.0. Conserved residues are marked with red background in white characters. The catalytic Cys residue is marked with ( ∗). The conserved motifs of Lambda GST [CP(F/Y)A] and DHAR GST (CPFC/S) are marked with black dashed box (Fig
adopted from Vaish et al. 2018)
Table 3.
Characteristics of Ser and Cys plant GSTs
| S. no | Class | Typical catalytic motif with active site residue | Average amino acid length | Oligomerization state | References | |
|---|---|---|---|---|---|---|
| 1 | Tau | 10/20 W(A/V)S(P/M) | 250 | Dimer | Sylvestre-Gonon et al. (2019) | |
| 2 | Phi | 12A(A/V)(C/N)P | 215 | Dimer | Pégeot et al. (2017) | |
| 3 | Theta | 10SQPS/C | 250 | Dimer | Sylvestre-Gonon et al. (2019) | |
| 4 | Zeta | 20SSCS/A | 225 | Dimer | Thom et al. (2001) | |
| 5 | Lambda | 40CPF/YA | 230 | Monomer | Lallement et al. (2014a, b) | |
| 6 | DHAR | 20CPFC/S | 220 | Monomer | Lallement et al. (2014a, b) | |
| 7 | TCHQD | ?SCIS | 265 | Monomer | Lallement et al. (2014a, b) | |
| 8 | mPGES-2 | 120CPYC | 310 | Dimer | Lallement et al. (2014a, b) | |
| 9 | GHR | 50CPWA | 330 | Dimer | Lallement et al. (2014a, b) | |
| 10 | GSTI | 120CPYC | 490 | ? | Lallement et al. (2014a, b) | |
| 11 | GSTH | 50CPF/YT | 510 | ? | Lallement et al. (2014a, b) | |
GSTs in biotechnological applications
The ongoing research on GSTs and demarcation of their varied function abilities have made them a striking tool in various biotechnological applications:
GSTs in stress-resilient crops
The salient functional features of GSTs like detoxification of foreign compounds, inter or intracellular chemical compounds, against several stresses and contribution in herbicide detoxification have fascinated the researchers to combine it with genetic engineering and plant breeding approaches and to develop herbicide-resistant, negative climatic factors such as drought, flood, high salt, and cold resilient crops. Many research groups have developed herbicide-tolerant plants that exhibit resistance to fluorodifen, oxyfluorfen, alachlor, atrazine, and other multiple herbicides (Benekos et al. 2010; Kissoudis et al. 2015). Additionally, transgenic tobacco plants were developed showing chilling tolerance, expressing the genes from J. regia (Yang et al. 2016). In a transgenic approach by Sharma et al. (2014), transgenic Arabidopsis plants were developed showing increased tolerance to salinity and oxidative stress. Fusarium is a causative agent of Fusarium head blight (FHB), a fungal disease, in the wheat crop. Due to this fungal disease, the wheat production gets hampered worldwide. In a recent approach by Wang et al. (2020), a FHB resistant gene, Fhb7, was cloned by accumulating the genome of Thinopyrum elongatum. The Fhb7 gene encodes a GST protein which functions by detoxifying trichothecenes through de-epoxidation reaction and provides tolerance against Fusarium. So Fhb7 gene can be a good option in plant breeding approaches to the development of the Fusarium resistant transgenic crops.
GSTs in nanotechnology
Nanotechnology is an emerging field of science and catching the attention of molecular biologists in the area of agricultural science for the engineering of nanomaterials with plant proteins such as GSTs. Bai et al. (2013) designed a self-assembled nanoring by exploiting the concept of metal-ion-chelating interactions and nonspecific protein − protein interactions and key structural topographies of Schistosoma japonicum GST protein (SjGST) as a building block. The SjGST protein is present naturally as a stable homodimer with two subunits which are linked to each other via noncovalent interactions and have twofold axis symmetry (C2) with His residue and Ni metal-chelating sites on the protein surface. Nanowires were also developed using the Ni ion directed and His tag arm (Zhang et al. 2012).
GSTs in developing biosensors
From the past few decades there has been extensive work in the development of biosensor as they have many advantages over traditional chemical experiments:quick to perform, accurate results, easy sample preparation, etc. By exploiting the ability of GSTs in conjugation reaction of GSH with xenobiotics, herbicides, pesticides have given an idea of developing biosensors that can be used in the detection of cancers, various pathogen infections, pesticides, and herbicides, etc. Safarpour et al. (2012), developed a quantum dot FRET-based biosensor for detecting the fungal infection in sugar beets. Anti-GST, the GST protein corresponding antibody, was conjugated to the quantum dots. Recently, an optical GST-based biosensor was designed for the detection of pesticide, i.e., α-endosulfan which is a toxic insecticide and also a Persistent Organic Pollutant (POP). A structure-based design method was implemented for generating a synthetic and an augmented GST mutant PvGmGSTUG, by taking the glutathione transferase from P. vulgaris and G. max (Chronopoulou et al. 2019).Additionally, the GST–glutathione (GSH) interaction system was also employed in the immobilization protein surface for developing the matrixes (Paternolli et al. 2002). Ki et al. (2013), by exploiting the affinity binding between GST and GSH, created a recombinant selection fused to GST (GST-SIL), by protein immobilization method. GST-SIL protein was then bound to the GSH coated glass plate and the matrix developed could be used in biosensing, biocatalyzing equipment, various diagnostic plates, and also in the bioactive tissue-culture framework. Recently, for the detection of the cancerous cells, an electrochemical biosensor was developed. For generating this kind of biosensor in which GSH is a biomarker for cancer cells, GSTs (as a probe) were immobilized on the MoS2 sheet. MoS2 is a two- dimensional graphene or transition metal dichalcogenides (TMDs), frequently used in developing highly sensitive biosensors (Rawat et al. 2020). Oliveira et al. (2013), developed a GST based biosensor to quantify molinate, a thiocarbamate herbicide commonly used in rice paddy fields worldwide to control weed overgrowth. The biosensor was prepared by immobilizing the GST on the glassy carbon electrode.
GSTs in environmental biotechnology
The application of GSTs is also widespread in metabolizing and bioremediation of the environmental pollutants such as TNT (2,4,6-trinitrotoluene), anthracene, and chloromequat chloride. Two tau class GST from Arabidopsis U24 and U25 exhibits detoxifying activity towards TNT and was found to be upregulated on exposure to TNT (Gunning et al. 2014). Additionally, a transgenic Arabidopsis plant expressing the GST gene from Drosophila melanogaster (DmGSTE6) was found tolerant to TNT and highly efficient in removing the environmental TNT and could be an attractive option for the biodegradation of such hazardous chemical compounds (Tanikawa et al. 2002). BphKLB400 protein of a bacteria Burkholderia xenovoransLB400, is a GST analog and catalyzes the dechlorination of toxic chlorinated organic compounds (chloromequat chloride). It can also be a potential natural tool in bioremediation (McGuinness et al. 2007). In a study by Dixit et al. (2011), a transgenic tobacco plant expressing a GST gene from fungi Trichoderma virens was involved in the destruction and remediation of a polyaromatic hydrocarbon- anthracene.
Concluding remarks and future perspectives
With the ongoing research plant GSTs have been identified and characterized diversely. Their extensive functional characterization reveals its versatility towards a wide range of functions from traditional glutathionylation to deglutathionylation, combatting against several biotic and abiotic stresses, plant growth and development, secondary metabolism, tetrapyrrole metabolism, etc. The evolutions of GSTs were reported through gene duplication, transposition, and exon shuffling of an ancestral GSH binding protein, yet a lot of work is needed to find the mystery of the evolution of GSTs. Cytosolic GSTs are most numerous and extensively characterized both structurally and functionally. The interacting surfaces of GSTs can be hydrophilic or hydrophobic and their interaction with the substrate ranges from salt bridges to polar. With the all-embracing research from the few last decades, many new classes of plant GSTs like iota, hemerythrin, GHR, Ure2p, and metaxin have also been reported. Due to its flexibility relating to its roles this protein has gained a lot of attention in the field of recombinant DNA technology to selectively modify GST expression concerning genetically engineering resistance against abiotic and biotic stresses. Specifically, the detoxification properties of tau and phi class GSTs have been exploited for the development of herbicide-tolerant (e.g., alachlor, fluorodifen) crops like rice and tobacco.
In the current review the key features of Plant GSTs which represent a multipurpose and resourceful tool have been shown. The constant analysis of this protein superfamily will undoubtedly reveal many other examples of their functional expansions like their role in plant physiology such as photosynthesis, respiration, etc. that will further excite the researchers to think and work out of the box. Although a plethora of research is been done and so far continue at different levels in plant GSTs, yet a wide range of queries are arising, such as activities like the evolution of GSTs is through gene duplication only? or any epigenetic change in related protein get evolved into a new class called as Glutathione-S transferase? Additionally, a series of questions are left to be answered such as the following: is there any new class of plant GSTs to be identified? Is there any organelle-specific plant GST? Are TATA-less promoters a feature of zeta class only? Do GSTs interact with substrates other than glutathione, xenobiotics, ROS, etc.? Reports suggested Theta, Zeta, and Omega GSTs as the most ancestral classes but why there is no report of omega GSTs in plants? What is the reason behind the extensive tandem duplication event in the GST gene family with the diverse kinetic property? If concentrating on the earlier and currently characterized GST gene family in different plants and crops it was found that the number of tau class GSTs are more than any other classes, exceptionally in wheat, so why the tau GSTs are the highest in number? Why is there no report of tau GST in P. patens which is an only bryophyte in which GST gene family has been identified and characterized? Additionally, why there is no report of iota and hemerythrin in higher plants except moss?
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Abbreviations
- GST
Glutathione S-Transferase
- GSH
Glutathione;
- TCHQD
Tetrachloro-hydroquinone dehalogenase
- EF1B γ
Elongation factor 1B gamma
- DHAR
Dehydroascorbate reductases
- GHRs
Glutathionyl-hydroquinone reductases
- mPGES-2 s
Microsomal prostaglandin E synthase type 2
- Cyt P450s
Cytochrome P450s
- SMV
Soybean Mosaic Virus
- BaMV
Bamboo Mosaic Virus
- ROS
Reactive Oxygen Species
- POD
Peroxidases
- SOD
Superoxide Dismutase
- GPOX
Glutathione Peroxidase
- Ka/Ks ratio
Nonsynonymous/ Synonymous Mutation
- AsA
Ascorbic Acid
- GSTU
Tau GST
- GSTF
Phi GST
Author contributions
All the authors have contributed for writing and discussing the manuscript; they all have seen and approved the content of the final manuscript.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Contributor Information
Swati Vaish, Email: swati.vaish88@gmail.com.
Divya Gupta, Email: guptadivya06@gmail.com.
Rajesh Mehrotra, Email: rmehrotra@pilani.bits-pilani.ac.in.
Sandhya Mehrotra, Email: sandhyam@goa.bits-pilani.ac.in.
Mahesh Kumar Basantani, Email: mkbasantani@gmail.com.
References
- Abdallah IB, Tlili N, Martinez-Force E, Pérez Rubio AG, Perez-Camino MC, Albouchi A, Boukhchina S. Content of carotenoids, tocopherols, sterols, triterpenes and aliphatic alcohols, and volatile compounds in six walnuts (Juglans regia L.) varieties. Food Chem. 2015;173:972–978. doi: 10.1016/j.foodchem.2014.10.095. [DOI] [PubMed] [Google Scholar]
- Ahmad L, Rylott EL, Bruce NC, Edwards R, Grogan G. Structural evidence for Arabidopsis glutathione transferase at GSTF2 functioning as a transporter of small organic ligands. FEBS Open Bio. 2017;7:122–132. doi: 10.1002/2211-5463.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Zhang DY, Xu ZL, Xu L, Yi JX, He XL, Huang YH, Liu XQ, Khan AA, Trethowan RM, Ma HX. Uncovering the salt response of soybean by unraveling its wild and cultivated functional genomes using tag sequencing. PLoS ONE. 2012;7:e48819. doi: 10.1371/journal.pone.0048819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial, and eukaryotic proteomes. J Mol Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- Armstrong RN. Mechanistic imperatives for the evolution of glutathione transferases. Current Op in Chem Biol. 1998;2:618–623. doi: 10.1016/s1367-5931(98)80093-8. [DOI] [PubMed] [Google Scholar]
- Axarli I, Dhavala P, Papageorgiou AC, Labrou NE. Crystallographic and functional characterization of the fluorodifen-inducible glutathione transferase from Glycine max reveals an active site topography suited for diphenylether herbicides and a novel L-site. J Mol Biol. 2009;385:984–1002. doi: 10.1016/j.jmb.2008.10.084. [DOI] [PubMed] [Google Scholar]
- Babu M, Gagarinova AG, Brandle JE, Wang A. Association of the transcriptional response of soybean plants with soybean mosaic virus systemic infection. J Gen Virol. 2008;89:1069–1080. doi: 10.1099/vir.0.83531-0. [DOI] [PubMed] [Google Scholar]
- Bai Y, Luo Q, Zhang W, Miao L, Xu J, Li H, Liu J. Highly ordered protein nanorings designed by accurate control of glutathione S-transferase self-assembly. J Am Chem Soc. 2013;135:10966–10969. doi: 10.1021/ja405519s. [DOI] [PubMed] [Google Scholar]
- Banday ZZ, Nandi AK. Arabidopsis thaliana GLUTATHIONE S-TRANSFERASE THETA 2 interacts with RSI1/FLD to activate systemic acquired resistance: GSTT2 interacts with FLD and regulates SAR. Mol Plant Pathol. 2018;19:464–475. doi: 10.1111/mpp.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basantani M, Srivastava A. Plant glutathione transferases: a decade falls short. Can J Bot. 2007;85:443–456. [Google Scholar]
- Basantani M, Srivastava A, Sen S. Elevated antioxidant response and induction of tau-class glutathione S-transferase after glyphosate treatment in Vigna radiata (L.) Wilczek. Pesti Biochem Physiol. 2011;99:111–117. [Google Scholar]
- Benekos K, Kissoudis C, Nianiou-Obeidat I, Labrou N, Madesis P, Kalamaki M, Makris A, Tsaftaris A. Overexpression of a specific soybean GmGSTU4 isoenzyme improves diphenyl ether and chloroacetanilide herbicide tolerance of transgenic tobacco plants. J Biotechnol. 2010;150:195–201. doi: 10.1016/j.jbiotec.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Board PG, Baker RT, Chelvanayagam G, Jermiin LS. Zeta, a novel class of glutathione transferases in a range of species from plants to humans. Biochem J. 1997;328:929–935. doi: 10.1042/bj3280929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board PG, Taylor MC, Coggan M, Parker MW, Lantum HB, Anders MW. Clarification of the role of key active site residues of glutathione transferase Zeta/maleylacetoacetate isomerase by a new spectrophotometric technique. Biochem J. 2003;374:731–737. doi: 10.1042/BJ20030625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairrao E, Couderchet M, Soares AMVM, Guilhermino L. Glutathione-S-transferase activity of Fucus spp. as a biomarker of environmental contamination. Aquatic Toxicol. 2004;70:277–286. doi: 10.1016/j.aquatox.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Chen IC, Huang IC, Liu MJ, Wang ZG, Chung SS, Hsieh HL. Glutathione S -transferase interacting with far-red insensitive 219 is involved in phytochrome A-mediated signaling in Arabidopsis. Plant Physiol. 2007;143:1189–1202. doi: 10.1104/pp.106.094185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Chiu MH, Cheng SF, Hsu YH, Tsai CH. The glutathione transferase of Nicotiana benthamiana NbGSTU4 plays a role in regulating the early replication of Bamboo mosaic Virus. New Phytol. 2013;199:749–757. doi: 10.1111/nph.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Huang H, Wei S, Huang Z, Wang X, Zhang C. Investigating the mechanisms of glyphosate resistance in goosegrass (Eleusine indica (L.) Gaertn.) by RNA sequencing technology. Plant J. 2017;89:407–415. doi: 10.1111/tpj.13395. [DOI] [PubMed] [Google Scholar]
- Chi Y, Cheng Y, Vanitha J, Kumar N, Ramamoorthy R, Ramachandran S, Jiang S. Expansion Mechanisms and Functional Divergence of the Glutathione S Transferase Family in Sorghum and Other Higher Plants. DNA Res. 2011;18:1–16. doi: 10.1093/dnares/dsq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- Chronopoulou E, Madesis P, Tsaftaris A, Labrou NK. Cloning and characterization of a biotic-stress-inducible glutathione transferase from phaseolus vulgaris. Planta. 2012;235:1253–1269. doi: 10.1007/s12010-013-0509-3. [DOI] [PubMed] [Google Scholar]
- Chronopoulou E, Ataya FS, Pouliou F, Perperopoulou F, Georgakis N, Nianiou-Obeidat I, et al. Glutathione in plant growth, development, and stress tolerance. London: Springer; 2017. Structure evolution and functional roles of plant glutathione transferases; pp. 195–213. [Google Scholar]
- Chronopoulou E, Georgakis N, Nianiou-Obeidat I, Madesis P, Perperopoulou F, Pouliou F, et al. Glutathione in Plant Growth, Development, and Stress Tolerance. London: Springer; 2017. Plant glutathione transferases in abiotic stress response and herbicide resistance; pp. 215–233. [Google Scholar]
- Chronopoulou EG, Vlachakis D, Papageorgiou AC, Ataya FS, Labrou NE. Structure-based design and application of an engineered glutathione transferase for the development of an optical biosensor for pesticides determination. Biochimica Biophysica Acta. 2019;1863:565–576. doi: 10.1016/j.bbagen.2018.12.004. [DOI] [PubMed] [Google Scholar]
- Cicero LL, Madesis P, Tsaftaris A, Piero ALR. Tobacco plants over-expressing the sweet orange tau glutathione transferases (CsGSTUs) acquire tolerance to the diphenyl ether herbicide fluorodifen and to salt and drought stresses. J Phytochem. 2015;116:69–77. doi: 10.1016/j.phytochem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Conn S, Curtin C, Zier AB, Franco C, Zhang W. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J Experimental Botany. 2008;59:3621–3634. doi: 10.1093/jxb/ern217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csiszar J, Horvath E, Vary Z, Galle A, Bela K, Brunner S, Tari I. Glutathione transferase supergene family in tomato: salt stress-regulated expression of representative genes from distinct GST classes in plants primed with salicylic acid. Plant Physiol Biochem. 2014;78:15–26. doi: 10.1016/j.plaphy.2014.02.010. [DOI] [PubMed] [Google Scholar]
- Cummins I, Cole DJ, Edwards R. Purification of multiple glutathione transferases involved in herbicide detoxification from wheat (Triticum aestivum L.) treated with the safener fenchlorazole-ethyl. Pestic Biochem Physiol. 1998;59:35–49. [Google Scholar]
- Cummins I, Cole DJ, Edwards R. A role for glutathione transferases functioning as glutathione peroxidases in resistance to multiple herbicides in black-grass. Plant J. 1999;18:285–292. doi: 10.1046/j.1365-313x.1999.00452.x. [DOI] [PubMed] [Google Scholar]
- Cummins I, Wortley DJ, Sabbadin F, He Z, Coxon CR, Straker HE, Sellars JD, Knight K, Edwards L, Hughes D, Kaundun SS, Hutchings SJ, Steel PG, Edwards R. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc Natl Acad Sci. 2013;110:5812–5817. doi: 10.1073/pnas.1221179110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca RR, Johnson WE, O’Brien SJ, Vasconcelos V, Antunes A. Molecular evolution and the role of oxidative stress in the expansion and functional diversification of cytosolic glutathione transferases. BMC Evol Biol. 2010;10:281. doi: 10.1186/1471-2148-10-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton DA, Boniface C, Turner Z, Lindah A, Kim HJ, Jelinek L, Govindarajulu M, Finger RE, Taylor CG. Physiological roles of glutathione S-transferases in soybean root nodules. Plant Physiol. 2009;150:521–530. doi: 10.1104/pp.109.136630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Wang A, Zhang X, Wu Y, Wang R, Cui H, Huang R, Luo Y. Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses. BMC Plant Biol. 2017;17:225. doi: 10.1186/s12870-017-1179-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit P, Mukherjee PK, Sherkhane PD, Kale SP, Eapen S. Enhanced tolerance and remediation of anthracene by transgenic tobacco plants expressing a fungal glutathione transferase gene. J Hazardous Mat. 2011;192:270–276. doi: 10.1016/j.jhazmat.2011.05.018. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases. J Biol Chem. 2009;284:21249–21256. doi: 10.1074/jbc.M109.020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Cummins I, Cole DJ, Edwards R. Glutathione-mediated detoxification systems in plants. Current Op in Plant Biol. 1998;1:258–266. doi: 10.1016/s1369-5266(98)80114-3. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases. Genome Boil. 2002;3:1–3004. doi: 10.1186/gb-2002-3-3-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Madesis P, Mudd EA, Day A, Edwards R. Binding and glutathione conjugation of porphyrinogens by plant glutathione transferases. J of Biolog Chem. 2008;283:20268–20276. doi: 10.1074/jbc.M802026200. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Hawkins T, Hussey PJ, Edwards R. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J Exp Bot. 2009;60:1207–1218. doi: 10.1093/jxb/ern365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Skipsey M, Edwards R. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry. 2010;71:338–350. doi: 10.1016/j.phytochem.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Sellars JD, Edwards R. The Arabidopsis phi class glutathione transferase AtGSTF2: binding and regulation by biologically active heterocyclic ligands. Biochem J. 2011;438:63–70. doi: 10.1042/BJ20101884. [DOI] [PubMed] [Google Scholar]
- Du J, Ren J, Ye X, Hou A, Fu W, Mei F, Liu Z. Genome-wide identification and expression analysis of the glutathione S-transferase (GST) family under different developmental tissues and abiotic stresses in Chinese Peer. J Preprints. 2018;6:62. [Google Scholar]
- Edward PL, Lee N, Watson D, Ray FR. Glutathion-S-transferase is present in a variety of microorganisms. Chemosphere. 1980;9:565–569. [Google Scholar]
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: with multiple functions in sickness and in health. Trends Plant Sci. 2000;5:193–198. doi: 10.1016/s1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- Frear DS, Swanson HR. Biosynthesis of S-(4-ethylamino-6-isopropylamino- 2-s-triazino) glutathione: partial purification and properties of a glutathione S-transferase from corn. Phytochemistry. 1970;9:2123–2132. [Google Scholar]
- Frova C. The plant glutathione transferase gene family: genomic structure, functions, expression and evolution. Physiol Plant. 2003;119:469–479. [Google Scholar]
- Gallé A, Csiszár J, Secenji M, Tari I, Györgyey J, Dudits D, Erdei L. Changes of glutathione S-transferase activities and gene expression in Triticum aestivum during polyethylene-glycol induced osmotic stress. Acta Biologica Szegediensis. 2005;49:95–96. [Google Scholar]
- Gallé Á, Czékus Z, Bela K, Horváth E, Ördög A, Csiszár J, Poór P. Plant glutathione transferases and light. Front Plant Sci. 2019;9:1944. doi: 10.3389/fpls.2018.01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Chen B, Lin H, Liu Y, Wei Y, Chen F, Li W. Identification and characterization of the glutathione S-Transferase (GST) family in radish reveals a likely role in anthocyanin biosynthesis and heavy metal stress tolerance. Gene. 2020;743:144484. doi: 10.1016/j.gene.2020.144484. [DOI] [PubMed] [Google Scholar]
- Georgakis N, Poudel N, Papageorgiou AC, Labrou NE. Comparative structural and functional analysis of phi class glutathione transferases involved in multiple-herbicide resistance of grass weeds and crops. Plant Physiol. Biochem. 2020;149:266–276. doi: 10.1016/j.plaphy.2020.02.012. [DOI] [PubMed] [Google Scholar]
- Gong H, Jiao Y, Hu WW, Pua EC. Expression of glutathione-S-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro, Plant Mol. Biol. 2005;57:53–66. doi: 10.1007/s11103-004-4516-1. [DOI] [PubMed] [Google Scholar]
- Gunning V, Tzafestas K, Sparrow H, Johnston EJ, Brentnall AS, Potts JR, Rylott EL, Bruce NC. Arabidopsis glutathione transferasesU24 andU25 exhibit arange of detoxification activities with the environmental pollutant and explosive, 2,4,6-trinitrotoluene. Plant Physiol. 2014;165:854–865. doi: 10.1104/pp.114.237180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han XM, Yang ZL, Liu YJ, Yang HL, Zeng QY. Genome-wide profiling of expression and biochemical functions of the Medicago glutathione S-transferase gene family. Plant Physiol et Biochem. 2018;126:126–133. doi: 10.1016/j.plaphy.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Herv’E C, Groisillier A, Thierry TT, Boyen C. New members of the glutathione transferase family discovered in red and brown algae. Biochem J. 2008;412:535–544. doi: 10.1042/BJ20071464. [DOI] [PubMed] [Google Scholar]
- Horváth E, Bela K, Papdi C, Gallé Á, Szabados L, Tari I, Jolán C. The role of Arabidopsis glutathione transferase F9 gene under oxidative stress in seedlings. Acta Biol Hungarica. 2015;66:406–418. doi: 10.1556/018.66.2015.4.5. [DOI] [PubMed] [Google Scholar]
- Hu B, Zhao J, Lai B, Qin Y, Wang H, Hu G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016;35:831–843. doi: 10.1007/s00299-015-1924-4. [DOI] [PubMed] [Google Scholar]
- Islam S, Sajib SD, Jui ZS, Arabia S, Islam T. Genome-wide identification of glutathione S-transferase gene family in pepper, its classification, and expression profiling under different anatomical and environmental conditions. Scientific Reports. 2019;9:9101. doi: 10.1038/s41598-019-45320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Ghanashyam C, Bhattacharjee A. Comprehensive expression analysis suggests overlapping and specific roles of glutathione S-transferases during development and stress responses in rice. BMC Genomics. 2010;11:73. doi: 10.1186/1471-2164-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson VJ, Lay DC, Holt SW, Bright J, Andrew J. Cloning and characterization of maize herbicide safener-induced cDNAs encoding subunits of glutathione S-transferase isoforms I. II and IV Plant Mol Bio. 1994;26:1855–1866. doi: 10.1007/BF00019498. [DOI] [PubMed] [Google Scholar]
- Jiang HW, Liu MJ, Chen IC, Huang CH, Chao LY, Hsieh HL. A Glutathione S-transferase regulated by light and hormones participates in the modulation of arabidopsis seedling development. Plant Physiol. 2010;154:1646–1658. doi: 10.1104/pp.110.159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampranis SC, Damianova R, Atallah M, Toby G, Kondi G, Tsichlis PN, Makris AM. A novel plant glutathione S-transferase/peroxidase suppresses Bax lethality in yeast. J Biol Chem. 2000;275:29207–29216. doi: 10.1074/jbc.M002359200. [DOI] [PubMed] [Google Scholar]
- Kayum MA, Nath UK, Park JI, Biswas MK, Choi EK, Song JY, Kim HT, Nou IS. Genome-wide identification, characterization, and expression profiling of glutathione S-transferase (GST) family in pumpkin reveals likely role in cold-stress tolerance. Genes. 2018;9:84. doi: 10.3390/genes9020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ki M, Yeo KB, Pack SP. Surface immobilization of protein via biosilification catalyzed by silicatein fused to glutathione S-transferase (GST) Bioprocess Biosyst Eng. 2013;36:643–648. doi: 10.1007/s00449-012-0818-x. [DOI] [PubMed] [Google Scholar]
- Kissoudis C, Kalloniati C, Pavli O, Flemetakis E, Labrou NE, Madesis P, Skaracis G, Tsaftaris A, Nianiou-Obeidat I. Stress inducible GmGSTU4 shapes transgenic tobacco plants metabolome towards increased salinity tolerance. Acta Physiol Plant. 2015;37:102. [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004;37:104–114. doi: 10.1046/j.1365-313x.2003.01943.x. [DOI] [PubMed] [Google Scholar]
- Kitamura S, Akita Y, Ishizaka H, Narumia I, Tanaka A. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in cyclamen. J Plant Physiol. 2012;169:636–642. doi: 10.1016/j.jplph.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Mushegian AR, Tatusov RL, Altschul SF, Bryant SH, Bork P, Valencia A. Eukaryotic translation elongation factor 1c contains a glutathione transferase domain - study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994;3:2045–2054. doi: 10.1002/pro.5560031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou M, Liu YJ, Li ZY, Zhang YG, Tang W, Yan H, Wang X, Chen XG, Su ZX, Arisha MH, Li Q, Ma DF. A novel glutathione S-transferase gene from sweetpotato, IbGSTF4, is involved in anthocyanin sequestration. Plant Physiol and Biochem. 2019;135:395–403. doi: 10.1016/j.plaphy.2018.12.028. [DOI] [PubMed] [Google Scholar]
- Kumar S, Trivedi PK. Glutathione S-transferases: role in combating abiotic stresses including arsenic detoxification in plants. Front Plant Sci. 2018;9:751. doi: 10.3389/fpls.2018.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Asif MH, Chakrabarty D, Tripathi RD, Dubey RS, Trivedi PK. Expression of a rice Lambda class of glutathione S-transferase, OsGSTL2, in Arabidopsis provides tolerance to heavy metal and other abiotic stresses. J Hazardous Materials. 2013;248–249:228–237. doi: 10.1016/j.jhazmat.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Labrou NE, Papageorgiou AC, Pavli O, Flemetakis E. Plant GSTome: structure and functional role in xenome network and plant stress response. Current Op in Biotechnol. 2015;32:186–194. doi: 10.1016/j.copbio.2014.12.024. [DOI] [PubMed] [Google Scholar]
- Lallement PA, Brouwer B, Keech O, Hecker A, Rouhier N. The still mysterious roles of cysteine-containing glutathione transferases in plants. Front Pharmacol. 2014;5:192. doi: 10.3389/fphar.2014.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallement PA, Meux E, Gualberto JM, Prosper P, Didierjean C, Saul F, Haouz A, Rouhier N, Hecker A. Structural and enzymatic insights into Lambda glutathione transferases from populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants. Biochem J. 2014;462:39–52. doi: 10.1042/BJ20140390. [DOI] [PubMed] [Google Scholar]
- Lan T, Yang ZL, Yang X, Liu YJ, Wang XR, Zeng QY. Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell. 2009;21:3749–3766. doi: 10.1105/tpc.109.070219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T, Wang XR, Zeng QY. Structural and functional evolution of positively selected sites in pine glutathione s-transferase enzyme family. J of Biol Chem. 2013;288:24441–24451. doi: 10.1074/jbc.M113.456863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer B, Böger P. Binding and protection of porphyrins by glutathione S-transferases of Zea mays L. Biochem Biophys Acta. 2003;1621:226–233. doi: 10.1016/s0304-4165(03)00073-4. [DOI] [PubMed] [Google Scholar]
- Li D, Xu L, Pang S, Liu Z, Wang K, Wang C. Variable levels of glutathione S-transferases are responsible for the differential tolerance to metolachlor between maize (Zea mays) shoots and roots. J Agric Food Chem. 2017;65:39–44. doi: 10.1021/acs.jafc.6b04129. [DOI] [PubMed] [Google Scholar]
- Licciardello C, Agostino N, Traini A, Recupero GR, Frusciante L, Chiusano ML. Characterization of the glutathione S-transferase gene family through ESTs and expression analyses within common and pigmented cultivars of Citrus sinensis (L) Osbeck. BMC Plant Biol. 2014;14:39. doi: 10.1186/1471-2229-14-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Han XM, Ren LL, Yang HL, Zeng QY. Functional divergence of the glutathione S-transferase supergene family in Physcomitrella patens reveals complex patterns of large gene family evolution in land plants. Plant Physiol. 2013;161:773–786. doi: 10.1104/pp.112.205815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HJ, Tang ZX, Han XM, Yang ZL, Zhang FM, Yang HL, Liu YJ, Zeng QY. Divergence in enzymatic activities in the soybean GST supergene family provides new insight into the evolutionary dynamics of whole-genome duplicates. Mol Biol Evol. 2015;32:2844–2859. doi: 10.1093/molbev/msv156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv F, Zhou J, Zeng L, Xing D. β-cyclocitral upregulates salicylic acid signalling to enhance excess light acclimation in Arabidopsis. J Exp Bot. 2015;66:4719–4732. doi: 10.1093/jxb/erv231. [DOI] [PubMed] [Google Scholar]
- Marrs KA. The functions and regulation of Glutathione S-transferases in plants. Annu Rev Plant Physiol . 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- Martin JL. Thioredoxin—a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- McGonigle B, Keeler SJ, Lau S-MC, Koeppe MK, O’Keefe DP. A genomics approach to the comprehensive analysis of the glutathione s-transferase gene family in soybean and maize. Plant Physiol. 2000;124:1105–1120. doi: 10.1104/pp.124.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness M, Mazurkiewicz V, Brennan E, Dowling D. Dechlorination of pesticides by a specific bacterial glutathione S -transferase, BphKLB400: Potential for Bioremediation. Eng Life Sci. 2007;7:611–615. [Google Scholar]
- Moons A. Osgstu3 and osgtu4, encoding tau class glutathione S-transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots. FEBS Lett. 2003;553:427–432. doi: 10.1016/s0014-5793(03)01077-9. [DOI] [PubMed] [Google Scholar]
- Moons A. Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs) Vitam And Horm. 2005;72:155–202. doi: 10.1016/S0083-6729(05)72005-7. [DOI] [PubMed] [Google Scholar]
- Morales A, O'Rourke JA, Van M, Mortel DE. Transcriptome analyses and virus induced gene silencing identify genes in the Rpp4-mediated Asian soybean rust resistance pathway. Funct Plant Biol. 2013;40:1029–1047. doi: 10.1071/FP12296. [DOI] [PubMed] [Google Scholar]
- Mueller S, Hilbert B, Dueckershoff K, Roitsch T, Krischke M, Mueller MJ, Berger S. General detoxification and stress responses are mediated by oxi- dized lipids through TGA transcription factors in Arabidopsis. Plant Cell. 2008;20:768–785. doi: 10.1105/tpc.107.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakka S, Jugulam M, Peterson D, Asif M. Herbicide resistance: Development of wheat production systems and current status of resistant weeds in wheat cropping systems. The Crop J. 2019;7:750–760. [Google Scholar]
- Nianiou-Obeidat I, Madesis P, Kissoudis C, Voulgari G, Chronopoulou E, Tsaftaris A, Labrou NE. Plant glutathione transferase-mediated stress tolerance: functions and biotechnological applications. Plant Cell Rep. 2017;36:791–805. doi: 10.1007/s00299-017-2139-7. [DOI] [PubMed] [Google Scholar]
- Oakley AJ. Glutathione transferases: new functions. Curr Opin Struct Biol. 2005;15:716–723. doi: 10.1016/j.sbi.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Oliveira TIS, Oliveira M, Viswanathan S, Barroso MF, Barreiros L, Nunes OC, Rodrigues JA, de Lima-Neto P, Mazzetto SE, Morais S, Delerue-Matos C. Molinate quantification in environmental water by a glutathione-S-transferase based biosensor. Talanta. 2013;106:249–254. doi: 10.1016/j.talanta.2012.10.074. [DOI] [PubMed] [Google Scholar]
- Öztetik E. A Tale of Plant Glutathione S-Transferases: Since 1970. Bot Rev. 2008;74:419–437. [Google Scholar]
- Oztetik E, Kockar F, Alper M, Iscan M. Molecular characterization of zeta class glutathione S-transferases from Pinus brutia Ten. J Genet. 2015;94:417–423. doi: 10.1007/s12041-015-0538-5. [DOI] [PubMed] [Google Scholar]
- Pandey T, Singh SK, Chhetri G, Tripathi T, Singh AK. Characterization of a highly ph stable Chi Class Glutathione S-Transferase from Synechocystis PCC 6803. PLoS ONE. 2015;10:e0126811. doi: 10.1371/journal.pone.0126811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternolli C, Ghisellini P, Nicolini C. Development of immobilization techniques of cytochrome P450-GST fusion protein. Colloids Surf B. 2002;23:305–311. [Google Scholar]
- Pégeot H, Koh CS, Petre B, Mathiot S, Duplessis S, Hecker A, Didierjean C, Rouhier N. The poplar Phi class glutathione transferase: expression, activity and structure of GSTF1. Front Plant Sci. 2014;5:712. doi: 10.3389/fpls.2014.00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pégeot H, Mathiot S, Perrot T, Gense F, Hecker A, Didierjean C, Rouhier N. Structural plasticity among glutathione transferase Phi members: natural combination of catalytic residues confers dual biochemical activities. FEBS J. 2017;284:2442–2463. doi: 10.1111/febs.14138. [DOI] [PubMed] [Google Scholar]
- Peragón J, Amores-Escobar MT. Olive tree glutathione S-transferase and its response against the herbicides oxyfluorfen and glyphosate. Sci Hortic. 2018;231:194–200. [Google Scholar]
- Plomion C, Aury J, Amselem J, et al. Oak genome reveals facets of long lifespan. Nature Plants. 2018;4:440–452. doi: 10.1038/s41477-018-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prade L, Hof P, Bieseler B. Dimer interface of glutathione S-transferase rom Arabidopsis thaliana: influence of the G site architecture on the dimer interface and implications for classification. Biol Chem. 1997;378:317–320. doi: 10.1515/bchm.1997.378.3-4.317. [DOI] [PubMed] [Google Scholar]
- Puglisi I, Lo Cicero L, Lo Piero AR. The glutathione S-transferase gene superfamily: an in silico approach to study the post translational regulation. Biodegradation. 2013;24:471–485. doi: 10.1007/s10532-012-9604-3. [DOI] [PubMed] [Google Scholar]
- Rawat B, Mishra KK, Barman U, Arora L, Pal D, Paily RP. Two-Dimensional MoS2-based electrochemical biosensor for highly selective detection of glutathione. IEEE Sensors J. 2020;1:1. doi: 10.1109/JSEN.2020.2978275. [DOI] [Google Scholar]
- Rezaei MK, Shobbar ZS, Shahbazi M, Abedini R, Zare S. Glutathione S-transferase (GST) family in barley: Identification of members, enzyme activity, and gene expression pattern. J Plant Physiol. 2013;170:1277–1284. doi: 10.1016/j.jplph.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Roxas VP, Smith RK, Allen ER, Allen RD. Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat Biotechnol. 1997;15:988–991. doi: 10.1038/nbt1097-988. [DOI] [PubMed] [Google Scholar]
- Safarpour H, Safarnejad MR, Tabatabaei M, Mohsenifar A, Rad F, Basirat M, Shahryari F, Hasanzadeh F. Development of a quantum dots FRET-based biosensor for efficient detection of Polymyxa betae. Biochem Cell Biol. 2012;2012(34):507–515. [Google Scholar]
- Sharma R, Sahoo A, Devendran R, Jain M. Over-expression of a rice tau class glutathione s transferase gene improves tolerance to salinity and oxidative stresses in Arabidopsis. PLoS ONE. 2014;9:e92900. doi: 10.1371/journal.pone.0092900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HY, Li ZH, Zhang YX, Chen L, Xiang DY, Zhang YF. Two pear glutathione S-transferases genes are regulated during fruit development and involved in response to salicylic acid, Auxin, and glucose signaling. PLoS ONE. 2014;9:e89926. doi: 10.1371/journal.pone.0089926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. Cytoprotective and regulatory functions of glutathione S-transferases in cancer cell proliferation and cell death. Cancer Chemother Pharmacol. 2015;75:1–15. doi: 10.1007/s00280-014-2566-x. [DOI] [PubMed] [Google Scholar]
- Singh BR, Shaw RW. Selective inhibition of oat glutathione-S-transferase activity by tetrapyrroles. FEBS Lett. 1988;234:379–382. doi: 10.1016/0014-5793(88)80120-0. [DOI] [Google Scholar]
- Singhal SS, Singh SP, Singhal P, Horne D, Singhal J, Awasthi S. Antioxidant role of glutathione S-transferases: 4-hydroxynonenal, a key molecule in stress-mediated signaling. Toxicol Appl Pharmacol. 2015;289:361–370. doi: 10.1016/j.taap.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitou K, Muleta AW, Papageorgiou AC, Chronopoulou E, Labrou NE. Catalytic features and crystal structure of a tau class glutathione transferase from glycine max specifically upregulated in response to soybean mosaic virus infections. Biochem Biophys Acta. 2015;1854:166–177. doi: 10.1016/j.bbapap.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Smith AP, Nourizadeh SD, Peer WA, Xu J, Bandyopadhyay A, Murphy AS, Goldsbrough PB. Arabidopsis AtGSTF2 is regulated by ethylene and auxin, and encodes a glutathione S-transferase that interacts with flavonoids. Plant J. 2003;36:433–442. doi: 10.1046/j.1365-313x.2003.01890.x. [DOI] [PubMed] [Google Scholar]
- Soranzo N, Gorla MS, Mizzi L, De TG, Frova C. Organisation and structural evolution of the rice glutathione S-transferase gene family. Mol Genet Genomics. 2004;271:511–521. doi: 10.1007/s00438-004-1006-8. [DOI] [PubMed] [Google Scholar]
- Su T, Xu J, Li Y, Lei L, Zhao L, Yang H, Feng J, Liu G, Ren D. Glutathione-indole-3- acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell. 2011;23:364–380. doi: 10.1105/tpc.110.079145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre-Gonon E, Law SR, Schwartz M, Robe K, Keech O, Didierjean C, Dubos C, Rouhier N, Hecker A. Functional, Structural and Biochemical Features of Plant Serinyl-Glutathione Transferases. Front Plant Sci. 2019;10:608. doi: 10.3389/fpls.2019.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre-Gonon E, Schwartz M, Girardet JM, Hecker A, Rouhier N. Is there a role for tau glutathione transferases in tetrapyrrole metabolism and retrograde signalling in plants? Soc: Phil. Trans. R; 2020. p. B37520190404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takusagawa F. Microsomal prostaglandin E synthase type2 (mPGES2) is a glutathione dependent heme protein, and di thiothreitol dissociates the bound heme to produce active prostaglandin E2 synthase in vitro. J Biol Chem. 2013;288:10166–10175. doi: 10.1074/jbc.M112.418475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Hoagland KD, Siegfried BD. Uptake and bioconcentration of atrazine by selected freshwater algae. Environ Toxicol Chem. 1998;17:1085–1090. [Google Scholar]
- Tanikawa N, Ohmiya Y, Ohkubo H, Hashimoto K, Kangawa K, Kojima M, Ito S, Watanabe K. Identification and characterization of a novel type of membrane- associated prostaglandin Esynthase. Biochem Biophys Res Commun. 2002;291:884–889. doi: 10.1006/bbrc.2002.6531. [DOI] [PubMed] [Google Scholar]
- Thom R, Dixon DP, Edwards R, Cole DJ, Lapthorn AJ. The structure of a zeta class glutathione S-transferase from Arabidopsis thaliana: characterisation of a GST with novel active-site architecture and a putative role in tyrosine catabolism. J Mol Biol. 2001;308:949–962. doi: 10.1006/jmbi.2001.4638. [DOI] [PubMed] [Google Scholar]
- Tiwari V, Patel MK, Chaturvedi AK, Mishra A, Jha B. Functional characterization of the Tau Class Glutathione-S-Transferases Gene (SbGSTU) promoter of Salicornia brachiata under salinity and osmotic stress. PLoS ONE. 2016;2:e0148494. doi: 10.1371/journal.pone.0148494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaish S, Awasthi P, Tiwari S, Tiwari SK, Basantani MK. In silico genome-wide identification and characterization of glutathione S-transferase gene family in Vigna radiata (L.) Wilczek. Genome. 2018;61:311–322. doi: 10.1139/gen-2017-0192. [DOI] [PubMed] [Google Scholar]
- Vijayakumar H, Thamilarasan SK, Shanmugam A, Natarajan SK, Jung HJ, Park JI, Kim H, Chung MY, Nou Ill S. Glutathione transferases superfamily: cold-inducible expression of distinct GST genes in Brassica oleracea. Int J Mol Sci. 2016;17:1211. doi: 10.3390/ijms17081211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sun S, Ge W, Zhao L, Hou B, Wang K, et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;368:6493. doi: 10.1126/science.aba5435. [DOI] [PubMed] [Google Scholar]
- Warner JR, Behlen LS, Copley SD. A trade-off between catalytic power and substrate inhibition in TCHQ dehalogenase. Biochemistry. 2008;47:3258–3265. doi: 10.1021/bi702431n. [DOI] [PubMed] [Google Scholar]
- Wongsantichon J, Ketterman AJ. Alternative splicing of glutathione S-transferases. Methods Enzymol. 2005;401:100–116. doi: 10.1016/S0076-6879(05)01006-2. [DOI] [PubMed] [Google Scholar]
- Xu XY, Fan R, Zheng R, Li CM, Yu DY. Proteomic analysis of seed germination under salt stress in soybeans. J Zhejiang Univ Sci B. 2011;12:507–517. doi: 10.1631/jzus.B1100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xing XJ, Tian YS, Peng RH, Xue Y, Zhao W, Yao QH. Transgenic arabidopsis plants expressing tomato glutathione S-transferase showed enhanced resistance to salt and drought stress. PLoS ONE. 2014;10:e0136960. doi: 10.1371/journal.pone.0136960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tian YS, Xing XJ, Xu ZS, Zhu B, Fu XY, Peng RH, Yao QH. Enhancement of phenol stress tolerance in transgenic Arabidopsis plants overexpressing glutathione S-transferase. Plant Growth Regul. 2017;82:37–45. [Google Scholar]
- Yamada T, Komoto J, Watanabe K, Ohmiya Y, Takusagawa F. Crystal structure and possible catalytic mechanism of microsomal prostaglandin E synthase type2 (mPGES-2) J Mol Bio. 2005;348:1163–1176. doi: 10.1016/j.jmb.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Yang HL, Zhao YR, Wang CL, Yang ZL, Zeng QY, Lu H. Molecular characterization of a dehydroascorbate reductase from Pinus bungeana. J Integr Plant Biol. 2009;51:993–1001. doi: 10.1111/j.1744-7909.2009.00848.x. [DOI] [PubMed] [Google Scholar]
- Yang Q, Liu YJ, Zeng QY. Biochemical functions of the glutathione transferase supergene family of Larix kaempferi. Plant Physiol and Biochem. 2014;7:99–107. doi: 10.1016/j.plaphy.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Yang G, Xu Z, Peng S, Sun Y, Jia C, Zhai M. In planta characterization of a tau class glutathione S-transferase gene from Juglans regia (JrGSTTau1) involved in chilling tolerance. Plant Cell Rep. 2016;35:681–692. doi: 10.1007/s00299-015-1912-8. [DOI] [PubMed] [Google Scholar]
- Zeng QY, Lu H, Wang XR. Molecular characterization of a glutathione transferase from Pinus tabulaeformis (Pinaceae) Biochimie. 2005;87:445–455. doi: 10.1016/j.biochi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Zettl R, Schell J, Palme K. Photoaffinity labeling of Arabidopsis thaliana plasma membrane vesicles by 5-azido-[7-3H] indole-3-acetic acid: identification of a glutathione S-transferase. Proc Natl Acad Sci. 1994;91:689–693. doi: 10.1073/pnas.91.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Luo Q, Miao L, Hou C, Bai Y, Dong Z, Xu J, Liu J. Self-assembly of glutathione S-transferase into nanowires. Nanoscale. 2012;4:5847–5851. doi: 10.1039/c2nr31244a. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang Y, Ma K, Shi C, Chen W, Liu D. A novel sigma class glutathione S-transferase gene in freshwater planarian Dugesia japonica: cloning, characterization and protective effects in herbicide glyphosate stress. Ecotoxicology. 2020;29:295–304. doi: 10.1007/s10646-020-02173-9. [DOI] [PubMed] [Google Scholar]
- Zheng K, Board PG, Fei X, Sun Y, Lv S, Yan G, Liu J, Shen J, Luo G. A novel selenium containing glutathione transferase zeta1-1, the activity of which surpasses the level of some native glutathione peroxidases. Int J Biochem Cell Biol. 2008;40:2090–2097. doi: 10.1016/j.biocel.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Zhu JH, Li HL, Guo D, Wang Y, Dai HF, Mei WL, Peng SQ. Transcriptome-wide identification and expression analysis of glutathione S-transferase genes involved in flavonoids accumulation in Dracaena cambodiana. Plant Physiol and Biochem. 2016;S0981–942830:182–186. doi: 10.1016/j.plaphy.2016.05.012. [DOI] [PubMed] [Google Scholar]