Highlights
-
•
B-cell epitopes is the antigen portion binding to the immunoglobulin or antibody.
-
•
T-Cell epitope play a crucial role in eliciting vigorous protective immune response.
-
•
Antigenicity refers to the capacity of virus to bind to specific antibody molecule.
Keywords: SARS-CoV-2, COVID-19, T-cell epitopes, B-cell epitopes, Bioinformatics
Abstract
The beginning of 2020 was marked as the emergence of a COVID-19 outbreak caused by a new coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Currently, there is no vaccine or approved treatment for this infectious virus so the invention of an efficient vaccine is certainly a high priority. Some studies have employed several techniques to facilitate the combination of the immunoinformatics approach and comparative genomic approach in order to determine the potential peptides for designing the T-cell epitope-based peptide vaccine using the 2019-nCoV envelope protein as a target. Via screening the bioimmunoinformatic SARS-CoV2 derived B-cell and T-cell epitopes within the basic immunogenic of SARS-CoV2 proteins, we presented a set of inferred B-cell and T-cell epitopes from the spike (S) and nucleocapsid (N) proteins with high antigenicity and without allergenic property or toxic effects. Our findings provide a screened set of epitopes that can be introduced as potential targets for developing peptide vaccines against the SARS-CoV-2 virus.
1. Introduction
The recent outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the Chinese seafood market town of Wuhan has led to a rapid transmission to 210 countries worldwide [1]. Worldwide, scientists struggle to understand this novel and rapidly spreading virus that causes SARS-CoV-2 with the aim of developing effective involvements for controlling and preventing Coronaviruses (CoVs) which cause various lethal diseases [2].
CoVs, which belong to Nidovirales order, are enveloped viruses with the largest single-stranded RNA and genome size of 26–32 kb in length [3]. The severe acute respiratory syndrome coronavirus (SARS-CoV-1) and Middle East respiratory syndrome coronavirus (MERS-CoV) are also beta coronaviruses that are zoonotic in origin and have been linked to potential fatal illness during the outbreaks in 2003 and 2012, respectively.
Due to the rapid increase of SARS-CoV-2 infections and affected countries, efforts for developing an effective SARS-CoV-2 vaccine are urgently required. CoVs encodes multiple structural and non-structural proteins. The structural proteins include the spike (S) protein, the envelope (E) protein, the membrane (M) protein, and the nucleocapsid (N) protein. With SARS-CoV-2 being discovered very recently, there is currently insufficient information about immunogenic epitopes eliciting antibody or T-cell responses. Introductory studies suggest that SARS-CoV-2 is rather similar to SARS-CoV-1, based on phylogenetic analysis, complete genome, and similarity in the mechanism of cell entry and binding of the human cell receptor [4], [5]. Concerning the similarity between the two viruses, previous investigations that provided a reflective protective immune response against SARS-CoV-1 can potentially encourage vaccine design for SARS-CoV-2. Research also has shown that the antibody responses generated against the S protein, the most exposed protein of SARS-CoV, can be protectective against infection in animal models [6]. Furthermore, numerous studies have shown that antibodies are produced against the SARS-CoV protein N, a highly immunogenic protein and widely expressed, during infection [7]. Moreover, studies have demonstrated that SARS-CoV-2 elicits a robust B cell response and this humoral immune response causes the clearance of SARS-CoV-2. T cells likewise play a fundamental role in viral infections for example CD4 T cells provide B cell-help for antibody production which leads to long lasting protection. Hence, development of vaccines against SARS-CoV-2 that can stimulate B and T cells is highly useful.
On the one hand, as Peptide-based vaccines do not need in vitro culture of pathogenic virus, they are safe and their selectivity allows accurate activation of immune responses whereas traditional vaccines are costly, allergenic, time-consuming, and also dangerious because they require in vitro culture [8]. On the other hand, antibody-dependent enhancement (ADE) of viral entry, vaccine development, and antibody-based drug therapy have been among major concerns for epidemiologists in dealing with many viruses. ADE of virus infection is a phenomenon in which virus-specific antibodies enhance the entry of virus and in some cases the replication of virus into immune cells through interaction with Fc and/or complement receptors. ADE has been observed in coronaviruses for decades and due to the recent progress toward understanding the receptor recognition and membrane fusion mechanisms of coronavirus spikes, coronaviruses represent an excellent model system for investigating ADE of viral infections. Concerning the literature on multiple SARS-CoV-1 and MERS-CoV vaccine efforts which have failed due to ADE in animal models, it is reasonable to hypothesize a similar ADE risk for SARS-CoV2 vaccine efforts unless they specifically target domains which will block virus-immune cell fusion.
Further, some studies have demonstrated that Neutralizing MAbs targeting other parts of viral spikes would be less likely to mediate ADE if they do not trigger the conformational changes of the spikes. Hence, to reduce the likelihood of ADE, spike-based subunit vaccines lacking the receptor-binding domain (RBD) can be designed to prevent viral infections.
Through this review, we have attempted to collect the information that is totally matching and contains no mutation in the available SARS-CoV-2 sequences. These epitopes have the potential to incite an effective response against SARS-CoV-2. The aim of this systematic review (SR) is to help researchers to produce vaccine by collecting data for the control and prevention of SARS-CoV-2 and the immune and bioinformatic identification of T-cell and B-cell epitopes.
2. Methods
The present review was conducted over the rearch studies published on immune and bioinformatics identification of T-cell and B-cell epitopes in the protein structure of SARS-CoV-2. In developing our systematic review (SR) protocol, preferred reporting elements for the SR statements, meta-analyses (PRISMA) and guidelines from the Cochrane Reviewer’s Handbook were used (http://www.prisma-statement.org/Extensions/InDevelopment.aspx) [9], [10], Noorimotlagh et al. [11].
2.1. Information sources and search strategy
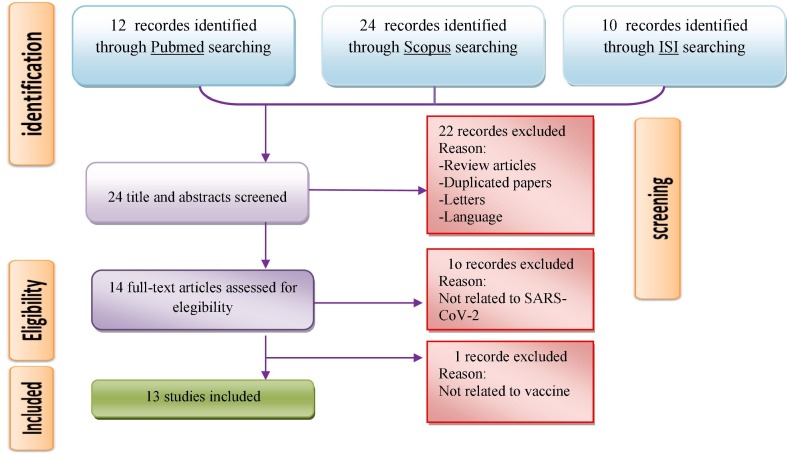
We performed a systematic bibliographic search during 2019–2020. The last search was conducted on April 24, 2020. Institute for Scientific Information (ISI) Web Science, Scopus, MEDLINE and Google Scholar databases were used to search using MeSH (Medical Subject Headings), free text words and all possible combination. The following proper keywords were used: (“nCov” OR “Novel Coronaviruses” OR “2019 Novel Coronavirus” OR “Covid-19” OR ”2019-nCoV“ OR ”Severe Acute Respiratory Syndrome- Coronaviruses-2” OR “SARS-COV-2”) AND (”B-cell“ OR ”T-cell“ OR ”Epitope“ OR ”Peptide“ OR ”Vaccine“, as illustrated in Fig. 1 .
Fig. 1.
Summary of a standard four-step protocol for literature review.
2.2. Inclusion/Exclusion criteria for the included studies
Articles were systematically reviewed and publications were selected based on the following criteria: articles needed to be a) written in English, b) original papers, c) electronicly available (online), and d) vaccine-focused research for SARS-CoV-2 and identification bioinformatics of T-cell and B-cell epitopes. The book review, book chapters, guidelines, review articles, duplicate articles, other languages (French, German, Italian, Spanish,…), letters to editors, short communications, oral presentation, conference documents and comments were considered as excludsion criteria in this systematic review. Overall, we examined articles which introduced epitopes that were identified by B and T cells and were extracted from viral antigenic proteins such as S, M, N, and E. Also, these selected epitopes needed to be analyzed in terms of antigenicity, allergenicity, and physiological properties and were presented as effective epitopes in inducing immune responses.
2.3. Data extraction
Two reviewers (SM & MA) independently investigated the titles, abstracts, and fulltexts from each database. Considering each selected study, items such as first author, country, year of publication, type of protein, B cell/T cell epitope, antigenicity, HLA Class I, HLA Class II, threshold, score and results were extracted.
3. Results
In this SR, 166 articles were identified through the initial searches. Based on title and abstract, 22 articles were excluded because they were either duplicates, reviews, letters or written in other languages. Eleven full-text papers were also excluded because they were unrelated to SARS-CoV-2 and vaccine. Finally, a total of 13 unique papers were included for the purpose of this research. Fig. 1 illustrates the flow diagram of the selection of ‘step-by-step’ studies and the number of studies identified for this review. Table 1 summarizes a detailed description of the 13 articles reviewed. Next, all the selected articles were stratified according to the characteristics of B and T cell epitopes identified in SARS-CoV-2 (Table 2, Table 3 ). Also, Table S1 contains more information about T-cell epitopes identified in 2019‐nCoV.
Table 1.
Summary of the main detailed of the included studies.
| Study ID | Type of Protein | B cell epitope | T cell epitope | Antigenicity |
HLA Class I | HLA Class II | Threshold |
Score |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B cell | T cell | B cell | T cell | B cell | T cell | ||||||
| (Grifoni et al., 2020), USA | Surface glycoprotein Nucleocapsid phosphoprotein Membrane glycoprotein sequences Orf3a protein Orf 1ab protein |
✓ | ✓ | – | – | ✓ | – | ✓ | ✓ | – | ✓ |
| (Baruah and Bose, 2020), India | Surface glycoprotein | ✓ | ✓ | ✓ | ✓ | ✓ | – | ✓ | ✓ | – | ✓ |
| (Kumar et al., 2020), India | Spike protein | – | ✓ | – | ✓ | – | – | – | – | – | ✓ |
| (Ahmed et al., 2020), China | Surface glycoprotein Nucleocapsid phosphoprotein |
✓ | ✓ | – | – | ✓ | ✓ | – | – | – | – |
| (Abdelmageed et al., 2020), Sudan | Envelope Protein | – | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | – | – |
| (Sarkar et al., 2020), Bangladesh | Nucleocapsid phosphoprotein Surface glycoprotein |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | – | – |
| (Li et al., 2020), China | Spike protein | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| (Saha and Prasad, 2020), India | Spike protein | – | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | – | ✓ |
| (Ismail et al., 2020), Pakistan | Spike protein | ✓ | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | ✓ | – |
| (Kalita et al., 2020), India | Nucleocapsid protein Membrane glycoprotein Surface spikeglycoprotein |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | – | ✓ | – |
| (Bhattacharya et al., 2020), Korea | spike protein | ✓ | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | – | – |
| (Bojin et al., 2020), Romania | Spike protein Membrane protein |
– | ✓ | – | – | ✓ | ✓ | – | – | – | ✓ |
| (Rehman et al., 2020), Pakistan | spike glycoprotein | ✓ | – | – | – | – | – | – | – | – | – |
Table 2.
Literature review of characteristics of B-cell epitopes identified in SARS-CoV-2.
| Study ID | linear B‐cell epitopes/ length | Antigenicity | Threshold | Score | Results |
|---|---|---|---|---|---|
| [12], USA |
Surface glycoprotein *287DAVDCALDPLSETKCTLKSFTVEKGIYQTSN317 / 31 *524VCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVI598 /74 *601GTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYTGS640 / 39 *802FSQILPDPSKPSKRSFIE819 /17 *888FGAGAALQIPFAMQMAYRFNGI909 / 21 Nucleocapsid phosphoprotein 1MADSNGTITVEELKKLLEQWNLVI24 /24 132PLLESELVIGAVILRGHLRI151 / 19 Membrane glycoprotein 42RPQGLPNNTASWFTALTQHGK62 / 20 153NNNAATVLQLPQGTTLPKGF172 / 19 355NKHIDAYKTFPPTEPKKDKKKKTDEAQPLPQRQKKQPTVTLLPAADM401 / 46 |
– | cutoff of 0.55 (corresponding to specificity greater than 0.81 and sensitivity below 0.3) |
– | SARS-CoV epitopes in conjunction with bioinformatic predictions points to specific regions of SARS-CoV-2 that have a high likelihood of being recognized by human immune responses. The observation that many B and T cell epitopes are highly conserved between SARS-CoV-2 and SARS-CoV is important |
| [13], India |
Surface glycoprotein CVNLTTRTQLPPAYTN / 16 NVTWFHAIHVSGTNGT / 16 SFSTFKCYGVSPTKLNDL / 18 |
1.38 0.84 1.06 |
0.51 | IFN‐γ epitope −0.92 −0.30 −0.16 |
Three sequential B cell epitopes in the viral surface glycoprotein can be potential candidates for the development of 2019‐nCoV vaccines. |
|
[14], China |
Surface glycoprotein * Nucleocapsidphosphoprotein DVVNQNAQALNTLVKQL / 17 *FFGMSRIGMEVTPSGTW / 17 EAEVQIDRLITGRLQSL / 17 *GLPNNTASWFTALTQHGK / 18 GAGICASY / 8 *GTTLPK / 6 AISSVLNDILSRLDKVE / 17 *IRQGTDYKHWPQIAQFA / 17 GSFCTQLN / 8 *KHIDAYKTFPPTEPKKDKKK / 20 ILSRLDKVEAEVQIDRL / 17 * KHWPQIAQFAPSASAFF / 17 KGIYQTSN / 8 *YNVTQAFGRRGPEQTQGNF / 19 AMQMAYRF / 8 *KTFPPTEPKKDKKKK / 15 KNHTSPDVDLGDISGIN / 17 *QLPQGTTLPKGFYAEGSRGGSQ / 22 MAYRFNGIGVTQNVLYE / 17 * LNKHIDAYKTFPPTEPK / 17 AATKMSECVLGQSKRVD / 17 *LPQGTTLPKG / 10 PFAMQMAYRFNGIGVTQ / 17 * PKGFYAEGSRGGSQASSR / 18 QALNTLVKQLSSNFGAI / 17 * QFAPSASAFFGMSRIGM / 17 QLIRAAEIRASANLAAT / 17 *QGTDYKHW / 8 QQFGRD / 6 *QLPQGTTLPKGFYAE / 15 RASANLAATKMSECVLG / 17 *QLPQGTTLPKGFYAEGSR / 18 RLITGRLQSLQTYVTQQ / 17 *TFPPTEPK / 8 EIDRLNEVAKNLNESLIDLQELGKYEQY / 28 * LLPAAD / 6 SLQTYVTQQLIRAAEIR / 17 * RRPQGLPNNTASWFT / 15 DLGDISGINASVVNIQK / 17 * SRGGSQASSRSSSRSR / 16 EVAKNLNESLIDLQELGGAALQIPFAMQMAYRFN / 34 *SQASSRSS / 8 |
– | – | – |
Only 23% and 16% of known SARS-CoV T cell and B cell epitopes map identically to SARS-CoV-2, respectively, and with no mutation having been observed in these epitopes among the available SARS-CoV-2 sequences set of SARS-CoV epitopes map identically to SARS-CoV-2 |
|
[15], Bangladesh |
Surface glycoprotein LTPGDSSSGWTAG / 13 VRQIAPGQTGKIAD / 14 YQAGSTPCNGV / 1 1 QTQTNSPRRARSV / 13 ILPDPSKPSKRS / 12 *Nucleocapsid phosphoprotein *MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRP QGLPNNTAS / 51 *KTFPPTEPKKDKKKKADETQALPQRQKKQQ / 30 *GTTLPKGFYAEGSRGGSQASSRSSSRSRNSSRNSTPGSSRGTSP ARMAGNGGD / 53 *SKMSGKGQQQQGQTVTKKSAAEASKKPRQKRTATKAYN / 38 *RIRGGDGKMKDL / 12 *TGPEAGLPYGANK / 13 |
Antigenic (0.534) Antigenic (0.709) |
Antigenicity (tumor model, threshold = 0.4) |
– |
Potential subunit vaccines were designed against the SARS-CoV-2 using various methods of reverse vaccinology and immunoinformatics |
|
[16], China |
Spike protein 407VRQIAPGQTGKIAD420 / 14 1033VLGQSKRVDFCGKG1046 / 14 545GLTGTGVLTESNKK558 / 14 417KIADYNYKLPDDFT431 / 14 |
1.2606 1.3582 1.0227 0.9567 |
0.9 | 0.9 | The B- and T-cell epitopes identified here may assist the development of potent peptide-based vaccines to address the SARS-CoV-2 |
| [17], Pakistan |
S protein (spike protein) SQCVNLTTRTQLPPAYTNSFTRGVY / 25 FSNVTWFHAIHVSGTNGTKRFDN / 23 DPFLGVYYHKNNKSWME / 17 MDLEGKQGNFKNL / 13 KHTPINLVRDLPQGFS / 16 TPGDSSSGWTA / 11 KSFTVEKGIYQTSNFRVQP / 19 FPNITNLCPFGEVFNATRFASVYAWNRKRISNCVA / 35 YNSASFSTFKCYGVSPTKLNDLCFT / 25 GDEVRQIAPGQTGKIADYNYKLP / 23 NLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN / 62 ELLHAPATVCGPKKSTNLVKN / 21 VNNSYECDIPI / 11 ASYQTQTNSPRRARSVASQ / 19 YTMSLGAENSVAYSNN / 16 VNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGI / 40 SCCKFDEDDSEPVLKG / 16 |
– | – | Linear B cell epitopes were mapped using Bepipred Linear Epitope Prediction 2.0 and those with score greater than 0.5 were subjected to T-cell epitopes identification step |
SARS-CoV-2 spike glycoprotein for antigenic peptides and proposed a multi-epitope peptide vaccine construct (MEPVC) by means of several computational immunological methods and biophysical calculations |
| [18], India |
Nucleocapsid protein TRRIRGGDGKMKDLSP / 16 KSAAEASKKPRQKRTA / 16 EGALNTPKDHIGTRNP / 16 Membrane glycoprotein RSMWSFNPETNILLNV / 16 SFRLFARTRSMWSFNP / 16 Surface spike glycoprotein YACWHHSIGFDYVYNP / 16 VVKIYCPACHNSEVGP / 16 TLKGGAPTKVTFGDDT / 16 TSRYWEPEFYEAMYTP / 16 |
0.987 1.053 1.0412 |
– | 0.94 0.93 0.93 0.89 0.88 0.96 0.96 0.95 0.94 |
A multi-epitope-based subunit vaccine was including highly antigenic epitopes from three virus proteins and had a good protective efficacy and safety against SARS-CoV-2 infection |
| [19], Korea |
Spike glycoprotein 22SQCVNLTTRTQLPPAYTNSFTRGVY46 / 25 68FSNVTWFHAIHVSGTNGTKRFDN90 / 23 106KS107 /2 147DPFLGVYYHKNNKSWME163 / 17 186MDLEGKQGNFKNL198 /13 215KHTPINLVRDLPQGFS230 / 16 259TPGDSSSGWTA269 / 11 302LDPL305 / 4 313KSFTVEKGIYQTSNFRVQP331 / 19 338FPNITNLCPFGEVFNATRFASVYAWNRKRISNCVA372 / 35 378YNSASFSTFKCYGVSPTKLNDLCFT402 / 25 413GDEVRQIAPGQTGKIADYNYKLP435 / 23 449NLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN510 / 62 525ELLHAPATVCGPKKSTNLVKN545 / 21 564SNKKFLPF571 / 8 589QTLE592 / 4 611TNTSN615 / 5 625NCTEVPVAIHADQLTPT641 / 17 643RVYSTGSNVFQ653 / 11 665VNNSYECDIPI675 / 11 681ASYQTQTNSPRRARSVASQ699 / 19 704YTMSLGAENSVAYSNN719/ 16 757E757 / 1 782EQDKNTQ788 / 7 795KQIYKTPPIKDFGGF809 15 816PDPSKPSK823 / 8 837LADAGFIKQYGDCLG851 / 15 997EAEVQ1001 / 5 1044GQSKRVDFC1052 / 9 1116RNFYEPQIITTD1127 / 12 1142VNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGI1181 / 40 1212LGKY1215 / 4 1261SCCKFDEDDSEPVLKG1276 / 16 1278K1278 / 1 |
– | – | – |
immunoinformatic analysis pointed out 13 MHC‐I and 3 MHC‐II epitopes within the spike glycoprotein of SARS‐COV‐2. These epitopes are the ideal candidate to formulate a multi‐epitopic peptide vaccine, not only because of being selected from the linear B‐cell epitopic region but also because of their antigenic property was confirmed |
| [20], Pakistan |
Spike glycoprotein 4FLVLLPLVSSQCVNL18 / 15 617QVAVLYQDV627 / 9 34RGVYYPDK41 / 8 644CTEVPVAIHAD653 / 11 44RSSVLHST51 / 8 667AGCLIGA674 / 7 53DLFLPFFS60 / 8 667GAGICASY674 / 8 65FHAIHV70 / 6 687VASQSII693 / 7 81NPVLPFN87 / 7 723TTEILPVS730 /8 115QSLLIVN121 / 7 735SVDCTMY741 /7 125NVVIKVCEFQ134 / 10 750SNLLLQYGSFCTQL763 /14 136CNDPFLGVYYH146 / 11 781VFAQVKQI788 /8 168FEYVSQP174 / 7 803SQILPD808 /6 210INLVRDL216 / 7 837YGDCLGD843 /7 223LEPLVDLP230 / 8 837RDLICAQ853 /7 239QTLLALHRSY248 / 10 858LTVLPPL864 / 7 263AAYYVGYL270 / 8 873YTSALLAG880 /8 288PRTFLLK295 / 7 959LNTLVKQL966 / 8 333AVDCALDP339 / 8 973ISSVLND979 / 7 359TNLCPFG371 / 7 1003SLQTYVTQQ1011 / 9 376SNCVADYSVLYNS385 / 13 1030SECVLGQS1037 / 8 430TFKCYGVSPT435 / 10 1057PHGVVFLHVTYVPA1070 / 14 488TGCVIA495 / 6 1079PAICHDG1085 / 7 505CYFPLQSY527 / 8 1123SGNCDVVIGI1132 / 10 592YQPYRVVVLSFELLHAPATVCGP599 / 23 1174ASVVNI1179 / 6 607FGGVSVIT615 / 8 1221IAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKF1256 / 36 |
– | – | – | Potential T-cells and B-cell epitopes (continuous) have been recognized from SARS-CoV-2 main protease, Nsp12 RNA polymerase, Spike and Nsp13 helicase proteins to design multi-epitope construct (MVC) by using adjuvants (hβ defensins) and appropriate linkers |
Table 3.
Literature review of characteristics of T-cell epitopes identified in SARS-CoV-2.
| Study ID | T cell epitope | MHC I & MHC II | Antigenicity | Threshold | Score | Results |
|---|---|---|---|---|---|---|
|
[12], USA |
S protein N protein *IRGWIFGTTLDSKTQSLL *ALNTPKDHI *CTFEYVSQPFLMD *LQLPQGTTL *QPFLMDLEGKQGN *GDAALALLLL *TRFQTLLALHRSYLTPGD *LALLLLDRL *KSFTVEKGIYQTSNFRVQ *LLLDRLNQL *SASFSTFKCYGVSPTKL *RLNQLESKM *KLPDDFTGCV *TKAYNVTQAF *NLDSKVGGNYNYLYRLFR *GMSRIGMEV *YLYRLFRKSNLKPFERDI *MEVTPSGTWL *KPFERDISTEIYQ *NFKDQVILL *QSIIAYTMSLGAENSVAY Orf 1ab protein *TECSNLLLQYGSFCTQL *WLMWLIINL *VKQIYKTPPIKDFGGFNF *ILLLDQALV *DSLSSTASALGKLQDVV *SACVLAAEC *ALNTLVKQL *SLPGVFCGV *VLNDILSRL *TLMNVLTLV *LITGRLQSL *SMWALIISV *QLIRAAEIRASANLAATK *CLEASFNYL *HWFVTQRNFYEPQII M protein *RLNEVAKNL *GLMWLSYFI *NLNESLIDL *HLRIAGHHL *FIAGLIAIV *TLACFVLAAV *SIIAYTMSL Orf 3a protein *RIFTIGTVTLKQGEI *TVTLKQGEI |
S | – |
HLA class II = . cutoff%20 HLA class I = cutoff ≥ 6%. |
RF ≥ 0.3 | Virus Parallel bioinformatic predictions identified a priori potential B and T cell epitopes for SARS-CoV-2. |
|
[13], India |
Surface glycoprotein *YLQPRTFLL *GVYFASTEK *EPVLKGVKL *VVNQNAQAL *WTAGAAAYY |
S | * (0.45)% * (0.71)% * (1.23)% * (0.47)% * (0.63)% |
0.5 | 0.83/0.64 0.58/0.98 0.73/0.61 0.77/0.78 0.82/0.54 |
One overlapping CTL epitope between MERS‐CoV and 2019‐nCoV with one gap and one mismatch |
|
[23], India |
Spike protein *RISNCVADY *CVADYSVLY * RSFIEDLLF * RVDFCGKGY * MTSCCSCLK * VLKGVKLHY |
S | *357 *361 *815 *1039 *1237 *1264 |
– | scores > 1.25 = highest sensitivity | Spike glycoprotein sequences of 2019-nCoV and SARS-CoV exhibits 76.2% identity, 87.2% similarity and 2% Gaps |
| [14], China |
N protein S protein *ILLNKHID *GAALQIPFAMQMAYRF *AFFGMSRIGMEVTPSGTW *MAYRFNGIGVTQNVLY *MEVTPSGTWL *QLIRAAEIRASANLAATK *GMSRIGMEV *FIAGLIAIV *ILLNKHIDA *ALNTLVKQL *ALNTPKDHI *LITGRLQSL *IRQGTDYKHWPQIAQFA *NLNESLIDL *KHWPQIAQFAPSASAFF *QALNTLVKQLSSNFGAI *LALLLLDRL *RLNEVAKNL *LLLDRLNQL *VLNDILSRL *LLNKHIDAYKTFPPTEPK *LQLPQGTTL *LQLPQGTTL *AQFAPSASAFFGMSR *AQFAPSASAFFGMSRIGM *RRPQGLPNNTASWFT *YKTFPPTEPKKDKKKK |
S | – | – | – |
Only 23% and 16% of known SARS-CoV T cell and B cell epitopes map identically to SARS-CoV-2, respectively |
| [8], Sudan |
MHC-1 Peptide MHC-II Peptide *YVYSRVKNL *KPSFYVYSRVKNLNS *SLVKPSFYV *VKPSFYVYSRVKNLN *SVLLFLAFV *LVKPSFYVYSRV *FLAFVVFLL *PSFYVYSRVKNLNSS *VLLFLAFVV *NIVNVSLVKPSFYVY *RLCAYCCNI *LLVTLAILTALRLCA *FVSEETGTL *SFYVYSRVKNLNSSR *LTALRLCAY *LVTLAILTALRLCAY *LVKPSFYVY *VTLAILTALRLCAYC *NIVNVSLVK *CNIVNVSLVKPSFYV |
S | 0.6025 | 0.4 | – | the following 10 peptide in MHC1 with the highest world population, coverage as a good candidate for vaccine, another 10 peptides in MHC2 as candidates for vaccine designed based on the world population percentage |
|
[15], Bangladesh |
MHC-I& N protein MHC-II&N protein *AGLPYGANK *LIRQGTDYKHWP *AADLDDFSK *RLNQLESKMSGK *QLESKMSGK *LDRLNQLESKM MHC-I&S protein *LNQLESKMSGKG *SVLNDILSR *QELIRQGTDYKH *GVLTESNKK MHC-II&S protein *RLFRKSNLK * SNFRVQPTESIV *QIAPGQTGK * LLIVNNATNVVI * TSNFRVQPTESI |
S | S | 0.4 | – | potential subunit vaccines were designed against the SARS-CoV-2 using various methods of reverse vaccinology and immunoinformatics |
| [16], China |
MHC-1 MHC-11 *VGYQPYRVVVLSFEL *FAMQMAYRFNGIGVT *NFTISVTTEILPVSM *LPIGINITRF *LVLLPLVSSQCVNLT *IAIVMVTIM *VVFLHVTYVPAQEKN *TNFTISVTTEILPVS *GYQPYRVVVLSFELL *PTNFTISVTTEILPV *ALQIPFAMQMAYRFN |
S | MHC-1 S MHC-II S |
MHC-1 1% MHC-II 10% |
MHC-1 = 3 MHC-II = 5 |
The B- and T-cell epitopes can potentially promote an immune response in the host via the vaccine against SARS-CoV-2 |
|
[21] India |
MHC-I MHC-II *TLDSKTQSL *IGINITRFQ *GKQGNFKNL *YGFQPTNGV *CYGVSPTKL *VLSFELLHA *KIADYNYKL *LQIPFAMQM *VVVLSFELL *IAIVMVTIM |
S | MHC-1 S MHC-II S |
0.4 | score > 1 | 5 MHC I and 5 MHC II epitopes with high antigenic potential and strong binding affinity within the S protein of 2019- nCoV |
| [17], Pakistan |
Spike protein *TRTQLPPAY *FPNITNLCP *RTQLPPAYT *FSTFKCYGV *TTRTQLPPA *PTKLNDLCF *FSNVTWFHA *GQTGKIADY *TWFHAIHVS *IAPGQTGKI *PRRARSVAS *FRKSNLKPF *YYHKNNKSW *NLKPFERDI *KQGNFKNLR *KSNLKPFER *KHTPINLVR *PKKSTNLVK *PGDSSSGWT *KKSTNLVKN *YQTSNFRVQ *PNITNLCPF *NSYECDIPI *YAWNRKRIS *FNATRFASV |
S | S | 0.5 | – | SARS-CoV-2 spike glycoprotein for antigenic peptides and proposed a MEPVC by means of several computational immunological methods and biophysical calculations |
| [18], India |
Nucleocapsid protein *RIAGHHLGR *GTWLTYTGAIKLD *LPKEI TVAT *AALALLLLDRLNQ Surface spike glycoprotein *LLLDRLNQL *ATRFASVYAWNRK *GMSRIGMEV *ITRFQTLLALHRSY *KSAAEASKK *EFVFKNIDGYFKIY *KTFPPTEPK *YLQPRTFLL *FPRGQGVPI *KIADYNYKL *KPRQKRTAT *RLFRKSNLK *Membrane Glycoprotei *GVYFASTEK *RLFARTRSM *SPRRARSVA *NRFLYIIKLIFLWLL *IPTNFTISV *GLMWLSYFI *FVLAAVYRI *LSYFIASFR |
S | Nucleocapsi d protein = 0.9871 Membrane Glycoprotein =1.0532 Surface spike glycoprotein =1.0412 |
– | – | Multi-epitope-based subunit vaccine has a probability to show good protective efficacy and safety against SARS-CoV-2 infection in humans |
|
[19] Korea |
Spike protein *SQCVNLTTR *NSYECDIPI *YTNSFTRGV *SPRRARSVA *GVYYHKNNK *LGAENSVAY *GKQGNFKNL *KQIYKTPPI *TPINLVRDL *FIKQYGDCL *GIYQTSNFR *RNFYEPQII *NLCPFGEVF *VNNTVYDPL *FASVYAWNR *ELDSFKEEL *ASFSTFKCY *FKNHTSPDV *VSPTKLNDL *DEDDSEPVL *KIADYNYKL *IHVSGTNGT *KVGGNYNYL *VYYHKNNKS *RLFRKSNLK *LVRDLPQGF *FERDISTEI *VFNATRFAS *EGFNCYFPL *YRLFRKSNL *ELLHAPATV *FERDISTEI *GPKKSTNLV *YQTQTNSPR *TEVPVAIHA *FKNHTSPDV *RVYSTGSNV |
S | S | 0.4 | – | pointed out 13 MHC‐I and 3 MHC‐II epitopes within the spike glycoprotein of SARS‐COV‐2 |
| [22], Romania |
MHC-I&S protein MHC-I&M protein *YLQPRTFLL *GLMWLSYFI *KIADYNYKL *FVLAAVYRI *FQFCNDPFL *KLLEQWNLV *SIIAYTMSL *ATSRTLSYY *VLNDILSRL *YFIASFRLF *LTDEMIAQY *SYFIASFRL *WTAGAAAYY *YANRNRFLY *NYNYLYRLF *VATSRTLSY *QYIKWPWYI *FAYANRNRF *IPFAMQMAY MHC-II&M protein *LPFNDGVYF *SYFIASFRLFARTRS *VASQSIIAY *AVILRGHLRIAGHH *FAMQMAYRF *EITVATSRTLSYYK *LGAENSVAY MHC-II&S protein *GNYNYLYRLFRKSN *IRAAEIRASANLAA *INLVRDLPQGFSAL |
S | – | – | S | Candidate peptides could counteract the novel China coronavirus by eliciting both CD4 + and CD8 + T cell responses |
4. Discussion
The outbreak of the new coronavirus (SARS-CoV-2) in China and its subsequent pandemic raised a great deal of global concerns. Confining this viral infection in human communities requires adequate information on its transmission routes, severity and complications, as well as access to appropriate therapeutics and vaccines [23]. Efforts to develop an effective vaccine against the SARS-CoV-2 aim to limit and then eradicate the infection from human societies. However, producing an effective SARS-CoV-2 vaccine is problematic due to the lack of sufficient information about its biological characteristics and immune responses against it [14]. Thus, it seems that the immunoinformatics-based approachs are required to investigate the immunogenic epitopes and vaccine design using data from proteins sequencing of the COVID-19.
The SARS-CoV-2 is an RNA virus with four structural proteins; S (Spike), E (Envelope), N (Nucleocapsid), and M (Membrane). These structural proteins play important roles in the entry of the virus into host cells and subsequent compartmentation of viral particles. Immune responses including antibody production against the structural proteins are essential to limit the spread of the infection. Therefore, efforts have been dedicated to design and produce peptide vaccines targeting immunogenic epitopes on the viral structural proteins to help immune cells to quickly identify these important viral epitopes [23], [16].
T and B lymphocytes are two important types of immune cells in the body [13], [15]. Upon entry of an antigen into the body, it is directed to helper T-cells through MHC-II molecules expressed by antigen-presenting cells (APC). While helper T-cells express the CD4 surface marker, another subset of lymphocytes, known as cytotoxic T-cells, express CD8. Antigens are presented to these cells though MHC-I molecules. On the other hand, activated helper T-cells induce B lymphocytes which ultimately secrete large amounts of neutralizing antibodies. Consequent antibody-dependent enhancement (ADE) of viral entry has been detected for many viruses. It was revealed that antibodies selected not only one serotype of viruses but also sub-neutralize additional, principal ADE of the latter viruses [24]. The original mechanism for ADE includes a neutralizing antibody binding to the virus-surface structure protein of coronaviruses like a viral receptor, activating a conformational modification of the spike, and facilitating viral entry into IgG-Fc-receptor-expressing cells through recognized viral-receptor-dependent pathways. In spite of the sequential relationship between the timings of severe SARS and antibody expansion, other possible elucidations for the detected pathology have not been scientifically proved. Previous antibody expansion could be determined by grooming effect from previous human CoV infections with no effect on pathogenesis regardless of the time-based coincidence with other severe diseases. Moreover, the kinetics of antibody response is also known to be influenced by viral load and innate immune reactions [25].
The helper cells also activate cytotoxic T-cells which directly kill infected cells [15]. Therefore, it is important to identify the SARS-CoV-2 virus epitopes that induce efficient T-helper, T-cytotoxic, and B-cells to develop vaccines against the virus. SARS-CoV-2 closest relative among human coronaviruses is SARS-CoV, with 79% genetic similarity. There is 72% similarity in the amino acid sequence of the RBDs of SARS-CoV and SARS-CoV-2, with highly similar 3D structures. Homology modelling and biophysical properties designate that the SARS-CoV-2 RBD domain binds to ACE2 with 10 to 20-fold higher affinity than that of SARS-CoV [26].
Many bioinformatics studies have been conducted in recent months to determine epitopes which are detected by T and B cells. We have presented some of these studies in Table 1 to Table 3. In these studies, the proteins of COVID-19 virus that have an essential role in virulence of the virus was selected and then its antigenic properties was examined. After the antigenic analysis of these proteins, goal proteins such as S or N proteins were selected for final analysis. The Immune Epitope Database or IEDB that contains a huge collection of experimental data on T-cell epitopes was used for the prediction of epitope of helper T-cell based on their percentile rank and the median inhibitory concentration (IC50) values. Also NetCTL 1.2 and NetMHC pan EL 4.0 servers were used for the prediction of CTL epitope [15]. Epitopes identified by T-cells are short peptide fragments that must be evaluated for their abilities to bind MHC-I and MHC-II molecules. The epitopes selected for developing vaccines should also have high affinity for MHC-I and II molecules and present the highest world population coverage [14]. Linear epitopes identified by B-cells are generally longer than those identified by T cells and can be parts of structural proteins. Such epitopes, if being immunogenic, can induce more effective humoral responses against viral infections. For the prediction of linear B-cell epitopes in SARS-CoV-2 surface glycoprotein, nucleocapsid phosphoprotein, and membrane glycoprotein, these studies used BepiPred 2.0 and ABCPred servers [27].
The designed vaccine candidate must represent high antigenic property to be able to stimulate effective immune responses against viral infections without the need for any adjuvant and also must not induce an allergic reaction to the body. So the epitopes selected in the previous step in these studies were examined in terms of antigenicity (using the VaxiJen server), allergenicity (using the AllerTOP server), and physicochemical parameters (using ProtParam server). Finally, epitopes with high antigenicity and without allergenic or toxic effects were introduced as potential targets for developing peptide vaccines against SARS-CoV-2 virus.
Advanced epitope maps (T-cell and B-cell tests) are required to decide on the potential of recognized epitopes to trigger a positive resistant reaction against SARS-CoV-2. This would offer assistance to help refine the detailed set of epitope, based on the immunogenicity inspected; an critical thought for the immunogenic plan. Overall, as the distinguished set of SARS-CoV epitopes is indistinctly distinguished from SARS-CoV-2, they show valuable potential candidacies for directing exploratory endeavors towards creating immunizations against SARS-CoV-2. More for the most part, our deliberate assistance highlights the potential importance of past clinical and exploratory considerations for SARS-CoV, and is used in conjunction with growing information for SARS-CoV-2, in the search for compelling immunizations to combat the COVID-19 outbreak. Characteristically all vaccines were created using live or weakened microorganisms or parts of them. However, the use of whole organisms, their components or the biological procedure for vaccine production has several weaknesses. The presence of immunologically dismissed biological components or biological impurities in such vaccines might cause main complications. All the unfavorable features of traditional vaccines might be incredulous via the development of entirely synthetic peptide-based vaccines. However, once only minimal antigenic epitopes are applied for immunisation, the immune responses are poor. The use of an adjuvant can overcome this hindrance; however, it may increase new malfunctions [28].
5. Conclusions
B and T cells are two important arms of the immune system against pathogens, including viruses. Activation of these cells plays an important role in the body's defenses. These cells are activated in both, natural active (pathogen entry) and artificial active (vaccine) forms. The vaccine is now being used to treat many infections. In the present study, we reviewed epitopes of structural proteins identified as effective and recognizable targets by immune (B&T) cells for SARS-CoV-2 virus. This can help early stages of vaccine development attempts to produce more effective peptide vaccines.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank the Ilam University of Medical Sciences, Ilam, Iran for the financially support (Grant No: 991002/8).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2020.106738.
Contributor Information
Sanaz Mami, Email: sani_vet@yahoo.com.
Mahdieh Azizi, Email: Mirzaee.seyyed@gmail.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Din M.A.U., Boppana L.K.T. An update on the 2019-nCoV outbreak. Am. J. Infect. Control. 2020 doi: 10.1016/j.ajic.2020.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong N.S., Zheng B.J., Li Y.M., Poon L.L.M., Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z., Kong W., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ying L.I.N., Xu S., Yang R.F., Li Y.X., Ji Y.Y., He Y.Y., De Shi M., Wei L.U., Shi T.L., Jin W. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13:141–145. doi: 10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdelmageed M.I., Abdelmoneim A.H., Mustafa M.I., Elfadol N.M., Murshed N.S., Shantier S.W., Makhawi A.M. Design of multi epitope-based peptide vaccine against E protein of human 2019-nCoV: An immunoinformatics approach. BioRxiv. 2020 doi: 10.1155/2020/2683286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohamoud Y.A., Mumtaz G.R., Riome S., Miller D., Abu-Raddad L.J. The epidemiology of hepatitis C virus in Egypt: a systematic review and data synthesis. BMC Infect. Dis. 2013;13:288. doi: 10.1186/1471-2334-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noorimotlagh Z., Mirzaee S.A., Martinez S.S., Rachoń D., Hoseinzadeh M., Jaafarzadeh N. Environmental exposure to nonylphenol and cancer progression Risk–A systematic review. Environ. Res. 2020;109263 doi: 10.1016/j.envres.2020.109263. [DOI] [PubMed] [Google Scholar]
- 11.Noorimotlagh Z., Mirzaee S.A., Ahmadi M., Jaafarzadeh N., Rahim F. The possible DNA damage induced by environmental organic compounds: The case of Nonylphenol. Ecotoxicol. Environ. Saf. 2018:158. doi: 10.1016/j.ecoenv.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 12.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020 doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar B., Ullah M.A., Johora F.T., Taniya M.A., Araf Y. The essential facts of Wuhan novel coronavirus outbreak in China and epitope-based vaccine designing against 2019-nCoV. BioRxiv. 2020 [Google Scholar]
- 16.Li L., Sun T., He Y., Li W., Fan Y., Zhang J. Epitope-based peptide vaccines predicted against novel coronavirus disease caused by SARS-CoV-2. BioRxiv. 2020 doi: 10.1016/j.virusres.2020.198082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ismail S., Ahmad S., Azam S.S. Immuno-informatics characterization SARS-CoV-2 spike glycoprotein for prioritization of epitope based multivalent peptide vaccine. bioRxiv. 2020 doi: 10.1016/j.molliq.2020.113612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.P. Kalita, A.K. Padhi, K.Y. Zhang, T. Tripathi, Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2, 2020. [DOI] [PMC free article] [PubMed]
- 19.Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Lee S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J. Med. Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.H.M. Rehman, M.U. Mirza, M. Saleem, M. Froeyen, S. Ahmad, R. Gul, M.S. Aslam, M. Sajjad, M.A. Bhinder, A putative prophylactic solution for COVID-19: development of novel multiepitope vaccine candidate against SARS-COV-2 by comprehensive immunoinformatic and molecular modelling approach, 2020. [DOI] [PMC free article] [PubMed]
- 21.Saha R., Prasad B.V.L.S. In silico approach for designing of a multi-epitope based vaccine against novel Coronavirus (SARS-COV-2) bioRxiv. 2020 [Google Scholar]
- 22.F. Bojin, O. Gavriliuc, M.-B. Margineanu, V. Paunescu, Design of an Epitope-Based Synthetic Long Peptide Vaccine to Counteract the Novel China Coronavirus (2019-nCoV), 2020.
- 23.Kumar S., Maurya V.K., Prasad A.K., Bhatt M.L.B., Saxena S.K. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) VirusDisease. 2020:1–9. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Alwis R., Chen S., Gan E.S., Ooi E.E. Impact of immune enhancement on Covid-19 polyclonal hyperimmune globulin therapy and vaccine development. EBioMedicine. 2020 doi: 10.1016/j.ebiom.2020.102768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., He L., Chen Y., Wu J., Shi Z. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020:94. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tay M.Z., Poh C.M., Renia L., Macary P.A., Ng L.F. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020:1–12. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu A., Sarkar A., Maulik U. Strategies for vaccine design for corona virus using Immunoinformatics techniques. bioRxiv. 2020 [Google Scholar]
- 28.Skwarczynski M., Toth I. Peptide-based synthetic vaccines. Chem. Sci. 2016;7:842–854. doi: 10.1039/c5sc03892h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.