Abstract
Papaver nudicaule L. (Iceland poppy) is widely used for ornamental purposes. A previous study demonstrated the alleviation of lipopolysaccharide-induced inflammation mediated by P. nudicaule extract through nuclear factor-kappa B and signal transducer and activator of transcription 3 inactivation. As isoquinoline alkaloids are chemical markers and bioactive constituents of Papaver species, the present study investigated the alkaloid profile of aerial parts of five P. nudicaule cultivars with different flower colors and a P. rhoeas cropped for two years. A combination of liquid chromatography high-resolution mass spectrometry and molecular networking was used to cluster isoquinoline alkaloids in the species and highlight the possible metabolites. Aside from the 12 compounds, including rotundine, muramine, and allocryptopine, identified from Global Natural Products Social library and reported information, 46 structurally related metabolites were quantitatively investigated. Forty-two and 16 compounds were proposed for chemical profiles of P. nudicaule and P. rhoeas, respectively. Some species-specific metabolites showed similar fragmentation patterns. The alkaloid abundance of P. nudicaule differed depending on the flower color, and the possible chemical markers were proposed. These results show that molecular networking-guided dereplication allows investigation of unidentified metabolites. The derived chemical profile may facilitate evaluation of P. nudicaule quality for pharmacological applications.
Keywords: Papaver nudicaule, alkaloids, LC-MS/MS chemical profiling, GNPS molecular networking
1. Introduction
Plants in the Papaver L. genus from Papaveraceae (commonly known as poppy) family have been used for traditional medicinal practices and ornamental purposes for a long time. Papaver rhoeas L. (Corn poppy) and Papaver nudicaule L. (Iceland poppy) are widely used for ornamentation owing to their multicolored flowers. Nudicaulins, which are flavonoid-derived indole alkaloids, along with pelargonidin, kaempferol, and gossypetin glycosides, are responsible for the white, red, orange, and yellow colored petals of different P. nudicaule cultivars [1,2]. P. nudicaule belongs to the Papaveraceae family, and is an annual herbaceous species. This plant can produce various alkaloids, particularly isoquinoline alkaloids (IAs) [3]. IAs from Papaver species possess potent pharmacological properties, including narcotic analgesic, antimicrobial, muscle relaxant, cough suppressant, and anticancer effects [4,5]. However, studies on chemical constituents and biological activities of Papaver species have mainly been performed on P. somniferum, which produces morphine alkaloids [6].
To understand the metabolites of other Papaver species and their biological activities, the metabolite profiling of IAs in P. rhoeas was previously performed using liquid chromatography (LC) coupled with quadrupole time-of-flight (Q-TOF) mass spectrometry (MS) [7]. P. nudicaule extracts have anti-inflammatory activity and decrease the inflammatory response induced by lipopolysaccharide by inhibiting the nuclear factor-kappa B (NF-κB) and signal transducer and activator of transcription 3 (STAT3) signaling pathway in the previous report [8]. The study proposed that IAs, such as allocryptopine, from P. nudicaule could be the active constituents and a representative subset for this species. However, IAs of P. nudicaule have been rarely reported, except for flavonols; indole alkaloid skeleton of yellow petals [9,10,11] and some alkaloids of aerial parts, including 8, 14-dihydroflavinantine, pseudoprotopine, allocryptopine, dihydroamuronine, amuronine, amurensinine N-oxide A, and (−)-amurensinine N-oxide B [12,13].
Alkaloid profiling of P. nudicaule is crucial to investigate its pharmaceutical potential. However, a major challenge for MS-based chemical profiling of P. nudicaule is the identification of alkaloids. To the best of our knowledge, the compound information and MS/MS spectra of P. nudicaule have been rarely reported. Our main interest is to highlight the species-specific metabolites and propose a reliable alkaloid profile. Therefore, this study utilized the Global Natural Products Social (GNPS) molecular networking platform and significantly accelerated the MS/MS-based clustering of alkaloids, even for compounds with no spectral matches in any public database or in-house MS/MS library [14,15].
This study aimed to identify IAs and investigate their abundance between P. nudicaule and P. rhoeas, and further analyze the chemical markers in specimens from aerial parts of five cultivars of P. nudicaule based on liquid chromatography high-resolution mass spectrometry (LC-HRMS) and molecular network analysis.
2. Results
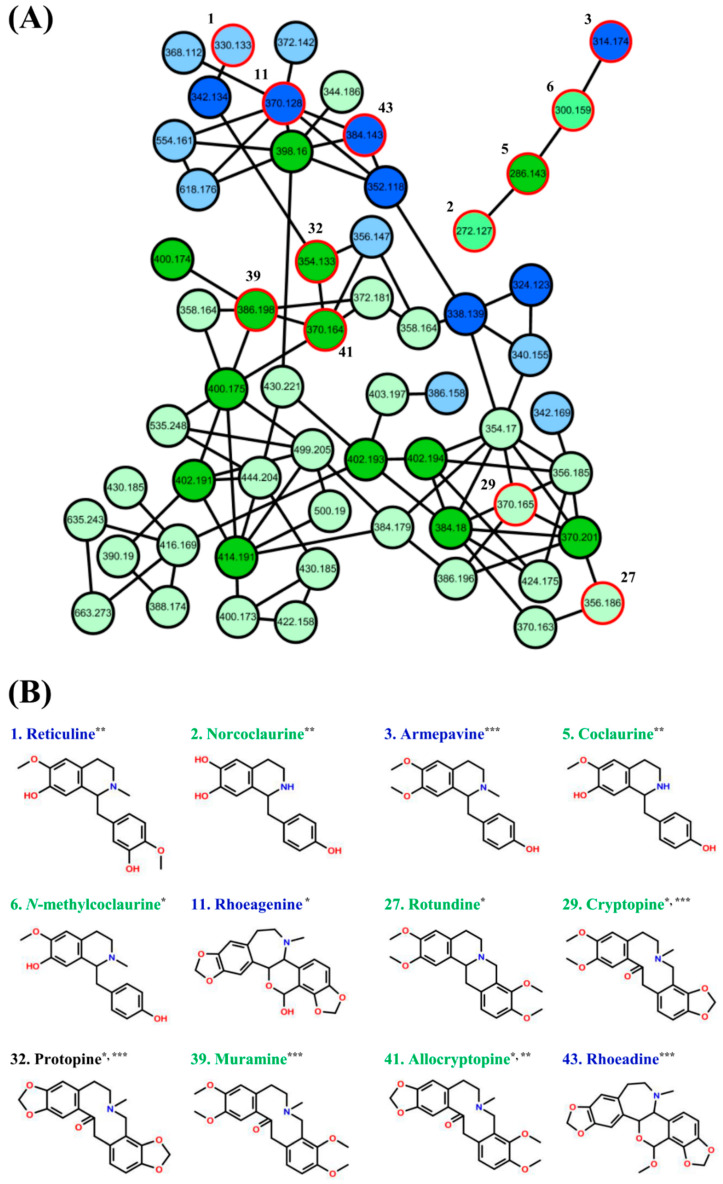
2.1. Molecular Network-Based Annotation of Species-Specific Metabolites
To organize the compound candidates for the alkaloid profiling of P. nudicaule and P. rhoeas, the LC-MS/MS data from 12 extracts was submitted to molecular networking through the GNPS web platform (http://gnps.ucsd.edu, ID: d38cc1e769524002ad2c2122f7375f08). The 12 extracts were prepared from the five cultivars of P. nudicaule with different flower colors (orange, purple, scarlet, white, and yellow), and a P. rhoeas sample was collected for two years (2017 and 2018). The extract from aerial parts of P. nudicaule with white bloom in 2017 is abbreviated as 17NW. The same format is followed for other extracts (i.e., orange, NO; purple, NP; scarlet, NS; and yellow, NY). The extract of aerial parts of P. rhoeas collected in 2018 is abbreviated as 18RA. The resulting molecular network comprised 2121 molecular features and 102 independent clusters with at least three features (Supplementary Figure S1). A total of six alkaloids were identified through MS/MS library matching of GNPS. The nodes belonged to two molecular clusters (Figure 1A). The two clusters comprising 58 molecular features were selected as species-specific alkaloids from the chemical profiles of P. nudicaule and P. rhoeas. This strategy was based on molecular networking using spectral similarity that allowed grouping of structurally related metabolites [16]. Although the molecular networking platform significantly increased structural annotations and clustering of Papaver species, the MS/MS library matching unfortunately failed to cover all metabolites. Indeed, 44 IAs were identified from P. rhoeas in the previous report [7], but the molecular network-guided profiling in this study missed some of these alkaloids. The unidentified alkaloids belong to other clusters that are not yet characterized are speculated.
Figure 1.
(A) Alkaloid-targeted molecular network generated from the ethanolic extracts of P. nudicaule and P. rhoeas. Colors inside nodes represent species-specific metabolites: green, P. nudicaule; blue, P. rhoeas. (B) Nodes with red outer circles represent ions dereplicated by GNP libraries (*), authentic compounds (**), and literature (***). Identified ions are presented with their common names and chemical structures.
Within the 58 metabolites, six alkaloids were further annotated by manually comparing their MS/MS spectra with the in-house MS/MS library and previous literature [17,18]. The molecular network-guided approach reduced this laborious step by narrowing down the possible MS/MS features. Twelve alkaloids were identified (Figure 1B), and 46 metabolites were proposed to be structurally related alkaloids. Table 1 summarizes the chromatographic information (retention times), high-resolution MS/MS-based information (observed mass, formulas, MS/MS fragment ions), and quantitative information (abundant species, fold changes in peak areas) of each metabolite.
Table 1.
Molecular network-based alkaloids of P. nudicaule and P. rhoeas by liquid chromatography (LC)- quadrupole time-of-flight (QTOF)-mass spectrometry (MS)/MS.
| Peak No. | Rt (min) | Observed Mass (Da) | Calculated Mass (Da) | Formula | Adduct Form | MS/MS Fragment Ions (m/z) | Species | Fold Change * |
|---|---|---|---|---|---|---|---|---|
| 1 | 4.68 | 330.1326 | 330.1342 | C19H24NO4 | [M + H]+ | 263.0700, 194.0804 *, 189.0688, 177.0769 *, 176.0703 * | P. rhoeas | −1.92 |
| 2 | 5.17 | 272.1274 | 272.1287 | C16H18NO3 | [M + H]+ | 256.1035 *, 238.0969, 162.0625, 108.0529 *, 107.0501 * | P. nudicaule | 0.41 |
| 3 | 5.73 | 314.1745 | 314.1756 | C19H24NO3 | [M + H]+ | 271.1322, 269.1156, 237.0897, 175.0758, 107.0499 * | P. rhoeas | −1.84 |
| 4 | 6.07 | 554.1616 | 554.1630 | C38H22N2O3 | [M + H]+ | 352.1170 *, 334.1059, 190.0855 * | P. rhoeas | −1.98 |
| 5 | 6.16 | 286.1437 | 286.1443 | C17H20NO3 | [M + H]+ | 270.1225 *, 238.0931, 210.0994, 108.0535 *, 107.0498 * | P. nudicaule | 1.10 |
| 6 | 6.27 | 300.1589 | 300.1600 | C18H22NO3 | [M + H]+ | 270.1193 *, 238.0928, 176.0773, 108.0514 *, 107.0482 * | P. nudicaule | 1.07 |
| 7 | 6.31 | 358.1643 | 358.1655 | C20H24NO5 | [M + H]+ | 277.0835, 191.0929, 190.0851 *, 151.0750 | P. nudicaule | 2.28 |
| 8 | 6.48 | 342.1335 | 342.1342 | C19H20NO5 | [M + H]+ | 263.0708, 235.0753, 206.0803, 176.0704 *, 165.0543 | P. rhoeas | −1.92 |
| 9 | 6.48 | 368.1123 | 368.1134 | C20H18NO6 | [M + H]+ | 350.1043, 332.0949, 261.0567, 188.0705 * | P. rhoeas | −1.67 |
| 10 | 6.64 | 342.1694 | 342.1705 | C20H24NO4 | [M + H]+ | 297.1128 *, 282.0899, 265.0851 *, 237.0906, 191.0858 | P. rhoeas | −0.77 |
| 11 | 6.67 | 370.1282 | 370.1291 | C20H20NO6 | [M + H]+ | 352.1191 *, 334.1087, 320.0927, 190.0865 *, 188.0710 * | P. rhoeas | −3.26 |
| 12 | 6.82 | 372.1424 | 372.1447 | C20H22NO6 | [M + H]+ | 354.1340, 322.1152, 204.1051 *, 192.1013 | P. rhoeas | −0.81 |
| 13 | 6.91 | 618.1764 | 618.1791 | C39H26N2O6 | [M + H]+ | 370.1305, 352.1171 *, 321.0745, 190.0835 | P. rhoeas | −2.60 |
| 14 | 6.94 | 535.2476 | 535.2444 | C30H35N2O7 | [M + H]+ | 518.2158, 504.2009 *, 320.1053, 220.0970 *, 205.0732 | P. nudicaule | 3.35 |
| 15 | 7.11 | 344.1864 | 344.1862 | C20H26NO4 | [M + H]+ | 298.1471, 267.1026, 191.0932, 190.0876 * | P. nudicaule | 0.56 |
| 16 | 7.12 | 358.1641 | 358.1655 | C20H24NO5 | [M + H]+ | 340.1564, 278.0934, 194.0830, 176.0704 * | P. nudicaule | 0.33 |
| 17 | 7.27 | 370.1635 | 370.1655 | C21H24NO5 | [M + H]+ | 338.1369, 238.0629, 192.1021 * | P. nudicaule | 2.07 |
| 18 | 7.38 | 372.1808 | 372.1811 | C21H26NO5 | [M + H]+ | 354.1692, 291.1015, 222.1118, 204.1017 *, 190.0853 | P. nudicaule | 2.20 |
| 19 | 7.39 | 386.1581 | 386.1604 | C21H24NO6 | [M + H]+ | 306.1193, 206.1168, 191., 190.0865 * | P. rhoeas | −1.67 |
| 20 | 7.78 | 356.1475 | 356.1498 | C20H22NO5 | [M + H]+ | 340.1547 *, 325.1345, 267.0990, 192.1015 *, 177.0782 | P. rhoeas | −1.26 |
| 21 | 7.93 | 430.1854 | 430.1866 | C23H28NO7 | [M + H]+ | 412.1746 *, 350.1138, 220.0961 *, 218.0798, 205.0839 | P. nudicaule | 2.25 |
| 22 | 8.00 | 402.1930 | 402.1917 | C22H28NO6 | [M + H]+ | 384.1809 *, 335.1278, 206.1175 *, 193.0857, 179.0701 | P. nudicaule | 3.25 |
| 23 | 8.01 | 390.1907 | 390.1917 | C21H28NO6 | [M + H]+ | 372.1808 *, 310.1199, 208.0969 *, 193.0724 | P. nudicaule | 2.85 |
| 24 | 8.05 | 356.1859 | 356.1862 | C21H26NO4 | [M + H]+ | 206.1178 *, 190.0868, 162.0911 | P. nudicaule | 2.92 |
| 25 | 8.07 | 340.1547 | 340.1549 | C20H22NO4 | [M + H]+ | 323.1095, 277.0838, 192.1016 *, 177.0790 | P. rhoeas | −1.67 |
| 26 | 8.30 | 352.1182 | 352.1185 | C20H18NO5 | [M + H]+ | 334.1082, 320.0924, 190.0863 * | P. rhoeas | −3.99 |
| 27 | 8.97 | 356.1857 | 356.1862 | C21H26NO4 | [M + H]+ | 325.1384, 249.1838, 192.1017 *, 177.0777 | P. nudicaule | 2.78 |
| 28 | 9.13 | 400.1745 | 400.1760 | C22H26NO6 | [M + H]+ | 382.1670, 341.1352, 282.1280, 204.1006 *, 165.0901 * | P. nudicaule | 2.08 |
| 29 | 9.19 | 370.1650 | 370.1655 | C21H24NO5 | [M + H]+ | 352.1534, 291.1014, 222.1122, 205.1094 *, 204.1018 * | P. nudicaule | 2.10 |
| 30 | 9.30 | 414.1901 | 414.1917 | C23H28NO6 | [M + H]+ | 220.0969 *, 205.0740 | P. nudicaule | 3.84 |
| 31 | 9.38 | 388.1750 | 388.1760 | C21H26NO6 | [M + H]+ | 370.1647 *, 352.1541 *, 336.1232, 322.1189, 308.1273 | P. nudicaule | 2.59 |
| 32 | 9.47 | 354.1338 | 354.1342 | C20H20NO5 | [M + H]+ | 275.0710, 247.0758, 206.0812, 189.0779 *, 188.0708 * | P. nudicaule | 0.82 |
| 33 | 9.47 | 403.1974 | 403.1995 | C22H29NO6 | [M + H]+ | 385.1845, 354.1717, 280.1043, 207.1207 *, 206.1171 * | P. nudicaule | 3.67 |
| 34 | 9.48 | 384.1797 | 384.1811 | C22H26NO5 | [M + H]+ | 352.1535, 325.1429, 206.1183 *, 190.0870 | P. nudicaule | 3.83 |
| 35 | 9.48 | 402.1935 | 402.1917 | C22H28NO6 | [M + H]+ | 384.1807 *, 353.1394, 325.1411, 206.1183 *, 190.0869 | P. nudicaule | 3.73 |
| 36 | 9.49 | 424.1749 | 424.1760 | C24H26NO6 | [M + H]+ | 384.1911, 214.0871, 206.1176 * | P. nudicaule | 2.78 |
| 37 | 9.59 | 400.1747 | 400.1760 | C22H26NO6 | [M + H]+ | 382.1643, 206.0815 *, 191.0586 | P. nudicaule | 3.57 |
| 38 | 9.60 | 422.1580 | 422.1604 | C24H24NO6 | [M + H]+ | 351.1177, 206.0833 *, 191.0590 * | P. nudicaule | 2.14 |
| 39 | 10.05 | 386.1980 | 386.1968 | C22H28NO5 | [M + H]+ | 368.1870, 306.1264 *, 222.1139, 204.1025 *, 190.0872 | P. nudicaule | 3.65 |
| 40 | 10.10 | 416.1691 | 416.1709 | C22H26NO7 | [M + H]+ | 398.1606, 222.0767 *, 205.0733 | P. nudicaule | 2.41 |
| 41 | 10.48 | 370.1643 | 370.1655 | C21H24NO5 | [M + H]+ | 290.0944 *, 206.0813, 188.0709 *, 181.0861 | P. nudicaule | 2.72 |
| 42 | 10.52 | 338.1385 | 338.1392 | C20H20NO4 | [M + H]+ | 277.0861, 249.0912, 190.0859 *, 149.0596 | P. rhoeas | −1.67 |
| 43 | 10.80 | 384.1433 | 384.1447 | C21H22NO6 | [M + H]+ | 352.1182 *, 334.1080, 320.0922, 190.0858 *, 188.0707 * | P. rhoeas | −4.37 |
| 44 | 11.04 | 370.2009 | 370.2018 | C22H28NO4 | [M + H]+ | 206.1178 *, 190.0868 | P. nudicaule | 4.08 |
| 45 | 11.16 | 324.1227 | 324.1236 | C19H18NO4 | [M + H]+ | 250.0942, 176.0707 *, 149.0596 * | P. rhoeas | −3.29 |
| 46 | 11.28 | 635.2425 | 635.2393 | C37H35N2O8 | [M + H]+ | 499.2079, 398.1601 *, 380.1486, 220.0967 * | P. nudicaule | 2.05 |
| 47 | 11.70 | 500.1908 | 500.1921 | C26H30NO9 | [M + H]+ | 456.2026, 397.1888, 220.0972 *, 205.0742 | P. nudicaule | 0.97 |
| 48 | 11.71 | 398.1595 | 398.1604 | C22H24NO6 | [M + H]+ | 382.1282 *, 364.1176, 336.1231 *, 193.0859 | P. nudicaule | 3.51 |
| 49 | 11.78 | 386.1957 | 386.1968 | C22H28NO5 | [M + H]+ | 222.1129 *, 161.0831 | P. nudicaule | 3.33 |
| 50 | 12.15 | 354.1695 | 354.1705 | C21H24NO4 | [M + H]+ | 338.1361, 190.0864 *, 149.0593 | P. nudicaule | 3.11 |
| 51 | 15.51 | 402.1907 | 402.1917 | C22H28NO6 | [M + H]+ | 384.1812 *, 322.1200, 220.0975 *, 205.0734 | P. nudicaule | 3.85 |
| 52 | 15.75 | 400.1748 | 400.1760 | C22H26NO6 | [M + H]+ | 382.1649, 320.1051, 220.0965 *, 205.0736 | P. nudicaule | 3.47 |
| 53 | 16.00 | 663.2720 | 663.2706 | C39H39N2O8 | [M + H]+ | 499.2075, 398.1603 *, 380.1482, 220.0972 * | P. nudicaule | 2.55 |
| 54 | 16.01 | 384.1793 | 384.1811 | C22H26NO5 | [M + H]+ | 322.1197, 220.0970 *, 205.0729 | P. nudicaule | 2.77 |
| 55 | 16.15 | 444.2040 | 444.2022 | C24H30NO7 | [M + H]+ | 384.1814 *, 322.1204, 220.0974 *, 205.0730 | P. nudicaule | 3.51 |
| 56 | 16.18 | 430.1852 | 430.1866 | C23H28NO7 | [M + H]+ | 370.1661 *, 322.1185, 206.0810 *, 191.0576 | P. nudicaule | 2.35 |
| 57 | 16.30 | 499.2038 | 499.2022 | C33H27N2O3 | [M + H]+ | 351.1147, 320.1046, 220.0969 *, 205.0733 | P. nudicaule | 2.94 |
| 58 | 16.36 | 430.2216 | 430.2230 | C24H32NO6 | [M + H]+ | 384.1808 *, 353.1377, 206.1173 *, 192.1008 | P. nudicaule | 2.21 |
* Relative fold changes between the two species were calculated from the ratio of the mean peak areas of P. nudicaule samples to that of P. rhoeas samples.
2.2. Characterized Alkaloids of P. nudicaule
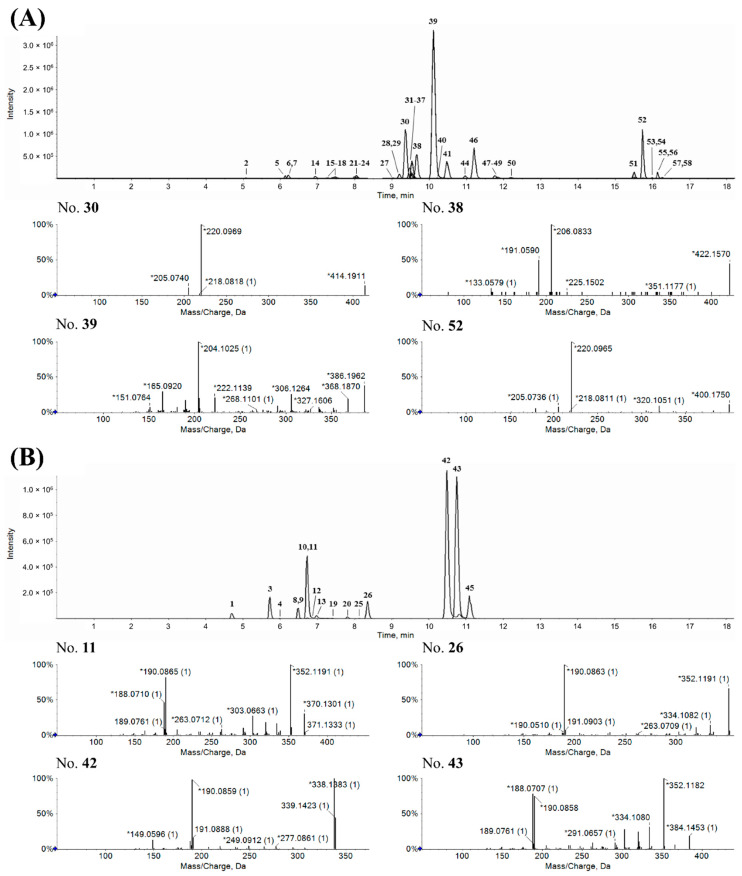
Representative chromatograms for the selected IAs are shown in Figure 2. The major metabolites of P. nudicaule revealed some specific molecular ions upon MS/MS fragmentation. As shown in Figure 2A, compounds 30 and 52 generated fragment ions at m/z 220.10, whereas compounds 38 and 39 generated fragment ions at m/z 206.08 and m/z 204.10, respectively. The molecular ions at m/z 204.10, m/z 206.08, m/z 206.12 (data not shown), and m/z 220.10 could be diagnostic ions of P. nudicaule, as most major metabolites generated one of the fragment ions (Table 1). However, the major metabolites of P. rhoeas (compounds 11, 26, 42, and 43) were mainly fragmented to molecular ions with m/z 190.09 (Figure 2B).
Figure 2.
Representative extracted ion chromatograms of P. nudicaule (A) and P. rhoeas (B) based on clustered alkaloids. Specific metabolites of each species were numbered according to their retention times, and the fragmentation patterns of major peaks are presented.
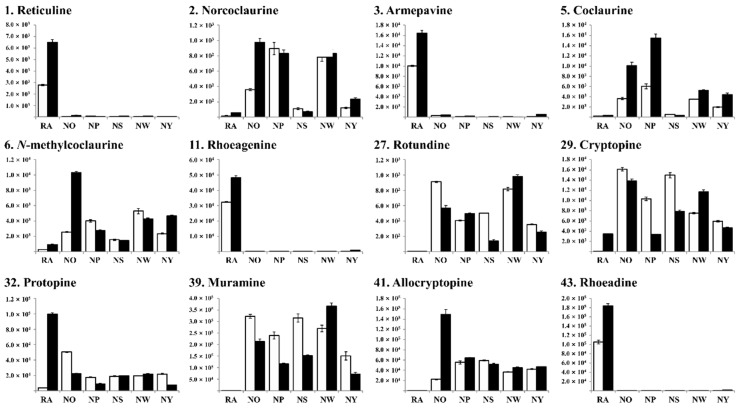
A relative quantification analysis was performed to explore the alkaloid abundance in P. nudicaule and P. rhoeas. Among the 58 compounds within two molecular clusters, 42 and 16 were abundant in P. nudicaule and P. rhoeas, respectively. As shown in Supplementary Table S1, the peak areas of each compound and the relative fold change between the two species were calculated. Considering the mean peak areas of P. nudicaule samples, species-specific alkaloids are summarized in Table 1 depending on fold change values. Furthermore, 12 identified alkaloids were quantitatively compared based on their peak areas. Figure 3 presents their abundance in 12 specimens harvested for two years (2017 and 2018) comprising five cultivars of P. nudicaule blooms and a P. rhoeas. Reticuline (1), armepavine (3), rhoeagenine (11), and rhoeadine (43) were more abundant in P. rhoeas. The levels of demethylcoclaurine (2), coclaurine (5), N-methylcoclaurine (6), rotundine (27), muramine (39), and allocryptopine (41) were much higher in P. nudicaule (Figure 3). Protopine (32) was detected in both species among protopine-type alkaloids.
Figure 3.
The abundance of the identified alkaloids in P. nudicaule and P. rhoeas, as determined by liquid chromatography high-resolution mass spectrometry (LC-HRMS). Bars with white and black colors represent specimens of 2017 and 2018, respectively. Reticuline (1), demethylcoclaurine (2), armepavine (3), coclaurine (5), N-methylcoclaurine (6), rhoeagenine (11), rotundine (27), cryptopine (29), protopine (32), muramine (39), allocryptopine (41), and rhoeadine (43).
2.3. Alkaloid-Based Multivariate Analysis
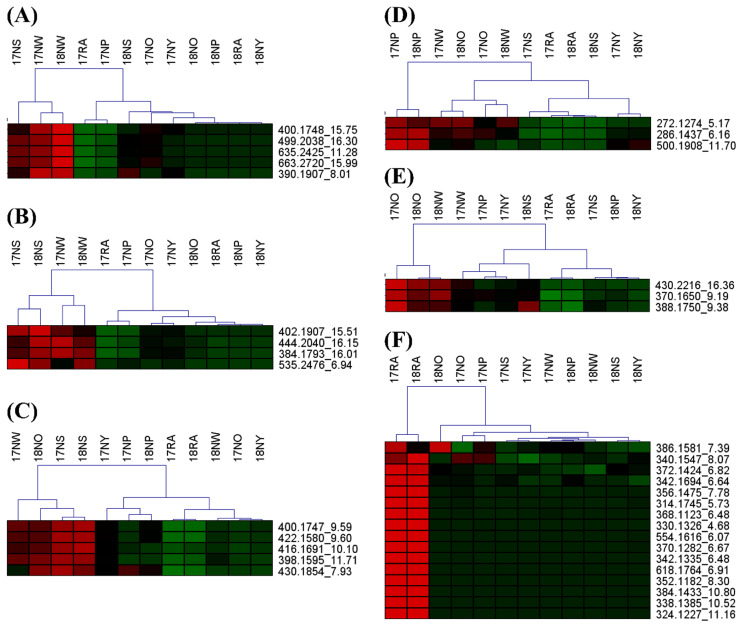
A multivariate analysis was performed for 12 specimens based on molecular network-guided IAs. Figure 4 presents the K-means clustering for 58 identified alkaloids. Representative compounds are proposed. Compounds 23, 46, 52, 53, and 57 were significantly abundant in NW (Figure 4A), while compounds 14, 51, 54, and 55 were abundant in both NW and NS (Figure 4B). Compounds 21, 37, 38, 40, and 48 in NS (Figure 4C); 2, 5, and 47 in NP (Figure 4D); and 29, 31, and 58 in NO (Figure 4E) could be representative metabolites. None was represented for the specimen with yellow colored flower. Sixteen metabolites were significantly abundant in P. rhoeas as compared with those in P. nudicaule (Figure 4F), suggesting that alkaloid production in the two species depends on different biosynthesis pathways and that, even within the same species, the abundance of IAs is very different for each P. nudicaule cultivar depending on flower color. These results further the understanding of the biosynthesis of alkaloids in P. nudicaule, and the quantitative variation may explain the difference in biological activities within the same species. Protopine detected in both P. rhoeas and P. nudicaule (Figure 3) is also one of the IAs found in Sanguinaria canadensis L., another species in the family Papaveraceae. Protopine has various pharmacological and biological activities, including anti-inflammatory, anti-infectious, neuroprotective, and antithrombotic effects [19]. Further investigations should address the correlation between their chemical composition and biological activities.
Figure 4.
Heatmaps of the molecular features illustrating K-means clustering for the 58 alkaloids of P. nudicaule and P. rhoeas derived from molecular network-guided analysis. Specimens from each analysis were hierarchically clustered. Representative metabolites were proposed for (A) NW, (B) NS and NW, (C) NS, (D) NP, (E) NO, and (F) RA.
3. Materials and Methods
3.1. Plant Material
The aerial parts of P. nudicaule L. and P. rhoeas L. were harvested by the Korea National Academy of Agricultural Science, Rural Development Administration. The five cultivars of P. nudicaule with different flower colors (orange, purple, scarlet, white, and yellow) and P. rhoeas samples collected for two years (2017 and 2018) were immediately frozen in liquid nitrogen and stored at −70°C in a deep freezer. For each specimen, experiments were performed in triplicate under the same conditions.
3.2. Reagents and Chemicals
The suppliers of the reference standard for IAs were previously described [5]. High-performance liquid chromatography (HPLC)-grade acetonitrile and methanol were purchased from Honeywell Burdick and Jackson (Morristown, NJ, USA). Formic acid and ammonium formate were procured from Sigma-Aldrich (St. Louis, MO, USA). Deionized water was obtained using a pure water purification system (Human Co., Seoul, Korea).
3.3. Sample Preparation
Aerial parts of lyophilized Papaver species were ground into a fine powder. Two grams of all samples were ultrasonicated in 5 mL of ethanol for 30 min. The supernatants were filtered through 0.2 μm polytetrafluoroethylene syringe filters and dried using a SAVANT SPD2010 SpeedVacTM concentrator (Thermo Scientific, Asheville, NC, USA). Ethanolic extracts were dissolved in HPLC-grade methanol to 2.0 mg/mL and either directly analyzed or stored at −70°C until analysis.
3.4. LC and MS Analysis
The LC-MS system comprised a Vanquish UHPLC system (Thermo Fisher Scientific, Sunnyvale, CA, USA) with an Acquity UPLC HSS T3 column (2.1 mm × 100 mm, 1.7 μm; Waters) and a Triple TOF 5600+ mass spectrometer system (Sciex, Foster City, CA, USA). The ultrahigh performance liquid chromatography (UHPLC) system used 0.05% formic acid and 2.5 mM ammonium formate in water as eluent A and acetonitrile as eluent B. The optimized elution program was as follows: 0–2.5 min (1% B), 2.5–3.0 min (1–10% B), 3.0–6.0 min (10–19% B), 6.0–9.0 min (19–22% B), 9.0–14.0 min (22–25% B), 14.0–17.0 min (25–70% B), 17.0–19.0 min (70–100% B), 19.0–22.0 min (100% B), and equilibration with 1% B for 3 min at a flow rate of 0.4 mL/min. The column was maintained at 40 °C, and the auto-sampler was held at 4 °C. The injection volume of each sample solution was 1 μL. The MS/MS data were acquired by an information-dependent acquisition scan at positive-ion mode, and the parameters were as follows: mass range 50–1500 m/z, ion spray voltage, 4.5 kV; source temperature, 450 °C; declustering potential, 50 V; nitrogen as nebulizer gas, 50 L/min; heater gas, 50 L/min; curtain gas, 25 L/min; and collision energy, 10 eV.
3.5. LC-MS/MS Data Processing
The acquired AB Sciex dataset (.wiff) was directly imported into MZmine 2.53 [20]. Each detection of ms1 and ms2 levels was filtered with ions showing a minimum height 1.0 × 103 and 5, respectively, and the extracted ion chromatograms (XICs) were built with ions showing a minimum time span of 0.02 min, a minimum height of 1.0 × 103, and an m/z tolerance of 0.002 Da (or 10.0 ppm). The chromatograms were deconvoluted by a baseline cut-off algorithm using the following parameters: minimum peak height 1.0 × 103, peak duration range 0.02–0.25 min, and baseline level 1.0 × 10. The deconvoluted peaks were deisotoped using an isotopic peaks grouper algorithm with an m/z tolerance of 0.002 Da (or 10.0 ppm) and a retention time tolerance of 0.1 min, and aligned together into a peak list using a join aligner module and following parameters: m/z tolerance at 0.005 Da (or 20.0 ppm), weight for m/z of 70, absolute retention time tolerance of 0.1 min, and weight for m/z of 30. The chromatogram was gap-filled by a peak finder module with an intensity tolerance of 10.0%, m/z tolerance of 0.002 Da (or 10.0 ppm), and absolute retention time tolerance of 0.2 min. The processed peak list was eventually exported to an mgf file using the GNPS-FBMN module for GNPS molecular networking [21].
3.6. Identification of Species-Specific Metabolites Using Molecular Network Processing
The MZmine-processed LC-MS/MS peak lists were window-filtered by choosing only the top six peaks in the ± 50 Da window throughout the spectrum. A network was created where edges were filtered to have a cosine score > 0.70 and more than four matched peaks. Further edges between two nodes were retained in the network only if each node appeared in every other’s respective top-10 most similar nodes. The spectra in the network were then searched against the spectral library of GNPS. The library spectra were filtered in the same manner as the input data. The molecular network was visualized using Cytoscape 3.7.2 [22].
3.7. Multivariate Analysis
Identified alkaloids and peak areas were scaled by unit variance based on the year of specimen collection. The scaled data were subjected to K-means clustering and hierarchical tree construction using Multi-experiment Viewer (MeV) version 4.9.0 [23]. The K-means clustering was performed using Pearson’s correlation with 12 cluster numbers and optimized order of molecular features, and the hierarchical trees were constructed using complete linkage clustering with optimized order of sample leaf.
4. Conclusions
In the present study, a combination of LC-HRMS and molecular networking was applied to cluster structurally related compounds in natural products and highlight the possible metabolites important for the chemical profiling of the species. As a case study, IAs from P. nudicaule and P. rhoeas were investigated. The chemical annotation was significantly increased using the molecular networking platform. A total of 58 metabolites were selected and quantitatively analyzed. Forty-two and 16 compounds were proposed as chemical profiles of P. nudicaule and P. rhoeas, respectively. The alkaloid abundance of P. nudicaule was different depending on the color of flowers. These results suggest that the LC-HRMS and molecular networking-guided dereplication method can be a powerful tool for the chemical investigation of unidentified metabolites and chemotaxonomic approaches in the phytomedicine field. Together, these results may contribute to the understanding of the biosynthesis of alkaloids in P. nudicaule and the evaluation of the quality of this plant for further pharmaceutical applications.
Acknowledgments
We would like to thank Editage (www.editage.co.kr) for English language editing.
Supplementary Materials
The following are available online, Figure S1: Molecular network from ethanolic extracts of Papaver nudicaule and Papaver rhoeas; Table S1: LC-MS/MS information for clustered metabolites.
Author Contributions
Conceptualization, K.S., J.-H.O., and I.J.H.; Methodology, K.S.; Software, K.S., M.Y.L.; Formal analysis, K.S., M.Y.L.; Investigation, K.S., J.-H.O., Resources, J.-H.O., S.-G.L., and I.J.H.; Data curation, K.S.; Writing–original draft preparation, K.S., J.-H.O.; Writing–review and editing, S.-G.L., I.J.H.; Supervision, S.-G.L., I.J.H.; Project administration, S.-G.L., I.J.H.; Funding acquisition, J.-H.O., S.-G.L., and I.J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-2018R1D1A1B07047610) and was carried out with the support of "Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01045502)" Rural Development Administration, Republic of Korea.
Conflicts of Interest
The authors have declared no conflict of interest.
Footnotes
Sample Availability: Samples of the examined materials are available from the authors.
References
- 1.Harborne J.B. Plant polyphenols—XV: Flavonols as yellow flower pigments. Phytochemistry. 1965;4:647–657. doi: 10.1016/S0031-9422(00)86234-2. [DOI] [Google Scholar]
- 2.Tatsis E.C., Böhm H., Schneider B. Occurrence of nudicaulin structural variants in flowers of papaveraceous species. Phytochemistry. 2013;92:105–112. doi: 10.1016/j.phytochem.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Cordell G.A. Fifty years of alkaloid biosynthesis in phytochemistry. Phytochemistry. 2013;91:29–51. doi: 10.1016/j.phytochem.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Hagel J.M., Facchini P.J. Benzylisoquinoline alkaloid metabolism: A century of discovery and a brave new world. Plant Cell Physiol. 2013;54:647–672. doi: 10.1093/pcp/pct020. [DOI] [PubMed] [Google Scholar]
- 5.Farrow S.C., Hagel J.M., Facchini P.J. Transcript and metabolite profiling in cell cultures of 18 plant species that produce benzylisoquinoline alkaloids. Phytochemistry. 2012;77:79–88. doi: 10.1016/j.phytochem.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Ziegler J., Facchini P.J., Geissler R., Schmidt J., Ammer C., Kramell R., Voigtlander S., Gesell A., Pienkny S., Brandt W. Evolution of morphine biosynthesis in opium poppy. Phytochemistry. 2009;70:1696–1707. doi: 10.1016/j.phytochem.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Oh J.-H., Ha I.J., Lee M.Y., Kim E.-O., Park D., Lee J.-H., Lee S.-G., Kim D.-W., Lee T.-H., Lee E.-J., et al. Identification and metabolites profiling of alkaloids in aerial parts of Papaver rhoeas by liquid chromatography coupled with quadrupole time-of-Flight tandem mass spectrometry. J. Sep. Sci. 2018;41:2517–2527. doi: 10.1002/jssc.201701402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh J.-H., Yun M., Park D., Ha I.J., Kim C.-K., Kim D.-W., Kim E.-O., Lee S.-G. Papaver nudicaule (Iceland poppy) alleviates lipopolysaccharide-Induced inflammation through inactivating NF-κB and STAT3. BMC Complement. Altern. Med. 2019;19:90. doi: 10.1186/s12906-019-2497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schliemann W., Schneider B., Wray V., Schmidt J., Nimtz M., Porzel A., Bohm H. Flavonols and an indole alkaloid skeleton bearing identical acylated glycosidic groups from yellow petals of Papaver nudicaule. Phytochemistry. 2006;67:191–201. doi: 10.1016/j.phytochem.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Tatsis E.C., Schaumloffel A., Warskulat A.C., Massiot G., Schneider B., Bringmann G. Nudicaulins, yellow flower pigments of Papaver nudicaule: Revised constitution and assignment of absolute configuration. Org. Lett. 2013;15:156–159. doi: 10.1021/ol303211w. [DOI] [PubMed] [Google Scholar]
- 11.Dudek B., Warskulat A.-C., Schneider B. The occurrence of flavonoids and related compounds in flower sections of Papaver nudicaule. Plants. 2016;5:28. doi: 10.3390/plants5020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Istatkova R., Philipov S., Yadamsurenghiin G.O., Samdan J., Dangaa S. Alkaloids from Papaver nudicaule L. Nat. Prod. Res. 2008;22:607–611. doi: 10.1080/14786410701605315. [DOI] [PubMed] [Google Scholar]
- 13.Philipov S., Istatkova R., Yadamsurenghiin G.O., Samdan J., Dangaa S. A new 8,14-dihydropromorphinane alkaloid from Papaver nudicaule L. Nat. Prod. Res. 2007;21:852–856. doi: 10.1080/14786410701494777. [DOI] [PubMed] [Google Scholar]
- 14.Kang K.B., Ernst M., Hooft J.J.J., Silva R.R., Park J., Medema M.H., Sung S.H., Dorrestein P.C. Comprehensive mass spectrometry-Guided phenotyping of plant specialized metabolites reveals metabolic diversity in the cosmopolitan plant family Rhamnaceae. Plant J. 2019;98:1134–1144. doi: 10.1111/tpj.14292. [DOI] [PubMed] [Google Scholar]
- 15.Olivier-Jimenez D., Chollet-Krugler M., Rondeau D., Beniddir M.A., Ferron S., Delhaye T., Allard P.-M., Wolfender J.-L., Sipman H.J.M., Lucking R., et al. A database of high-Resolution MS/MS spectra for lichen metabolites. Sci. Data. 2019;6:294. doi: 10.1038/s41597-019-0305-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst M., Kang K.B., Caraballo-Rodriquez A.M., Nothias L.-F., Wandy J., Chen C., Wang M., Rogers S., Medema M.H., Dorrestein P.C., et al. MolNetEnhancer: Enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites. 2019;9:144. doi: 10.3390/metabo9070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong E.-K., Lee S.Y., Yu S.M., Park N.H., Lee H.-S., Yim Y.-H., Hwang G.-S., Cheong C., Jung J.H., Hong J. Identification of structurally diverse alkaloids in Corydalis species by liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2012;26:1661–1674. doi: 10.1002/rcm.6272. [DOI] [PubMed] [Google Scholar]
- 18.Tsugawa H., Nakabayashi R., Mori T., Yamada Y., Takahashi M., Rai A., Sugiyama R., Yamamoto H., Nakaya T., Yamazaki M., et al. A cheminformatics approach to characterize metabolomes in stable-Isotope-Labeled organism. Nat. Methods. 2019;16:295–298. doi: 10.1038/s41592-019-0358-2. [DOI] [PubMed] [Google Scholar]
- 19.Croaker A., King G.J., Pyne J.H., Anoopkumar-Dukie S., Liu L. Sanguinaria canadensis: Traditional Medicine, Phytochemical Composition, Biological Activities and Current Uses. Int. J. Mol. Sci. 2016;17:1414. doi: 10.3390/ijms17091414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M.X., Carver J.J. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M. TM4: A free, open-Source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.