Abstract
Microcrystal electron diffraction (MicroED) was first coined and developed in 2013 at the Janelia Research Campus as a new modality in electron cryomicroscopy (cryoEM). Since then, MicroED has not only made important contributions in pushing the resolution limits of cryoEM protein structure characterization but also of peptides, small-organic and inorganic molecules, and natural-products that have resisted structure determination by other methods. This review showcases important recent developments in MicroED, highlighting the importance of the technique in fields of studies beyond protein structure determination where MicroED is beginning to have paradigm shifting roles.
Current Opinion in Structural Biology 2020, 64:51–58
This review comes from a themed issue on Cryo electron microscopy
Edited by Sara Sandin and Daniela Rhodes
For a complete overview see the Issue and the Editorial
Available online 28th June 2020
https://doi.org/10.1016/j.sbi.2020.05.018
0959-440X/© 2020 Elsevier Ltd. All rights reserved.
Introduction
MicroED [1•,2••] has been pushing the limits of cryoEM in determining structures of macromolecular protein assemblies, peptides, and chemical compounds [1•,3,4••,5•]. Prior the technological advancements in detectors [6, 7, 8, 9] and software [10, 11, 12, 13] that made the ‘cryoEM resolution revolution [14]’ possible, structure determination of biological assemblies, peptides, and chemical compounds has been dominated X-ray crystallography. To date, there are close to 150 000 depositions in the Protein Data Bank (PDB), comprised of roughly 90% X-ray, 8% NMR, and 2% EM structures. The first near atomic resolution structure by cryoEM reported in 2005 [15] set the stage for the highest growth in transmission electron microscopy (TEM) structure deposition that occurred in the last five years.
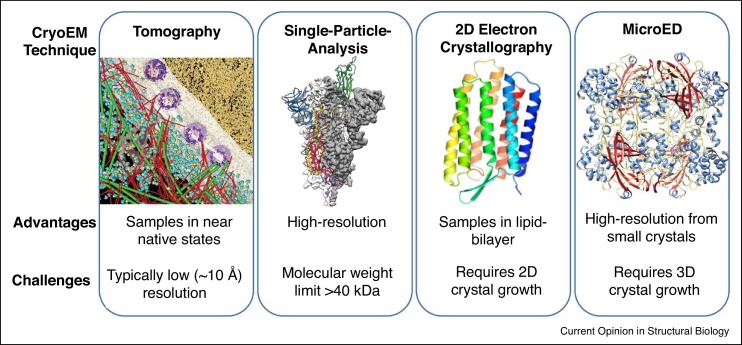
The field of cryoEM includes at least four major techniques: cryo-electron tomography (cryoET) [16,17], single-particle-analysis (SPA) [18, 19, 20], 2-dimenstional (2D) electron crystallography [21,22], and MicroED [1•,2••] (Figure 1 ). All of these cryoEM techniques exploit the advantage that electrons interact orders of magnitude more strongly with materials than X-rays, allowing the application of samples that are not tractable by other methods [23]. While CryoET and SPA use imaging, the crystallographic cryoEM methods of 2D electron crystallography and MicroED also take advantage of electron diffraction. 2D-electron crystallography is typically used for structure determination of moleucles in 2D arrays, which traditionally have been of membrane proteins that are crystallized within the native environment of the lipid bilayer [22]. In contrast, MicroED uses 3D crystals and data collection by continuous rotation to yield structures of a wide range of samples including soluble and membrane proteins, peptides, small organic and inorganic molecules, semi-conductors, and natural-products [5•].
Figure 1.
The four major modalities of cryoEM.
From left to right. A model of the HeLa cell nuclear periphery by Cryo-ET (Reprint from Ref. [24] with permission from AAAS). Model of COVID-19 spike protein by SPA (Reprint from Ref. [25] with permission from AAAS). The structure of bacteriorhodopsin determined by electron crystallography (Reprint from Ref. [21]). The structure of catalase determined by MicroED (Reprint from Ref. [26]).
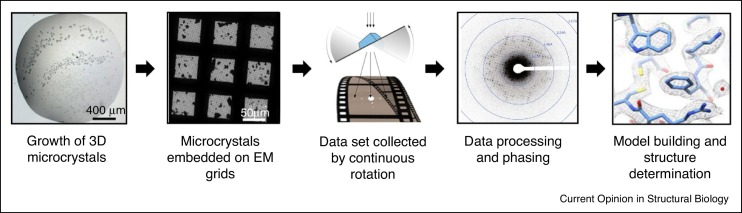
During MicroED experiments, crystals are harvested and prepared in a number of ways for embedment on EM grids (discussed below) [1•] (Figure 2 ). Biological samples, which are more sensitive to radiation, are typically vitrified to protect from radiation damage and to withstand the high-vacuum within the electron microscope [1•,27]. Once well-diffracting crystals are detected, MicroED datasets are collected by exposure of the sample to an electron beam in diffraction mode during continuous rotation of the stage [2••] (Figure 2). MicroED data are then collected on a fast camera as a movie, where each frame contains a diffraction pattern that represents a wedge of the reciprocal space [27]. Because continuous rotation for MicroED is analogous to the rotation method in X-ray crystallography, the data collected can be directly processed by existing standard X-ray crystallography software such as Mosflm [28], XDS [29], DIALS [30], SHELX [31] and HKL2000 [32] (Figure 2).
Figure 2.
MicroED workflow.
From left to right. Crystals are grown, harvested, and placed directly on EM grids. Manual or automated screening to assess for electron diffraction of the crystals. When a well-diffracting crystal is identified, a dataset is collected by continuous rotation. The diffraction dataset is then processed using standard crystallography software to allow model building and structure refinement.
After data-processing, the phases are determined and structures are built using the electron density maps [1•]. Given its wide-application and ability to extract structural information from nanocrystals, often a billionth the volume of those needed for X-ray crystallography, there are a growing number of structures determined by MicroED. Since its inception in 2013, there are close to 100 PDB entries produced by MicroED, with the highest growth in the just the past two years. To date, several laboratories have published MicroED studies and the number of practitioners are growing. Still, there are several challenges that lay ahead for MicroED including additional methodologies for sample preparation and technological developments for data collection that currently limit widespread usage of the technique. Below we discuss the most recent developments and strategies to expand the use of the MicroED.
Protein structure determination by MicroED
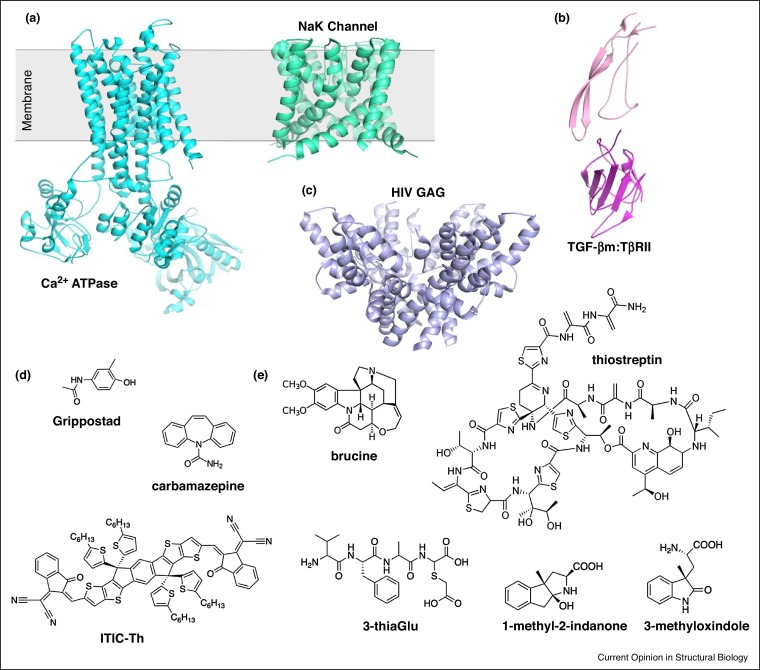
Formation of large crystals continue to be the most challenging and time-consuming step for X-ray crystallography, especially for membrane proteins and protein complexes [33]. The small crystals that are typically formed by membrane proteins and protein complexes can often diffract electrons using very low exposures to minimize radiation damage (0.01 e−/Å2) [1•]. One of the earliest membrane protein structures determined by MicroED was the Ca2+ ATPase (PDB 3J7T/U) [34••] (Figure 3 a). The Ca2+ ATPase structure illustrated the utility of MicroED for generating Coulomb potential (charge density) maps to detail information about the charged-states of amino-acid sidechains, cofactors, metals, and ligands [34••]. Since Ca2+ ATPase, there have been several important structures determined by MicroED including the non-selective sodium-potassium (NaK) channel (PDB 6CPV) [35••] and the complex of the transforming growth factor beta paired type II (TGF-βm:TβRII) (PBD 5TY4) [36] (Figure 3a,b). The MicroED structure of NaK is similar to those previously determined X-ray crystallography [37]. However, like Ca2+ ATPase, the structure of NaK by MicroED allowed generation of Coulomb potential maps to unambiguously place Na+ within the channel and to visualize a new transient state of the channel [35••]. The heterodimeric complex between TGF-βm and TβRII plays essential roles in the adaptive immune response and maintenance of the extracellular matrix [38]. Unlike Ca2+ ATPase and NaK, which formed nanocrystals, the structure of TGF-βm:TβRII was obtained from fragmentation of large, imperfect crystals (discussed below) [36]. Compared with the imperfect, large parent crystals, this approach led to better MicroED data from well-ordered microcrystal fragments that ultimately yielded atomic-resolution structures [36]. This study expanded the application of MircoED to include a wider range of crystal sizes.
Figure 3.
Representative MicroED structures of membrane proteins, protein complexes, small-molecules, and natural-products.
(a) MicroED structures of the membrane proteins rendered as cyan and green ribbon for Ca2+ ATPase (PDB 3J7T/U) [34••] and NaK (PDB 6CPV) [35••], repectively. (b) MircoED structure of the TGF-βm:TβRII protein–protein complex (PBD 5TY4) [36]. Protein rendered as pink and magenta ribbon for TGF-βm and TβRII, respectively. (c) MircoED structure of the HIV-GAG-bevirimat rendered as blue ribbon (PDB 6N3U) [39•]. (d) A galley of small-molecules with structures determined by MircoED. (e) Examples of natural-products with structures determined by MircoED.
MicroED in drug discovery
MicroED has already made important contributions to drug discovery by determining structures of protein–drug complexes and supra-resolution of small-molecules and natural-products, often directly from powders, bypassing crystallization experiments. The MicroED structure of HIV-GAG, which plays important roles in the life-cycle of HIV, was solved in complex with the antiviral drug, bevirimat (PDB 6N3U) [39•] (Figure 3c). The HIV-GAG-bevirimat complex provided important information about the antiviral drug mechanism and was the first demonstration of drug discovery using MicroED.
MicroED was originally intended for studying protein assemblies [26], however, it was rapidly recognized that this technique is a powerful tool for the characterization of small-molecules and natural-products. In 2016, the structure of the sodium channel blocker carbamezapine was determined to ∼1 Å resolution [40] (Figure 3d). In 2018, a method for small-molecule sample preparation using a "powder to structure" pipeline was described for carbamezapine [33] and later expanded to several small organic molecules [4••]. The structure of MBBF4 [41•], a methylene blue derivative with wide medical applications including its activity as a photo-activatable antimicrobial agent [42], was also solved. Since then, several structures of small-molecules have been reported by MicroED. These MicroED structures include Grippostad [41•], an antiviral drug for the treatment of the common cold and the flu [43], and a recent example of the non-fulleren acceptor (NFA) semi-conductive material ITIC-Th (Figure 3d).
MicroED has also proven its usefulness for the structure characterization of several natural-products that have previously been challenging or, in some cases, impossible to determine by other techniques. Unlike their synthetic small-molecules counterparts, biosynthesized natural-products are typically larger, structurally dynamic, obtained in small amounts, and difficult to crystallize, posing considerable challenges for X-ray studies. Even when natural-products form lattices, these crystals are often too small and are not useful for X-ray diffraction [44••,45••]. Brucine is an alkaloid toxin currently being tested for its anticancer properties [46] (Figure 3e). The MicroED structure of brucine at 0.9 Å resolution allowed for definitive assignment of its two chiral centers, key for understanding its toxicity and anticancer properties [4••] (Figure 3). Brucine, while large compared to small-molecules, is relatively small compared to amino-acid derived natural-products called ribosomally synthesized and post-translationally modified peptides (RiPPs), including 3-thiaGlu [44••] and thiostreptin [47] (Figure 3e). Glutamylated thiols, similar to the peptide modification on 3-thiaGlu, have been shown to block jasmonate and ethylene signaling pathways [48]. Thiostreptin is an antibiotic currently used in veterinary medicine [47] (Figure 3e). When efforts failed by X-ray crystallography, MicroED readily provided a 0.9 Å resolution of the 3-thiaGlu peptide (PDB 6PO6) [44••]. While thiostreptin has been studied by NMR [44••] and X-ray crystallography [49] previously, the ease of its characterization speaks to the robustness of MicroED for structure determination of large, flexible natural-products (Figure 3e). Like the difficulties encountered for 3-thiaGlu, the structures of 3-substituted oxindole derivatives (Figure 3e), that contain a new stereocenter at the γ carbon installed by an enzyme through directed-evolution, was only solved with the application of MicroED [45••]. These recent studies demonstrate that MicoED provides an additional strategy for more complete analysis of absolute configuration based on internal markers [44••,45••].
Strategies for crystal preparation for MicroED
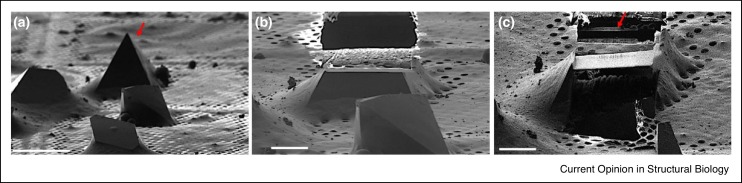
Electrons interact much more strongly with material than X-ray [23]. This phenomenon, however, results in high absorption and, thus, electrons can only penetrate very thin materials. Crystals that are greater than 500 nm in thickness must be thinned before MicroED data can be collected [36]. There are two strategies for trimming large crystals to thicknesses suitable for MicroED diffraction including mechanical fragmentation (typically by sonication, vigorous pipetting, or vortexing) [36] and milling with a focused ion beam (FIB) [50,51,52•]. Mechanical fragmentation has been successful for determining protein structures from large crystals of lysozyme, TGF-βm TβRII, xylanase, thaumatin, trypsin, proteinase K, thermolysin, and a segment of the protein tau [36]. Moving forward, the most current and promising technique for trimming large crystals for MicroED is FIB milling [50,51,52•]. During FIB milling, a crystal is repeatedly exposed a gallium beam to trim away the surrounding materials and generate lamellas with controllable thicknesses. As proof of principle, the structures of several proteins, including lysosome and proteinase K, have been determined by FIB mill and MicroED [50,51,52•,53,54] (Figure 4 ). Currently, FIB milling crystals is a relatively slow process but,even then, about ten crystal lamellas can be prepared per day.
Figure 4.
FIB milling of crystals for MicroED.
(a) Image of select proteinase K crystals at high magnification before milling. The arrow indicates the crystal that was milled. (b) FIB image after milling the top of the crystal. (c) FIB image after milling and cleaning both the top and bottom of the crystal leaving a lamella indicated by an arrow (Reprint from Ref. [52•]).
Outrunning radiation damage and phasing MicroED data
Radiation damage in structural studies continue to be a major challenge leading to poor processing statistics and map quality [55,56]. When electrons penetrate materials, they deposit energy that can deteriorate the samples, a process referred to as radiation damage. Radiation damage can be categorized into two forms: global and site-specific. Global radiation damage typically results in the disruption of the crystal lattice which can be detected during data-processing when decreases in overall diffraction intensities and increases in B-factors are observed [57,58]. On the other hand, site-specific radiation damage is not uniform, is not typically detected during data-processing, and observable only during examination of the real-space map [59]. The degree of radiation damage depends, among other things, on the content of the sample, the surrounding solution, and is proportional to the amount of energy used during diffraction studies. For MicroED, site-specific radiation damage has been illustrated to occur on specific amino-acids including cysteines, glutamates, and aspartic acids [60•]. To curb the effects of radiation damage, samples are often vitrified [61]. However, even the combination of vitrification and exposure to extremely low doses of electrons (0.01 e−/Å2/s) during MicroED experiments can still lead to detectable radiation damage [60•].
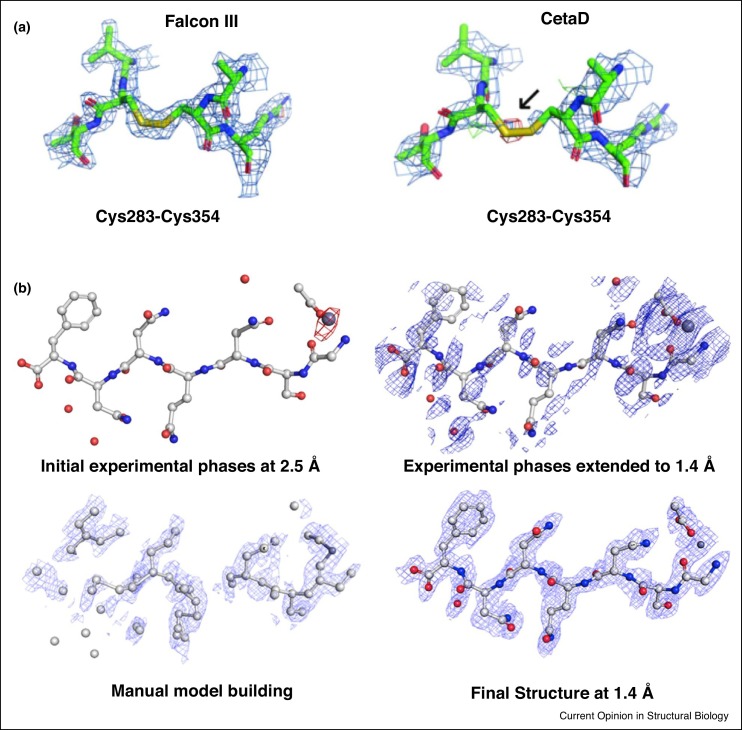
TEMs for cryo-EM studies are typically equipped with highly-sensitive direct-electron detectors designed for imaging [9,14,62]. These highly sensitive cameras, however, have not been used extensively for MicroED because of concerns of damage to the sensors. As such, MicroED data are typically collected on indirect-electron detectors such as the complementary metal oxide semiconductor (CMOS)-based CetaD and TVIPS TemCam-F416 cameras. This strategy, however, limits the availability of MicroED because most facility TEMs are typically outfitted with top-of-the-line direct-electron detectors for imaging and not CMOS cameras. Recently, the Falcon III direct-electron camera was tested for MicroED data collection [63••]. This study demonstrates that MicroED data collected at lower electron exposure, to avoid camera damage of Falcon III, lead to greater mean completeness relative to CMOS detectors and to higher quality maps. As proof of principle, examination of the maps of proteinase K from data collected on the Falcon III camera preserved the disulfide bonds,which are highly susceptible to radiation damage [63••] (Figure 5 a). A similar approach has been used recently with a Gatan K2 direct-electron detector in counting mode with an exposure 25 times less than for the Falcon III detector and a seemingly damage-free structure of Proteinase K has been determined.
Figure 5.
MicroED radiation damage and experimental phasing.
(a) Disulfide bonds of the proteinase K structures determined from data collected on Falcon III and CetaD. The 2mFo-DFc densities (blue meshes) are contoured at 1.5 σ (Reprint of Ref. [63••]). (b) Fourier difference maps between the damaged and undamaged structure of a peptide, contoured at 3 σ (Top left). Maps of the experimental phases of the peptide extended to 1.4 Å (Top right). Maps of an intermediate model-building step (Bottom left) to generate the final peptide structure at 1.4 Å (Bottom right) (Reprint from Ref. [64••]).
Outrunning radiation damage using the direct electron detector Falcon III has been instrumental in determining the structures of samples that are highly susceptible to damage [44••,45••] and we believe that many more such examples would be forthcoming. Radiation damage was recently exploited to establish a pipeline for phasing MicroED data [64••] (Figure 5b). A low-damage followed by a high-damage data sets were taken from the same crystal. A difference Patterson was calculated and allowed for generation of initial phases. Following cycles of manual model building and refinement, the structure of a peptide was determined (Figure 5b). This study demonstrates the ability to extract meaningful phase information using radiation damage in MicroED.
Concluding remarks
MicroED is proving to be an important new tool in structural biology not only in determining structures of proteins but also of peptides, small organic and inorganic molecules, and natural-products. Continuous rotation MicroED is paradigm shifting because it has proven to be a robust, fast, and efficient method for structure determination of small molecules and natural products, even without crystallization and directly from mixtures [4••]. To our knowledge, no other structural biology method is capable of determining atomic resolution structures directly from mixtures, making MicroED a useful and powerful tool for an array of problems that are yet to be explored. Moving forward, the application of FIB-milling and fast-cameras will certainly expand MicroED for structure determination of varying types of samples with a wide-range of crystal sizes and to facilitate time resolved studies. These advancements could ultimately be applied to establish automated pipelines that mirror those for X-ray crystallography to facilitate obtaining MicroED structures. Early examples of automation in MicroED has already reported in which several hundred data sets could be collected overnight to demonstrate similar throughput as synchrotrons [65].
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We would like to thank the Gonen laboratory and all collaborators who worked with us on MicroED applications. The Gonen lab is funded by the Howard Hughes Medical Institute and the National Institutes of Health P41-GM136508.
References
- 1•.Shi D., Nannenga B.L., Iadanza M.G., Gonen T. Three-dimensional electron crystallography of protein microcrystals. eLife. 2013;2:1–17. doi: 10.7554/eLife.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]; These studies mark the development and launch of MicroED from Janelia Research Campus for protein structure determination. Using lysozyme as a model system, the first MicroED structure of this protein was solved to 2.9 Å resolution.
- 2••.Nannenga B.L., Shi D., Leslie A.G.W., Gonen T. High-resolution structure determination by continuous-rotation data collection in MicroEDED. Nat Methods. 2014;11:927–930. doi: 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]; Important accompanying studies to the launch of MicroED in 2013 that illustrates continuous-rotation data collection. This work demonstrates that continuous-data collection for MicroED leads to better processing statistics, which can be achieved using already existing X-ray crystallography software.
- 3.Rodriguez J.A. Structure of the toxic core of α-synuclein from invisible crystals. Nature. 2015;525:486–490. doi: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.Jones C.G. The CryoEM method MicroED as a powerful tool for small molecule structure determination. ACS Cent Sci. 2018;4:1587–1592. doi: 10.1021/acscentsci.8b00760. [DOI] [PMC free article] [PubMed] [Google Scholar]; After the successful application of MicroED for proteins, this techniques was applied for structure determination of small-molecules and natural-products. These studies tout a ‘powder to structure’ pipeline where structures of small-molecules and natural-products were obtained minutes after sample preparation and MicroED data collection. Further, this work illustrates the precision of MicroED in obtaining structures from polymorphic mixtures of small-molecules and natural-products.
- 5•.Nannenga B.L., Gonen T. The cryo-EM method microcrystal electron diffraction (MicroED) Nat Methods. 2019;16:369–379. doi: 10.1038/s41592-019-0395-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review outlines the development of MicroED form 2013 to 2019, highlighting important achievements that establish MicroED as an important addition to the field of cryoEM.
- 6.Bai X.C., Fernandez I.S., McMullan G., Scheres S.H.W. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2013:2–13. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herzik M.A., Wu M., Lander G.C. Achieving better-than-3-Å resolution by single-particle cryo-EM at 200 keV. Nat Methods. 2017;14:1075–1078. doi: 10.1038/nmeth.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myasnikov A., Zheng S., Bulkley D., Cheng Y., Agard D. K3 - a first look at the new direct electron detection camera from Gatan company. Microsc Microanal. 2018;24:890–891. [Google Scholar]
- 10.Wagner T. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol. 2019;2:1–13. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zivanov J. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:1–22. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tegunov D., Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 14.Kühlbrandt W. The resolution revolution. Science (80-.) 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 15.Gonen T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komeili A., Li Z., Newman D.K., Jensen G.J. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science (80-.) 2006;311:242–246. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 17.Murphy G.E., Jensen G.J. Electron cryotomography of the E. coli pyruvate and 2-oxoglutarate dehydrogenase complexes. Structure. 2005;13:1765–1773. doi: 10.1016/j.str.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Frank J. vol 1. 1975. pp. 159–162. (The Cavendish Laboratory, Free School Lane, Cambridge CB2 3RQ, UK). [Google Scholar]
- 19.Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 20.Taylor D.W. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science (80-.) 2015;348:581–586. doi: 10.1126/science.aaa4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wisedchaisri G., Reichow S.L., Gonen T. Advances in structural and functional analysis of membrane proteins by electron crystallography. Structure. 2011;29:1381–1393. doi: 10.1016/j.str.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson R. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 23.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- 24.Mahamid J. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science (80-.) 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 25.Wrapp D. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-.) 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nannenga B.L., Shi D., Hattne J., Reyes F.E., Gonen T. Structure of catalase determined by MicroED. eLife. 2014;3 doi: 10.7554/eLife.03600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi D. The collection of microED data. Nat Protoc. 2016;11:895–904. doi: 10.1038/nprot.2016.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Battye T.G.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr Sect D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabsch W. XDS. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waterman D.G. Diffraction-geometry refinement in the DIALS framework. Acta Crystallogr Sect D Struct Biol. 2016;72:558–575. doi: 10.1107/S2059798316002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheldrick G.M., Gould R.O. Structure solution by iterative peaklist optimization and tangent expansion in space group P1. Acta Crystallogr Sect B. 1995;51:423–431. [Google Scholar]
- 32.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Fromme P., Spence J.C.H. Femtosecond nanocrystallography using X-ray lasers for membrane protein structure determination. Curr Opin Struct Biol. 2011;21:509–516. doi: 10.1016/j.sbi.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Yonekura K., Kato K., Ogasawara M., Tomita M., Toyoshima C. Electron crystallography of ultrathin 3D protein crystals: atomic model with charges. Proc Natl Acad Sci U S A. 2015;112:3368–3373. doi: 10.1073/pnas.1500724112. [DOI] [PMC free article] [PubMed] [Google Scholar]; These studies successfully launch the application of MicroED for structure determination of membrane proteins. Further, Coulomb maps generated from electron diffraction allow assignment of charges in Ca2+ ATPase.
- 35••.Liu S., Gonen T. MicroED structure of the NaK ion channel reveals a Na+ partition process into the selectivity filter. Commun Biol. 2018;1:1–6. doi: 10.1038/s42003-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; These studies firmly establish the application of MicroED for structure determination of membrane proteins. Using nanocrystals, MicroED captured two new conformations of Nak and visualization of the sodium within the channel, important missing links in the study of this family of proteins.
- 36.De La Cruz M.J. Atomic-resolution structures from fragmented protein crystals with the cryoEMEM method MicroEDED. Nat Methods. 2017;14:399–402. doi: 10.1038/nmeth.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alam A., Jiang Y. Structural analysis of ion selectivity in the NaK channel. Nat Struct Mol Biol. 2009;16:35–41. doi: 10.1038/nsmb.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morikawa M., Derynck R., Miyazono K. TGF- β and the TGF-β family: context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Purdy M.D. MicroED structures of HIV-1 Gag CTD-SP1 reveal binding interactions with the maturation inhibitor bevirimat. Proc Natl Acad Sci U S A. 2018;115:13258–13263. doi: 10.1073/pnas.1806806115. [DOI] [PMC free article] [PubMed] [Google Scholar]; The structure of HIV-GAG bound the anti-viral drug bevirimat provided important insights into its mechanism of action. Using MicroED for structures determination of protein–drug complexes establishes its application for structures-based drugs design.
- 40.Van Genderen E. Ab initio structure determination of nanocrystals of organic pharmaceutical compounds by electron diffraction at room temperature using a Timepix quantum area direct electron detector. Acta Crystallogr Sect A Found Adv. 2016;72:236–242. doi: 10.1107/S2053273315022500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Gruene T. Rapid structure determination of microcrystalline molecular compounds using electron diffraction. Angew Chem Int Ed. 2018;57:16313–16317. doi: 10.1002/anie.201811318. [DOI] [PMC free article] [PubMed] [Google Scholar]; These studies also establish that MicroED is a powerful tool for structure determination using powders of small molecules. These studies illustrate the expanded use of MicroED in the field biology and chemistry.
- 42.Wainwright M. Methylene blue derivatives - suitable photoantimicrobials for blood product disinfection? Int J Antimicrob Agents. 2000;16:381–394. doi: 10.1016/s0924-8579(00)00207-7. [DOI] [PubMed] [Google Scholar]
- 43.Koytchev R. Evaluation of the efficacy of a combined formulation (Grippostad®-C) in the therapy of symptoms of common cold: a randomized, double-blind, multicenter trial. Int J Clin Pharmacol Ther. 2003;41:114–125. doi: 10.5414/cpp41114. [DOI] [PubMed] [Google Scholar]
- 44••.Ting C.P. Use of a scaffold peptide in the biosynthesis of amino acid-derived natural products. Science (80-.) 2019;365:280–284. doi: 10.1126/science.aau6232. [DOI] [PMC free article] [PubMed] [Google Scholar]; Crystals of the natural product 3-thiaglu, a RiPP with a novel peptide modification with biological activities, failed to generate a structure by X-ray crystallography. MicroED achieved a 0.9 Å resolution of 3-thiaglu to allow assignment of the absolute configuration of its stereocenters, providing important insights into the mechanism for the installation of this unique modification.
- 45••.Dick M., Sarai N.S., Martynowycz M.W., Gonen T., Arnold F.H. Tailoring tryptophan synthase TrpB for selective quaternary carbon bond formation. J Am Chem Soc. 2019;141:19817–19822. doi: 10.1021/jacs.9b09864. [DOI] [PMC free article] [PubMed] [Google Scholar]; Crystals of 3-substituted oxindole derivatives, which contain a new stereocenter at the γ carbon that were installed by the TrpB enzyme through directed-evolution, failed to generate structures by X-ray crystallography. MicroED achieved structure determination of several of these 3-substituted oxindole derivatives at supra-resolution, allowing complete assignment of their stereocenters.
- 46.Qin J. Antitumor effects of brucine immuno-nanoparticles on hepatocellular carcinoma in vivo. Oncol Lett. 2018;15:6137–6146. doi: 10.3892/ol.2018.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kutscher A.H., Seguin L., Zegarelli E.V., Piro J.D. Antimicrobial activity of thiostrepton: tube dilution studies. J Am Dent Assoc. 1959;59:715–720. doi: 10.14219/jada.archive.1959.0207. [DOI] [PubMed] [Google Scholar]
- 48.Arrebola E., Cazorla F.M., Perez-García A., de Vicente A. Chemical and metabolic aspects of antimetabolite toxins produced by Pseudomonas syringae pathovars. Toxins (Basel) 2011;3:1089–1110. doi: 10.3390/toxins3091089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson B., Hodgkin D.C., Viswamitra M.A. The structure of thiostrepton. Nature. 1970;225:233–235. doi: 10.1038/225233a0. [DOI] [PubMed] [Google Scholar]
- 50.Martynowycz M.W. Collection of continuous rotation MicroED data from ion beam- milled crystals of any size. Structure. 2019;27:545–548. doi: 10.1016/j.str.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duyvesteyn H.M.E. Machining protein microcrystals for structure determination by electron diffraction. Proc Natl Acad Sci U S A. 2018;115:9569–9573. doi: 10.1073/pnas.1809978115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Martynowycz M.W., Zhao W., Hattne J., Jensen G.J., Gonen T. Qualitative analyses of polishing and precoating FIB milled crystals for MicroED. Structure. 2019;27:1594–1600.e2. doi: 10.1016/j.str.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; FIB milling has been establishes as a tool to expand MicroED for structure determination of large crystals. Here, these studies applied an additional step called ‘polishing’ to improve the MicroED diffraction quality of crystals from FIB milling.
- 53.Li X., Zhang S., Zhang J., Sun F. In situ protein micro-crystal fabrication by cryo-FIB for electron diffraction. Biophys Rep. 2018;4:339–347. doi: 10.1007/s41048-018-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou H., Luo Z., Li X. Using focus ion beam to prepare crystal lamella for electron diffraction. J Struct Biol. 2019;205:59–64. doi: 10.1016/j.jsb.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Carugo O., Carugo K.D. When X-rays modify the protein structure: radiation damage at work. Trends Biochem Sci. 2005;30:213–219. doi: 10.1016/j.tibs.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 56.Baker L.A., Rubinstein J.L. Radiation damage in electron cryomicroscopy. Methods Enzymol. 2010;481 doi: 10.1016/S0076-6879(10)81015-8. Elsevier Masson SAS. [DOI] [PubMed] [Google Scholar]
- 57.Kmetko J., Husseini N.S., Naides M., Kalinin Y., Thorne R.E. Quantifying X-ray radiation damage in protein crystals at cryogenic temperatures. Acta Crystallogr Sect D Biol Crystallogr. 2006;62:1030–1038. doi: 10.1107/S0907444906023869. [DOI] [PubMed] [Google Scholar]
- 58.Ravelli R.B.G., Theveneau P., McSweeney S., Caffrey M. Unit-cell volume change as a metric of radiation damage in crystals of macromolecules. J Synchrotron Radiat. 2002;9:355–360. doi: 10.1107/s0909049502014541. [DOI] [PubMed] [Google Scholar]
- 59.Weik M. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc Natl Acad Sci U S A. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60•.Hattne J. Analysis of global and site-specific radiation damage in Cryo-EM. Structure. 2018;26:759–766.e4. doi: 10.1016/j.str.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first comprehensive study to decipher the effects of radiation damage during MicroED experiments with implications to all cryoEM studies. These studies found both global and site-specific radiation damage during MicroED studies.
- 61.Karuppasamy M., Karimi Nejadasl F., Vulovic M., Koster A.J., Ravelli R.B.G. Radiation damage in single-particle cryo-electron microscopy: effects of dose and dose rate. J. Synchrotron Radiat. 2011;18:398–412. doi: 10.1107/S090904951100820X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu S., Armache J.P., Cheng Y. Single-particle cryo-EM data acquisition by using direct electron detection camera. Reprod Syst Sex Disord. 2016;65:35–41. doi: 10.1093/jmicro/dfv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63••.Hattne J., Martynowycz M.W., Penczek P.A., Gonen T. MicroED with the Falcon III direct electron detector. IUCrJ. 2019;6:921–926. doi: 10.1107/S2052252519010583. [DOI] [PMC free article] [PubMed] [Google Scholar]; These studies applied the first usage of a FalconIII direct electron detector for mitigating radiation damage from MicroED studies. The use of the Falcon III detector collected data with higher completeness and lower radiation damage as compared to CMOS detectors.
- 64••.Martynowycz M.W., Hattne J., Gonen T. Experimental phasing of MicroED data using radiation damage. Structure. 2020;28:458–464.e2. doi: 10.1016/j.str.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates the use of radiation damage for ab initio phasing with MicroED.
- 65.de la Cruz M.J., Martynowycz M.W., Hattne J., Gonen T. MicroED data collection with SerialEM. Ultramicroscopy. 2019;201:77–80. doi: 10.1016/j.ultramic.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]