Abstract
Objective
Klebsiella pneumoniae, one of the clinical superbugs, causes diverse infections because of its variable capsular antigens. This study focused on K. pneumoniae and aimed to assess any correlation between capsular serotype, capsule-associated virulence genes, and evaluate its resistance to conventional antibiotics in order to gain insight into any regional differences.
Materials and Methods
A total of 61 K. pneumoniae collected from various clinical specimens were confirmed genotypically. Clinical and demographic data for all patients were reviewed. All isolates were subjected to antimicrobial susceptibility tests. Capsular serotyping and capsule-associated virulence genes were studied using the molecular method.
Results
All typeable isolates were typed into K5, K20, and K54 serotypes, and among them, K54 was observed to be predominant. The most common capsule-associated virulence genes comprised uge (93.4%), ycfM (91.8%), and wabG (88.5%), while wcaG (29.5%) and rmpA (21.3%) were noted at much lower prevalence rates. The gene wcaG was significantly associated with K54 positive isolates (p = 0.001), while rmpA was associated with K20 positive isolates (p = 0.01).
Conclusion
Serotype K54 had a high frequency in isolates collected from patients with pulmonary diseases, while serotype K20 was associated with burn patients. Carbapenems and levofloxacin were the best therapeutic options for the treatment of infections with serotypes K20 and K54.
Keywords: Klebsiella pneumoniae, capsular serotype, virulence factor, extended-spectrum beta-lactamase, multidrug-resistance, multiplex-PCR
Introduction
Klebsiella pneumoniae is an upcoming superbug of clinical concern.1,2 The organism possesses a pronounced capsule, described as a K type, which provides a mucoid phenotype to the isolate3 and is an important virulence factor. The organism has been discriminated into 79 capsular serotypes.4,5 Of these, serotypes K1 and K2 are associated with bacteremia and are reported to be correlated with high mortality rates in Taiwan, Europe, and North America.6,7 Capsule-associated genes are a prime cause of pathogenicity in K. pneumoniae isolates. Virulence factors such as wabG (responsible for the biosynthesis of core lipopolysaccharide), uge (uridine diphosphate galacturonate 4-epimerase), and ycfM (the outer membrane lipoprotein) are involved in capsule production and promote infection by resistance to phagocytosis.8 The plasmid gene rmpA (regulator of mucoid phenotype A) provides a hypermucoviscous phenotype to K. pneumoniae by enhancing capsular polysaccharide production.9 The virulence gene wcaG is also responsible for K. pneumoniae capsule biosynthesis. The presence of this gene boosts the ability of bacteria to evade phagocytosis by macrophages.3,10 It is well known that bacteria undergo several modifications depending on the geographical region and with respect to the time, leading to changes in their characteristics. This study aimed to investigate the clinical features, capsular types, and any correlation between the presence of capsular serotypes with capsule-associated genes and antibiotic resistance in clinical K. pneumoniae isolates.
Materials and Methods
Bacterial Isolates
A total of 61 K. pneumoniae were isolated from various clinical specimens and the pertinent information on any underlying disease and other demographic data were collected from the medical records. Bacterial isolation and identification were carried out following standard cultural and biochemical techniques such as reactions in TSI (triple sugar iron), MR-VP test, SIM (sulfate/indole ⁄motility) and citrate agar.1 The hypervirulent phenotype was defined when a colony touched with the loop and lifted vertically from the surface of the agar plate, produced a string-like growth between the loop and the surface of the plate.11 The K. pneumoniae was later confirmed genotypically using primers as depicted in Table S1.9 The identified isolates were stored in trypticase soy broth containing 20% glycerol at −80°C for further use.
Definition
Hospital-acquired strains or nosocomial-associated bacteria refers to any bacteria contracted by a patient in a hospital at least 72 hours after being admitted, and if the infection was not obviously associated with the clinical conditions of the patient at the time of admission. Otherwise, infection was considered to be community-acquired.12
Antibiotic Susceptibility Pattern
Disk diffusion testing was performed and analyzed as per Clinical Laboratory Standards Institute (CLSI) guidelines.13 The resistance pattern of K. pneumoniae isolates was analyzed and is presented in Table 1. For confirmation of Extended-Spectrum Beta-Lactamase (ESBL) production, a double-disk test method utilizing cefotaxime and ceftazidime with and without clavulanic acid disks was used.13 Escherichia coli ATCC 25922 was used as quality control for antibiotic susceptibility testing. The isolates that were resistant to at least one antimicrobial agent in three or more of the categories were considered as MDR (multidrug resistance).14
Table 1.
Antibiotic Pattern of K. pneumoniae Isolates
| No. of Profiles | Resistance Profiles | No. of AB | No. of Isolatesa |
|---|---|---|---|
| I | PTZ, NI | 2 | 2 |
| II | SXT, NI | 2 | 2 |
| III | CP, SXT, CAZ, CTX, NI | 5 | 3 |
| IV | SXT, CAZ, CTX, NI, LEV | 5 | 2 |
| V | CP, SXT, CAZ, CTX, PTZ, NI | 6 | 2 |
| VI | GM, CP, SXT, CAZ, CTX, PTZ, NI | 7 | 2 |
| VII | GM, CP, SXT, CAZ, CTX, PTZ, NI | 7 | 2 |
| VIII | GM, CP, AN, SXT, CAZ, CTX, PTZ, NI | 8 | 5 |
| IX | GM, CP, AN, IMI, MEM, SXT, CAZ, CTX, PTZ, NI, LEV | 11 | 2 |
Note: aProfile of two or more similar isolates was included.
Abbreviations: GM, gentamicin; CP, ciprofloxacin; AN, amikacin; IMI, imipenem; MEM, meropenem; SXT, co-trimoxazole; CAZ, ceftazidime; CTX, cefotaxime; PTZ, piperacillin-tazobactam; NI, nitrofurantoin; LEV, levofloxacin; AB, antibiotic disks.
DNA Extraction and Multiplex PCR
DNA was extracted from K. pneumoniae isolates using the commercial DNA extraction kit (Stratec Biomedical systems, Birkenfeld, Germany). Briefly, 1mL of bacterial suspension matched equivalent to 0.5 McFarland was prepared from an overnight culture and then centrifuged. DNA was extracted as per the instructions provided in the kit from the pellet and finally resolved in 100µL TE buffer. Table S1 depicts the primers for serotyping (K1, K2, K5, K20, K54, and K57),9 confirmation of K. pneumoniae (K. pneumoniae 16S–23S ITS)9 and capsule-associated virulence genes (rmpA, wcaG, uge, ycfM, and wabG).10 Amplification of the respective genes was carried out as a multiplex PCR performed as a 25μL reaction mixture with 5pmol of each primer and 2μL of DNA added to the Master PCR mixture (Yekta Tajhiz Azma®, Iran). Two separate Multiplex PCRs were carried out with the same thermal cycling conditions for detecting the capsular serotypes and capsule-associated genes as described previously.9,10 The amplified products were electrophoresed on 1% agarose gel (Yekta Tajhiz Azma®, Iran) and stained with Cyber safe stain (Yekta Tajhiz Azma®, Iran).
Data Analysis
The logistic regression model was performed for the multivariate analysis to identify risk factors for mortality. We used the X2 test and the Fisher exact test (if necessary) to find the relationship between serotypes and other variables. P-value <0.05 was considered significant statistically. The data were analyzed with SPSS statistics (Version 20) program (IBM Corporation).
Ethical Approval
This study was approved by the Regional Ethics Committee of Tabriz (Tabriz University of Medical Sciences, Tabriz, Iran, No. IR.TBZMED.REC.1397.058).
Results
Among 468 bacterial isolates obtained from various clinical specimens, 61 (13.03%) isolates were identified as K. pneumoniae by traditional biochemical tests. Urine was the most common [n=31; (50.8%)] clinical specimen from which K. pneumoniae was isolated, followed by wound [n=15; (24.6%)], blood [n=8; (13.1%)], endotracheal aspirate [n=4; (6.6%)], and other body fluids [n=3; (4.9%)]. All 61 isolates were positive for the internal transcribed spacer region (K. pneumoniae 16S–23S) and were confirmed at the molecular level as K. pneumoniae.
Serotyping
Of the 61 K. pneumoniae isolates, 36 (59%) were found to be typeable with K5, K20, and K54 primers, while 25 (41%) were non-typeable. Serotype K54 was the most prevalent [n=18; 29.5%] followed by K20 and K5, which accounted for 13 (21.3%) and 5 (8.1%) isolates, respectively (Table 2).
Table 2.
Relation of K-Serotypes with Capsule-Associated Virulence Factors in Various Clinical Wards
| Number of | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wards | K. pneumoniae N (%) | K-Serotypes | Virulence Factors | ||||||
| K54 (n=18) | K20 (n=13) | K5 (n=5) | wcaG (n=18) | rmpA (n=13) | uge (n=57) | ycfM (n=56) | wabG (n=54) | ||
| Burn ICU | 7 (11.5) | 2 | 5* | 0 | 2 | 4* | 7 | 7 | 7 |
| General ICU | 6 (9.8) | 1 | 0 | 0 | 1 | 0 | 6 | 6 | 6 |
| Infectious ICU | 4 (6.6) | 4 | 1 | 0 | 4 | 0 | 3 | 4 | 4 |
| Internal ICU | 3 (4.9) | 0 | 0 | 1 | 0 | 0 | 2 | 2 | 2 |
| Surgery ICU | 2 (3.3) | 1 | 0 | 1 | 1 | 0 | 2 | 2 | 2 |
| Internal | 8 (13.1) | 2 | 0 | 2 | 2 | 0 | 8 | 7 | 7 |
| Burn | 6 (9.8) | 1 | 4* | 0 | 1 | 5* | 6 | 5 | 5 |
| Urology | 6 (9.8) | 2 | 0 | 0 | 2 | 1 | 5 | 5 | 4 |
| Surgery | 2 (3.3) | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 2 |
| Emergency | 2 (3.3) | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 |
| Infectious | 4 (6.6) | 1 | 2 | 0 | 1 | 0 | 4 | 4 | 3 |
| Out-patients | 11 (18) | 2 | 1 | 1 | 2 | 1 | 10 | 10 | 10 |
Note: *P-value<0.05, calculated by Chi-square test or Fisher exact test.
Abbreviations: ICU, intensive care unit; K, capsular polysaccharide (K antigen).
Clinical Wards
Considering the source, 43 (70.5%) isolates were obtained from in-patients admitted to five different intensive care units (ICU) and six other wards (Table 2). K. pneumoniae was the causative agent of nosocomial infection in ICU patients with a high prevalence [n= 22; 36.2%], and the most common isolates were obtained from the burn ICU [n=7; 11.5%]. The hospital lengths of stay of patients from whom K. pneumoniae were isolated ranged from 7 to 60 days. One patient admitted to the infectious ICU had the longest duration of hospitalization, equaling approximately 205 days. The distribution of capsular serotypes, and capsule-associated virulence genes among the various clinical wards of the hospital is depicted in Table 2. The K20 serotype and rmpA gene were prevalent in patients admitted to the burn ICU and burn wards (p < 0.05, Table 2).
Clinical Data
Table 3 shows that 28 (45.9%) and 33 (54.1%) of isolates were isolated from males and females, respectively. Thirty-four isolates (55.6%) were collected as hospital-acquired and 27 (44.4%) were community-acquired. Eighteen (29.5%) patients were residents of rural areas. The ages of patients ranged from 3 to 89 years (mean ± SD 56.7 ± 23.42 yrs.). Almost 50% of infections occurred in elderly patients (≥60 years). When patients were studied for any underlying medical condition or had undergone medical modality, it was found that most of the patients had undergone either radiography [n=34; 55.7%] or ultrasonography [n=21; 34.4%]. Other medical interventions included previous surgery [n=18; 29.5%] and a need for mechanical ventilation [n=9; 14.8%]. Table 3 also shows the patients afflicted with various types of K. pneumoniae-associated diseases. K54 was the most common serotype significantly (p = 0.001) compared to patients with pulmonary diseases [8 (72.7%) of the 11 patients afflicted with pulmonary disease], while K20 correlated (p = 0.003) with burn patients who had undergone skin grafting [7 (70%) of 10 infected burn patients]. When the typeability of K. pneumoniae was compared between the two hospital settings, 24 of 34 (71%) hospital-acquired isolates were typeable in comparison to only 12 of 27 (45%) community-acquired isolates (p = 0.03). Urinary tract infection (UTI), renal disease, diabetes mellitus, and bacteremia with frequencies of 13 (21.3%), 7 (11.4%), 5 (8.1%), and 3 (4.9%) patients, respectively, were the common diseases observed from community-acquired K. pneumoniae infections in this study. One patient suffered from both diabetes mellitus and renal disease. There was no statistical relation between the three common capsular serotypes and community-acquired infections.
Table 3.
Demographic and Clinical Data of 61 K. pneumoniae-Infected Patients and Relation with Serotypes
| Variables | Number (%) | Statistically Significant Association with Capsular Serotypes |
|---|---|---|
| Gender | ||
| Male | 28 (45.9) | – |
| Female | 33 (54.1) | K20* |
| Infection setting | ||
| Hospital-acquired | 34 (55.6) | K5, K20, K54* |
| Community-acquired | 27 (44.4) | - |
| Age (mean ± SD) | 56.70 ± 23.42 years | - |
| Accommodation | ||
| Urban | 43 (70.5) | - |
| Rural | 18 (29.5) | - |
| Underlying medical conditions | ||
| ICU stay | 22 (36.2) | - |
| Bed-ridden | 43 (70.5) | - |
| Radiography | 34 (55.7) | - |
| Ultrasonography | 21 (34.4) | - |
| CT scan | 18 (29.5) | - |
| Tracheostomy | 5 (8.2) | - |
| Bronchoscopy | 4 (6.6) | - |
| Cystoscopy | 4 (6.6) | - |
| Skin graft | 11(18) | K20* |
| Mechanical ventilation | 9 (14.8) | - |
| Pneumonia | 7 (11.5) | - |
| Surgery procedure | 18 (29.5) | - |
| Prior antibiotic usage | 43 (70.5) | - |
| Diseases | ||
| Pulmonary | 11 (18) | K54* |
| Renal diseases and UTI | 20 (32.7) | - |
| Gastrointestinal | 7 (11.4) | - |
| Infectious disease | 7 (11.4) | - |
| Ulcer and abscess | 6 (9.7) | - |
| Burn | 10 (16.4) | K20* |
| Diabetes mellitus | 5 (8.2) | - |
| DJ procedure | 5 (8.2) | - |
| Bacteremia | 3 (4.9) | - |
| Cardiovascular | 2 (3.3) | - |
| Hyperplasia of prostate | 2 (3.3) | - |
| Syndromes | ||
| Kimmel-Wilson syndrome | 4 (6.6) | - |
| Respiratory distress syndrome | 2 (3.3) | - |
| Outcome | ||
| Survived | 50 (82) | - |
| Died | 11 (18) | - |
Note: *P-value<0.05, calculated by Chi-square test or Fisher exact test.
Abbreviations: SD, standard deviation; ICU, intensive care unit; CT, computed tomography; UTI, urinary tract infection; DJ, double J; K, capsular polysaccharide (K antigen).
Prevalence of Capsule-Associated Genes in Predominant Serotypes
Overall, uge was the most commonly detected putative virulence gene [n=57; (93.4%)], followed by ycfM [n=56; (91.8%)] and wabG [n=54; (88.5%)]. The wcaG [n=18; (29.5%)] and rmpA [n=13; (21.3%)] genes had lower distributions among various K-serotypes. The presence of wcaG was highly associated with K54 positive isolates [18/18 (100%); p = 0.001], while rmpA had a higher prevalence among K20 positive isolates [8/13 (61.5%); p = 0.01]. The source of both wcaG and rmpA positive isolates was the hospital [p < 0.05] (Table 4).
Table 4.
Distribution of Putative Capsule-Associated Virulence Genes in K. pneumoniae in Predominant Capsular Serotypes
| Capsule-Associated Genesa | ||||||
|---|---|---|---|---|---|---|
| rmpA (n=13) | wcaG (n=18) | uge (n=57) | ycfM (n=56) | wabG (n=54) | ||
| Hospital-acquired isolates | 12* | 13* | 32 | 31 | 31 | |
| Capsular serotypes | K5 (n=5) | 0 | 2 | 5 | 5 | 5 |
| K20 (n=13) | 8* | 3 | 12 | 12 | 12 | |
| K54 (n=18) | 6 | 18* | 18 | 17 | 18 | |
Note: aAs the capsule-associated genes were common among two serotypes, thus, the numbers cannot add up to numbers mentioned for each of them. *P-value<0.05, calculated by Chi-square test or Fisher exact test.
Abbreviation: K, capsular polysaccharide (K antigen).
Prevalence of Serotypes and Capsule-Associated Genes in Both Genders
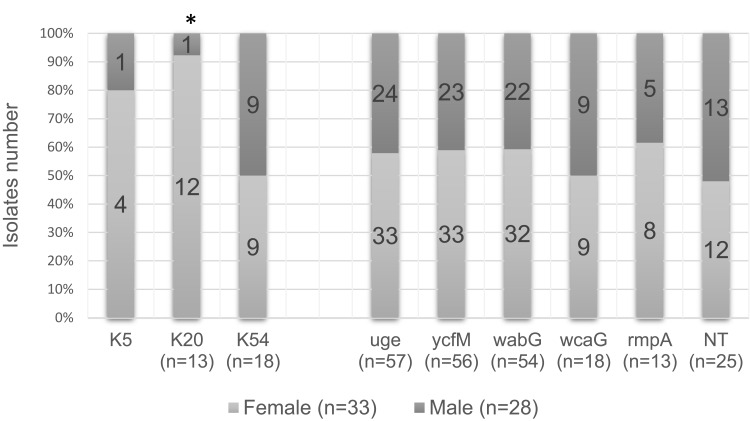
Figure 1 displays the distribution of capsular serotypes with capsule-associated genes in K. pneumoniae. K20 serotype had a higher prevalence in clinical specimens collected from females than males (P = 0.02). In contrast, the distribution of K5, and K54 serotypes with five capsule-associated virulence factors was not different between genders.
Figure 1.
Distribution of capsular serotypes and capsule-associated virulence genes among the isolates collected from male and female specimens.
Note: *Higher prevalence of K20 serotype was found in the clinical specimens collected from females in comparison to males (p-value = 0.02). NT: the isolates that did not belong to the K1, K2, K5, K20, K54, or K57 serotype.
Abbreviations: K, capsular polysaccharide (K antigen); NT, non-typeable.
Mucoid Phenotype
The mucoid phenotype detected by the string test was observed in 16 (26.22%) isolates, and the sources of these isolates comprised 7 (43.7%) from urine, 5 (31.2%) from wounds, 2 (12.5%) from blood, and 2 (12.5%) from other body fluid.
Table 5 depicts that of the 16 mucoid phenotype isolates, 14 were typeable [87.5%; p=0.01]. Six of the 18 K54 positive isolates had a mucoid phenotype (p = 0.304), while such correlation was observed in 10 of the 13 K20 positive isolates (p < 0.001) and in only 1 of the K5 positive isolates (p = 0.606). Among the 13 rmpA positive K. pneumoniae isolates, 10 (76.9%; p = 0.003) had mucoid phenotype.
Table 5.
Correlation of Mucoid Phenotype with K-Serotypes, Capsule-Associated Virulence Genes, and ESBL Among K. pneumoniae Isolates
| Number (%) of | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mucoid phenotype | Typeable isolate (n=36) | Capsular serotypes | NTa isolate (n=25) | Capsule-associated virulence factors | Extended-spectrum beta-lactamase | |||||||||||
| K5 (n=5) | K20 (n=13) | K54 (n=18) | rmpA (n=13) | wcaG (n=18) | uge (n=57) | ycfM (n=56) | wabG (n=54) | ESBL isolates (n=36) | K5, ESBLb (n=2) | K20, ESBLb (n=9) | K54, ESBLb (n=12) | rmpA, ESBLc (n=12) | Non-ESBL isolates (n=25) | |||
| Total=16 (26.22%) | 14 (87.5)* | 1 (6.25) | 10 (62.5)* | 3 (18.7) | 2 (12.5) | 10 (62.5)* | 3 (18.7) | 15 (93.7) | 15 (93.7) | 15 (93.7) | 10 (62.5) | 0 | 7 (43.7)* | 3 (18.7) | 7 (43.7)* | 6 (37.5) |
Note: aNT, the isolates that were not found to be typeable with K1, K2, K5, K20, K54, and K57 primers. bIsolates with both K5/K20 serotype and ESBL positivity. cIsolates with both rmpA gene and ESBL positivity (the only virulence factor with a significant association with mucoid phenotype). *P-value<0.05, calculated by Chi-square test or Fisher exact test.
Abbreviations: NT, non-typeable; ESBL, extended-spectrum beta-lactamase; K, capsular polysaccharide (K antigen).
Among 36 ESBL-positive isolates, only 10 were found to be associated with mucoid phenotypes (p = 0.490). In two isolates (with both serotype K5 and ESBL positivity), no mucoid phenotype was detected, while among nine isolates with both serotype K20 and ESBL positivity, 7 were observed to be associated with mucoid phenotypes (p < 0.001). Similarly, among 12 isolates with serotype K54 and ESBL producers, 5 were found to be associated with mucoid phenotypes. In 12 isolates with both rmpA and ESBL positivity, 7 were observed to be associated with mucoid phenotypes (p < 0.001). Comparatively, among 25 non-ESBL isolates, only 6 were found to have the mucoid phenotype. Among the K. pneumoniae isolates obtained from in-patients, 9 (75%) had mucoid phenotype. The one isolate obtained from an out-patient also had the same morphology.
Antibiotic Resistance
The outcome of the antibiotic resistance pattern is depicted in Table 1. All K. pneumoniae isolates could be typed into nine different antibiotypes, and among these resistance patterns, profiles number VIII (with 5 isolates) and III (with 3 isolates) had the highest frequencies.
Resistance to the tested antibiotics was as follows: cefotaxime 78.8%, ceftazidime 75.4%, ciprofloxacin 68.9%, nitrofurantoin 68.9%, co-trimoxazole 67.2%, piperacillin-tazobactam 57.4%, gentamicin 45.9%, amikacin 39.3%, imipenem 24.6%, meropenem 24.6%, and levofloxacin 24.6%. Among the K. pneumoniae isolates studied, 36 (59%) of them were found to be ESBL producers, while 47 (77%) produced MDR. Twenty-four of these ESBL-positive isolates were related to hospital-acquired infections (p = 0.036). Serotype K20 was significantly associated with resistance to amikacin and gentamicin [n=9 (69%), p = 0.01], while serotype K54 was found to be significantly associated with resistance to ciprofloxacin [16 of 18 [88%], p = 0.02]. Serotype K54 was a prevalent serotype with MDR [17 of 18 (94.4%), p = 0.02]. Among the capsule-associated virulence genes, rmpA had a high frequency in ESBL-producing isolates [12 of 13 (92.3%), p = 0.01], while wcaG had a high prevalence in MDR-producing isolates [17 of 18 (94.4%), p = 0.02] and was also related to ciprofloxacin resistance [16 of 18 (88.8%), p = 0.03]. Other capsule-associated virulence factors, uge, ycfM, and wabG, had high prevalence rates and were similarly distributed among all antibiotic-resistant isolates. Interestingly, resistance to imipenem, meropenem, and levofloxacin was law and was found to be the best choices for treatment of these serotypes.
When hospital-acquired ESBL K. pneumoniae strains were compared for capsule-associated genes, 11 of the 13 rmpA positive isolates had a significant (p < 0.001) relation with hospital-acquired ESBL strains. Conversely, 6 of 18 wcaG positive, 21 of 53 wabG positive, 21 of 55 ycfM positive, and 21 of 56 uge positive isolates were not related to hospital-acquired ESBL strains [(p = 0.500), (p = 0.451), (p = 0.642), and (p = 0.498), respectively].
Risk Factor for Mortality
In the current study, the crude mortality rate was 18%. By multivariate logistic regression analysis, some variables, like old age [≥60 years] (p=0.02), infection with MDR (p=0.04), long-term usage of amikacin (p=0.01), infectious diseases [septicemia, tetanus, peritonitis, and wound infections] (p=0.03), and lung disease (p=0.01), were associated with a significantly higher mortality rate. Among the three predominant serotypes in this investigation, the K54 serotype was associated with higher mortality and variables such as old age (p=0.04), infection with MDR (p=0.03), and pulmonary diseases (p=0.02).
Discussion
In the present investigation, six capsular serotypes and capsule-associated virulence factors were assessed in 61 K. pneumoniae isolates obtained from various clinical sources. The rate of hospital-acquired K. pneumoniae infections was 55.7%, which is higher than other hospital-acquired reports in Vietnam (29.5%)15 and in ICUs in Southern Europe, Turkey, and Iran (23.5%),16 which may be due to the type of infections seen in medical practices at various places.
The mortality rate of K. pneumoniae-associated bacteremia varies from 20% to 70% depending on the treatment regimen and disease severity.17–19 In two studies which dealt with bloodstream infections (BSI) caused by K. pneumoniae, the overall mortality ranged from 35.6% to 36.7%.20,21 By multivariate analysis, some variables were recognized as predictors of mortality associated with K. pneumoniae infections such as isolates being MDR, resistance to amikacin, presence of infectious diseases, and involvement of the pulmonary system. The total mortality rate in the current study was 18%, which is lower than the mortality rates reported earlier.22 Because all kinds of infections caused by Klebsiella pneumoniae were assessed in this study, the mortality rate was low compared with specifically mentioned infections such as bacteremia.19,20 Concerning community-acquired infections, two available studies from India23 and New York24 reported that K. pneumoniae infections are often associated with pyogenic liver abscess. In contrast, research data from the present investigation showed a higher prevalence of hospital-acquired infections and the absence of serotypes K1, K2, and K57. Moreover, this study did not include any patients with liver abscess.
ESBL production and MDR in K. pneumoniae are major problems in patient care globally.20 According to the current results, MDR isolates were more related to mortality outcomes, perhaps because of delays at the beginning of the appropriate therapy. Similar to the study by Tsay Ren-Wen et al,12 high mortality was correlated to the presence of lung infection.
Of the 61 K. pneumoniae isolates, 36 (59%) were typeable, distributed in the three serotypes K5, K20, and K54. Though research studies performed elsewhere have shown more than 85% of isolates as being typeable with vast distributions,25,26 other investigations with high numbers of isolates could not wholly type the isolates.12,27,28 Most of the serological studies done on K. pneumoniae have been related to pyogenic liver abscess syndrome and its relationship with capsular serotypes. K. pneumoniae isolates from liver abscess specimens have similar characteristics, like the prevalence of serotype K1 or K2 and genomic heterogeneity.28,29
A recent study reported from Iran30 found K54 as the most frequent (68%) capsular serotype while K1 (8%) was the lowest frequency. However, contrary to our findings, a higher frequency of serotype K5 (60%) was reported.30 Prevalence of K5, K20, and K54 serotypes was significantly lower in Taiwan and Europe25,31 in comparison to our study. In a study similar to the present study by Turton et al9 from the UK in 2010, reported a higher incidence of serotype K54 compared to other capsular types. However, contrary to the results of the present study, the K2 serotype had the highest frequency. The differences in the frequency of capsular serotypes may be due to differences in the types of samples collected in other studies. Only serotyping was performed in this investigation; thus, we cannot confirm an outbreak. Secondly, as the isolates were obtained from different wards (though same hospital sources), it can be an outbreak of three predominant serotypes. Nevertheless, precise typing molecular methods are required to confirm this.
The present results demonstrated the presence of uge (in 93.4% of isolates), ycfM (91.8%), and wabG (88.5%) capsule-associated genes among the K. pneumoniae isolates encoding capsule lipoprotein, external membrane protein, and capsule, respectively. Two other studies from Turkey and Iraq10,32 reported the prevalence of these genes in more than 80% of their K. pneumoniae isolates. These genes are also involved in lipopolysaccharide biosynthesis and promote infection by resistance to phagocytosis.8 Therefore, because uge, ycfM, and wabG were commonly found in K. pneumoniae isolates, they seem to be at the basis of the pathogenicity of K. pneumoniae. Earlier uge and wabG genes have been identified from patients with invasive and serious infections.33
According to previous studies,34,35 the plasmid-borne gene rmpA is related to capsule production. It has also been associated with 6 PLA (pyogenic liver abscess)-related capsular types (K1, K2, K5, K54, K57, and KN1).34,35 In the current investigation, the absence of a liver abscess specimen may be the main reason for the absence of K1 and K2 capsular serotypes. The results showed that rmpA had a high frequency with K20 serotype (p < 0.05). Moreover, 61.53% of rmpA genes and 76.9% of K20 serotypes were associated with the mucoid phenotype. Two other research studies previously performed in Taiwan showed that a 180-kilobase plasmid holding rmpA (regulator of mucoid phenotypes) was associated with the mucoid phenotype.36,37 Another putative virulence factor which may encode capsular fucose synthesis in K. pneumoniae and assist the bacterial evasion from the phagocytosis is the presence of wcaG,34,35 a gene involved in capsule production.38 Turton et al demonstrated that the presence of wcaG in their isolates was correlated with K1, K16, K54, and K58 capsular serotypes.9 The current results showed that wcaG was associated with the K54 serotype (p < 0.05). Earlier, K. pneumoniae isolates possessing wcaG were isolated from patients with serious and invasive diseases.9 The outcomes of two researches indicated wcaG had a high effect on K. pneumoniae virulence.9,39 Of the five capsule-associated genes studied in the present investigation, wcaG was correlated with serotype K54, while rmpA was correlated with K20. According to the present findings, rmpA had a high prevalence in ESBL-producing isolates, and wcaG was found to be the predominant capsule-associated gene in MDR isolates. Serotype K20 and rmpA positive isolates were associated with a mucoid phenotype. Moreover, isolates with resistance to gentamicin and amikacin were correlated with serotype K20, while ciprofloxacin-resistant isolates were concomitantly related to serotype K54. These antibiotic consequences are significant in therapeutic spheres, where physicians can prescribe either carbapenems or medications from other drug groups apart from aminoglycosides or fluoroquinolones for K. pneumoniae infections in our region; however, this necessitates the confirmation of antibiotic susceptibility towards carbapenems at the molecular level.
In this study, it was found that K5 and K54 had higher prevalence rates in females compared with males. As the sample size was small, no further conclusions cannot be made on such type of associations. Not many publications are available on the relationship of K-serotypes with gender or age. However, Liu and Guo (2019) in their investigation on bacteremic patients collected 175 K. pneumoniae isolates but found no significant differences in gender or age between the two groups in terms of serotype prevalence.40 Similarly, Lee et al (2010) studied 91 K. pneumoniae from various clinical disorders and found K1/K2 serotypes; however, none of them were correlated with the male gender.41
The distribution of prevalent capsular serotypes in various medical wards was also assessed, and serotype K20 was found to be associated with infections in the burn ICU and burn wards, especially among patients who developed skin infections after grafting. Serotype K54 was found to be associated with pulmonary diseases. Other studies have assessed the association of serotypes K1 and K2 with diseases only. For example, Chi-Tai Fang et al from Taiwan found serotype K1 to be an emerging pathogen capable of causing central nervous system complications and catastrophic septic ocular.31 The current study is the first of its kind to focus on any association between capsular serotypes, various medical wards, and antibiotic resistance and to show the relationship of serotypes K20 and K54 with the above-mentioned diseases.
Conclusion
The current study found that diverse capsular serotypes are involved in K. pneumoniae-associated infections. Serotype K54 had a high frequency in pulmonary diseases, while serotype K20 was associated with burn infections. Of the five capsule-associated genes studied, wcaG was correlated with serotype K54, while rmpA was correlated with K20. Serotype K20 and rmpA positive isolates were associated with a mucoid phenotype. Carbapenems and levofloxacin could be recommended for the treatment of infections with serotypes K20 and K54. Information about the distribution of capsular serotypes in specific diseases and medical wards with a special pattern of antibiotic resistance could aid physicians in prescribing appropriate treatments.
Acknowledgments
The authors would like to thank Tabriz University of Medical Sciences, Faculty of Medicine for providing the expertise that greatly assisted. Also, we thank Mrs. Leila Dehghani for her technical assistance in the collection of clinical isolates. This study was supported by Immunology Research Center, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, I. R. Iran. This report is part of a database of MSc thesis of the second author registered in the Tabriz University of Medical Sciences (Thesis No-58558).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ko W-C, Paterson DL, Sagnimeni AJ, et al. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis. 2002;8(2):160–166. doi: 10.3201/eid0802.010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woldu M. Klebsiella pneumoniae and its growing concern in healthcare settings. Clin Exp Pharmacol Physiol. 2016;6(199):2161. doi: 10.4172/2161-1459.1000199 [DOI] [Google Scholar]
- 3.Derakhshan S, Najar Peerayeh S, Bakhshi B. Association between presence of virulence genes and antibiotic resistance in clinical klebsiella pneumoniae isolates. Lab Med. 2016;47(4):306–311. doi: 10.1093/labmed/lmw030 [DOI] [PubMed] [Google Scholar]
- 4.Brisse S, Passet V, Haugaard AB, et al. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51(12):4073–4078. doi: 10.1128/JCM.01924-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Y-J, Lin T-L, Chen C-T, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep. 2015;5(1):15573. doi: 10.1038/srep15573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turton JF, Englender H, Gabriel SN, et al. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J Med Microbiol. 2007;56(Pt5):593–597. doi: 10.1099/jmm.0.46964-0 [DOI] [PubMed] [Google Scholar]
- 7.Jian-li W, Yuan-yuan S, Shou-yu G, et al. Serotype and virulence genes of Klebsiella pneumoniae isolated from mink and its pathogenesis in mice and mink. Sci Rep. 2017;7(1):17291. doi: 10.1038/s41598-017-17681-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortés G, Borrell N, de Astorza B, et al. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect Immun. 2002;70(5):2583–2590. doi: 10.1128/IAI.70.5.2583-2590.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turton JF, Perry C, Elgohari S, et al. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59(5):541–547. doi: 10.1099/jmm.0.015198-0 [DOI] [PubMed] [Google Scholar]
- 10.Candan ED, Aksöz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62(4):867–874. doi: 10.18388/abp.2015_1148 [DOI] [PubMed] [Google Scholar]
- 11.Victor LY, Hansen DS, Ko WC, et al. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis. 2007;13(7):986. doi: 10.3201/eid1307.070187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsay R-W, Siu L, Fung C-P, et al. Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch Intern Med. 2002;162(9):1021–1027. doi: 10.1001/archinte.162.9.1021 [DOI] [PubMed] [Google Scholar]
- 13.Wayne P. Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing. 2017;142–158. [Google Scholar]
- 14.Magiorakos AP, Srinivasan A, Carey R, et al. Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 15.Phu VD, Wertheim HF, Larsson M, et al. Burden of hospital acquired infections and antimicrobial use in Vietnamese adult intensive care units. PLoS One. 2016;11(1):e0147544. doi: 10.1371/journal.pone.0147544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erdem H, Inan A, Altindis S, et al. Surveillance, control and management of infections in intensive care units in Southern Europe, Turkey and Iran – a prospective multicenter point prevalence study. J Infect. 2014;68(2):131–140. doi: 10.1016/j.jinf.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 17.Daikos GL, Tsaousi S, Tzouvelekis LS, et al. Carbapenemase-producing Klebsiella pneumoniae bloodstream infections: lowering mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob Agents Chemother. 2014;58(4):2322–2328. doi: 10.1128/aac.02166-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machuca I, Gutierrez-Gutierrez B, Gracia-Ahufinger I, et al. Mortality associated with bacteremia due to colistin-resistant klebsiella pneumoniae with high-level meropenem resistance: importance of combination therapy without colistin and carbapenems. Antimicrob Agents Chemother. 2017;61(8):e00406–e00417. doi: 10.1128/aac.00406-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tumbarello M, Viale P, Viscoli C, et al. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase–producing K. pneumoniae: importance of combination therapy. Clin Infect Dis. 2012;55(7):943–950. doi: 10.1093/cid/cis588 [DOI] [PubMed] [Google Scholar]
- 20.Kang C-I, Kim S-H, Park WB, et al. Bloodstream infections due to extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for mortality and treatment outcome, with special emphasis on antimicrobial therapy. Antimicrob Agents Chemother. 2004;48(12):4574–4581. doi: 10.1128/AAC.48.12.4574-4581.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tumbarello M, Spanu T, Sanguinetti M, et al. Bloodstream infections caused by extended-spectrum-β-lactamase-producing Klebsiella pneumoniae: risk factors, molecular epidemiology, and clinical outcome. Antimicrob Agents Chemother. 2006;50(2):498–504. doi: 10.1128/AAC.50.2.498-504.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18. doi: 10.1186/s12941-017-0191-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shankar C, Veeraraghavan B, Nabarro LEB, et al. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 2018;18(1):6. doi: 10.1186/s12866-017-1148-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cryz S, Mortimer P, Mansfield V, et al. Seroepidemiology of Klebsiella bacteremic isolates and implications for vaccine development. J Clin Microbiol. 1986;23(4):687–690. doi: 10.1128/JCM.23.4.687-690.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh K-M, Kurup A, Siu L, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol. 2007;45(2):466–471. doi: 10.1128/JCM.01150-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y-T, Lai Y-C, Tan M-C, et al. Prevalence and characteristics of pks genotoxin gene cluster-positive clinical Klebsiella pneumoniae isolates in Taiwan. Sci Rep. 2017;7(1):43120. doi: 10.1038/srep43120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y, Bao C, Liu J, et al. Microbiological characterisation of Klebsiella pneumoniae isolates causing bloodstream infections from five tertiary hospitals in Beijing, China. J Glob Antimicrob Resist. 2018;12:162–166. doi: 10.1016/j.jgar.2017.10.002 [DOI] [PubMed] [Google Scholar]
- 29.Yeh KM, Lin JC, Yin FY, et al. Revisiting the importance of virulence determinant magA and its surrounding genes in Klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. J Infect Dis. 2010;201(8):1259–1267. doi: 10.1086/606010 [DOI] [PubMed] [Google Scholar]
- 30.TAVAKOL M, MOMTAZ H. Molecular characterization of serotypes and capsular virulence genes in cps gen group of Klebsiella pneumonia isolated from Tehran hospitals. JMW. 2017;10:18–25. [Google Scholar]
- 31.Fang C-T, Lai S-Y, Yi W-C, et al. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45(3):284–293. doi: 10.1086/519262 [DOI] [PubMed] [Google Scholar]
- 32.Aljanaby AAJ, Alhasani AHA. Virulence factors and antibiotic susceptibility patterns of multidrug resistance Klebsiella pneumoniae isolated from different clinical infections. Afr J Microbiol Res. 2016;10(22):829–843. doi: 10.5897/AJMR2016.8051 [DOI] [Google Scholar]
- 33.Regué M, Hita B, Piqué N, et al. A gene, uge, is essential for Klebsiella pneumoniae virulence. Infect Immun. 2004;72(1):54–61. doi: 10.1128/IAI.72.1.54-61.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng H, Chen Y, Wu C, et al. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. 2010;192(12):3144–3158. doi: 10.1128/JB.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu C-R, Lin T-L, Chen Y-C, et al. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiol. 2011;157(12):3446–3457. doi: 10.1099/mic.0.050336-0 [DOI] [PubMed] [Google Scholar]
- 36.Lin H-A, Huang Y-L, Yeh K-M, et al. Regulator of the mucoid phenotype A gene increases the virulent ability of extended-spectrum beta-lactamase-producing serotype non-K1/K2 Klebsiella pneumonia. J Microbiol Immunol Infect. 2016;49(4):494–501. doi: 10.1016/j.jmii.2014.08.023 [DOI] [PubMed] [Google Scholar]
- 37.Yu W-L, Ko W-C, Cheng K-C, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42(10):1351–1358. doi: 10.1086/503420 [DOI] [PubMed] [Google Scholar]
- 38.Shu H-Y, Fung C-P, Liu Y-M, et al. Genetic diversity of capsular polysaccharide biosynthesis in Klebsiella pneumoniae clinical isolates. Microbiol. 2009;155(12):4170–4183. doi: 10.1099/mic.0.029017-0 [DOI] [PubMed] [Google Scholar]
- 39.Yeh K-M, Lin J-C, Yin F-Y, et al. Revisiting the importance of virulence determinant magA and its surrounding genes in Klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. J Infect Dis. 2010;201(8):1259–1267. doi: 10.1086/606010 [DOI] [PubMed] [Google Scholar]
- 40.Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob. 2019;18(1):4. doi: 10.1186/s12941-018-0302-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee C-H, Liu J-W, Su L-H, et al. Hypermucoviscosity associated with Klebsiella pneumoniae-mediated invasive syndrome: a prospective cross-sectional study in Taiwan. Int J Infect Dis. 2010;14(8):e688–e92. doi: 10.1016/j.ijid.2010.01.007 [DOI] [PubMed] [Google Scholar]