Figure 1.
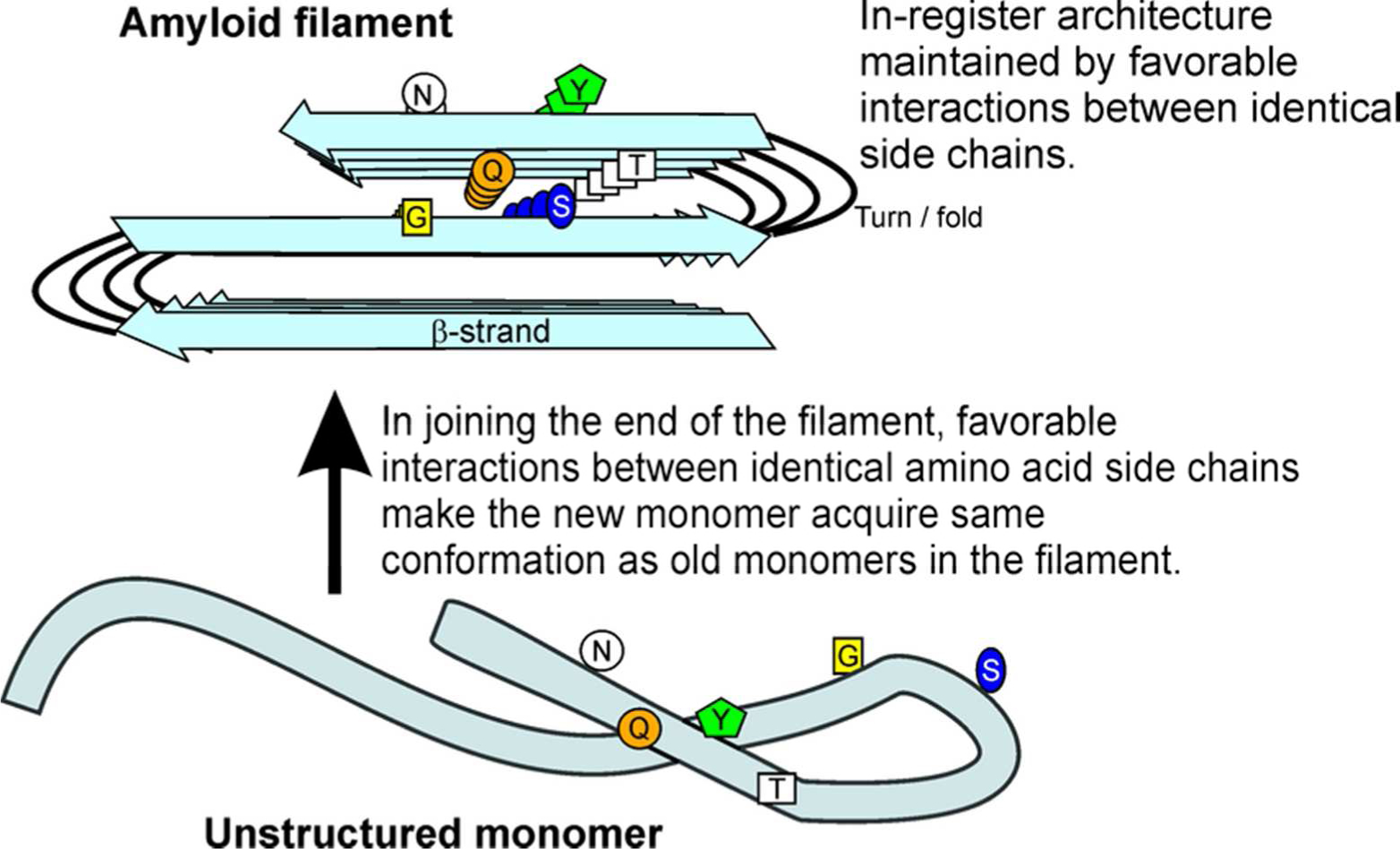
End-on view of an amyloid filament. The folded, parallel in-register β-sheet architecture of infectious amyloids of the prion domains of Sup35p, Ure2p and Rnq1p may explain the ability of prion variants to propagate the amyloid conformation. Prion variants are proposed to differ in the location of the folds of the β-sheet. Favorable interactions among identical amino acid side chains (H-bonding or hydrophobic interactions) require that adjacent molecules be aligned (in-register). This forces molecules joining the ends of the filaments to have their turns (= folds of the sheet) in the same locations as molecules already in the sheet. This constitutes templating of conformation, and can explain how each prion variant is stably propagated (2). Modified from ref. (2).
