Abstract
Cognitive decline is a feared aspect of growing old. It is a major contributor to lower quality of life and loss of independence in old age. We investigated the genetic contribution to individual differences in non-pathological cognitive ageing in five cohorts of older adults. We undertook a genome-wide association analysis using 549 692 single nucleotide polymorphisms (SNPs) in 3511 unrelated adults in the Cognitive Ageing Genetics in England and Scotland (CAGES) project. These individuals have detailed longitudinal cognitive data from which phenotypes measuring each individual’s cognitive changes were constructed. One SNP—rs2075650, located in TOMM40—had a genome-wide significant association with cognitive ageing (p = 2.5 × 10−8). This result was replicated in a meta-analysis of three independent Swedish cohorts (p = 2.41 × 10−6). An APOE haplotype (adjacent to TOMM40), previously associated with cognitive ageing, had a significant effect on cognitive ageing in the CAGES sample (p = 2.18 × 10−8; females, p = 1.66 × 10−11; males, p = 0.01). Fine SNP-mapping of the TOMM40/APOE region identified both APOE (rs429358; p = 3.66 × 10−11) and TOMM40 (rs11556505; p = 2.45 × 10−8) as loci that were associated with cognitive ageing. Imputation and conditional analyses in the discovery and replication cohorts strongly suggest that this effect is due to APOE (rs429358). Functional genomic analysis indicated that SNPs in the TOMM40/APOE region have a functional, regulatory non protein-coding effect. The APOE region is significantly associated with non-pathological cognitive ageing. The identity and mechanism of one or multiple causal variants remain unclear.
Keywords: GWAS, genetics, cognitive ageing, APOE, TOMM40
Introduction
As human life expectancy has increased, addressing the causes of bodily ageing and age-related diseases has become a higher priority in research. Like aspects of physical function, many cognitive abilities decline with age. Cognitive decline can begin in the 20s or 30s, though it is gradual in most people, and even from the 50s onwards the change can be modest in those who do not have dementia or a physical illness that affects cognitive functions1,2. By old age most people will have cognitive decrements compared with their earlier capabilities, but there are individual differences in the severity of this change, even in people who do not have dementia or mild cognitive impairment3. Age-related cognitive impairment is an increasing problem for society and one that carries with it high personal, social and economic burdens4,5. As the global population aged over 60 is set to rise threefold to 2 billion by 2050 the burden of age-related cognitive impairment on society will increase6. Research must address the causes of the early stages of cognitive ageing in order to preserve healthy brains and help delay or prevent dementia, and allow older individuals to enjoy a more independent and active old age.
Identifying risk factors for individual differences in age-related cognitive decline is, therefore, amongst the greatest challenges facing the healthcare of older people. Twin studies have attempted to identify genetic influences on cognitive ageing; however, until recently, the proportion of the variance in lifetime cognitive change explained by genetic and environmental influences remained unknown, not least because it is rare to find appropriate samples with cognitive abilities tested repeatedly over a sufficiently long period7–10. Deary et al., using three of the cohorts included in the present study, utilised population–based genetic analyses to quantify the genetic and environmental contribution to stability and changes in intelligence differences for most of the human lifespan11. Our estimate of the lower limit of the narrow-sense heritability of lifetime (age 11 to old age) cognitive change was 0.24 (±0.20), and the genetic correlation between childhood and old age intelligence was estimated to be 0.62 (±0.22). These results suggest that genetic factors may have a role in cognitive change between childhood and old age.
A replicated finding in non-pathological, age-related cognitive change is a small contribution from variation in the Apolipoprotein E (APOE) gene; other suggested individual genetic contributions are largely unreplicated12,13. A meta-analysis indicated that cognitively healthy carriers of the APOE e4 allele, previously associated with late onset Alzheimer’s disease (LOAD), performed worse on several domains of cognitive ability and this effect increased with age13. A recent genome-wide association study (GWAS) of age-related cognitive decline also reported a genome-wide significant finding for APOE; however, the discovery and replication cohorts included a large proportion of individuals with mild cognitive impairment or with Alzheimer’s disease, with just 60% of the discovery cohort being classified as cognitively normal14. APOE is located in a gene-dense region of chromosome 19, the association of this region with AD has been known for many years and has been the subject of many studies15,16. The APOE e2 allele has been found to be positively associated with survival and longevity in older adults and appears to have a protective effect in LOAD13,17,18. There is evidence that APOE e3 carriers with a long poly-T repeat (rs10524523 or ‘523’) in the neighbouring Translocase of the Outer Mitochondrial Membrane 40 homolog gene (TOMM40) gene have an earlier age of onset of LOAD than those with shorter ‘523’ repeats19. Other variants in TOMM40 have also been associated with LOAD (http://www.alzgene.org/), but few studies have examined the influence of TOMM40 on non-pathological cognitive decline. A small study showed that APOE e3 homozygotes with long ‘523’ repeats have decreased memory abilities and gray matter volumes compared to those with shorter repeats20. In a longitudinal study across most of the ninth decade, both APOE e4 and ‘523’ repeats were associated with age-related cognitive decline, but there was no interaction between the two genes21. Ideally, longitudinal cognitive data should be collected over more extensive periods of time to discover any genetic causes of cognitive ageing, because each is likely to have a small effect.
Here we report a GWAS of non-pathological cognitive ageing. We investigate the genetic contribution to individual differences in cognitive change by performing a GWAS using five UK cohorts of older adults (N = 3511) with longitudinal cognitive ability data, and by attempting replication in three independent Swedish cohorts (N = 1367).
Materials and Methods
Cohort Descriptions
The five UK cohorts described below make up the Cognitive Ageing Genetics in England and Scotland (CAGES) consortium (Supplementary Table 1). These are the discovery samples. Three Swedish cohorts are the replication samples (Supplementary Table 1).
Discovery cohorts
The Lothian Birth Cohort 1921 (LBC1921)
The LBC1921 is a longitudinal study of cognitive ageing conducted at the University of Edinburgh. Individuals recruited into the LBC1921 were all born in 1921 and most had completed the Moray House Test (MHT) No. 12 assessment of general intelligence in the Scottish Mental Survey 1932 at a mean age of 11 years22,23. A total of 550 individuals (234 men and 316 women) were recruited and tested at a mean age of 79.1 years (SD = 0.6)24. The recruitment and re-testing of these individuals in old age has been described previously23,24. Following informed consent, venesected whole blood was collected for DNA extraction. Ethical approval was obtained from Lothian Research Ethics Committee.
The Lothian Birth Cohort 1936 (LBC1936)
The LBC1936 is a longitudinal study of cognitive ageing conducted at the University of Edinburgh. Individuals recruited into the LBC1936 were all born in 1936 and most had completed the MHT, in the Scottish Mental Survey 1947, at a mean age of 11 years23,25. A total of 1091 participants (548 men and 543 women) were recruited and tested individually at a mean age of 69.5 years (SD = 0.8). The recruitment and re-testing of these individuals in old age has been described previously26. Following informed consent, venesected whole blood was collected for DNA extraction. Ethical approval was obtained from Scotland’s Multicentre Research Ethics Committee and Lothian Research Ethics Committee.
Aberdeen Birth Cohort 1936 (ABC1936)
The ABC1936 is a longitudinal study of cognitive ageing conducted at the University of Aberdeen. Individuals recruited into the ABC1936 were all born in 1936 and most had completed the MHT in the Scottish Mental Survey 1947 at a mean age of 11 years23,25. The recruitment and re-testing of these individuals in old age has been described previously24. A total of 498 relatively healthy participants (243 men, 255 women) were traced and tested at mean age 64.6 years (SD = 0.9). Following informed consent, venesected whole blood was collected for DNA extraction. Ethical approval was obtained from the Grampian Research Ethics Committee.
Manchester and Newcastle Longitudinal Studies of Cognitive Ageing Cohorts
The University of Manchester Age and Cognitive Performance Research Centre (ACPRC) programme has documented longitudinal trajectories in cognitive function in a large sample of older adults in the North of England whilst examining its predictors and their confounders27. At the outset of the study, 6063 volunteers were available (1825 men, 4238 women), with a median age of 65 years and a range from 44 to 93 years. The Dyne Steel DNA Archive for Ageing and Cognition was established following invitation to all participating volunteers between 1999 and 2004. Approximately 2000 volunteers attended the Universities (or, in some cases, were visited at home). These volunteers had undergone biennial cognitive test sessions with two alternating different task batteries. The average number sessions participated in by volunteers for battery A=4 and battery B=3. The median interval between first and last test session was 14 years, range 12–18 years. Following informed consent, venesected whole blood was collected for DNA extraction. Ethical approval was obtained from University of Manchester.
Replication cohorts
Three related twin studies from the Swedish Twin Registry were used in the replication analyses28.
The Swedish Adoption/Twin Study of Aging (SATSA)
SATSA includes a matched sample of same-sex twin pairs reared apart with those reared together29,30. In-person testing (IPT) sessions include those 50 years and older and were conducted at three-year intervals with the exception of a gap at IPT4 which encompassed a telephone interview only. DNA was extracted from blood samples collected at the third or subsequent IPT sessions. Cognitive data and marker information were available for those without prevalent dementia for up to 633 individuals (58.8% female) among 372 pairs and with up to five waves (IPT1-IPT3, IPT5, IPT6) of data. The average age at baseline was 67.6 years (SD = 8.6). On average, SATSA twins participated in 3.3 IPT waves (SD = 1.2).
The Sex Differences in Health and Aging Study (Gender)
Gender is a completed study of 498 opposite-sex twins, born between 1906 and 1925, entailing three IPT waves31. Blood samples for DNA extraction were taken at the first IPT occasion. Cognitive data and marker information were available among those without prevalent dementia for up to 385 individuals (49.0% female) among 230 opposite-sex twin pairs and on up to three occasions of measurement. The baseline average age was 74.4 years (SD = 2.6). On average, Gender twins participated in 1.7 IPT waves (SD = 0.8).
The Study of Origins of Variance in the Oldest-Old (OCTO-Twin)
OCTO-Twin is a completed five-wave study of 702 twins born during or prior to 191332. Blood for DNA extraction was taken at the first or second IPT. Cognitive data and marker information were available among those without prevalent dementia for up to 349 individuals (64.2% female) among 246 pairs and for up to 5 waves. The average age at baseline was 82.8 years (SD = 2.4). On average, twins participated in 2.9 IPT waves (SD = 1.5).
Phenotype construction
For fluid-type intelligence in old age, principal components analyses (PCA) were used in the following cohorts to derive a general intelligence factor (strictly, PCA does not produce ‘factors’, but this is common usage): the LBC1921 and LBC1936, and the ABC1936. In each case, the scores on a number of fluid-type cognitive tests were subjected to PCA. Fluid cognitive tests typically show age-related decrements. In all cases a single component was indicated (using analysis of the scree slope and the Eigenvalues-greater-than-one criterion), and was extracted. Individuals’ scores on the first unrotated principal component were used as the indicator of general fluid-type intelligence (gf). The tests used to form the gf factor in the LBC1921 were the Moray House Test23, Raven’s Standard Progressive Matrices33, Logical Memory34, and Verbal Fluency35. The tests used to form the gf factor in the LBC1936 were six tests from the Wechsler Adult Intelligence Scale (WAIS)-IIIUK 36: Digit Symbol Coding, Block Design, Matrix Reasoning, Digit Span Backward, Symbol Search, and Letter-number Sequencing. The tests used to form the gf factor in the ABC1936 were Raven’s Standard Progressive Matrices33, Digit Symbol37, Uses of Common Objects38, and Auditory Verbal Learning Test35. The gf factors for each cohort were then adjusted for prior cognitive ability using the Moray House Test scores at age 11, thus providing a quantitative measure of cognitive change from age 11 to old age. Both gf and age 11 Moray House Test scores were adjusted for age in days at time of testing prior to the creation of the cognitive change measure. These measures were extracted and standardized independently for males and females.
A fluid intelligence variable (gf) for the Manchester and Newcastle cohorts was derived from the Alice Heim 4 and Cattell Culture Fair tests overall total correct score27. In all cases men and women were examined and scores standardized separately. Growth curve models were estimated that took the 0 point on the age scale as age 70, and measured variation about that in units of 10 years. Data were available for up to four occasions of measurement. The fixed part of the models that described the overall pattern of change in the construct for the sample as a whole included linear and quadratic age terms. In addition, to account for possible artifactual improvement due to practice effects (a single step function), each test score was allowed to increment between the first and subsequent occasions in which each particular test was taken. Individual differences were accounted for by allowing subject-specific random effects for the intercept, describing the within-sample variation in performance at age 70, and for a linear growth term, describing the individual differences in the trend of cognitive decline over the period of follow-up. The random effects were assumed to be bivariate normally distributed and the models estimated in gllamm by maximum likelihood using adaptive quadrature (www.gllamm.org). Incomplete data observations were included under an assumption that the missing scores were missing at random (MAR), allowing attrition to be selective with respect to age, sex, and observed test scores. Subject-specific intercepts and linear trends were estimated using empirical Bayes’s methods. The above fully describes the model in the circumstance where a single repeated cognitive test was available. Where multiple tests were available at each occasion then a factor growth model was estimated as illustrated in Supplementary Figure 1 for the two-measure case. Different tests were allowed different means, scales and error variances but the tests were assumed to reflect a single underlying construct with a common trend.
The cognitive outcome for the combined SATSA, Gender, and OCTO-Twin analyses included the 1st unrotated principal component from a PCA of four tests in common across the three studies spanning crystallized/verbal (Synonyms), fluid/spatial (Block Design), perceptual speed (Symbol Digit) and episodic memory (Thurstone Picture Memory) domains39. PCA scores across waves were constructed by applying standardized scoring coefficients to test scores standardized against the first in-person occasion means and SDs, and transformed into t-score units within gender (Mean = 50, SD = 10 at the first occasion within gender). The combined sample size was 1,367 (582 men, 785 women).
In the Swedish cohorts, for individuals who developed incident dementia, cognitive scores were set to missing for the data collected after age of dementia onset. Growth curve models were fitted centering on age 70, with change parameters in units of 10 years. The fixed effect model included intercept, linear, and quadratic age terms. In addition, a retest effect accounted for possible improvement due to practice effects (a single step function between the first and subsequent testing occasions). The random effects model allowed for individual variation for the intercept (performance at age 70), a linear growth term (estimating the linear trend at age 70 years), and nonlinear quadratic change across age. Dependency among pairs was accounted for by including between- and within-pair random effects. The growth models were fitted in SAS PROC Mixed by full maximum likelihood estimation. Those with incomplete data were included in the model-fitting.
Genotyping and Quality Control
Discovery cohorts
Genomic DNA was isolated by standard procedure at the Wellcome Trust Clinical Research Facility (WTCRF) Genetics Core, Western General Hospital, Edinburgh (for the LBC1936 and the ABC1936), and the Medical Research Council Technology, Western General Hospital, Edinburgh (for the LBC1921). The UK DNA Banking Network was used for the Manchester and Newcastle Dyne-Steel samples. In total, 3802 samples (LBC1936 N = 1042; LBC1921 N = 526; ABC1936 N = 456; Manchester N = 901; and Newcastle N = 877) were genotyped at the WTCRF Genetics Core using the Illumina610-Quadv1 chip (Illumina, Inc., San Diego, CA, USA). These samples were then subjected to the quality control (QC) procedures described below.
Individuals were excluded from further analysis if there was a disagreement between genetic and reported gender. Relatedness between subjects was investigated and, for any related pair of individuals, one was removed (PI_HAT [proportion of IBD] > 0.25). Samples with a call rate ≤ 0.95, and those showing evidence of non-Caucasian descent by multidimensional scaling, were also removed. SNPs were included in the analyses if they met the following conditions: call rate ≥ 0.98, minor allele frequency ≥ 0.01, and Hardy-Weinberg Equilibrium test with p ≥ 0.001. After QC, 3511 samples remained (LBC1936 N = 1005; LBC1921 N = 517; ABC1936 N = 426; Manchester N = 805; and Newcastle N = 758). The final number of SNPs included in the study was 549 692.
Replication cohorts
Genotyping of rs2075650 was available for the Swedish cohorts from the Cardio-MetaboChip (Illumina, San Diego, CA, USA) that provided coverage across the genome as well as rare variants implicated in metabolic and cardiovascular disease traits. Genotyping was performed by the Uppsala University SNP technology platform (http://www.medsci.uu.se/molmed/snpgenotyping/methods.htm).
APOE genotyping
For LBC1936 and ABC1936, APOE SNPs rs7412 and rs429358 were genotyped using TaqMan® technology (Applied Biosystems, Carlsbad, CA, USA) by the Wellcome Trust Clinical Research Facility Genetics Core, Western General Hospital, Edinburgh. For LBC1921, APOE haplotype was determined by polymerase chain reaction (PCR) amplification of a 227-bp fragment of the APOE gene containing two polymorphic sites (rs7412 and rs429358) that account for the three alleles, e2, e3, and e4, followed by restriction digest with Cfo1, and electrophoresis in 4% NuSieve gels40. The Manchester and Newcastle cohorts were genotyped by Sequenom using the iPLEX method. This method has been described previously41. For replication cohorts, the APOE SNPs, rs7412 and rs429358, were assayed using Illumina GoldenGate assays (Illumina, San Diego, CA, USA).
Genotyping of TOMM40/APOE region
To follow up the significant GWAS result located in the TOMM40 gene (see below), 62 SNPs were tested which covered the TOMM40/APOE region of chromosome 19. SNPs were identified from several sources (HapMap, 1000 Genomes project (NCBI dbSNP Build 131), and The Duke Bryan Alzheimer’s Research Center cohort42) with the criterion that they had previously been validated in a Caucasian population. These SNPs were then genotyped in the CAGES cohorts using an OpenArray® Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA). Six SNPs were genotyped using TaqMan® technology (Applied Biosystems, Carlsbad, CA, USA), because it was not possible to genotype them using the OpenArray method. Genotyping was performed at the WTCRF Genetics Core, Western General Hospital, Edinburgh. Quality control was performed on these SNPs and 54 were retained for analysis. SNPs were excluded if they failed to achieve a 90% call rate. Minor allele frequencies and Hardy-Weinberg equilibrium were checked but no exclusions were made.
Imputation of TOMM40/APOE region
The GWAS and fine mapping data were combined to provide the most detailed genotyping available for the region of interest. 2684 SNPs were imputed, using the Minimac software, in a 200kb region surrounding TOMM40/APOE. The 1000 Genomes v2 was used as the reference sample. An imputation quality cut off of 0.3 and a MAF < 0.005 filter were applied to the data prior to analysis. After QC 594 SNPs remained.
Polymorphic poly-T variant
A polymorphic poly-T variant, rs10524523 (hereinafter ‘523’), in the TOMM40 gene was previously genotyped in LBC1921 by Polymorphic DNA technologies (Alameda, CA, USA)21. For this study, LBC1936, and the Manchester and Newcastle Cohorts were genotyped by the Roses laboratory using a method described previously43,44. Briefly, TOMM40 ‘523’ polyT genotypes were determined based on length variation. The ‘523’ region of each genomic DNA sample was PCR-amplified using a fluorescently labeled primer. Genotypes were determined on an ABI 3730 DNA Analyzer using GeneMapper, version 4.0 software (Applied Biosystems, Foster City, CA) for allelic size assessment. The ‘523’ allele was assigned according to the length of the PCR product. The convention established by Roses et al. for determining alleles was used: Short (S), ≤19; Long (L) - 20–29; Very Long (VL) ≥3019.
Statistical Analysis
Genome-wide association analyses (GWAS) of cognitive ageing phenotypes were initially carried out for each of the CAGES cohorts, split by gender, using linear regression under an additive model, in PLINK45. Meta-analyses were performed to combine the males and females for each cohort using the program METAL (www.sph.umich.edu/csg/abecasis/Metal). All association analyses were adjusted for four MDS components to correct for any population stratification, as described previously46. An inverse variance weighted meta-analysis of all of the CAGES cohorts was performed (www.sph.umich.edu/csg/abecasis/Metal). Gene-based tests for association were performed on the meta-analysis results using VEGAS47. Published candidate genes previously associated with Alzheimer’s disease (http://www.alzgene.org/) were examined for association with cognitive ageing using this gene-based method.
To investigate the effect of APOE on cognitive ageing, individuals were firstly classified according to APOE e4 allele carrier status due to previously-reported associations of the e4 allele with cognitive variables13. Association of the APOE e4 carrier status variable with cognitive ageing was assessed using a linear regression analysis. All GWAS analyses described above were repeated with APOE e4 allele dosage fitted as a covariate. A second APOE analysis was performed using APOE e2 allele carrier status to investigate the previously-reported protective effect of the e2 allele on LOAD17. Association of the APOE e2 carrier status variable with cognitive ageing was assessed using a linear regression analysis.
Biological pathway analysis was performed using WebGestalt248,49. A list of suggestive findings from the VEGAS analysis (p < 0.01) was used in this analysis. An enrichment analysis for the Gene Ontology (GO) categories was performed using the hypergeometric test. The resulting p-values were corrected for multiple testing using the Benjamini-Hochberg method50. The significance threshold was p < 0.05 and the minimum number of genes for a category was two. Functional annotation analysis was carried out using DAVID, a data-mining integrated environment of bioinformatics resources51. Suggestive findings from the VEGAS analysis were used and functional annotation of this list was performed using GO terms, pathways, protein domains, disease associations and functional categories. The highest clustering stringency was used as this generates fewer functional groups, with more tightly associated genes in each group. An enrichment score of 1.3 or greater indicates a cluster which should be examined in further detail; this is equivalent to p = 0.05.
Replication analyses
In the Swedish cohorts, tests of marker effects proceeded in the context of simultaneous growth model fitting. The rs2075650 marker was recoded into number of rare alleles. A baseline growth model was fitted including the retest covariate. A second model added rs2075650 as a predictor of performance level and change. Model comparisons were conducted by calculating difference chi-square tests to evaluate the association of rs2075650 with the three cognitive trajectory parameters (intercept, linear and quadratic change). Where indicated, analyses included APOE entered as a two-SNP set in the baseline model (rs429358, rs7412 each recoded into number of rare alleles), and subsequently rs2075650 was added to determine its unique contributions. Tests of individual growth parameter effects were based upon t-statistics and associated p-values calculated by the parameter estimate divided by its asymptotic standard error.
Fine mapping
Genotype-phenotype association analyses were initially carried out on the fine mapping data for each of the CAGES cohorts, split by gender, using linear regression under an additive model in PLINK45. An inverse variance weighted meta-analysis of all of the CAGES cohorts was performed using METAL (www.sph.umich.edu/csg/abecasis/Metal). Conditional analyses were performed to test for independence of the top hits. Conditional analysis fits the allelic dosage of the specified SNP as a covariate in a linear regression analysis. SNPs remaining significant are assumed to have an independent effect on the phenotype with respect to the conditioned SNP. Allelic dosage of the top SNP from each locus of interest was fitted as a covariate in a genotype-phenotype analysis. These analyses, both genotype-phenotype association and conditional, were repeated for the female subjects using imputed data, this analysis was performed using the MACH2 QTL software52,53.
The TOMM40 ‘523’ variants were first classified into three categories, as described previously19,21. The classifications were: ‘Short’ (S, T≤19), ‘Long’ (L, 20≤T≤29) and ‘Very Long’ (VL, T≥30). Each individual was then assigned to one of three groups: short homozygotes, short heterozygotes, and non-short. Univariate analysis of variance was performed to assess the association between ‘523’ repeat length and cognitive ageing.
Functional genomic analysis
Data mining of publicly available databases was performed by comparing existing and uploaded tracks on the UCSC genome browser 2009 human reference sequence (GRCh37 hg19)54; see Supplementary Note Appendix 1, Table A1 for details of the tracks used. As regulatory features show both spatial/temporal and state differences between cell lines and tissues, the most appropriate analysis for this study was to investigate adult brain tissue or neuronal/glial cell datasets. Where possible, data mining was therefore restricted to CNS relevant cell lines or tissues. We analyzed the genome-wide distribution of CpG islands (CPGI) using data from the UCSC genome browser CPGI track (hg19, cpgIslandExt) and protein coding gene models obtained from ENSEMBL (release 64)55,56. The genomic overlap between CPGI and gene models was computed using in-house scripts. This analysis was applied to the SNPs which reached suggestive significance from the fine mapping analysis (see Supplementary Note).
Results
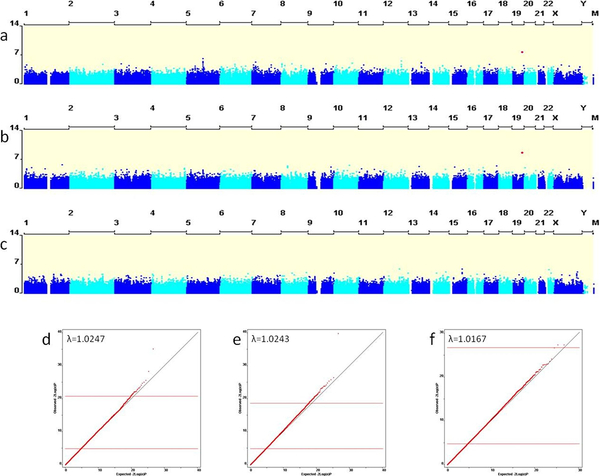
The meta-analysis of the GWAS of the CAGES cohorts identified a single SNP (rs2075650), on chromosome 19, that achieved a genome-wide significant association with cognitive ageing (p = 2.47 × 10−8) (Figure 1 and Table 1). The SNP is located in the TOMM40 gene. The meta-analyses split by gender revealed genome-wide significance in the females (p = 2.43 × 10−9), but not the males (p = 0.15) (Figure 1, Table 1 and Supplementary Figure 2). Further results from these meta-analyses are shown in Supplementary Tables 2–4, in which all SNPs that reached a suggestive significance threshold of p < 1 × 10−5 are reported. The meta-analysis of the CAGES females identified a SNP in the PLXNA2 gene which reached suggestive significance (p = 1.87 × 10−6). A SNP in the ARHGAP19 gene was found to be of suggestive significance (p = 8.84 × 10−6) in the meta-analysis of the CAGES cohorts.
Figure 1. Meta-analytic genome-wide association results for all five samples in the Cognitive Ageing Genetics in England and Scotland consortium.
Manhattan plots show meta-analysis results for all of the CAGES individuals (a), females (b) and males (c). The −log10 p values (y axis) of 549 692 SNPs are presented based on their chromosomal position (x axis). The red point (b) indicates a p value of genome-wide significance (p<5×10−8). Quantile-quantile plots of the meta-analysis p-values for all of the CAGES individuals (d), females (e) and males (f). The red dots represent the observed data; the black line is the expectation under the null hypothesis of no association.
Table 1.
Results for the SNP rs2075650 (G allele) in each of the discovery cohorts and the replication cohorts. Meta-analysis results for the discovery and replication datasets are also shown. N indicates the sample size, beta is the effect size, SE is the standard error of the beta, and AF is the allele frequency of the effect allele.
| All | Females | Males | |||||
|---|---|---|---|---|---|---|---|
| Beta (SE) | P (N) | Beta (SE) | P (N) | Beta (SE) | P (N) | AF | |
| ABC1936 | 0.05 (0.11) | 0.64 (347) | −0.062 (0.16) | 0.70 (168) | 0.16 (0.15) | 0.30 (179) | 0.14 |
| LBC1936 | −0.20 (0.06) | 0.002 (932) | −0.25 (0.09) | 7.7 × 10−3 (457) | −0.15 (0.09) | 0.08 (475) | 0.16 |
| LBC1921 | −0.26 (0.10) | 0.007 (453) | −0.39 (0.13) | 1.81 × 10−3 (268) | −0.06 (0.15) | 0.69 (185) | 0.15 |
| Manchester | −0.30 (0.08) | 1.9 × 10−4 (805) | −0.35 (0.10) | 3.62 × 10−4 (571) | −0.14 (0.17) | 0.41 (234) | 0.15 |
| Newcastle | −0.21 (0.08) | 0.006 (753) | −0.25 (0.10) | 8.71 × 10−3 (535) | −0.1 (0.14) | 0.48 (218) | 0.13 |
| CAGES meta-analysis | −0.20 (0.04) | 2.47 × 10−8 (3290) | −0.27 (0.05) | 2.43 × 10−9 (1999) | −0.08 (0.06) | 0.15 (1291) | 0.14 |
| Swedish pooled-analysis | −1.30 (0.27) | 2.41 × 10−6 (1367) | −1.05 (0.38) | 5.85 × 10−3 (785) | −1.54 (0.38) | 6.47 × 10−5 (582) | 0.15 |
Gene-based tests of association resulted in no significant findings based on a Bonferroni threshold (p < 2.8 × 10−6). Suggestive gene-based findings (p < 0.01) are detailed in Supplementary Tables 5–7.
The genome-wide significant finding, rs2075650, is an intronic SNP within TOMM40. This result was replicated in a pooled-analysis of three later-life Swedish cohorts with cognitive ageing measures (Table 1). In the Swedish analysis, the rs2075650 marker predicted variation in cognitive trajectories in both men (χ2 (3) = 15.7, p = 1.31 × 10−3) and women (χ2 (3) = 9.4, p = 2.44 × 10−2) with a consistent effect of the marker on linear decline at age 70 (pmen = 6.47 × 10−5, pwomen = 5.85 × 10−3). The Swedish pooled-analysis is the only analysis that finds a significant association of rs2075650 with cognitive ageing in men (Table 1).
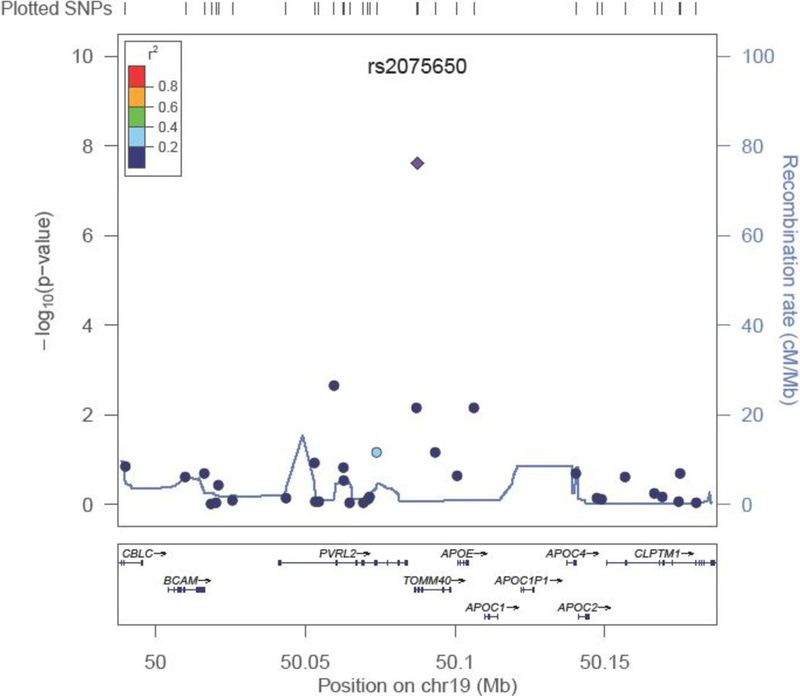
Figure 2 demonstrates the GWAS results for the CAGES females for a 100kb region surrounding the TOMM40 gene. From the plot it can be seen that there are several genes in this region; however, the region is poorly covered by SNPs on the genotyping array used in this study. In close proximity to TOMM40 is APOE which has been previously linked to cognition in later life57,58. Although not on the genotyping array used in this study, APOE e2/e3/e4 genotyping was available on the CAGES individuals and was included in some of the analyses. Regression analyses were performed to investigate the associations between the APOE e4 and e2 alleles and cognitive ageing (Table 2 and Supplementary Table 8). The APOE e4 allele is significantly associated with a greater decline in cognitive ability in the entire CAGES sample (p = 2.18 × 10−8) and the male and female subgroups; however, the effect appears to be greater in females (pwomen = 1.66 × 10−11; pmen = 0.01). The APOE e2 allele shows a nominally significant association with less cognitive decline in the females (p = 0.05). Supplementary Figure 3 shows the Manhattan and Quantile-Quantile plots for the CAGES GWAS analyses adjusted for APOE e4 allele dosage. No SNPs were significantly associated with cognitive ageing. When adjusted for APOE e4 allele dosage, rs2075650 does not achieve even a suggestive significance threshold (pall = 0.69; pwomen = 0.37; pmen = 0.65).
Figure 2. Region plot showing detailed results for a 200kb region on chromosome 19 flanking the TOMM40 gene.
The circles represent each genotyped SNP with the colour indicating pairwise LD in relation to the top hit (calculated from 1000 Genomes June 2010 CEU), −log10 p values are also indicated (y axis). The purple diamond represents the genome-wide significant SNP. This figure also shows the poor coverage of the region on the genotyping array. The solid blue line indicates the recombination rate.
Table 2.
Association of APOE e4 allele with cognitive ageing. A negative beta indicates that the e4 allele is associated with a greater decline in cognitive ability. SE is the standard error of the beta. AF is the allele frequency of the e4 allele.
| All | Females | Males | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cohort | Beta | SE | P | Beta | SE | P | Beta | SE | P | AF |
| ABC1936 | −0.19 | 0.12 | 0.11 | −0.27 | 0.17 | 0.1 | −0.11 | 0.16 | 0.52 | 0.14 |
| LBC1936 | −0.18 | 0.06 | 0.004 | −0.23 | 0.10 | 0.016 | −0.14 | 0.09 | 0.1 | 0.16 |
| LBC1921 | −0.34 | 0.10 | 6.89 × 10−4 | −0.44 | 0.13 | 7.27 × 10−4 | −0.19 | 0.16 | 0.25 | 0.13 |
| Manchester | −0.36 | 0.08 | 1.18 × 10−5 | −0.4 | 0.09 | 2.08 × 10−5 | −0.22 | 0.17 | 0.19 | 0.14 |
| Newcastle | −0.22 | 0.07 | 0.002 | −0.27 | 0.09 | 0.002 | −0.11 | 0.13 | 0.41 | 0.14 |
| Meta-analysis | −0.22 | 0.04 | 2.18 × 10−8 | −0.31 | 0.05 | 1.66 × 10−11 | −0.15 | 0.06 | 0.01 | 0.14 |
In the pooled analysis of the Swedish samples, the two APOE SNPs are significant predictors of the cognitive trajectories in both men (omnibus p = 3.30E-05) and women (omnibus p = 1.62E-03). When rs2075650 is added to a model containing the APOE SNPs, the effect of rs2075650 is not significant via the omnibus test in either men (p > 0.53) or women (p > 0.18). The linear effect is also not significant in either men or women; however, there is a trend in the intercept effect for women (p = 0.0552).
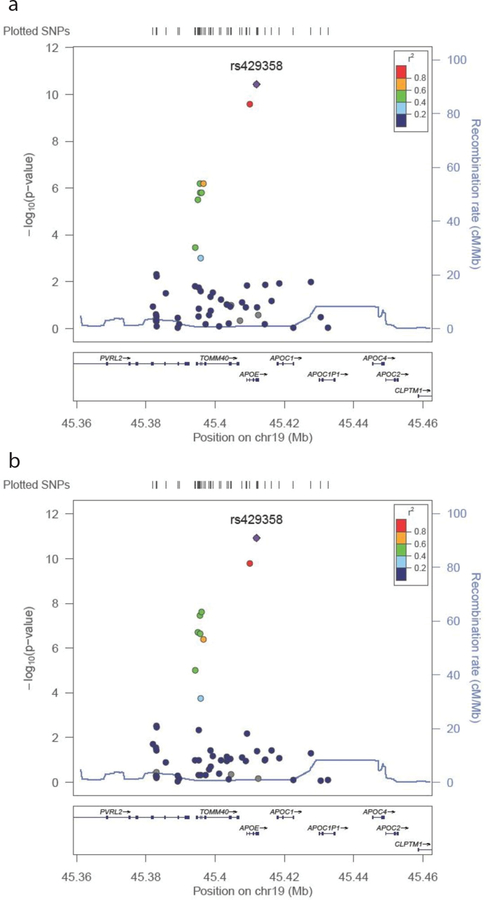
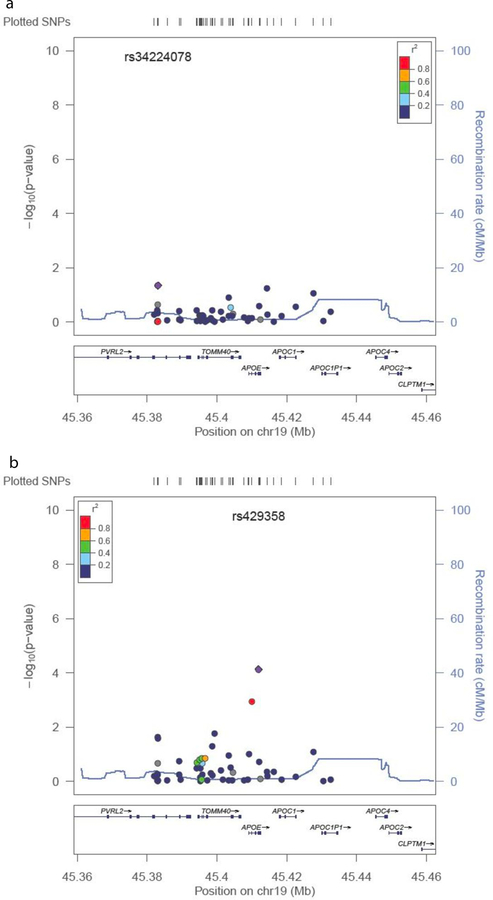
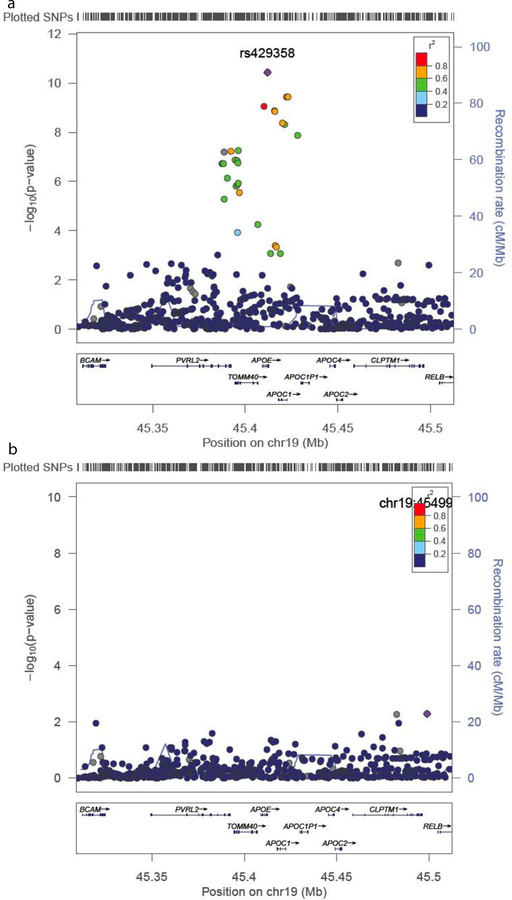
The results of the association analyses from the fine-mapping genotype data in the CAGES sample are shown in Table 3, Figure 3, and Supplementary Table 9. Results for all SNPs which reached a suggestive significance level of p < 1 × 10−5 in the meta-analysis of the females are presented in Table 3. Four SNPs achieved genome-wide significance in the females, two of which also reached genome-wide significance in the whole sample. Results from both the meta-analysis of the CAGES cohorts and of the females are shown in Figure 3. The top two SNPs in each analysis are located in APOE. The further seven suggestive SNPs in the females are located in TOMM40, five of which also reached suggestive significance in the whole sample. No significant results were observed from the analyses of the males. The pairwise linkage disequilibrium (LD), calculated from the CAGES samples, between the top two SNPs, rs429358 and rs769449, is r2 = 0.795, and between rs429358 and rs11556505 is r2 = 0.539. Because these SNPs are in moderately high LD and APOE has been previously linked to cognitive phenotypes, conditional analyses were performed to test the independence of these SNP effects (Figure 4). This analysis was only carried out on the female sample. When the analysis was conditioned on rs429358, no significant associations were observed. However, when the analysis was conditioned on rs10556505, rs429358 remained significant (p = 7.38 × 10−5) at a Bonferroni corrected significance threshold (p < 9.26 × 10−4). The results from the analysis of the imputed data in the female subjects are shown in Figure 5. Again, the meta-analysis results (Figure 5A) appear to highlight two loci of interest, APOE and TOMM40. However, when the analysis is conditioned on the top APOE SNP (rs429358), none of the other SNPs remain significantly associated. This suggests that the effects of APOE and TOMM40 on cognitive ageing are not statistically independent effects and that the association is driven by APOE-based variation. For the APOE SNP rs429358 a positive beta is associated with the T allele; therefore, the presence of a T allele is associated with less cognitive decline. The T allele is associated with the absence of an e4 allele, which is consistent with previous findings linking e4 with increased cognitive decline13.
Table 3.
Top hits from APOE/TOMM40 region fine-mapping analysis. SNPs which achieve suggestive significance (p < 1 × 10−5) in the female only meta-analysis. AF is the allele frequency of the effect allele in the entire CAGES sample. Beta is the effect size. SE is the standard error of the beta. The values for rs2075650 differ slightly from those presented in Table 1 as genotype data were available for more individuals for this analysis.
| All | Females | Males | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Effect Allele | AF | Beta | SE | p | Beta | SE | p | Beta | SE | p |
| rs429358 | T | 0.86 | 0.26 | 0.04 | 3.66 × 10−11 | 0.35 | 0.05 | 1.19 × 10−11 | 0.14 | 0.06 | 0.03 |
| rs769449 | A | 0.12 | −0.26 | 0.04 | 2.51 × 10−10 | −0.34 | 0.05 | 1.62 × 10−10 | −0.14 | 0.06 | 0.03 |
| rs11556505 | T | 0.15 | −0.18 | 0.04 | 1.56 × 10−6 | −0.28 | 0.05 | 2.45 × 10−8 | −0.05 | 0.06 | 0.40 |
| rs2075650 | G | 0.86 | −0.19 | 0.04 | 6.10 × 10−7 | −0.27 | 0.05 | 3.47 × 10−8 | −0.07 | 0.06 | 0.25 |
| rs184017 | T | 0.80 | 0.16 | 0.03 | 3.13 × 10−6 | 0.23 | 0.04 | 1.96 × 10−7 | 0.06 | 0.05 | 0.29 |
| rs157581 | T | 0.79 | 0.16 | 0.03 | 1.56 × 10−6 | 0.23 | 0.04 | 2.28 × 10−7 | 0.07 | 0.05 | 0.19 |
| rs59007384 | T | 0.20 | −0.17 | 0.03 | 6.26 × 10−7 | −0.22 | 0.04 | 4.06 × 10−7 | −0.09 | 0.05 | 0.08 |
| rs71352238 | A | 0.85 | 0.12 | 0.04 | 4.72 × 10−3 | 0.22 | 0.05 | 9.99 × 10−6 | 0.02 | 0.06 | 0.79 |
Figure 3. Meta-analytic results for the TOMM40/APOE region.
Results from both the meta-analysis of all subjects (a) and the meta-analysis of female subjects (b) are shown. The circles represent each genotyped SNP with the colour indicating pairwise LD in relation to the top hit (calculated from 1000 Genomes June 2010 CEU); −log10 p values are also indicated (y axis). The purple diamond represents the top hit, rs429358. The solid blue line indicates the recombination rate.
Figure 4. Results from the conditional analyses of the TOMM40/APOE region.
Two analyses were performed, one conditioning on the top GWAS hit rs429358 (a), and secondly on rs11556505 (b) which is the best SNP from the TOMM40 locus. The circles represent each genotyped SNP with the colour indicating pairwise LD in relation to the top hit (calculated from 1000 Genomes June 2010 CEU); −log10 p values are also indicated (y axis). The purple diamond represents the top hit. The solid blue line indicates the recombination rate.
Figure 5. Meta-analytic results for the 1000 Genomes imputation of the TOMM40/APOE region.
Results from the meta-analysis of the female subjects (a) and a conditional analysis conditioning on the top GWAS hit rs429358 (b). The circles represent each genotyped SNP with the colour indicating pairwise LD in relation to the top hit (calculated from 1000 Genomes June 2010 CEU); −log10 p values are also indicated (y axis). The purple diamond represents the top hit, rs429358. The solid blue line indicates the recombination rate.
Data on the TOMM40 ‘523’ variant, rs10524523, were available for the Lothian Birth Cohorts and the Manchester and Newcastle cohorts. Results for the females are shown in Table 4. Only one significant association was observed, in the Newcastle females (p = 0.045). No significant associations were observed with the males in any of the cohorts.
Table 4.
Associations between TOMM40 rs10524523 genotype and cognitive ageing in females. The estimates for the TOMM40 genotype represent the associations between TOMM40 variants and cognitive ageing, with the TOMM40 short-short genotype as the reference category. A positive beta indicates a greater decline in cognitive ability. TOMM40, translocase of outer mitochondrial membrane 40 homolog; 95% CI, confidence interval.
| LBC1921 | LBC1936 | Manchester | Newcastle | |||||
|---|---|---|---|---|---|---|---|---|
| Parameter | Estimate (95% CI) | p | Estimate (95% CI) | p | Estimate (95% CI) | p | Estimate (95% CI) | p |
| TOMM40 genotype (overall) | ||||||||
| Short heterozygotes | 0.035 (−0.29; 0.36) | 0.83 | −0.029 (−0.29; 0.23) | 0.82 | −0.08 (−0.36; 0.21) | 0.6 | 0.11 (−0.13; 0.35) | 0.35 |
| Non-short | −0.009 (−0.35; 0.33) | 0.96 | −0.47 (−0.32; 0.23) | 0.74 | 0.14 (−0.15; 0.44) | 0.34 | 0.26 (0.005; 0.51) | 0.045 |
To date, APOE and TOMM40 have been predominantly linked with Alzheimer’s disease (AD). Because AD affects individuals in later life and is known to affect cognitive functioning, a candidate gene list was created from the AD literature and the association with cognitive ageing explored using the results from the gene-based analyses. Table 5 indicates that the only genes showing a significant association signal with cognitive ageing in the CAGES cohorts are APOE and TOMM40.
Table 5.
The association of candidate genes, previously identified in the literature as associated with Alzheimer’s disease, using a gene-based test. The genes were chosen from the Alzgene (www.alzgene.org) top results ranking. Alzgene only includes findings from meta-analyses with p<0.00001 and all results are assessed for their epidemiological credibility. Please note that due to LD the p-values for APOE and TOMM40 are not independent.
| Chromosome | Gene | Number of SNPs in gene | p-value All | p-value Females | p-value Males |
|---|---|---|---|---|---|
| 19 | APOE | 19 | 0.0017 | 0.001 | 0.29 |
| 2 | BIN1 | 48 | 0.55 | 0.30 | 0.95 |
| 8 | CLU | 39 | 0.90 | 0.77 | 0.55 |
| 19 | ABCA7 | 34 | 0.15 | 0.13 | 0.42 |
| 1 | CR1 | 44 | 0.44 | 0.088 | 0.99 |
| 11 | PICALM | 44 | 0.74 | 0.33 | 0.92 |
| 11 | MS4A6A | 10 | 0.70 | 0.15 | 0.72 |
| 19 | CD33 | 26 | 0.80 | 0.55 | 0.98 |
| 19 | TOMM40 | 19 | 0.0021 | 0.0014 | 0.32 |
| 6 | CD2AP | 24 | 0.53 | 0.82 | 0.52 |
Biological pathway analysis was performed on the whole CAGES sample (188 genes) and on males (171 genes) and females (215 genes) separately. The results from these analyses are presented in Supplementary Figures 4 and 5, Supplementary Tables 10 and 11. Seven GO categories were found to be nominally associated with cognitive ageing in the entire sample. Five of these categories involved lipoproteins and triglycerides; APOE featured in each of these categories. The remaining two categories were linked to the Major Histocompatibility Complex (MHC). The analysis of the males yielded some significant results under molecular function. The analysis of the females produced no significant results at the p < 0.05 threshold.
The functional annotation analysis produced some significantly enriched annotation clusters (Supplementary Tables 12–14). Lipoproteins and triglycerides featured again here with enrichment scores of 1.49 in the whole sample and 1.63 in the females. However the individual categories mostly did not survive a Bonferroni correction for multiple testing of the category specific terms. Two MHC categories in the whole sample, and one cholesterol linked category in the females remained significant; no categories remained significant in the males.
Functional genomic analysis of the eight suggestively significant SNPs and rs7412 from the fine mapping analysis produced some evidence to support that these variants have a functional non protein-coding effect. One SNP in particular (rs71352238) lies within a genomic region of open chromatin containing functional regulatory elements in CNS relevant cells and brain tissue, and thus is a clear candidate to exert a functional effect on gene expression; see the Supplementary Note for details. Five additional SNPs are also housed within genomic regions indicated to be transcriptionally active. DNA methylation analysis also supports that three of the variants (rs184017, rs429358 and rs7412) may play a role in DNA methylation-mediated modification of gene expression. Of these, the two APOE e4 encoding variants were found to have similar methylation states and are indicated to be within a region that shows differential methylation status. This finding provides support for the argument that epigenetic modification of these nucleotides may contribute to the APOE e4 disease risks.
Discussion
The study found genome-wide significant association of the APOE region of chromosome 19 with non-pathological cognitive ageing in older adults and, more particularly, in older females. This result was replicated in a pooled-analysis of three independent Swedish cohorts of older adults, in which the result was also significant in males. A fine mapping approach enabled this result to be dissected further and significant associations were identified with SNPs in both the TOMM40 and APOE genes. This region contains multiple genes and exhibits moderate to strong LD, and the conditional analyses indicate that APOE is driving the observed association. This is discussed in more detail below. Pathway analysis and functional annotation, whilst not producing highly significant results, also indicated APOE-related pathways involving lipoproteins, triglycerides and cholesterol. Associations between APOE and cognitive phenotypes, including cognitive ageing, in some of the CAGES cohorts have been reported previously21,57–59, but this study included the first meta-analyses of these data. Functional genomic analysis of SNPs in these loci provided evidence to support that variants in this region may have a functional non-protein-coding effect.
A recent paper by De Jager et al. presented results from a GWAS of age-related cognitive decline14. They, too, identified an association between the TOMM40/APOE region and cognitive decline. However, this study included a large number of individuals with mild cognitive impairment (23.5%) and with Alzheimer’s disease (24.9%) and the average time of follow up was nine years in the discovery sample14. Therefore, the results of the De Jager et al. study may have been driven by the pathological cognitive decline of a significant number of individuals. Although we cannot rule out the possibility that individuals in our study were suffering from pre-clinical dementia, all were living independently in the community and none were diagnosed with Alzheimer’s disease or other dementia. Therefore, the present study is a GWAS of mostly non-pathological cognitive ageing. We acknowledge, however, that the separation we have made into non-pathological and pathological cognitive ageing is not based on biological criteria, and that the biology of the two entities will overlap. Within the range of cognitive ageing we have studied there are many individuals who will never proceed to dementia, and there will be some who are in the early stages of pathological cognitive ageing. It is valuable to explore the genetic bases of this milder variation in cognitive change because, being more common than dementia, it affects a greater proportion of the population and, as we discussed previously, the genetic effects on dementia cannot account for all of the effects of, for example, APOE on non-pathological cognitive ageing58.
There was a suggestive finding in the PLXNA2 gene, which has been associated with schizophrenia60 and anxiety61 both of which have been linked with cognitive decline62,63. PLXNA2 encodes the plexin-A2 protein, a member of the plexin-A family of semaphorin coreceptors. Semaphorins are a large family of secreted or membrane-bound proteins that mediate repulsive effects on axon pathfinding during nervous system development. Plexins have been implicated in nervous system development and adult neurogenesis. This requires further work in order to replicate the finding; it will be of interest to follow this up in both clinical and population based cohorts.
It is not unexpected to find that APOE and TOMM40 have a role in cognitive ageing, as there are multiple studies describing associations of these genes with Alzheimer’s disease19,64–66, cognitive phenotypes in old age14,21,58,67,68 and exceptional longevity69. Previous studies involving the LBC1921 cohort reported that individuals with at least one APOE e4 allele declined more in intelligence functions across two periods of the lifecourse: from childhood to old age, and within the ninth decade21,58. However, two studies of the LBC1936 cohort reported that APOE e4 did not influence memory abilities at 70 years of age70 but did have an effect on general cognitive ability, non-verbal cognitive ability, and choice reaction time57. Pendleton et al. reported no association between cognitive decline and APOE in a subset of the Manchester and Newcastle cohorts used in the present study59. It appears that the association with APOE in the CAGES cohorts may be associated with change in the oldest individuals and that the effect is greater in females. The direction of the effects observed for both APOE e2 and APOE e4 were consistent with the literature. The association between APOE and age was not followed up in this study as three of the cohorts are narrow range birth cohorts and thus have little age variation and the remaining cohorts would lack the power required to detect such effects. One obvious cause of this apparent age and sex difference would be if the samples contained considerably more older female individuals; however, if we take LBC1921 as an example we see that a greater effect is observed in the females. There is little age variation and the sample comprises 60% females. Also significant effects were observed in the Swedish males; however, the combined Swedish cohorts have a mean age of 82.8 years, which is considerably older than a large proportion of the CAGES individuals. From the earlier APOE studies described above it is apparent that the individual cohorts at times lacked power to detect this APOE effect and that by combining these cohorts to create the CAGES project—and focusing on a phenotype of cognitive ageing—we have increased the sample size, and thus the power, to enable the discovery of such effects on cognitive ageing.
A recent study identified a polymorphic poly-T variant in the TOMM40 gene that provides greatly-increased precision in the estimation of age of onset of late onset Alzheimer’s disease (LOAD) for APOE e3 carriers19. Previously, APOE studies have predominantly focused on the e4 allele as this was identified originally as the AD risk allele. However, this study by Roses et al. suggests that the association between this region and cognitive ageing and age-related disease appears to be more complex than solely the APOE e4 risk allele. A subsequent paper found this Poly_T variant to be highly variable in diverse populations41. Data on this ‘523’ repeat were available for the Lothian Birth Cohorts and the Manchester and Newcastle cohorts. A significant result was observed in the Newcastle cohort females (p = 0.045). No significant associations were seen with the ‘523’ variant and the other cohorts.
The results described here along with the close physical proximity of TOMM40 and APOE, the level of LD observed, and previous associations reported in this region suggest that there is a significant association with cognitive ageing in this region. It has not been possible to dissect this association further, although the conditional analyses suggest that APOE is driving the observed association and this is the most parsimonious explanation of the results. As in any SNP-trait association, statistical association alone is not sufficient to infer causality. Also, we cannot be certain that we have analysed all potentially causal variants in the region.
A recent study describing a functional analysis of the APOE locus, including TOMM40 and APOC1, reported that multiple APOE locus cis-regulatory elements influence both APOE and TOMM40 promoter activity71. This suggests that there is a complex transcriptional regulatory structure modulating regional gene expression at this locus. Functional genomic analysis, performed as part of the present study, of TOMM40/APOE variants that were significantly associated at a suggestive level with cognitive change also produced some evidence indicating that these variants have regulatory non protein-coding effects. Of note, the potential for DNA methylation modifications of the APOE e4 nucleotides supports the hypothesis that epigenetic mechanisms may contribute to the phenotype of cognitive decline. Deep sequencing of this region in larger cohorts combined with functional work may identify the causal variants involved in this association and elucidate the relationship between APOE and the ageing brain.
A limitation of the present study is the potential for phenotypic heterogeneity, because the time between measurements is different between the English and Scottish cohorts. However, the later-life change documented in the English cohorts will also have occurred in the Scottish cohorts; therefore, any associations linked specifically to later-life change should be significant in the joint analyses. Previous studies of cognitive change have often suffered from a poor phenotype, for example being studied over short periods of time with insufficient measurements7. The strength of this study lies in the rare phenotypes available from the CAGES cohorts. Very few cohorts have measured cognitive change over such an extended time period as those which were the follow up studies of the Scottish Mental Surveys. The cognitive change phenotypes for the Scottish cohorts were calculated over 53 to 68 year intervals. We note that—given our recent estimate of the additive genetic contribution to lifetime cognitive change being about .2411—the present results leave most of this heritability still missing, most likely because the effect sizes of remaining variants that are in LD with the common SNPs are too small to be detected with the sample size of our discovery cohorts.
In conclusion, we report results of the first GWAS on non-pathological cognitive ageing in older individuals. We show an association of variants within the APOE region with cognitive ageing and have replicated this finding in three independent Swedish cohorts. The association appears to be greater in females and more significant in cohorts with the oldest individuals. This study also highlights that the candidate variants within this region may have a cis-regulatory role on gene transcription, and that noncoding epigenetic mechanisms may contribute to the well-established APOE e4 risk effect. Deep sequencing and in-depth functional genomics studies will be required to dissect further this association with the APOE region.
Supplementary Material
Acknowledgements
We thank the cohort participants who contributed to these studies. We thank Martha Pollard for Lothian Birth Cohort 1921data collection. Genotyping of the CAGES cohorts and the analyses conducted here were supported by the UK’s Biotechnology and Biological Sciences Research Council (BBSRC). Phenotype collection in the Lothian Birth Cohort 1921 was supported by the BBSRC, The Royal Society, and The Chief Scientist Office of the Scottish Government. Phenotype collection in the Lothian Birth Cohort 1936 was supported by Research Into Ageing (continues as part of Age UK’s The Disconnected Mind project). Phenotype collection in the Aberdeen Birth Cohort 1936 was supported by BBSRC, the Wellcome Trust, and the Alzheimer’s Research Trust. Phenotype collection in the Manchester and Newcastle Longitudinal Studies of Cognitive Ageing cohorts was supported by Social Science Research Council, Medical Research Council, Economic and Social Research Council, Research Into Ageing, Wellcome Trust and Unilever plc. We gratefully acknowledge the support of the Swedish Research Council (2009-2298), National Institute on Aging (AG 04563, AG10175, AG08861, AG08724, AG028555). The Australian-based researcher acknowledges support from the Australian Research Council and the National Health and Medical Research Council. The work was undertaken in The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (G0700704/84698). Funding from the BBSRC, EPSRC, ESRC and MRC is gratefully acknowledged.
This work was funded by the Biotechnology and Biological Sciences Research Council, The Royal Society, The Chief Scientist Office of the Scottish Government, Research Into Ageing, Age UK, the Wellcome Trust, the Alzheimer’s Research Trust, Social Science Research Council, Medical Research Council, Economic and Social Research Council, Unilever plc, and the Engineering and Physical Sciences Research Council. Work with the Swedish samples was supported by the Swedish Research Council (2009-2298), and the US National Institute on Aging (AG04563, AG10175, AG08861, AG08724, AG028555). PMV is supported by the Australian National Health and Medical Research Council.
Footnotes
Conflict of interest
Allen D Roses is the CEO and only stock-holder of Zinfandel Pharmaceuticals, Inc., a company in an Alliance with Takeda Pharmaceuticals, Inc., to perform the prospective qualification of the TOMM40 marker for age of onset distribution of Alzheimer’s Disease. For this study, Zinfandel Pharmaceuticals, Inc. paid for the TOMM40 assays to be performed for medical research, not as a clinical diagnostic.
Supplementary information is available at Molecular Psychiatry’s website.
References
- 1.Rabbitt P, and Lowe C. Patterns of cognitive ageing. Psychol Res 2000; 63: 308–316. [DOI] [PubMed] [Google Scholar]
- 2.Salthouse TA. Major issues in cognitive aging. Oxford University Press: New York, NY, 2010. [Google Scholar]
- 3.Brayne C The elephant in the room – healthy brains in later life, epidemiology and public health. Nat Rev Neurosci 2007; 8: 233–239. [DOI] [PubMed] [Google Scholar]
- 4.Tannenbaum C, Mayo N, Ducharme F. Older women’s health priorities and perceptions of care delivery: results of the WOW health survey. Can Med Assoc J 2005; 173: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comas-Herrera A, Wittenberg R, Pickard L, Knapp M. Cognitive impairment in older people: future demand for long-term care services and the associated costs. Int J Geriatr Psych 2007; 22: 1037–1045. [DOI] [PubMed] [Google Scholar]
- 6.World Population Prospects 1950–2050. UN Publications, 2001. [Google Scholar]
- 7.Lee T, Henry JD, Trollor JN, Sachdev PS. Genetic influences on cognitive functions in the elderly: a selective review of twin studies. Brain Res Rev 2010; 64: 1–13. [DOI] [PubMed] [Google Scholar]
- 8.Reynolds CA, Finkel D, McArdle JJ, Gatz M, Berg S, Pedersen NL. Quantitative genetic analysis of latent growth curve models of cognitive abilities in adulthood. Dev Psychol 2005; 41: 3–16. [DOI] [PubMed] [Google Scholar]
- 9.Finkel D, Reynolds CA, McArdle JJ, Hamagami F, Pedersen NL. Genetic variance in processing speed drives variation in aging of spatial and memory abilities. Dev Psychol 2009; 45: 820–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGue M, Christensen K. Social activity and healthy aging: a study of aging Danish twins. Twin Res Hum Genet 2007; 10: 255–265. [DOI] [PubMed] [Google Scholar]
- 11.Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, Liewald D et al. Genetic contributions to stability and change in intelligence from childhood to old age. Nature 2012; doi: 10.1038/nature10781. [DOI] [PubMed] [Google Scholar]
- 12.Harris SE, Deary IJ. The genetics of cognitive ability and cognitive ageing in healthy older people. Trends Cogn Sci 2011; 15: 388–394. [DOI] [PubMed] [Google Scholar]
- 13.Wisdom NM, Callahan JL, Hawkins KA. The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiol Aging 2011; 32: 63–74. [DOI] [PubMed] [Google Scholar]
- 14.De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol Aging 2011; doi: 10.1016/j.neurobiolaging.2011.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schellenberg GD, Deeb SS, Boehnke M, Bryant EM, Martin GM, Lampe TH et al. Association of an apolipoprotein CII allele with familial dementia of the Alzheimer type. J Neurogenet 1987; 4: 97–108. [PubMed] [Google Scholar]
- 16.Chartier-Harlin MC, Parfitt M, Legrain S, Perez-Tur J, Brousseau T, Evans A et al. Apolipoprotein E, e4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: analysis of the 19q13.2 chromosomal region. Hum Mol Genet 1994; 3: 569–574. [DOI] [PubMed] [Google Scholar]
- 17.Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 1994; 7: 180–184. [DOI] [PubMed] [Google Scholar]
- 18.Corder EH, Lannfelt L, Viitanen M, Corder LS, Manton KG, Winblad B. Apolipoprotein E genotype determines survival in the oldest old (85 years or older) who have good cognition. Arch Neurol 1996; 53: 418–422. [DOI] [PubMed] [Google Scholar]
- 19.Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS et al. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J 2009; 10: 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SC, La Rue A, Hermann BP, Xu G, Koscik RL, Jonaitis EM et al. The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOE ε3/ε3 genotype. Alzheimer’s & Dementia 2011; 7: 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiepers OJG, Harris SE, Gow AJ, Pattie A, Brett CE, Starr JM et al. APOE E4 status predicts age-related cognitive decline in the ninth decade: longitudinal follow-up of the Lothian Birth Cohort 1921. Mol Psychiatry 2011; e-pub ahead of print 25 January 2011; doi: 10.1038/mp.2010.137. [DOI] [PubMed] [Google Scholar]
- 22.Scottish Council for Research in Education. The Intelligence of Scottish Children: A National Survey of an Age-group. University of London Press: London, UK, 1933. [Google Scholar]
- 23.Deary IJ, Whalley LJ, Starr JM. A Lifetime of Intelligence: Follow-up Studies of the Scottish Mental Surveys of 1932 and 1947. American Psychological Association: Washington, D.C., 2009. [Google Scholar]
- 24.Deary IJ, Whiteman MC, Starr JM, Whalley LJ, Fox HC. The impact of childhood intelligence on later life: following up the Scottish Mental Surveys of 1932 and 1947. J Pers Soc Psychol 2004; 86: 130–147. [DOI] [PubMed] [Google Scholar]
- 25.Scottish Council for Research in Education. The Trend of Scottish Intelligence. University of London Press: London, UK, 1949. [Google Scholar]
- 26.Deary IJ, Gow AJ, Taylor MD, Corley J, Brett C, Wilson V et al. The Lothian Birth Cohort 1936: a study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatrics 2007; 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabbitt PMA, McInnes L, Diggle P, Holland F, Bent N, Abson V et al. The University of Manchester longitudinal study of cognition in normal healthy old age, 1983 through 2003. Aging Neuropsychol Cogn 2004; 11: 245–279. [Google Scholar]
- 28.Pedersen NL, Lichtenstein P, Svedberg P. The Swedish Twin Registry in the third millennium. Twin Res 2002; 5:427–432. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen NL, McClearn GE, Plomin R, Nesselroade JR, Berg S, DeFaire U. The Swedish Adoption Twin Study of Aging: an update. Acta Geneticae Medicae et Gemellologiae: Twin Research 1991; 40: 7–20. [DOI] [PubMed] [Google Scholar]
- 30.Finkel D, & Pedersen NL. Processing Speed and Longitudinal Trajectories of Change for Cognitive Abilities: The Swedish Adoption/Twin Study of Aging. Aging, Neuropsychology, and Cognition 2004; 11(2–3): 325–345. [Google Scholar]
- 31.Gold CH, Malmberg B, McClearn GE, Pedersen NL, Berg S. Gender and health: a study of older unlike-sex twins. J Gerontol B Psychol Sci Soc Sci 2002; 57: S168–176. [DOI] [PubMed] [Google Scholar]
- 32.McClearn GE, Johansson B, Berg S, Pedersen NL, Ahern F, Petrill SA et al. Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science 1997; 276: 1560–1563. [DOI] [PubMed] [Google Scholar]
- 33.Raven JC, Court JH, Raven J. Manual for Raven’s Progressive Matrices and Vocabulary Scales. HK Lewis: London, UK, 1977. [Google Scholar]
- 34.Wechsler D Wechsler Memory Scale–Revised. Psychological Corporation: San Antonio, TX, 1987. [Google Scholar]
- 35.Lezak MD, Howieson DB, Loring DW. Neuropsychological Assessment. 4th edn Oxford University Press: Oxford, UK, 2004. [Google Scholar]
- 36.Wechsler D WAIS-III UK Administration and Scoring Manual. Psychological Corporation: London, UK, 1998. [Google Scholar]
- 37.Wechsler D Wechsler Adult Intelligence Scale-Revised. Psychological Corporation: New York, 1981. [Google Scholar]
- 38.Nelson HE, Willison JR. National Adult Reading Test (NART) Test Manual. 2nd edn NFER-Nelson: Windsor, UK, 1991. [Google Scholar]
- 39.Pedersen NL, Plomin R, Nesselroade JR, McClearn GE. A quantitative genetic analysis of cognitive abilities during the second half of the life span. Psychol Sci 1992; 3: 346–353. [Google Scholar]
- 40.Deary IJ, Whiteman MC, Pattie A, Starr JM, Hayward C, Wright AF et al. Apolipoprotein E gene variability and cognitive functions at age 79: follow up of the Scottish Mental Survey 1932. Psychology and Aging 2004; 19: 367–371. [DOI] [PubMed] [Google Scholar]
- 41.Ghebranious N, Ivacic L, Mallum J, Dokken C. Detection of ApoE E2, E3 and E4 alleles using MALDI-TOF mass spectrometry and the homogeneous mass-extend technology. Nucleic Acids Research 2005; 33: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Wetten S, Li L, St. Jean PL, Upmanyu R, Surh L et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer’s disease. Arch Neurol 2008; 65: 45–53. [DOI] [PubMed] [Google Scholar]
- 43.Linnertz C, Saunders AM, Lutz MW, Crenshaw DM, Grossman I, Burns DK et al. Characterization of the Poly-T Variant in the TOMM40 Gene in Diverse Populations. PLoS ONE 2012; 7: e30994 e-pub 16 February 2012; PMID:22359560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayden KM, McEvoy JM, Linnertz C, Attix D, Kuchibhatla M, Saunders AM et al. A homopolymer polymorphism in the TOMM40 gene contributes to cognitive performance in aging. Alzheimer’s and Dementia 2012; In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol. Psychiatry 2011; 16: 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM et al. A Versatile Gene-Based Test for Genome-wide Association Studies. Am J Hum Genet 2010; 87: 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duncan D, Prodduturi N, Zhang B. WebGestalt2: an updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC Bioinformatics 2010; 11. [Google Scholar]
- 49.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005; 33: W741–W8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 1995; 57: 289–300. [Google Scholar]
- 51.Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc 2009; 4(1): 44–57. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Willer CJ, Ding J, Scheet P and Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Willer CJ, Sanna S and Abecasis GR. Genotype Imputation . Annu Rev Genomics Hum Genet 2009; 10: 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM et al. The human genome browser at UCSC. Genome Res. 2002; 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gardiner-Garden M & Frommer M. CpG islands in vertebrate genomes. J Mol Biol 1987; 196: 261–282. [DOI] [PubMed] [Google Scholar]
- 56.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D et al. Ensembl 2012. Nucleic Acids Research 2012; 40: D84–D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luciano M, Gow AJ, Harris SE, Hayward C, Allerhand M, Starr JM et al. Cognitive ability at age 11 and 70 years, information processing speed and APOE variation: the Lothian Birth Cohort 1936 Study. Psychol Aging 2009; 1: 129–138. [DOI] [PubMed] [Google Scholar]
- 58.Deary IJ, Whiteman MC, Pattie A, Starr JM, Hayward C, Wright AF et al. Cognitive change and the APOE e4 allele. Nature 2002; 418: 932. [DOI] [PubMed] [Google Scholar]
- 59.Pendleton N, Payton A, van den Boogerd EH, Holland F, Diggle P, Rabbitt PM et al. Apolipoprotein E genotype does not predict decline in intelligence in healthy older adults. Neurosci Lett 2002; 324: 74–76. [DOI] [PubMed] [Google Scholar]
- 60.Mah S, Nelson MR, DeLisi LE, Reneland RH, Markward N, James MR et al. Identification of the semaphorin receptor PLXNA2 as a candidate for susceptibility to schizophrenia. Mol Psychiatry 2006; 11: 471–478. [DOI] [PubMed] [Google Scholar]
- 61.Wray NR, James MR, Mah SP, Nelson M, Andrews G, Sullivan PF et al. Anxiety and comorbid measures associated with PLXNA2. Arch Gen Psychiat 2007; 64: 318–326. [DOI] [PubMed] [Google Scholar]
- 62.Green MF. Cognitive impairment and functional outcome in schizophrenia and bipolar disorder. J Clin Psychiat 2006; 67(10):e12. [PubMed] [Google Scholar]
- 63.Beaudreau SA, O’Hara R. Late-life anxiety and cognitive impairment: a review. Am J Geriat Psychiat 2008; 16: 790–803. [DOI] [PubMed] [Google Scholar]
- 64.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GA et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993; 261: 921–923. [DOI] [PubMed] [Google Scholar]
- 65.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS et al. Apolipoprotein E: High avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer’s disease. Proc Nat Acad Sci 1993; 90; 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harald D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 2009; 41: 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Small BJ, Rosnick CB, Fratiglioni L, Backman L. Apolipoprotein E and cognitive performance: A meta-analysis. Psychol Aging 2004; 19: 592–600. [DOI] [PubMed] [Google Scholar]
- 68.Reynolds CA, Prince JA, Feuk L, Gatz M, & Pedersen NL. Longitudinal memory performance during normal aging: twin association models of APOE and other Alzheimer candidate genes. Behav Genet 2006; 36: 185–94. [DOI] [PubMed] [Google Scholar]
- 69.Sebastiani P, Solovieff N, DeWan AT, Walsh KM, Puca A, Hartley SW et al. Genetic Signatures of Exceptional Longevity in Humans. PloS One 2012; 7: e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luciano M, Gow AJ, Taylor MD, Hayward C, Harris SE, Campbell H et al. Apolipoprotein E is not related to memory abilities at 70 years of age. Behav Genet 2009; 39: 6–14. [DOI] [PubMed] [Google Scholar]
- 71.Bekris LM, Lutz F, & Yu C. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J Hum Genet 2011; 57: 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.