Abstract
Drosophila has been extensively used to model the human blood-immune system, as both systems share many developmental and immune response mechanisms. However, while many human blood cell types have been identified, only three were found in flies: plasmatocytes, crystal cells and lamellocytes. To better understand the complexity of fly blood system, we used single-cell RNA sequencing technology to generate comprehensive gene expression profiles for Drosophila circulating blood cells. In addition to the known cell types, we identified two new Drosophila blood cell types: thanacytes and primocytes. Thanacytes, which express many stimulus response genes, are involved in distinct responses to different types of bacteria. Primocytes, which express cell fate commitment and signaling genes, appear to be involved in keeping stem cells in the blood. Furthermore, our data revealed four novel plasmatocyte subtypes (Ppn+, CAH7+, Lsp+ and reservoir plasmatocytes), each with unique molecular identities and distinct predicted functions. We also identified cross-species markers from Drosophila hemocytes to human blood cells. Our analysis unveiled a more complex Drosophila blood system and broadened the scope of using Drosophila to model human blood system in development and disease.
Keywords: Drosophila, blood, single-cell RNA-seq, thanacyte, primocyte, plasmatocyte
INTRODUCTION
Drosophila has been used extensively to study the human blood-vascular system. At first glance both systems might seem to have little in common; however, at the molecular level the systems are highly conserved (Evans et al., 2003). For example, they share specific transcription factors and signaling pathways during development. And, in both systems the terminally differentiated blood cell lineages are derived from common progenitor cells. Moreover, at the functional level Drosophila blood cells demonstrate phagocytosis, innate immunity, wound healing, engulfing of large particles (e.g., wasp infestation), and sensing of environmental gasses like oxygen levels, similar to the human myeloid blood cell system.
Decades of research have revealed a complex and numerous cell system in human blood, comprising erythrocytes, leukocytes (neutrophils, T lymphocytes and B lymphocytes), natural killer (NK) cells, macrophages and thrombocytes. In contrast, to date, only three terminally differentiated blood cell types have been described in Drosophila: plasmatocytes, crystal cells and lamellocytes.
Plasmatocytes are the most numerous (~90–95% of hemocytes). During embryogenesis plasmatocytes constitute a major source of extracellular matrix proteins which are essential for embryonic renal tubule morphogenesis (Artero et al., 2006), further they engulf apoptotic cells by endocytosis (Kurucz et al., 2007a). Within the immune system they provide phagocytic and antimicrobial functions to remove any invading particles, similar to human macrophages. Plasmatocytes are known to express the free radical scavenging enzyme Peroxidasin (Pxn) (Nelson et al., 1994), and several cell surface molecules involved in phagocytosis, including Nimrod C1 (NimC1; P1 antigen) (Kurucz et al., 2007a) and Eater receptors (Kocks et al., 2005).
Crystal cells make up a much smaller portion of Drosophila hemocytes (~2–5%). They are named for their distinct crystalline structures, and these inclusions contain Prohenoloxidase (ProPO) enzymes that mediate melanization in response to injury. Further, crystal cells facilitate innate immunity and the hypoxic response, not unlike human platelet and granulocyte functions. Early markers include lozenge (lz) and pebbled (peb), while later markers include the ProPO enzymes PPO1 and PPO2.
Lamellocytes are generally very rare in healthy flies; however, their numbers increase rapidly in response to wasp infection. They arise from trans-differentiation of plasmatocytes. Lamellocyte morphology stands out as they form large flat disc-shaped cells, ideal for enveloping intrusions. They often contain more lysosomes and phagocytic vacuoles than plasmatocytes but lamellocytes do not exhibit phagocytic activity. Their human equivalents are the multinucleated giant cells that arise when monocytes or macrophages fuse together during an infection. Marker expression includes PPO3, atilla, Integrin alphaPS4 subunit (ItgaPS4), misshapen (msn), puckered (puc), L6 or L2 antigens, and myospheroid (mys; encodes β subunit of the integrin dimer). The latter is also expressed by hemocyte progenitors and plasmatocytes.
Cells, including those from the Drosophila blood system, have traditionally been classified based on their morphology and the expression of a limited number of defined protein markers. However, single-cell RNA sequencing (scRNA-seq) technology can assay gene expression of an individual cell at a genome-wide scale, for thousands of cells in a single experiment (Saliba et al., 2014), and has provided many new insights into the richness of cell type variety that makes up different tissues and organs.
In this manuscript we applied scRNA-seq technology to study the cellular heterogeneity of the total circulating blood cells in Drosophila wandering third instar larvae. Our analysis revealed four previously unknown plasmatocytes subtypes: Ppn+ plasmatocytes, CAH7+ plasmatocytes, Lsp+ plasmatocytes and reservoir plasmatocytes. And, we describe new markers that uniquely distinguish crystal cells and lamellocytes among hemocytes. Our findings also uncovered two new Drosophila blood cell types: thanacytes and primocytes, which display distinct gene expression profiles with unique markers. By silencing Tep4, a thanacyte-specific gene, we showed that thanacytes are responsible for the distinct response to different type of bacterial infection. We further compared the expression profiles of these newly identified cells to those of the human blood system and identified cross-species markers linking fly and human blood cell types. Our analysis unveiled a more complex Drosophila blood system and broader scope for using Drosophila to model human blood system.
RESULTS
Drosophila blood cell diversity revealed by single-cell RNA-seq
We performed scRNA-seq on circulating hemocytes from wandering third instar larvae (w1118, n = 18; Fig. 1A) to uncover the molecular heterogeneity of Drosophila blood cells. After removing cell doublets and debris, 3,424 cells were left and included in cluster analysis (Table S1; Fig. S1). Interestingly, tSNE identified two unique cell clusters in addition to the three cell types described to date. All five clusters showed expression of pan-hemocyte marker srp (Fig. 1C). Each cluster also showed unique sets of differentially expressed genes (Fig. 1C). Plasmatocytes (cluster 1) make up the majority of the blood cells (89.5%) (Fig. 1B), which is consistent with previous reports of around 95% of total hemocytes (Kemp et al., 2013). Crystal cells (cluster 2) and lamellocytes (cluster 3) account for 0.3% and 1.4% respectively under our experimental conditions. Moreover, they are clearly separated from the other cell clusters, which further demonstrates the sensitivity and objectivity of our clustering and classification.
Fig. 1.
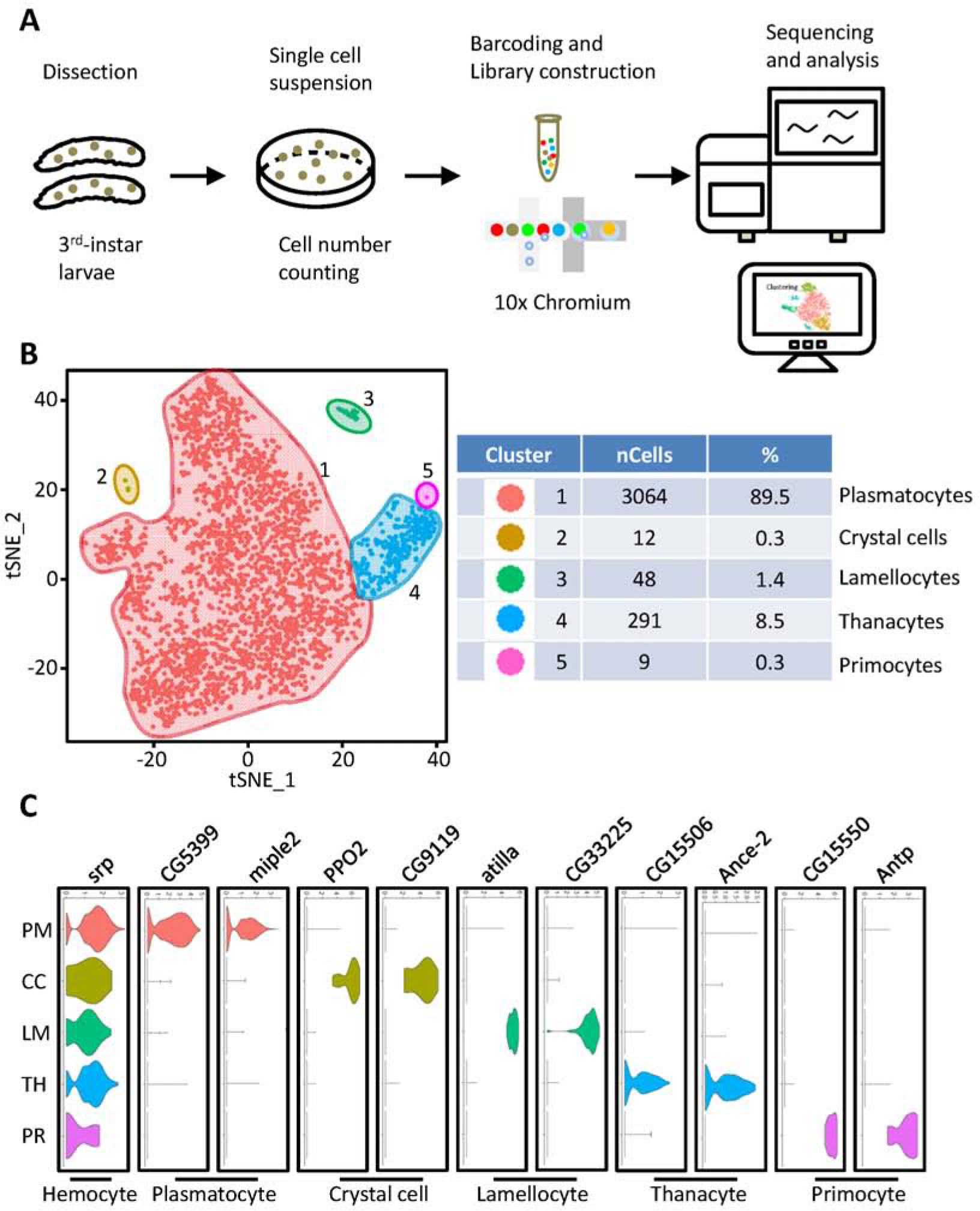
Cell diversity in Drosophila blood cells delineated by single-cell transcriptomic analysis. A: Schematic representation of scRNA-seq workflow of blood cells from L3 stage larvae by 10x Chromium platform (10x Genomics, CA, USA). B: tSNE feature plot representing Drosophila blood scRNAseq data (left). Panel (right) shows cell numbers and percentage of cells/total for each tSNE cluster. C: Violin plots show expression of indicated genes across the different tSNE cell clusters: srp (pan-hemocyte) and representative marker genes for each of the five hemocyte clusters. X-axis: log scale normalized read count. Y: PM, plasmatocytes; CC, crystal cells; LM, lamellocytes; TH, thanacytes; PR, primocytes.
Defining the heterogeneous population of Drosophila plasmatocytes
Differences in morphology among plasmatocytes have been described before; however, classification into subtypes has been hampered by lack of specific markers. Therefore, we performed unbiased sub-cluster analysis on this cluster (cluster 1), which. further categorized the plasmatocytes into four sub-groups, which we based on their foremost expressed genes: Papilin positive plasmatocytes (Ppn+ PM), Carbonic anhydrase 7 positive plasmatocytes (CAH7+ PM), Larval serum protein positive plasmatocytes (Lsp+ PM) and reservoir plasmatocytes (reservoir PM) (Fig. 2A). Among these reservoir plasmatocytes are the most prominent (1815 cells, 59.24% of plasmatocytes), followed by CAH7+ PM (646 cells; 21.08% of plasmatocytes), Ppn+ PM (381 cells; 12.43% of plasmatocytes) and Lsp+ PM (222 cells; 7.25% of plasmatocytes). These data demonstrate the transcriptional heterogeneity of plasmatocytes and reveal the presence of defined subtypes.
Fig. 2.
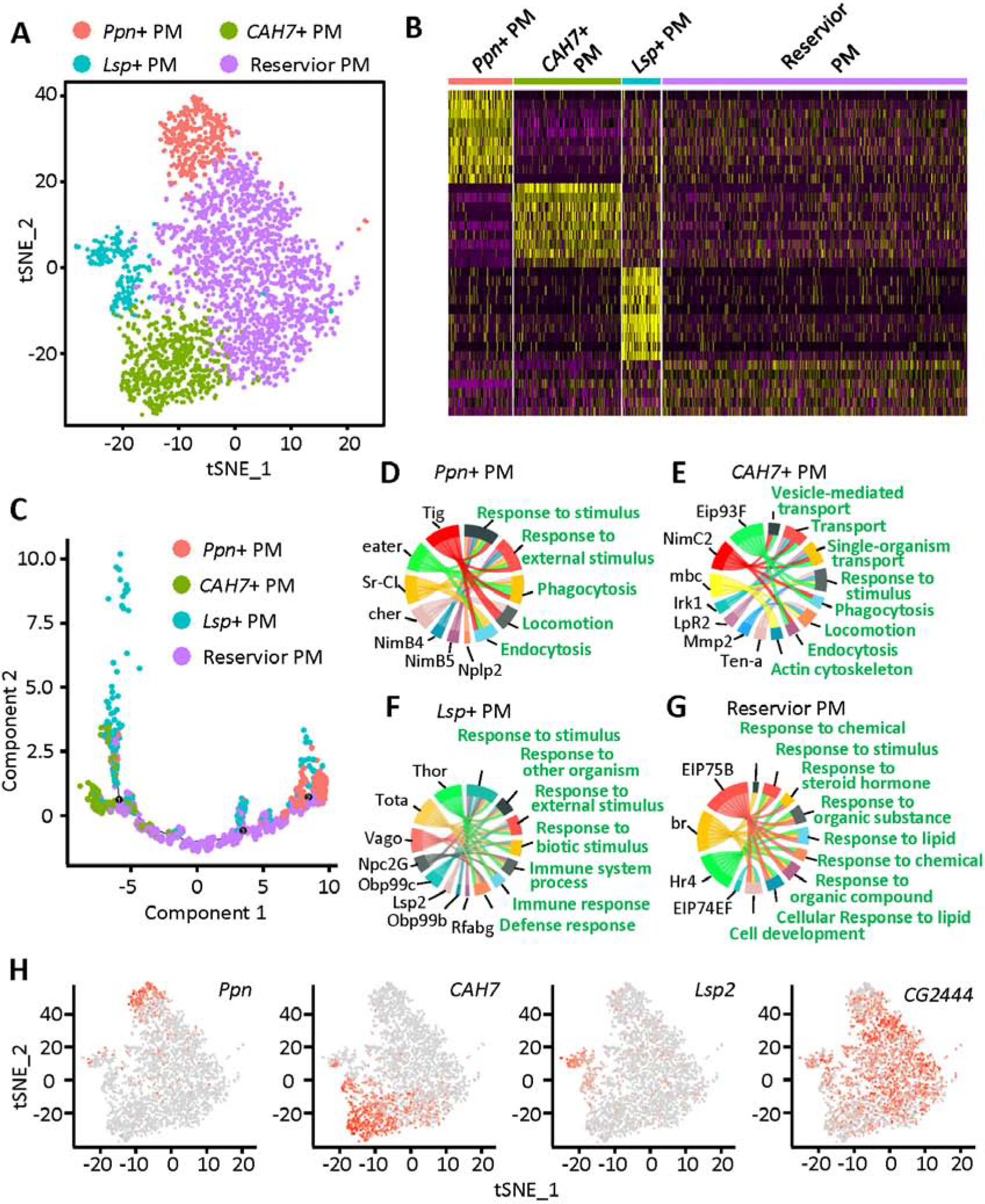
Characterization of plasmatocyte subtypes: Ppn+ PM, CAH7+ PM, Lsp+ PM and reservoir PM. A: tSNE plot of the plasmatocyte subclusters visualized by Seurat (R package). B: Heatmap representing the differentially expressed genes for each of the plasmatocyte subclusters. C: Single-cell trajectory for the four subtypes of plasmatocytes, reconstructed using Monocle. D – G: GO pathway analysis using the genes most differentially expressed in the Ppn+ PM (D), CAH7+ PM (E), Lsp+ PM (F) and reservoir PM (G) clusters. H: Feature plots showing the expression of marker genes specific to the Ppn+ PM, CAH7+ PM, Lsp+ PM and reservoir PM cell clusters. PM, plasmatocyte.
All sub-clusters highly expressed known pan-plasmatocyte markers srp, Hemese (He), Pxn and croquemort (crq) (Fig. S3); however, each of the four plasmatocyte subtypes also showed a distinctive gene expression profile (Fig. 2B). Unlike Ppn+ PM, CAH7+ PM and Lsp+ PM, the reservoir PM could not be defined by expression of unique marker genes (Fig. 2H). Ppn, a member of the metzincin superfamily of metalloproteases, is specifically expressed in Ppn+ PM. CAH7 with both carbonate dehydratase activity and zinc ion binding capability is highly expressed in CAH7+ PM, although some Lsp+ PM cells also showed its expression. It is interesting and important to observe that Lsp2, a gene previously considered to be expressed only in larvae fat body (Benes et al., 1990), showed specific expression in the Lsp+ PM subset of plasmatocytes (Fig. 2H). This cell cluster is different from fat body for they express all known hemocyte markers including srp, He, Pxn and crq (Fig. S3). Although no specific markers have been identified in reservoir PM, the gene CG2444 showed higher expression in reservoir PM than the other three plasmatocyte subtypes (Fig. 2H). CG2444 encodes a “Single domain Von Willebrand factor type C” (SVWC) containing protein which has been implicated to respond to environmental challenges, such as bacterial infection, anti-viral immunity, as well as nutritional status.
We used Monocle to carry out single-cell trajectory analysis, in an unbiased assessment (Qiu et al., 2017). Monocle placed reservoir PM at the root of the trajectory tree, and the other three plasmatocyte subtypes were placed near the end of the branches (Fig. 2C). These results imply that reservoir PM may represent a plastic type of plasmatocytes, which could differentiate or mature into other plasmatocyte types depending on exposure to certain biological processes, such as bacterial infection.
Pathway analysis of the differentially expressed genes in each cell type, enabled us to predict the function of a specific cell type. Ppn+ PM and CAH7+ PM represent two types of differentiated “killer” cells, based on enrichment of phagocytosis and endocytosis pathways in these clusters (Fig. 2D and E). Ppn+ PM showed high expression of eater, which encodes a well-known transmembrane receptor of the Nimrod family specifically expressed in hemocytes and required for phagocytosis of bacteria (Kocks et al., 2005). NimB4 and NimB5 members of the Nimrod family are also highly enriched in this cell cluster. Neuropeptide-like precursor 2 (Nplp2), another gene implicated in defense response (Uttenweiler-Joseph et al., 1998) and humoral immune response (Verleyen et al., 2006), is also specifically expressed in Ppn+ PM. CAH7+ PM appears to be a dynamic cell type that may transform, engulf and transport pathogens, as evident by the high levels of expression of genes involved in cytoskeleton regulation and transportation, including myoblast city (mbc) and NimC2. CAH7+ PM cells display high expression of matrix metalloproteinase 2 (Mmp2), which encodes a proteinase that cleaves proteins in the extracellular matrix and contributes to tissue remodeling (Page-McCaw et al., 2003).
Lsp+ PM on the other hand, also show the ability to respond to external stimuli and are involved in the immune system process (Fig. 2F). However, these cells do not seem to express the “weapons” needed to fight external pathogens, as such they may be involved in the regulation of the immune response in a regulatory or signaling capacity. Pathway analysis for the reservoir PM indicates a key task is response to different stimuli (Fig. 2G), ranging from lipids to steroid hormones. The data suggest reservoir PM might respond to chemical and signaling molecules in addition to a direct immune response to external pathogens. Further support comes from the high expression level of ecdysone-induced protein 75B (Eip75B) and ecdysone-induced protein 74EF (Eip74EF) in reservoir PM. Eip75B encodes a nuclear receptor and Eip74EF encodes a transcription factor, which respond to NO and 20-hydroxyecdysone, respectively. Taken together, these data suggest reservoir PM might represent cells capable of plasticity.
Detection of plasmatocyte subtypes using newly identified markers
In order to validate the gene expression-based classifications, we looked for available antibodies and transgenic Gal4-driven/GFP fly lines for the genes with expression unique to specific cell types (Tables S2 and S3). We confirmed 13 of 25 antibodies, and 6 of 25 tested transgenic fly lines. We used mCD8-GFP driven by Ppn-Gal4 to successfully visualize Ppn+ PM in Drosophila blood in vivo, which located Ppn expression to the cell bodies (Figs. 2H, 3A and 3B). Ecdysone receptor (EcR) is largely restricted to Ppn+ PM (Fig. 3C), with around 21% of blood cells showing detectable EcR transcription in the scRNA-seq data. EcR encodes a protein that binds to ecdysone response elements in conjunction with co-activators and co-repressors to regulate transcription of target genes. In line with the scRNA-seq findings, immunofluorescent staining using antibody against EcR protein labeled a subset of hemocytes accounting for 37% of total blood cells (Fig. 3D). EcR antibody staining is detected adjacent to the nuclear membrane, which is consistent with its function in gene regulation. Enabled (ena) was expressed highest in CAH7+ PM (Fig. 3E), although sporadic cells with lower levels of expression were observed in Lsp+ PM and reservoir PM. Ena encodes a processive actin polymerase to regulate actin cytoskeleton organization, which is consistent with pathway analysis results for CAH7+ PM that indicate this group of cells is morphologically dynamic.
Fig. 3.
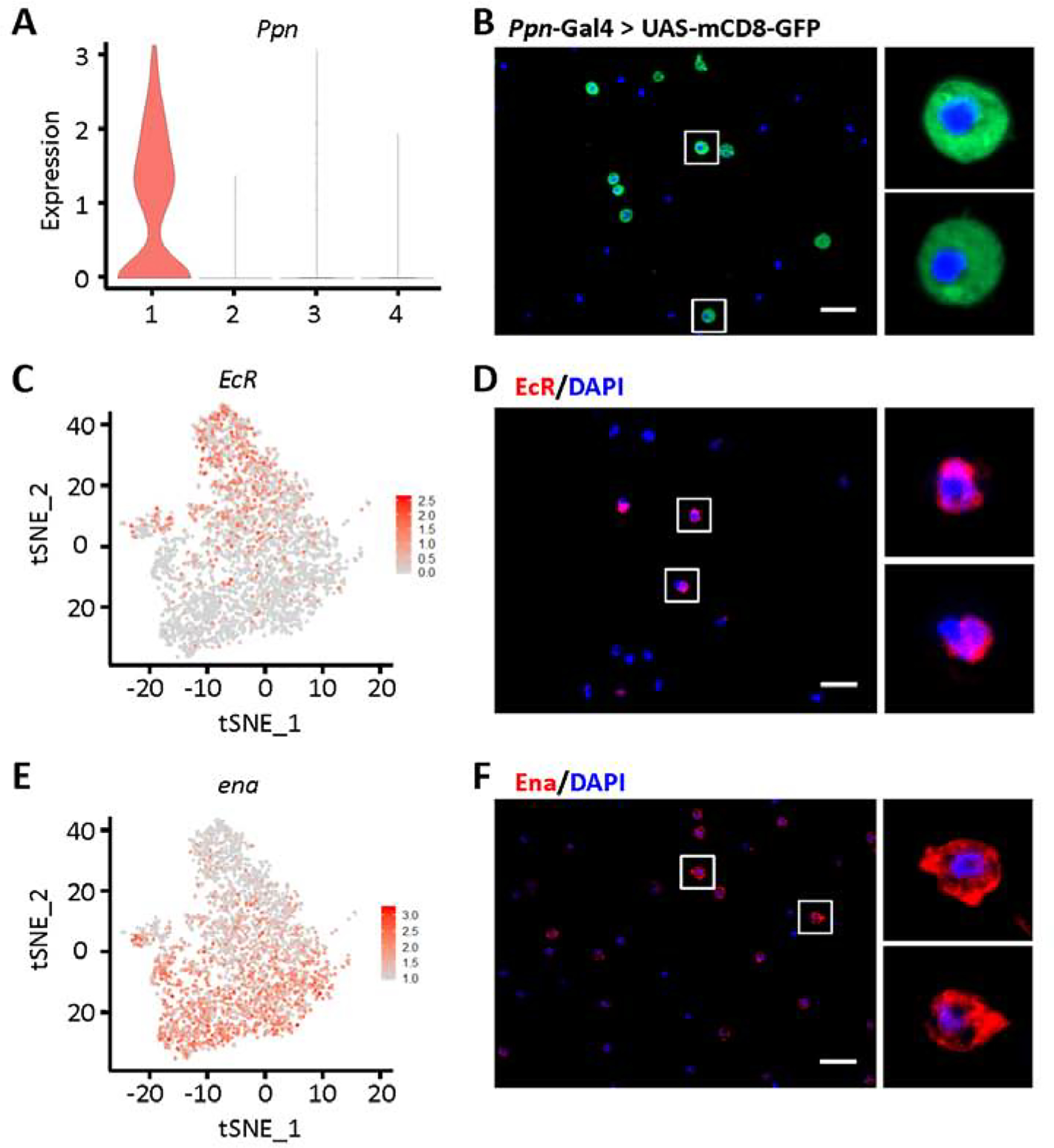
Labeling of Ppn+ and CAH7+ plasmatocytes by differentially expressed markers. A: Violin plot showing Ppn expression in plasmatocyte subclusters. Y-axis: log scale normalized read count. X-axis: 1: Ppn+ PM; 2: CAH7+ PM; 3: Lsp+ PM; 4: reservoir PM. B: Representing Apotome image of Ppn expression (green) in blood cells from Drosophila mCD8-GFP driven by Ppn-Gal4. Nuclear counterstain with DAPI (blue). Scale bar: 20 μm. C and E: Feature plots representing expression (level and distribution) in plasmatocytes for EcR (C) and ena (E). D and F: Representing Apotome images EcR (D; red) and Ena (F; red) protein expression in Drosophila blood cells, by immunofluorescence. Nuclear counterstain with DAPI (blue). Scale bars: 20 μm.
Identification of novel cell types in Drosophila blood
We uncovered two novel distinct cell clusters in our scRNA-seq data (Fig. 1B). Each of these showed unique molecular signatures and expresses cell type-specific markers (Figs. 1C, 4A and 4D); we named these two novel cell types primocytes and thanacytes.
Fig. 4.
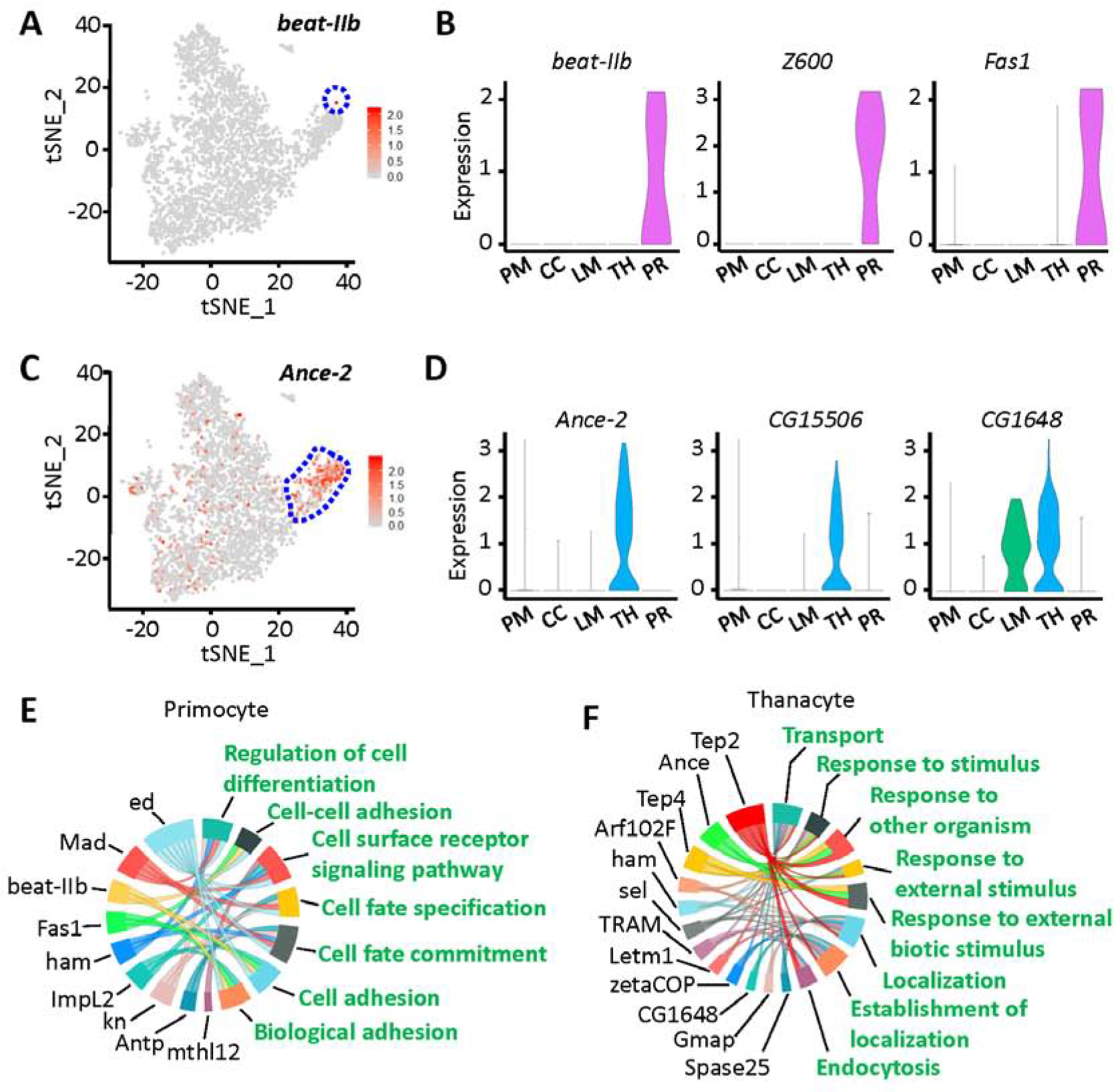
Identification of two novel cell types in Drosophila blood: thanacytes and primocytes. A: Feature plot representing beat-IIb gene expression in hemocytes (scRNA-seq data). Primocyte cell cluster bounded in blue dashed line. B: Violin plots showing the expression for beat-IIb, Z600 and Fas1 genes in Drosophila blood scRNA-seq data. C: Feature plot representing Ance-2 gene expression in hemocytes (scRNA-seq data). Thanacyte cell cluster bounded in blue dashed line. D: Violin plots for Ance-2, CG15506 and CG1648 expression in Drosophila blood scRNA-seq data. X-axis: PM, plasmatocytes; CC, crystal cells; LM, lamellocytes; TH, thanacytes; PR, primocytes. Y-axis: log scale normalized read count. E and F: GO pathway analysis of differentially expressed genes for primocyte (E) and thanacyte (F) cell clusters. Differentially expressed genes are shown in black, and enriched GO pathways are shown in green.
Primocytes are rare in Drosophila blood, accounting for only 0.3% of total hemocytes (Fig. 1B). They express a very unique set of marker genes (Figs. 1C, 4A and 4B), including the early hemocyte marker srp but have no detectable levels of more mature hemocyte markers like He, Pxn and crq (Fig. S3). They also express asrij (Kulkarni et al., 2011), a pan-hemocyte gene that has been implicated in maintaining stem cell potency and hematopoiesis (Fig. S4A). Beaten path IIb (beat-IIb) and Fasciclin 1 (Fas1), both encode cell adhesion molecules, are highly and specifically expressed in primocytes (Fig. 4A and B). GO pathway analysis indicates primocytes are mainly involved in cell adhesion/recognition, and cell fate commitment (Fig. 4E). These cells are likely a group of hematopoietic precursor cells, for they express a series of transcription factors which regulate cell fate and cell differentiation, including Antennapedia (Antp), knot (kn), hamlet (ham), Mothers against Dpp (Mad) and Z600 (Figs. 1C, 4B, 4E and S4B). We stained circulating hemocytes with anti-Antp antibody and found it labeled a small subset (<1%) of circulating hemocytes, presumably primocytes (Fig. S4C).
Thanacytes make up the other new cell cluster and are more abundant (8.5% of hemocytes; Fig. 1B) than crystal cells, lamellocytes and primocytes combined. Although thanacytes express multiple hemocyte markers srp, He, Pxn and crq (Fig. S3), they are distinct from plasmatocytes because they show no detectable expression of the plasmatocyte marker NimC1 (Kurucz et al., 2007a) (Fig. 5B). Interestingly, CG1648 is expressed in thanacytes and lamellocytes clusters only (Fig. 4B), and is known to be induced by bacterial (Irving et al., 2005) and viral (Cordes et al., 2013; Kemp et al., 2013) infections. Angiotensin converting enzyme 2 (Ance-2) and CG15506 are highly and specifically expressed in thanacytes (Fig. 4C and D). Ance-2 is predicted to have exopeptidase activity, and CG15506 is a yet uncharacterized protein-coding gene. Moreover, GO pathway analysis indicated thanacytes are involved in the defense response (Fig. 4F). Two genes encoding Thioester-containing proteins, Tep2 and Tep4, showed high expression in thanacytes, and previously have been demonstrated to control defense response to microbial pathogens via regulation of Toll pathway, metabolism and inflammation (Dostalova et al., 2017; Shokal et al., 2018).
Fig. 5.
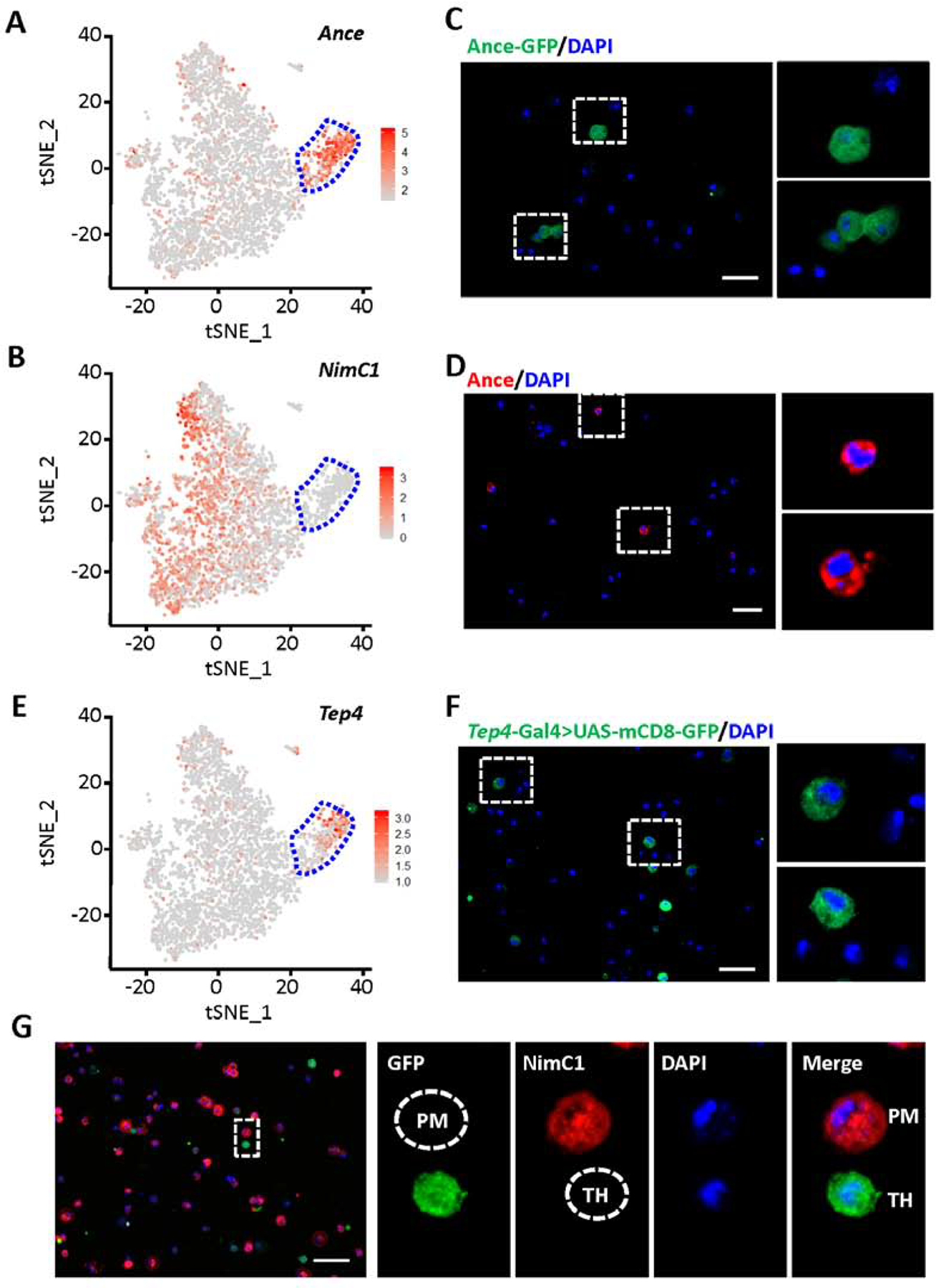
Labeling of thanacytes in Drosophila blood. A, B and E: tSNE feature plots of Drosophila blood scRNA-seq data. Thanacyte cell cluster bounded by blue dashed line. A: tSNE plot representing expression of Ance. B: tSNE plot representing expression of NimC1. E: tSNE plot representing expression of Tep4. C, D, F and G: Fluorescent images captured by Apotome microscopy. Scale bars: 20 μm. C: Ance protein expression (GFP; green) in Drosophila blood cells from Ance-GFP fly line. D: Ance protein expression (red) detected by antibody in Drosophila blood cells. F: Expression of Tep4 (green) in blood cells from a Drosophila Tep4-Gal4-driven UAS-mCD8-GFP line. G: NimC1 (P1), plasmatocyte marker, protein expression (red), and expression of Tep4 (green) in blood cells from Tep4-Gal4>UAS-mCD8-GFP Drosophila line. PM, indicates a GFP negative, NimC1 positive plasmatocyte; and, TH, indicates a GFP positive, NimC1 negative thanacyte.
Labeling thanacytes using existing genetic resources for marker genes identified by scRNA-seq
Single-cell RNA-seq data showed Ance and Tep4 are highly expressed in thanacytes (Fig. 5A and E). We used various methodologies to visualize the expression of Ance and Tep4 in Drosophila blood cells. We found Ance, which encodes an extracellular glycosylated metallopeptidase (Hurst et al., 2003), is expressed at high levels in nearly every thanacyte. We identified a fly line which expresses GFP under the Ance promoter and an anti-Ance antibody, to visually identify Ance gene or protein (~18%) expressing cells (Fig. 5C and D). Tep4 is also highly expressed in thanacytes (Fig. 5E). We used a Tep4-Gal4 line to drive UAS-mCD8-GFP, a membrane-targeted GFP line (Fig. 5F). Findings were consistent with scRNA-seq, and the percentage of mCD8-GFP positive cells (18.02 ± 2.43 %) was very close to the number of Ance-GFP cells (18.18 ± 1.37 %) in Drosophila blood. Double immune labeling showed that while the majority of hemocytes labeled NimC1 positive (plasmatocyte marker), the smaller population of Tep4+ (a proxy for thanacytes) are negative for NimC1 (P1) (Fig. 5G). These data collectively demonstrate that thanacytes are a unique subset of blood cells.
Thanacytes are involved in distinct responses to different types of bacteria
In order to examine the function of thanacytes, we used Tep4-Gal4 to drive UAS-Reaper to induce apoptosis to thanacytes. However, these flies die very early, suggesting that Tep4 function is required in other tissues during early development. To analyze thanacyte-specific role of Tep4, we combined Tep4-Gal4 with UAS-Tep4-RNAi to silence this thanacyte-specific gene, and infected these flies with different types of bacteria including Escherichia coli (E. coli), Photorhabdus luminescens (P. luminescens), Photorhabdus asymbiotica (P. asymbiotica), Micrococcus luteus (M. luteus), Listeria monocytogenes (L. monocytogenes), and Staphylococcus aureus (S. aureus) (Fig. 6). We found that both control and Tep4-Gal4>Tep4-RNAi flies survived well. Interestingly, thanacyte-specific silencing of Tep4 led to higher resistance to P. asymbiotica (Fig. 6A) and L. monocytogenes (Fig. 6B) when compared with control flies. At 36 hrs after infection by P. asymbiotica, only 10% flies survived in the control group, while ~36% flies were still alive in Tep4-silenced flies (Fig. 6A). After infection by L. monocytogenes, 35% control flies were alive at 36 h, while ~69% of Tep4-silenced flies were still alive, this trend continued at 60 h (Fig. 6B). These data indicate that thanacytes, through Tep4 expression, are involved in distinct responses to different types of bacteria.
Fig. 6.
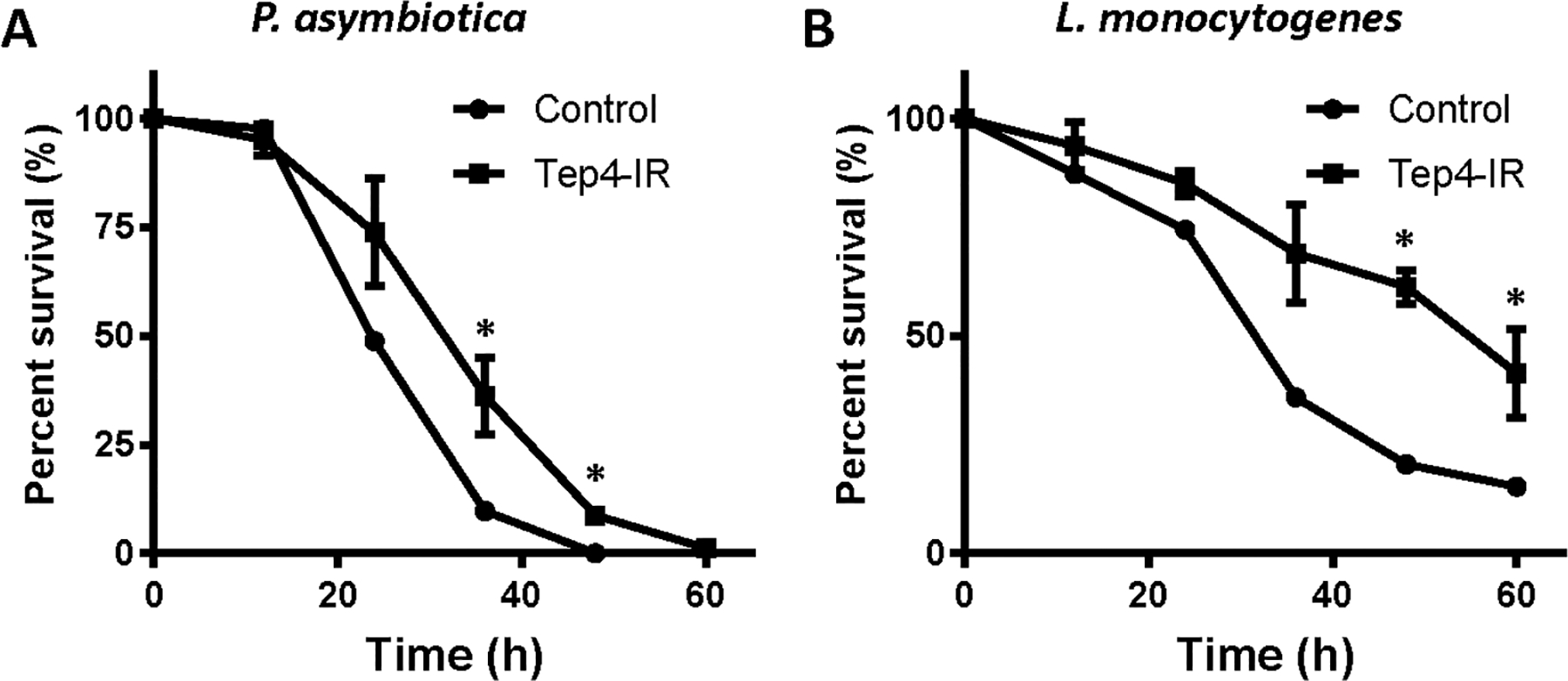
Silencing of Tep4 expression in thanacytes led to distinct responses to different types of bacteria. A and B: Survival plots for control flies (w1118: circle) and Tep4-IR flies (Tep4-Gal4>Tep4-IR: square) after intrathoracic injection with different types of bacteria. Whereas there is no difference observed between these two groups when injecting Escherichia coli (E. coli), Photorhabdus luminescens (P. luminsescens), Micrococcus luteus (M. luteus), or Staphylococcus aureus (S. aureus), injection of Photorhabdus asymbiotica (P. asymbiotica) (A) or Listeria monocytogenes (L. monocytogenes) (B) showed significant different survival curves. *P-value < 0.05. Graphs depict survival of 40 flies per experimental group, monitored for 60 h at 12-hour intervals.
Single-cell RNA-seq disentangles differentially expressed genes with low expression in conventional RNA-seq
Conventional RNA-seq is typically performed on so-called “bulk tissue”. However, averaging gene expression across thousands to millions of cells lowly expressed genes might be masked. For example, read counts for CG31174 (crystal cell), ItgaPS4 (lamellocytes) and Leucine zipper and EF-hand containing transmembrane protein 1 (Letm1) (thanacytes) were relatively low and would most likely not pass the detection threshold in conventional RNA-seq (Fig. S5A). However, scRNA-seq data revealed these genes were highly expressed in specific blood cell types (Fig. S5B–D). For example, kn showed high and specific expression in primocytes (Fig. S5E), which account for only 0.3% of hemocytes. These data exemplify how scRNA-seq analyses increase sensitivity and enable more defined gene expression profiles for specific cell populations.
Identification of new marker genes for crystal cells and lamellocytes
Crystal cells are a group of cells which contain crystalline inclusions required for humoral melanization. Prophenoloxidase (PPO) is produced and stored by crystal cells and is involved in the melanization reaction upon wounding. Our scRNA-seq results showed PPO2 is highly expressed in crystal cells (Fig. 1C). In addition to PPO2, we identified several new marker genes unique to crystal cells, including vav guanine-nucleotide exchange factor (Vav), Ninjurin B (NijB), and sulfotransferase 3 (St3) (Fig. S6A). Vav encodes a GDP/GTP exchange factor for the product of Rac1. NijB is predicted to localize to the integral component of membranes and to be involved in cell adhesion. Its human homolog Ninj1 has been implicated in Toll-like receptor signaling and inflammation (Jennewein et al., 2015). St3 encodes an enzyme involved in sulfation, xenobiotic metabolism and the defense response to bacteria. GO analysis demonstrated the crystal cells are mainly involved in the immune response, melanization and wound healing (Fig. S7A), which is consistent with their previously described functions.
Lamellocytes are large blood cells that rapidly increase in number in response to an immune invasion. Atilla is a known lamellocyte marker (Honti et al., 2009; Kurucz et al., 2007b). Our scRNA-seq data are consistent with previous reports that atilla is highly and specifically expressed in lamellocytes (Fig. 1C). Moreover, we identified additional lamellocyte-specific genes, including Cbl-associated protein (CAP), short stop (shot), and methuselah-like 4 (mthl4) (Fig. S6B). Interestingly, CAP and shot are both known to play a role in the cytoskeleton dynamics and to interact with actin and/or microtubules (Bharadwaj et al., 2013), which are critical for the engulfment function of lamellocytes. GO pathway analysis revealed that lamellocytes are involved in external stimulus, cytoskeleton organization and locomotion (Fig. S7B), which is consistent with their known function in localizing and engulfing parasitoid wasp eggs (Rizki and Rizki, 1992; Williams, 2007).
Therefore, scRNA-seq results from Drosophila blood not only confirmed previously reported marker genes for crystal cells and lamellocytes, but also identified new markers.
Conserved functions between Drosophila and human blood cells
Previous studies have shown blood development in Drosophila shares several interesting features with hematopoiesis in vertebrates, including spatiotemporal regulation, and the use of similar transcriptional regulators and signaling pathways (Evans et al., 2003). We searched for known human homologs of the fly genes differentially expressed between cell types, then examined their expression in a publicly available dataset of human peripheral blood mononuclear cells (PBMCs) (Zheng et al., 2017). Interestingly, we found several human homologs with specific expression (Fig. 7A). Aldehyde dehydrogenase (Aldh) is expressed in Ppn+ plasmatocytes (Fig. 7B), while its human homolog ALDH2 is expressed in CD14+ monocytes and dendritic cells (Fig. 7E). Drosophila Aldh and human ALDH2 showed highly similar protein sequences (Fig. S8A). Two protein-coding Drosophila genes, CG30088 and CG30090, are highly expressed in thanacytes (Fig. 7C and D), while their human homologs, Granzyme B (GZMB) and GZMH, are specifically expressed in NK cells and CD8+ (cytotoxic) T cells (Fig. 7F and G). Both CG30088 and CG30090 encode trypsin-like serine proteases of the S1A family, which are endopeptidases for cleavage of amide substrates following Arginine or Lysine at the P1 position. Similarly, GZMB and GZMH are part of the peptidase S1 family of serine proteases, which are secreted by NK cells and CD8+ (cytotoxic) T lymphocytes (Sedelies et al., 2004). Therefore, evidence that the key trypsin-like serine protease domains (smart00020: Tryp_SPc) show high conservation among these proteins (Fig. S7B and C) supports the notion that these genes are functional homologs from flies to humans. Our data demonstrate that Drosophila hemocytes exhibit traces of conservation to human blood cells.
Fig. 7.
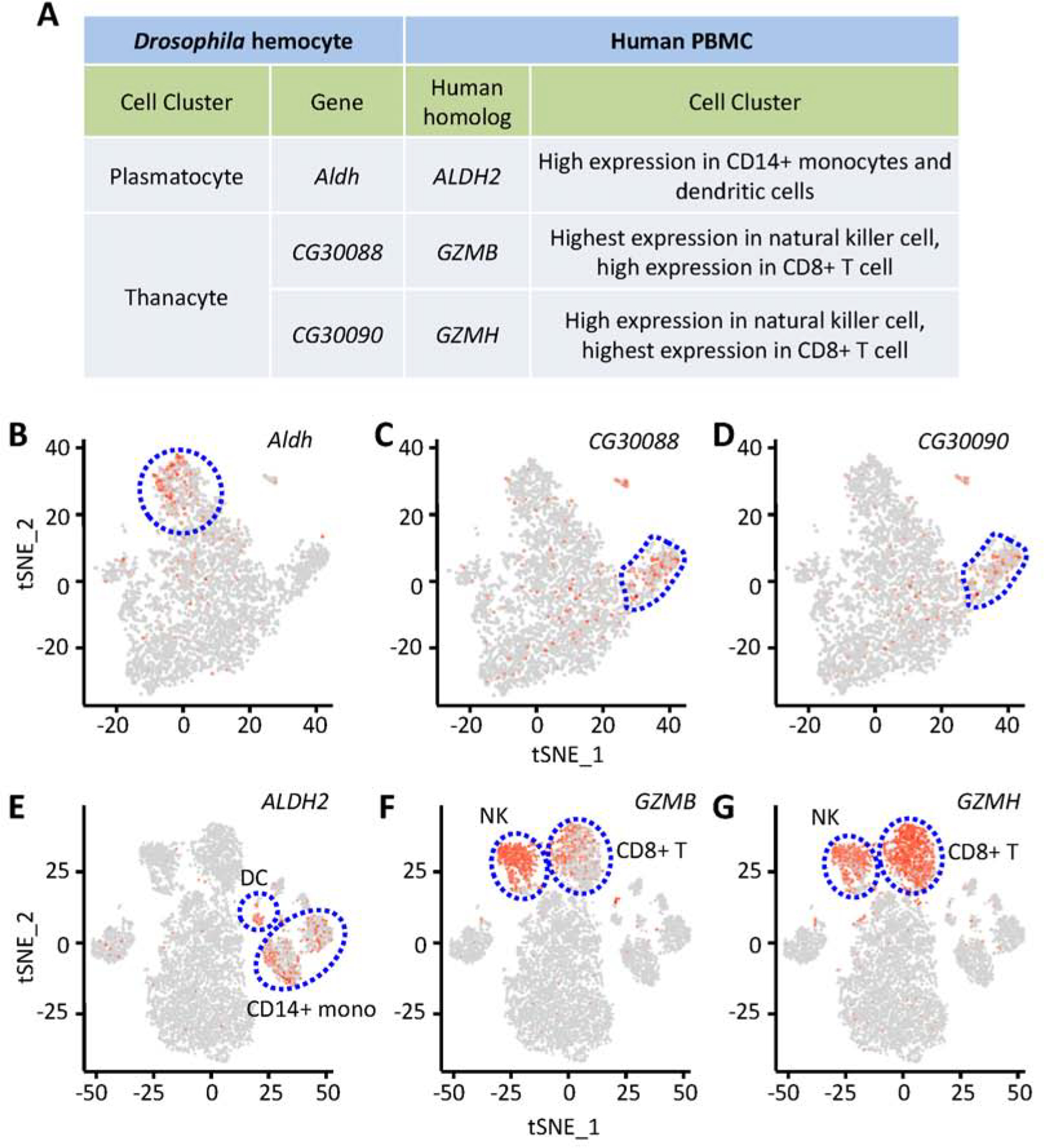
Functional conservation between Drosophila and human blood cells. A: Summary of expression of Drosophila genes and their human homologs. B–D: tSNE feature plots of Drosophila blood scRNA-seq data. E–G: tSNE feature plots of human blood scRNA-seq data (peripheral blood mononuclear cells (Zheng et al., 2017)). B and E: Representing expression of Aldh (B), fly plasmatocyte cluster bounded by dashed blue line; and, its human homolog ALDH2 (E), CD14+ monocytes (CD14+ mono) and dendritic cells (DC) clusters indicated. C, D, F and G: Representing expression of CG30088 (C) and CG30090 (D), fly thanacytes cluster bounded by dashed blue line; and, their human homologs GZMB (F) and GZMH (G), respectively, natural killer cell and CD8+ (cytotoxic) T cell clusters indicated.
DISCUSSION
The immune system forms the first line of defense against any foreign materials of an organism. Indeed, the human blood system is versatile and many different cell types, each with specialized functions, have been identified. Drosophila has proven to be a valuable model to study the human immune system; however, to date only three types of blood cells have been described in the fly: plasmatocytes, crystal cells and lamellocytes. The advent of technological advances now enables us to study transcriptomes at the single-cell level. We leveraged this technique to study the composition of Drosophila blood by sequencing all circulating hemocytes. This unbiased approached allowed us to identify a very small population of a previously unknown cell type, as well as subpopulations of known cell types. These findings would likely have been lost in conventional (“bulk”) RNA-seq data (Fig. S5).
Uncovering plasmatocyte diversity
We identified four plasmatocyte subtypes (Ppn+, CAH7+, Lsp+, and reservoir plasmatocytes), each with a unique expression profile and pathways that indicate specialized functions (Fig. 2). Interestingly, these sub types appear to fulfill defined roles in the immune response system (Fig. 2 D–G). For example, pathway analysis indicates phagocytic activity is specific to the Ppn+ PM and CAH7+ PM, suggesting they are at the front line of eliminating pathogens and cell debris.
Reservoir PM make up the majority of plasmatocytes (60%). They show a very broad response to immune and non-immune stimuli and unique molecular cell markers to define this group seem to be missing, suggesting they might represent a plastic plasmatocyte cell state. For example, similar to the human naïve T cells, which are mature T cells but have not yet been exposed to an antigen upon which their molecular profile changes drastically (Carter et al., 1998). This is further supported by pseudotemporal trajectory tracing, which places the reservoir plasmatocytes at the root with the other PM subtypes branching off based on differing transcriptional profiles (Fig. 2C). Of note, lamellocyte numbers are known to rapidly increase in response to wasp infection, arising from trans-differentiating plasmatocytes (Anderl et al., 2016). Our findings suggest that reservoir PM might be the source for these immune-induced lamellocytes. Further studies are needed to tease out the functional responsibilities and contributions of the individual blood cell types and subtypes, and how these might change in response to an immune challenge.
Expanding the variety of Drosophila blood cell types
The scRNA-seq data provided a wealth of information at the single-cell level, and revealed five distinct cell types, including the known plasmatocytes, crystal cells and lamellocytes. The two additional cell types expressed the pan-hemocyte marker srp, but expression of established blood cell type-specific markers did not rise above the detection threshold. Based on their unique gene expression profiles, we identified them as novel Drosophila blood cell types. The first is a small, quite distinct cell cluster as it only expresses select early pan-hemocyte genes. Further analysis shows pathways of cell fate commitment and regulation of differentiation are particularly active, indicating a hitherto undescribed circulating progenitor cell type in Drosophila blood, which we named primocytes. Primocytes have high level of expression of transcription factors that regulate cell fate commitment, including Antp, kn, Mad, and ham. Interestingly, Antp and Kn are also highly expressed in the Posterior Signaling Center (PSC) of lymph gland primary lobe (Makki et al., 2010). This role of the PSC is reminiscent of the “niche”, the micro-environment of hematopoietic stem cells in vertebrates. Therefore, we speculate that the primocytes might be involved in keeping stem cell fate in the Drosophila blood. The PSC cells in lymph gland only consist of a small number of cells, consistent with the small percentage of primocytes in the circulating hemocytes. Thanacytes, on the other hand, express common hemocyte markers, but show no expression of pan-plasmatocyte markers above threshold. Tep4 is highly expressed in thanacytes, but nearly absent in all other hemocyte cell types (Fig. 5E). These Tep4+ cells show a striking contrast with plasmatocytes in that they do not express NimC1 (P1, marker of plasmatocytes) at detectable levels (Fig. 5G), which further supports our scRNA-seq findings that these cells represent a novel blood cell type. Tep4 is known to play a role in the Drosophila cellular immune response to certain Gram-negative bacteria (Shokal and Eleftherianos, 2017a; Shokal et al., 2018). We used Tep4 expression as a proxy for thanacytes and carried out an immune response assay. We found that the thanacyte-specific Tep4 expression, if silenced, could lead to distinct responses to certain types of bacteria (Fig. 6). This finding suggests that thanacytes have distinct responses to different types of bacteria. We also found that thanacytes express several proteases that might aid to remove infected cells, similar to those found in cytoplasmic granules of cytotoxic T cells and NK cells. Further studies are warranted to deduce the exact nature and limitations of the thanacyte response.
Does a more elaborate Drosophila blood system better model its human equivalent
Drosophila models have long been used to study the human blood-vascular system. The prevailing view has been that the mammalian hematopoietic cells differentiate into lymphoid and myeloid lineages. Hemocytes in Drosophila, and other invertebrates, are considered to be restricted to the myeloid lineage, even though they show strong conservation of genetic homology with human immune cells. In line with these thoughts we found Drosophila Aldh is uniquely expressed in Ppn+ PM, while its human homolog ALDH2 is specific to CD14+ monocytes and dendritic cells of the myeloid lineage. CD14+ monocytes make up 2–10% of all leukocytes, while Ppn+ PM make up ~12% of all plasmatocytes. Both CD14+ monocytes and dendritic cells are part of the mammalian innate immune system, they have three main functions phagocytosis, antigen presentation, and cytokine production. Interestingly, Ppn+ PM in fly are one of two blood cell subtypes in Drosophila, in which we denoted phagocytic activity.
Further we uncovered data that might blur the invertebrate-mammalian division and expand the notion that the mammalian myeloid and lymphoid blood cells at times diverge from their traditional lineages, especially under challenging conditions (Doulatov et al., 2010; Hartenstein and Mandal, 2006; Kawamoto and Katsura, 2009). For example, CAH7, expression of which is specific to the CAH7+ PM, encodes a carbonic anhydrase. An enzyme essential for homeostasis of oxygen-hemoglobin binding, which was first discovered in red blood cells (Meldrum and Roughton, 1933). Drosophila has no known red blood cell equivalent. Furthermore, the human homologs to two prominent thanacyte-specific molecular markers, GZMB (Drosophila GC30088) and GZMH (Drosophila GC30090), both encode members of the granzyme family. These serine proteases are released by cytotoxic T cells and NK cells via cytoplasmic granules, and induce targeted lysis of infected cells through endocytosis (Trapani, 2001). Thanacytes express several proteases and show enriched endocytosis pathways, suggesting they might operate in a similar fashion.
The transcriptome-wide molecular profiles obtained by scRNA-seq uncovered new cell types and sub cell types and identified new markers to aid in future studies of their functions. Already our analysis unveiled a much more complex Drosophila blood system than described to date. Moreover, it provides hints of potential non-myeloid activity in Drosophila blood cells that might blur the lines of conventional lineage restriction. Studies directly comparing the molecular profiles of human and Drosophila blood cell types, as well as their dynamics in response to immune challenges will be fundamental. The data presented here contribute knowledge and identified resources (Tables S2 and S3) towards an increased understanding of how both systems relate, thereby widening the scope of the Drosophila model to study the human blood system in development, health and disease.
METHODS
Fly strains
All fly stocks were maintained at 25°C on standard diet. The w1118, Ance [MB09828] (#27809), MIC-Ance [MI05748] (#42109), Ppn-Gal4 (#77733), atilla [MB03539] (#23540), alphaTub85E (#60267), Drs (#67099), Drs(#55707), CG30090 (#65315), Hr4 (#38655), Eip78C (#38637), Eip74EF (#38636), crq-Gal4 (#25041), LpR2(#60219), lz(#43954), Tep4-Gal4 (#76750), UAS-mCD8-GFP (32185), lines were all from the Bloomington Drosophila Stock Center (Bloomington, USA).
Sample preparation and single-cell RNA sequencing
Circulating hemocytes from 18 wandering third instar larvae of w1118 were pooled and resuspended in 30 μL ice-cold Schneider’s medium. Cell numbers were counted with a Neubauer chamber. Single-cell suspensions were assayed a Chromium Single Cell Controller (10x Genomics, USA). Barcoding and cDNA synthesis were performed according to the manufacturer’s instructions using GemCode Technology to add Unique Molecular Identifiers (UMIs) to each PCR product for library construction. cDNA amplicon size was optimized by enzymatic fragmentation and size selection using Beckman Coulter (USA) SPRIselect Reagent prior to library construction. Quality of cDNA/libraries was tested using the Agilent Bioanalyzer High Sensitivity assay (Agilent, USA) and Qubit dsDNA BR Assay (Invitrogen, USA). For quantification, Illumina Library Quantification Kit (KAPA Biosystems, Cat# KK4824, USA) was used. Libraries were sequenced on an Illumina NextSeq (Illumina, USA) with 2×150 paired-end kits using the following read length: 26 bp Read1 for cell barcode and UMI, 8 bp I7 index for sample index and 98 bp Read2 for transcript. Cell Ranger (http://10xgenomics.com) was used to process Chromium single-cell 3′ RNA-seq output. The data matrix was generated by Cell Ranger (10x Genomics).
Data preparation, dimensionality reduction and cell clustering
Once the gene-cell data matrix was generated, cells with less than 200 or more than 2500 unique genes expressed genes (potentially cell duplets) were excluded. Only genes expressed in three or more cells were used for further analysis. Remaining data were log-normalized and cell-cell variation was regressed out by the number of detected molecules per cell (UMIs). We performed Principal Component Analysis (PCA) on the scaled data for dimensionality reduction (Seurat R package; version 2.3.4). Statistical significance of PCAs was based on strong enrichment of genes with low P-values and used for downstream clustering analysis. We used the JackStrawPlot function to compare the distribution of P-values for each PC with a uniform distribution. FindClusters function was applied to cluster the cells based on previously identified PCs. Non-linear dimensional reduction (t-distributed stochastic neighbor embedding (tSNE)) was performed to visualize the distribution of these cells in two dimensions based on expression signatures of the variable genes.
Differentially expressed genes and gene ontology (GO) analysis
Differentially expressed genes of each cluster (both directions) were identified using the Seurat R package (version 2.3.4). Expression heatmaps for a given cell cluster represents marker genes with the top 10 markers (or all markers if less than 10). GO enrichment analysis was performed using DAVID (https://david.ncifcrf.gov/summary.jsp). Adjusted P (< 0.05) was used for terms ranking and selection.
Cell trajectory analysis of plasmatocytes using Monocle
To perform pseudotemporal analysis of plasmatocytes, the normalized data of the clustered cells were passed directly into Monocle2 (version 2.14.0) (Qiu et al., 2017). Outliers were removed, and after dimensionality reduction, the top 1000 most significant differentially expressed genes were used to perform trajectory analysis. Cells were ordered along the trajectory without bias.
Immunofluorescence
Hemocytes from wandering third instar larvae were collected in Schneider medium or artificial hemolymph. Cells were fixed in 4% formaldehyde (10 min), and processed using standard immunochemistry protocol. Antibody against Ance was kindly provided by Dr. Elwyn Isaac University of Leeds (UK) (Hurst et al., 2003), and all other primary antibodies were purchased from Developmental Studies Hybridoma Bank (DSHB, USA). Secondary antibodies (AlexaFluor 568 anti-mouse or anti-rabbit, Thermo Fisher Scientific, USA). Images were obtained with an Apotome microscope (Zeiss, Germany).
Bacterial infection and survival experiments
Procedures were performed as described previously (Shokal and Eleftherianos, 2017b). Bacteria M. luteus (#4698), S. aureus (#12600) and L. monocytogenes (#19115) were purchased from ATCC (USA). Bacteria P. asymbiotica, P. luminescens and M. luteus were cultured in sterile Luria-Bertani (LB) broth for 18–22 h at 30°C on a shaker at 225 rpm. E. coli, M. luteus, S. aureus and L. monocytogenes were cultured at 37°C on a shaker at 225 rpm. For infections, bacterial concentrations were adjusted to an optical density (OD, 600 nm) of 0.1 for P. luminescens, 0.25 for P. asymbiotica, M. luteus, S. aureus and L. monocytogenes, and 0.015 for E. coli, as measured by spectrophotometer (NanoDropTM 2000c, Thermo Fisher Scientific). Adult flies of seven- to ten-day-old (40 / condition) were anesthetized with CO2 and injected in the thorax with 20 nL of bacterial suspension or sterile 1X PBS (injury control). Flies were kept at 25°C, and survival was monitored for 96 h after injection.
Expression of human homologs of the differentially expressed Drosophila genes in PBMCs
We checked for human homologs of differentially expressed genes in our Drosophila scRNA-seq data using the DIOPT - DRSC Integrative Ortholog Prediction Tool (version 7.1) maintained by Harvard Medical School. Next, we studied the expression and distribution of the human genes in peripheral blood mononuclear cells (PBMCs) (Zheng et al., 2017). Both human and Drosophila protein sequences were downloaded from Uniprot database and were aligned with CLUSTAL O (1.2.4) maintained by EMBL-EBI (UK).
Statistical analysis
Digital image processing and analysis were performed with Fiji (http://fiji.sc/) and ZEN 2 (Zeiss, Germany). Statistical data are expressed as mean ± SEM, and Student t test was applied to determine the significance of the difference by GraphPad Prism 7.00 software. A value of P < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Bloomington Drosophila Stock Center for fly stocks, and the University of Iowa Developmental Studies Hybridoma Bank for antibodies. We thank Dr. Elwyn Isaac (University of Leeds, UK) for kindly providing Ance antibody and Dr. Ioannis Eleftherianos for providing bacteria P. asymbiotica and P. luminescens (George Washington University, USA). This work was supported by grants from NIH to Z.H. (R01-HL134940, R01-DK098410).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderl I, Vesala L, Ihalainen TO, Vanha-Aho LM, Ando I, Ramet M, Hultmark D, 2016. Transdifferentiation and proliferation in two distinct hemocyte lineages in Drosophila melanogaster larvae after wasp infection. PLoS pathog. 12, e1005746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artero RD, Monferrer L, Garcia-Lopez A, Baylies MK, 2006. Serpent and a hibris reporter are co-expressed in migrating cells during Drosophila hematopoiesis and Malpighian tubule formation. Hereditas. 143, 117–122. [DOI] [PubMed] [Google Scholar]
- Benes H, Spivey DW, Miles J, Neal K, Edmondson RG, 1990. Fat-body-specific expression of the Drosophila Lsp-2 gene. SAAS Bull Biochem Biotechnol. 3, 129–133. [PubMed] [Google Scholar]
- Bharadwaj R, Roy M, Ohyama T, Sivan-Loukianova E, Delannoy M, Lloyd TE, Zlatic M, Eberl DF, Kolodkin AL, 2013. Cbl-associated protein regulates assembly and function of two tension-sensing structures in Drosophila. Development. 140, 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter LL, Zhang X, Dubey C, Rogers P, Tsui L, Swain SL, 1998. Regulation of T cell subsets from naive to memory. J Immunother. 21, 181–187. [DOI] [PubMed] [Google Scholar]
- Cordes EJ, Licking-Murray KD, Carlson KA, 2013. Differential gene expression related to Nora virus infection of Drosophila melanogaster. Virus Res. 175, 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostalova A, Rommelaere S, Poidevin M, Lemaitre B, 2017. Thioester-containing proteins regulate the Toll pathway and play a role in Drosophila defence against microbial pathogens and parasitoid wasps. BMC Bio. 15, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE, 2010. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 11, 585–593. [DOI] [PubMed] [Google Scholar]
- Evans CJ, Hartenstein V, Banerjee U, 2003. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev Cell. 5, 673–690. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Mandal L, 2006. The blood/vascular system in a phylogenetic perspective. BioEssays. 28, 1203–1210. [DOI] [PubMed] [Google Scholar]
- Honti V, Kurucz E, Csordas G, Laurinyecz B, Markus R, Ando I, 2009. In vivo detection of lamellocytes in Drosophila melanogaster. Immunol Lett. 126, 83–84. [DOI] [PubMed] [Google Scholar]
- Hurst D, Rylett CM, Isaac RE, Shirras AD, 2003. The Drosophila angiotensin-converting enzyme homologue Ance is required for spermiogenesis. Dev Bio. 254, 238–247. [DOI] [PubMed] [Google Scholar]
- Irving P, Ubeda JM, Doucet D, Troxler L, Lagueux M, Zachary D, Hoffmann JA, Hetru C, Meister M, 2005. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell Microbiol. 7, 335–350. [DOI] [PubMed] [Google Scholar]
- Jennewein C, Sowa R, Faber AC, Dildey M, von Knethen A, Meybohm P, Scheller B, Drose S, Zacharowski K, 2015. Contribution of Ninjurin1 to Toll-like receptor 4 signaling and systemic inflammation. Am J Resp Cell Mol. 53, 656–663. [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Katsura Y, 2009. A new paradigm for hematopoietic cell lineages: revision of the classical concept of the myeloid-lymphoid dichotomy. Trends Immunol. 30, 193–200. [DOI] [PubMed] [Google Scholar]
- Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, Pfeffer S, Hoffmann JA, Imler JL, 2013. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol. 190, 650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, Strom C, Conto SL, Hetru C, Stuart LM, Stehle T, Hoffmann JA, Reichhart JM, Ferrandon D, Ramet M, Ezekowitz RA, 2005. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 123, 335–346. [DOI] [PubMed] [Google Scholar]
- Kulkarni V, Khadilkar RJ, Magadi SS, Inamdar MS, 2011. Asrij maintains the stem cell niche and controls differentiation during Drosophila lymph gland hematopoiesis. PLoS One. 6, e27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurucz E, Markus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, Udvardy A, Krausz I, Lukacsovich T, Gateff E, Zettervall CJ, Hultmark D, Ando I, 2007a. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. CB 17, 649–654. [DOI] [PubMed] [Google Scholar]
- Kurucz E, Vaczi B, Markus R, Laurinyecz B, Vilmos P, Zsamboki J, Csorba K, Gateff E, Hultmark D, Ando I, 2007b. Definition of Drosophila hemocyte subsets by cell-type specific antigens. Acta Biol Hung. 58 Suppl, 95–111. [DOI] [PubMed] [Google Scholar]
- Makki R, Meister M, Pennetier D, Ubeda JM, Braun A, Daburon V, Krzemien J, Bourbon HM, Zhou R, Vincent A, Crozatier M, 2010. A short receptor downregulates JAK/STAT signalling to control the Drosophila cellular immune response. PLoS Biol. 8, e1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldrum NU, Roughton FJ, 1933. Carbonic anhydrase. Its preparation and properties. J Physiol. 80, 113–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RE, Fessler LI, Takagi Y, Blumberg B, Keene DR, Olson PF, Parker CG, Fessler JH, 1994. Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J. 13, 3438–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A, Serano J, Sante JM, Rubin GM, 2003. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev Cell. 4, 95–106. [DOI] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C, 2017. Single-cell mRNA quantification and differential analysis with Census. Nat methods. 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki TM, Rizki RM, 1992. Lamellocyte differentiation in Drosophila larvae parasitized by Leptopilina. Dev Comp Immunol. 16, 103–110. [DOI] [PubMed] [Google Scholar]
- Saliba AE, Westermann AJ, Gorski SA, Vogel J, 2014. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 42, 8845–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelies KA, Sayers TJ, Edwards KM, Chen W, Pellicci DG, Godfrey DI, Trapani JA, 2004. Discordant regulation of granzyme H and granzyme B expression in human lymphocytes. J Biol Chem. 279, 26581–26587. [DOI] [PubMed] [Google Scholar]
- Shokal U, Eleftherianos I, 2017a. The Drosophila Thioester containing Protein-4 participates in the induction of the cellular immune response to the pathogen Photorhabdus. Dev Comp Immunol. 76, 200–208. [DOI] [PubMed] [Google Scholar]
- Shokal U, Eleftherianos I, 2017b. Thioester-Containing Protein-4 Regulates the Drosophila Immune Signaling and Function against the Pathogen Photorhabdus. J Innate Immun. 9, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokal U, Kopydlowski H, Harsh S, Eleftherianos I, 2018. Thioester-Containing Proteins 2 and 4 Affect the Metabolic Activity and Inflammation Response in Drosophila. Infection and immunity 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani JA, 2001. Granzymes: a family of lymphocyte granule serine proteases. Genome Biol. 2, reviews3014.1–reviews3014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P, 1998. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc Natl Acad Sci U S A. 95, 11342–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verleyen P, Baggerman G, D’Hertog W, Vierstraete E, Husson SJ, Schoofs L, 2006. Identification of new immune induced molecules in the haemolymph of Drosophila melanogaster by 2D-nanoLC MS/MS. J Insect Physiol. 52, 379–388. [DOI] [PubMed] [Google Scholar]
- Williams MJ, 2007. Drosophila hemopoiesis and cellular immunity. J Immunol. 178, 4711–4716. [DOI] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


