Significance
Our work identifies regulatory mechanisms between ferroptosis and autophagy. We discovered that PE-binding protein-1 (PEBP1) is pivotal for dynamic interactions between the ferroptotic cell death program and prosurvival autophagy in asthmatic/Type 2 stimulated airway epithelial cells and that concurrent activation of autophagy protects cells from ferroptotic death and mitochondrial DNA release. Similar findings are observed in Type 2 Hi asthmatic epithelial cells where high 15-lipoxygenase-1 (15LO1)–PEBP1 and light chain-3 II (LC3-II) levels associate with low bronchoalveolar lavage fluid mitochondrial DNA and severe disease. Our findings of concomitant activation of ferroptosis and autophagy by 15LO1–PEBP1 complexes and their hydroperoxy-phospholipids reveal a pathobiologic pathway relevant to asthma and amenable to therapeutic targeting.
Keywords: autophagy, ferroptosis, asthma
Abstract
Temporally harmonized elimination of damaged or unnecessary organelles and cells is a prerequisite of health. Under Type 2 inflammatory conditions, human airway epithelial cells (HAECs) generate proferroptotic hydroperoxy-arachidonoyl-phosphatidylethanolamines (HpETE-PEs) as proximate death signals. Production of 15-HpETE-PE depends on activation of 15-lipoxygenase-1 (15LO1) in complex with PE-binding protein-1 (PEBP1). We hypothesized that cellular membrane damage induced by these proferroptotic phospholipids triggers compensatory prosurvival pathways, and in particular autophagic pathways, to prevent cell elimination through programmed death. We discovered that PEBP1 is pivotal to driving dynamic interactions with both proferroptotic 15LO1 and the autophagic protein microtubule-associated light chain-3 (LC3). Further, the 15LO1–PEBP1-generated ferroptotic phospholipid, 15-HpETE-PE, promoted LC3-I lipidation to stimulate autophagy. This concurrent activation of autophagy protects cells from ferroptotic death and release of mitochondrial DNA. Similar findings are observed in Type 2 Hi asthma, where high levels of both 15LO1–PEBP1 and LC3-II are seen in HAECs, in association with low bronchoalveolar lavage fluid mitochondrial DNA and more severe disease. The concomitant activation of ferroptosis and autophagy by 15LO1–PEBP1 complexes and their hydroperoxy-phospholipids reveals a pathobiologic pathway relevant to asthma and amenable to therapeutic targeting.
Harmonized epithelial cell communities depend on the fidelity of their individual members to eliminate metabolic and/or toxic consequences to individual cells that subsequently endanger the health of the entire population. Several cell death programs have been identified as instruments of this sacrificial behavior, beginning with apoptosis and more recently expanded into other contextually regulated necrotic platforms (e.g., necroptosis, pyroptosis) (1–3). Ferroptosis—the response to redox disbalance between the prooxidant enzymatically driven reactions of lipid peroxidation and their thiol-dependent control by glutathione (GSH) peroxidases—is a recent addition to these programmed cell death pathways (4); 15-lipoxygenase (15LO)-initiated peroxidation of arachidonoyl-phosphatidylethanolamines (AA-PEs) to hydroperoxy-products distinguishes the characteristic features of ferroptotic cell demise (5). We discovered that the selectivity and specificity of the peroxidation reaction are modulated by the complex of 15LO with a promiscuous protein, PE-binding protein 1 (PEBP1) (6). Generally, PEBP1 is bound to and acts as an inhibitor of Raf-1 kinase [hence, its alternate name, Raf-1 kinase inhibitory protein (7)]. However, in Type 2 immune conditions, dramatically increased expression of 15LO1 creates conditions whereby PEBP1 avidly binds 15LO1, changing its catalytic competence from free arachidonic acid(AA) to AA-PE and specifically initiating the generation of the ferroptotic hydroperoxy-phospholipid, 15-hydroperoxyeicosaetetranoic acid (15-HpETE-PE) (6, 8). While 15LO1–PEBP1 complex formation has been reported ex vivo in asthmatic human airway epithelial cells (HAECs) (6, 8), the consequences of these interactions to epithelial death, survival, and human disease are less clear.
Because of the irrevocability of cell death, a variety of protective processes can be triggered to prevent and/or eliminate the “damage” by activating repair mechanisms (1–3, 9). Among them is autophagy (9–11). For successful autophagy to proceed, the essential autophagy protein, microtubule light chain protein-3 I (LC3-I), must conjugate with the ferroptosis-linked phospholipid phosphatidylethanolamine (PE) to form membrane-bound lipidated LC3-II (12). Current understanding of the cross-talk between autophagy and ferroptotic cell death is controversial. However, the majority of studies suggest autophagy contributes to enhancement or even execution of ferroptotic cell death, through pathways including ferritinophagy and chaperone-mediated autophagy (13, 14). However, no specific biochemical links have been identified. Interestingly, in addition to complexing with 15LO1 to activate ferroptosis, PEBP1 binds LC3-I, suggesting potential biochemical cross-talk between the pathways at the level of PEBP1, as well as through PE (15). The consequences of this promiscuous PEBP1 partnering with LC3 to ferroptosis are unknown.
With this background, we hypothesized that 15LO1–PEBP1 interactions play two seemingly opposing, yet complementary roles: generating 15-HpETE-PE, which can induce ferroptotic cellular damage and death, while simultaneously inhibiting PEBP1 binding with LC3-I and promoting its subsequent lipidation, which activates autophagic prosurvival pathways to limit cell destruction. Using fresh and cultured primary HAECs, we demonstrate that under Type 2 high interleukin-13 (IL-13) conditions, dynamic interactions of PEBP1 with both LC3 and 15LO1 engage protective autophagic cell survival in the face of potential ferroptotic damage. These in vitro findings are mirrored ex vivo in asthmatic lungs, where concordant increases in both 15LO1 (in relation to PEBP1) and LC3-II associate with greater asthma severity.
Results
LC3-II Is Induced by 15LO1-Dependent Mechanisms in HAECs In Vitro under IL-13 Stimulation.
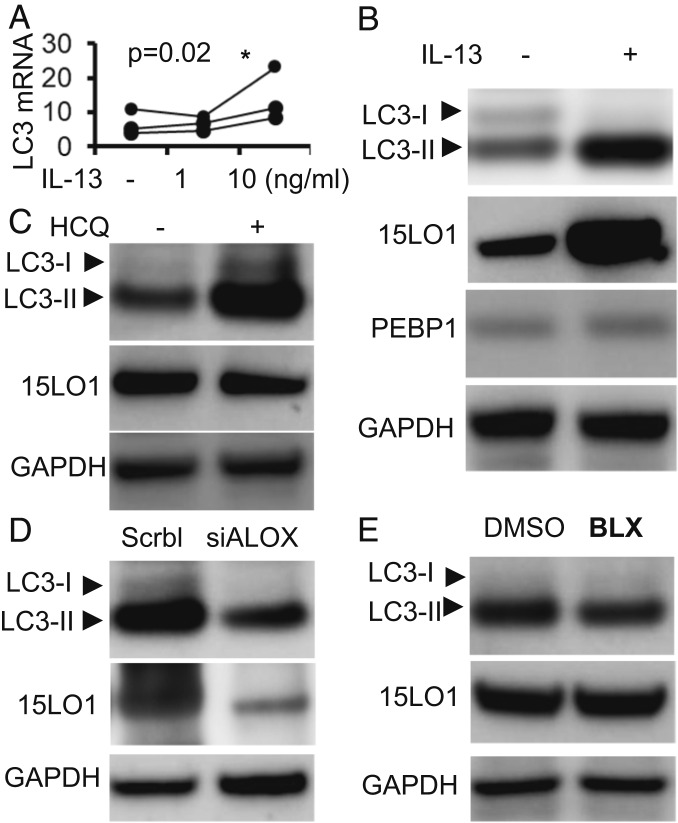
15LO1, elevated in asthmatic HAECs ex vivo, is induced by IL-13 in vitro, where it forms a complex with PEBP1 and generates the unstable ferroptosis-inducing oxidized phospholipid 15-HpETE-PE (6, 16). While the necessity of PEBP1–15LO1 complex for ferroptosis has been previously documented (6), we additionally confirmed this using CRISPR-CasP9 knockout (KO) protocol. We found that PEBP1 KO HT22 cells demonstrated very high resistance to RSL3-induced ferroptosis compared with wild-type control cells (SI Appendix, Fig. S1). Similar to ferroptosis, the autophagic cell survival pathway also engages membrane phospholipids, particularly PE, to form new phospholipid membranes, which protect cells by recycling damaged cell organelles under conditions of stress. Although multiple proteins are associated with autophagy, lipidation of microtubule-associated light chain-3 (LC3-I) to form LC3-II via conjugation with PE is accepted as an essential stage of autophagosome formation (17). Given the common utilization of PE by both autophagic and ferroptotic processes, as well as the reported binding of LC3 with PEBP1, we explored a potential link between the three proteins, 15LO1, PEBP1, and LC3, as well as the canonical ferroptotic mediator, 15-HpETE-PE, in primary HAECs under air–liquid interface (ALI) culture conditions stimulated with IL-13 (6, 16). Under IL-13 conditions, LC3 messenger RNA (mRNA) expression was induced in a dose-dependent manner with optimal expression at 10 ng/mL (Fig. 1A), supporting general enhancement of de novo synthesis. In addition to this overall increase, IL-13 stimulation markedly (and disproportionately) increased lipidated LC3-II, as detected by western blot, while LC3-I decreased (Fig. 1B and SI Appendix, Fig. S2A). Supporting wider autophagy activation, IL-13 also induced p62 expression (SI Appendix, Fig. S2B). As previously reported, IL-13 induced higher 15LO1 expression without altering PEBP1 (Fig. 1B). Addition of the autophagosome-progression inhibitor, hydroxychloroquine (HCQ; 10 μM, overnight), further increased IL-13–induced LC3-II and p62, confirming the inhibition of autophagic flux and presence of autophagy (Fig. 1C and SI Appendix, Fig. S2C).
Fig. 1.
LC3 and LC3-II are induced by 15LO1-dependent mechanisms in HAECs in vitro under IL-13 stimulation. (A) IL-13 induced LC3 mRNA expression in a dose-dependent manner. HAECs under ALI were stimulated with IL-13 1 and 10 ng/mL for 7 d. LC3 mRNA was analyzed by RT-PCR with beta-glucuronidase (GUSB) as the internal control (*as compared to no-IL-13 control). (B) Representative western blot showing IL-13–induced LC3-II protein expression in HAECs in vitro. HAECs under ALI were stimulated with IL-13 for 7 d. Total cell lysate protein was collected for western blot analysis. (C) Representative western blot showing HCQ further increased IL-13–induced LC3-II. HAECs were all stimulated with IL-13 for 7 d before treatment with or without HCQ (10 μM, overnight). (D) 15LO1 siRNA (siALOX15) transfection suppressed IL-13–induced LC3-II protein expression. HAECs transfected with siALOX15 were all stimulated with IL-13 for 5 d, with scramble siRNA as the negative control. (E) The 15LO1 inhibitor BLX2477 (BLX) suppresses IL-13–induced LC3-II. HAECs were all stimulated with IL-13 for 7 d before treatment with BLX2477 (2 μM, overnight), with DMSO as the control. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
These data suggest that chronic IL-13 stimulation induces autophagic flux in HAECs, which under steady-state conditions, does not lead to “net” p62 destruction (18–23). Given the similar directionality and general appreciation that LC3 is a central autophagy biomarker, the remaining studies focused on LC3-I and -II (17).
To explore the regulation of LC3 expression and/or lipidation by 15LO1 presence or activation, Dicer-substrate short interfering RNAs (DsiRNA) knockdown of 15LO1 (DsiALOX15) was performed. DsiALOX15 modestly decreased LC3 mRNA expression (SI Appendix, Fig. S2D) but robustly decreased lipidated LC3-II following IL-13 stimulation (Fig. 1D and SI Appendix, Fig. S2E). Confirming these results, a specific 15LO1 chemical inhibitor BLX2477 (24) also decreased LC3-II (Fig. 1E and SI Appendix, Fig. S2F). Together, these data confirm that 15LO1 promotes both the expression and the lipidation of LC3-II under Type 2/IL-13 conditions in vitro. The parallel decrease seen with the enzyme inhibitor suggests that a 15LO1-generated mediator is responsible for this effect. Under IL-13 conditions, these are primarily 15-HpETE-PE and its metabolite 15-HETE-PE (6, 25). Given the critical importance of 15LO1 binding with PEBP1 to generate 15-HpETE-PE (6) and the recent report of PEBP1 binding with LC3, we then determined whether PEBP1 served as a potential intermediary between ferroptosis and autophagy.
Sequestration of LC3 by PEBP1 Prevents Its Lipidation and Subsequent Activation of Autophagy.
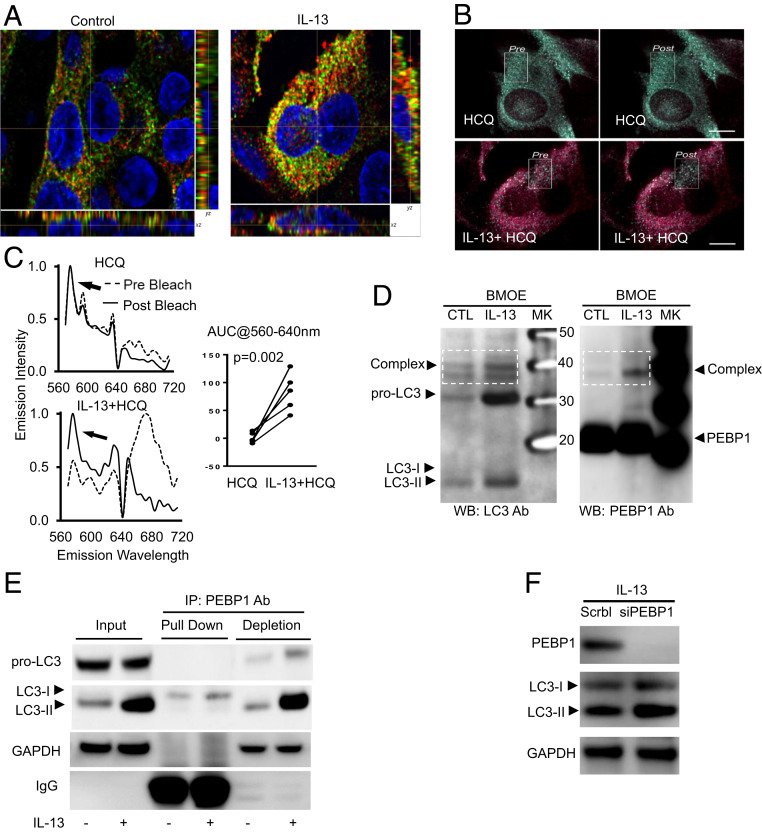
PEBP1 is a promiscuous protein known to bind 15LO1 (6, 8), Raf-1, GRK2 (26), and IkKB (27). It was also reported to bind LC3 (15). To determine whether PEBP1, alone and/or through interactions with 15LO, regulates LC3/autophagy, we first confirmed the colocalization of PEBP1 with LC3 by dual object-based colocalization analysis of HAECs using IF/confocal microscopy in vitro. Mirroring the overall increase in LC3 mRNA and protein, IL-13 increased the numbers of LC3 puncta colocalized with PEBP1 as compared with unstimulated cells (Fig. 2A and SI Appendix, Fig. S3A). LC3 and PEBP1 binding with IL-13 stimulation was confirmed by fluorescence resonance energy transfer (FRET). No/minimal FRET was detected in HCQ-only control conditions as shown by the low baseline emissions of the FRET acceptor (cy5, λEM ∼ 640 to 700 nm) (Fig. 2 B, Upper and C, Upper Left), whereas stimulation with IL-13 dramatically increased the acceptor emissions (Fig. 2 B, Lower and C, Lower Left). FRET was confirmed by acceptor photobleaching and the unquenching of the donor emissions (cy3, λEM ∼ 570 nm). Using HCQ to maximize detection of autophagosomes in all cells, quantitation analysis of area under curve showed a significant increase of FRET induced by IL-13 as compared with HCQ alone (Fig. 2 C, Right). PEBP1 binding with LC3 was further confirmed by protein cross-linking with bis-maleimidoethane (BMOE) followed by separately detecting LC3 and PEBP1 on two identical western blot membranes. IL-13 increased a complex at ∼40 kDa identified by both LC3 and PEBP1 antibodies (Fig. 2D). Consistent with the overall increase in LC3 synthesis, IL-13 also increased an LC3 band at about 32 kDa (Fig. 2D), presumably the full-length precursor pro-LC3 as previously reported (17). However, this form was not detected by PEBP1 antibody.
Fig. 2.
Sequestration of LC3 by PEBP1 prevents its lipidation and subsequent activation of autophagy. HAECs were stimulated with/without IL-13 for 7 d, and IF/confocal microscopy and object-based localization were performed. (A) Representative images of object-based colocalization showing IL-13 increased total number of LC3 puncta colocalized with PEBP1 as compared with unstimulated cells. Red, PEBP1; green, LC3; yellow, PEBP1/LC3 colocalization. (B and C) FRET showing IL-13–induced PEBP1–LC3 interactions. HAECs were stimulated with IL-13 for 7 d. Cells were then treated overnight with or without HCQ (10 μM). Images were collected at 60× (14 NA) on a Nikon A1 equipped with a spectral detector. Scale bar, 10 microns. Images are colored based on the wavelength of the peak emissions for each pixel of the image, following the selective excitation of the donor fluorophore (cy3, 540 nm). Green pixels represent the emissions of the donor, and red pixels represent the emissions from the acceptor (cy5) and are indicative of FRET. The absence of red pixels in the untreated samples suggests that minimal FRET occurred, whereas the strong emission from the acceptor in the IL-13–treated samples supports the presence of FRET (and thereby, interaction of PEBP1 and LC3), which was confirmed by acceptor photobleaching within the white boxed region, and the unquenching of the donor emissions (the appearance of green pixels). (D) Cross-linking of total protein with BMOE showing IL-13 increased PEBP1–LC3 complex formation. Total cell lysate protein was incubated with BMOE (0.2 mM) for 2 h at 4 °C, and the reaction was stopped by directly boiling in 2× SDS-DTT sample buffer. Samples were divided and loaded onto two identical SDS/PAGE gels for western blot for LC3 and PEBP1, respectively. (E) Co-IP/WB showing IL-13–induced PEBP1 binding with LC3-I but not with pro-LC3 and LC3-II. (F) siPEBP1 transfection increased IL-13–induced LC3-II protein levels. HAECs transfected with siPEBP1 were all stimulated with IL-13 for 5 d, with scramble siRNA as the negative control. NA, numerical aperture; AUC, area under curve; CTL, control; SDS, sodium dodecyl sulfate; DTT, dithiothreitol; PAGE, polyacrylamide gel electrophoresis; MK, maker; IP, immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; WB, western blot.
Since traditional immunofluorescence (IF)/confocal, FRET, and cross-link/western blot cannot distinguish between binding of PEBP1 with lipidated or nonlipidated LC3, coimmunoprecipitation (Co-IP) followed by western blot of control and IL-13–treated HAECs was performed using anti-PEBP1 antibody (pull down) on total cell lysates. Both the immunoprecipitation (pull-down fraction) and supernatant (antibody-depleted fraction) were collected for LC3 detection by western blot. As shown in Fig. 2E, IL-13 increased expression of all forms of LC3 (pro-LC3, LC3-I, and LC3-II) (input in Fig. 2E), but only LC3-I coimmunoprecipitated with PEBP1 (pull down in Fig. 2E), while pro-LC3 and LC3-II were only detectable in the antibody-depleted supernatant fraction (depletion in Fig. 2E). These results confirm the previously reported binding of PEBP1 with LC3-I (15) and further show enhancement of this binding by IL-13, at least partially due to overall increases in LC3 expression with IL-13 stimulation. Since the full-length precursor protein pro-LC3 (17) did not interact with PEBP1 in both cross-linking and Co-IP results, the remaining studies focused on LC3-I and LC3-II.
To determine whether PEBP1 functionally limits LC3-I lipidation (even in the presence of 15LO1), DsiRNA PEBP1 knockdown (siPEBP1) was performed. siPEBP1 increased IL-13–induced LC3-II as compared with scramble small interfering RNA (siRNA) control, with only modest effects on LC3-I, consistent with more unbound LC3-I (Fig. 2F and SI Appendix, Fig. S3B). Thus, under IL-13 conditions (with high 15LO1 and LC3 levels), PEBP1 binding to LC3 still functions to limit LC3 lipidation. However, the effect is inadequate, and LC3-II levels, paradoxically, increase. Thus, we next explored the concurrent interactions of PEBP1 with 15LO1, which could explain this.
15LO1 Outcompetes LC3 for Binding to PEBP1 Promoting Its Lipidation.
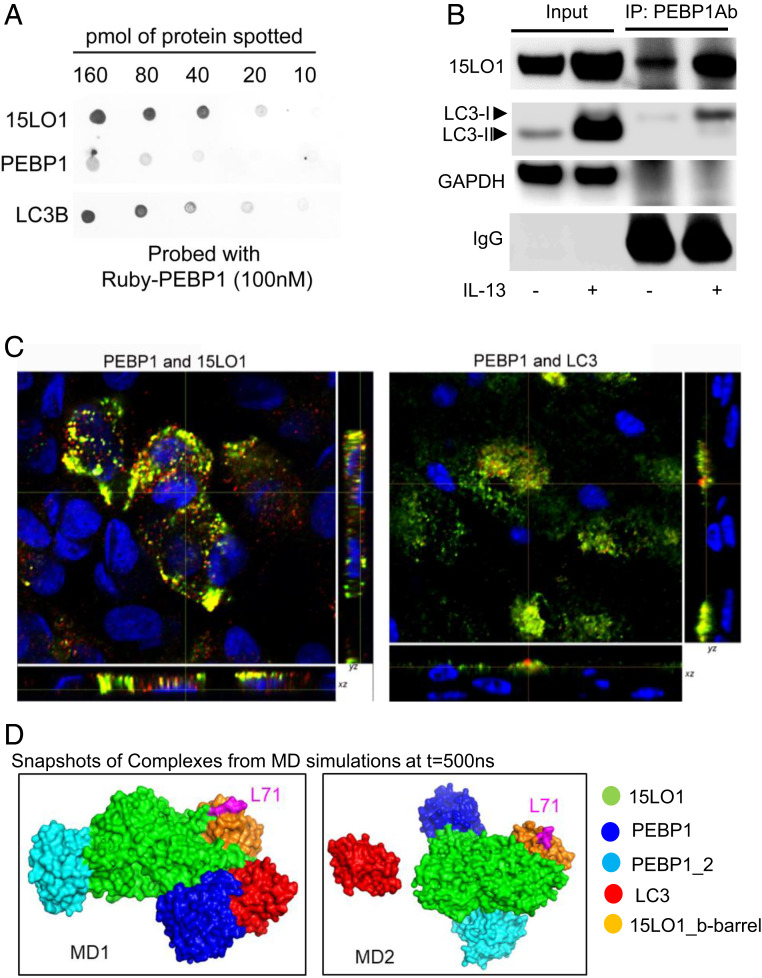
Despite this brake of PEBP1 on LC3 lipidation, following IL-13 stimulation (to induce high 15LO1 conditions), lipidated LC3-II paradoxically increases (Fig. 1 A and B). To initially explore whether the high LC3-II observed under 15LO1 conditions is due to 15LO1 outcompeting LC3-I for binding to PEBP1 (hence reducing its overall inhibitory capacity on LC3 lipidation), a dot blot with equal amounts of recombinant 15LO1, PEBP1, or LC3 was performed and then probed with mRuby2-PEBP1 protein. Both 15LO1 and LC3 bind with PEBP1 (Fig. 3A and SI Appendix, Fig. S4A). Quantification analysis shows there is relative preference for PEBP1 to interact with 15LO1 over LC3 (SI Appendix, Fig. S4A). To confirm this biochemical observation in vitro, HAECs were simulated with IL-13 for 5 d, and total protein was harvested for Co-IP to determine binding of 15LO1 and PEBP1, as compared with LC3 and PEBP1 under IL-13 conditions. IL-13 markedly increased PEBP1–15LO1 complex formation as previously reported (8), when compared with the proportionately smaller increase in PEBP1–LC3-I complexes (Fig. 3B). Quantitative IF/confocal analysis of HAECs stimulated with IL-13 confirmed more colocalization of PEBP1 with 15LO1 than with LC3 in response to IL-13 stimulation (Fig. 3C and SI Appendix, Fig. S4B).
Fig. 3.
15LO1 outcompetes LC3 for binding to PEBP1 promoting its lipidation. (A) PEBP1 has greater binding affinity toward 15LO1 as compared with LC3 using dot-blot approach. The indicated quantities of purified recombinant 15LO1, PEBP1, and LC3B were applied to nitrocellulose, probed with Ruby-tagged PEBP1, and binding visualized by fluorescence at 585 nm, indicating that PEBP1 was able to bind 15LO1 and LC3B. (B) IL-13 induced more 15LO1–PEBP1 complexes as compared with PEBP1–LC3-I complexes (by Co-IP/western blot). (C) IL-13 induced more 15LO1–PEBP1 colocalization as compared with PEBP1–LC3 in HAECs by IF staining and colocalization analysis. Cells were all stimulated with IL-13 for 7 d. Green, PEBP1; red, LC3 or 15LO1; yellow, colocalization. (D) Intermolecular interactions observed in coarse-grained simulations. PEBP1 exhibited a higher propensity to form a complex with 15LO1 than with LC3. Snapshots of the complex formed at the end (t = 500 ns) of two independent runs are displayed. The interacting proteins are colored as per the key on the right, and L71 (magenta) on b barrel of 15LO1 is indicated for orientation purposes. IP, immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
These preferential interactions between PEBP1, 15LO1, and LC3 were further supported by molecular modeling and simulations, which also showed that 15LO1–PEBP1 complex formation was favored as compared with PEBP1–LC3 complexes (Fig. 3D and SI Appendix, Fig. S4C). Intermolecular interactions in multicomponent systems composed of a randomly positioned 15LO1 molecule, together with two PEBP1 molecules and one LC3 molecule, were simulated in a solvated (water plus ions) environment to examine the time evolution of complex formation between these molecules, if any. Three trajectories of 500 ns each were generated using the molecular dynamics (MD) simulation protocol described in Materials and Methods. Heterooligomeric complex formations were observed in two of the runs, termed MD1 and MD2 (Fig. 3D and SI Appendix, Fig. S4C). Detailed time evolution of the association and dissociation of 15LO1, PEBP1, and LC3 molecules is presented in SI Appendix, Fig. S4 C–F. PEBP1 exhibited preferential binding to 15LO1 in both cases, with both PEBP1 molecules associating with 15LO1 (Fig. 3D and SI Appendix, Fig. S4 C, D, Upper, and E, Upper). The LC3 molecules in MD1 then associated with a PEBP1 molecule, which was already bound to 15LO1 (Fig. 3D and SI Appendix, Fig. S4 D, Lower and E, Lower). In the third simulation (SI Appendix, Fig. S4F), no complexes were observed. These results demonstrate that with 15LO1 in the vicinity, PEBP1 would bind 15LO1 at the expense of its potential interaction with LC3. Both PEBP1–LC3 and PEBP1–15LO1 complexes remained associated for the remainder of the simulation period, after a complex was formed (SI Appendix, Fig. S4C).
Since PEBP1 expression does not change with IL-13 (Fig. 1B), we assumed that the increase in LC3-II is a result from the elevated affinity of PEBP1 for 15LO1 and/or higher level of 15LO1 which both enhance the expression of LC3-I for lipidation reaction (6).
Ferroptotic Hydroperoxy-Phospholipids Generated by 15LO1–PEBP1 Complexes Induce Compensatory Autophagic Responses.
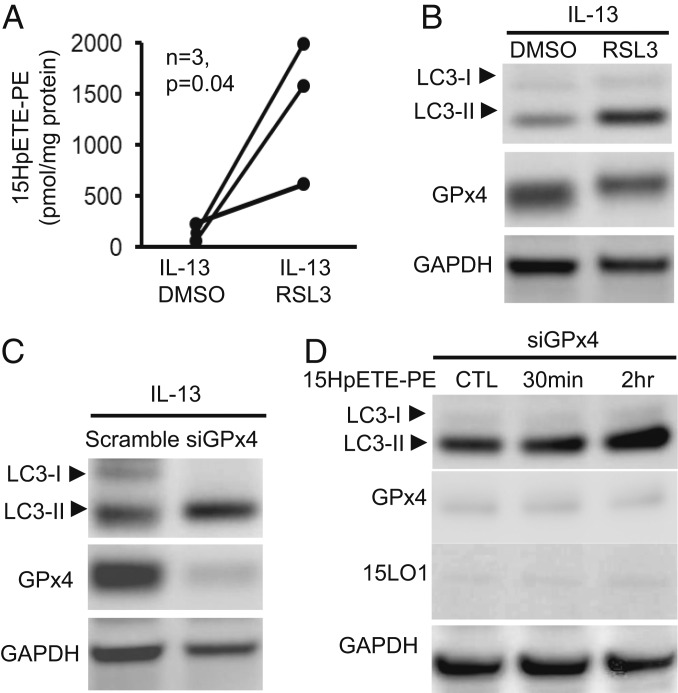
15LO1 binds PEBP1 to generate ferroptotic hypdroperoxy-phospholipids, including 15-HpETE-PE (6). Previous studies suggested lipoxygenase (LOX)-derived oxidized PE-containing phospholipids could serve as substrates for LC3 lipidation, although ferroptotic hydroperoxy-PL was not evaluated (28). To investigate whether “protective” autophagic responses occurred in response to generation of proferroptotic 15-HpETE-PE in HAECs, we first investigated whether increased autophagic activity would be observed under conditions of glutathione peroxidase 4 (GPX4) inactivation where 15-HpETE-PE levels are increased. GPX4 is the only GPX capable of metabolizing 15-HpETE-PE to its inert metabolite 15-HETE-PE (29, 30). HAECs were stimulated with IL-13 for 5 d to induce high levels of both 15LO1 and LC3 before overnight treatment with the specific GPX4 inhibitor, RSL3; 15-HpETE-PE increased as expected following RSL3 treatment of IL-13–stimulated HAECs (Fig. 4A). With RSL3/GPX4 inhibition and in the presence of high 15-HpETE-PE, LC3-II levels increased further as compared with dimethyl sulfoxide (DMSO) control (sufficient GPX4) conditions, while LC3-I levels were minimally impacted (Fig. 4B and SI Appendix, Fig. S5A). These effects were confirmed by GPX4 knockdown (DsiGPX4 for 5 d), which similarly increased LC3-II expression under IL-13 conditions, while minimally deceasing LC3-I (Fig. 4C and SI Appendix, Fig. S5B). To determine whether increases in LC3-II after GPX4 inhibition were directly induced by increased 15-HpETE-PE, exogenous 15-HpETE-PE was added to cultured HAECs in the absence of IL-13 (hence no 15LO1), as well as in the presence of DsiGPX4 knockdown to prevent its further metabolism. LC3-II increased as early as 30 min postaddition of exogenous 15-HpETE-PE, further increasing up to 2 h (Fig. 4D and SI Appendix, Fig. S5C). No significant changes in 15LO1 or GPX4 were observed. Overall, these data show that 15-HpETE-PE is not only capable of inducing ferroptosis but concomitantly of stimulating lipidation of LC3 to initiate cell-protective autophagy. Supporting these LC3-II increases, p62 was also enhanced by both RSL3 and erastin treatment (SI Appendix, Fig. S5 D and E).
Fig. 4.
Ferroptotic hydroperoxy-phospholipids generated by 15LO1–PEBP1 complexes induce compensatory autophagic responses. (A) HPLC/MS analysis showing RSL3 induced 15-HpETE-PE in HAECs stimulated with IL-13. HAECs were stimulated with IL-13 for 5 d before RSL3 treatment (10 μM, overnight), with DMSO as control. (B) RSL3 enhanced IL-13–induced LC3-II expression measured by western blot. HAECs were stimulated with IL-13 for 5 d before treatment with RSL3 (10 μM, overnight), with DMSO as control. (C) siGPx4 transfection increased IL-13–induced LC3-II as measured by western blot. HAECs transfected with siGPX4 were all stimulated with IL-13 for 5 d, with scramble siRNA as the negative control. (D) Exogenous 15-HpETE-PE induced LC3-II expression as measured by western blot in HAECs. HAECs with siGPX4 transfection were cultured under ALI (without IL-13 stimulation) for 5 d before treatment with 15-HpETE-PE (10 μg/mL), with DMPE as the control. CTL, control; HPLC, high performance liquid chromatography; MS, mass spectrometry; DMPE, dimyristoyl-sn-glycero-3-phosphoethanolamine; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
15LO1–PEBP1 Complex-Driven LC3-II Generation Limits Ferroptotic Cell Death and Extracellular Mitochondria Release.
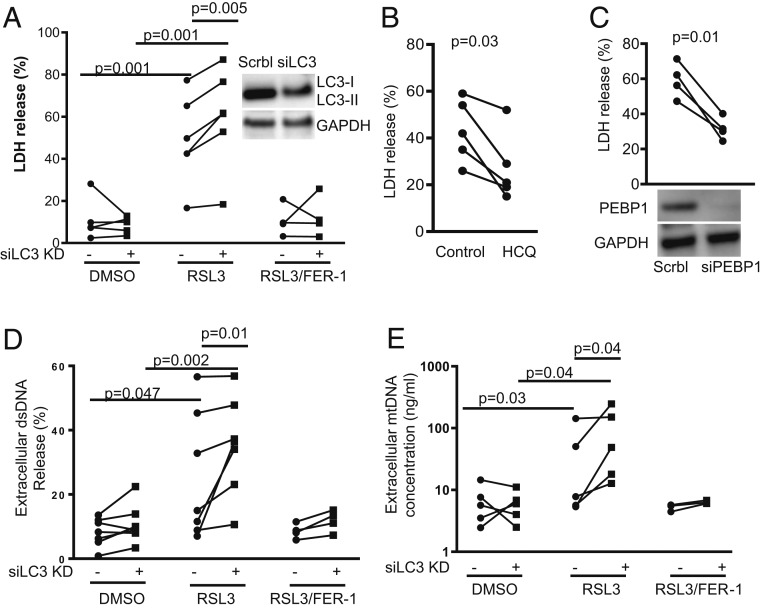
To determine whether the increased LC3-II generation, which occurs concurrently with 15LO1–PEBP1-induced generation of ferroptotic hydroperoxy phospholipids, limits overall ferroptotic cellular damage, siRNA knockdown (KD) of LC3 (DsiLC3) was performed in the presence and absence of traditional RSL3-induced ferroptosis and ferrostatin (FER-1) rescue (31). Lactate dehydrogenase (LDH) release, as a measure of ferroptotic cell death, was compared between DsiLC3 KD and scramble control. IL-13 alone did not induce cell death as indicated by low LDH release (Fig. 5A, DMSO condition). However, inhibition of GPX4 by RSL3 triggered significant LDH release as compared with DMSO control, which was rescued by FER-1 (Fig. 5A). A protective effect of LC3 was confirmed as DsiLC3 KD significantly enhanced LDH release induced by RSL3 (Fig. 5A). To confirm these results, ferroptotic cell death was further induced by erastin stimulation, which lowers intracellular GSH levels to reduce GPX4 activity. Similar to RSL3, LC3 KD enhanced LDH release induced by erastin (SI Appendix, Fig. S6A). Each of these increases in LDH was prevented by FER-1, supporting ferroptotic cell death. Similarly, HCQ and PEBP1 KD, previously shown to increase LC3-II expression (Figs. 1C and 2F), also decreased RLS3-induced LDH release (Fig. 5 B and C). To further confirm a protective effect, the human bronchial epithelial cell line was used. Unlike primary HAECs, these cells require serum for survival and proliferation. In contrast to LC3 KD, serum starvation [which induces autophagy (32, 33)] decreased erastin-induced cell death as measured by propidium iodide staining using flow cytometry, as an alternative measurement of cell death (6) (SI Appendix, Fig. S6B). These results strongly confirm a protective role of LC3-II in ferroptotic cell death.
Fig. 5.
15LO1–PEBP1 complex-driven LC3-II generation limits ferroptotic cell death and extracellular mitochondria release. (A) RSL3 induced extracellular LDH release in HAECs stimulated with IL-13, which was further increased by siLC3 transfection. All cells with/without siRNA transfection were stimulated with IL-13 for 5 d before treatment with/without RSL3 (10 μM, overnight) and FER-1 (1 μM, overnight). DMSO served as vehicle control. Cell-free culture supernatants and cell lysates were collected for LDH measurement, respectively. The results are presented as percentage released as described in Materials and Methods. (B) HCQ decreased RSL3-induced LDH release. HAECs were stimulated with IL-13 for 5 d before treatment with RSL3 (10 μM, overnight) and with/without HCQ (10 μM, overnight). The results are presented as the percentage released as described in Materials and Methods. (C) siPEBP1 transfection decreased RSL3-induced LDH release. All cells with siRNA transfection were stimulated with IL-13 for 5 d before treatment with RSL3 (10 μM, overnight). The results are presented as the percentage released as described in Materials and Methods. (D) RSL3 increased total extracellular dsDNA measured by Picogreen assay in HAECs stimulated with IL-13, which was further increased by siLC3 transfection. All cells with/without siRNA transfection were stimulated with IL-13 for 5 d before treatment with/without RSL3 (10 μM, overnight). Cell-free culture supernatants and cell lysates were collected for dsDNA measurement, respectively. The results are presented as the percentage released as described in Materials and Methods. (E) RSL3 increased extracellular MtDNA release, measured by qPCR in culture supernatants as compared with DMSO control, and siLC3 knockdown further increased extracellular MtDNA release induced by RSL3. All cells with/without siRNA transfection were stimulated with IL-13 for 5 d before treatment with RSL3 (10 μM, overnight). Cell-free culture supernatants were collected for MtDNA measurement, and the results are presented as nanograms per milliliter culture supernatants as assessed from standard curve. siLC3, LC3 siRNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
It is conceivable that the known release of Raf-1 from PEBP1 in the presence of 15LO1 and subsequent activation of extracellular signal-regulated kinase (ERK) could also drive cell death and/or autophagic survival. To investigate this, we confirmed that PEBP1 KD [previously reported to decrease cell death (6)] was associated with increased phospho-ERK (SI Appendix, Fig. S6C). We then further determined that the ERK inhibitor, PD89059, had no effect on RSL3-induced cell death, as shown by flow cytometry using propidium iodide staining and establishing that PD89059 did not act as a ferroptosis inhibitor (SI Appendix, Fig. S6 D and E). Similarly, PD89059 was ineffective in changing RSL3-induced ferroptotic response when PEBP1 was pharmacologically dissociated from Raf-1 by Sorafenib (SI Appendix, Fig. S6 D–F).
The mechanisms by which ferroptosis causes cell death are unclear, with little data to suggest disruption of nuclear membranes. However, intracellular membranes and organelles, including endoplasmic reticulum and mitochondrial membranes, may be affected as numerous studies report mitochondrial damage in cells undergoing ferroptosis (34, 35). Any insult that sufficiently damages the inner and the outer mitochondrial membranes could lead to extracellular release of mitochondrial DNA (MtDNA) (36). To determine whether ferroptotic death also increases extracellular MtDNA, extracellular double-stranded DNA (dsDNA) and specific MtDNA (MT-ND1) were measured following induction of ferroptosis in HAECs by RSL3. Similar to LDH release, RSL3 treatment increased extracellular dsDNA (measured by Picogreen dsDNA assay; Invitrogen) (Fig. 5D) and MtDNA (measured by PCR) (Fig. 5E) as compared with DMSO baseline and consistent with mitochondrial damage. Mirroring the effect on cell death/LDH, DsiLC3 KD significantly enhanced RSL3-induced extracellular dsDNA (Fig. 5D) and MtDNA (Fig. 5E) as compared with scramble control, with each rescued by FER-1. Release of extracellular histone-associated (nuclear) DNA fragments [measured by Cell death Detection ELISA; Sigma-Aldrich (37)] did not differ between conditions (SI Appendix, Fig. S6G), confirming the specific release of MtDNA in the setting of ferroptotic death.
Elevated 15LO1 in Relation to PEBP1 Associates with High LC3-II in T2-High Severe Asthmatic HAECs and Low Bronchoalveolar Lavage Fluid MtDNA.
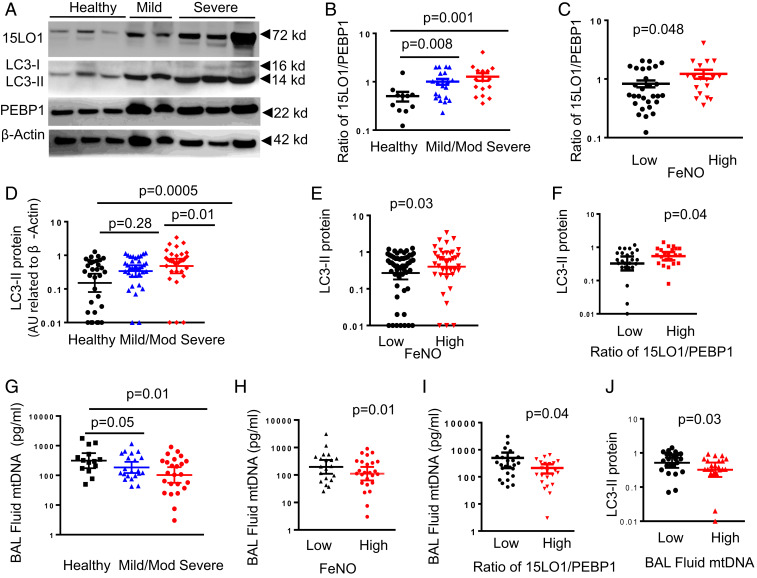
15LO1 complexes with PEBP1 in freshly brushed asthmatic HAECs, with more complexes observed in association with biomarkers of Type 2 inflammation (6). This binding promotes the generation of the ferroptotic mediator, 15-HpETE-PE. Thus, the abundance of 15LO1 relative to PEBP1 (as indicated by the ratio of 15LO1 to PEBP1) was determined in freshly brushed asthmatic HAECs by western blot and compared with healthy controls (HCs) (SI Appendix, Table S1 shows subject demographics). As expected, 15LO1 was elevated in asthmatic HAECs (Fig. 6A), while PEBP1 varied minimally across subject groups (Fig. 6A and SI Appendix, Fig. S7 A and B). The ratio of 15LO1 to PEBP1 was also higher in asthma (Fig. 6B), higher with increasing severity, and associated with higher levels of the Type 2 biomarker FeNO (Fig. 6C).
Fig. 6.
Elevated 15LO1 in relation to PEBP1 associates with high LC3-II in T2-high severe asthmatic HAECs and low BAL fluid MtDNA. Freshly brushed HAECs from asthmatic and HC participants were collected for protein detection by western blot, while BAL fluid was collected for MtDNA measurement using qPCR. (A) Representative western blots showing elevated 15LO1 and LC3-II expression in asthmatic compared with HC participants, without significant change in PEBP1 across groups. (B) Western blot densitometry (presented as arbitrary unit [AU] related to housekeeping gene expression) showing higher ratios of 15LO1 to PEBP1 in freshly obtained HAECs from severe as compared with mild/moderate (Mild/Mod) asthma and HC ex vivo and (C) higher ratios of 15LO1 to PEBP1 in freshly obtained HAECs ex vivo from subjects with high FeNO (>30 ppb) as compared with subjects with low FeNO (<30 ppb). (D) Higher LC3-II in freshly obtained HAECs from severe as compared with Mild/Mod asthma and HC ex vivo and (E) higher LC3-II in HAECs with high FeNO (>30 ppb) and (F) high 15LO1–PEBP1 ratio. (G) Lower MtDNA in BAL fluid (by qPCR) from severe as compared with Mild/Mod asthma and HC participants and (H) lower MtDNA in BAL fluid from subjects with high FeNO (>30 ppb) and (I) high 15LO1–PEBP1ratio. (J) Lower MtDNA in BAL fluid (by qPCR) from participants with higher LC3-II in HAECs ex vivo.
As our in vitro data suggested that 15LO1–PEBP1-mediated ferroptosis simultaneously induced compensatory autophagic responses, the levels of LC3-I and -II ex vivo were similarly determined by western blot. While both LC3-I and LC3-II were detectable (Fig. 6A), LC3-II was higher in asthmatic HAECs as compared with HCs, associating with greater disease severity (Fig. 6D), higher FeNO (Fig. 6E), and higher 15LO1 to PEBP1 ratios in HAECs (Fig. 6F). These results mirror the in vitro findings reflecting an environment conducive to both ferroptotic mediator generation and autophagic/mitophagic survival.
Since our in vitro results confirm that 15LO1-induced autophagy/mitophagy limits ferroptotic release of MtDNA, dsDNA and MtDNA levels in bronchoalveolar lavage (BAL) fluid were also measured. Low levels of dsDNA were detected in BAL fluid, which did not differ across groups or by FeNO (SI Appendix, Fig. S7 C and D). In contrast, abundant MtDNA was detected in HC BAL fluid, which was significantly higher than BAL fluid levels in asthmatic and particularly severe asthmatic patients (Fig. 6G). Moreover, MtDNA negatively associated with FeNO and 15LO1 to PEBP1 ratios (Fig. 6 H and I). Similar to in vitro IL-13 conditions, higher HAEC LC3-II levels associated with lower BAL fluid MtDNA (Fig. 6J), consistent with concomitant autophagic/mitophagy effects to limit MtDNA release (36). These data suggest asthmatic epithelial cells exist in a state of unhealthy balance between autophagic survival and ferroptosis, which could contribute to asthma and its severity.
Discussion
Effective aerobic life and catalysis are maintained by myriads of well-coordinated metabolic reactions occurring in perilous environments. Inevitably, this leads to accumulation of aberrant toxic products. Coping with these “life-threatening” issues is a special task fulfilled by a number of elimination catabolic processes, including autophagy. Although autophagy has been linked to a specific form of cell death (autophagic cell death), it primarily serves to eliminate damaged proteins and organelles to prevent cell death under stressed conditions. Despite this, previous studies suggested that activation of autophagy during ferroptosis promoted cell death, rather than prevented it (13, 38, 39). In contrast, data presented here identify autophagy as an antagonist of ferroptosis under Type 2/IL-13 conditions through complex interplay between three proteins, 15LO1, PEBP1, and LC3, and one hydroperoxy-phospholipid 15-HpETE-PE. The promiscuous scaffold protein PEBP1 can both inhibit autophagy by sequestering LC3 and promote ferroptotic mediator generation (15-HpETE-PE) when bound to 15LO1. However, under high 15LO1 conditions, the enzyme outcompetes LC3-I and binds to PEBP1, thus facilitating the generation of both the ferroptotic signal, 15-HpETE-PE, and lipidation of the autophagic LC3-I to LC3-II. Rather than worsening ferroptiotic cell death, or increasing a cell’s susceptibility to it, this increase in LC3-II/autophagy serves to protect the cell (and perhaps its mitochondria) from ferroptotic cell death.
Although generation of ferroptotic phospholipids drives an efficient form of oxidatively induced cell death, cells have developed protective mechanisms that promote survival rather than death. GPX4 is considered the penultimate inhibitor of ferroptotic death when it metabolizes 15-HpETE-PE to its neutral alcohol, 15-HETE-PE (40, 41). Recently, GPX4-independent protective pathways against ferroptosis have been identified. A ferroptosis suppressor protein-1 has been shown to recycle Coenzyme Q10 (CoQ10) back to its reduced state and prevent the accumulation of phospholipid hydroperoxides (42, 43).
In contrast to these molecular/biochemical inhibitors of ferroptosis, and even unexpectedly, previous studies suggested autophagy does not inhibit but rather enhances ferroptosis. Ferritinophagy, or the sequestration of ferritin by specific autophagic processes, lowers intracellular levels of the iron-binding protein ferritin, increasing intracellular iron levels and enhancing ferroptosis (44). Ferroptosis was also suggested to be promoted by loss of GPX4 through chaperone-mediated autophagy (13). Finally, the autophagic protein beclin−1 was shown to inhibit the cystine transporter System Xc– (SLC7A11/SLC3A2) (45). This inhibition then lowers intracellular GSH, which induces ferroptosis. Thus, in contrast to its primary prosurvival function, autophagy has been viewed as a proferroptotic/procell death pathway.
There is emerging understanding of central molecular mechanisms engaged by cells to determine which particular death or survival pathway to execute. Caspase-8 is increasingly recognized as such a molecule, which both initiates apoptosis and inhibits necroptosis (46, 47). Thus, the previous report that PEBP1, the protein critical to generation of ferroptotic lipids by LOXs, also bound (and sequestered) the quintessential autophagic protein LC3 (15) suggests novel mechanisms by which ferroptosis and autophagy might intersect. Our observations of concurrent ferroptotic mediator generation and autophagic activity in IL-13 stimulated HAECs further supported PEBP1 as a likely candidate for regulation of the interactions.
While the autophagy-related (ATG) pathway is considered the canonical pathway to generate autophagic membranes, many additional proteins have now been identified, including PEBP1 through its hypothesized inhibition of LC3 lipidation (15, 48). The data presented here expand on this report by showing intracellular binding of nonlipidated LC3-I to PEBP1 using co-IP, immunofluorescence, and finally, FRET. Despite this, IL-13 even more dramatically up-regulates 15LO1, which then outcompetes LC3 for PEBP1 binding, preventing it from being sequestered and promoting its availability for lipidation and activation of autophagy. Supportively, computer modeling and biochemical studies both show preferential avidity of PEBP1 for 15LO1 compared with LC3. Thus, binding of 15LO1 to PEBP1 overwhelms the inhibitory effect of PEBP1 on autophagy, increasing overall LC3 expression and freeing LC3 for lipidation under conditions of proferroptotic 15-HpETE-PE generation.
Execution of autophagy requires the lipidation of LC3 (ATG8), traditionally through the formation of a covalent bond with PE. This lipidation process is highly sensitive to membrane curvature and is strongly enhanced if the membrane bilayer contains lipid packing defects caused by cone-shaped lipids such as PE (49). Given that oxidatively modified lipids can also disrupt the bilayer organization (50), it is conceivable that oxidized PE species (15-HpETE-PE, 15-HETE-PE) may facilitate the LC3 lipidation process (28). Our results favor the involvement of HpETE-PE—generated by the 15LO1–PEBP1 complex and accumulating further with GPX4/GSH deficiency—in the LC3 lipidation process. This may be due to the conversion of HpETE-PE into oxidatively truncated species with a higher bilayer-destabilizing capacity (50) as well as their potent electrophilicity and covalent attack on the nucleophilic LC3 sites. Thus, the same hydroperoxy-phospholipid, which induces cell death (15-HpETE-PE), could simultaneously promote autophagic self-survival by serving as the phospholipid donor for LC3. Interestingly, similar to LC3-II, IL-13 also induces p62 expression in HAECs, which is further enhanced by HCQ. This suggests that rather than degradation of p62 when LC3-I is lipidated to LC3-II as seen in classic autophagy (18), p62 increases as LC3-II increases under IL-13 stimulation. These data suggest that chronic IL-13 simulation of HAECs promotes continuous expression and parallel degradation of p62 leading to an autophagic equilibrium. In fact, multiple studies have observed similar parallel increases (20–23).
None of these findings determine whether concurrent activation of autophagy enhances or protects from ferroptotic death. After GPX4 is inhibited (by RSL3), a significant increase in LDH release occurs (indicative of cell death), which is fully rescued by FER-1. In the presence of LC3 KD, the release of LDH was significantly greater than scramble control, supporting a protective effect of LC3 in this setting. Given this protective effect of autophagy in this system, and the described impact of ferroptosis on mitochondria (34, 35), we then asked whether the protective effect of autophagy might be through an effect on mitophagy. Consistent with an effect on both inner and outer mitochondrial membranes, extracellular MtDNA increased under proferroptotic (RSL3) conditions and was rescued by FER-1. This is also in line with earlier reports of the presence of 15LO1 in the outer leaflet of mitochondrial membranes (51, 52). LC3 KD significantly increased extracellular MtDNA levels, consistent with a protective effect of LC3. These results suggest that mitophagy may be the pathway by which LC3 is protecting cells from ferroptotic cell death.
Importantly, these in vitro findings are recapitulated ex vivo. Increases in LC3-II are observed ex vivo in fresh asthmatic HAECs, corresponding with increased 15LO1 expression and more severe but currently stable T2-Hi asthma. We speculate this combination of high 15LO1 and LC3-II observed in severe asthma is indicative of the same coactivation of ferroptotic phospholipid generation and autophagic, perhaps mitophagic, processes observed in vitro under IL-13 conditions. This evidence for autophagy/mitophagy is associated with lower BAL fluid MtDNA observed in asthmatic compared with healthy BAL fluid. While these lower levels could be controlled by multiple factors, the relationship to LC3-II is supportive of a possible role of autophagy/mitophagy in slowing epithelial cell turnover, as compared with healthy epithelium (53, 54). Given that excessive extracellular MtDNA released into BAL fluid during acute lung injury and pneumonia functions as a damage-associated molecular pattern (55–57), an autophagic effect in stable asthma could serve to control excessive inflammation. However, factors that increase oxidative stress would deactivate GPX4, increase ferroptotic 15-HpETE-PE, and overwhelm autophagy. Release of high amounts of MtDNA could then increase the inflammation and promote asthma exacerbations.
In conclusion, the data presented here identify fundamental dynamic interactions in vitro and ex vivo between three proteins (15LO1, PEBP1, LC3) and the hydroperoxy-phospholipid 15-HpETE-PE, the balance of which determines epithelial cell survival and death. Up-regulation of these pathways ex vivo may represent a precarious balance between cell survival and death in the asthmatic epithelium. Development of 15LO1–PEBP1-targeted interventions to normalize these processes could potentially improve asthma outcomes.
Materials and Methods
Details are in SI Appendix, SI Materials and Methods.
Reagents, Antibodies, and Primers.
Antibodies against LC3 (rabbit immunoglobulin G [IgG]) were purchased from Sigma-Aldrich; 15LO1 (rabbit IgG) was from Abnova, and PEBP1 (mouse and rabbit IgG1) was from Santa Cruz. All other bodies and reagents used are described in SI Appendix, SI Materials and Methods.
Sources of HAEC for In Vitro and Ex Vivo Studies.
HAECs were obtained by bronchoscopic brushing of asthmatic and HC airways as previously described (58). The study was approved by the University of Pittsburgh Institutional Review Board and all participants gave informed consent. HAECs were cultured in ALI under serum-free condition as previously described (8, 59). Details are in SI Appendix, SI Materials and Methods.
CRISPR Knockout of PEBP1.
PEBP1 KO cell line was established in HT22 cells. Details are in SI Appendix, SI Materials and Methods.
Coarse-Grained MD Simulations.
The web-based graphical interphase Chemistry at Harvard Macromolecular Mechanics-Graphical User Interface (CHARMM-GUI) was used to generate the molecular simulation system. Details are in SI Appendix, SI Materials and Methods.
Statistical Analysis.
Statistical analysis was performed using JMP SAS software. Data that were normally distributed were represented as means ± SEM. Log transformation was done on MtDNA in BAL fluid that was heavily skewed. Each “n” identifies the number of biologically replicated experiments from different donors. For each donor condition, technical replicates were generally run in triplicate, except for western blots, which were done in singlets due to limited sample volume. Differences between conditions and subject groups were analyzed by ANOVA. P values of <0.05 were considered statistically significant.
Data Availability.
All data are presented and available in the text and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants P01 HL114453 (to V.E.K.), R01 AI145406 (to V.E.K. and S.E.W.), P01 AI106684 (to S.E.W.), and U01 HL109086 (to S.E.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.R.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921618117/-/DCSupplemental.
References
- 1.Tait S. W., Ichim G., Green D. R., Die another way: Non-apoptotic mechanisms of cell death. J. Cell Sci. 127, 2135–2144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buja L. M., Eigenbrodt M. L., Eigenbrodt E. H., Apoptosis and necrosis. Basic types and mechanisms of cell death. Arch. Pathol. Lab. Med. 117, 1208–1214 (1993). [PubMed] [Google Scholar]
- 3.Galluzzi L. et al., Molecular mechanisms of cell death: Recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 25, 486–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W. S., Stockwell B. R., Ferroptosis: Death by lipid peroxidation. Trends Cell Biol. 26, 165–176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kagan V. E. et al., Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 13, 81–90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenzel S. E. et al., PEBP1 wardens ferroptosis by enabling lipoxygenase generation of lipid death signals. Cell 171, 628–641.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeung K. et al., Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401, 173–177 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Zhao J. et al., 15-Lipoxygenase 1 interacts with phosphatidylethanolamine-binding protein to regulate MAPK signaling in human airway epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 14246–14251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B. et al., Quantitative assessment of cell fate decision between autophagy and apoptosis. Sci. Rep. 7, 17605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu L., Chen Y., Tooze S. A., Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14, 207–215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Youle R. J., Narendra D. P., Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabeya Y. et al., LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Wu Z. et al., Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 116, 2996–3005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou W. et al., Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 12, 1425–1428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noh H. S. et al., PEBP1, a RAF kinase inhibitory protein, negatively regulates starvation-induced autophagy by direct interaction with LC3. Autophagy 12, 2183–2196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J. et al., Interleukin-13-induced MUC5AC is regulated by 15-lipoxygenase 1 pathway in human bronchial epithelial cells. Am. J. Respir. Crit. Care Med. 179, 782–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabeya Y. et al., LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima N., Yoshimori T., Levine B., Methods in mammalian autophagy research. Cell 140, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang P., Mizushima N., LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods 75, 13–18 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Zhu Z. D., Yu T., Liu H. J., Jin J., He J., SOCE induced calcium overload regulates autophagy in acute pancreatitis via calcineurin activation. Cell Death Dis. 9, 50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng X. H., Chen B., Zhang J. P., Intracellular insulin and impaired autophagy in a Zebrafish model and a cell model of type 2 diabetes. Int. J. Biol. Sci. 13, 985–995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farah B. L. et al., β-Adrenergic agonist and antagonist regulation of autophagy in HepG2 cells, primary mouse hepatocytes, and mouse liver. PLoS One 9, e98155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aisa Z. et al., Effect of autophagy on myocardial infarction and its mechanism. Eur. Rev. Med. Pharmacol. Sci. 21, 3705–3713 (2017). [PubMed] [Google Scholar]
- 24.Pelcman B. et al., 3-Substituted pyrazoles and 4-substituted triazoles as inhibitors of human 15-lipoxygenase-1. Bioorg. Med. Chem. Lett. 25, 3024–3029 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Zhao J. et al., Preferential generation of 15-HETE-PE induced by IL-13 regulates goblet cell differentiation in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 57, 692–701 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenz K., Lohse M. J., Quitterer U., Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature 426, 574–579 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Yeung K. C. et al., Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation. Mol. Cell. Biol. 21, 7207–7217 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan A. H. et al., A novel role for 12/15-lipoxygenase in regulating autophagy. Redox Biol. 4, 40–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seiler A. et al., Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 8, 237–248 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Ordoñez C. L. et al., Mild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expression. Am. J. Respir. Crit. Care Med. 163, 517–523 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Dixon S. J. et al., Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J. S. et al., Autophagy plays an essential role in cigarette smoke-induced expression of MUC5AC in airway epithelium. Am. J. Physiol. Lung Cell. Mol. Physiol. 310, L1042–L1052 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Chen R. et al., The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J. Cell Biol. 206, 173–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neitemeier S. et al., BID links ferroptosis to mitochondrial cell death pathways. Redox Biol. 12, 558–570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C. et al., Induction of ferroptosis and mitochondrial dysfunction by oxidative stress in PC12 cells. Sci. Rep. 8, 574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bueno M. et al., PINK1 attenuates mtDNA release in alveolar epithelial cells and TLR9 mediated profibrotic responses. PLoS One 14, e0218003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heiserman J. P. et al., TLR4 mutation and HSP60-induced cell death in adult mouse cardiac myocytes. Cell Stress Chaperones 20, 527–535 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buccarelli M. et al., Inhibition of autophagy increases susceptibility of glioblastoma stem cells to temozolomide by igniting ferroptosis. Cell Death Dis. 9, 841 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang R., Tang D., Autophagy and ferroptosis–What’s the connection? Curr. Pathobiol. Rep. 5, 153–159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang W. S. et al., Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedmann Angeli J. P. et al., Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doll S. et al., FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Bersuker K. et al., The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mancias J. D., Wang X., Gygi S. P., Harper J. W., Kimmelman A. C., Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song X. et al., AMPK-mediated BECN1 phosphorylation promotes ferroptosis by directly blocking system Xc(-) activity. Curr. Biol. 28, 2388–2399.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fritsch M. et al., Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature 575, 683–687 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Newton K. et al., Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature 574, 428–431 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Yang Y., Qin M., Bao P., Xu W., Xu J., Secretory carrier membrane protein 5 is an autophagy inhibitor that promotes the secretion of α-synuclein via exosome. PLoS One 12, e0180892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dancourt J., Melia T. J., Lipidation of the autophagy proteins LC3 and GABARAP is a membrane-curvature dependent process. Autophagy 10, 1470–1471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sankhagowit S., Lee E. Y., Wong G. C., Malmstadt N., Oxidation of membrane curvature-regulating phosphatidylethanolamine lipid results in formation of bilayer and cubic structures. Langmuir 32, 2450–2457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Leyen K., Duvoisin R. M., Engelhardt H., Wiedmann M., A function for lipoxygenase in programmed organelle degradation. Nature 395, 392–395 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Brinckmann R. et al., Membrane translocation of 15-lipoxygenase in hematopoietic cells is calcium-dependent and activates the oxygenase activity of the enzyme. Blood 91, 64–74 (1998). [PubMed] [Google Scholar]
- 53.Boers J. E., Ambergen A. W., Thunnissen F. B., Number and proliferation of clara cells in normal human airway epithelium. Am. J. Respir. Crit. Care Med. 159, 1585–1591 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Boers J. E., Ambergen A. W., Thunnissen F. B., Number and proliferation of basal and parabasal cells in normal human airway epithelium. Am. J. Respir. Crit. Care Med. 157, 2000–2006 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Grazioli S. et al., Mitochondrial alarmins are tissue mediators of ventilator-induced lung injury and ARDS. PLoS One 14, e0225468 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szczesny B. et al., Mitochondrial DNA damage and subsequent activation of Z-DNA binding protein 1 links oxidative stress to inflammation in epithelial cells. Sci. Rep. 8, 914 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simmons J. D. et al., Mitochondrial DNA damage associated molecular patterns in ventilator-associated pneumonia: Prevention and reversal by intratracheal DNase I. J. Trauma Acute Care Surg. 82, 120–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu H. W. et al., Transforming growth factor-beta2 induces bronchial epithelial mucin expression in asthma. Am. J. Pathol. 165, 1097–1106 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albano G. D. et al., IL-13 desensitizes beta2-adrenergic receptors in human airway epithelial cells through a 15-lipoxygenase/G protein receptor kinase 2 mechanism. J. Allergy Clin. Immunol. 135, 1144–1153–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented and available in the text and SI Appendix.