Significance
Neuron-to-neuron communication is based on synapse activity where presynapses release neurotransmitters, which activate postsynaptic neurotransmitter receptors. The various families of glutamate receptors at the excitatory synapses are responsible of the fast synaptic transmission as well as the regulation of the long-term signaling implicated in information storage and memory. The organization at the nanometer scale of these postsynaptic receptors is a key determinant for synaptic transmission efficiency. Here, we combined dual-color superresolution imaging with electrophysiology and modeling to determine how the various glutamate receptors are co-organized at the nanoscale and to what extent this organization regulates the receptor activation by a single vesicle release.
Keywords: synaptic transmission, glutamate receptors, superresolution microscopy
Abstract
The nanoscale co-organization of neurotransmitter receptors facing presynaptic release sites is a fundamental determinant of their coactivation and of synaptic physiology. At excitatory synapses, how endogenous AMPARs, NMDARs, and mGluRs are co-organized inside the synapse and their respective activation during glutamate release are still unclear. Combining single-molecule superresolution microscopy, electrophysiology, and modeling, we determined the average quantity of each glutamate receptor type, their nanoscale organization, and their respective activation. We observed that NMDARs form a unique cluster mainly at the center of the PSD, while AMPARs segregate in clusters surrounding the NMDARs. mGluR5 presents a different organization and is homogenously dispersed at the synaptic surface. From these results, we build a model predicting the synaptic transmission properties of a unitary synapse, allowing better understanding of synaptic physiology.
The discrete chemical signals mediated by glutamate release at presynapses are computed into various pathways such as rapid depolarization induced by AMPA receptors (AMPARs), calcium entry through NMDA receptors (NMDARs), and activation of long-term signaling pathways via metabotropic receptors (mGluRs). The ability of the postsynapse to integrate the presynaptic message is thought to be part of the memory storage mechanism. However, due mainly to technical limitations, glutamate receptor organization and activation profiles have not been clearly described yet. For example, most studies on the functional relationship between AMPAR and NMDAR have been performed with electrophysiology by stimulating a large number of axons, thereby mixing per se the presynaptic release probabilities, synapses where AMPARs are absent (1), and variations in postsynaptic properties.
The general view, based on imaging, electrophysiology, and biochemical experiments, is that a spine contains between 50 and 100 AMPARs (2, 3). However, only 20 to 25% of this pool of AMPARs is activated by a single vesicle release, revealing the nonsaturation of synaptic AMPARs by glutamate (4). Similar experiments on NMDARs concluded that among the 20 to 30 NMDARs present inside a spine (5), only 2 to 3 are activated by a vesicle release in the absence of magnesium block (6–8).
The nonsaturation of AMPARs and NMDARs has been explained by modeling the glutamate gradient, which rapidly fades away (9, 10). While a glutamate concentration of 4 mM is reached at the vesicle fusion pore, it decreases to 0.5 mM 100 nm away from it 100 µs after release. For this reason, the nanoscale organization of the receptors with respect to the release site shapes the amplitude of synaptic transmission (11, 12). This is particularly true for AMPARs, which have a low affinity for glutamate and whose activation is thus exquisitely sensitive to their location in the gradient. The emergence of superresolution imaging has improved our understanding of the nanoscale organization of glutamate receptors. Around half of the synaptic AMPARs are clustered in domains of around 80 nm, with the rest diffusing freely (3). This organization is stabilized by subdomains of the scaffolding protein PSD95 present inside the PSD (3, 13, 14). These AMPAR domains are co-organized with the presynaptic glutamate release site via transsynaptic adhesion proteins like neurexin and neuroligin (11, 15). Such a tight molecular organization improves the synaptic transmission efficiency, and modeling has suggested that a 100 nm shift in this prepost alignment decreases synaptic transmission by more than 30% (11).
More recently, the intimate synaptic organization of NMDARs was imaged (16, 17). They are distributed in small clusters inside the PSD, although their co-organization with respect to the release sites has not been established. Some data are available regarding the co-organization of AMPARs and NMDARs; electron microscopy studies have demonstrated a slightly higher density of NMDARs at the center of the PSD, while AMPARs are enriched in a ring at the periphery (18–20). In parallel, several studies have reported the subsynaptic organization of metabotropic receptors. In 1994, Nusser and colleagues demonstrated that AMPAR clusters are more in the center of the synapse, while mGluR1, belonging to the same subgroup as mGluR5, is found more at the periphery (21). Other electron microscopy studies also reported this perisynaptic localization of metabotropic type I family members (22).
Despite attempts to decipher the nanoscale organization of the various glutamate receptors, a clear vision of their co-organization and coactivation is still missing. Here, we combined dual-color superresolution imaging with electrophysiology and modeling to determine how the various glutamate receptors are organized at the nanoscale and to what extent they are activated by a single vesicle release.
Results
NMDARs and AMPARs Are Clustered in Spines, While mGluR5 Is Homogeneously Distributed.
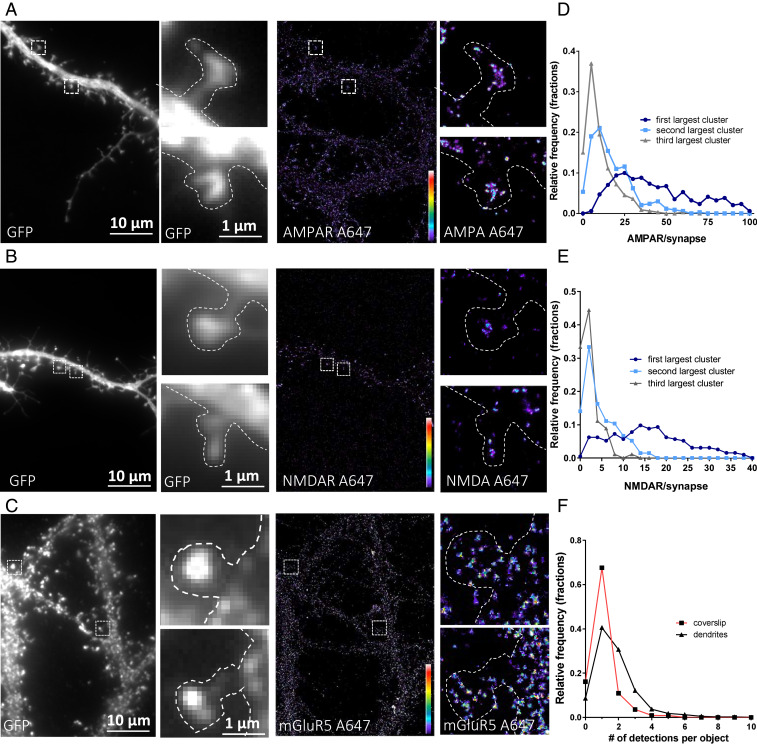
We first determined the nanoscale synaptic organization of each receptor type in hippocampal neuronal cultures using single-color immunocytochemistry and direct stochastic optical reconstruction microscopy (dSTORM). In live cells, we labeled endogenous GluA2 to visualize AMPAR (Fig. 1A), NR1 for NMDAR (Fig. 1B), and mGluR5 for mGluR (Fig. 1C) with monoclonal primary antibodies, then with secondary antibodies tagged with Alexa-647. Both GluA2- and NR1-containing receptors were organized in clusters, while mGluR5 was homogeneously distributed in the entire synapse.
Fig. 1.
Epifluorescence (Left) and dSTORM images (Right) of endogenous glutamate receptors: GluA2-containing AMPAR (A), NR1-containing NMDAR (B), and mGluR5-containing mGluR (C). Intensity is color-coded, and the scale goes from 1 (purple) to 100 (white) detections per pixel. Average spine density (D and E) represents the distribution of the estimated number of receptors per nano-object inside the synapse. Only the three largest clusters per spine are taken into account, with the others being mainly single receptors. (F) Comparison between the intensity of nano-objects on the coverslip (single secondary antibody) and on the neuron (primary and secondary antibodies on the mGluR5-containing mGluR). Both AMPAR and NMDAR present a clustered organization, while mGluR5 seems more evenly distributed.
In contrast to epifluorescence microscopy images which are blurred by diffraction, dSTORM images are more punctate and each punctum, called a nano-object, varies in size and intensity as a function of its fluorophore content. To resolve the nanoscale organization of the various receptors at the synapse, we quantified the receptor content of each synaptic nano-object from intensity-based image reconstructions (SI Appendix, Fig. 1.1A). Briefly, we first estimated the number of localizations per single receptor on the dendrite. On dendrites, there is a clear peak of distribution of individual nano-objects that, we assume, represents an individual receptor. Then, we extracted all of the individual nano-objects by image segmentation using a threshold of five localizations per 25 × 25 nm pixels to extract noise (SI Appendix, Fig. 1.1A). Finally, the number of detections in individual nano-objects was divided by the average number of detections emitted by the average smallest nano-object, which we consider to be a single receptor (11).
The distribution of the total number of GluA2-containing AMPAR receptors per spine (SI Appendix, Fig. 1.1B) presents a peaked shape with a median of 108 ± 3 GluA2-containing receptors but does not present a normal distribution, probably due to the variations in PSD size and the existence of spines with multiple PSDs (23). We then determined the receptor content for all individual nano-objects, single and clustered receptors, per synapse. This distribution revealed that more than 60% of nano-objects are composed of one or two receptors (SI Appendix, Fig. 1.1B, Inset). To further refine our analysis, we then classified the nano-object populations within each individual synapse from the one containing the highest number of receptors to the lowest. Fig. 1D represents the distribution of the largest (dark blue), second-largest (light blue), and third-largest (gray) clusters per spine. The other ones were mainly singles or pairs of receptors, as illustrated (SI Appendix, Fig. 1.1A).
The distribution of the different synaptic AMPAR clusters classified from largest to smallest (Fig. 1D) revealed that each synapse has a main domain containing between 15 and 40 receptors (average of 36 ± 1) with an average full width at half maximum (FWHM) of 97 ± 3 nm (SI Appendix, Fig. 1.1C). These main synaptic clusters correspond to the AMPAR nanodomains previously described in Nair et al. (3). The second-largest clusters contained around 12 AMPARs (12.6 ± 0.5), the third has 7 ± 0.3, and the others were isolated or distributed in clusters containing fewer than 5 AMPARs.
Concerning NMDARs, spines contained mainly between 10 and 40 NR1-containing NMDARs (median of 32 ± 3, SI Appendix, Fig. 1.1D) organized in various clusters. The largest cluster presented a FWHM of 110 ± 2 nm and was composed of about 15 ± 2 receptors. Unlike AMPARs, no significant secondary clusters were observed (Fig. 1E), with the other NMDARs being organized in objects containing few receptors. For example, the second-largest cluster contained 4 ± 0.3, and the third had 2 ± 0.2.
mGluR5 was organized differently; between 5 and 30 receptors were found per spine, with a median of 21 ± 2, and they presented a more homogeneous distribution (SI Appendix, Fig. 1.1F). Indeed, comparison of the nearest-neighbor distance between mGluR from experimental data and from simulated data (SI Appendix, Fig. 1.1G) reveals that mGluR5 distribution has a random distribution. Interestingly, the number of detections per object in the dendrites and spines (1.5 ± 0.3) was 50% larger than that of the secondary antibodies attached to the coverslip (1 ± 0.2). This could be due to either the presence of mGluR5 dimers or to the fact that multiple secondary antibodies labeled a single monoclonal antibody.
mGluR5 is widely known to interact inside the synapse with the protein Homer, which aggregates at the PSD (24). To understand the discrepancy between the previously reported role of Homer as a scaffolding protein of the PSD and the spread and homogeneous organization of mGluR5, we performed dSTORM experiments on Homer to characterize its nanoscale organization and single-particle tracking photoactivated localization microscopy (sptPALM) experiments on mGluR5 to measure its membrane diffusion properties (SI Appendix, Figs. 1.2 and 1.3). Homer displayed a highly clustered organization in disks measuring approximately the size of the PSD (area of 0.1 ± 0.003 µm2). sptPALM experiments on mGluR5, performed by expressing a protein fusion of mGluR5 with the photoactivatable fluorophore mEOS2, demonstrated a high mobility in both the dendrites and the spines, revealing no long-term trapping at the synapse. Step analysis performed to identify potential transient immobilization did not reveal any particular restriction in the diffusive behavior of mGluR5 in the vicinity of the PSD.
Hence, AMPARs, NMDARs, and mGluR5 display widely different organizations in synapses, with AMPARs displaying clusters of different sizes, while NMDARs display mainly a single cluster and mGluR5 presents a more diffuse distribution.
NMDAR and AMPAR Clusters Do Not Colocalize but Are Co-organized in a 300-nm Area.
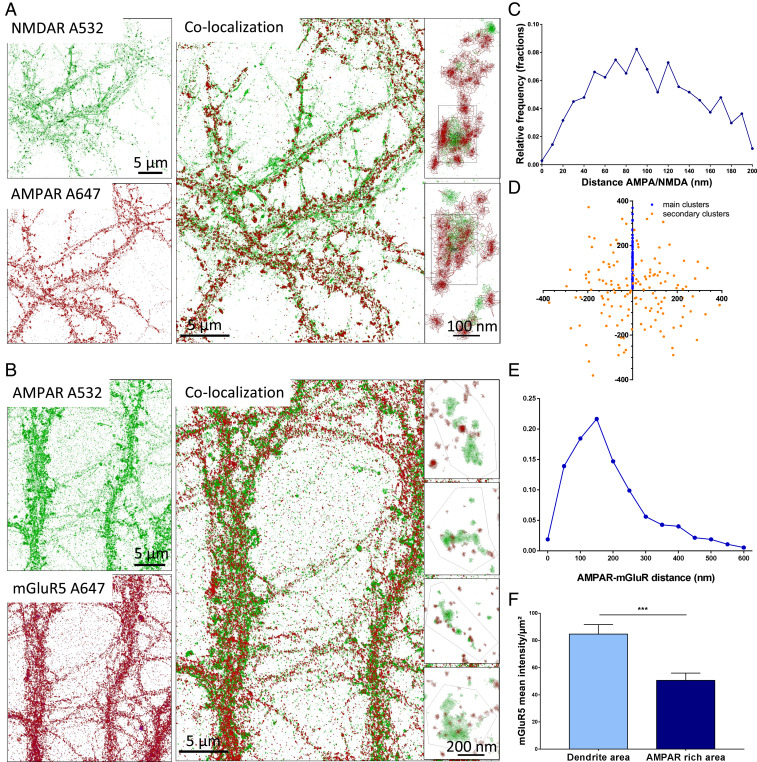
We then assessed the co-organization of the various glutamate receptors. As we failed to obtain a third color allowing superresolution imaging, we separately analyzed AMPAR/NMDAR and AMPAR/mGluR co-organization by using secondary antibodies labeled, respectively, with Alexa 647 nm and Alexa 532 nm. In our experimental conditions, we measured an average pointing accuracy of 12 nm with Alexa 647 nm dye and 28 nm with Alexa 532 nm [based on nearest-neighbor localization precision (25)]. Chromatic aberrations were corrected by using a correction matrix calculated before each experiment by taking an image of TetraSpeck beads adsorbed on the coverslip, as described in (26). The average distance over the entire field of view between the same beads in the two colors was 146 nm (interquartile range [IQR] 138–163) before correction and reached 12 nm (IQR 8–16) after correction.
To evaluate our precision in measuring the co-organization, we initially performed colabeling of endogenous GluA2/GluA1. These two AMPAR subunits are present in the same nano-objects because they mainly belong to the same tetrameric receptor (SI Appendix, Fig. 2.2 and ref. 27). Using dual-color tessellation-based colocalization analysis (26), we measured the distance between the center of the closest cluster of each color. This center is determined as a function of the repartition of the density of the detection and not only as a function of the shape of the cluster. This, centroid-to-centroid distance between GluA2 and GluA1 nanoclusters ranged from 10 to 110 nm, with an average of 58 ± 2.3 nm.
We then determined the distances between the main synaptic clusters of endogenous GluA2-containing AMPARs and NR1-containing NMDARs (Fig. 2A), considering only the clusters containing more than five receptors. The cluster-to-cluster distance distribution displays a bell-shaped curve, with an average of 100 ± 1.5 nm. Only a few clusters were closer than 40 nm, and none were farther than 200 nm. Moreover, the distribution of the cluster area overlap showed that only 20% of the clusters colocalized over more than 40% of their area, revealing a separation between the two types of clusters (SI Appendix, Fig. 2.2F). No such organization could be observed on dendrites, which can present some AMPAR clusters that are less dense and present almost no NMDAR clusters.
Fig. 2.
Co-organization of the various glutamate receptors. Dual-color dSTORM imaging is represented with Coloc-Tesseler software of NR1-containing NMDAR and GluA2-containing AMPAR (A) and mGluR5-containing mGluR and GluA2-containing AMPAR (B). For A and B, the Upper Left panel represents the 532 nm channel, and the Lower Left panel represents the 647 nm channel; the overlay is shown at the center, and synapses are zoomed in the Right. In A it appears clearly that the NMDAR cluster (in green) is surrounded by one to two AMPAR clusters (in red). C and E represent the distribution of the cluster centroid–to–cluster centroid distance for AMPAR/NMDAR (C) and AMPAR/mGluR (E). D is the distribution of the AMPAR domains around the NMDAR clusters (located at the origin of the graph). The x and y coordinates are expressed in nanometers. The larger AMPAR cluster distance is projected by rotation on the positive y axis (blue dots); the other cluster coordinates are obtained by applying similar rotation vectors (orange dots). Data represent 91 individuals synapses and 325 AMPAR clusters. (F) Because mGluR does not present clustering, its overlapping with AMPAR is not relevant, so we represent the density of mGluR single objects on the dendrite and in the area enriched in AMPAR. ***P < 0.001.
To report the relative position between AMPAR and NMDAR clusters, we computed the NMDAR–AMPAR centroid-to-centroid distances and display their spatial distribution with respect to the axis defined between the largest AMPAR cluster and the NMAR cluster (blue dots) for each synapse (Fig. 2D). We observed that secondary clusters containing more than five receptors (orange dots) are homogenously distributed around NMDAR clusters (Fig. 2D).
The distance distribution between the AMPAR clusters and the mGluR5 objects ranged between 100 and 600 nm, with an average of 187 ± 6 nm (Fig. 2E). Interestingly, there were 50% fewer mGluR5 objects inside the AMPAR domains than in the other part of the spines or inside the dendrites (Fig. 2F).
Determining the Activation of NMDAR and AMPAR by Glutamate Release during Miniature Excitatory PostSynaptic Currents (EPSCs).
After characterizing the co-organization of the glutamate receptors, we measured their activation by single vesicle release. To this aim, we initially bathed the cell in classical Tyrode’s solution deprived of Mg2+ and in the presence of tetrodotoxin (TTX). The miniature currents of AMPARs were recorded for 1 to 2 min; we then perfused 2,3-dioxo-6-nitro-7-sulfamoyl-benzo[f]quinoxaline (NBQX) (10 µM) to block them. Five micromolar glycine was added to activate the NMDARs, and the miniature currents were recorded for 1 to 2 min. The obtained NMDAR currents were very noisy; hence, to verify their specificity, we perfused them with (2R)-amino-5-phosphonovaleric acid (APV) (50 µM) to block NMDAR activity (examples of traces in Fig. 3A). The Detection Mini software was used to extract the properties of the AMPAR and NMDAR miniature currents. Fig. 3C shows an average amplitude of 14.9 ± 0.22 pA for AMPAR miniature currents and 9.5 ± 0.1 pA for NMDAR ones.
Fig. 3.
Example of AMPAR and NMDAR miniature currents. (A) From top to bottom, TTX and bicuculline are present but Mg2+ is absent (first trace), then NBQX is added to block AMPARs (second trace), glycine is added to favor activation of NMDARs (third trace), and, finally, APV is added to block NMDARs and estimate the noise level (fourth trace). (B) Average trace of AMPAR (Left) and NMDAR (Right) miniature currents. (C) Distribution of the AMPAR (black line) and NMDAR (red line) miniature current amplitudes (pA). (D) Miniature frequency (in Hz) of AMPARs (black) and NMDARs (red) on the same neurons, showing that only half of the frequency of AMPAR currents can be detected when recording NMDAR currents.
To establish the number of activated NMDARs in a single miniature EPSC, we first estimated the single-channel conductance. To this aim, single-conductance analysis was applied to currents recorded either in the presence of TTX + NBQX + glycine or when APV was added to this mixture. SI Appendix, Fig. 3.1 shows that APV application suppressed a conductance of around 4 pA. Such currents due to a channel having approximately a conductance of 50 pS and being blocked by APV can be attributed to NMDARs. To obtain the number of activated NMDARs per miniature current, we divided the distribution of NMDAR miniature currents by the single-receptor current (SI Appendix, Fig. 3.1D). In agreement with previously reported findings, we observed that an average of 2.5 ± 0.02 NMDARs were activated by a single vesicle release. This value is probably overestimated; indeed, when we compared, on the same neurons, the frequency of AMPAR- and NMDAR-detected miniatures, we obtained 4.5 ± 0.9 Hz for AMPAR miniatures and 2.3 ± 0.2 Hz for NMDAR miniatures. As the release frequency is quite stable all along a recording, this suggests that almost half of the glutamate releases either trigger NMDAR miniatures which are below the detection threshold or do not even activate any NMDARs.
Modeling of Receptor Co-organization and Coactivation with the MCell Model.
To improve our understanding of the impact of the glutamate receptor nanoscale organization on synaptic transmission, we created a model using the MCell/CellBlender software. The synaptic architecture was based on three-dimensional reconstruction of electron microscopy (EM) (3DEM) data, and the localization and the size of the PSD were determined on the basis of 3DEM images (SI Appendix, fig. 1 and ref. 28). The simulations were divided in two sequences. The purpose of the first sequence was to initialize the steady-state organization of proteins inside the synapse for 10 s with a simulation time step of 1 ms (SI Appendix, Fig. 4.1 A and B). The second sequence simulated the activation of the various receptors upon glutamate release for 200 ms with a simulation time step of 1 µs (SI Appendix, Fig. 4.1 C and D).
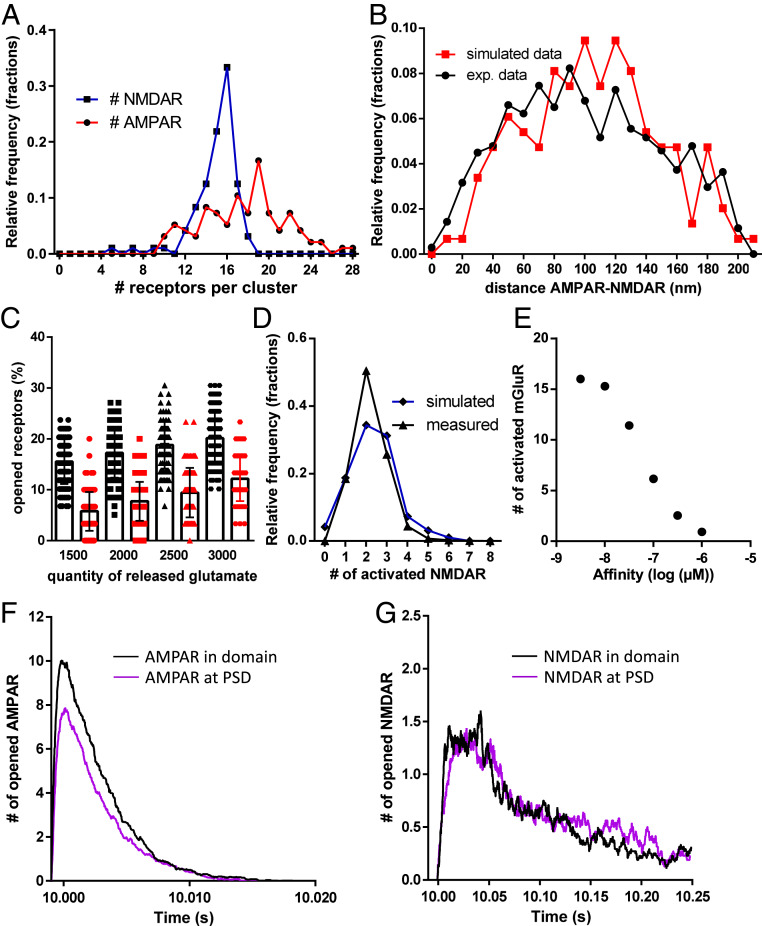
For initial conditions, the average numbers of AMPARs, NMDARs, and mGluRs experimentally measured with dSTORM were released at the virtual synapse, i.e., 60 AMPARs, 30 NMDARs, and 20 mGluRs. All receptors were set to diffuse freely on the surface at 0.1 µm2/s as classically observed with single-particle tracking experiments. Inside the PSD area, NMDARs and AMPARs were trapped by predefined spots in randomly spaced clusters. The affinity of the receptors for the traps was adjusted to obtain an average receptor content per domain similar to that observed in dSTORM (around 18 for AMPARs and 15 for NMDARs, see Fig. 4A). Fig. 4B shows the centroid-to-centroid distance of AMPAR to NMDAR clusters from experimental (black line) and simulated (red line) data. The simulated random organization between the two cluster types matched the experimental data.
Fig. 4.
(A) Simulated data representing the distribution of the accumulation of AMPARs (red line) and NMDARs (blue line) inside synaptic nanoclusters. (B) Distribution of simulated (red line) and measured (black line) data of the centroid-to-centroid distance between AMPAR and NMDAR clusters. (C) Evolution of the number of activated AMPARs (black line) and NMDARs (red line) as a function of the quantity of released glutamate. (D) Comparison between the distribution of simulated (blue line) and measured (dark line) activated NMDARs. (E) Dose–response curve of the simulated number of mGluRs activated per synapse when 2,500 glutamate molecules are released as a function of the mGluR affinity. (F and G) Simulated traces of AMPAR (F) and NMDAR (G) currents when the same number of receptors is accumulated either in nanodomains or for the entire PSD.
After 10 s of simulation, the proper steady-state organization of the various receptors was established, and glutamate was released to estimate the number of activated receptors. The release site was obtained by aligning the presynaptic site in front of the centroid of the AMPAR cluster. Between 1,500 and 3,000 glutamate molecules were released, and the number of activated AMPARs and NMDARs is shown in Fig. 4C.
The release of 2,000 glutamate molecules activated 17.5% of AMPARs and 7% of NMDARs, i.e., about 10 AMPARs and 2.5 NMDARs, as obtained experimentally. Interestingly, each step increase of 500 glutamate molecules per vesicle triggered the additional activation of 1 AMPAR and 0.5 NMDAR on average, revealing the relatively low impact of glutamate content per vesicle on synaptic currents.
To complete the validation of the model, we compared the experimentally measured distribution of the number of activated NMDARs per miniature with the simulated one, when released vesicles contained 2,000 glutamate molecules. Both distributions had comparable averages, 2.2 ± 0.02 for measured values and 2.32 ± 0.1 for simulated ones (Fig. 4D).
The similarities between the simulated and experimental data validated our model and allowed us to estimate the number of bound mGluR5 during glutamate release. Fig. 4E shows the number of activated mGluR5 as a function of their affinity for glutamate when 2,000 glutamate molecules were released. In these conditions, half of the mGluRs were activated if they had a 50 nM affinity for glutamate. With an affinity of 1 µM as described in the literature, only one receptor was activated on average (29).
Finally, we used the model to determine the efficiency of receptor activation depending of their nanoscale organization (Fig. 4 F and G). We compared the AMPAR and NMDAR activation when the same number of receptors was either clustered in a nanodomain (black lines) or randomly accumulated at the PSD (purple lines). Interestingly, AMPARs and NMDARs do not behave similarly. For AMPARs, their random accumulation at the entire PSD causes a 22% decrease in their activation compared to a nanocluster, while such organization of NMDARs does not affect NMDAR activation, meaning that NMDARs are less sensitive to their nanoscale organization than AMPARs.
In conclusion, the nanoscale clusterization of NMDARs is less important for their activation than their total number at the PSD, while for AMPAR,, the clustering clearly improves their activation and thus the synaptic response efficiency.
Discussion
Nanoscale Organization of AMPARs, NMDARs, and mGluRs.
Using dSTORM, we deciphered the nanoscale organization of the three main glutamate receptor subtypes, individually and relative to each other. Endogenous GluA2, NR1, and mGluR5 were labeled, and their quantity and nanoscale organization inside spines were characterized. As described previously, GluA2-containing AMPARs are unevenly distributed in the synapse; 50% of the identified objects are composed of one to two receptors, representing probably the mobile fraction of the receptors. The other 50% are organized in domains, with a main domain containing around 25 receptors. Other smaller domains were found to contain 5 to 15 receptors. It is not clear whether these secondary domains play a physiological role and are co-organized with presynaptic release sites or whether they form a reserve pool of receptors inside the synapse. Given the relatively low affinity of AMPARs for glutamate, in the millimolar range, it is unlikely that release in front of one AMPAR domain can activate a significant number of AMPARs in a neighboring domain (11). The larger synapses present multiple domains, but their detailed characterization is rendered difficult by the high density of receptors. They may correspond to synapses containing multiple PSDs, as described previously (23).
Concerning the organization of NR1-containing NMDARs, their organization in clusters is similar to that recently described (30), with a single main cluster and surrounding isolated receptors. Quantitative analysis revealed a single main domain composed of half of the synaptic NMDARs (∼15 receptors), with the rest being mainly in the form of single or pairs of receptors. These isolated single receptors might represent the pool of mobile receptors already reported in the literature (30, 31).
Interestingly, mGluR5 was not organized in clusters but was distributed more homogeneously, probably due to the rapid and constant lateral diffusion, as observed with sptPALM technique. This is somewhat in contradiction to the measured strong interaction with Homer 1b and 1c proteins, which are considered scaffolding proteins. This paradox could be due to a dual role of Homer, first as an interactor of mGluRs acting as a regulator of mGluR signaling (32) and, in parallel, as a scaffolding protein inside the PSD, involved in the trapping of other types of proteins. We cannot rule out that the overexpression of mGluR5–mEos used for sptPALM limited our ability to detect the few trapped receptors. However, the nonclustered organization of endogenous receptors found with dSTORM experiments is in favor of the hypothesis that mGluR5 is not tightly attached to the PSD.
Co-organization of Glutamate Receptors.
AMPAR and NMDAR co-organization was already studied with classical fluorescent microscopy techniques and electron microscopy, revealing a colocalization inside the PSD (18, 20). Here, we demonstrate that AMPAR and NMDAR clusters barely colocalize at the nanometric scale, their centroid-to-centroid distance ranging from 50 to 250 nm. This maximum distance of 200 nm could correspond to the presence of both clusters inside the PSD. Moreover, NMDAR clusters were localized mainly centrally and were surrounded by one or more AMPAR clusters. We used simulations to reproduce the bell-shaped distribution of the AMPAR–NMDAR distance. A similar curve was obtained when NMDAR clusters were placed at the center of the PSD and after placing the AMPAR clusters randomly inside the PSD (Fig. 4B).
Physiologically, this pattern of organization would be obtained if NMDARs create the initial aggregation of scaffold proteins and then AMPARs are recruited at the PSD. In that scenario, the release site would be fixed subsequently at the top of the AMPAR clusters, as previously reported (11, 15, 33). This is just one hypothesis that can be formulated to explain this random AMPAR/NMDAR colocalization inside the PSD. In any case, even if both receptor types interact with similar scaffolding proteins (mainly PSD95), they do not present colocalization. This indicates that synapses are physically able to discriminate the trapping sites for AMPAR and NMDAR. This is in line with previous experiments indicating that AMPAR and NMDAR binding to PSD95 can be differentially completed (34). It is also in line with recent results indicating that sequences upstream of the extreme PDZ ligand in AMPAR auxiliary proteins can control their binding to PSD95 (35), hence opening the possibility of differential binding of various PSD95 interactors.
The different types of organization between AMPARs and mGluRs, with AMPARs clustered and mGluRs highly mobile, clearly account for the random distribution of the distance between the AMPAR cluster centroids and the closest mGluR5. None of these receptor types presents any form of co-organization. We even observed an exclusion of mGluR5 outside of the AMPAR clusters (Fig. 2F). As this exclusion is only partial, it might simply present a form of steric exclusion due to the highly dense cluster of AMPAR complexes. Interestingly, the previously reported enrichment in mGluRs at the perisynapse could be explained by this exclusion. Indeed, exclusion of mGluRs outside of the PSD would mimic an accumulation at the perisynapse.
In conclusion, the present findings point to the central position of a unique NMDAR cluster inside the PSD with AMPAR clusters surrounding it, while mGluR5 diffuses rapidly inside the entire synapse.
Coactivation of the Various Receptor Types.
The specific organization of glutamate receptor subtypes with respect to the release site determines the signature of their distinctive activation properties. By combining electrophysiological data and simulations, we were able to estimate the number of receptors activated by the release of glutamate. Electrophysiologically, we found that each release triggers a current of 14 pA for AMPARs and 9 pA for NMDARs. The single-channel current driven by the glutamate receptor is around 1 pA for AMPARs and 4 pA for NMDARs. This means that a single glutamate release activates between 10 and 15 AMPARs and around 2 NMDARs. Such values, which are in agreement with published values (6–8), are validated by the results of simulations based on well-defined single-channel gating properties and the observed nanoscale organization.
The relatively low activation of NMDARs is partly due to the nonalignment of NMDAR clusters with glutamate release sites. Indeed, when we simulated a release in front of a NMDAR cluster, we obtained around six to seven activated NMDARs (24 to 28 pA of current), representing 20 to 25% of the total number of synaptic NMDARs. As such a current was not observed experimentally, this indicates that miniature release does not occur in front of NMDAR clusters.
Interestingly, the activation of NMDAR is more sensitive to their total number at the PSD than to their nanoscale organization. Indeed, modeling shows that the same number of NMDARs is activated whether they are inside a nanodomain or trapped at the PSD. However, similar experiments with AMPARs triggers a more than 20% decrease in the number of activated AMPARs.
In view of our model which accurately predicts AMPAR and NMDAR activation, we evaluated the number of mGluRs bound to glutamate according to their affinity. Even if their exact affinity is difficult to measure, it is in the micromolar range (29). This would lead to the low activation of mGluRs by miniature release, in the range of zero to two receptors.
Through this work, we describe the nanoscale co-organization and activation of three glutamate receptor types. While the synapse seems efficient at activating AMPARs, the organization observed in this study seems inefficient to properly activate both NMDARs and mGluR5. Their full activation would necessitate the summation of a train of inputs. Since this synaptic model seems to be able to reflect receptor activation obtained on hippocampal cell culture, it would now be interesting to determine whether such a form of organization is common to all glutamatergic synapses or is specific to this synaptic type. Simulation demonstrated that varying from 1,500 to 3,000 glutamate molecules per vesicle does not strongly affect the amplitude of the response, reinforcing the idea that the quantum value of the synaptic response is not solely dependent on the quantity of glutamate but more on the quantity of receptors and their nanoscale organization. Out of 2,000 released glutamate molecules, only 60 to 80 are used for postsynaptic glutamate receptor activation, so it could be important to determine to what extent the remaining 95% are involved in undefined signaling, such as activation of the astrocytes, feedback on the presynapse, and other pathways.
Material and Methods
Hippocampal Neuron Culture and Transfection.
Sprague–Dawley pregnant rats (Janvier Labs) were killed according to the European Directive rules (2010/63/EU). Primary hippocampal cultures were prepared according to the Banker protocol. For imaging, neurons were transfected with GFP at days in vitro 9. For sptPALM experiments, neurons were transfected with an mEOS3.2–mGluR5 construct following a Ca–phosphate protocol [described in Haas et al. (11)].
dSTORM Experiments.
For dSTORM imaging, primary neuronal cultures were labeled with monoclonal antibodies: mouse anti-GluA2 IgG2b isotype antibody, mouse anti-GluN1 IgG1 isotype antibody (provided by E. Gouaux, Vollum Institute and Howard Hughes Medical Institute, Oregon Health and Science University, Portland, Oregon), rabbit anti-GluA1 antibody (Agrobio), and rabbit anti-mGluR5 (AB5675, Merck Millipore). AMPAR and NMDAR labeling is realized on living cells at DIVs 14 to 16 by a 7 min incubation at 37 °C before paraformaldehyde fixation. mGluR5 labeling necessitated fixation and then permeabilization with Triton X-100 (0.2%) before labeling. Primary antibodies were revealed with 45 min incubation with goat anti-mouse IgG2b Alexa 647 or 532 for AMPARs (A21242, Thermo Fisher Scientific), goat anti-mouse IgG1 Alexa 532 or 647 for NMDARs (Thermo Fisher Scientific, with coupling done at the laboratory), and goat anti-rabbit Alexa 647 for mGluR5 (A21244, Thermo Fisher Scientific).
dSTORM imaging was performed as described in Haas et al. (11). Multicolor fluorescent microspheres (TetraSpeck, Invitrogen) were used as fiducial markers to register long-term acquisitions and correct for lateral drifts.
Single-molecule localization was achived using WaveTracer software operating as a plugin of MetaMorph software (36). Intensity-based drift-corrected superresolution images were reconstructed with a 25 nm pixel size, using home-made PALMTracer software operating as a plugin of MetaMorph.
Dual-Color Analysis.
Dual-color dSTORM images were analyzed using the tessellation-based colocalization analysis software (26). Two color images (532 and 647 nm) were acquired with the same dichroic mirror, and chromatic aberrations were corrected using fiduciary markers (TetraSpeck beads) using PALMTracer software.
For each color, clusters were segmented using local density factors computed from the polygons embedding each localization. Nanoclusters were segmented by thresholding the local density parameter. To compare clusters similar to the ones identified Fig. 1 on intensity-based images (of five detections per 25 nm pixel), we used a threshold density factor of 1.5, corresponding approximatively to 8,000 detections/µm2 (SI Appendix, Fig. 2.1). Only AMPAR or NMDAR nanoclusters containing more than five receptors (number of localizations of 80 for Alexa 647 nm and 35 for Alexa 532 nm, calibrated from the intensity-based images analysis) were kept for centroid-to-centroid colocalization analysis. With these conditions, we succeed at analyzing comparable domains between intensity-based and tessellation-based images (SI Appendix, Fig. 2.1).
Electrophysiology.
Miniature excitatory postsynaptic current recordings in neuronal culture were performed as described in Haas et al. (11). Extracellular recording solution was composed of the following (in mM): 110 NaCl, 5 KCl, 2 CaCl2, 10 Hepes, 10 d-glucose, 0.0005 tetrodotoxin, 0.02 bicuculline, 0.01 NBQX, 0.005 glycine, and 0.05 d-APV (pH 7.4; ∼256 mOsm/L). The pipettes were filled with intracellular solution composed of the following (in mM): 100 K gluconate, 10 Hepes, 1.1 ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid), 3 adenosine triphosphate, 0.3 guanosine triphosphate, 0.1 CaCl2, 5 MgCl2 (pH 7.3; 230 mOsm). Unless specified otherwise, all chemicals were purchased from Sigma-Aldrich, except for drugs, which were from Tocris Bioscience, and miniature EPSC analysis was performed using software developed by Michel Goillandeau (Detection Mini). Briefly, the principle of the detection used is the median filter. The program takes a window with a width set by the experimenter. For each point of the biological signal, the software calculates the median of values in the window before and after the point. The detection is not made on the biological signal but on another signal (called the detection signal), calculated from the difference between the filtered signal and the baseline signal. For further analysis, only detected events with an amplitude between 5 and 50 pA are taken into account.
Single-Channel Analysis.
Data were analyzed with the software Igor Pro-8. Current records were first corrected for baseline drift. The baseline was estimated by smoothing the data (Smooth-Loes, Smoothing = 0.2) and was subtracted from the original data. To reduce high-frequency noise, the records were then digitally filtered at a 200 Hz cutoff. Histograms were acquired using a bin width of 0.1 to 0.18 pA to obtain ∼200 bins. Histograms were fitted with sums of Gaussian using the multipeak fit module of Igor Pro-8.
Modeling.
Computer modeling was performed using the MCell/CellBlender simulation environment with MCell version 3.4. The realistic model of glutamatergic synaptic environment was constructed from 3DEM of hippocampal area CA1 neuropil as described in (28–37). The AMPAR chemical kinetic properties were obtained from the well-established model published in Jonas et al. (38), and the kinetic parameters were adjusted to fit with the recorded mEPSCs (see ref. 11). The NMDAR kinetics were obtained from Vargas-Caballero and Robinson (39). All values are reported in SI Appendix, Fig. 4.1.
Two surface properties were defined: the synapse and the PSD (identified on EM data). According to the literature and to our dSTORM data, 200 PSD95 molecules, 60 AMPARs, 30 NMDARs, and 20 mGluRs were released. They freely diffused at the synapse. Inside the PSD, PSD95 was reversibly palmitoylated (pPSD95) at a defined rate (association constant [kon] = 35, dissociation constant [koff] = 0.7).
A clusterization point called “L” was set at the center of the PSD. pPSD95 aggregates in contact with L (kon = 7, koff = 1) to form a domain. NMDARs interacted with this domain and are trapped in an NMDAR cluster (kon = 10, koff = 1).
A “G” molecule was released inside the PSD and was immobilized when it randomly interacted with a pPSD95. After immobilization, the G molecule recruited the insertion of a presynaptic neurotransmitter release site into the presynaptic membrane at the point closest to the location of G. At the same time, G clustered pPSD95 (kon = 100, koff = 1) which, in turn, clustered AMPARs (kon = 10, koff = 1). The detailed interactions with their Kd (affinity constant) and the kinetics of accumulation are reported SI Appendix, Fig. 4.2.
All this organization of PSD95, AMPARs, NMDARs, and mGluRs at the synapse was simulated with a time step of 1 ms for 10,000 iterations (10 s), until reaching a steady state (as illustrated in SI Appendix, Fig. 4.1). It is important to note that the means employed here to achieve receptor organization are only intended to give the desired final organization in our model and are not intended to model the physiological mechanisms by which this occurs in real synapses. After reaching the desired organization, the simulations were switched to a time step of 1 µs for 250,000 iterations to model the AMPAR, NMDAR, and mGluR activation when the glutamate was released at the presynaptic level, in front of G.
Sampling and Statistics.
Statistics are presented as mean ± SEM. Statistical significance tests were performed using GraphPad Prism software. At least 10 cells from three independent neuronal preparations are acquired per set of data.
Ethical Approval.
All experiments were approved by the Regional Ethical Committee on Animal Experiments of Bordeaux.
Data Availability.
The model is accessible at www.mcell.cnl.salk.edu/models/hippocampus-glutamate-receptor-organization-2020-1. All other data and associated protocols used for this study are available in the main paper and the SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge E. Gouaux for the anti-GluA2 and anti-NR1 antibodies. The Bordeaux Imaging Center, part of the France-BioImaging national infrastructure (Grant ANR-10INBS-04-0) is thanked for support in microscopy. We thank the Initiative for Neurosciences cell biology core facilities (Laboratoire d'Excellence Bordeaux Region Aquitaine [Grant ANR-10-LABX-43]), in particular, C. Breillat, E. Verdier, and N. Retailleau, for cell culture and plasmid production and Jorge Aldana from the Salk Institute for computing support. We thank Corey Butler for his advice on superresolution microscopy and colocalization analysis. This work was supported by funding from the Ministère de l’Enseignement Supérieur et de la Recherche (Grant AMPAR-trapping [ANR AMPAR-T]), Fulbright Program and the Philippe Foundation to E.H. and D.C., Agence Nationale pour la Recherche (NewOptogeneticTools, Grant ANR-15-CE11-0029-01; Laboratory of Excellence, Ion Channel Science and Therapeutics, Grant ANR-11-LABX-0015-01) to M.V., Grant ANR-16-CE13-0018 to J.-B.S., and Centre National de la Recherche Scientifique, European Research Council Grant ADOS (339541) and DynSynMem Grant (787340) to D.C.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922563117/-/DCSupplemental.
References
- 1.Bekkers J. M., Stevens C. F., NMDA and non-NMDA receptors are co-localized at individual excitatory synapses in cultured rat hippocampus. Nature 341, 230–233 (1989). [DOI] [PubMed] [Google Scholar]
- 2.Fukazawa Y., Shigemoto R., Intra-synapse-type and inter-synapse-type relationships between synaptic size and AMPAR expression. Curr. Opin. Neurobiol. 22, 446–452 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Nair D. et al., Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J. Neurosci. 33, 13204–13224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu G., Choi S., Tsien R. W., Variability of neurotransmitter concentration and nonsaturation of postsynaptic AMPA receptors at synapses in hippocampal cultures and slices. Neuron 22, 395–409 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Shinohara Y., Quantification of postsynaptic density proteins: Glutamate receptor subunits and scaffolding proteins. Hippocampus 22, 942–953 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Mainen Z. F., Malinow R., Svoboda K., Synaptic calcium transients in single spines indicate that NMDA receptors are not saturated. Nature 399, 151–155 (1999). [DOI] [PubMed] [Google Scholar]
- 7.McAllister A. K., Stevens C. F., Nonsaturation of AMPA and NMDA receptors at hippocampal synapses. Proc. Natl. Acad. Sci. U.S.A. 97, 6173–6178 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nimchinsky E. A., Yasuda R., Oertner T. G., Svoboda K., The number of glutamate receptors opened by synaptic stimulation in single hippocampal spines. J. Neurosci. 24, 2054–2064 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghavachari S., Lisman J. E., Properties of quantal transmission at CA1 synapses. J. Neurophysiol. 92, 2456–2467 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Rusakov D. A., Kullmann D. M., Extrasynaptic glutamate diffusion in the hippocampus: Ultrastructural constraints, uptake, and receptor activation. J. Neurosci. 18, 3158–3170 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas K. T. et al., Pre-post synaptic alignment through neuroligin-1 tunes synaptic transmission efficiency. eLife 7, e31755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarusawa E. et al., Input-specific intrasynaptic arrangements of ionotropic glutamate receptors and their impact on postsynaptic responses. J. Neurosci. 29, 12896–12908 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukata Y. et al., Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J. Cell Biol. 202, 145–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacGillavry H. D., Song Y., Raghavachari S., Blanpied T. A., Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron 78, 615–622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang A. H. et al., A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210–214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellermayer B. et al., Differential nanoscale topography and functional role of GluN2-NMDA receptor subtypes at glutamatergic synapses. Neuron 100, 106–119.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Ladepeche L. et al., Single-molecule imaging of the functional crosstalk between surface NMDA and dopamine D1 receptors. Proc. Natl. Acad. Sci. U.S.A. 110, 18005–18010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X. et al., PSD-95 family MAGUKs are essential for anchoring AMPA and NMDA receptor complexes at the postsynaptic density. Proc. Natl. Acad. Sci. U.S.A. 112, E6983–E6992 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kharazia V. N., Weinberg R. J., Tangential synaptic distribution of NMDA and AMPA receptors in rat neocortex. Neurosci. Lett. 238, 41–44 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Racca C., Stephenson F. A., Streit P., Roberts J. D., Somogyi P., NMDA receptor content of synapses in stratum radiatum of the hippocampal CA1 area. J. Neurosci. 20, 2512–2522 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nusser Z., Mulvihill E., Streit P., Somogyi P., Subsynaptic segregation of metabotropic and ionotropic glutamate receptors as revealed by immunogold localization. Neuroscience 61, 421–427 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Shigemoto R. et al., Immunohistochemical localization of a metabotropic glutamate receptor, mGluR5, in the rat brain. Neurosci. Lett. 163, 53–57 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Hruska M., Henderson N., Le Marchand S. J., Jafri H., Dalva M. B., Synaptic nanomodules underlie the organization and plasticity of spine synapses. Nat. Neurosci. 21, 671–682 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fagni L., Ango F., Perroy J., Bockaert J., Identification and functional roles of metabotropic glutamate receptor-interacting proteins. Semin. Cell Dev. Biol. 15, 289–298 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Endesfelder U., Malkusch S., Fricke F., Heilemann M., A simple method to estimate the average localization precision of a single-molecule localization microscopy experiment. Histochem. Cell Biol. 141, 629–638 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Levet F. et al., A tessellation-based colocalization analysis approach for single-molecule localization microscopy. Nat. Commun. 10, 2379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y., Chen S., Swensen A. C., Qian W. J., Gouaux E., Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science 364, 355–362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartol T. M. et al., Computational reconstitution of spine calcium transients from individual proteins. Front. Synaptic Neurosci. 7, 17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholler P. et al., HTS-compatible FRET-based conformational sensors clarify membrane receptor activation. Nat. Chem. Biol. 13, 372–380 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Willems J. et al., ORANGE: A CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLoS Biol. 18, e3000665 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groc L. et al., Differential activity-dependent regulation of the lateral mobilities of AMPA and NMDA receptors. Nat. Neurosci. 7, 695–696 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Suh Y. H., Chang K., Roche K. W., Metabotropic glutamate receptor trafficking. Mol. Cell. Neurosci. 91, 10–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mondin M. et al., Neurexin-neuroligin adhesions capture surface-diffusing AMPA receptors through PSD-95 scaffolds. J. Neurosci. 31, 13500–13515 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bard L. et al., Dynamic and specific interaction between synaptic NR2-NMDA receptor and PDZ proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 19561–19566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng M. et al., Phase separation-mediated TARP/MAGUK complex condensation and AMPA receptor synaptic transmission. Neuron 104, 529–543.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kechkar A., Nair D., Heilemann M., Choquet D., Sibarita J. B., Real-time analysis and visualization for single-molecule based super-resolution microscopy. PLoS One 8, e62918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinney J. P. et al., Extracellular sheets and tunnels modulate glutamate diffusion in hippocampal neuropil. J. Comp. Neurol. 521, 448–464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonas P., Major G., Sakmann B., Quantal components of unitary EPSCs at the mossy fibre synapse on CA3 pyramidal cells of rat hippocampus. J. Physiol. 472, 615–663 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vargas-Caballero M., Robinson H. P., Fast and slow voltage-dependent dynamics of magnesium block in the NMDA receptor: The asymmetric trapping block model. J. Neurosci. 24, 6171–6180 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The model is accessible at www.mcell.cnl.salk.edu/models/hippocampus-glutamate-receptor-organization-2020-1. All other data and associated protocols used for this study are available in the main paper and the SI Appendix.