Significance
While studies of domestication reveal the process of evolution under human influence, many works focused on identifying single genes showing large phenotypic effects. In this work we assembled the chromosome-level genome of bitter gourd (Momordica charantia) and investigated the genomic changes under domestication. Domestication in this species appears to be a complex process, where distinct human preferences among countries prevented the fixation of major mutations responsible for trait evolution, at the same time resulting in large phenotypic differences between geographically structured cultivar groups. Beyond strong directional selection on Mendelian traits, this work highlights the importance of other factors, such as balancing selection within and divergent selection among cultivar groups, in shaping crop diversity.
Keywords: Momordica charantia, genome assembly, domestication, artificial selection, population genetics
Abstract
The genetic architecture of quantitative traits is determined by both Mendelian and polygenic factors, yet classic examples of plant domestication focused on selective sweep of newly mutated Mendelian genes. Here we report the chromosome-level genome assembly and the genomic investigation of a nonclassic domestication example, bitter gourd (Momordica charantia), an important Asian vegetable and medicinal plant of the family Cucurbitaceae. Population resequencing revealed the divergence between wild and South Asian cultivars about 6,000 y ago, followed by the separation of the Southeast Asian cultivars about 800 y ago, with the latter exhibiting more extreme trait divergence from wild progenitors and stronger signs of selection on fruit traits. Unlike some crops where the largest phenotypic changes and traces of selection happened between wild and cultivar groups, in bitter gourd large differences exist between two regional cultivar groups, likely reflecting the distinct consumer preferences in different countries. Despite breeding efforts toward increasing female flower proportion, a gynoecy locus exhibits complex patterns of balanced polymorphism among haplogroups, with potential signs of selective sweep within haplogroups likely reflecting artificial selection and introgression from cultivars back to wild accessions. Our study highlights the importance to investigate such nonclassic example of domestication showing signs of balancing selection and polygenic trait architecture in addition to classic selective sweep in Mendelian factors.
Domestication involves human actively modifying organismal traits and is considered a good model to study the process of evolution (1). Classic examples include the TEOSINTE BRANCHED 1 (TB1) gene generating nonbranching of maize (2), the QTL of seed shattering in chromosome 1 (qSH1) gene for nonshattering in both Asian and African rice (3, 4), as well as many others. Intriguingly, these classic examples involve strong directional selection on novel mutations of Mendelian traits, which left strong signatures of hard selective sweep. On the other hand, in many plants, domestication inevitably involves the enlargement of seeds or fruits, likely highly polygenic traits where selection may only slightly alter the allele frequencies of standing variations. In some plants, the domesticated forms, semiwild forms, and wild progenitors were all utilized by humans, and the continuum of phenotypic divergence is not as discrete as in many other crops. The situation may be further complicated by the parallel selection in different countries, resulting in different sets of “domestication genes” for the same phenotype in cultivars of diverse genetic backgrounds. Therefore, to understand the process of domestication and how human might have shaped the genomes of plants, studies on these nonclassic cases are necessary. Here we focus on bitter gourd (Mormordica charantia, 2n = 2x = 22) (5).
Bitter gourd is a vegetable and medicinal plant of the family Cucurbitaceae, cultivated in tropical and subtropical Asia and characterized by its spiny skin pattern and bitter taste. Bitter gourd fruits are rich in vitamin C, minerals, and carotenes (6). The pharmacological effect of bitter gourd has been widely investigated (7), especially in type 2 diabetes. Bitter gourd fruits contain substances with the antidiabetic effect such as charatin, vicine, and polypeptide-p, which may improve insulin sensitivity and decrease blood glucose level (8). Furthermore, bitter gourd resides in a distinct clade far from all other assembled Cucurbitaceae genomes (9), providing a valuable resource for investigating genome evolution in Cucurbitaceae. While a good reference genome is strongly needed, the chromosome-level genome of M. charantia based on long-read sequencing is not yet available. The most recent publicly available M. charantia genome is a short-read scaffold-level assembly (6) as well as another short-read based assembly connected by linkage map (European nucleotide archive PRJEB24032).
Previous studies have investigated the patterns of genetic variation of M. charantia: Five clusters were identified in the collection of India cultivars (10), and three clusters were found using accessions from east and southeast Asia and 160 SSR markers (11). The most recent study, using 50 SSR markers and 114 accessions, identified three major subgroups: India, Philippines, and Thailand (12). However, all population genetics study available to date used low-density markers. To fully investigate the demographic history of domestication as well as the genetic architecture underlying domestication traits and their patterns of selection, a population genomics study from resequencing diverse genomes is needed.
Here we report the long-read genome assembly of M. charantia, currently one of the most complete assemblies among publicly available genomes in Cucurbitaceae. With population resequencing, we also investigated the genetic structure, demographic history, genomewide patterns of selection, as well as the traces of selection on specific fruit traits.
Results
Genome Architecture.
We used PacBio contigs and two linkage maps to construct the chromosome-level genome of M. charantia. In total, 2,366,274 subreads with 10,725-bp read on average, equivalent to 25.3 Gb, were acquired. The genome was assembled into 302.99 Mb in 221 contigs with 96.4% BUSCO completeness (13–15). The previous Illumina RNA-sequencing (RNA-seq) data have close to 97% of reads mapped to this reference genome with 90% properly paired, and the average read mapping rate for diverse accessions (used for population genetics analyses below) is higher than 99% with 97% properly paired.
For chromosome map development, two independent linkage maps were reconstructed using previously analyzed restriction-site associated DNA sequence (RAD-seq) data of two F2 populations (6, 16). After imputing missing marker genotypes in F2 populations, we identified 12 linkage groups from the OHB61-5 × OHB95-1A cross (6) and 10 linkage groups from the K44 × Dali-11 cross (16). The final set of 11 chromosomes was identified by comparison between the two linkage maps. By mapping de novo assembly against this chromosome map, 96.27% of the sequences (291.7 Mb, including 39 long contigs) can be anchored on chromosomes, with 28 gaps in total.
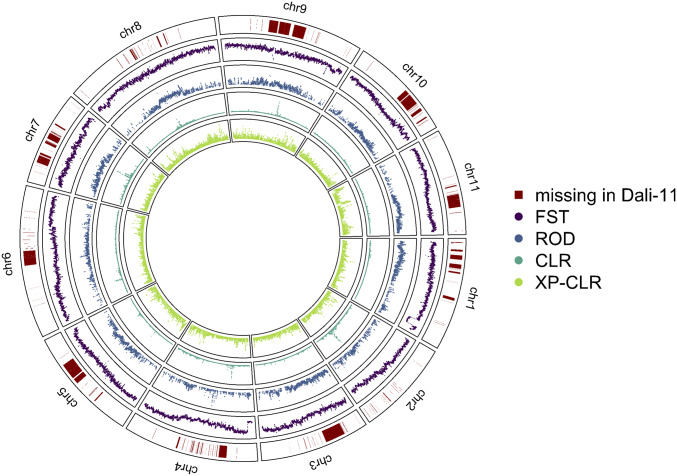
Comparing among all published Cucurbitaceae genomes (including a recent Illumina-based M. charantia assembly of the Dali-11 accession, European nucleotide archive PRJEB24032), our assembly has the highest contig N50 (close to 10 Mb), including a recently improved watermelon genome (17) (SI Appendix, Table S1). Comparison between our long-read and the recent short-read assemblies revealed that much of the centromeric regions in the long-read assembly is absent from the short-read assembly (Fig. 1 and SI Appendix, Fig. S1).
Fig. 1.
Features of the Goya v2 assembly. Shown are regions missed in the Dali-11 assembly, FST, reduction of diversity (ROD), composite likelihood ratio (CLR), and cross-population composite likelihood ratio (XP-CLR) test results.
We identified 159-Mb repetitive elements (REs), representing 52.52% of the genome (SI Appendix, Table S2). Using the same repeat annotation pipeline, we found the repeat coverage in our assembly is higher than the Dali-11 M. charantia assembly (45.43%), demonstrating the better assembly of repetitive regions. Compared to Dali-11, long-terminal repeats (LTRs), representing about 24% of the genome and 46% of all REs, are largely responsible for the higher RE proportion in our assembly (SI Appendix, Table S3). Gypsy and Copia subfamilies constitute most of the LTRs (25.6% and 15.8% of REs).
We further plotted the genome-wide distribution of each type of repeats. LTR, DNA transposons, and unknown repeats are enriched near the centromeric regions, representing the major improvement of long-read over the short-read assembly (SI Appendix, Fig. S2 A–C). For other repeat categories, short interspersed nuclear elements (SINEs) and simple repeats have similar distribution patterns to genes (SI Appendix, Fig. S2 D and E), and rRNAs had six unique clusters in the genome (SI Appendix, Fig. S2F). Interestingly, while LTRs are concentrated near the centromere, DNA transposons and long interspersed nuclear elements (LINEs) have a more pericentromeric distribution pattern (SI Appendix, Fig. S2 B and G).
Our genome assembly also allows the synteny comparison between M. charantia and six other Cucurbitaceae species (SI Appendix, Fig. S3). Between the bitter gourd and other cucurbit genomes, in general there is not a one-to-one relationship in chromosomes, indicating that these Cucurbitaceae species do not have similar karyotype to bitter gourd, consistent with the fact that the genus Momordica is in a different clade from most published Cucurbitaceae genomes (9). It is worth noting that in our assembly, the repeat-rich pericentromeric regions often have little match on other genomes, again demonstrating we have assembled regions that were previously difficult for short-read genomes. Highly conserved synteny between bitter gourd and melon (Cucumis melo) could be observed in two pairs of chromosomes (chrs) (M. charantia chr1 to C. melo chr8, M. charantia chr3 to C. melo chr12, SI Appendix, Fig. S3). Particularly, according to dotplots (SI Appendix, Fig. S4), more than 8 Mb of euchromatic region in the end of bitter gourd chr1 showed conserved synteny with chromosomes in all analyzed Cucurbitaceae plants, while inversions were sometimes observed. In Cucurbita maxima and Cucurbita moschata, bitter gourd chr1 is in syntenic to their chr3 and chr7, reflecting a known allo-tetraploidization event specific to Cucurbita species (18) but not in other Cucurbitaceae species (19).
Demographic History.
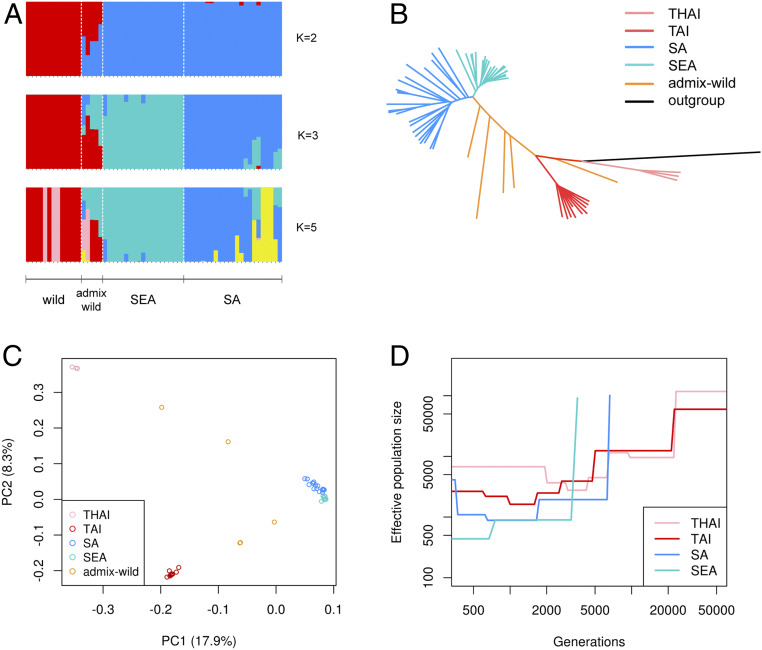
We sampled 42 cultivars, 18 wild accessions, and an outgroup (Momordica cochinchinensis) (20). Population genetic analyses from ADMIXTURE (Fig. 2A), phylogenetic tree (Fig. 2B), and principal component analysis (PCA, Fig. 2C) consistently identified four genetic groups, including two cultivar groups from South Asia (SA) and Southeast Asia (SEA) as well as wild genetic groups from Taiwan (TAI) and Thailand (THAI). These methods give largely consistent results, with ADMIXTURE K = 2 first separated wild and cultivar groups, followed by K = 3 separating the two cultivar groups. Under K = 5, the two wild groups as well as a small subgroup, Bangladesh within the SA group, were further separated. Correspondingly, the ADMIXTURE models had lower cross-validation errors under K = 3 or 5 (SI Appendix, Fig. S5). We did not observe any admixed individual between TAI and THAI wild groups probably due to the discontinuous spatial sampling, and the cultivar-wild admixture accessions (admix-wild) consistently possess introgressions from wild groups of the same geographic area.
Fig. 2.
Population structure and demographic history of M. charantia. Shown are the (A) population structure, (B) phylogenetic tree, (C) principal component analysis, and (D) demographic history of different wild (THAI and TAI) and cultivar (SA and SEA) groups.
As expected, the wild group has the most rapid decay of linkage disequilibrium (LD) among the unadmixed groups, reaching low LD (r2 = 0.25) at about 10 kb. The pattern is seconded by SA (at about 670 kb) and SEA cultivars (about 1 Mb) (SI Appendix, Fig. S6). Consistent with the patterns of LD decay, the wild group has highest mean pairwise nucleotide distance and number of heterozygous sites among all three groups while SEA has the lowest (SI Appendix, Figs. S7 and S8), suggesting the SEA cultivars represent a more recent split from the SA cultivars. Finally, the admix-wild group consisting of admixed accessions between wild and cultivars contains the highest variation and heterozygosity, consistent with their hybrid origin.
We used SMC++ (21) to infer the divergence time among these groups. Assuming one generation per year, the cultivars diverged from the wild groups at about 6,100 y ago, and the divergence between SA and SEA cultivars happened much more recently, roughly 800 y ago (Fig. 2D and SI Appendix, Fig. S9).
Genetic Architecture of Fruit Traits.
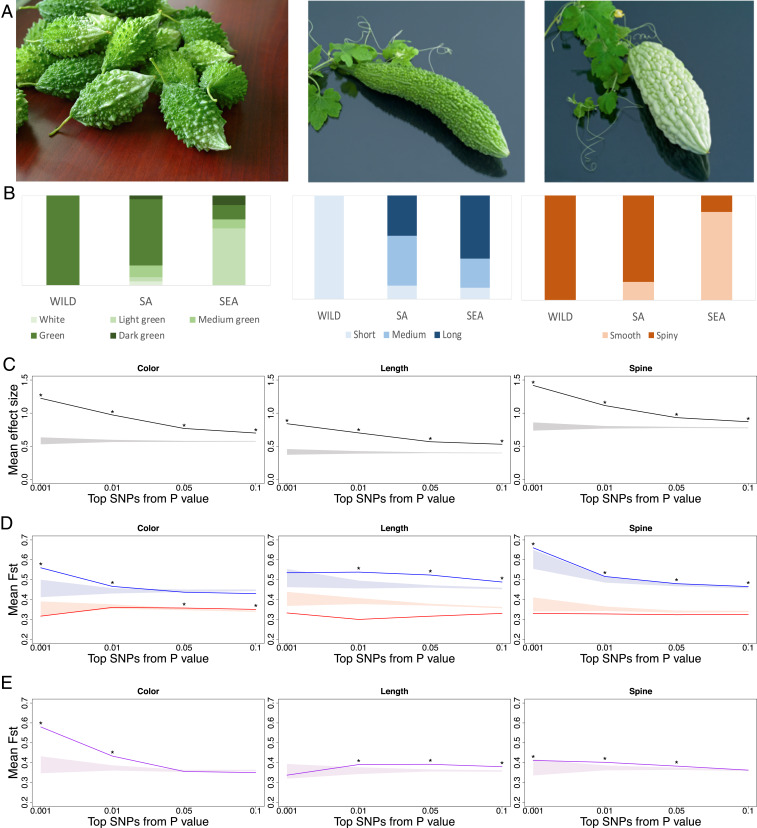
From wild to SA to SEA groups, in general the fruits became lighter in color, larger, and less spiny (Fig. 3A). Despite both being widely consumed cultivar groups, SA has higher genetic variation and more rapid linkage disequilibrium decay (SI Appendix, Figs. S6 and S7) than SEA and phenotypic characteristics more similar to wild accessions, while SEA appears to be a relatively recently derived population with extreme trait values.
Fig. 3.
Genetic architecture of selection in bitter gourd fruit traits. (A) Pictures (not to scale) showing the typical phenotypes of wild (Left, short, green, and spiny) and cultivar accessions. The typical weight of wild fruits is less than 30 g, whereas cultivar fruits could be more than 500 g. (B) Phenotypic distributions among genetic groups for fruit color, length, and presence of spines. (C–E) Relationship among top trait-associated SNPs and their mean (C) effect size, (D) FST between cultivar and wild groups, and (E) FST between the SA and SEA cultivar groups. The horizontal axes show SNPs with different magnitudes of association with traits (0.1, 1, 5, and 10% SNPs with the lowest P values). Solid lines represent mean test statistic for target SNPs, and shaded areas represent the 95% range of 1,000 resampled background SNP sets. Asterisks above solid lines represent values higher than the top 5% of background values. Blue represents WILD-SEA, red represents WILD-SA, and purple represents the SA-SEA comparisons.
To test whether the extreme trait differences of the SEA group were caused by selection or consequences of genetic drift and to investigate their genetic architecture, we calculated the association between single-nucleotide polymorphisms (SNPS) and fruit color, fruit length, and skin pattern while controlling for population structure. While we recognize the relatively lower sample size may not allow a formal genome-wide association study (GWAS), here we do not aim to identify specific GWAS peaks but instead focus on the broad patterns of SNPs’ trait association and magnitude of differentiation among populations, aiming to deduce the evolutionary pattern of these traits in different cultivar groups. We first show that, as we focused on the top 0.1%, 1%, 5%, and 10% SNPs with highest trait association (smallest P values), their mean effect sizes decline gradually as expected but are still significantly larger than the 95% range of genomic background (Fig. 3C, following a novel method in ref. 22) (Materials and Methods). In other words, even with 10% of SNPs (pruned for linkage disequilibrium), their relatively small effect sizes are still significantly larger than those possibly confounded by population structure, suggesting these traits were not controlled by only a few genomic locations with large effect sizes.
Following the same resampling method (22), we investigated the magnitude of divergence (FST) of trait-associated SNPs between wild and cultivar groups. If strong selection was driving the divergence of fruit-related traits, we expect to observe higher wild-cultivar FST in trait-associated SNPs than background SNPs. In general, after controlling for genomic background, trait-associated SNPs have significant divergence between SEA and WILD but not much so between SA and WILD (Fig. 3D), consistent with SEA’s higher magnitude of trait divergence from WILD and suggesting they were under stronger selection despite only differentiated from the SA cultivars less than 1,000 y ago (Fig. 2D). We further made the same comparison between the SA and SEA cultivars, and the results are highly consistent with those between WILD and SEA (Fig. 3E), demonstrating that the signals of selection between WILD and SEA mainly resulted from the selection during the SA–SEA divergence stage, not caused by the gradual and cumulative changes from WILD to SA to SEA.
Interestingly, these traits differ in their genetic architecture of selection. For example, SEA fruits mostly have smooth surface, and SNPs ranging from high (top 0.1%) to moderate association (top 10%) all have significantly higher differentiation with WILD than neutral expectation. On the other hand, the much lighter fruit color of SEA seems to be driven mainly by larger-effect variants (up to top 1%) while SNPs with more moderate trait association have no significant allele frequency difference from WILD or SA accessions. For fruit size, while trait divergence appears to be associated with SNPs having moderate effects (top 1%, 5%, and 10%), the top 0.1% SNPs do not have significantly higher FST than genomic background, suggesting alleles with large and opposite effects are still segregating within both cultivar populations. This pattern may partly explain why heterosis was often observed in crosses between the SEA and SA cultivars.
While further studies are required to validate the SNP-trait association, we identified potential candidate genes enriched for the top 0.1% SNPs with highest trait association (SI Appendix, Table S4). For example, for fruit length we found ANT, an ethylene-responsive transcription factor required for the development of female gametophyte, ovule integument, and gynoecium marginal tissues (23–25). For the presence of spines, we identified TRN2, a protein related with auxin transportation and involved in shoot apical meristem patterning in the peripheral zone as well as leaf patterning process (26, 27). For fruit color, APRR2 was identified, which was shown to be associated with fruit pigmentation in several species (28–30).
Signatures of Selection in the Gynoecy Locus.
We employed four methods to investigate signatures of selection during domestication: the composite likelihood ratio test (CLR) within the cultivars and the fixation index (FST), reduction of diversity (ROD), and cross-population composite likelihood ratio test (XP-CLR) between wild accessions and cultivars (Fig. 1). While these methods have individually identified putative regions with signatures of directional selection, in general we did not observe strong agreements among these methods in most regions. From each method we further chose the top 1% regions and investigated the enrichment of gene ontology (GO) functional groups. GO terms associated with metabolic processes, especially for macromolecule and organonitrogen compounds, are enriched in the genomic regions with top scores of these selection tests, suggesting the wild and cultivar groups may be differentiated in metabolism-related traits, likely associated with the unique tastes of bitter gourd fruits (SI Appendix, Fig. S10).
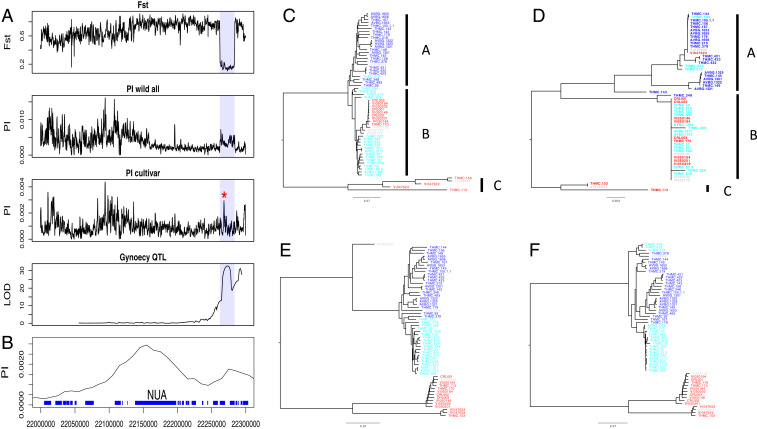
Given the high divergence between wild and cultivar groups, the baseline FST is too high to show obvious peaks. On the other hand, we observed two regions with exceptionally low FST, one near the end of chromosome 1 and the other at the beginning of chromosome 4 (Fig. 1), suggesting forces preventing the divergence between wild and cultivar groups in these regions. Interestingly, the end of chromosome 1 harbors a locus for gynoecy, affecting the ratio of male to female flowers in this monoecious species. The locus was identified in a cross between Japanese accessions OHB61-5 and OHB95-1A (31), and our reanalyses identified two closely linked quantitative trait loci (QTL) in this region, where the QTL with larger effect (with logarithm of the odds [LOD] score >30) completely overlapped this low-FST region (Fig. 4A). QTL in the same region were also identified in an independent cross between Chinese accessions Dali-11 and K44 (16), demonstrating the polymorphism conferring different flower sex ratios was shared among populations. While increasing the proportion of female flowers is the focus of continuous breeding efforts in the accession level, in the population level this locus might be under negative frequency dependent selection as either the fixation or loss of female-biased allele results in overall lower population fitness. As expected from balancing selection, levels of polymorphism in this low-FST region are high in both wild and cultivar groups (Fig. 4A). Under the high-polymorphism peak in cultivars we identified a gene NUA (NUCLEAR PORE ANCHOR) (Fig. 4B). Most of this 60-kb gene is intron, and the coding sequence constitutes 6,240 bp. Given that BLAST search identified full-length NUA genes in many other dicots and its homolog in Arabidopsis thaliana (AT1G79280) has a 6,345 bp coding sequence, we do not consider this exceptionally long gene as annotation error. In A. thaliana, mutants of NUA and ESD4 (EARLY IN SHORT DAYS 4) greatly reduce stamen length and anther size (32), suggesting NUA’s potential role in bitter gourd flower sex ratio.
Fig. 4.
The region with low differentiation between and high polymorphism within wild and cultivar groups in chromosome 1. (A) This region in chromosome 1 is labeled in shade, showing low FST, high PI, and the colocalization with the gynoecy QTL. The left half of chromosome 1 is repeat-rich centromeric regions, and the red asterisk labels the region enlarged for (B). (B) The high-polymorphism region in cultivars contains a candidate gene NUA likely affecting flower sex ratio. (C) Phylogenetic tree of the 1.77-Mb shaded area in A. The phylogeny is mostly consistent with genomewide phylogeny, except some wild accessions have obvious introgression from cultivars. (D) Phylogenetic tree of the NUA coding region, showing several balanced allelic groups with very low diversity within and high difference between groups. (E and F) Phylogenetic trees of 500-kb flanking regions upstream and downstream of the 1.77-Mb low-FST region. For C to D, tip label colors represent the population assigned based on genomewide SNPs. Red, wild accessions; gray, wild-cultivar admixed accessions; blue, South Asian cultivars; and cyan, Southeast Asian cultivars.
Phylogenetic reconstruction of this 1.77-Mb low-FST region and the coding region of NUA show distinct patterns. The 1.77-Mb low-FST region (Fig. 4C) generally follows the genomewide pattern of neutral divergence (Fig. 2B), with the exception that some wild accessions obviously harbor the allele recently introgressed from cultivars. In this region, the relative divergence between the two cultivar haplogroups versus the divergence between cultivar and original wild haplogroups (Fig. 4C, Dxy between clade A and B/Dxy between clade AB and C = 0.149) is similar to genomewide average (Fig. 2B, Dxy between SA and SEA cultivars/Dxy between cultivar and wild groups = 0.151). For the NUA gene, we observed two highly diverged haplogroups, both containing accessions belonging to the SA cultivars, SEA cultivar, and wild accessions (Fig. 4D). The relative divergence between the haplogroups is roughly equal to their divergence to the true wild haplogroup (Fig. 4D, Dxy between clade A and B/Dxy between clade AB and C = 1.072), suggesting these were highly balanced alleles that existed at least since the split between wild and cultivar groups. In the coding region of this gene, very low variation within haplogroup B and part of haplogroup A was also observed (Fig. 4D), and many cultivars and the wild accessions possess identical coding region sequences, suggesting a rapid expansion of these alleles in the cultivars and introgression back to some wild accessions. Finally, given the abrupt change of FST at the boundaries of this 1.77-Mb segment (Fig. 4A) instead of a typical valley reflecting continuous recombination, we hypothesize this could be a structural rearrangement between wild and cultivar groups, where a new structural variant was introgressed from cultivars into wild accessions, elevating the polymorphism of the whole 1.77-Mb region in wild accessions. As a support, the 500-Kb flanking regions immediately upstream and downstream of this region have phylogenetic trees reflecting the genomewide pattern (Fig. 4 E and F).
In summary, we observed complex patterns in the candidate gene NUA: The balanced distribution of highly differentiated haplogroups among populations is consistent with patterns of balancing selection, and the exceptionally low variation within haplogroups suggests recent selective sweep. Introgressions from cultivars back to wild accessions were also observed, which rapidly increased the frequency of a linked 1.77-Mb chromosomal segment, causing the high variation within wild accessions and low differentiation between wild and cultivar accessions. While most studies reported how domestication efforts left strong signatures of directional selection in the genome, here we report a more complicated case likely involving balancing selection, selective sweep, and introgression of a chromosomal segment from cultivars back to wild progenitors.
Discussion
Unlike classic examples of domestication where progressive evolution of key traits have been observed from wild progenitors to landraces to elite cultivars, the direction of selection in bitter gourd is not ubiquitous: Consumers from different cultures have their own preferences. South Asians like highly bitter fruits with smaller size (although still larger than wild progenitors) and spiny, dark green features. Southeast Asians like less bitter fruits with light green (or white) and smooth skin (12). Considerable phenotypic variations therefore exist for M. charantia cultivars. Further, some cultures may also value and directly cultivate the wild progenitors for consumption, increasing the chance of introgression between wild and cultivar accessions, a situation also observed in the pepper (Capsicum) species in Latin America (33). Under such situation, it is therefore conceivable that it would be difficult to identify classic signatures of selective sweep and strong Mendelian genes, given that the most obvious target of selection is fruit size, likely a polygenic trait, and even for fruit size we did not identify strong signs of selection in the SA cultivar group (Fig. 3D, the fruit-size-associated SNPs actually have lower FST than neutral expectation between SA and WILD). Therefore, the process of selection may be slower in bitter gourd, with introgressions between wild and cultivar groups preventing the strong and rapid fixation of domestication genes. Such situation, on the other hand, suggests potential for the further improvement of bitter gourd, as we have shown that the top 0.1% SNPs associated with fruit size are not yet highly diverged between cultivar and wild accessions. The improvement of bitter gourd cultivars is a current focus of the World Vegetable Center to provide more nutritious and climate/pest/disease resistant vegetables for the developing countries.
Traditionally, bitter gourd was separated into two “varieties” where M. charantia var. charantia refers to cultivars and M. charantia var. muricata refers to wild accessions. Such distinction, however, is mostly based on fruit size without much genetic information (34). While our samples collected from the wild environment all have muricata-type morphology, among the “cultivar accessions” conserved by the stock center, THMC113, THMC153, and THMC170 (SI Appendix, Table S5) have the typical muricata fruits and are genetically close to the wild groups. These accessions therefore represent good examples where people still directly cultivate and consume wild accessions and treat them as “cultivars.” Of particular interest is accession THMC113, originally recorded as a cultivar collected in Belize but is genetically close to the TAI wild group. As Central America does not appear to be the native range of M. charantia, this accession may be a more recent introduction of an old-world wild progenitor into the new world as a cultivar. At the same time, we recognize that our samples of wild accessions may only cover a small portion of the native range, and a more thorough expedition is required to investigate the global diversity of wild M. charantia.
In bitter gourd, the SA and SEA cultivar groups exhibit very different patterns: The SA cultivar group first diverged from wild progenitors at about 6,000 y ago, with higher diversity, faster LD decay, and phenotypes slightly closer to the wild progenitor. The SEA group later separated at about 800 y ago, with much lower diversity, slower LD decay, and highly distinct phenotypes. For trait-associated SNPs, we also identified significantly higher FST than background SNPs in SEA-WILD and SEA-SA but not much so in the SA-WILD comparison. Taken together, unlike classic Mendelian examples such as the loss of branching in maize (2) or the loss of shattering in rice (3, 4) where large phenotypic and genetic changes occurred during the domestication process from wild progenitor to landraces, our results suggest that the SA and SEA cultivar groups may represent two different stages of domestication, with SA being relatively closer to wild accessions and the SEA group further exhibiting low diversity, extreme trait values, as well as highly differentiated SNPs with trait association.
Interestingly, such two-stage patterns of quantitative trait evolution have also been observed in maize (35), watermelon (17), and tomato (36). In maize, the signatures of selection appear to be stronger in the domestication (from wild progenitor to landrace) rather than the improvement (from landrace to modern inbred lines) stage (35). For the increase of fruit size and sugar content in watermelon, the largest phenotypic changes and signs of selection were also observed during the domestication rather than the improvement stage (17). Different from these examples, the larger magnitudes of phenotypic changes in bitter gourd do not exist between wild progenitors and cultivars but likely between two geographically distinct cultivar groups, partly due to the aforementioned cultural preferences, providing an interesting case in domestication studies.
In contrast to classic examples of selective sweep, we found one region with very low divergence between wild and cultivar groups colocalizing with the locus conferring gynoecy. Despite being a continuous focus of breeding efforts (31), we observed two highly diverged haplogroups balanced within the cultivars. While we identified potential signs of rapid spread of alleles within each haplogroup, this region did not show an overall sign of selective sweep in most tests due to the existence of balanced haplogroups in both cultivar groups as well as wild accessions. Taken together, our investigations showed that the bitter gourd may provide a valuable nonclassic model of domestication, where the intermittent weaker selection with different directions and polygenic genetic architecture precludes the identification of strong single candidate genes, and the directional artificial selection for gynoecy cannot overwhelm the force of balancing selection in nature.
Materials and Methods
Genome Assembly.
High molecular weight genomic DNA was extracted from the leaves of M. charantia OHB3-1 accession following the protocol provided from Pacific Biosciences with modification. Briefly, genomic DNA was extracted from the leaf tissue using Carlson lysis buffer containing cetyl trimethyl ammonium bromide (CTAB) and precipitated by ethanol after chloroform/isoamyl alcohol extraction. RNase- and proteinase-treated genomic DNA was purified using Genomic-tip (Qiagen). SMART library was prepared from high molecular weight genomic DNA (>50 kbp) and applied to sequencing by PacBio Sequel.
Subreads from PacBio sequencing were corrected and assembled using Canu 1.7 with default settings for PacBio (13, 14). The obtained contigs were polished by pilon 1.23 using paired-end Illumina HiSeq2500 reads (250b × 2) from the same genomic DNA (15).
Restriction-site associated DNA sequence (RAD-seq) data were obtained from two F2 crosses: 97 F2 individuals from a cross in Japan (6) and 423 F2 individuals from a cross in China (16). In order to solve the low coverage and high missing-data problem in RAD-seq data, we employed a window-based method to define marker genotypes (37). Briefly, the genome was cut into 100-kb windows, and the parental genotype of each F2 individual within each window was called based on the proportion of parental reads within the window. SNPs with allele depth (AD) <3 and maternal allele proportion ≥95% or ≤5% across all samples were excluded. For 100-kb windows used for linkage map construction, if the depth of a sample in a window is lower than 5, we called it missing, and a window was excluded if the proportion of missing individuals is higher than 60%.
MSTmap (38) was used for constructing linkage maps, and filtering of genotyping errors and data imputation were applied. We identified 12 linkage groups from the Japanese cross (6) and 10 linkage groups from the Chinese cross (16). The final set of 11 linkage groups was identified by comparison between the two linkage maps. JCVI-ALLMAPS v0.8.12 (39) was used to combine the two linkage maps and produce the chromosome-level assembly. Scaffolds smaller than 10 kb were excluded from the construction. We set the weight of Japan linkage map to 1.5 and Chinese map to 1 since our genome accession was genetically closer to parents in the Japan cross.
Synteny blocks were identified between the genomes of bitter gourd and other Cucurbitaceae species. Sequences of pseudomolecules and generic feature format (GFF) files for the predicted genes in Cucumis melo (40), Cucumis sativus (41), Citrullus lanatus (42), C. maxima, C. moschata (18), and Lagenaria siceraria (19) were applied to the analysis by SyMAP 4.2 (43) with default settings.
Gene Annotation.
We performed repeat annotation by RepeatMasker 1.332 (44) with a de novo repeat library constructed by RepeatModeler 1.0.11 (45) and Repbase (46). We used ab initio gene prediction and RNA-seq data for gene annotation. RNA-seq data from three tissues, root (SRR3535149), leaf (SRR3535138), and flower (SRR3535137) were mapped to the genome by HISAT 2 2.1.0 (47) and subsequently assembled and merged by StringTie 1.3.5 (48). We used TransDecoder 5.5.0 (49) to predict the ORF based on assembled transcripts, followed by the use of parameter “retain_blastp_hits” to validate the result using blastp 2.8.1 (50) on UniProt (51) database. Ab initio gene prediction by AUGUSTUS 3.3.2 (52) was performed with the repeat-masked genome with “-species Arabidopsis” option. The species parameters of AUGUSTUS were trained by genome mode BUSCO 3.0.2 (53) with eudicotyledons_odb10 database.
The ab initio predictions, RNA-seq alignments, and ORF predictions were submitted to Evidencemodeler 1.1.1 (EVM) (54) to identify consensus gene model. The weight of ab initio and ORF prediction is 1, and the weight of RNA-seq data is 10, based on the recommendation of EVM. The gene set from EVM was sent to BUSCO for assessing the completeness with eudicotyledons_odb10 database.
The complete gene set was loaded into blast2go 5.2.5 (55) and compared with UniProtKB/Swiss-prot (51) database using local blastx. Blast E-expectation value (E-value) cutoff was set to 0.001 and word size to 6. Moreover, we mapped the genes annotated by blastx to the GO database. The mapped GO terms were further evaluated by GO evidence codes, which indicated the experimental and computational evidence of GO terms. GO enrichment analysis of genomic regions with signatures of selection was implemented with Fisher’s exact test.
Plant Materials and Population Genetics Analyses.
A total of 60 M. charantia accessions were used for population genomics analyses (20). Our samples consist of M. charantia var. muricata (small-fruit) type accessions collected from wild environments (CRL and VI accessions in SI Appendix, Table S5) as well as cultivars with mixed fruit sizes from the World Vegetable Center collections (AVBG and THMC accessions in SI Appendix, Table S5). Many of the accessions used here were collected by indigenous projects focusing on landraces and wild accessions (56). As we have discussed, some people directly cultivate and consume wild accessions, and therefore some accessions originally classified as cultivars by the stock center might actually be phenotypically and genetically close to wild accessions. The original wild-cultivar distinction therefore does not necessarily agree with population genetics results, and we chose to use the genetic groups separated by genetic data for all following analyses. The phenotypic data were received from the World Vegetable Center East and Southeast Asia, Thailand. All of them are categorical and graded data (SI Appendix, Table S5). The estimation method of the phenotypes had been reported in a previous study (12). The outgroup, M. cochinchinensis, were obtained from a horticulture market in Taiwan, and its species identity was validated with chloroplast MaturaseK gene (MatK) markers (SI Appendix, Table S6) (57).
The genomic DNA was extracted from leaves using DNeasy Plant Mini Kit (Qiagen) with 100 mg of leaf tissue, and DNA quality and concentration were estimated with gel electrophoresis and Qubit. NEBNext Ultra II DNA Library Prep Kit was used to construct the illumina library, and the libraries were sequenced with 150 bp paired-end using Illumina HiSeq X-ten.
Reads were trimmed base on sequence quality by SolexaQA++ v3.1.7.1 (58), and adaptor sequences were removed by cutadapt 1.14 (59). Reads were mapped to the reference genome by BWA 0.7.15 (60). The duplicated reads produced by PCR were marked with Picard Tools 2.9.0-1 (http://broadinstitute.github.io/picard). SNP genotypes were called following GATK 3.7 (61) best practice. Variant sites were then filtered with vcftools v0.1.13 (62) by keeping biallelic SNP sites only, QUAL >30, missing rate <10%, and minor allele frequency (MAF) >1%. Sites with depth among all samples lower or higher than 3 SDs of genome‐wide average were filtered out. The first step of filtering resulted in 9,743,755 SNPs including the outgroup.
PLINK v1.90b4.5 (63) was used to perform SNP LD pruning in 50-kb windows, 5 kb between each step, and r2 threshold of 0.5. This results in 1,159,323 SNPs for the following population structure analyses. The neighbor-joining tree was reconstructed with TASSEL 5.0 (64). PCA was performed by PLINK (63) with default settings. Ancestral proportion analysis was performed by ADMIXTURE 1.3.0 (65), and the admixture Q matrix was plotted by R package pophelper 2.2.5 (66).
For LD day, nucleotide diversity estimation, demography, and following analyses testing for traces of selection, we used the original SNP dataset without pruning for linkage disequilibrium nor filtering for minor allele frequency. In total this dataset without the outgroup species contains 6,135,286 SNPs, with different number of SNPs being used for analyses specific to each population. LD decay was calculated and plotted with PopLDdecay 3.40 (67). We removed the admixed individuals identified by ADMIXTURE before LD estimation within each genetic group. Nucleotide diversity was calculated in 50-kb windows with 10-kb steps by vcftools. Heterozygosity of each individual was counted by vcftools with “–het” option. We used SMC++ v1.15.2 (21) to estimate the demographic history of M. charantia. SMC++ had two advantages: 1) It required only unphased genomes, which was suitable for nonmodel organisms; and 2) multiple samples could be included in the analysis for constructing recent history. The admixed individuals in each group were excluded before analyses. Historical population sizes of four genetic groups, THAI, TAI, SA, and SEA were separately estimated with the “estimate” option, and their pairwise divergence times were estimated by “split” option. After summarizing the mutation rates frequently used for eudicots, the mutation rate per generation was set as 2 × 10−8.
The wild group we used in selection models was the Taiwan wild group since it was genetically closer to the cultivars. The fixation index (FST) between wild and cultivar populations was calculated in 50-kb windows with 10-kb step size by vcftools. Reduction of diversity (ROD) was calculated in 50-kb windows with 10-kb step size between the wild and cultivar populations. The formula was: log10 (πwild/πcultivar). CLR (68) was performed within cultivars by SweeD 3.0 (69), where each chromosome was separated into 2,000 bins. XP-CLR (70) was estimated between wild and cultivar populations in 50-kb windows with 10-kb step size.
To investigate the genetic architecture of fruit traits, we estimated the association between SNPs and traits while controlling for the PCA values of genomic background. To ensure relative independence among SNPs, all following analyses were performed with SNPs further pruned for LD, and SNPs with minor allele frequency less than 0.1 were excluded, resulting in about 154,000 SNPs. For each trait separately, we obtained the top 0.1%, 1%, 5%, and 10% of LD-pruned SNPs with lowest P values and estimated their mean effect sizes (in units of trait SD) as well as the FST values between genetic groups. To test whether the observed test statistics (effect size and FST) of the target SNPs deviate significantly from genomewide average, we employed a novel resampling method (22). Specifically, SNPs were separated into 400 grids, consisted of 20 bins based on local LD by 20 bins based on MAF. For a specific set of target SNPs (for example, the 1% SNPs with the lowest P values for fruit length), the number of these SNPs in each of the 400 grids were first calculated, and equal amounts of background SNPs were sampled from the same grids and the test statistic was calculated. The process was repeated 1,000 times, resulting in the distribution of 1,000 genomewide mean test statistics with the same patterns of local LD and MAF as the target SNPs.
Data Availability.
The assembled genome is available under DNA Data Bank of Japan, accession number BLBB01000001-BLBB01000193. The PacBio reads were submitted under DNA Data Bank of Japan, accession number DRA009109. The Illumina reads of the OHB3-1 genome accession was submitted under DNA Data Bank of Japan, accession number DRA009106. Population re-sequencing Illumina reads were submitted under NCBI BioProject PRJNA578358.
Supplementary Material
Acknowledgments
We thank Chia-Yu Chen, Pei-Min Yeh, Jo-Wei Hsieh, and Jo-Yi Yen for assistance; Maarten van Zonneveld for valuable comments; and those who devoted their time collecting and conserving bitter gourd germplasm. We are grateful for the support from National Taiwan University’s Computer and Information Networking Center for their high-performance computing facilities and College of Life Science Technology Commons for their molecular biology facilities. This work was supported by Taiwan Ministry of Science and Technology Grants 107-2636-B-002-004 and 108-2636-B-002-004, Japan Society for the Promotion of Science, Grant-in-Aid for Scientific Research Grants JP17K07601 and 16H06279 (Platform for Advanced Genome Science), and long-term strategic donors to the World Vegetable Center: Republic of China (Taiwan), UK aid from the UK government, US Agency for International Development, Australian Centre for International Agricultural Research, Germany, Thailand, Philippines, Korea, and Japan.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The assembled genome is available under DNA Data Bank of Japan, accession nos. BLBB01000001–BLBB01000193. The PacBio reads were submitted under DNA Data Bank of Japan, accession no. DRA009109. The Illumina reads of the OHB3-1 genome accession was submitted under DNA Data Bank of Japan, accession no. DRA009106. Population resequencing Illumina reads were submitted under NCBI BioProject PRJNA578358.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921016117/-/DCSupplemental.
References
- 1.Meyer R. S., Purugganan M. D., Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Wang R.-L., Stec A., Hey J., Lukens L., Doebley J., The limits of selection during maize domestication. Nature 398, 236–239 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Konishi S. et al., An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Wang M. et al., The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46, 982–988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaman M. Y., Alam S. S., Karyotype diversity in three cultivars of Momordica charantia L. Cytologia 74, 473–478 (2009). [Google Scholar]
- 6.Urasaki N. et al., Draft genome sequence of bitter gourd (Momordica charantia), a vegetable and medicinal plant in tropical and subtropical regions. DNA Res. 24, 51–58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan S. P., Kha T. C., Parks S. E., Roach P. D., Bitter melon (Momordica charantia L.) bioactive composition and health benefits: A review. Food Rev. Int. 32, 181–202 (2016). [Google Scholar]
- 8.Krawinkel M. B., Keding G. B., Bitter gourd (Momordica Charantia): A dietary approach to hyperglycemia. Nutr. Rev. 64, 331–337 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Renner S. S., Schaefer H., Phylogeny and Evolution of the Cucurbitaceae. Genetics and Genomics of Cucurbitaceae, (Springer, 2016), pp. 13–23. [Google Scholar]
- 10.Gaikwad A. B. et al., Amplified fragment length polymorphism analysis provides strategies for improvement of bitter gourd (Momordica charantia L.). HortScience 43, 127–133 (2008). [Google Scholar]
- 11.Saxena S. et al., Development of novel simple sequence repeat markers in bitter gourd (Momordica charantia L.) through enriched genomic libraries and their utilization in analysis of genetic diversity and cross-species transferability. Appl. Biochem. Biotechnol. 175, 93–118 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Dhillon N. P. S., Sanguansil S., Schafleitner R., Wang Y.-W., McCreight J. D., Diversity among a wide Asian collection of bitter gourd landraces and their genetic relationships with commercial hybrid cultivars. J. Am. Soc. Hortic. Sci. 141, 475–484 (2016). [Google Scholar]
- 13.Matsumura H., Hsiao M.-C., Toyoda A., Taniai N., Miyagi N., Tarora K., Urasaki N., Lee C.-R., Momordica charantia DNA contig, BLBB01000001-BLBB01000193, DDBJ Annotated/Assembled Sequences database. http://getentry.ddbj.nig.ac.jp/getentry/na/BLBB010000001/ to http://getentry.ddbj.nig.ac.jp/getentry/na/BLBB010000193/. Deposited 7 Nov 2019.
- 14.Matsumura H., Momordica charantia PacBio Sequel sequencing, DRA009109, DDBJ Sequence Read Archive. http://trace.ddbj.nig.ac.jp/DRASearch/submission?acc=DRA009109. Deposited 17 October 2019.
- 15.Matsumura H., Momordica charantia Illumina sequencing, DRA009106, DDBJ Sequence Read Archive. http://trace.ddbj.nig.ac.jp/DRASearch/submission?acc=DRA009106. Deposited 15 October 2019.
- 16.Cui J. et al., A RAD-based genetic map for anchoring scaffold sequences and identifying QTLs in bitter gourd (Momordica charantia). Front Plant Sci 9, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo S. et al., Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat. Genet. 51, 1616–1623 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Sun H. et al., Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant 10, 1293–1306 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Wu S. et al., The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring-spot virus resistance locus. Plant J. 92, 963–975 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Hsiao M.-C., Anand S. S., Schafleitner R., Lee C.-R., Population whole genome sequencing of Momordica charantia, PRJNA578358, National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov/bioproject/578358. Deposited 18 October 2019.
- 21.Terhorst J., Kamm J. A., Song Y. S., Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49, 303–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo J. et al., Global genetic differentiation of complex traits shaped by natural selection in humans. Nat. Commun. 9, 1865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z., Franks R. G., Klink V. P., Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12, 1879–1892 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klucher K. M., Chow H., Reiser L., Fischer R. L., The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizukami Y., Fischer R. L., Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 942–947 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cnops G. et al., The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. Plant Cell 18, 852–866 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu W.-H., Chandler J., Cnops G., Van Lijsebettens M., Werr W., Mutations in the TORNADO2 gene affect cellular decisions in the peripheral zone of the shoot apical meristem of Arabidopsis thaliana. Plant Mol. Biol. 63, 731–744 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Oren E. et al., The multi-allelic APRR2 gene is associated with fruit pigment accumulation in melon and watermelon. J. Exp. Bot. 70, 3781–3794 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan Y. et al., Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 161, 1476–1485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao G. et al., A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 51, 1607–1615 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Matsumura H. et al., Mapping of the gynoecy in bitter gourd (Momordica charantia) using RAD-seq analysis. PLoS One 9, e87138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X. M. et al., NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19, 1537–1548 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Zonneveld M. et al., Screening genetic resources of Capsicum peppers in their primary center of diversity in Bolivia and Peru. PLoS One 10, e0134663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behera T. K. et al., Bitter gourd: Botany, horticulture, breeding. Hortic. Rev. (Am. Soc. Hortic. Sci.) 37, 101–141 (2010). [Google Scholar]
- 35.Hufford M. B. et al., Comparative population genomics of maize domestication and improvement. Nat. Genet. 44, 808–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin T. et al., Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46, 1220–1226 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Lee C.-R. et al., Young inversion with multiple linked QTLs under selection in a hybrid zone. Nat. Ecol. Evol. 1, 119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y., Bhat P. R., Close T. J., Lonardi S., Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4, e1000212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang H. et al., ALLMAPS: Robust scaffold ordering based on multiple maps. Genome Biol. 16, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Argyris J. M. et al., Use of targeted SNP selection for an improved anchoring of the melon (Cucumis melo L.) scaffold genome assembly. BMC Genom. 16, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q. et al., A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). Gigascience 8, giz072 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo S. et al., The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 45, 51–58 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Soderlund C., Bomhoff M., Nelson W. M., SyMAP v3.4: A turnkey synteny system with application to plant genomes. Nucleic Acids Res. 39, e68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarailo-Graovac M., Chen N., Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinf. Chapter 4, Unit 4 10 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Smit A. F. A., Hubley R., RepeatModeler Open-1.0 (2008–2015). http://www.repeatmasker.org. Accessed 29 May 2019.
- 46.Bao W., Kojima K. K., Kohany O., Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pertea M. et al., StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas B. J. et al., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 51.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanke M., Morgenstern B., AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, W465–W467 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterhouse R. M. et al., BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haas B. J. et al., Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Götz S. et al., High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engle L. M., Faustino F. C., Conserving the indigenous vegetable germplasm of southeast Asia. Acta Hortic 752, 55–60 (2007). [Google Scholar]
- 57.Ka O., Endo Y., Yokoyama J., Murakami N., Useful primer designs to amplify DNA fragments of the plastid gene matK from angiosperm plants. J. Jpn. Bot. 70, 328–331 (1995). [Google Scholar]
- 58.Cox M. P., Peterson D. A., Biggs P. J., SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinf. 11, 485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10 (2011). [Google Scholar]
- 60.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenna A. et al., The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Danecek P. et al.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purcell S. et al., PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bradbury P. J. et al., TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Francis R. M., pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17, 27–32 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Zhang C., Dong S. S., Xu J. Y., He W. M., Yang T. L., PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Zhu L., Bustamante C. D., A composite-likelihood approach for detecting directional selection from DNA sequence data. Genetics 170, 1411–1421 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pavlidis P., Živkovic D., Stamatakis A., Alachiotis N., SweeD: Likelihood-based detection of selective sweeps in thousands of genomes. Mol. Biol. Evol. 30, 2224–2234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen H., Patterson N., Reich D., Population differentiation as a test for selective sweeps. Genome Res. 20, 393–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled genome is available under DNA Data Bank of Japan, accession number BLBB01000001-BLBB01000193. The PacBio reads were submitted under DNA Data Bank of Japan, accession number DRA009109. The Illumina reads of the OHB3-1 genome accession was submitted under DNA Data Bank of Japan, accession number DRA009106. Population re-sequencing Illumina reads were submitted under NCBI BioProject PRJNA578358.