Significance
Despite decades of research, the question of how single cells regulate their size remains unclear. Here we demonstrate that a widely supported molecular model for the fundamental origin of size control in budding yeast is inconsistent with a set of experiments testing the model’s key prediction. We therefore conclude that the problem of cell-size control in budding yeast remains largely unsolved. This work highlights the need for rigorous testing of future models of size control in order to make progress on this fundamental question.
Keywords: cell-size control, budding yeast, single-cell time-lapse microscopy, Start, Whi5
Abstract
Cells must couple cell-cycle progress to their growth rate to restrict the spread of cell sizes present throughout a population. Linear, rather than exponential, accumulation of Whi5, was proposed to provide this coordination by causing a higher Whi5 concentration in cells born at a smaller size. We tested this model using the inducible GAL1 promoter to make the Whi5 concentration independent of cell size. At an expression level that equalizes the mean cell size with that of wild-type cells, the size distributions of cells with galactose-induced Whi5 expression and wild-type cells are indistinguishable. Fluorescence microscopy confirms that the endogenous and GAL1 promoters produce different relationships between Whi5 concentration and cell volume without diminishing size control in the G1 phase. We also expressed Cln3 from the GAL1 promoter, finding that the spread in cell sizes for an asynchronous population is unaffected by this perturbation. Our findings indicate that size control in budding yeast does not fundamentally originate from the linear accumulation of Whi5, contradicting a previous claim and demonstrating the need for further models of cell-cycle regulation to explain how cell size controls passage through Start.
Cells from all domains of life display size control (1–4), coupling their cell size and cell-cycle progression to reduce the variability in size observed throughout a population. We refer readers to recent reviews in the following references for further discussion: refs. 5–8. Despite this widespread behavior, the parameters that cells monitor as proxies for their size and the mechanisms by which these parameters control progress through the cell cycle remain unclear. In the budding yeast Saccharomyces cerevisiae, studies of the cell cycle have produced several hypotheses for the mechanism of size control (9–14). However, no consensus has yet been reached that favors one model above all others. Several models include a role for Whi5, a transcriptional inhibitor that delays progress through Start (15–17), the transition that commits yeast cells to replicating their DNA and then dividing. Here we test how cell-size regulation depends on the expression dynamics of Whi5.
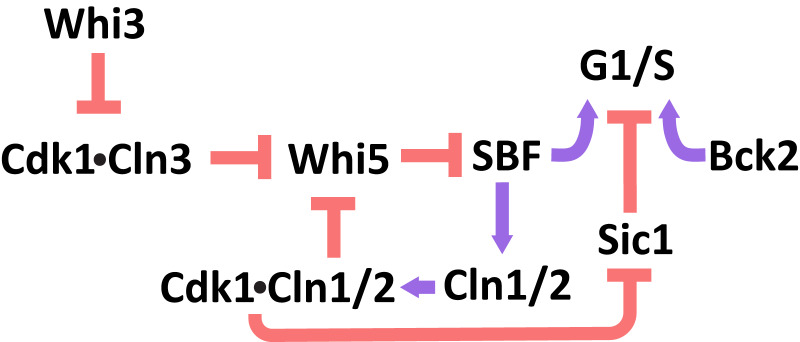
Budding yeast divides asymmetrically, with size control acting in the first cell cycle of the smaller, newborn daughter cells. During their first cell cycle, small daughters have a longer G1 phase, which delays Start relative to the cell cycle of larger daughter cells (3, 18, 19). In contrast, the timing of the budded portion of the cell cycle is roughly independent of cell size (18, 20). The core regulatory network that controls passage through Start has been studied extensively and is outlined in Fig. 1 (15, 16, 21–28).
Fig. 1.
Genetic regulation of passage through Start.
At the end of mitosis, Whi5 enters the nuclei of both daughter and mother cells, where it remains for much of the subsequent G1 phase (15). Whi5’s nuclear localization allows the protein to bind to and inhibit the heterodimeric transcription factor SBF, preventing it from inducing the expression of genes required for passage through Start (15, 16). The G1 cyclin Cln3 forms a heterodimer with the cyclin-dependent kinase Cdk1, which phosphorylates both Whi5 and SBF at multiple sites, causing Whi5 to unbind from SBF (25, 15, 16, 29, 30). SBF and another transcription factor, MBF, induce genes that drive passage through Start including two other G1 cyclins, Cln1 and Cln2. The expression of Cln1 and Cln2 completes a positive feedback loop that commits cells to passage through Start and leads to the nuclear export of phosphorylated Whi5 (31, 32). The differential lengthening of G1 in small daughter cells occurs prior to Whi5’s nuclear exit, demonstrating that cell size must influence the rate of activation of this positive feedback loop (33).
The genetic circuit that regulates Start also includes Bck2, a protein responsible for Cdk1-indpendent activation of genes that promote passage through Start (27, 34–36). The parallel Bck2- and Cln3-dependent induction of gene expression is revealed by the G1 arrest of cln3Δ bck2Δ cells, a behavior not seen in either single mutant (27, 35).
A range of different hypotheses for how cell size could regulate this genetic network have been proposed. We highlight two paradigms of size control: inhibitor dilution and activator accumulation (37, 38). Recent observations support an inhibitor dilution model enacted through Whi5, wherein the growth-mediated dilution of Whi5 relative to a roughly constant concentration of Cln3 during G1 increases the rate of passage through Start as cells grow larger (12, 39). Since the concentration of Whi5 is negatively correlated with daughter-cell volume at birth, this dilution mechanism could explain the longer G1 of small daughter cells; smaller cells would need to grow proportionally more to dilute their higher initial concentration of Whi5. This negative correlation between Whi5 concentration at birth and cell volume was proposed to originate from a volume-independent synthesis rate of Whi5 during the budded portion of the cell cycle, with only a small fraction of Whi5 synthesis occurring during the G1 phase. This contrasts with the synthesis rate of Cln3 which, like most other proteins, scales with cell volume. Here we focus on the predictions of the Whi5 dilution model.
Results
Perturbing the Dynamics of Whi5 Expression Does Not Alter the Cell-Size Distribution.
We tested whether the details of how Whi5 accumulates determine the success of cell-size regulation. The negative correlation between Whi5 concentration at birth and cell volume is essential for the Whi5 dilution model to regulate cell size (12). We perturbed this correlation by expressing Whi5 proportionally to cell volume, decoupling Whi5 concentration at birth from cell volume at birth. Our approach also eliminates the periodic nature of Whi5 synthesis, meaning that a synthesis rate proportional to volume would cause Whi5 concentration to reach a constant value, independent of cell size. Since the rate of passage through Start is proposed to decrease with increasing Whi5 concentration, cells would delay Start significantly if the steady-state Whi5 concentration was higher than that of newborn wild-type (WT) daughters, and the perturbed cells would become larger with each cell cycle. Conversely, if the perturbed Whi5 level was lower than that of WT cells, the cells with altered Whi5 expression would become progressively smaller. This behavior is illustrated in SI Appendix, Fig. S1, with simulations validating these predictions (SI Appendix, Methods). However, these extremes are not seen experimentally; cells lacking Whi5 are indeed smaller than WT cells, but these cells still display size control, rather than the steadily decreasing cell size that the simple, Whi5-dependent model predicts (15, 16, 40, 41).
We perturbed Whi5 synthesis by placing the WHI5 gene under the control of the GAL1 promoter (PGAL1-WHI5). This promoter has previously been used to show that overexpressing Whi5 in an otherwise WT background increases mean cell size (15, 42). It is, however, difficult to achieve finely graded control of Whi5 expression in these cells: expression from the GAL1 promoter is bistable at intermediate galactose concentrations, and cells metabolize galactose, altering its concentration throughout an experiment. Deleting GAL1 blocks galactose phosphorylation and metabolism, and placing GAL3 under the control of a strong promoter (PACT1-GAL3) makes expression from PGAL1 vary smoothly with the concentration of galactose in the medium (42). To quantify Whi5 expression, we generated a fusion to Whi5 using the fast-maturing fluorescent protein mVenNB (43). SI Appendix, Fig. S2, demonstrates that overexpressing Whi5 by exposing these PGAL1-WHI5-mVenNB cells to high levels of exogenously added galactose generates cells that are larger than PWHI5-WHI5-mVenNB cells. For brevity, these cell types will henceforth be abbreviated as PGAL1-WHI5 and PWHI5-WHI5. SI Appendix, Fig. S2, further shows that, at a high galactose concentration, these PGAL1-WHI5 cells also maintain a concentration of Whi5 in G1 that is several times larger than that of PWHI5-WHI5 cells (measured by fluorescence microscopy using fluorescence intensity as a proxy for protein concentration). Despite this, these large cells still maintain a reproducible characteristic cell size. These findings are consistent with previous observations (15, 16), but are inconsistent with the inhibitor dilution model’s prediction that these unusually high levels of Whi5 expression will lead to an unconstrained increase in average cell size over time (SI Appendix, Fig. S1).
Since the predictions of an inhibitor dilution model were not validated by overexpressing Whi5, we decided to characterize the cell-size control displayed by our PGAL1-WHI5 cells in greater detail to assess the effect of decoupling Whi5 concentration at birth from cell volume at birth. Tuning the exogenous galactose concentration to 125 μM generated cells the average size of which was identical to that of PWHI5-WHI5 cells (SI Appendix, Fig. S3). Synthesizing Whi5 proportionally to cell volume should eliminate size control during G1 by removing the negative correlation between cell size and Whi5 concentration at birth. If the rate of passage through Start couples to cell size through the Whi5 concentration, as has been claimed (12), the rate of passage through Start will be uncorrelated with cell volume. Because the primary size control in budding yeast occurs during G1, the loss of this control is predicted to generate a substantial broadening of the cell-size distribution. Indeed, in the absence of compensatory mechanisms, a constant Whi5 concentration is expected to generate arbitrarily broad distributions of cell size, as demonstrated through simulations in SI Appendix, Fig. S1 G–I. SI Appendix, Fig. S3, shows that in bulk cultures we observed no such broadening of the cell-size distribution, which is inconsistent with the predictions of the Whi5 inhibitor dilution model. Because the Whi5 dilution model was initially tested in cells lacking Bck2, we also compared the size of PWHI5-WHI5, bck2∆ and PGAL1-WHI5, bck2∆ cells at 125 μM galactose (12). As expected, bck2∆ cells were larger than BCK2 cells, but neither the mean cell size nor the SD in cell size was significantly different between the PWHI5-WHI5, bck2∆ and PGAL1-WHI5, bck2∆ cells. Aside from these measurements of asynchronous populations of cells (SI Appendix, Fig. S3), our data were obtained from BCK2 cells.
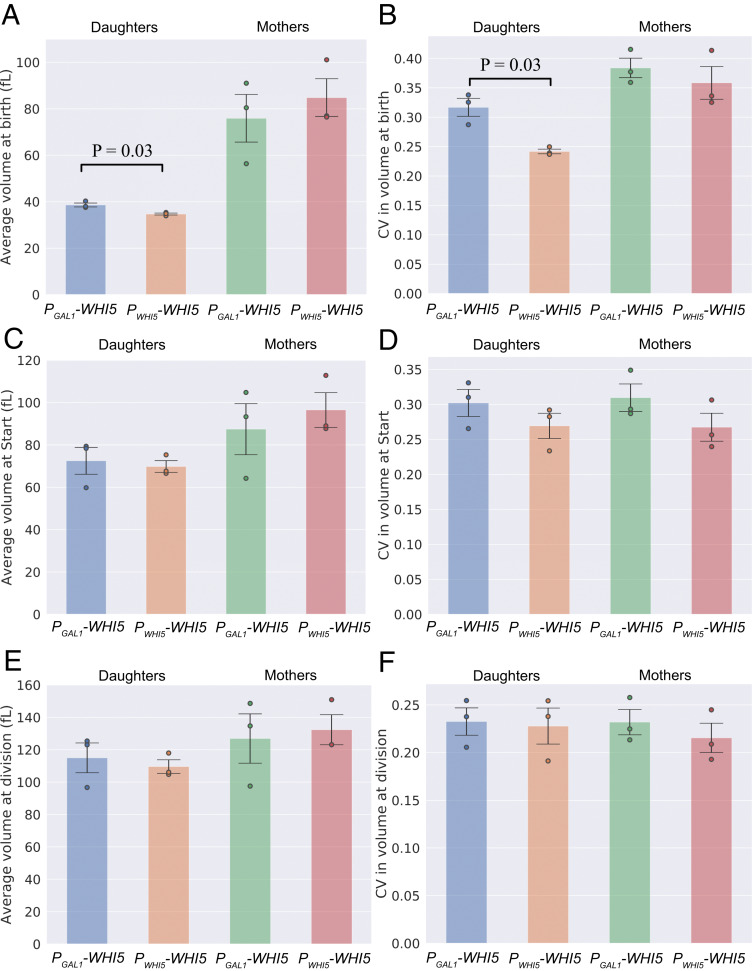
To study the distributions of cell size at specific points in the cell cycle, we used a microfluidic device to track the growth of immobilized cells in a flow chamber when exposed to 125 μM galactose. We extracted information on 3,581 individual cell cycles using a custom-designed algorithm (Methods). Fig. 2 shows the average cell size and coefficient of variation (CV = SD/mean) in cell size measured at birth, at Start, and at division for daughter and mother cells separately. These observations show a statistically significant, although minor, increase in the average and CV of volume at birth for PGAL1-WHI5 daughter cells relative to PWHI5-WHI5 cells. We did not observe statistically significant differences in CV in cell volume between PGAL1-WHI5 cells and PWHI5-WHI5 cells at Start or at division. SI Appendix, Fig. S3, shows cell-size statistics for asynchronous populations, measured both by using a Coulter Counter and by imaging large numbers of cells from exponentially growing cultures at a single time point. Fluorescence imaging in the second approach allowed us to focus on cells in G1 based on the localization of fluorescently tagged Whi5: unbudded cells with Whi5 in the nucleus have not passed Start, whereas those with Whi5 in the cytoplasm have passed Start. However, we were unable to distinguish between mother and daughter cells using this approach. Our measurements of G1 cells using single-time-point microscopy and over the full cell cycle using a Coulter Counter showed a spread in cell size that was independent of whether Whi5 was expressed from the PGAL1 or PWHI5 promoter; both populations had a similar SD and CV. We note, however, that the “smearing” effect of measuring cell size in an asynchronous population may obscure any small increase in the CV for volume at birth. Thus, the only statistically significant difference in cell size between our PGAL1-WHI5 cells and our PWHI5-WHI5 cells is a modest increase in the CV in volume at birth in newborn daughter cells. Notably, this finding is inconsistent with the substantially broader cell-size distribution predicted by the Whi5 dilution model (SI Appendix, Fig. S1).
Fig. 2.
Perturbing Whi5 expression using a galactose-inducible promoter has a minimal effect on the spread in cell size, generating a modest increase in CV at birth, which is not observable at Start or cell division. Cell-size distributions are compared for PGAL1-WHI5 and PWHI5-WHI5 cell types for volume at birth, at Start, and at division. Volumes were measured via time-lapse microscopy and plotted by cell type (inducible and noninducible, mothers and daughters). (A) Average size at birth. (B) CV in size at birth. (C) Average size at Start. (D) CV in size at Start. (E) Average size at division. (F) CV in size at division. Values represent the mean across three biological replicates of the relevant statistic (average or CV). Data are compiled from three experiments for each cell type with a total of 347 PGAL1-WHI5 daughters, 800 PGAL1-WHI5 mothers, 853 PWHI5-WHI5 daughters and 1,581 PWHI5-WHI5 mothers. Error bars represent the SD taken across three biological replicates. Black lines correspond to statistically significant differences with two-tailed P values less than 0.05 quoted, calculated by comparing daughters and mothers separately between cell types using a Welch’s t test across biological replicates. Dots correspond to values for individual biological replicates.
Another potential difference between the PGAL1 promoter and the PWHI5 promoter is in the stochasticity of gene expression. SI Appendix, Fig. S2, demonstrates based on time-point microscopy that the CV in the concentration of Whi5-mVenNB within PGAL1-WHI5 cells is greater than that of PWHI5-WHI5 cells. In contrast, our measurements of the CV in cell size at distinct points in the cell cycle, shown in Fig. 1, show little difference between our PGAL1-WHI5 and PWHI5-WHI5 strains. This observation indicates that any difference in the noisiness of gene expression between these promoters does not lead to a notably broader size distribution. We note that both the magnitude and relative size of these CVs at birth, Start, and division are comparable to previous measurements of cells in similar growth conditions (19). If Whi5 concentration is a crucial element in sensing cell size, then greater variability in expression might be expected to lead to a broader size distribution than that seen in WT cells. This contrasts with the unchanged CVs in cell size that we observe and is consistent with our conclusion that the dynamics of Whi5 expression are unimportant in coupling size to passage through Start.
The Dynamics of Whi5 Accumulation Depend on the Promoter Driving Its Expression.
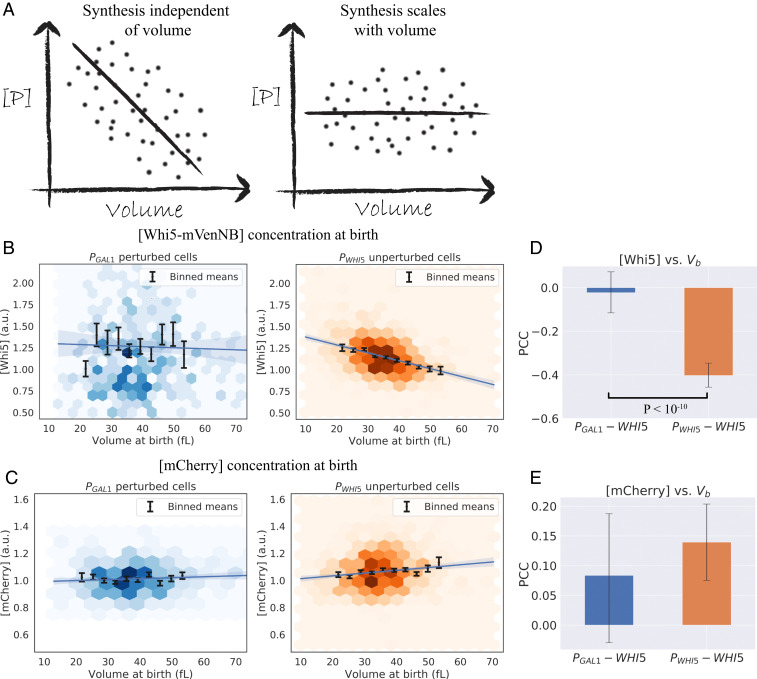
We used fluorescence microscopy to confirm that the scaling of Whi5 synthesis with cell volume was perturbed in PGAL1-WHI5 cells. In the Whi5 dilution model, a linear accumulation of Whi5 leads to a negative correlation between Whi5 concentration and cell volume in newly born cells (12), which was proposed to be essential for controlling the distribution of cell sizes. We therefore measured the correlation between these two quantities in our strains with time-lapse microscopy, using average fluorescence intensity as a proxy for the concentration of Whi5-mVenNB protein at cell birth at the galactose concentration (125 μM) studied in Fig. 2. For PWHI5-WHI5 cells, we confirmed the previously observed negative correlation between Whi5 concentration at birth and cell volume, as shown in Fig. 3B for daughter cells. Fig. 3C shows the concentration profile for the expression of PACT1-mCherry in the same cells, with this red fluorescent protein driven by the promoter of the gene ACT1. Unlike Whi5-mVenNB, the concentration of mCherry does not decline in larger cells. The PGAL1-WHI5 cells showed a different pattern of Whi5 accumulation, with no statistically significant correlation between Whi5 concentration and cell volume at birth. Comparing Fig. 3 B and C demonstrates that inducing Whi5 synthesis with the GAL1 promoter brings the correlation between Whi5 concentration at birth and cell volume at birth closer to that observed for our PACT1-mCherry construct. Because individual time-lapse measurements showed greater variability in their correlations and contained fewer cells, we performed our statistical tests on time-lapse data aggregated from at least three experimental replicates. SI Appendix, Fig. S4, shows that PWHI5-WHI5 cells recapitulate the previously observed decrease in Whi5 concentration throughout G1 (12), whereas the Whi5 concentration remains roughly constant throughout G1 in PGAL1-WHI5 cells. SI Appendix, Fig. S5, shows that the average level of both Whi5-mVenNB and mCherry is indistinguishable between cells that express Whi5 from the WHI5 or GAL1 promoters.
Fig. 3.
Expressing Whi5 from the GAL1 promoter alters the relationship between Whi5 concentration and cell size. (A) Illustrations of the predicted correlation of protein concentration with cell size for a gene the production rate of which scales linearly with cell volume (53), contrasted with a gene the synthesis of which does not scale with cell volume. (B and C) Concentration of fluorescent proteins at cell birth vs. volume at birth (Vb), grouped by cell type (PWHI5-WHI5 “unperturbed” cells, and PGAL1-WHI5 “perturbed” cells) and derived from time-lapse experiments to monitor cell growth. The fluorescence intensity averaged over the cell is used as a proxy for protein concentration. Colored hexagons represent a 2D histogram of data points, with darker hexagons showing increased local density of data points. Black lines correspond to averages of the same data binned with respect to Vb, with error bars showing the SEM. Blue lines correspond to linear regression fits with 95% confidence intervals. Data are compiled from three experiments for each cell type with a total of 347 PGAL1-WHI5 daughters, 800 PGAL1-WHI5 mothers, 853 PWHI5-WHI5 daughters and 1,581 PWHI5-WHI5 mothers. (B) Whi5 signal. PWHI5-WHI5 cells (orange) show a negative correlation between Whi5 concentration at birth and cell volume at birth. PGAL1-WHI5 cells (blue) lose this negative correlation, consistent with Whi5 synthesis being proportional to cell volume. (C) pACT1-mCherry signal. Daughter cells display a weak positive correlation between [mCherry] and cell volume at birth. The origin of this correlation is unknown, although it is consistent between PGAL1-WHI5 and PWHI5-WHI5 cell types. (D and E) Pearson correlation coefficients (PCC) measured for the datasets plotted in B and C. Error bars correspond to 95% confidence intervals inferred by bootstrapping analysis. Black lines correspond to statistically significant differences with P values less than 0.05 quoted, calculated using a Fisher’s z-transformation on both datasets. (D) PCC values for [Whi5] measured at birth vs. Vb for daughter cells show a statistically significant difference between cell types with P < 10−10. (E) PCC values for [mCherry] measured at birth vs. Vb for daughter cells shows no statistically significant difference between the two cell types.
Because time-lapse microscopy can alter cell cycle timing and fluorescent proteins show photobleaching after repeated illumination, we also measured the correlation of fluorescence intensity with cell volume for G1 cells in asynchronous cultures. SI Appendix, Fig. S6, confirms our observations from time-lapse microscopy: the concentration of Whi5-mVenNB expressed from PWHI5 falls with increasing cell volume, whereas the concentration of Whi5-mVenNB expressed from PGAL1 rises with cell volume. This rise is qualitatively similar to the positive correlation observed for mCherry expressed from PACT1. We conclude that expressing Whi5 from PGAL1 strongly perturbs the correlation between the volume of G1 cells and their Whi5 concentration relative to cells expressing Whi5 from its own promoter, bringing it closer to the behavior observed for expression from the PACT1 promoter. These findings, combined with those of previous sections, show that the distribution of cell size is only modestly affected by the difference between a linear and exponential accumulation of Whi5 over time, invalidating a key prediction of the Whi5 dilution model.
Correlations in Cell-Cycle Timing and Cell Volume.
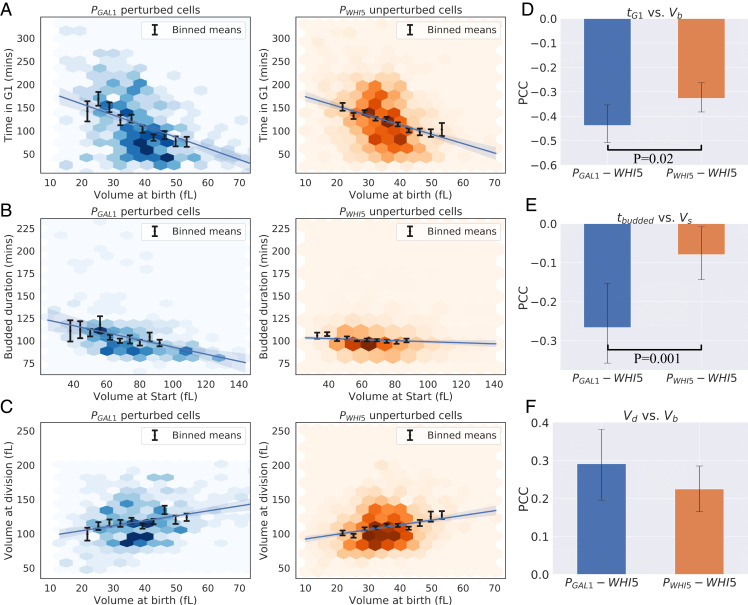
Perturbing the dynamics of Whi5 synthesis failed to change the cell-size distribution in asynchronous populations and showed only a modest increase in CV in volume at birth. To study the effects of this perturbation on size control in greater detail, we examined the dependence of cell-cycle timing on cell volume in PGAL1-WHI5 cells. We defined time the between birth and Start as the duration of Whi5 nuclear localization, while the rest of the cell cycle was defined as the period when Whi5 was excluded from the nucleus. This metric for G1 duration has been validated in previous studies of cell-size regulation (33). SI Appendix, Fig. S7, compares the time spent in the distinct portions of the cell cycle for PGAL1-WHI5 and PWHI5-WHI5 cells. The average durations of the entire cell cycle and the pre- and post-Start intervals are statistically indistinguishable between the two populations. Fig. 4A shows that the negative correlation between G1 timing and cell volume is seen in both PGAL1-WHI5 and PWHI5-WHI5 cells, with PGAL1-WHI5 cells showing a slightly stronger negative correlation (Fisher z-transformation). The correlation between the duration of the budded portion of the cell cycle (tbudded) and cell volume at Start has a slightly stronger negative correlation for PGAL1-WHI5 cells than for PWHI5-WHI5 cells (Fisher z-transformation). This finding is consistent with weak size control acting in the budded portion of the cell cycle, but since size control during G1 remains functional in our perturbed cell type, the interpretation of this difference is unclear (Fig. 4B). Furthermore, there is no statistically significant difference in the correlation between cell volume at birth and division [a common indicator for the mode of cell-size control (8)], when compared between the two cell types. In order to determine whether these differences in the correlations of cell-cycle timing with cell volume translated to effects on cell volume, we studied the correlations between the cell volume added in different phases of the cell cycle and cell volume at the start of those phases. SI Appendix, Fig. S8, shows these correlations for volume added in G1 as a function of volume at birth, volume added during budding as a function of volume at Start, and volume added over the full cell cycle as a function of volume at birth. These datasets show no statistically significant differences between PGAL1-WHI5 and PWHI5-WHI5 cells, consistent with size control being conserved between these cell types.
Fig. 4.
PGAL1-WHI5 cells retain size control during the G1 phase, in addition to weak size control in the budded portion of the cell cycle. (A–C) Cell-cycle correlations for daughter cells. See Fig. 3 for details on plotting features for A–C. Data are compiled from three experiments for each cell type with a total of 347 PGAL1-WHI5 daughters, 800 PGAL1-WHI5 mothers, 853 PWHI5-WHI5 daughters and 1,581 PWHI5-WHI5 mothers. (A) Plot of time spent in G1 phase (determined by nuclear localization of Whi5) vs. cell volume at birth (Vb) for PGAL1-WHI5 and PWHI5-WHI5 cells. (B) Plot of time spent in the budded phases (the sum of S-phase, G2, and mitosis, determined by nuclear exclusion of Whi5) vs. cell volume at Start (Vs) for PGAL1-WHI5 and PWHI5-WHI5 cells. (C) Plot of Vb vs. volume at division (Vd). (D–F) PCCs measured for the datasets plotted in A–C. Error bars show 95% confidence intervals inferred by bootstrapping analysis. Black lines correspond to statistically significant differences with P values less than 0.05 quoted, calculated using a Fisher’s z-transformation on both datasets. (D) PCC values for G1 duration vs. Vb for daughter cells show a statistically significant difference between cell types (P = 0.02). This difference is consistent with stronger size control occurring during the G1 phase for PGAL1-WHI5 cells, not weaker as would be predicted by the inhibitor dilution model. (E) PCC values for budded duration measured at birth vs. Vb for daughter cells shows a statistically significant difference between the two cell types with P = 0.001. This difference corresponds to the presence of weak size control during the budded portion of the cell cycle. (F) PCC values for Vb vs. Vd for daughter cells show no statistically significant difference between the two cell types.
Finally, we tested the dependence of G1 duration on Whi5 concentration at birth. SI Appendix, Fig. S9, demonstrates that the duration of G1 retains a positive correlation with Whi5 concentration at birth in both PGAL1-WHI5 and PWHI5-WHI5 cells, with no statistically significant difference between the correlation coefficients measured for these two cell types. This observation is consistent with our findings and those of others that overexpressing Whi5 leads to an increase in cell size by delaying Start (12, 15, 16).
Cell-Size Control Does Not Depend on the Dynamics of Cln3 Expression.
Previous studies have focused on the roles of Cln3, as an activator, and Whi5, as an inhibitor, in coupling passage through Start to cell-size control (9, 11, 12, 44, 45). To explore whether the details of Cln3 accumulation influence cell-size control, we constructed strains where the endogenous copy of CLN3 was replaced by a galactose-inducible version. As expected, increasing Cln3 expression by increasing the galactose concentration decreased cell size, in contrast to the increased cell size caused by increasing Whi5 expression. Tuning the galactose concentration to 200 μM yielded an average cell size corresponding to that of WT cells. In this condition, our PGAL1-CLN3 cells show no statistically significant difference in mean, SD, or CV of their size from that of WT cells (SI Appendix, Fig. S10). This finding indicates that, at the level of an asynchronous population, perturbing the details of Cln3 synthesis does not lead to any observable broadening of the cell-size distribution.
Discussion
We investigated the role of Whi5 in controlling cell size in the budding yeast S. cerevisiae. Like previous groups, we find that overexpressing Whi5 makes cells bigger and that preventing its expression makes them smaller. Titrating the expression of Whi5 from the galactose-inducible GAL1 promoter produces populations of cells the mean size and spread in cell size of which are indistinguishable from those of cells that express Whi5 from its endogenous promoter. Because expressing Whi5 from PGAL1 makes the rate of Whi5 synthesis scale linearly, rather than sublinearly, with cell volume, this result is inconsistent with the model that the sublinear scaling of Whi5 synthesis with cell volume plays a critical role in controlling the distribution of cell size. Minor differences in the CV in volume at birth were observed in time-lapse microscopy; however, this modest increase in CV for PGAL1-WHI5 cells remains inconsistent with the perturbations predicted by an inhibitor dilution model. To confirm that cell size was still being controlled by the length of the interval between cell birth and Start, we verified that this timing retains its strong negative correlation with cell volume at birth when Whi5 is expressed from PGAL1. Additionally, regardless of whichever promoter drives Whi5 expression, we see a positive correlation between Whi5 concentration and the interval between birth and Start within a population of cells, consistent with the observations that led to the proposal of the Whi5 dilution model (12).
Our results show that the concentration of Whi5 influences the size at which the cell population passes through Start but the dynamics of Whi5 accumulation have only a modest effect on the spread in size. This indicates that Whi5 dilution is not the dominant mechanism constraining the spread in the cell-size distribution in WT cells. Our results therefore invalidate a key prediction of the Whi5 dilution model and reveal that this model is insufficient to explain cell-size control in budding yeast. We note, however, that, even if the Whi5 dilution model is not the principal way of controlling the cell-size distribution, this mechanism may still play a redundant role, the individual effect of which is too small to be statistically significant within our experiments. It remains possible that Whi5 dilution may act in a complementary fashion to other, presently unknown, mechanisms of size control. Such a size-control mechanism is unlikely to rely exclusively on Bck2, since our experiments in bulk cultures show that the cell-size distribution is unaffected by perturbing Whi5 expression in cells lacking Bck2.
The observations that led to the Whi5 dilution model were made of cells grown in 2% glycerol plus 1% ethanol as a carbon source in order to generate small daughter cells with strong size control. This growth medium differs from the 2% Raffinose carbon source used here. Although both are nonfermentable carbon sources that generate a small average cell size, we cannot rule out the possibility that this difference in carbon source has contributed to the difference between the conclusions of the two studies. However, both studies observed a correlation between Whi5 concentration and G1 phase duration in WT cells, and our study reproduces the negative correlation between Whi5 concentration and cell volume at birth in G1 that lies at the heart of the inhibitor dilution model. Additionally, SI Appendix, Table S2, shows that the duration of cell-cycle phases measured in our growth medium is comparable to those published previously, displaying an extended G1 phase but very similar interdivision times (33). Dorsey et al. (13) used microscopy to infer absolute protein copy number as cells passed through the cell cycle. This work did not observe any reduction of the Whi5 concentration during G1, after controlling for photobleaching effects, which contrasts with our own and earlier work (12). However, their measurements were acquired using glucose or glycerol as carbon sources. Another recent study proposed that this discrepancy may arise from differences in growth media rather than from photobleaching effects, since the study’s observations showed Whi5 dilution in the G1 phase to be more pronounced for cells grown in media with longer doubling times (44). Our single-time-point microscopy experiments for cells in G1 demonstrate that correlation between birth cell volume and Whi5 concentration (Fig. 3) that we used as a basis for invalidating the Whi5 dilution model is not a result of photobleaching. We cannot, however, rigorously exclude the effects of photobleaching in our recapitulation of Whi5 dilution during the G1 phase (SI Appendix, Fig. S4) (12).
Are there viable alternatives to the Whi5 dilution model? A variety of other models have been proposed. They propose the accumulation, throughout G1, of a component that is limiting for passage through Start, such as Cln3 or the SBF subunit Swi4 (9, 13), or the integration of Cln3 activity throughout G1 through its ability to phosphorylate and inhibit Whi5 (11). Attempts to measure Cln3 levels have been limited by its rapid degradation, causing authors to use stabilized Cln3 mutants rather than WT Cln3 (11, 12). To address this concern, a recent study used a self-cleaving linker to allow a fluorescent protein to report on Cln3 translation without affecting Cln3 function or accumulation (45). These authors observed that WT cells experience a pulse of overall protein synthesis in late G1, leading to an increase in Cln3 concentration which drives cells through Start. How this burst couples passage through Start to cell size remains unclear. Cln3 translation is hindered by its long 5′ untranslated region (5′ UTR) (46), an effect which may be mediated by the binding of Whi3 (an inhibitor of Cln3 translation) to key motifs in Cln3 messenger RNA (mRNA) transcripts (47). Collectively, these observations have led to the suggestion that the translation of Cln3 may play a role in sensing cell size at the G1/S transition, for example, by having the rate of Cln3 translation increase nonlinearly with cell volume during G1 (41). Our measurements on asynchronous populations do not support such a model. SI Appendix, Fig. S10, demonstrates that replacing the 5′ UTR of the Cln3 mRNA with that of Gal1 mRNA and adjusting Cln3 expression from an inducible promoter to achieve the same mean cell size as WT cells did not cause any statistically significant increase in the spread of cell size, as would be expected if the details of Cln3 synthesis were key elements in cell-size regulation. We acknowledge, however, that there may be more subtle effects on the spread in cell size that are unobservable in these measurements on asynchronous populations.
There is clear phenomenological evidence for mechanisms that regulate the distribution of cell sizes: newborn daughters spend longer in G1 than their larger mothers, and very large cells have a smaller exponential growth constant than cells at the median size (48). But what are the molecular mechanisms that produce these correlations? One common theme in the models described above is their focus on the role of single genes in regulating cell size. Although deleting the genes invoked by these hypotheses either increases (cln3Δ, swi4Δ) or decreases (whi3Δ, whi5Δ) cell size, none of these perturbations produces the dramatic broadening of the cell-size distribution expected if a single gene controlled the spread of cell size (15, 16, 49). In contrast, similar work in the fission yeast Schizosaccharomyces pombe has revealed a number of candidate genes the deletion or inactivation of which leads to an increase in the spread in cell size, such as the protein Pom1 or the double mutant wee1-50 cdc25Δ (50, 51). Our interpretation of our own and earlier work is that an understanding of how budding yeast cells couple their size to the cell cycle remains elusive. Although the field has identified proteins the expression of which alters the average cell size, no model has succeeded in explaining how these proteins collectively prevent the cell-size distribution from growing broader over time. We see three classes of model that could close the gap. The first is that there is a single, as yet unidentified, protein that regulates passage through Start, the abundance or concentration of which is controlled by cell size in a way that controls the distribution of cell size. The second is that there are multiple proteins that activate passage through Start and multiple proteins that inhibit it, with the synthesis rate of the activators rising with cell size more steeply than that of the inhibitors, so that, as cells grow, the activators eventually prevail and drive cells through Start (52). In this model, size control is a collective exercise, and it would require the removal of many activators or inhibitors to broaden the cell-size distribution. Finally, there are likely to be passive mechanisms that regulate cell size in addition to these active mechanisms. As an example, if the ratio between protein synthesis rate and cell volume has an optimal value over a modest range, but then falls in larger or smaller cells, the cells that are too big or too small would replicate more slowly. This would set a limit to the steady-state distribution of cell size, even in the absence of an active control linking cell-cycle progress to cell size. Indeed, recent evidence suggests that the growth of large cells slows because transcription reaches a maximal rate (48, 53). Further measurements of how cell growth rate is affected by cell size will be useful in understanding the constraints on the spread of cell size that may be imposed by such a passive size-control mechanism in the absence of other active size-control strategies (54).
Methods
See SI Appendix, Methods, for a full description of the methods used in this text.
Yeast Strains and Plasmids.
The strains used in this study were congenic with W303 and are listed in full in SI Appendix, Table S1. All strains were constructed using standard genetic methods.
Image Processing.
We performed brightfield image segmentation and cell tracking using the open source CellStar algorithm (55). Cell volume was inferred based on this brightfield segmentation by fitting an ellipse to the two-dimensional (2D) mask and assuming a three-dimensional prolate spheroid shape. We designed a custom, semiautomated image-processing pipeline to incorporate fluorescence data and compile measurements of individual cell cycles. Cell-cycle progression was assessed based on Whi5 nuclear localization in accordance with previous approaches (33). All relevant code is available at https://github.com/AWMurrayLab/image_processing_cellstar_public.
Live-Cell Microscopy.
For time-lapse microscopy, cells were loaded into a CellASIC microfluidics flow chamber and imaged using a Nikon Eclipse Ti spinning disk confocal microscope. Growth medium with the appropriate carbon source and galactose concentration was made to flow from two wells at a pressure of 1 pound per square inch using the ONIX microfluidics system. Our single time-point imaging used agar pads with a concentration between 1 and 2%, made using the appropriate growth medium.
Cell Culture.
All experiments were performed in 2× complete synthetic medium (56) with various carbon sources. Cells were taken from exponentially growing cultures with a culture density between 1 and 5 × 106 cells/mL and were grown in the relevant growth medium for a minimum of 20 h prior to measurement.
Data Availability.
The datasets used to generate the figures within this publication are available for download at https://data.4tu.nl/repository/uuid:c3dcc24a-ac07-42bb-8bc2-3824077738fc. All code required to analyze these datasets is available at https://github.com/AWMurrayLab/image_processing_cellstar_public.
Supplementary Material
Acknowledgments
F.B. was supported by the William Georgetti Trust, a Harvard Graduate Merit Award, a Harvard Quantitative Biology Initiative Student Award, The Milton Fund and The Volkswagen Foundation while conducting this research; A.A. was supported by NSF CAREER Award 1752024; and A.W.M. was supported by NIH Grant RO1-GM43987 and the NSF-Simons Center for Mathematical and Statistical Analysis of Biology at Harvard (NSF-Simons Grants 1764269 and 594596). We thank Naama Barkai, Ilya Soifer, Angelika Amon, Bruce Futcher, and Jan Skotheim for helpful suggestions in writing this manuscript. F.B. thanks Zachary Niziolek, Jeffery Nelson, and the staff at the Harvard Bauer Core Facility for their help with the Coulter counter instrument.
Footnotes
The authors declare no competing interest.
Data deposition: The datasets used to generate the figures within this publication are available for download from the Center for Research Data at https://data.4tu.nl/repository/uuid:c3dcc24a-ac07-42bb-8bc2-3824077738fc. All code required to analyze these datasets is available in GitHub at https://github.com/AWMurrayLab/image_processing_cellstar_public.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001255117/-/DCSupplemental.
References
- 1.Donachie W. D., Relationship between cell size and time of initiation of DNA replication. Nature 219, 1077–1079 (1968). [DOI] [PubMed] [Google Scholar]
- 2.Nurse P., Genetic control of cell size at cell division in yeast. Nature 256, 547–551 (1975). [DOI] [PubMed] [Google Scholar]
- 3.Johnston G. C., Pringle J. R., Hartwell L. H., Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105, 79–98 (1977). [DOI] [PubMed] [Google Scholar]
- 4.Eun Y.-J. et al., Archaeal cells share common size control with bacteria despite noisier growth and division. Nat. Microbiol. 3, 148–154 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Amodeo A. A., Skotheim J. M., Cell-size control. Cold Spring Harb. Perspect. Biol. 8, a019083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osella M., Tans S. J., Cosentino Lagomarsino M., Step by step, cell by cell: Quantification of the bacterial cell cycle. Trends Microbiol. 25, 250–256 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Facchetti G., Chang F., Howard M., Controlling cell size through sizer mechanisms. Curr. Opin. Syst. Biol. 5, 86–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho P.-Y., Lin J., Amir A., Modeling cell size regulation: From single-cell-level statistics to molecular mechanisms and population-level effects. Annu. Rev. Biophys. 47, 251–271 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Carey L. B., Cai Y., Wijnen H., Futcher B., Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoS Biol. 7, e1000189 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrezuelo F. et al., The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat. Commun. 3, 1012 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Liu X. et al., Reliable cell cycle commitment in budding yeast is ensured by signal integration. eLife 4, e03977 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmoller K. M., Turner J. J., Kõivomägi M., Skotheim J. M., Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size. Nature 526, 268–272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorsey S. et al., G1/S transcription factor copy number is a growth-dependent determinant of cell cycle commitment in yeast. Cell Syst. 6, 539–554.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Heldt F. S., Lunstone R., Tyson J. J., Novák B., Dilution and titration of cell-cycle regulators may control cell size in budding yeast. PLOS Comput. Biol. 14, e1006548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costanzo M. et al., CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117, 899–913 (2004). [DOI] [PubMed] [Google Scholar]
- 16.de Bruin R. A. M., McDonald W. H., Kalashnikova T. I., Yates J. III, Wittenberg C., Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117, 887–898 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Charvin G., Oikonomou C., Siggia E. D., Cross F. R., Origin of irreversibility of cell cycle start in budding yeast. PLoS Biol. 8, e1000284 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartwell L. H., Unger M. W., Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J. Cell Biol. 75, 422–435 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soifer I., Robert L., Amir A., Single-cell analysis of growth in budding yeast and bacteria reveals a common size regulation strategy. Curr. Biol. 26, 356–361 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Allard C. A. H., Decker F., Weiner O. D., Toettcher J. E., Graziano B. R., A size-invariant bud-duration timer enables robustness in yeast cell size control. PLoS One 13, e0209301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beach D., Durkacz B., Nurse P., Functionally homologous cell cycle control genes in budding and fission yeast. Nature 300, 706–709 (1982). [DOI] [PubMed] [Google Scholar]
- 22.Cross F. R., DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 4675–4684 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nash R., Tokiwa G., Anand S., Erickson K., Futcher A. B., The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7, 4335–4346 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyers M. et al., Characterization of G1 and mitotic cyclins of budding yeast. Cold Spring Harb. Symp. Quant. Biol. 56, 21–32 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Tyers M., Tokiwa G., Futcher B., Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12, 1955–1968 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider B. L., Yang Q.-H., Futcher A. B., Linkage of replication to start by the Cdk inhibitor Sic1. Science 272, 560–562 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Wijnen H., Futcher B., Genetic analysis of the shared role of CLN3 and BCK2 at the G(1)-S transition in Saccharomyces cerevisiae. Genetics 153, 1131–1143 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Garí E., Vergés E., Gallego C., Aldea M., Recruitment of Cdc28 by Whi3 restricts nuclear accumulation of the G1 cyclin-Cdk complex to late G1. EMBO J. 23, 180–190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cross F. R., Blake C. M., The yeast Cln3 protein is an unstable activator of Cdc28. Mol. Cell. Biol. 13, 3266–3271 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner M. V. et al., Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS One 4, e4300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cross F. R., Tinkelenberg A. H., A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell 65, 875–883 (1991). [DOI] [PubMed] [Google Scholar]
- 32.Skotheim J. M., Di Talia S., Siggia E. D., Cross F. R., Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454, 291–296 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Talia S., Skotheim J. M., Bean J. M., Siggia E. D., Cross F. R., The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448, 947–951 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Epstein C. B., Cross F. R., Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol. Cell. Biol. 14, 2041–2047 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Como C. J., Chang H., Arndt K. T., Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol. Cell. Biol. 15, 1835–1846 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrezuelo F., Aldea M., Futcher B., Bck2 is a phase-independent activator of cell cycle-regulated genes in yeast. Cell Cycle 8, 239–252 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Fantes P. A., Grant W. D., Pritchard R. H., Sudbery P. E., Wheals A. E., The regulation of cell size and the control of mitosis. J. Theor. Biol. 50, 213–244 (1975). [DOI] [PubMed] [Google Scholar]
- 38.Barber F., Ho P.-Y., Murray A. W., Amir A., Details matter: Noise and model structure set the relationship between cell size and cell cycle timing. Front. Cell Dev. Biol. 5, 92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmoller K. M., Skotheim J. M., The biosynthetic basis of cell size control. Trends Cell Biol. 25, 793–802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soifer I., Barkai N., Systematic identification of cell size regulators in budding yeast. Mol. Syst. Biol. 10, 761 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jonas F., Soifer I., Barkai N., A visual framework for classifying determinants of cell size. Cell Rep. 25, 3519–3529.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingolia N., Bistability and Positive Feedback in Genetic Networks, (Harvard University, Cambridge, MA, 2006). [Google Scholar]
- 43.Balleza E., Kim J. M., Cluzel P., Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods 15, 47–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu Y. et al., Cell cycle inhibitor Whi5 records environmental information to coordinate growth and division in yeast. Cell Rep. 29, 987–994.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Litsios A. et al., Differential scaling between G1 protein production and cell size dynamics promotes commitment to the cell division cycle in budding yeast. Nat. Cell Biol. 21, 1382–1392 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Danaie P., Altmann M., Hall M. N., Trachsel H., Helliwell S. B., CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E. Biochem. J. 340, 135–141 (1999). [PMC free article] [PubMed] [Google Scholar]
- 47.Cai Y., Futcher B., Effects of the yeast RNA-binding protein Whi3 on the half-life and abundance of CLN3 mRNA and other targets. PLoS One 8, e84630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neurohr G. E. et al., Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell 176, 1083–1097.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jorgensen P., Nishikawa J. L., Breitkreutz B.-J., Tyers M., Systematic identification of pathways that couple cell growth and division in yeast. Science 297, 395–400 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Wood E., Nurse P., Pom1 and cell size homeostasis in fission yeast. Cell Cycle 12, 3228–3236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sveiczer A., Novak B., Mitchison J. M., The size control of fission yeast revisited. J. Cell Sci. 109, 2947–2957 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Chen Y., Zhao G., Zahumensky J., Honey S., Futcher B., Differential scaling of gene expression with cell size may explain size control in budding yeast. Mol. Cell 78, 359.e6-370.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin J., Amir A., Homeostasis of protein and mRNA concentrations in growing cells. Nat. Commun. 9, 4496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goranov A. I. et al., The rate of cell growth is governed by cell cycle stage. Genes Dev. 23, 1408–1422 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Versari C. et al., Long-term tracking of budding yeast cells in brightfield microscopy: CellStar and the evaluation platform. J. R. Soc. Interface 14, 20160705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Synthetic complete (SC) medium. Cold Spring Harbor Protoc. 2016, pdb.rec090589 (2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used to generate the figures within this publication are available for download at https://data.4tu.nl/repository/uuid:c3dcc24a-ac07-42bb-8bc2-3824077738fc. All code required to analyze these datasets is available at https://github.com/AWMurrayLab/image_processing_cellstar_public.