Significance
The correlation of cryogenic superresolution fluorescence imaging, which features single-molecule sensitivity and specific labelling, with the molecular-scale cellular context of cryogenic electron tomography is a powerful approach for observing the subcellular arrangement of biomolecules. This approach is still in its infancy, and numerous challenges need to be overcome to make this approach routine. This manuscript describes a four-stage workflow using cryogenic single-molecule localizations to annotate cryogenic electron tomography images. The workflow is applied to three different proteins in Caulobacter crescentus and is used to visualize the relative subcellular organization of two proteins, SpmX and PopZ, whose interactions play a key regulatory role in the life cycle of C. crescentus.
Keywords: CLEM, superresolution, cryogenic electron tomography, correlative microscopy, CIASM
Abstract
Superresolution fluorescence microscopy and cryogenic electron tomography (CET) are powerful imaging methods for exploring the subcellular organization of biomolecules. Superresolution fluorescence microscopy based on covalent labeling highlights specific proteins and has sufficient sensitivity to observe single fluorescent molecules, but the reconstructions lack detailed cellular context. CET has molecular-scale resolution but lacks specific and nonperturbative intracellular labeling techniques. Here, we describe an imaging scheme that correlates cryogenic single-molecule fluorescence localizations with CET reconstructions. Our approach achieves single-molecule localizations with an average lateral precision of 9 nm, and a relative registration error between the set of localizations and CET reconstruction of ∼30 nm. We illustrate the workflow by annotating the positions of three proteins in the bacterium Caulobacter crescentus: McpA, PopZ, and SpmX. McpA, which forms a part of the chemoreceptor array, acts as a validation structure by being visible under both imaging modalities. In contrast, PopZ and SpmX cannot be directly identified in CET. While not directly discernable, PopZ fills a region at the cell poles that is devoid of electron-dense ribosomes. We annotate the position of PopZ with single-molecule localizations and confirm its position within the ribosome excluded region. We further use the locations of PopZ to provide context for localizations of SpmX, a low-copy integral membrane protein sequestered by PopZ as part of a signaling pathway that leads to an asymmetric cell division. Our correlative approach reveals that SpmX localizes along one side of the cell pole and its extent closely matches that of the PopZ region.
Visualizing the subcellular spatial organization of specific biomolecules is of paramount importance to understanding their function in vivo. This subcellular organization is often studied using fluorescence microscopy, which has the ability to image specific fluorescently labeled biomolecules of interest in the crowded cellular environment. However, the diffraction limit of visible light obscures nanoscale details. This limitation is particularly detrimental in the study of bacterial systems, where cellular length scales are on the order of the diffraction limit itself. The development of superresolution techniques in the mid-2000s has enabled the study of subcellular structures formed from specifically labeled biomolecules with information well below the diffraction limit (1–3). These superresolved images provide a detailed map of where labeled biomolecules are localized with ∼20-nm precision, but often in the absence of the key cellular context provided by other unlabeled structures. Cellular context, such as knowledge of subcellular features surrounding a biomolecule of interest or the presence of nearby interaction partners, can enhance understanding of the target feature. While the number of simultaneously labeled and superresolved structures is ever-increasing (4–6), most superresolved images still lack this key cellular context.
Another approach to observe the subcellular organization of biomolecules is cryogenic electron tomography (CET). This approach generates cellular images of frozen-hydrated samples with nanometer-scale resolution, where the process of plunge freezing in liquid ethane rapidly preserves the native cellular state in vitreous ice (7). An ongoing challenge for CET is the specific labeling of biomolecules of interest with electron-dense markers (8–10). Intracellular labeling with antibody-based markers, such as immunogold, disrupts the structural organization of the cell and the high-resolution information collected. In the absence of specific labeling, the recognizable features in a CET experiment are often restricted to large or periodic structures whose native contrast is sufficient for either template matching, where a volume is searched for all orientations of a template structure (11–13), or annotation, a process of manual labeling of visible structures. More recently, this annotation process has been assisted by the use of neural networks (14). Movie S1 gives an example of the results obtained with typical neural network-assisted annotation applied to a CET reconstruction of Caulobacter crescentus, a model bacterium investigated in this work.
Both superresolution fluorescence imaging and CET are powerful techniques for uncovering the subcellular organization of biomolecules. Each approach has unique strengths and limitations. The combination of superresolution imaging, with its noninvasive and specific labels, and CET, with its high resolution, would enable the visualization of biomolecules of interest in the presence of their cellular context. However, correlating these two techniques relies on development of superresolution fluorescence imaging of samples at cryogenic temperatures. There are numerous approaches to superresolution fluorescence imaging, each with their own challenges when adapting them to cryogenic temperatures. Here, we will focus specifically on single-molecule active control microscopy (SMACM). This method requires precise localization of single molecules in a situation where molecular overlap is removed by using some active control mechanism to ensure only a subset of molecules are emitting in any one imaging frame. In this approach, resolution beyond the diffraction limit arises from a pointillist reconstruction of the single-molecule positions. Retaining active control mechanisms under cryogenic conditions remains a challenge to adapting SMACM for cryogenic temperatures due to the reduced thermal energy available for stochastic processes and/or restricted molecular conformations. Methods of controlling the emissive states of fluorophores at room temperature often rely on diffusional processes or large conformational changes (15, 16), both of which are limited in frozen-hydrated samples. Recent work has identified several photoactivatable fluorescent proteins that maintain their ability to photoactivate at cryogenic temperatures (17–20), and other methods have been shown to induce stochastic blinking of some fluorescent dyes when much higher excitation powers are used (20). Here, we demonstrate an efficient workflow for correlative cryogenic SMACM and CET using the red photoactivatable protein PAmKate (18). The limited number of PAmKate molecules that can be activated and imaged means that densely sampled structures remain a future goal. The primary utility of the single-molecule fluorescence localizations at the moment is to provide single-molecule ground truth annotations of specific molecules in CET reconstructions. We therefore call this technique correlative imaging by annotation with single molecules, or CIASM, pronounced “chasm.” We employ the CIASM workflow to investigate three different proteins in C. crescentus.
C. crescentus undergoes an asymmetric cell division to produce a motile “swarmer” daughter cell and a sedentary “stalked” daughter cell (21) (Fig. 1). Visualizing the subcellular organization of structural and regulatory proteins in these cells is crucial to understanding their life cycle. The cellular asymmetry and distinct characteristics of the two daughter cells are driven by the underlying asymmetric localization of critical signaling proteins. We have chosen to investigate three proteins whose asymmetric localization varies as a function of the cell cycle. The first, methyl-accepting chemotaxis protein A (McpA), is a member of the methyl-accepting chemotaxis proteins (MCPs). MCPs are anchored in the inner cell membrane and sense external stimuli through periplasmic ligand-binding domains. Upon binding, conformational changes occur which regulate the activity of a histidine kinase that forms a hexagonal lattice with the cytoplasmic tips of MCPs (22–24). Ultimately, the activity of the histidine kinase is used to regulate the flagellar motor machinery, resulting in changes in flagellum rotation upon stimulation (25). The chemoreceptor array forms on the dorsal surface of the inner membrane near the flagellated swarmer pole. The location of the array close to the base of the flagellum is crucial to its role in providing feedback to the flagellum motor machinery. The second protein, polar organizing protein Z (PopZ), forms space-filling microdomains at both the stalked and flagellated poles (26, 27). These PopZ microdomains are membraneless organelles that play a crucial role in selectively sequestering proteins to enhance chemical reactions and robustly maintain chemical gradients within the small volume of a bacterial cell (27, 28). The complement of proteins recruited to these microdomains differ at the two poles leading to their asymmetric sequestration in the daughter cells upon cell division. One such protein, SpmX, is the third protein investigated here. SpmX is sequestered to the nascent stalked pole PopZ microdomain during the swarmer to stalked cell transition (29). It is an intrinsically disordered integral membrane protein situated at the inner membrane with an N-terminal lysozyme homology region. SpmX is responsible for recruiting the histidine kinase DivJ, which contributes to the activity of scores of genes involved in cell cycle progression (27).
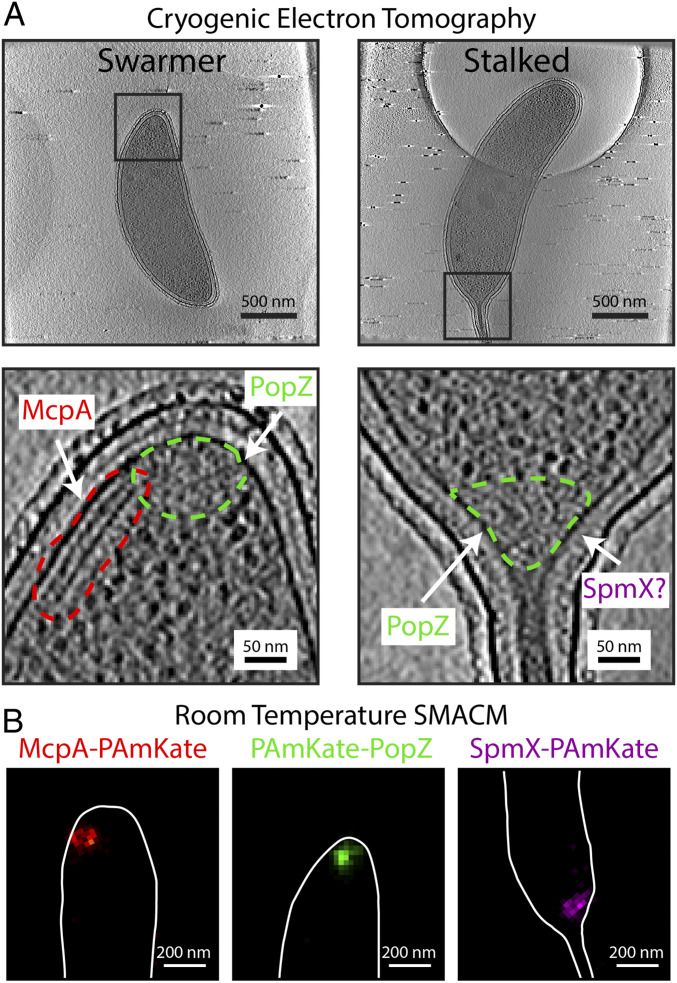
Fig. 1.
Three proteins, McpA, PopZ, and SpmX, in C. crescentus imaged with two different imaging modalities. (A, Top) Tomographic slices of CET data showing the two different cell types, motile swarmer and sessile stalked, of C. crescentus. (A, Bottom) Zoom-in of the Top regions outlined in black. Different protein structures are outlined in dashed colored lines. The parallel lines of density (red dashed line) show the chemoreceptor array containing McpA, the ribosome excluded regions (green dashed line) at the poles of the bacterium are the inferred position of PopZ, and the position of SpmX is undetermined in CET alone. (B) Single-color room temperature SMACM histogram reconstructions of the three proteins each separately fused to PAmKate. Cell outline manually added from bright-field images of the same region.
Slices from electron-tomographic reconstructions of C. crescentus (Fig. 1A) show the three different proteins in situ. Only in the case of McpA is a well-defined structure visible in the CET reconstructions, while the other two proteins, PopZ and SpmX, are not directly discernable. The chemoreceptor array, which contains McpA, is clearly visible under these imaging conditions. In this orientation the chemoreceptor array appears as two parallel lines in a given tomographic slice, but in a reconstructed volume is shown to be two parallel plates ∼12 to 15 nm apart (SI Appendix, Fig. S1). In this example, the plates are ∼190 × 100 nm. The assignment of McpA to this array structure has been well established. Briegel et al. (24) showed that an overexpression of McpA–mCherry resulted in an increased production of these array structures and that under this overexpression the arrays could be found at either cell pole. Furthermore, diffraction-limited correlative light and electron microscopy was used to assign McpA–mCherry to the array structure (24). The array structures observed in C. crescentus are similar to chemoreceptor arrays found across numerous bacteria phyla (23). Because the chemoreceptor array is so clearly visible in CET, we used McpA to validate the cryogenic CIASM workflow before applying the workflow to PopZ and SpmX.
PopZ (19 kDa) and SpmX (46 kDa) are positioned at the cell pole in multiprotein complexes. Neither protein has been observed to form well-ordered structures in situ, making them difficult to identify with the current capabilities of CET. While PopZ is not directly discernable in CET, the amorphous polar microdomain it forms in C. crescentus can be inferred in CET reconstructions by the absence of electron-dense ribosomes in the polar region of the cell (27, 30). There is no means of inferring the position of SpmX from CET alone; however, previous fluorescence data have demonstrated that SpmX localizes in the polar region of the cell on one side of the stalk (31), but its exact location in relation to PopZ has not yet been visualized. In contrast to CET data, all three proteins are clearly visible in the room temperature SMACM reconstructions when fused to PAmKate (Fig. 1B). Despite the high precision of the SMACM localizations (∼13 nm), the superstructures formed by the three proteins have similar size and shape in the SMACM reconstructions. Without the complexity of multicolor fluorescence, the typical cellular context provided by the optical microscopy is a simple cell outline provided by manually annotating bright-field images of the fluorescently imaged bacterial cell. The combination of SMACM localizations and CET using CIASM will provide the best of both imaging modalities. It will clearly assign McpA positions to the chemoreceptor array and will reveal the subcellular locations and organization of SpmX in relation to PopZ and the neighboring cellular environment.
Results
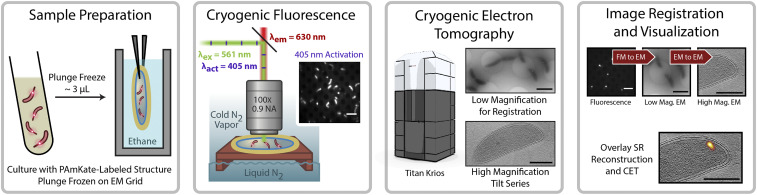
The process of correlating cryogenic SMACM localizations and CET can be divided into the four-stage workflow shown in Fig. 2: sample preparation, cryogenic single-molecule fluorescence imaging, CET, and image registration and visualization. Sample preparation and CET, with slight exceptions (Methods), follow established protocols (7). The cryogenic fluorescence imaging as well as the registration and visualization methods have aspects unique to this publication and will be described in detail below.
Fig. 2.
Workflow for CIASM. C. crescentus cultures with cells expressing PAmKate fusion proteins are plunge-frozen on electron microscopy grids. The grids are loaded onto a cryogenic microscope stage for single-molecule fluorescence imaging. The same grids are removed from the cryogenic stage and transported to a cryogenic electron microscope for CET. Following CET, a series of image transformations registers the fluorescence and CET data, resulting in the localizations of single molecules being overlaid with tomographic slices.
To prepare the sample, a small amount of bacterial cell culture containing cells expressing PAmKate fusion constructs is deposited on a holey carbon electron microscopy grid and plunge frozen. Next, the grid is loaded onto a cryogenic fluorescence microscope stage mounted on a home-built upright microscope. PAmKate molecules are photoactivated with 405-nm light, and the resulting active fluorophores are excited with 561-nm illumination at a low intensity (50 to 100 W/cm2) to avoid sample devitrification (SI Appendix, Fig. S2). The reduced quantum yield of photobleaching at cryogenic temperatures improves localization precision by permitting the total collection of an average of ∼11,800 photons per emitter (SI Appendix, Fig. S3), but this reduction in photobleaching paired with the low damage thresholds for devitrification at 77 K leads to long experiment times of ∼3 h (18). During these long experiments, axial and lateral drift caused by the large thermal gradients inherent in the imaging system as well as the replenishing of liquid nitrogen to the cryogenic stage pose a challenge. In order to avoid manual drift correction and simplify data acquisition, the point spread function (PSF) has been made astigmatic with the addition of a cylindrical lens prior to the image plane. Custom LabVIEW software runs a real-time fitting routine of a user-selected PSF generated from 40-nm-diameter fluorescent polystyrene beads and automatically adjusts a mechanical stage holding the objective to keep the PSF in focus (18). Despite best efforts, the reduced quantum yield for photobleaching and the limited 561-nm excitation intensities available invariably lead to some overlapping emitters, and not all PAmKate molecules are activated and imaged due to experimental time limits.
Despite these challenges, cryogenic SMACM can provide accurate and precise localizations for proteins of interest in a manner compatible with CET, yielding annotations of the CET volume with single-molecule positions from fluorescence. There are two primary errors that impact the ability to annotate CET reconstructions with fluorescence localizations: localization error and registration error. The first, localization error, is a random error for each emitter and describes the precision with which any emitter can be localized. The second, registration error, is a systematic error for all localizations within a registered field of view and impacts the accuracy between the fluorescence annotations and the CET reconstruction. We will first describe how precise localizations are obtained from the fluorescence data and then how those localizations are accurately registered with the CET reconstructions.
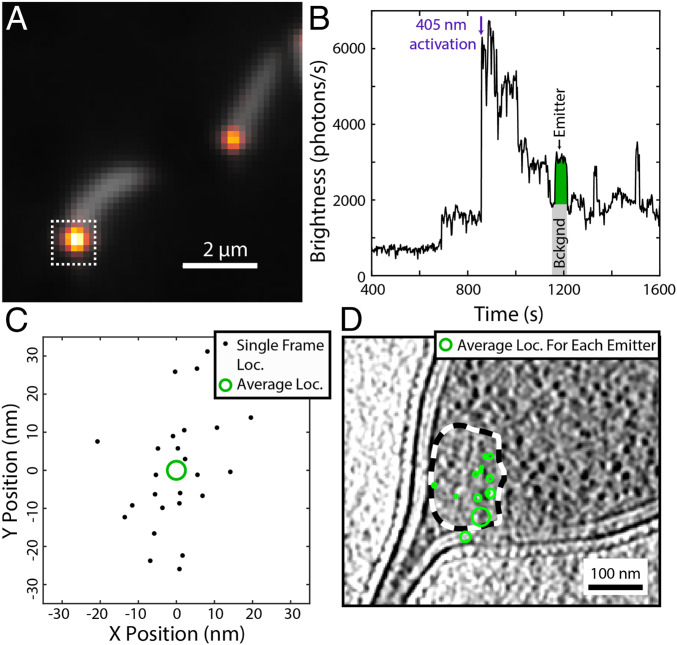
Fig. 3 shows representative cryogenic SMACM data from C. crescentus cells expressing inducible PAmKate–PopZ fusion constructs. The integrated fluorescence intensity from one cell pole (Fig. 3B) exhibits stepwise dynamics indicative of single emitters. From the intensity trace, it is clear that there are often overlapping emitters within a diffraction-limited region. However, due to the long on-times, fluorescence from a single emitter will span numerous 1-s imaging frames, and this temporal information can be leveraged to separate fluorescence contributions from spatially overlapping emitters. To localize an emitter of interest, frames that are either just prior to the emitter of interest’s activation event or just after the emitter of interest’s photobleaching event are manually identified. These preframes or postframes are averaged to estimate the background image generated from other emitters active in the same frame. This background image is then subtracted from the frames in question, thus isolating the single emitter’s PSF, similar to some approaches reported earlier (32–34). Each frame where the emitter of interest has been isolated provides a localization as calculated by ThunderSTORM (35), which is an estimate of the position of the emitter (Fig. 3C and SI Appendix, Fig. S4). Each of these frame localizations must be merged to give a single higher-precision estimate of the true emitter’s position. The precision of that estimate is the SEM of the frame localizations and is represented as the radius of a circle centered on the mean position. This process of isolating the contributions from single emitters of interest can be repeated for the entire SMACM time series, and following registration to CET data, the merged localizations provide single-molecule annotations, which can be overlaid onto a tomographic slice (Fig. 3D).
Fig. 3.
Representative cryogenic SMACM data from PAmKate–PopZ fusion constructs in C. crescentus. (A) Overlay of diffraction-limited fluorescence from the average of the 405-nm photoactivation frames (gray) showing cellular autofluorescence and the average of the 561-nm excitation frames (heat map) showing the overlapping diffraction-limited locations of PAmKate–PopZ molecules. (B) Fluorescence intensity trace from the integrated region indicated by the white dashed box in A. The fluorescence contributions from one emitter (green area) are obtained by subtracting a background (gray area) estimated by taking the average of several frames prior to the activation of the emitter. (C) Localizations from the single emitter shown in B across multiple frames (black dots). Each frame provides a noisy estimate for the true emitter’s position. These frame localizations are merged into a single estimate of the emitter’s location (green circle) with a precision given by the SE on the mean of the frame localizations represented as the radius of the green circle. (D) Overlay of the PAmKate–PopZ merged localizations (green) on a tomographic slice from their corresponding cell with the ribosome excluded region outlined with a dashed black and white line. Each green circle identifies the position and uncertainty in position of a different PAmKate–PopZ emitter.
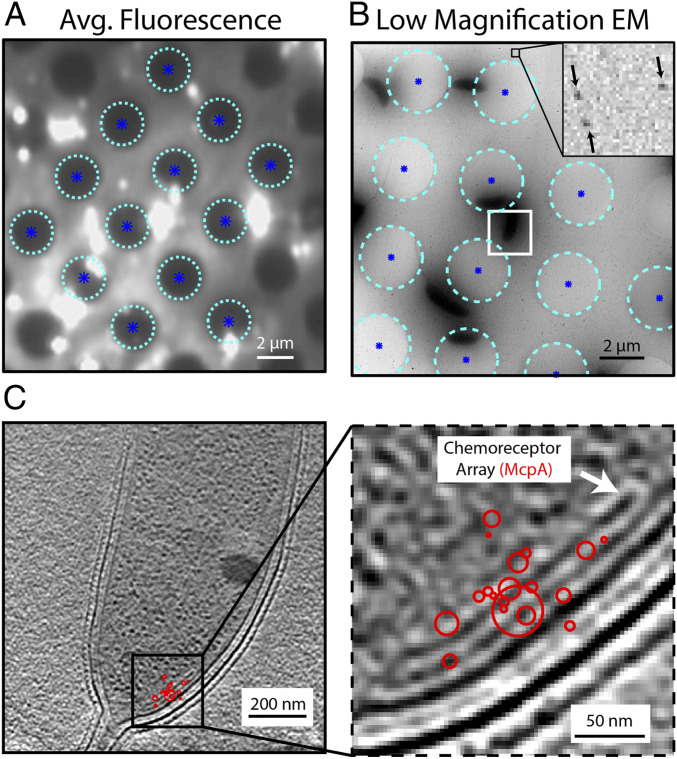
Registration of the SMACM localizations and the CET data are achieved through a series of image transformations starting with mapping the fluorescence to a low-magnification electron micrograph (3,600×, 38.8 Å/pixel) centered on the region of CET data collection. This low-magnification micrograph bridges the fluorescence and CET data. It is important that both the holes in the holey carbon grid and the gold beads used for CET alignment are clearly visible in this low-magnification micrograph, as both the holes and gold beads will be used in the registration process. First, the fluorescence data are registered with the low-magnification micrograph using the locations of 12 to 16 holes as control points for a projective transformation using custom Matlab software (Fig. 4A and SI Appendix, Note S1). We found this approach to be superior to using fluorescent beads as control points because it was challenging to achieve sufficiently high bead concentrations for accurate registration while simultaneously avoiding overlap with the biological structures of interest. Several automated methods for the identification of hole centers were explored, including template matching using an averaged hole image, centroid estimates, and Hough transformations that were used previously (36). Ultimately, software-assisted manual identification of hole centers proved to be the most robust method (SI Appendix, Fig. S5 and Note S2). Following registration of the fluorescence data to the low-magnification electron micrograph, the micrograph was registered with the higher-magnification CET data using the gold beads. To do this, a projection of the CET data was taken across the axial dimension following tomographic reconstruction, and 15 to 25 beads were manually selected using custom Matlab software. Then, using these bead locations as control point pairs, a similarity transformation that only allows for rotation and scaling was calculated. Once completed, the merged fluorescence localizations can be carried to the CET image space first by the application of the projective transformation and then the similarity transformation. In order to assess the quality of the registration process, a complete set of CET data were collected on a 40-nm polystyrene fluorescent bead (SI Appendix, Fig. S6). The distance between the center of the bead as measured in the CET data and the bead localization by fluorescence after registration is ∼30 nm. Further validation is provided by registering McpA–PAmKate localizations because the chemoreceptor array, which is partially composed of the labeled McpA, is visible in the CET data (Fig. 4B). It is clear from the overlay that SMACM localizations are superior for identifying and labeling a structure to traditional diffraction-limited correlative light and electron microscopy, which for this red fluorophore and imaging system would yield a fluorescent spot ∼350 nm in diameter (Fig. 4C and SI Appendix, Fig. S7).
Fig. 4.
Registration of SMACM localizations and CET data. (A) Heavily saturated average fluorescence data showing the identified centers (dark blue asterisks) of the holes (cyan circles) in the holey carbon grid. These hole centers will be used as control points for a projective transformation to register fluorescence and electron microscopy images. (B) Low-magnification electron micrograph of the same region imaged in A with the same holes and hole centers identified. The Inset shows 15-nm gold beads labeled with black arrows. Gold beads neighboring the cell of interest outlined in white are used as control points for a similar coordinate transformation that will register this low-magnification micrograph with the high magnification CET data. (C) Resulting CET and McpA–PAmKate SMACM localizations (red) for the cell of interest outlined in white in B. The agreement between the position of the chemoreceptor array and the single-molecule localizations validates the image registration process.
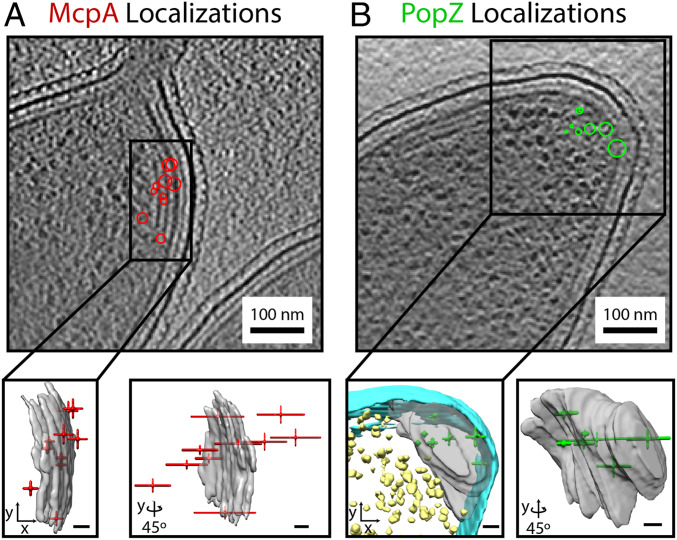
While the holey carbon substrate provides control points for the lateral registration of the fluorescence and CET data, our approach currently lacks control points for axial registration between the two imaging modalities. However, with the addition of astigmatism to the fluorescence microscope for axial drift correction during acquisition, there is axial localization information in the single-molecule PSFs. The resulting axial precision of these localizations is worse than expected by approximately a factor of 2 (SI Appendix, Fig. S8). This reduced precision is attributed to the residual effects of overlapping emitters. Despite lacking axial control points, absolute positioning of the SMACM localizations relative to the reconstructed tomograms can be performed by aligning the average position of the features observed in each imaging modality (Fig. 5 and Movie S2). Assuming that the axial registration error is dominated by the sparse SMACM localizations and that the SMACM localizations randomly sample the structure observed in CET, the error in the accuracy of the registration will follow SE statistics and decrease with an increased number of localizations. For example, the SD of the PAmKate–PopZ axial localizations shown in Fig. 5B is 90 nm. This results in a 27-nm SEM for the 11 localizations. Once axial registration has been performed, the SMACM localizations can be viewed in the full three-dimensional (3D) context provided by CET. While the axial registration error using this approach is comparable to that of the lateral registration error, this approach only works for structures that are visible directly in the CET data, somewhat defeating the purpose of correlation. Improvements in fiducials for axial registration are needed to make 3D correlations generalizable, and this is an area of ongoing development.
Fig. 5.
SMACM localizations in 3D cellular context. (A, Top) McpA–PAmKate localizations (red) overlaid onto a single tomographic slice. (A, Bottom) The same localizations represented as crosshairs where the length of the arm of the crosshairs is the SE on the mean position in the axial and lateral dimensions. The SMACM localizations in red are overlaid with the annotated chemoreceptor array shown in gray as visualized with the Chimera software. (B, Top) Same as A, but displaying PAmKate–PopZ localizations (green). (B, Bottom) Same as A, but with PAmKate–PopZ localizations shown in the 3D context of the annotated inner membrane (cyan), ribosomes (yellow), and ribosome excluded region (gray). (Scale bar: Bottom, 20 nm in all cases.)
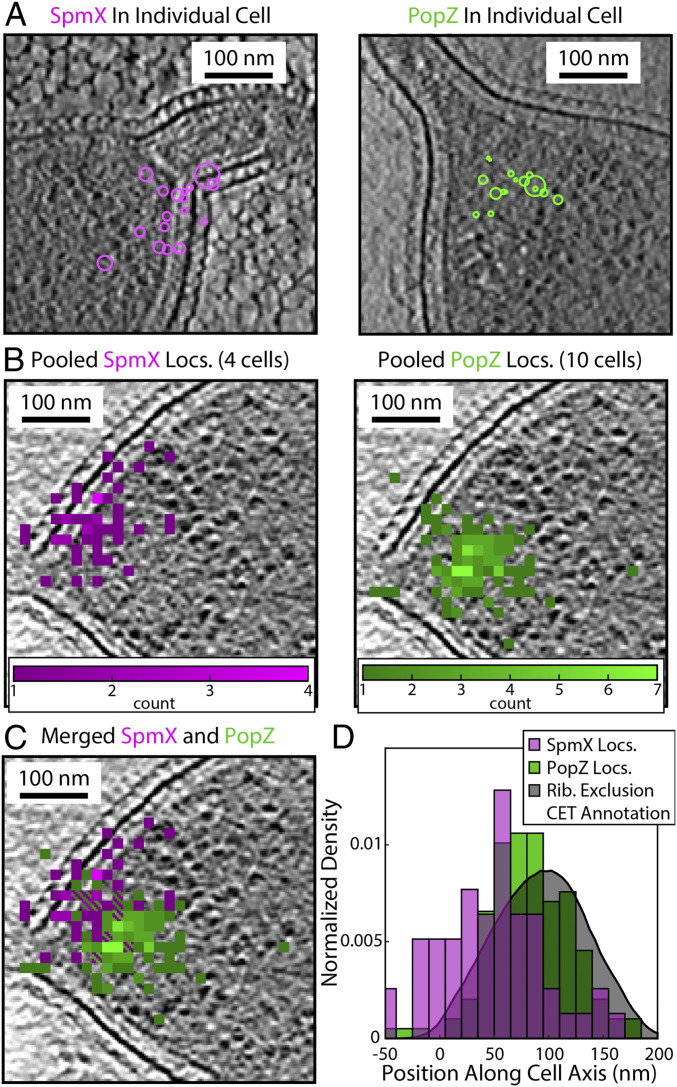
With the successful registration of SMACM localizations and CET for biomolecules of general interest in the two lateral dimensions, we turned our attention to the interaction between PopZ and SpmX by visualizing their subcellular organization. Fig. 6A shows the SpmX and PopZ localizations for individual cells. To overcome the sparsity of localizations recovered from individual cells, the localizations from all cells for which there are correlative data can be aligned based on the position of the base of the stalk and outer membrane, which are clearly observable in CET (SI Appendix, Fig. S9). Once aligned, a more detailed SMACM reconstruction can be overlaid by pooling all localizations onto a representative tomographic slice (Fig. 6B). Interestingly, while SpmX is localized to the stalked pole of the bacterium, it is not localized symmetrically about the stalk. Rather, the localizations are clustered on either the dorsal or ventral side of the bacterium when viewed from the top. This pattern of asymmetric localizations has been previously reported (31), albeit at a lower resolution, and is seen in all stalked poles for which correlative CET data were collected. The resulting visualization of both SpmX and PopZ shows SpmX is localized to the base of the PopZ microdomain (Fig. 6C). The extent of SpmX along the inner membrane closely matches that of PopZ. This shared extent of PopZ and SpmX is visible in the aligned cell data shown in Fig. 6C as well as in individual cells (SI Appendix, Fig. S9). A histogram of localizations along the cell body axis from the aligned cell data shows this arrangement of proteins, with the distribution of SpmX molecules being centered closer to the base of the stalk than PopZ (Fig. 6D and SI Appendix, Fig. S10). The spatial extent of PopZ and SpmX can be quantified in relation to observables from the CET data, such as the manually annotated ribosome exclusion region that normally defines the PopZ microdomain. We see good agreement between the CET and SMACM localizations, suggesting that PopZ is uniformly distributed throughout the ribosome exclusion volume as was previously assumed.
Fig. 6.
Subcellular organization of SpmX and PopZ revealed by CIASM. (A) SMACM localizations of SpmX–PAmKate (magenta) and PAmKate–PopZ (green) from individual cells registered with a tomographic slice from CET data. (B) The pooled SMACM localizations from several cells for either the SpmX–PAmKate or PAmKate–PopZ strains. Cells were aligned and the localizations pooled based on their correlative CET data and overlaid onto a representative tomographic slice. For representation, localizations were binned to produce a histogram with 14.5-nm pixels. (C) Merged data from B showing the relative positions of SpmX and PopZ. (D) Distributions of SpmX and PopZ localizations along the cell axis, where zero is defined as the base of the stalk as identified in CET. The shaded gray curve is the result of manual annotation of the ribosome exclusion region from the CET data, which shows good agreement with the PAmKate–PopZ localizations.
Discussion
We have established a four-stage workflow that correlates SMACM localizations with CET reconstructions to provide ground truth annotations of specifically labeled biomolecules of interest. This workflow achieves localizations with an average precision below 10 nm, resulting from the large number of photons being collected from each emitter due to the reduced quantum yield of photobleaching at cryogenic temperatures. These large numbers of photons collected over multiple frames make it essential that localizations be merged into a single estimate for an individual emitter both to prevent overcounting of the number of emitters and to achieve the highest possible localization precision. For these precise localizations to be useful as annotations for correlative imaging, they must be accurately registered with the CET data. Our workflow accomplishes this by using the centers of the holes in holey carbon grids as fiducials visible in both imaging modalities and achieves registration with an accuracy of ∼30 nm as assessed by the registration of a fluorescent polystyrene bead. These precise and accurate localizations identify the subcellular locations of the three proteins investigated in this study more clearly than conventional diffraction-limited correlative light and electron microscopy, room temperature SMACM, or CET.
The correlative imaging of McpA arrays provides a strong validation of this workflow due to the visibility of the chemoreceptor array in CET. This validation gives confidence in the assignment of both PopZ and SpmX localizations. In the case of PopZ, our results show good agreement between the ribosome exclusion region visible in CET and the registered PAmKate–PopZ localizations. The uniform distribution of PopZ throughout this excluded region strongly supports the model of PopZ being a space-filling plug in the polar region of the cell (27). Furthermore, SpmX, which forms no visible structure in CET, was found to localize on one side of the cell near the base of the stalk and extend along the inner membrane to the edge of the PopZ microdomain. This shared boundary of the microdomain formed by PopZ and the extent of membrane-bound SpmX is in agreement with data establishing that SpmX is recruited to the microdomain by its affinity for PopZ (37). However, the asymmetry of SpmX localizations within the polar microdomain, now clearly visible with the superior resolution in our experiments, is intriguing and raises the possibility that there may be other factors affecting the localization of SpmX within the PopZ microdomain. One possibility is that the localized pattern of SpmX could be a result of its self-association as has been demonstrated via biochemical experiments (37). Alternatively, it has been recently shown that the localization of stalked pole specific factors such as bactofilins and peptidoglycan binding proteins is mediated through SpmX in Asticcacaulis biprosthecum, a close relative of C. crescentus, and so interactions with other proteins may be driving SpmX’s asymmetric location within the PopZ microdomain (38). Understanding how SpmX oligomerization and interactions with other proteins lead to its subcellular organization and ultimately its biological function remains an exciting and active area of research in our laboratory.
The results presented here provide the clearest picture to date of the subcellular localization of the PopZ–SpmX signaling complex in C. crescentus, and a method for correlating CET and fluorescently tagged proteins in any cell. Currently, the most restrictive limitation in the quality of correlative images produced by our approach is the incomplete photophysical control over the emissive states of molecules at low temperatures using low excitation power. While PAmKate can be efficiently photoactivated at cryogenic temperatures, it is not possible to efficiently switch the emitters back to an off-state. Instead, we rely on stochastic processes to either permanently photobleach emitters or shelve them into long-lived dark states. These stochastic processes have very low quantum yields at cryogenic temperatures, and while these low yields permit the collection of more photons than room temperature studies, thus improving localization precision, the long on-times are actually limiting resolution by restricting the number of emitters that can be localized. Improved photophysical control over the fluorescent labels could possibly be obtained through new photochemistry that is robust to cryogenic temperatures or by exploiting the rugged cryogenic energy landscape to produce long-lived dark states not observed at room temperature. No matter how it is achieved, the ability to both activate and inactivate emitters efficiently is necessary for improved cryogenic superresolution imaging compatible with CET. Other aspects of this method that limit the quality of registered images include the axial precision, axial registration methods, and devitrification at low excitation powers.
None of these challenges is the result of fundamental physical limitations; rather, improvements in all of these areas can be obtained with additional engineering and experimentation. Given the importance of combined single-molecule sensitivity and specificity with high-resolution cellular context, we are confident that these challenges will be overcome and that CIASM has a bright future. The strengths and weaknesses of the SMACM and CET complement each other such that the resulting images are more informative than either technique is individually. We expect that this method will be useful for highlighting the large number of cellular protein complexes that cannot be directly discerned in CET due to either their small size or lack of periodic structure (such as the locations of the members of the divisome), and for identifying proteins in electron-dense structures of unknown composition.
Materials and Methods
Generation of C. crescentus PAmKate Fusion Strains.
McpA–PAmKate.
To construct C-terminal PAmKate fusions in a single step under a native locus, we used the vector pYFPC-2 (or under the xylose locus, pXyl-YFPC-2) backbones. The YFP gene was cut out using EcoRI and NheI enzymes, and the resulting backbone was purified by separation on an agarose gel. The PAmKate gene was amplified from Addgene plasmid #32691 (39) and inserted into the above backbones using Gibson assembly, resulting in the pPAmKateC-2 and pXylPAmKateC-2 vectors. The McpA coding region was amplified with appropriate overhangs from genomic DNA from NA1000 cells and gel purified. The pXPAmKateC-2 vector was linearized by digesting with NdeI, and the McpA gene was inserted using Gibson assembly. The linker used for the McpA fusion was GTLSRPENSNVHRS.
PAmKate–PopZ.
To construct an N-terminal fusion construct, PAmKate and PopZ genes were PCR amplified with a –GGGSGGGS– linker and cloned into pXyl-YFPC-2 that was digested by NdeI/NheI using Gibson Assembly (40, 41). The ligation was transformed into Escherichia coli DH5α cells and selected on LB-kan plates. Sequence-verified plasmid was then transformed into NA1000 cells via electroporation.
SpmX–PAmKate.
In order to produce the plasmid for the SpmX–PAmKate C-terminal fusion on the native promoter, the region encoding SpmX and the 500 base pairs upstream were amplified from genomic DNA from NA1000 cells and gel purified. This gene was then inserted at the EcoRI site on the linearized pPAmKateC-2 backbone. The linker used for the SpmX fusion was PAGALINMHGTLRSRAPENSNVTRHRSAT.
Room Temperature Superresolution Imaging.
C. crescentus cells were induced with 0.3% xylose for 3 h (McpA–PAmKate, PAmKate–PopZ) or cultured with endogenous expression (SpmX–PAmKate) and placed on agarose pads. These molecules either form relatively fixed structures or do not move dramatically during live cell imaging; highly mobile molecules are rejected as bad fits. Imaging was performed with a custom epifluorescence microscope. Fluorescence emission under 680 W/cm2 of 561 nm excitation (Coherent Sapphire) was collected using an oil immersion, supercorrected objective (Olympus PLANON60xOSC, 60×/N.A. 1.4) mounted in a Nikon Diaphot 200 microscope and imaged onto an EMCCD camera (Andor iXon) at 20 Hz. Occasional pulses (<1 s) of 400 W/cm2 of 405-nm photoactivation light (Coherent OBIS) were used to maintain sufficient concentrations of active emitters. In total, 7,000 to 9,000 frames were collected for each dataset. The acquired data were processed with ThunderSTORM (35) by fitting identified particles with a standard Gaussian model and filtering out bad fits (σ of Gaussian, >150 nm; intensity, <200 photons/frame; or uncertainty, >18 nm; <10% of localizations were removed by these filters).
Correlative Imaging: Sample Preparation.
McpA–PAmKate and PAmKate–PopZ cell cultures were induced with 0.3% xylose for 3 h prior to plunge freezing. The SpmX–PAmKate strain was endogenously expressed. A small aliquot of the bacterial cultures expressing PAmKate fusion constructs was mixed with 30% (vol/vol) ethylene glycol, which serves as a cryoprotectant, and with 50% (vol/vol) of a suspension of 15-nm gold beads and 40-nm fluorescent polystyrene beads. The mixture was then deposited on holey carbon electron microscopy finder grids (Quantifoil R2/2) and plunged into liquid ethane (CP3 Gatan). The frozen samples were stored in liquid nitrogen for several days to allow the ethane to sublime and then loaded onto the cryogenic fluorescence stage (CMS196 Linkam Scientific) for imaging.
Correlative Imaging: Cryogenic Fluorescence Microscopy.
Cryogenic imaging was performed on a homebuilt upright microscope that has been described previously (18). The tip of a long working distance objective (Nikon CFI Tu Plan Apo 100×/N.A. 0.9) was immersed in the cold nitrogen vapor in the sample chamber of a cryogenic fluorescence stage (CMS196 Linkam Scientific) just above the plunge frozen grid. The cryogenic stage holds the sample grid at cryogenic temperatures through strong thermal contact between the grid and a copper bridge that is partially submerged in a liquid nitrogen reservoir. Fluorescence was excited using an illumination intensity of ∼50 to 100 W/cm2 at the image plane from a 561-nm laser (CrystaLaser). Excitation intensities were kept low to avoid devitrification, which above this intensity occurs rapidly and locally to the area of excitation. Photoactivation was achieved using short pulses (50 to 500 ms) of ∼5 to 15 W/cm2 of 405-nm light. Both the intensity and duration of the 405-nm pulses were adjusted to achieve sufficient activation while avoiding excess emitter overlap as much as possible. Fluorescence emission from 561-nm excitation was filtered from scattered excitation light using a dichroic mirror (Semrock Di03-R561-t1-25x36) as well as a bandpass and a notch filter (FF01-607/70-25 and Chroma ZET561NF). A 1-m focal length cylindrical lens was mounted just prior to the camera (Andor iXon) to generate an astigmatic PSF that was used for axial localization and for live drift correction using custom Labview acquisition software (18). Drift correction was performed by fitting the PSF of 40-nm fluorescent polystyrene beads. Fluorescence frames were collected at 1 Hz, and ∼10,000 frames were acquired for each SMACM dataset. These frames were drift corrected using cross-correlation and then binned in time by a factor of 2 to improve the signal-to-noise ratio and reduce data volume. These drift-corrected frames are then used for subsequent data analysis including the localization process described in Fig. 3 and accompanying text.
Correlative Imaging: Cryogenic Electron Tomography and Microscopy.
CET data were collected using a 300-keV electron microscope (Titan Krios Thermo Fisher) with a direct detector (K2 Gatan) and an energy filter (Bioquantum Gatan). The tilt series was acquired with either 3° steps for PopZ imaging or 1° steps for SpmX and McpA imaging. The tilts were acquired in a bidirectional manner (42) beginning with the sample at 21°. For tilt series with 1° steps the stage positions were [21, 20, 19 … −45, 22, 23, 24… 45], and for 3° steps the stage positions were [21, 18, 15 … −45, 24, 27, 30 …45]. The total dose for each tilt series was 100 e-/Å2. Smaller step sizes enabled easier alignment of the tilt series during reconstruction using the Etomo package in IMOD (43). Tilts were acquired with a pixel size of 7.28 Å and 10 μm of defocus. These imaging parameters were chosen so that an entire C. crescentus cell could be acquired in a single tomogram. At these relatively low magnifications, resolving high-resolution structures would not be feasible, and so the defocus was set to improve low-resolution contrast. The additional low-magnification image required for image registration was taken following CET data collection and was centered on the cell in the CET data. This low-magnification micrograph had a pixel size of 38.8 Å. At this magnification, a sufficient number of holes, typically 12 to 16, were visible for registration to the fluorescence data, and the 15-nm gold beads used for alignment to the CET data were also visible (Fig. 4).
Correlative Imaging: Visualization.
CET data were reconstructed using the Etomo package from IMOD (43) using 15-nm gold beads as fiducials. The resulting tomograms were binned by four and, in the case of annotation, were annotated by hand or with the assistance of a neural network in EMAN2 (14). Fluorescence data in most cases were displayed as merged single-emitter localizations. The pooled reconstructions shown in Fig. 6 were displayed as histograms of the merged localizations with a pixel size of 14.6 nm.
Data Availability Statement.
Tomographic reconstructions and single-molecule localizations have been made publicly available in the Electron Microscopy Data Bank (accession no. EMD-21706) (44).
Supplementary Material
Acknowledgments
This work was supported in part by the National Institute of General Medical Sciences Grants R35GM118067 (to W.E.M.), R35GM118071 (to L.S.), and P01NS092525 (to W.C.), and Department of Energy Grant BER FWP100463 (to W.C.). A.M.S. is a William and Lynda Steere Bio-X Stanford Interdisciplinary Graduate Fellow, and J.W. is a Mona M. Burgess Bio-X Stanford Interdisciplinary Graduate Fellow. L.S. is a Chan Zuckerberg Biohub Investigator, W.C. is a Chan Zuckerberg Biohub Intercampus Research Award Investigator, and W.E.M. is a Chan Zuckerberg Biohub Intercampus Research Award Collaborator.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Tomographic reconstructions and single-molecule localizations have been deposited in the Electron Microscopy Data Bank (accession no. EMD-21706).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001849117/-/DCSupplemental.
References
- 1.Betzig E. et al., Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Hess S. T., Girirajan T. P. K., Mason M. D., Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rust M. J., Bates M., Zhuang X., Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schueder F. et al., Multiplexed 3D super-resolution imaging of whole cells using spinning disk confocal microscopy and DNA-PAINT. Nat. Commun. 8, 2090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hershko E., Weiss L. E., Michaeli T., Shechtman Y., Multicolor localization microscopy and point-spread-function engineering by deep learning. Opt. Express 27, 6158–6183 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Lehmann M., Lichtner G., Klenz H., Schmoranzer J., Novel organic dyes for multicolor localization-based super-resolution microscopy. J. Biophotonics 9, 161–170 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Thompson R. F., Walker M., Siebert C. A., Muench S. P., Ranson N. A., An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods 100, 3–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke N. I., Royle S. J., FerriTag is a new genetically-encoded inducible tag for correlative light-electron microscopy. Nat. Commun. 9, 2604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morphew M. K. et al., Metallothionein as a clonable tag for protein localization by electron microscopy of cells. J. Microsc. 260, 20–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q., Mercogliano C. P., Löwe J., A ferritin-based label for cellular electron cryotomography. Structure 19, 147–154 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Frangakis A. S. et al., Identification of macromolecular complexes in cryoelectron tomograms of phantom cells. Proc. Natl. Acad. Sci. U.S.A. 99, 14153–14158 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ortiz J. O., Förster F., Kürner J., Linaroudis A. A., Baumeister W., Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition. J. Struct. Biol. 156, 334–341 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Rigort A. et al., Automated segmentation of electron tomograms for a quantitative description of actin filament networks. J. Struct. Biol. 177, 135–144 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Chen M. et al., Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat. Methods 14, 983–985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subach F. V. et al., Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states. Proc. Natl. Acad. Sci. U.S.A. 106, 21097–21102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha T., Tinnefeld P., Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 63, 595–617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Y. W. et al., Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat. Methods 11, 737–739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahlberg P. D. et al., Identification of PAmKate as a red photoactivatable fluorescent protein for cryogenic super-resolution imaging. J. Am. Chem. Soc. 140, 12310–12313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuijtel M. W., Koster A. J., Jakobs S., Faas F. G. A., Sharp T. H., Correlative cryo super-resolution light and electron microscopy on mammalian cells using fluorescent proteins. Sci. Rep. 9, 1369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman D. P. et al., Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367, eaaz5357 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lasker K., Mann T. H., Shapiro L., An intracellular compass spatially coordinates cell cycle modules in Caulobacter crescentus. Curr. Opin. Microbiol. 33, 131–139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briegel A. et al., New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry 53, 1575–1585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briegel A. et al., Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. U.S.A. 106, 17181–17186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briegel A. et al., Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol. Microbiol. 69, 30–41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazelbauer G. L., Falke J. J., Parkinson J. S., Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem. Sci. 33, 9–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gahlmann A. et al., Quantitative multicolor subdiffraction imaging of bacterial protein ultrastructures in three dimensions. Nano Lett. 13, 987–993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowman G. R. et al., Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol. Microbiol. 76, 173–189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasker K. et al., Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat. Microbiol. 5, 418–429 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowman G. R. et al., Oligomerization and higher-order assembly contribute to sub-cellular localization of a bacterial scaffold. Mol. Microbiol. 90, 776–795 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebersbach G., Briegel A., Jensen G. J., Jacobs-Wagner C., A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134, 956–968 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang C., Brown P. J., Ducret A., Brun Y. V., Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature 506, 489–493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon M. P., Ha T., Selvin P. R., Single-molecule high-resolution imaging with photobleaching. Proc. Natl. Acad. Sci. U.S.A. 101, 6462–6465 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park H., Hoang D. T., Paeng K., Kaufman L. J., Localizing exciton recombination sites in conformationally distinct single conjugated polymers by super-resolution fluorescence imaging. ACS Nano 9, 3151–3158 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Qu X., Wu D., Mets L., Scherer N. F., Nanometer-localized multiple single-molecule fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 101, 11298–11303 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ovesný M., Křížek P., Borkovec J., Svindrych Z., Hagen G. M., ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson K. L., Page C., Swift M. F., Hanein D., Volkmann N., Marker-free method for accurate alignment between correlated light, cryo-light, and electron cryo-microscopy data using sample support features. J. Struct. Biol. 201, 46–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez A. M. et al., A localized complex of two protein oligomers controls the orientation of cell polarity. MBio 8, e02238-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caccamo P. D., Jacq M., VanNieuwenhze M. S., Brun Y. V., A division of labor in the recruitment and topological organization of a bacterial morphogenic complex. bioRxiv:10.1101/847335 (20 November 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunewardene M. S. et al., Superresolution imaging of multiple fluorescent proteins with highly overlapping emission spectra in living cells. Biophys. J. 101, 1522–1528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thanbichler M., Iniesta A. A., Shapiro L., A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35, e137 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibson D. G. et al., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Hagen W. J. H., Wan W., Briggs J. A. G., Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197, 191–198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Dahlberg P. D. et al., Cryogenic Single-Molecule Fluorescence Annotations for Electron Tomography Reveal In Situ Organization of Key Proteins in Caulobacter. https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-21706. Accessed 17 April 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tomographic reconstructions and single-molecule localizations have been made publicly available in the Electron Microscopy Data Bank (accession no. EMD-21706) (44).