Significance
In 1840, Justus von Liebig proposed the theory of mineral plant nutrition, through the invention of the Haber–Bosch process, leading to the industrialization of chemical fertilizer (inorganic nitrogen) to feed the human population. Because the excessive use of chemical fertilizer has led to numerous environmental problems, understanding the agroecosystem that contains plants, microbes, and soils is necessary for sustainable agriculture. We revealed the network structure of an agroecosystem established with different management practices and identified that organic nitrogen is a key component contributing to crop yield under the condition of soil solarization, even in the presence of inorganic nitrogen. Our finding provides a potential solution to make crop production more sustainable by utilizing organic nitrogen induced by soil solarization.
Keywords: agroecosystem, multiomics, soil solarization, organic nitrogen, Brassica rapa
Abstract
Both inorganic fertilizer inputs and crop yields have increased globally, with the concurrent increase in the pollution of water bodies due to nitrogen leaching from soils. Designing agroecosystems that are environmentally friendly is urgently required. Since agroecosystems are highly complex and consist of entangled webs of interactions between plants, microbes, and soils, identifying critical components in crop production remain elusive. To understand the network structure in agroecosystems engineered by several farming methods, including environmentally friendly soil solarization, we utilized a multiomics approach on a field planted with Brassica rapa. We found that the soil solarization increased plant shoot biomass irrespective of the type of fertilizer applied. Our multiomics and integrated informatics revealed complex interactions in the agroecosystem showing multiple network modules represented by plant traits heterogeneously associated with soil metabolites, minerals, and microbes. Unexpectedly, we identified soil organic nitrogen induced by soil solarization as one of the key components to increase crop yield. A germ-free plant in vitro assay and a pot experiment using arable soils confirmed that specific organic nitrogen, namely alanine and choline, directly increased plant biomass by acting as a nitrogen source and a biologically active compound. Thus, our study provides evidence at the agroecosystem level that organic nitrogen plays a key role in plant growth.
Agroecosystems are among the most inextricable self-organizing systems, with different levels such as plants, microbes, and soils interacting with each other and responding to different management practices, but they remain largely unexplored at the system level (1, 2). Although factors contributing to crop production have been pursued on an individual biotic and abiotic basis, the interaction between different levels in an agroecosystem carries unpredictability regarding agriculturally relevant crop phenotypes (3). In addition to plant responses against biotic and abiotic environmental factors, microbial function depends on their microhabitats, and plants modulate microbial community profile using phytohormones and metabolites (4–6). These complex interactions in agroecosystems give the capacity to self-organize through a series of continuous negative biological feedback loops (7). Thus, agroecosystems show dynamic changes governed by complex interactions via external signals.
Current agricultural management systems predominantly hold the simplified view centered on inorganic nitrogen based on the theory of mineral plant nutrition proposed by Justus von Liebig in 1840. This theory implies that plants acquire nutrients mostly in the inorganic form. Since nitrogen availability is a major factor determining plant growth, von Liebig’s vision transformed agriculture with the invention of the Haber–Bosch process, which chemically converts nitrogen gas into ammonia, and the industrialization of chemical fertilizer, eventually feeding the world population (8). However, current concern about environmental burdens of chemical fertilizer has forced us to revisit sustainable agriculture utilizing natural nutrient cycles and biological feedback mechanisms in soils (9, 10). Therefore, the comprehensive understanding of the multilevel interactions in view of agroecosystems is vitally important.
As an environmentally friendly farming method, soil solarization (SS) uses nature’s inexhaustible source of solar energy to disinfest soil-borne plant pathogens and remove weeds without using chemicals, and is also reported to improve plant growth, a phenomenon known as the increased growth response (IGR) (11). The SS-induced IGR has been reported in numerous field trials with various types of soils including silty clay and sandy loam, and with a broad range of crops such as tomato, eggplant, Chinese cabbage, and okra (11–17); therefore, SS can be a widely applicable agricultural crop production method (18, 19). Several hypotheses for the mechanisms explaining the SS-induced IGR have been proposed to date, such as changes in macro- and microelements in the soil such as nitrogen-containing compounds and several minerals, the elimination of minor pathogens, the release of growth regulator-like substances, and the stimulation of beneficial microorganisms. However, key components contributing to IGR are still unknown.
Recent development of multiple omics tools has enabled the generation of large datasets that can be used to understand complex agroecosystems (1). These datasets allow us to determine exactly how soil metabolic, mineral, and microbial components are organized into a network structure, but such an approach has not been undertaken to date to the authors’ knowledge. Here, we explored the responsive structure of an agroecosystem engineered by several farming methods, leveraging several omics technologies such as metabolome, ionome, microbiome, and phenome as well as integrated informatics. We revealed the network module structure behind the agroecosystem and identified SS-induced organic nitrogen as a key component contributing to crop yield.
Results
SS-Induced IGR in a Crop Grown on an Agricultural Field.
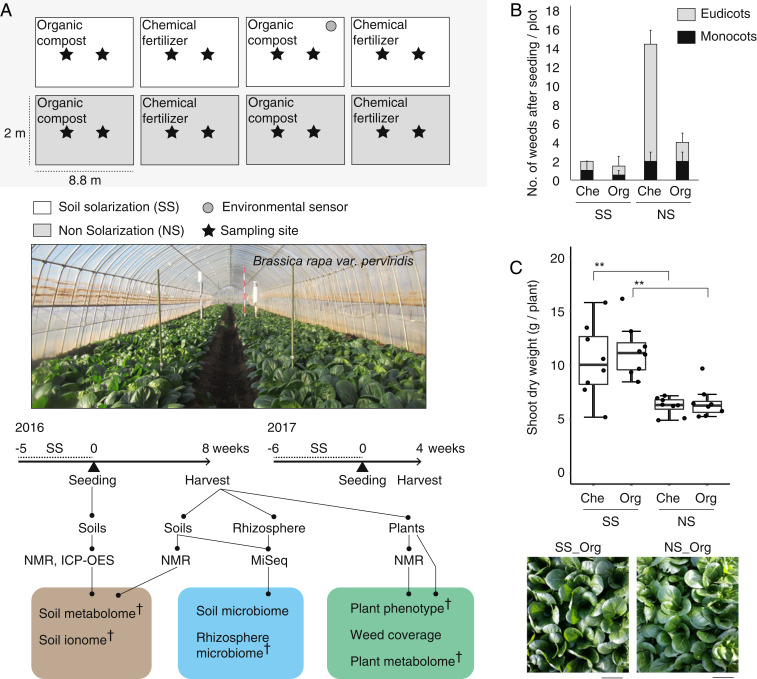
To dissect agroecosystems engineered by different farming methods, we grew Japanese mustard spinach (Brassica rapa var. perviridis) in a split-plot designed agricultural field in which the main effect was SS and the subeffect was fertilizer, monitored by several environmental sensors (Fig. 1A and SI Appendix, Fig. S1). Day accumulated soil temperature was over 1,000 °C during SS, and ∼200 mV soil redox potential (Eh) was maintained after SS at 10 cm soil depth (SI Appendix, Fig. S1), where facultative anaerobic bacteria prefer to propagate (20). Consistent with previous observations (16), SS decreased weed coverage, while we also detected that organic compost decreased weed coverage even in the nonsolarization (NS) plot (Fig. 1B and SI Appendix, Fig. S2). SS increased dry weight of plant shoots irrespective of the type of fertilizer (Fig. 1C), confirming the SS-induced IGR effect (11, 14, 15). Given that the crop phenotypic quality (i.e., metabolite profile, leaf shape traits, photosynthesis rate, sugar content, leaf chlorophyll, and taste index) was not significantly different between SS and NS treatments (SI Appendix, Table S1), SS likely induced additional nutrient release in the soil and stimulated physiological and morphological changes in plants to increase nutrient acquisition, rather than modifying the growth/immunity trade-off in plant resource allocation. In addition, we observed a robust IGR effect on crop yield (fresh weight [g]/m2) even after swapping the locations of SS and NS plots in the following year (SI Appendix, Fig. S3).
Fig. 1.
SS-induced IGR is detected in the crop grown on an agricultural field. (A) Split-plot designed field for B. rapa var. perviridis cultivation and experimental design for integrating omics approach. Analysis methods denoted with a dagger were used for integrated network analysis. (B) Number of weeds in each plot on harvest. Eudicots (gray) and monocots (black) were separately displayed. (C) Shoot dry weight (n = 8 plants) and leaf images at harvest stage (**P < 0.01 as analyzed by one-way ANOVA followed by a Tukey’s post hoc test). (Scale bars: 10 cm.) SS, soil solarization; NS, nonsolarization; Che, chemical fertilizer; Org, organic compost.
Soil Metabolic, Mineral, and Bacterial Dynamics in the Agroecosystem.
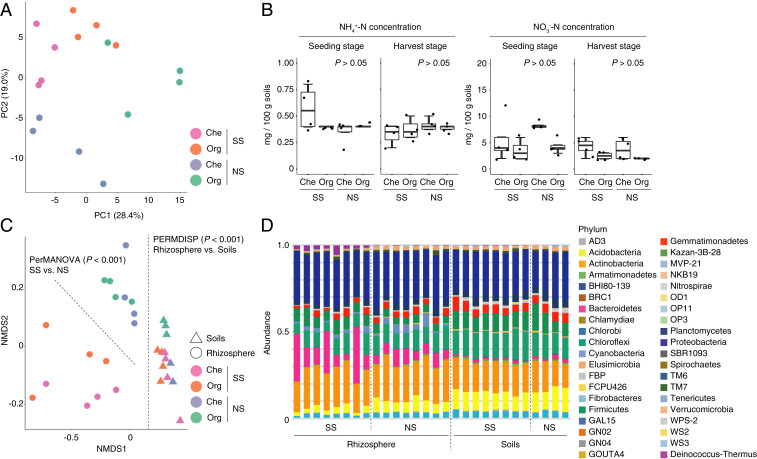
To detect changes in soil metabolic, mineral, and bacterial profiles in the agroecosystem where the SS-induced IGR was observed, we performed NMR-based metabolomics, ICP-OES-based ionomics, and bacterial 16S rRNA gene amplicon sequencing (Fig. 1A). Principal component analysis (PCA) of all soil metabolome and ionome data indicated that soil samples were grouped separately from each other depending on each experimental plot, validating our experimental treatments (Fig. 2A). Unexpectedly, the concentrations of soil inorganic nitrogen (NH4+-N and NO3−-N) were not significantly different between experimental treatments at either seeding or harvest stages (Fig. 2B), indicating that the SS-induced IGR effect observed in this study might be caused by the increased flux rates of nitrogen (i.e., rate of uptake by plants) in SS soils while maintaining equilibrium of decomposition of organic forms into inorganic forms among the treatments, or different from the prevailing idea that inorganic nitrogen is the major nitrogen source for agricultural crops. Consistent with a previous observation that SS increases soluble organic matter (15), an abundance test using a false discovery rate (FDR) of <0.05 between SS and NS treatments showed that organic nitrogen was transiently depleted at seeding stage but eventually enriched at harvest stage in the SS plot (SI Appendix, Table S2 and Figs. S4 and S5).
Fig. 2.
SS-induced soil nutrient cycle through metabolic, mineral, and bacterial dynamics. (A) Soil metabolome and ionome profile analyzed by PCA. (B) Concentration of inorganic nitrogen in soils at seeding and harvest stages. No significant differences were observed between treatments within each condition (P > 0.05, unequal-variance t test). (C) Microbiome profiles in rhizosphere and soils analyzed by NMDS using the Bray–Curtis distance matrix. Significance test was performed by PerMANOVA and PERMDISP. (D) Microbiome compositions in rhizosphere and soils displayed as stacked bar plot at phylum level. SS, soil solarization; NS, nonsolarization; Che, chemical fertilizer; Org, organic compost.
As reported in previous studies with Arabidopsis and rice microbiome (21, 22), nonmetric multidimensional scaling (NMDS) with the Bray–Curtis dissimilarity (β diversity) using family-level operational taxonomic units (OTUs) allows us to detect the differentiation of soil and rhizosphere bacterial community structure (permutational multivariate analysis of variance [PerMANOVA], df = 1, Fmodel = 16.5, P < 0.001). The detected differentiation might be caused by the heterogeneity of dispersions between soils and rhizosphere (permutational analysis of multivariate dispersions [PERMDISP], df = 1, F = 15.9, P < 0.001). Interestingly, the effect of SS was detected only on the community structure of rhizosphere bacteria but not of soil bacteria (PerMANOVA, df = 1, Fmodel = 8.25, P < 0.001; and df = 1, Fmodel = 1.45, P > 0.05, respectively; Fig. 2C). The PERMDISP analysis indicated that the differentiation of rhizosphere bacterial community structure induced by SS reflected change in the bacterial profile and not in the heterogeneity of dispersions (df = 1, F = 4.38, P > 0.05). Drastic differences in the proportions of phyla among SS and NS treatments were detected: the phyla Firmicutes and Deinococcus-Thermus were enriched, while Verrucomicrobia, Planctomycetes, Spirochaetes, Acidobacteria, and WS2 were depleted, in the rhizosphere bacterial profile in SS plots compared to NS plots (FDR < 0.05; Fig. 2D and SI Appendix, Fig. S6). In addition, we found that the genera Paenibacillus (phylum Firmicutes) and Pseudomonas (Proteobacteria), known as plant growth-promoting rhizobacteria (23, 24), are highly enriched in the rhizosphere bacterial profile in the SS plot (SI Appendix, Fig. S6), and indeed these bacteria can be isolated from plant roots grown in SS-treated soils (25). These data demonstrate that SS assembles and maintains the specific community profile of rhizosphere bacteria at the phylum level although the profile of soil bacteria was disrupted during plant cultivation.
Integrated Network Identifies Organic Nitrogen as a Key Component in the Agroecosystem.
In the agroecosystem we investigated, the SS treatment and not the type of fertilizer mainly explained the variation in the integrated all-omics data including plant phenome, plant metabolome, soil metabolome, soil ionome, and rhizosphere microbiome (SI Appendix, Fig. S7; principal component 1 explains 18.9% of the variance). To gain an insight into the responsive network structure in the agroecosystem, we constructed an unsigned correlation network using the integrated all-omics data. The resultant network has 9 major modules including more than 20 nodes determined by the Fast–Greedy modularity optimization algorithm (Fig. 3A and SI Appendix, Table S3). This indicates that the complex interactions seen in the agroecosystem show multiple network modules represented by plant traits heterogeneously associated with soil metabolites, minerals, and bacteria, which share a key structural similarity to other biological network systems. The module M7, which includes shoot dry weight, has soil organic nitrogen as highly interconnected hub metabolites, in addition to minerals and rhizosphere bacteria such as thermophilic bacteria, reflecting the pattern of SS treatment (Fig. 3 B and C). The M7 was connected with other modules (M1, M4, M5, and M8) having traits related to crop quality such as taste, brix, plant sodium, and plant nitrogen, but not the module M6 having inorganic nitrogen NO3−-N in soils (Fig. 3A and SI Appendix, Fig. S8). This allows us to hypothesize that soil organic nitrogen detected in the M7 directly affects plant growth and is a key component induced by SS during crop production.
Fig. 3.
Integrated network identifies the rhizosphere microbes and organic nitrogen sources correlating with the crop yield. (A) Integrated network constructed based on unsigned correlation network with a Fast–Greedy modularity optimization algorithm. Nodes and edges represent omics measurements and correlation-based interactions between omics measurements, respectively. Edge width indicates the weight of interaction. The nine major modules (from M1 to M9) are represented by different colored nodes. (B) The correlation network of integrated data categorized into M7 module. The size and color coding of nodes indicate log strength and strength, respectively, whose value sums up the weights of the adjacent edges for each node. (C) Heatmap visualization of M7 module. SS, soil solarization; NS, nonsolarization. Organic nitrogen sources denoted by an asterisk were used for the in vitro assay.
SS-Induced Organic Nitrogen Directly Increases Crop Yield.
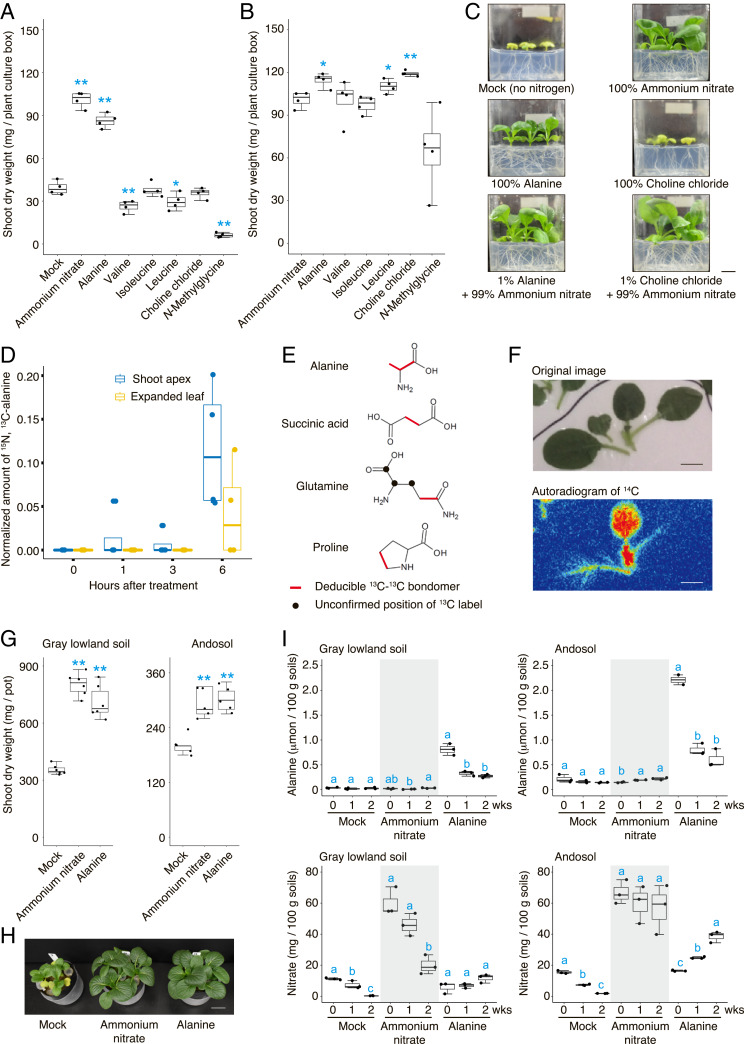
We tested this hypothesis by application experiments of the SS-induced organic nitrogen identified from the integrated network analysis. To exclude the possibility that bacteria and soil enzymatic activity could mineralize organic nitrogen, we established an in vitro germ-free B. rapa var. perviridis system. Shoot biomass of B. rapa was similar when grown with equal amounts (5 mM nitrogen) of inorganic nitrogen or alanine, while N-methylglycine, valine, and leucine inhibited plant growth (Fig. 4 A and C and SI Appendix, Fig. S9). This suggests that organic nitrogen such as alanine can increase plant shoot biomass as a nitrogen source. In addition, supplemental application of organic nitrogen (1% of the total nitrogen amount) with inorganic nitrogen revealed that the application of choline, alanine, or leucine with inorganic nitrogen increased the shoot biomass of B. rapa compared with only inorganic nitrogen under the equivalent nitrogen concentration (Fig. 4 B and C and SI Appendix, Fig. S9). Given that choline stimulates the photosynthetic activity in protoplasts (26, 27) and foliar application of choline was reported to enhance the growth of grass species (28), choline could function as a biologically active compound to enhance plant growth. This was confirmed by the fact that the supplemental application of choline (0.17% of the total nitrogen) with sufficient inorganic nitrogen (30 mM nitrogen) still enhanced the growth of B. rapa (SI Appendix, Fig. S10).
Fig. 4.
Organic nitrogen can be utilized to increase crop yield. (A) Evaluation of organic nitrogen as a nitrogen source (5 mM nitrogen concentration) for plant shoot growth (n = 4 plant culture boxes; n = 3 plants grown in each plant culture box). (B) Evaluation of organic nitrogen as supplemental application (1% of the total nitrogen amount) with inorganic nitrogen for plant shoot growth (n = 4 plant culture boxes; n = 3 plants grown in each plant culture box). (C) Images represent 14-d-old B. rapa seedlings for each treatment. (Scale bar: 1 cm.) (D) Incorporation of dual-labeled alanine into shoot tissue analyzed by LC-MS. (E) The 13C-labeled compounds (i.e., alanine, succinic acid, glutamine, and proline) metabolized from the absorbed alanine detected by NMR. Deducible 13C-13C bondomers are indicated by red lines, and unconfirmed positions of 13C label are indicated by black dots. (F) Accumulation of 14C in the shoot traced with labeled alanine. (Scale bars: 1 cm.) (G) Evaluation of alanine by pot experiments using field soils (n = 6 and 5 pots; n = 3 plants grown in each pot) for gray lowland soil and andosol, respectively (*P < 0.05 and **P < 0.01, unequal-variance t test compared with mock and ammonium nitrate for A and G and B, respectively). (H) Images represent 18-d-old B. rapa seedlings grown on gray lowland soil for each treatment. (Scale bar: 3 cm.) (I) Changes in concentrations of alanine and nitrate in soils during the pot experiments (0, 1, and 2 wk). Different letters indicate significant differences determined by one-way ANOVA followed by a Tukey’s test compared within each treatment (P < 0.05).
On the contrary, our in vitro assay suggests that alanine could be utilized not only as a nitrogen source but also as a biologically active compound. To reveal how a plant can uptake and utilize alanine without the presence of bacteria and soil enzymatic activity, we once again used our germ-free B. rapa system with the application of dual-labeled (15N, 13C) alanine into roots to detect the unique spectrum of the dual-labeled alanine in the shoot using LC-MS. Our data clearly showed that intact, labeled alanine was transported into young shoot tissue within 1 h, and subsequently into mature shoot tissue for up to 6 h (Fig. 4D). In addition, the 13C derived from the dual-labeled alanine treated in the root were detected in the alanine, succinic acid, glutamine, and proline in the shoot by NMR (Fig. 4E and SI Appendix, Fig. S11), suggesting that absorbed alanine is metabolized into other amino acids through the tricarboxylic acid (TCA) cycle within 6 h. The autoradiograph obtained using labeled alanine (14C-alanine) confirmed that the 14C derived from alanine accumulated in the proliferative region of young leaves at the shoot apex (Fig. 4F and SI Appendix, Fig. S12). These data suggest that B. rapa can absorb and utilize alanine as a nitrogen source, possibly bypassing the common mineralization pathway to save metabolic energy of converting absorbed inorganic nitrogen into amino acids (29, 30).
Finally, to reproduce the effect of alanine in the agricultural field, we performed a pot experiment using two different types of arable soils. Consistent with our in vitro assay, the application of alanine increased the growth of B. rapa in both gray lowland soil and andosol to the same extent as that of inorganic nitrogen (Fig. 4 G and H). The concentration of alanine decreased and the concentration of nitrate tended to increase in soils during plant cultivation (Fig. 4I), suggesting that alanine in soils can be directly absorbed by plants as well as decomposed to inorganic nitrogen. Taken together, our data suggest that SS could lead to the accumulation of organic nitrogen that is likely to function as direct and indirect nitrogen sources as well as biologically active compounds to increase crop production.
Discussion
Our study detected a heterogenous module structure among the complex interactions observed in an agroecosystem under different management practices (Fig. 2). This suggested that components in the different soil parameters interact with each other to orchestrate a complex agroecosystem or “black box,” which has hindered attempts to explain the link between “input” from different management practices and “output” in the form of plant phenotype. Based on the detected module structure, we assessed what soil components are associated with plant phenotype without any biases. Surprisingly, we found that organic nitrogen and not inorganic nitrogen is an important component to covary with crop performance in the SS-induced IGR effect (Fig. 3), which is confirmed by in vitro assay and pot experiment using arable soils (Fig. 4). On the contrary, soil inorganic nitrogen positively correlates with sugar content in the plant (SI Appendix, Fig. S8) owing to the fact that supply of inorganic nitrogen stimulates photosynthesis but does not always increase yield because of the complex internal regulation within plants (31). In addition to soil organic nitrogen, the module M7 has several rhizosphere bacteria including Paenibacillaceae and Thermaceae (Fig. 3). These bacteria might have specific functions in crop production and engage in the construction of a representative microbiome type in the agroecosystem (1). Notably, alanine and choline were detected by the integrated network analysis but not by the pair-wise comparison. Using only metabolome data, PCA can find that the contributed metabolites distinguishing the SS and NS treatments included alanine and choline (SI Appendix, Fig. S13), whereas the integrated network analysis using multiomics data can detect the correlation between these soil metabolites and crop yield. This indicates that our integrated omics approach has predictive power to detect multilevel interactions between plants, microbes, and soils and identify key components in agricultural field research.
Despite intensive studies on organic nitrogen in plant nutrition, the contribution of organic nitrogen to the crop yield is still unclear and controversial (32–37). Agricultural crops are clearly able to absorb amino acids even in the presence of inorganic nitrogen (33–35, 37). In addition, an amino acid transporter is involved in the uptake of amino acid in the field (36). However, the fraction of uptake of intact amino acids is lower compared with the uptake of inorganic nitrogen in the agricultural field, while tree species show higher rates of uptake of organic nitrogen (33). Thus, direct evidence that organic nitrogen contributes a significant amount of nitrogen to plant nutrition in an agricultural field is lacking (30). Our study assessing the responsive interactions in the agroecosystem demonstrated that organic nitrogen, and not inorganic nitrogen, covaries with crop yield in the agricultural field. Our in vitro assay using artificial media and a pot experiment using arable soils revealed that the application of amino acids led to an increase in crop yield, although the active nitrogen forms remain unclear. Taken together, this study shows a clear case that the presence of organic nitrogen in soils play a vital role in plant nitrogen nutrition.
We found a decrease in the soil C/N ratio during plant cultivation, but could not detect any differences in the C/N ratio among SS and NS treatments at the seedling and harvest stages (SI Appendix, Table S2). Although higher-resolution measurements of nitrogen concentration are necessary, this suggests that the SS treatment might not be affecting the whole process of decomposition of organic nitrogen but rather produces a specific type of organic nitrogen. Our network analysis indicates that the module correlated with crop yield includes rhizosphere bacteria in addition to organic nitrogen. Therefore, there is a possibility that these bacteria might have some functions in the decomposition of specific type of organic nitrogen. Since alanine stimulates the transport activity of alanine in rice (38) and choline increased root biomass even in the presence of inorganic nitrogen (SI Appendix, Figs. S9 and S10), SS-induced organic nitrogen might reprogram plants to increase the acquisition of organic nitrogen. Our findings imply that organic nitrogen can contribute to plant nutrition, with the potential to decrease the usage of chemical fertilizer in future crop production.
Materials and Methods
Experimental Design and Sampling.
The experiments were performed in a plastic greenhouse (∼5 m × 40 m) located in Chiba Prefecture, Japan (35.6°N, 140.3°E), where the type of soil was andosol and no plants were cultivated in the previous year. A split-plot designed agricultural field was prepared with SS treatments as the main effect and fertilizers as the subeffect, i.e., SS with organic compost, SS with chemical fertilizer, NS with organic compost, and NS with chemical fertilizer. There were two plots prepared for each treatment (a total of eight plots; size, 2.2 × 8 m per plot). Organic composts (18% C, 2% N measured by CN analyzer [2400 CHNS/O Series II System; Perkinelmer Japan]; estimated available nitrogen supply rate 50%) made from sorghum, cow dung, pig feces, and chicken manure were applied at 4 kg m−2, and chemical fertilizer (14% CH4N2O, 14% P2O5, 14% K2O; CAINZ) was applied at 0.3 kg m−2. The application amount was determined to equalize available nitrogen among organic and chemical fertilization. Organic compost and chemical fertilizer were applied on September 6, 2016. All plots were plowed after the application and moistened by sprinkler irrigation. Soil solarization was carried out by mulching soils with a plastic sheet for a total of 36 d from September to October. The temperature and moisture content of soil at a depth of 10 cm during the solarization period were collected by an environmental sensor (Vegetalia) in the field. The plastic sheet was removed on October 19, 2016, and Japanese mustard spinach (B. rapa var. perviridis) was sown on October 20, 2016. The Japanese mustard spinach was harvested on December 15, 2016. Soils at seeding and harvest stages were collected at a depth of 10 cm for soil metabolomics and ionomics, while soils at a depth of 10 cm between plants and rhizosphere including root tissues (3 cm from shoot/root junction) at harvest stage were collected using RNAlater stabilization solution (Thermo Fisher Scientific) for bacterial microbiome analysis, with two replicates for each experimental plot. These samples were kept at −20 °C until sample preparation. Plants at harvest stage were collected with two replicates for each experimental plot and kept at 4 °C. After transferring plants to the laboratory, the phenotypic traits (described later) were measured within 1 week. Soils and plants were collected from the middle two places of each plot to minimize the border effect. In 2017, the plots of SS and NS treatment were swapped in the same field, and the experiment was repeated. Organic compost and chemical fertilizer were applied on July 5, and soil solarization was carried out for a total of 47 d from July to August. Japanese mustard spinach was sown on August 23, and the harvest was carried out on September 25, 2017.
Phenotypic Trait Measurement.
Chlorophyll content, brix, acid, and photosynthesis traits were measured with the largest true leaf from each individual plant. SPAD values (proportional to the chlorophyll content) were measured by a chlorophyll meter spectrophotometer (SPAD-502Plus; Konica Minolta Japan). Brix and acid values (percent) were quantified with a digital brix/acid refractometer (PAL-BX/ACID3; ATAGO). Chlorophyll fluorescence was measured using a Pulse Amplified Modulation fluorometer (JUNIOR-PAM; Walz). The maximum photochemical quantum yield of photosystem II (PSII; Fv/Fm) and effective quantum yield of PSII [Y(II)] were calculated (39). Major nutritional quality traits of the crop (brix [%], antioxidation [TE mg/100 g], vitamin C [mg/100 g], NO3 [mg/L], and taste index [five-point scale]) in whole shoot were measured by the outsourced service provided by Delica Foods. The above traits would be changed during storage, but the measurements were performed for all samples at the same time. Also, the phenotypic traits are important in terms of postharvest shelf life. Morphological phenotypic traits such as shoot height, root length, and number of leaves (count >1 cm true leaves) were measured. To assess dry weight for a leaf (the largest true leaf) and whole shoot, they were dried at 80 °C for 5 d and then weighed. Detailed leaf traits such as leaf area, length, width, and leaf index (length/width) were measured for the largest true leaf in ImageJ (40). Amounts of weed and insect damage in each plot were investigated at 16, 40, and 55 d after seeding.
Ionomics and Metabolomics Analysis.
To determine the concentration of water-soluble inorganic nitrogens such as ammonium and nitrate ions, these compounds were extracted with water (soil-to-water ratio of 1:5). Reflectoquant test strips (Merc) were soaked in the extract, and the concentration of each compound was determined by a RQflex reflectometer with standard calibration (Merc). For ionomic analysis, 50 mg of the sieved soils and the powdered plant samples were extracted according to a previous study (41) with only a slight modification. The extracted elements were measured on SPECTROBLUE EOP (SPECTRO) with previously reported operating parameters (42). In addition, samples extracted from soil with 1 M ammonium acetate (soil-to-ammonium acetate ration, 1:5) were measured by inductively coupled plasma-optical emission spectrometry (ICP-OES, Perkinelmer Japan). To obtain the metabolome data showing an unbiased profile of major metabolites that can be used for plants, we performed the NMR analysis because NMR data are highly reproducible and quantitative over a wide dynamic range to directly detect the profile of metabolites with less bias (43). For metabolomic analysis, 15 g of soil and 50 mg of plant material were extracted with water solvent according to previous studies (44, 45). The extracted metabolites were measured on an NMR spectrometer (AVANCE II 700; Bruker Biospin) with a Bruker standard pulse program “jresgpprqf” using the parameters of 8 scans, 16 dummy scans, 32 (f1) and 16 K (f2) data points, and 50 Hz (f1) and 16 ppm (f2) spectral widths. The obtained spectra were preprocessed and peak-picked based on regions of interest (ROIs) as per a previous study (46). Soil extracts were also measured with “hsqcetgpsisp2.2” pulse program using the parameters of 128 scans, 16 dummy scans, 256 (f1) and 1,024 (f2) data points, and 140 (f1) and 14 (f2) ppm spectral widths. Metabolite annotations were performed using SpinAssign (47) (dmar.riken.jp/spinassign/) and SpinCouple (48) (dmar.riken.jp/spincouple/) programs, and annotated metabolites were further validated with reference to HMDB (49) (https://hmdb.ca/). Since the metabolite annotations using only software-based analysis cannot distinguish choline and acetylcholine in our data set, we performed an additional spiking experiment using reference standards including choline chloride (Wako Pure Chemical) and acetylcholine (FUJIFILM Wako Pure Chemical) to assess the annotation. Choline or acetylcholine was added to the extracted metabolites from soil at the concentration of 0.5 mM. The spiked mixtures were measured with a Bruker standard pulse program “jresgpprqf” using the parameters of 16 scans, 16 dummy scans, 16 K (f2) data points, and 16 ppm (f2) spectral width. The obtained spectra were converted by skyline projection to one-dimensional data. To quantify carbon and nitrogen, dried soil was wrapped in tin capsules and measured by CN analyzer (2400 CHNS/O Series II System; Perkinelmer Japan).
Microbiome Analysis.
DNA was extracted from the collected soils and rhizosphere using our custom protocol (50). Samples were transferred from RNAlater stabilization solution into a chilled mortar by liquid nitrogen, removing extra solution, and homogenized using a pestle. The ground sample powder was transferred into the lysis/binding buffer, and the DNA was extracted (50). DNA samples were eluted in 10 mM Tris⋅HCl and used for bacterial community profiling. The DNA were used in a two-step PCR amplification protocol. In the first PCR, the V4 region of bacterial 16S rRNA was amplified with 515f and 806rB frame-shifting primers fused with Illumina sequencing primers (forward primer, 5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG-[3–6-mer Ns]–GTG YCA GCM GCC GCG GTA A-3′; reverse primer, 5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA G [3–6-mer Ns]-GGA CTA CNV GGG TWT CTA AT-3′) (51, 52). Each sample (1 μL 100-fold diluted DNA) was amplified in a 10-μL reaction volume containing 0.2 U KOD FX Neo DNA polymerase (TOYOBO), 2× PCR buffer (TOYOBO), 0.4 mM dNTPs (TOYOBO), and 0.2 μM forward and reverse primers as well as 1 μM blocking primers (mPNA and pPNA; PNA BIO) to avoid contamination of mitochondrial and chloroplast 16S rRNA sequences (52). PCR was performed using the following temperatures: 94 °C/2 min followed by 35 cycles at 98 °C/10 s, 78 °C/10 s, 55 °C/30 s, 68 °C/50 s, and a final extension at 68 °C/5 min (ramp rate, 1 °C/s). To purify the PCR product, 2 μL ExoSAP-IT Express (Thermo Fisher Scientific) was added in 5 μL of the PCR product and incubated at 37 °C for 4 min, and enzymes were deactivated at 80 °C for 1 min. To complete P5/P7 Illumina adaptors with 8-mer index sequences, the second PCR was performed with the following primers: forward primer, 5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC AC- [8-mer index]-TCG TCG GCA GCG TC-3′; reverse primer, 5′-CAA GCA GAA GAC GGC ATA CGA GAT-[8-mer index]-GTC TCG TGG GCT CGG-3′ (53). Each sample (0.8 μL of purified first PCR product) was amplified in a 10-μL reaction volume containing 0.2 U KOD FX Neo DNA polymerase (TOYOBO), 2× PCR buffer (TOYOBO), 0.4 mM dNTPs (TOYOBO), and 0.3 μM forward and reverse primers as well as 1 μM blocking primers (mPNA and pPNA; PNA BIO). PCR was performed as follows: 94 °C/2 min followed by eight cycles at 98 °C/10 s, 78 °C/10 s, 55 °C/30 s, 68 °C/50 s, and a final extension at 68 °C/5 min (ramp rate, 1 °C/s). The PCR products were purified with 0.6 (vol/vol) of AMPure XP beads (Beckman Coulter). DNA concentration was fluorescently determined, and equal concentrations of the barcoded amplicon library were pooled. Paired-end Illumina sequencing was performed using the MiSeq sequencer (Illumina). Raw sequencing reads were processed using the Qiime software package (ver. 1.8.0) (54). Briefly, sequence pairs were assigned to each sample according to their barcodes. Low-quality reads were eliminated, and the remaining sequences were clustered into operational taxonomic units (OTUs) using USEARCH (55) at 97% identity. Taxonomy of representative OTUs was assigned using Ribosomal Database Project (RDP) classifier (56) trained on the Greengenes database (57). Bray–Curtis distance between samples was calculated using vegan 2.4–4. The differences in OTU compositions were examined by the permutational analysis of variance (PERMANOVA, 10,000 permutations) and multivariate homogeneity of dispersions (PERMDISP) and visualized with nonmetric multidimensional scaling (NMDS) using the Bray–Curtis distance matrix as an input.
Integrated Omics Analysis.
To integrate all omics datasets, data were filtered such that at least four of the samples should have >0 value and merged into one data matrix along with sample IDs. For the pairwise abundance test, data were log2-transformed and subjected to unequal-variance t test to compare between SS and NS, and an adjusted P value was calculated using BH methods (FDR, false discovery rate) (58). For principal component analysis (PCA) and correlation network analysis, data were z-score–normalized (R scale function). PC values were calculated based on the normalized data across samples (R prcomp function). Unsigned correlation network was constructed using the weighted gene correlation network analysis (WGCNA) package version 1.60 (59). The soft-thresholding power was chosen based on a scale-free topology with fit index 0.9. An adjacency matrix with the selected soft-thresholding power was calculated and transformed into a topological overlap matrix (TOM). Using the TOM, network properties such as strength were calculated, and the network was visualized using the igraph package version 1.0.1 (60). A Fast–Greedy modularity optimization algorithm was selected to define modules in the integrated network.
In Vitro Germ-Free B. rapa var. perviridis System.
To prevent microbes from decomposing organic nitrogen such as amino acids, an in vitro cultivation of B. rapa var. perviridis from germination to developmental stages was established under sterile condition. Seeds of B. rapa were sterilized in 1% sodium hypochlorite solution for 1 min, followed by five washes with sterile water. Seeds imbibed in sterile water were germinated on agarose medium for 1 d. For the nutritional application experiment, inorganic nitrogen was compared to seven different sources of organic nitrogen (L-alanine, L-valine, L-isoleucine, L-leucine, choline, N-methylglycine). The concentration of nitrogen in each treatment was 5 mM. A total of 50 mM of each nitrogen solution, L-alanine, L-valine, L-isoleucine, L-leucine, choline chloride, N-methylglycine, and ammonium nitrate, was sterilized by filtering through a 0.2-μm cellulose nitrate filter (Whatmann). N-free half-strength modified Hoagland’s nutrient solution (3 mM K, 2 mM Ca, 1 mM P, 0.5 mM S, 0.5 mM Mg) including 0.3% Gellan gum (Wako Pure Chemical) was placed in a vessel (Magenta vessel; Sigma-Aldrich Japan) and sterilized at 121 °C for 20 min. Sterilized nitrogen solutions were added to be equivalent to 5 mM nitrogen concentration into the N-free nutrient solution before solidifying Gellan gum. For the supplemental application experiment to evaluate organic nitrogen as a supplemental source, the nitrogen solutions in each treatment were 0.05 mM nitrogen concentration for each organic nitrogen and 4.95 mM nitrogen concentration for ammonium nitrate. All in vitro cultivations were carried out in a final volume of a 100-mL medium. Three germinated seeds were transferred to the aforementioned media and cultured in a controlled chamber for 13 d at 25 °C under cycles of 16 h of light and 8 h of dark. The experiments were conducted with four biological replicates. Moreover, three germinated seeds (2 d after sowing) were cultured in a half-strength Murashige–Skoog nutrient medium (30 mM N, 10 mM K, 0.63 mM P, 1.5 mM Ca, 0.75 mM Mg, 0.75 mM S, 1% sucrose, 0.8% Gellan gum, pH 5.8) including 0, 0.05, 0.5, and 5 mM concentrations of choline acetate (Agro Kanesho) in a controlled chamber for 12 d at 23 °C in cycles of 16 h of light and 8 h of dark. These experiments were conducted with five biological replicates. To ensure gas exchange for photoautotrophic growth, a hole about 8 mm in diameter was drilled in the top of the cover, and PTFE membrane filter with a 0.45-μm pore size (Milliseal; Merck) was attached. The above-ground material was sampled and then dried at 70 °C for 1 d and weighed.
Isotope Tracer Experiment.
To evaluate direct absorption of alanine, germinated B. rapa were cultured in half-strength Hoagland solution for 14 d under sterile conditions and immersed for 1, 3, and 6 h in the culture solution containing 5 mM double-labeled L-alanine [13C3, 99% and 15N, 99%; *CH3*CH(*NH2)*COOH; Cambridge Isotope Laboratories]. The expanded leaves and shoot apexes were sampled separately. These samples were kept at −80 °C until they were analyzed by LC-MS and NMR. LC-MS analysis can distinguish between double-labeled L-alanine (MW 93.07) and internal L-alanine (MW 89.09); this analysis was conducted by the outsourced service provided by Anatech. To capture the metabolization of dual-labeled L-alanine, NMR analysis was conducted as follows: 30 mg of plant material was extracted with 600 µL MeOD solvent and measured with “hsqcetgpsisp2.2” pulse program using the parameters of 16 scans, 16 dummy scans, 400 (f1) and 1,024 (f2) data points, and 40 (f1) and 14 (f2) ppm spectral widths. The two-dimensional data were processed by SINE (f1) and QSINE (f2) window function with 5.0 and 1.0 line-broadening factors, respectively. The SI parameter (zero filling) on Bruker TopSpin software was also set to 2,048 for f1 before Fourier transformation. Four individual plants were used for each experiment, and the experiment was repeated four times. To evaluate the carbon distribution of absorbed alanine in the shoot, B. rapa cultured for 14 d were immersed in the culture solution containing 5 mM alanine with 14C-alanine [5.9 kBq/mL, *CH3*CH(NH2)*COOH] for 1, 3, and 6 h. The above-ground material was cut and placed in contact with an imaging plate (IP; BAS IP TR; GE Healthcare) for autoradiogram of 14C. The IP was exposed at −80 °C for 12 h. Radiation images recorded in the IP were scanned with an image analyzer (FLA5000; Fujifilm) at a resolution of 100 µm.
Pot Experiment Using Arable Soils.
To mimic an agriculturally relevant experimental system, the pot experiments were carried out using two different types of field soils, andosol (pH 5.9, EC 0.14 dS/m, exchangeable Ca 386 mg CaO/100 g, exchangeable Mg 69.7 mg MgO/100 g, exchangeable K 38.8 mg K2O/100 g, CEC 34.0 mEq/100 g, NH4-N <0.1 mg/100 g, NO3-N 4.53 mg/100 g, Truog-P 6.36 mg P2O5/100 g, phosphate absorption coefficient 2,623 mg/100 g) and gray lowland soil (pH 7.0, EC 0.17 dS/m, exchangeable Ca 389 mg CaO/100 g, exchangeable Mg 106 mg MgO/100 g, exchangeable K 42.3 mg K2O/100 g, CEC 18.8 mEq/100 g, NH4-N 0.20 mg/100 g, NO3-N 1.89 mg/100 g, Truog-P 46.4 mg P2O5/100 g, phosphate absorption coefficient 429 mg/100 g). A total of 250 g of soil was mixed with 0.5 g of nitrogen in the form of ammonium nitrate or alanine and put into a plastic pot (upper diameter 8 cm, depth 7 cm). A total of 0.33 g of superphosphate was also applied to the Andosol (64 mg P2O5/100 g) to avoid phosphate deficiency. A treatment without nitrogen was provided as a control. Nine seeds of B. rapa per pot were sown, thinned out until three geminated seedlings at 5 d after sowing, and then cultivated in a growth chamber (16 h light/8 h dark, 25 °C) for 16 d for andosol or 13 d for gray lowland soils. Pots were regularly irrigated with deionized water. The experiments were conducted with five biological replicates in andosol and six biological replicates in gray lowland soil. The plant shoots were harvested and washed with deionized water. After drying at 80 °C for 2 d, the dry weight was measured. To measure the concentration of alanine and inorganic nitrogen in soils during the cultivation period, 15 g of each soil was collected at 0, 7, and 14 d after sowing and stored at −20 °C. To quantify the concentrations of hot water-soluble alanine and nitrate in soil, 2 g of soils were added to 8 mL of ultra-pure water containing 100 μM of [13C3,15N]-L-alanine (soil-to-water ratio, 1:2 wt/vol) and then heat-treated in an aluminum block thermostat for 30 min at 110 °C with occasional mixing. After centrifugation (1,000 × g, 10 min at room temperature), supernatant was filtered with a membrane filter (pore size, 1 μm). Aliquot of the extracts of 50 mg of soil was used for the quantification of alanine and nitrate concentration. The concentration of alanine (nmol per g of soil) was determined by using gas chromatography/time-of-flight mass spectrometry (LECO), and the concentration of nitrate was determined by ion chromatography (LC-20AD).
Data Accessibility.
Nucleotide sequence data reported are available in the DDBJ Sequence Read Archive under the accession number DRA007555.
Supplementary Material
Acknowledgments
We thank T. Yamagishi, M. Koiwai, and K. Watanabe for coordinating researchers and farmers; M. Ichikawa and S. Ichikawa for maintaining the agricultural field; Dr. T. Asakura, Dr. S. Masuda, Dr S. Hamamoto, Mr. R. Nakasone, Mr. K. Sakata, Mr. T. Kurokawa, and Ms. T. Nakatani for technical assistance. This work was supported by funding from the PRESTO, Japan Science and Technology Agency (JPMJPR15Q2), and the Cabinet Office, Government of Japan, Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for Smart Bio-industry and Agriculture” (funding agency: Bio-oriented Technology Research Advancement Institution, NARO) to Y.I.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Nucleotide sequence data reported are available in the DDBJ Sequence Read Archive (https://www.ddbj.nig.ac.jp/dra/index-e.html) under the accession number DRA007555.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917259117/-/DCSupplemental.
References
- 1.Toju H. et al., Core microbiomes for sustainable agroecosystems. Nat. Plants 4, 247–257 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Busby P. E. et al., Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 15, e2001793 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandey P., Irulappan V., Bagavathiannan M. V., Senthil-Kumar M., Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front Plant Sci 8, 537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang A. C. et al., A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364, eaau6389 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Hacquard S. et al., Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 7, 11362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castrillo G. et al., Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chellemi D. O., Gamliel A., Katan J., Subbarao K. V., Development and deployment of systems-based approaches for the management of soilborne plant pathogens. Phytopathology 106, 216–225 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Antonkiewicz J., Labetowicz J., Chemical innovation in plant nutrition in a historical continuum from ancient Greece and Rome until modern times. Chem. Didact. Ecol. Metrol. 21, 29–43 (2017). [Google Scholar]
- 9.Reganold J. P., Wachter J. M., Organic agriculture in the twenty-first century. Nat. Plants 2, 15221 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Stevens C. J., Nitrogen in the environment. Science 363, 578–580 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Chen Y., Katan J., Effect of solar heating of soils by transparent polyethylene mulching on their chemical properties. Soil Sci. 130, 271–277 (1980). [Google Scholar]
- 12.Grünzweig J. M., Katan J., Ben-Tal Y., Rabinowitch H. D., The role of mineral nutrients in the increased growth response of tomato plants in solarized soil. Plant Soil 206, 21–27 (1999). [Google Scholar]
- 13.Seman-Varner R., Mcsorley R., Gallaher R. N., Soil nutrient and plant responses to solarization in an agroecosystem utilizing an organic nutrient source. Renew. Agric. Food Syst. 23, 149–154 (2008). [Google Scholar]
- 14.Gamliel A., Katan J., Involvement of fluorescent pseudomonads and other microorganisms in increased growth-response of plants in solarized soils. Phytopathology 81, 494–502 (1991). [Google Scholar]
- 15.Chen Y., Katan J., Gamliel A., Aviad T., Schnitzer M., Involvement of soluble organic matter in increased plant growth in solarized soils. Biol. Fertil. Soils 32, 28–34 (2000). [Google Scholar]
- 16.Katan J., Greenberger H., Alon H., Grinstein A., Solar heating by polyethylene mulching for the control of diseases caused by soil-borne pathogens. Phytopathology 66, 683–688 (1976). [Google Scholar]
- 17.Stapleton J. J., Quick J., Devay J. E., Soil solarization: Effects on soil properties, crop fertilization and plant growth. Soil Biol. Biochem. 17, 369–373 (1985). [Google Scholar]
- 18.Stapleton J. J., Soil solarization in various agricultural production systems. Crop Prot. 19, 837–841 (2000). [Google Scholar]
- 19.Gamliel A., Katan J., Eds., Soil Solarization: Theory and Practice, (The American Phyutopathological Society, 2017). [Google Scholar]
- 20.Ray B., Ed., FUNDAMENTAL FOOD MICROBIOLOGY, (CRC press, 2014). [Google Scholar]
- 21.Bulgarelli D. et al., Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Edwards J. et al., Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grady E. N., MacDonald J., Liu L., Richman A., Yuan Z. C., Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 15, 203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beneduzi A., Ambrosini A., Passaglia L. M. P., Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35 (suppl. 4), 1044–1051 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tjamos E. C., Tsitsigiannis D. I., Tjamos S. E., Antoniou P. P., Katinakis P., Selection and screening of endorhizosphere bacteria from solarized soils as biocontrol agents against Verticillium dahliae of solanaceous hosts. Eur. J. Plant Pathol. 110, 35–44 (2004). [Google Scholar]
- 26.Hyeon S. B. et al., Effects of choline chloride and its analogues on photosynthesis in wheat protoplasts. Agric. Biol. Chem. 51, 917–919 (1987). [Google Scholar]
- 27.Che F. S. et al., Stimulation of photosynthesis and growth of photoautotrophically cultured plant cells by choline and its analogs. Plant Cell Rep. 12, 691–697 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi Y. et al., Promotive effect of choline salts on growth of Manilagrass (Zoysiamatrella Merr.) and bentgrass (Agrostis stronifera L. cv. Pencross). J. Japanese Soc. Turfgrass Sciense 19, 15–22 (1990). [Google Scholar]
- 29.Bloom A. J., Sukrapanna S. S., Warner R. L., Root respiration associated with ammonium and nitrate absorption and assimilation by barley. Plant Physiol. 99, 1294–1301 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Näsholm T., Kielland K., Ganeteg U., Uptake of organic nitrogen by plants. New Phytol. 182, 31–48 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Lawlor D. W., Carbon and nitrogen assimilation in relation to yield: Mechanisms are the key to understanding production systems. J. Exp. Bot. 53, 773–787 (2002). [PubMed] [Google Scholar]
- 32.Yamagata M., Ae N., Nitrogen uptake response of crops to organic nitrogen. Soil Sci. Plant Nutr. 42, 389–394 (1996). [Google Scholar]
- 33.Näsholm T., Huss-Danell K., Högberg P., Uptake of organic nitrogen in the field by four agriculturally important plant species. Ecology 81, 1155–1161 (2000). [Google Scholar]
- 34.Näsholm T., Huss-Danell K., Högberg P., Uptake of glycine by field grown wheat. New Phytol. 150, 59–63 (2001). [Google Scholar]
- 35.Chapin F. S., Moilanen L., Kielland K., Preferential use of organic nitrogen for growth by a non-mycorrhizal arctic sedge. Nature 361, 150–153 (1993). [Google Scholar]
- 36.Ganeteg U. et al., Amino acid transporter mutants of Arabidopsis provides evidence that a non-mycorrhizal plant acquires organic nitrogen from agricultural soil. Plant Cell Environ. 40, 413–423 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Czaban W. et al., Direct acquisition of organic N by white clover even in the presence of inorganic N. Plant Soil 407, 91–107 (2016). [Google Scholar]
- 38.Nihei N., Masuda S., Rai H., Nakanishi T. M., Imaging analysis of direct alanine uptake by rice seedlings. Radioisotopes 57, 361–366 (2008). [Google Scholar]
- 39.Genty B., Briantais J. M., Baker N. R., The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta, Gen. Subj. 990, 87–92 (1989). [Google Scholar]
- 40.Abràmoff M. D., Magalhães P. J., Ram S. J., Image processing with imageJ. Biophoton. Int. 11, 36–42 (2004). [Google Scholar]
- 41.Date Y. et al., In vitro evaluation method for screening of candidate prebiotic foods. Food Chem. 152, 251–260 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Shima H. et al., Exploring the impact of food on the gut ecosystem based on the combination of machine learning and network visualization. Nutrients 9, 1307 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markley J. L. et al., The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 43, 34–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogura T. et al., Improvement of physical, chemical, and biological properties of aridisol from Botswana by the incorporation of torrefied biomass. Sci. Rep. 6, 28011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida S., Date Y., Akama M., Kikuchi J., Comparative metabolomic and ionomic approach for abundant fishes in estuarine environments of Japan. Sci. Rep. 4, 7005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei F., Sakata K., Asakura T., Date Y., Kikuchi J., Systemic homeostasis in metabolome, ionome, and microbiome of wild yellowfin goby in estuarine ecosystem. Sci. Rep. 8, 3478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chikayama E. et al., Statistical indices for simultaneous large-scale metabolite detections for a single NMR spectrum. Anal. Chem. 82, 1653–1658 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Kikuchi J. et al., SpinCouple: Development of a web tool for analyzing metabolite mixtures via two-dimensional J-resolved NMR database. Anal. Chem. 88, 659–665 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Wishart D. S. et al., HMDB: The human metabolome database. Nucleic Acids Res. 35, D521–D526 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ichihashi Y., Fukushima A., Shibata A., Shirasu K., High impact gene discovery: Simple strand-specific mrna library construction and differential regulatory analysis based on gene co-expression network. Methods Mol. Biol. 1830, 163–189 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Caporaso J. G. et al., Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 (suppl. 1), 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lundberg D. S., Yourstone S., Mieczkowski P., Jones C. D., Dangl J. L., Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Toju H., Baba Y. G., DNA metabarcoding of spiders, insects, and springtails for exploring potential linkage between above- and below-ground food webs. Zoological Lett. 4, 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso J. G. et al., QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar R. C., Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Wang Q., Garrity G. M., Tiedje J. M., Cole J. R., Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeSantis T. Z. et al., Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995). [Google Scholar]
- 59.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Csardi G., Nepusz T., The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9 (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nucleotide sequence data reported are available in the DDBJ Sequence Read Archive under the accession number DRA007555.