Significance
The mechanism that controls recombination between homoeologous chromosomes in allopolyploids, also known as homoeologous exchange (HE), is not well understood. We found that HE has similar hallmarks as homologous recombination, occurring in subtelomeric regions and at recombination hotspot motifs. However, it differs by a significant aspect: HE is mostly restricted to gene bodies. This feature was shown through sequence analysis of a synthetic wheat allotetraploid and was also detected in other plant allopolyploids. An outcome of this intragenic recombination is the formation of novel transcripts and protein fusion variants. HE thus provides a mechanism for neo- and subfunctionalization of genes.
Keywords: allopolyploidy, homoeologous exchange, gene fusion, wheat
Abstract
Recombination between homeologous chromosomes, also known as homeologous exchange (HE), plays a significant role in shaping genome structure and gene expression in interspecific hybrids and allopolyploids of several plant species. However, the molecular mechanisms that govern HEs are not well understood. Here, we studied HE events in the progeny of a nascent allotetraploid (genome AADD) derived from two diploid progenitors of hexaploid bread wheat using cytological and whole-genome sequence analyses. In total, 37 HEs were identified and HE junctions were mapped precisely. HEs exhibit typical patterns of homologous recombination hotspots, being biased toward low-copy, subtelomeric regions of chromosome arms and showing association with known recombination hotspot motifs. But, strikingly, while homologous recombination preferentially takes place upstream and downstream of coding regions, HEs are highly enriched within gene bodies, giving rise to novel recombinant transcripts, which in turn are predicted to generate new protein fusion variants. To test whether this is a widespread phenomenon, a dataset of high-resolution HE junctions was analyzed for allopolyploid Brassica, rice, Arabidopsis suecica, banana, and peanut. Intragenic recombination and formation of chimeric genes was detected in HEs of all species and was prominent in most of them. HE thus provides a mechanism for evolutionary novelty in transcript and protein sequences in nascent allopolyploids.
Allopolyploidization—that is, interspecific hybridization followed by whole-genome duplication—is a major driving force in genome evolution and speciation of higher plants (1–5). Allopolyploidization is known to induce an array of genetic, epigenetic, and gene-expression changes (6–11). These changes can occur rapidly, as soon as in somatic cells of interspecific F1 hybrids, or in the generations following genome doubling. It has been proposed that this rapid response contributes to the initial stabilization and establishment of nascent allopolyploids toward new species (1, 8, 12, 13). These changes include transposable element (TE) activation (14, 15), sequence elimination (16, 17), changes in cytosine methylation (17), or small RNA profiles (18) that may lead to gene silencing or activation (19). The merging of divergent genomes may also lead to altered patterns of homoeologous gene expression, including transinteractions, subgenome dominance, and regional partitioning in response to developmental and environmental cues (20, 21).
One mechanism whereby rapid genomic change can be generated in nascent allopolyploids is via recombination between homoeologous chromosomes. Such homoeologous exchanges (HEs) are generally suppressed by dedicated loci, such as Ph1 in polyploid wheat, and with a weaker effect PrBn in Brassica (22, 23) or because of intrinsic parental sequence divergence, thus largely maintaining meiotic bivalent pairing and preventing multivalent formation that would otherwise lead to chromosome instability and sterility. Nevertheless, HEs do occur and can be tolerated as evidenced by several reports in allopolyploid crops (including wheat) and wild species (24–28). A recent study showed that HEs have played an important role in the evolution of polyploid Brassica species, being the main contributor of duplication-absence sequence polymorphism (29). Similarly, HEs were proposed to have played an important role in peanut domestication (30).
Because HEs generate alterations of the otherwise 2:2 homoeolog ratio, an immediate consequence is dosage-dependent changes in homoeologous transcripts of the HE-related genes (31). For those genes that are functionally differentiated between the diploid progenitor species, HEs are likely to have physiological and phenotypic consequences, as shown in several cases in Brassica allotetraploids (31–34). In addition, it was shown recently that HEs may sustain and even amplify allopolyploidization-induced DNA methylation repatterning, which in turn causes further changes in gene expression (35).
The wheat group provides a well-established paradigm of speciation via allopolyploidization in nature as well as under domestication (36–38). Approximately 0.5 to 0.9 Mya (39–41) allotetraploidization between diploid wheat, Triticum urartu (AA), and an unknown species related to Aegilops speltoides (genome SS) that contributed the B subgenome, led to the formation of tetraploid wild emmer wheat, Triticum turgidum, ssp. dicoccoides (BBAA). Then, ∼9,000 y ago, bread wheat was formed (Triticum aestivum, genome BBAADD) through allohexaploidization between a domesticated T. turgidum form (genome BBAA) and Aegilops tauschii (genome DD) (8, 40, 42).
A salient feature of tetraploid and hexaploid wheats, which differs sharply from most allopolyploid species, is the near absence of HEs due to the presence of the Ph1 locus, which efficiently prevents meiotic pairing and recombination between homoeologous chromosomes (22, 43), and due to divergence between subgenomes following allopolyploidization (16, 38). However, as shown in a recent report (28), the suppression of HE in wheat is not absolute, and its prior reported absence could be due to lack of high-resolution sequence-based analyses. Our previous studies on synthetic wheats indicate that while the combination of SSAA (BBAA), analogous to natural tetraploid wheat (T. turgidum), is chromosomally largely stable, combinations of SSDD and AADD, both lacking Ph1 and having no natural corresponding species, are characterized by widespread chromosome instabilities, including both HEs and aneuploidy (44, 45). In particular, we karyotyped a large number of progeny cohorts of AADD and assessed the impact of chromosomal variations on phenotypic manifestation at the individual and populations levels (45). We reasoned that this synthetic tetraploid wheat is a suitable material to pinpoint mechanisms of HE formation and its genetic consequences.
In this study, we have performed a detailed comparison of the genome sequence of nascent allotetraploids and of their parents, providing insight into the mechanism leading to HE formation. HEs occurred at various generations in the nascent synthetic allotetraploid wheat (AADD). Frequency was also variable, with HEs occurring in only 5 of 11 individuals of three lineages, mostly in the distal chromosome region and with greater propensity for some chromosomal groups. Most HE events were nonreciprocal, leading to either doubling or loss-of-gene copy number in large chromosomal segments. Finally, a detailed analysis of HE junctions at the molecular level lead to the discovery that most HE events take place within gene bodies, at sequence motifs typical for homologous recombination (HR). As a result, we show that HEs generate novel hybrid transcripts that can give rise to new protein variants. We confirmed and extended these findings by analyzing HE events in several other plant allopolyploid species that have quality genome sequences enabling the analyses, such as Brassica, peanut, rice, banana, and Arabidopsis suecica (25, 30, 35, 46, 47). We conclude that HE contributes to evolutionary novelty in allopolyploid plant species, not only by altering gene dosage of large chromosomal segments, but also via an overlooked yet likely generic mechanism, namely, the making of intergenomic “recombinant” proteins.
Results
Characterization of HEs by Karyotypic Analysis.
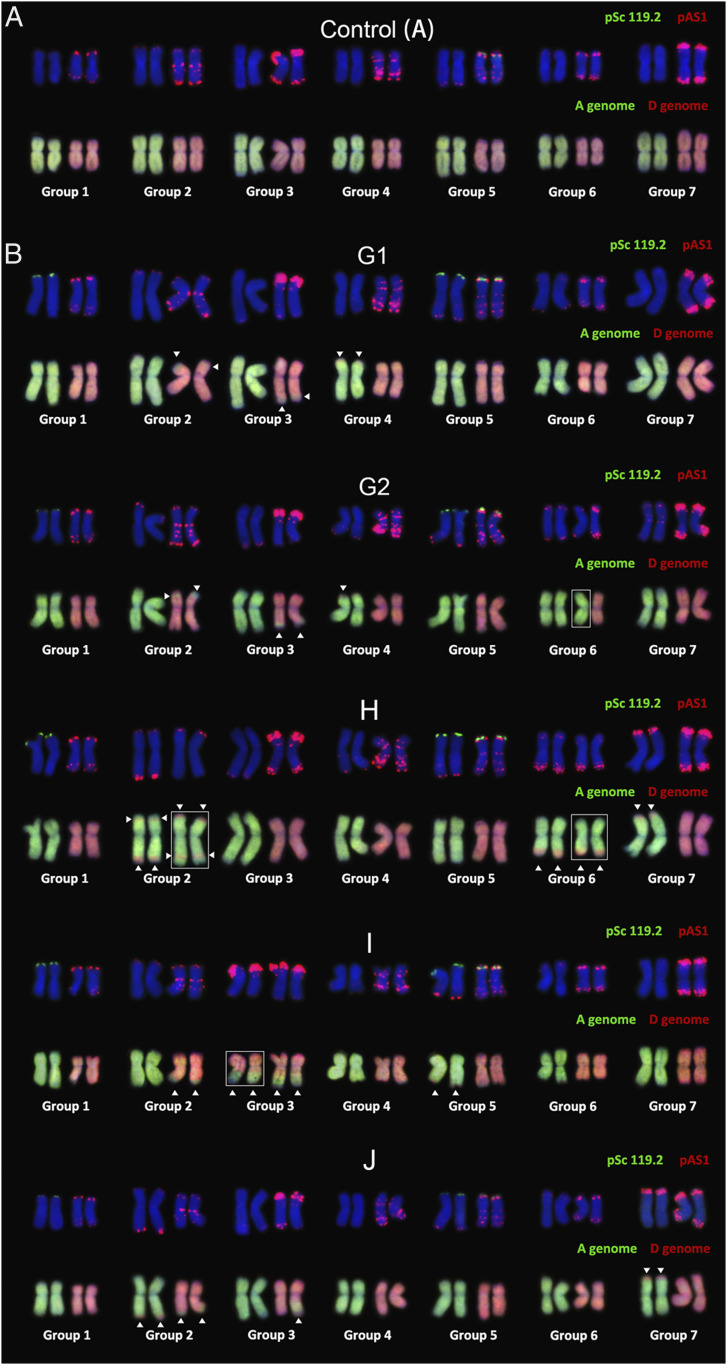
The pedigree of the 11 individual plants analyzed is shown in SI Appendix, Fig. S1, which represents 3 distinct lineages descended from a single founder plant at the first-selfed generation (S1) but separated at S2. One lineage (SI Appendix, Fig. S1, empty circles) contained plants (A to F) from three generations (S6, S9, and S12), while the other two lineages contained plants (G, H, I, and J) at the S9 generation (SI Appendix, Fig. S1). Together, these 11 plants should be representative for assessing both heritable (vertical transmission from earlier to later generations) and ongoing de novo genomic changes associated with allopolyploidization in the synthetic tetraploid wheat (AADD). Karyotyping of these plants was performed as described by Gou et al. (45). Six individuals did not show evidence for HEs, suggesting that they are bona fide euploids (Fig. 1A) or that the chromosomal segments involved in HE are submicroscopic. Among the five plants showing HEs (Fig. 1B), a variety of events were seen, including whole chromosomal exchanges, as in plant G2 concerning homoeologous group 6, which appears to be trisomic for chromosome 6A and monosomic for 6D. Most HE events seen here did not involve whole chromosomes, but rather consisted of segmental exchanges, which were generally located in the distal part of the chromosome. These segments varied in size, from approximately half a chromosomal arm (plant I, group 3) to small segments as in plant H (groups 6 and 7) or plant G2 (group 3). In most cases HE events were nonreciprocal, leading to a doubling of dosage of one parental segment and absence of the corresponding homoeologous segments. Only in one instance, in plant J (group 2), was a reciprocal HE seen at the tip of the chromosome. Group1 did not show any evidence for HE based on karyotypic analysis.
Fig. 1.
Karyotype illustration of HE events in synthetic allotetraploid wheat (AADD). (A) Standard karyotype image of the control tetraploid in S6 generation. FISH imaging was done using using two repetitive DNA probes, pSc119.2 (green) and pAS1 (red) (Upper). The same complement was probed by GISH of subgenome A (green) and subgenome D (red) (Lower). (B) FISH/GISH-based standard karyotypes of five tetraploids in S9 generation. The white arrows indicate visible HE events that occurred on corresponding chromosomes (small-scale chromosome replacement by homoeologous counterpart). Chromosomes in white boxes indicate almost whole chromosomes or chromosome arms that were replaced by their homoeologous counterparts.
Sequence-Based Identification of HEs.
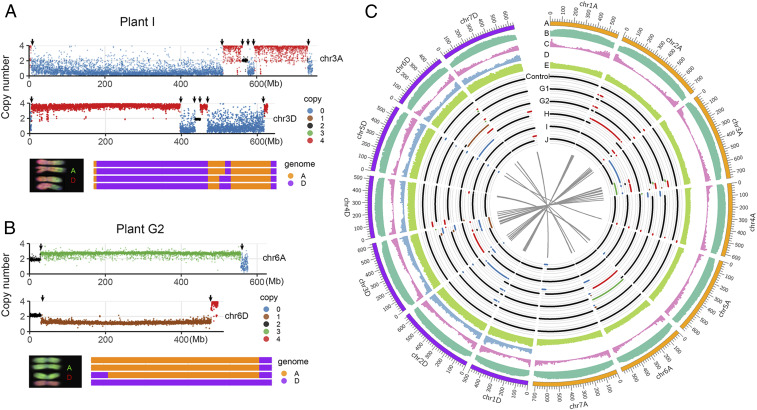
In order to validate the karyotypic observations, analyze the HE events at a sequence-based resolution, and gain insight into the possible underlying mechanism for HE events, we resequenced the diploid progenitors of the AADD allotetraploid. We compared the sequence of the 11 lines to an in silico AADD control that was constructed by concatenation of the whole-genome sequencing data of the two diploid parents (AA and DD) (SI Appendix, SI Materials and Methods). In addition, plant A in the S6 generation, which did not show any sign of HEs at the cytological level, was also treated as a tetraploid control for further analysis (Fig. 1A and SI Appendix, Fig. S1). To identify HE junctions, a series of bioinformatic steps were performed based on the whole-genome resequencing data (on average 10× coverage among individuals). A copy-number–dependent method was used to search original HE junction candidates and an SNP-based method was performed for further validation in each tetraploid individual (SI Appendix, SI Materials and Methods). In nonreciprocal HE events, these junctions correspond to the transition of regions ranging from 2 to 4 copies or 2 to 0; 0 to 4 or 4 to 0; 3 to 1 or 1 to 3 (Fig. 2 A and B). Note that reciprocal HE events cannot be detected by this copy number method; therefore, our focus here is on nonreciprocal HEs. Considering that only one reciprocal event was identified by karyotypic analysis, we assume that we missed only a small fraction of such HE events. A summary of all of the HE events identified through copy number is shown in Fig. 2C. Notably, transitions from 0 to 4 or 4 to 0 doses (Fig. 2A) ruled out possibilities of template switching as a PCR artifact that might have generated false junctions, as no alternative template is available.
Fig. 2.
Illustration of HE events based on whole-genome resequencing data. (A and B) Two examples of HE junctions in A and D chromosomes. The first and second tracks represent the profile of genomic copy number in A and D chromosomes, respectively. Blue, brown, black, green, and red dots represent genomic copy number from 0 to 4, respectively. Arrows indicate the position of HE junctions. The third track is a schematic summary of the chromosome constitution with corresponding karyotype diagram. (C) Summary of HE events and related genomic features. The feature density for each track was calculated based on a 10-Mb interval. Track A, the 14 chromosomes (orange and purple indicate chromosomes derived from the A and D subgenome, respectively); track B, TE density; track C, gene density; track D, density of SNP between A and D subgenomes; track E, CCN repeat motif; track Control, genomic copy number of control sample (the other five euploid tetraploids shared an identical HE event with the control sample, including a homoeolog translocation between chromosomes 4A and 5D; see main text for explanation); track G1-J, genomic copy number of five compensated euploidy plants. Blue, brown, black, green, and red lines represent genomic copy number from 0 to 4, respectively. The gray linkers in inner track indicate homoeologous HE junction pairs.
We also prepared Integrative Genomics Viewer (IGV) graphs based on both dosage and homoeolog state transition patterns (flanking 100-kb regions of each HE junction) (SI Appendix, Fig. S14) that fully supported authenticity of the HEs. In total, 74 HE junctions corresponding to 37 pairs of HE events (37 from A subgenome and 37 from D subgenome) were identified, which included two that occurred between chromosomes 4A and 5D. These two translocations were treated as HEs because they occurred in regions including an ancient translocation event between the long arms of chromosomes 4A and 5A in T. urartu (48). Note that during the process of HE junction identification, we could identify HE junctions with high confidence in the high-quality D subgenome. For the A subgenome, we used sequence data from the diploid A-genome T. urartu reference genome together with the sequences of the A subgenome of wild emmer wheat (T. turgidum ssp. dicoccoides) and of durum wheat (T. turgidum ssp. durum) in order to obtain high-confidence localization of the junctions (SI Appendix, Fig. S2).
Detailed analysis of the junctions showed various patterns. The most common and expected pattern is that an HE event gives rise to two pairwise junctions, one in A and one in D, that are precisely aligned to each other. In total, 23 pairs of HE junctions (of the 37) fulfilled this criterion (SI Appendix, Table S1, matched). In 14 HE junctions from the D subgenome, 6 were not aligned to the corresponding HE junctions in the A subgenome, but did align nearby (SI Appendix, Table S1, shift), possibly due to the divergence of the genome A parental sequence from the reference genomes that were used. The remaining eight HE pairs were not aligned to any genomic regions (SI Appendix, Table S1, no match, six junctions) or alignment was ambiguous due to repeats (SI Appendix, Table S1, low res., two junctions). Notably, all karyotypic nonreciprocal HE junctions were confirmed by the resequencing data, with additional cases that were not detected by cytology analysis because of their relatively small scale (Fig. 2B) (such as the terminal of 2AL, 3DL, and 6AL in samples G1 and G2; the terminal of 3AS and 3DL in sample I; and the proximal telomere region of 2AL). The HE rate cannot be accurately estimated due to the small number of resequenced lines in S9. Nonetheless, the rate can be estimated from data of a previous cytological analysis (45) using 1,426 individuals from the same genetic lineage as the 11 plants sequenced here, from the S12 generation in which 3,551 HE events were identified. The HE rate is thus 3,551/1,426 = 2.5 HE events per S12 plant on average. Assuming a constant rate per generation this corresponds to ∼0.1 HE event per meiosis (2.5/12/2, male or female). This rate is an underestimate given that small HE segments and interhomoeolog conversion would be cryptic at the cytological level.
Features of HE Events.
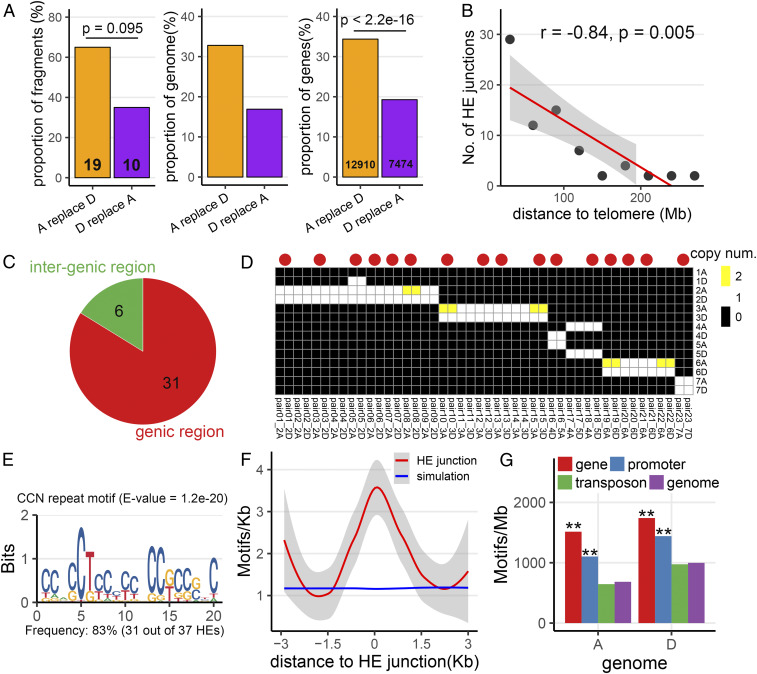
We carried out an analysis of various features of the 37 HE events. We found that chromosomal segments of A replaced segments of D in 19 instances compared to 10 instances for the opposite. While this was not significant from the expected 50% probability of A replacing D or vice versa (Fig. 3A) (χ2 test; P = 0.095), when considering the size of duplicated fragments and the gene content, the bias in favor of A became significant (Fig. 3A) (32.8% vs. 16.9%; chi-square test, P < 2.2e-16). The 74 HE junctions showed a nonrandom distribution along the chromosomes: Density increased on both arms along the centromere–telomere axis, correlating with the distance to the telomere (Fig. 3B) (r = −0.84; Pearson’s product-moment correlation, P = 0.005). Nonrandom occurrence of HEs was also observed among homoeologous groups, as already evident in the karyotypic analysis: That is, HE junctions were mainly occurring in chromosomes of groups 2 and 3 and were depleted in group 1 (Fig. 1B and SI Appendix, Table S2).
Fig. 3.
Features of HE events in the AADD wheat genome. (A) Nonreciprocal HE events lead to replacement of chromosomal segments of one subgenome by the other. This is measured for cases when A replaces D (orange column) or D replaces A (purple column) as percent of fragments (Left), percent of the genome (Center), and percent of genes (Right) in HE-involved chromosomal segments. The absolute number is shown at the bottom of the column and the χ2 test P value of the deviation from the randomly expected 50% ratio is above the columns. (B) Correlation between the number of HE junctions and their distance to telomeres (Pearson’s product-moment correlation, P = 0.005). The red line represents the trendline based on generalized linear model regression. (C) Genomic feature of HE junctions including genic and intergenic regions in the D subgenome. (D) Copy number of pairwise HE junction comparisons on all AADD chromosomes. Each HE junction was aligned to the AADD genome using Minimap2. Different colors represent different copy numbers to which the junction could align. Red circles above the cells indicates those pairwise HE junctions that were associated with homoeologous gene pairs. (E) Sequence logos of CCN repeat motif enriched in HE junctions based on the MEME Suite result. (F) The CCN repeat motif density around all HE junctions (red) and random genomic regions (blue, 1,000 replicates simulation), respectively. The trendline based on Loess regression are also shown. (G) Genomic features of motifs. Motif density in HE junctions, gene bodies, promoter regions, transposon regions, and whole-genome level are shown. The asterisks indicate significant enrichment (P < 2.2e-16) of motifs in corresponding genomic features.
Genomic feature analysis of 37 HE junctions from the D subgenome was performed. Interestingly, ∼83% (31 of 37) HE junctions were located either within gene-body regions, namely the region between the transcription start and termination sites, or their immediate adjacency. Considering that gene regions represent only 10% of the entire genome (Fig. 3C), our results indicate preferential occurrence of HEs in genic regions (χ2 test; P = 2.2e-16). When focusing on the 23 matched HE junction pairs (with clear syntenic alignment between homoeologs, 23 of 37), further analysis showed that HEs tended to preferentially occur in single-copy genomic regions (17 of 23) (Fig. 3D), even for those located in intergenic regions. Overall, HEs were enriched in gene-rich, low-copy genomic regions.
We searched for DNA motifs overrepresented in the 37 HE junctions of the D subgenome without a priori assumptions, using MEME Suite (49). The CCN repeat motif (E-value = 1.2e-20) was enriched in our HE junction set (Fig. 3E). This motif was previously described as being associated with crossovers between homologs in common wheat and Arabidopsis (50–52). In total, 83% of HEs (31 of 37) were associated with CCN repeat motifs. We then scanned the flanking regions of the above motif to validate if their distribution was specific to the HE junctions (SI Appendix, SI Materials and Methods). As shown in Fig. 3F, compared with simulation data, the density of the CCN repeat motif was found to decrease gradually with increasing distance to HE sites, suggesting motif-specific effects rather than regional effects occurring at HE junctions. In both subgenomes, the distribution of all motifs was consistent with HE patterns, with a higher proportion in the two distal regions (Fig. 2C and SI Appendix, Fig. S3). The abundance of the CCN repeat motif in different genomic features exhibited similar patterns in both subgenomes: It tended to be enriched in genic and promoter regions (Fig. 3G) (chi-square test, P < 2.2e-16), and depleted in TE regions. The distribution of the CCN repeat motif showed an inverse correlation with the pattern of CG DNA methylation in the gene-related region that is depleted in the gene body near the transcription start and transcription terminal site regions (SI Appendix, Figs. S4 and S5). Notably, the peak of the CCN repeat motif overlapped with that of chromatin accessibility (by ATAC-seq) (SI Appendix, Fig. S5). These results suggest that HE hotspot motifs correspond to genomic regions with lower DNA methylation level and higher chromatin accessibility.
HE as a Mechanism for Homoeologous Gene Fusion.
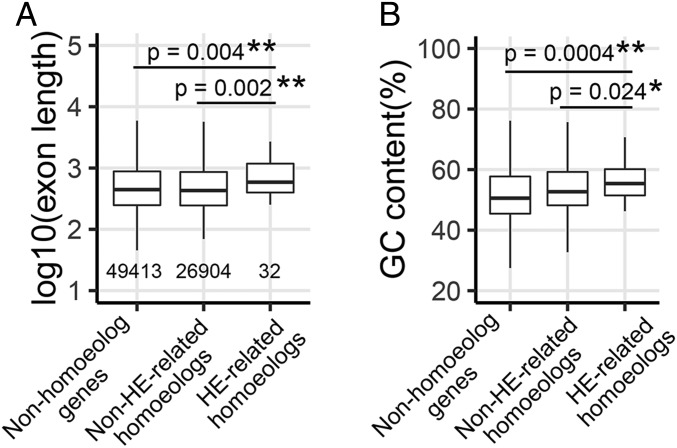
As shown above, most of HE junctions are located in genic regions. This raised the possibility that new chimeric genes are generated via HEs, combining the promoter of one parent with the coding region of the other, or that a new chimeric transcript is generated that could code for a new protein, assuming that the two homoeologous proteins have diverged in their amino acid sequences. To test these scenarios, we gathered 32 HE-associated genes (16 homoeologous pairs) to investigate their genomic features and expression profile. We compared the genomic features of genes associated with HEs vs. non-HE–related homoeologs and nonhomoeologous genes. As shown in Fig. 4, HE-related homoeologs possessed longer exon length than non-HE–related homoeologs (Fig. 4A) (median: 662 vs. 428 bp; mean: 915 vs. 699 bp; Mann–Whitney U test, P = 0.002). Exons of HE-related homoeologs were also longer than nonhomoeologous genes (median: 662 vs. 443 bp; mean: 915 vs. 720 bp; P = 0.004). Furthermore, the GC content of HE-related homoeologs was slightly higher than both non-HE–related homoeologs (Fig. 4B) (median: 56.3% vs. 52.7%; mean: 56.6% vs. 53.9%, P = 0.024) and nonhomoeologous genes (median: 56.3% vs. 50.6%; mean: 56.6% vs. 51.8%, P = 0.0004). These results suggest that longer exons and higher GC content may provide better target regions for pairing and recombination between homoeologous genes.
Fig. 4.
Comparison of genomic features of HE-related homoeologs with non-HE–related homoeologs and nonhomoeologous genes. (A) Exon length and (B) GC content among three types of gene sets. The asterisks indicate significant differences between HE-related homoeologs and non-HE–related homoeologs or nonhomoeologous genes (Mann–Whitney U test; *P < 0.05; **P < 0.01).
Next, we tested if HEs affected gene expression in situ or if it generated new fusion transcripts. Genes that were expressed in young leaves were included in the analysis. In total, 16 HE-related genes (8 homoeolog pairs) were expressed in both parents and in the corresponding recombinant HE tetraploid samples. Generally, fusion transcripts were expressed at similar levels as the control plants, with a few exceptions that could be due to trans effects from other genomic regions (SI Appendix, Fig. S6).
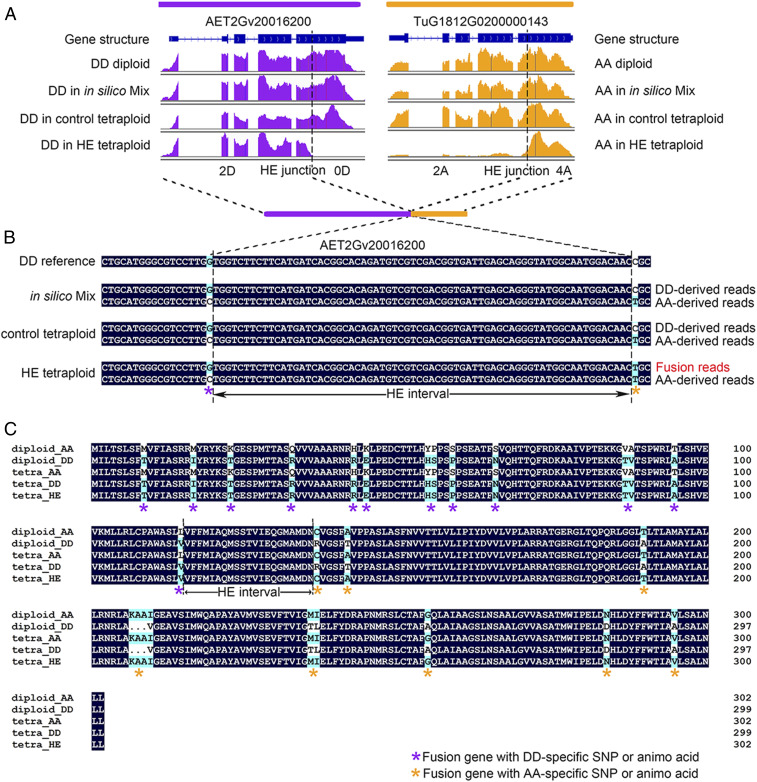
Regarding gene structure, all eight gene pairs that underwent HE retained the expected exons and reading frame of the parental proteins (Fig. 5A and SI Appendix, Fig. S7). The parental origin of exons could be determined based on SNPs from the RNA-sequencing (RNA-seq) data. Interestingly, several reads spanned the HE junctions (i.e., contained SNPs from both subgenomes), validating the presence of recombinant transcripts (Fig. 5B and SI Appendix, Fig. S7). Furthermore, RT-PCR and Sanger sequencing were performed for five genes to validate the full-length coding sequence and predict the completeness of ORF of fusion transcripts. An example is shown in Fig. 5C for one of the HE-related genes, validating that the full-length transcripts contained complete ORF regions, which matched well with RNA-seq and DNA data. The predicted amino acid sequences of the recombinant proteins indicated that new variants of the original protein sequences were obtained after HE.
Fig. 5.
Gene fusion between HE-related homoeologous gene pairs. (A) IGV illustration of a homoeologous fusion event based on RNA-seq reads depth. Each track (from up to down) represents the transcript structure and gene expression profile in diploid, in silico tetraploid, control tetraploid, and HE-related tetraploid, respectively. Orange and purple indicate the profile of the A and D subgenome gene, respectively. The position of the HE junctions were marked as dashed lines. The combination of number and letter flanking the HE junction represent the copy number of a given subgenome in HE-related tetraploid (2A indicating two copies of A subgenome). Note the HE-related tetraploid exhibits truncated expression pattern. (B) Validation of fusion transcript based on diploid-specific SNP method. The tracks represent the consensus sequencing reads in in silico mix tetraploid, control tetraploid, and HE-related tetraploid. Nucleotides without black background indicate high-confident SNPs between AA and DD diploid and were used for genotyping the sequence reads. The asterisks in different colors under SNP positions indicate the origin of SNP (orange for A subgenome and purple for D subgenome) in fusion read. Regions between adjacent asterisks of different colors represent HE junction intervals. (C) Predicted amino acid sequence of fusion transcript by ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) based on Sanger sequencing. Amino acids without black background indicate high-confidence variants between AA and DD diploids. The asterisks in different colors indicate the parental origin of the amino acid involved in the HE region (orange for A and purple for D) in the new predicted fusion protein. The region between adjacent asterisks of different colors represents the HE junction interval. Other similar cases of HE-related gene fusion events are shown in SI Appendix, Fig. S7.
Comparative Analysis of HE Junctions in Different Plant Allopolyploids.
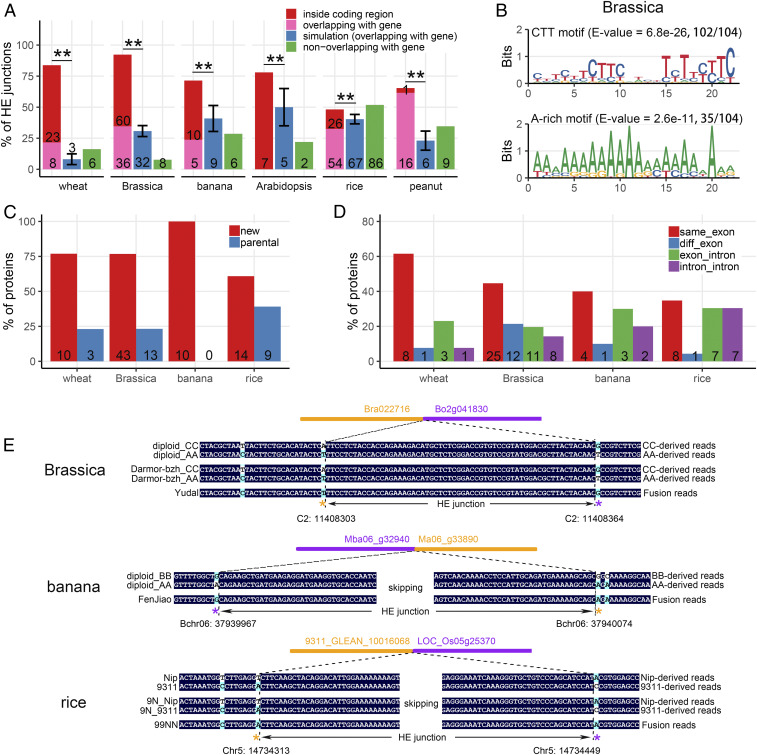
To test whether the salient feature of HE unraveled in the synthetic wheat (i.e., preferential occurrence within gene body and ready generation of novel, and potentially functional genic sequences) is a generic property of allopolyploidy, we analyzed several additional allopolyploid species with available quality genome sequence data. These species include Brassica (Brassica napus) (25), banana (Musa species) (47), Arabidopsis (A. suecica) (46), peanut (Arachis hypogaea) (30), and rice (Oryza sativa, synthetic tetraploid) (35). We interrogated the HE events in these species and compared the results with those of wheat. High-confidence HE junctions were identified in these species, ranging from 9 in Arabidopsis to 166 in rice, using both copy number- and SNP-based methods (SI Appendix, Tables S4–S8). IGV graphs based on both dosage and homoeolog state transition patterns for each HE junction were generated for these polyploid species (SI Appendix, Figs. S15–S18), fully supporting the authenticity of the HEs. The median resolution of HE junctions, based on SNP analysis, was from 76 bp in Arabidopsis to 6,963 bp in peanut, depending on SNP density between subgenomes (SI Appendix, Fig. S8). The HE junctions were significantly enriched in genic regions in all analyzed species: 92.3% in Brassica, 71.4% in banana, 78% in Arabidopsis, 48.2% in rice, and 65.3% in peanut (Fig. 6A) (χ2 test, P < 2.2e-16). Moreover, the proportion of HE junctions located within the gene body was significantly overrepresented in Brassica (57.7%, 60 of 104), banana (47.6%, 10 of 21), and Arabidopsis (78%, 7 of 9) and was comparable to that of wheat (62.2%, 23 of 37). In peanut, although 17 HE junctions were detected in genic regions (Fig. 6A), only 1 could be located precisely within the coding region due to low resolution of the sequencing data (Fig. 6A and SI Appendix, Fig. S8), precluding a statistical test.
Fig. 6.
HE features in Brassica, banana, Arabidopsis, rice, and peanut. (A) Proportion of HE junctions located inside a coding region (red), overlapping with a gene (pink), and not overlapping with a gene (green) in different polyploid species. Proportion of simulated HE junctions overlapping with the gene (blue) are also shown, assuming random occurrence of HEs in the genome. Error bars indicate the SD based on 1,000 simulations. The asterisks indicate significant differences between observed and simulated proportion of HE junction (χ2 test, P < 2.2e-16) in all species. The number of HE junctions are shown at the bottom of each bar. (B) HE-enriched motif logos in Brassica. Similar HE-enriched motif logos for rice and peanut are shown in SI Appendix, Fig. S10. (C) Percent of predicted HE-related fusion proteins based on the parental genome annotation. New (red) and parental (blue) protein represent a fusion protein with and without amino acid variations compared with parental proteins, respectively. (D) Distribution of a border of the HE junction that located within coding regions. The number of HE junction borders in the same exon (red), in different exon (blue), in exon–intron (green), and in intron–intron (purple) is shown. (E) validation of fusion transcripts based on diploid-specific SNPs in Brassica, banana, and rice. Tracks represent the consensus sequencing reads in diploid parents, different subgenomes in control tetraploid (Brassica) or control hybrid (rice, except banana), and HE-related polyploid. Nucleotides without black background indicate high-confidence SNPs between parental diploids that were used for genotyping the sequence reads. The asterisks in different colors under SNP positions indicate the origin of SNPs in the fusion read. The region between adjacent asterisks with different colors represents the HE junction interval. Other similar cases of HE-related gene fusion events in Brassica and banana are shown in SI Appendix, Figs. S12 and S13, respectively.
In the synthetic rice tetraploids, while 48.2% of HE junctions were in genic regions, only 15.7% (26 of 166) were in the coding region (Fig. 6A). This is not totally surprising given that rice tetraploids were parented by two closely related subspecies (japonica and indica); as such, there might be enough sequence similarity beyond the coding region for recombination to take place. Indeed, HEs in the rice synthetic tetraploids were preferentially mapped to up- and downstream regulatory regions (SI Appendix, Fig. S9), which is typical for HR (52, 53), but included also intragenic events (Fig. 6 C and D).
We searched for enrichment of HE-related motifs. In Brassica, we identified two motifs typical of interhomologs meiotic recombination hotspots (50, 53): The CTT repeat motif (E-value = 6.8e-26; present in 102 of 104 HE junctions, 98.1%) and the A-rich motif (E-value = 2.6e-11; present in 35 of 104 HE junctions, 33.7%) (Fig. 6B). Similarly, in peanut, both the CTT repeat motif (E-value = 8.6e-31, present in 26 of 26 junctions) and A-rich motif (E-value = 7.2e-17; present in 26 of 26 HE junctions) were enriched in HE junctions (SI Appendix, Fig. S10). In banana and Arabidopsis, the samples were too small for motif analysis. In rice, as discussed above, it is not clear if japonica and indica chromosomes can be considered as bona fide homoeologs due to their limited divergence; we found some variants of the Brassica CTT and wheat CCN repeat motif and the A-rich motif was also conserved (SI Appendix, Fig. S10). When looking at specific gene features (exons, introns) we excluded peanut from the analysis due to its low resolution of junction mapping. HE-related homoeologs were found to possess slightly longer exon length than non-HE–related homoeologs, although differences were not statistically significant (SI Appendix, Fig. S11) (Mann–Whitney U test, P > 0.05) but the trend was similar to that found in wheat (Fig. 4A).
Next, we analyzed the likelihood of HEs that were located within gene-body regions to generate potentially new functional coding sequences in these allopolyploids. We identified 76.8% (35 of 51), 100% (10 of 10), and 60.9% (14 of 23) such HEs in Brassica, banana, and rice, respectively, that generated in-frame, full-length coding sequences. These ratios are similar to those found in the synthetic wheat (76.9%, 10 of 13) (Fig. 6C).
Analysis of HE junction borders showed that they mainly located within the same exon (Fig. 6D) (44.6%, 25 of 56 in Brassica; 40%, 4 of 10 in banana; 34.8%, 8 of 23 in rice), similar to wheat (61.5%, 8 of 13). In wheat, 3 (3 of 13) HE events had one border in an exon, the second one in an intron, and only 1 of 13 events had exon–intron boundaries and intron–intron boundaries (Fig. 6D). In Brassica, 12 HE borders were located in different exons, 11 in exon–intron and 8 in intron–intron (Fig. 6D). In banana, only one HE border was located in different exons, three in exon–intron, and two in intron–intron (Fig. 6D); and in rice, only one HE border was located in different exons, and seven in both exon–intron and intron–intron. The general enrichment for borders within a single exon may be attributed to the longer length of sequence similarity (Fig. 4A and SI Appendix, Fig. S10).
When analyzing genes for which RNA-seq data were available, we found five transcript fusion events in B. napus cv. “Darmor-bzh” (five cases of nine expressed genes) and one in cv. “Yudal” (one case of two expressed genes) based on Illumina paired-end sequencing reads (Fig. 6E and SI Appendix, Fig. S12). Similarly, we also found two fusion cases in banana cv. “Fenjiao” (two cases of three expressed genes) (Fig. 6E and SI Appendix, Fig. S13) and one fusion case in rice (one case of two expressed genes of HE junction shorter than 300 bp) (Fig. 6E). The analysis was not performed in peanut due to low resolution of HE junctions, nor in Arabidopsis due to lack of corresponding RNA-seq data.
Discussion
In this work we have carried out a detailed analysis of HEs in the progenies of a nascent allotetraploid parented by two diploid progenitor species of hexaploid bread wheat. In particular, we have analyzed the junctions at HE sites in order to get a better understanding of the homoeologous recombination mechanism and its genetic consequences. A comparative analysis with several additional plant allopolyploids suggests that a general mechanism is at work. A series of evidence was used to validate HE junctions, including cytological analysis, transition in SNP dosage and heterozygosity, analysis of junctions from genomic data and from RNA transcripts (for expressed genes), and display of IGV graphs in the region flanking HE junctions. Taken together these data consistently support the robustness of HE junction identification.
HEs in Wheat.
We report here on the analysis of 37 HE events that took place progressively in different generations in the lineage of a nascent synthetic allopolyploid (genome AADD), raising questions regarding the role of HEs in wheat genome evolution. These findings are consistent with recent studies showing that the D genome contains segments from different diploid parents, suggesting that interspecific hybridization and HE took place during its formation (54). On the other hand, HE has not been widely reported in natural wheat polyploids. One possibility to reconcile these contradicting observations is that most studies on natural wheat allopolyploids involved domesticated tetraploid (genome BBAA) or hexaploid (genome BBAADD) wheat in the background of the 5B-encoded Ph1 locus, which suppresses homoeologous recombination (22, 43) and which was lacking in the synthetic allopolyploid studied here (genome AADD). However, the suppressive effect of Ph1 is not absolute as HEs have been documented in bread wheat (28). Moreover, the occurrence of HEs might have been underestimated in wheat, because large nonreciprocal HEs probably reduce fertility and were selected against in natural domesticated wheat as seen in Brassica (55), or because of HEs involving small segments, due to interhomoeologs crossover or conversion, which are harder to detect. Therefore, the role of HE in wheat evolution might turn out to be more important than previously thought.
HE Mechanism in Allopolyploids.
The role of HE in the evolution and domestication of several allopolyploids has been well documented, first in Brassica (56) and in recent years in several other species (26, 30, 35, 47, 57). We have performed a careful analysis of HE junctions in the synthetic allotetraploid wheat studied here, using data produced in the course of this work, as well as in Brassica, peanut, banana, rice, and A. suecica using published data. This analysis has enabled us to better understand the HE mechanism. Several hallmarks of HR hotspots were found for homoeologous recombination.
HE events were enriched in subtelomeric regions (Fig. 3B) as for HR (52, 58). Motifs enriched in HR hotspots—for example, CCN repeats, CTT repeats, and A-rich motifs (50, 53, 59, 60)—were also enriched at HE junctions (Figs. 3 and 6B and SI Appendix, Fig. S10). Such HE-related motifs shown here in wheat, Brassica, rice, and peanut had already been noticed in B. napus (61). Furthermore, as with other species, recombination was enriched in genic features (62). However, unlike crossover between homologous chromosomes, which showed a preference for promoters and terminators in several species, including wheat (51, 53, 63, 64), HE events reported here were strongly biased for gene-body regions (Figs. 3C and 6A). Interestingly, the A-rich motif, which was found for wheat HR hotspots (51), was not significantly enriched here in wheat HEs while it was enriched in Brassica and rice (SI Appendix, Table S9). This sequence motif, enriched in promoters and introns, is depleted of nucleosomes and thus provides high accessibility for homologous exchanges. The lack of significance for the A-rich motif enrichment in wheat HE regions might be due to higher sequence divergence in promoters and introns compared to exons.
An intriguing question that remains to be answered is whether the A-rich motif, which was shown to be a hotspot for SPO11-1–mediated double-strand break formation in Arabidopsis (65), has the same function in the species studied here. The HE-associated CCN repeat motif found here in wheat and in rice at HE sites (Fig. 3E) marks H3K4me3 chromatin modification and gene body (66). This, together with the higher GC content in regions involved in HE events, is consistent with HE preference for gene-body regions. The CTT repeat motif also found in Arabidopsis recombination hotspots (50, 59, 60) is enriched immediately downstream of the transcription start sites in nucleosomes with H2A.Z or H3K4me3 euchromatin marks. Its effect thus spreads through promoters, UTRs, and exons, as shown here in Brassica (SI Appendix, Table S9). The higher exon length at HE sites (Fig. 4A and SI Appendix, Fig. S11) provides some clues as to why recombination between homoeologs is driven to gene-body regions: Considering the high divergence between homoeologs, in particular in nongenic regions (67, 68), it is reasonable to assume that poor pairing and the mismatch repair machinery (69) are suppressing recombination in noncoding regions. In contrast, long stretches of DNA (as in long exons) with a high DNA sequence similarity, increase the chances for homoeologs to recombine. In most cases, given SNPs availability constraints, HEs could be located to a single (long) exon. The rice case also supports that HE is driven to gene body due to sequence divergence in other regions: In the tetraploid rice, the two parental subgenomes of indica and japonicum diverged only 200,000 to 400,000 y ago (70), compared to the wheat subgenomes, which diverged ∼2 to 3 Mya from a common progenitor (71). Therefore, even though intragenic HE events were identified, recombination between the subgenomes was more similar to classic HR than HE.
We conclude that HE between divergent genomes is probably using the standard HR machinery, but is restricted to regions of high similarity, such as gene-body regions. Similarity constraints being less stringent for HR, it tends to occur in high-accessibility regions, such as promoters that are AT-rich and have low nucleosome occupancy.
HE Generates New Chimeric Genes.
While changes in gene dosage for hundreds or thousands of genes involved in nonreciprocal HEs is the obvious and major impact of HEs on genome structure and function (31, 55), we wish to highlight here a particularly interesting outcome of HEs for the structure of genes at the junctions of HEs. The enrichment of HEs within coding regions together with the relatively high divergence between the homoeologous genes gave rise to novel “recombinant” genes in wheat, Brassica, banana, and rice (Fig. 6E). Overall, we show that HE provides a mechanism for generating new chimeric transcripts containing the promoter of one species and the coding region of another species, as well as a mechanism for generating new protein variants due to in-frame fusion of homoeologous genes of parental species. This reshuffling within genes could give rise to new patterns of gene expression or to new protein functions. While this is not different from what standard HR can do, the limited divergence between homologs, together with the tendency for recombination out of the coding region, make the formation of new proteins less likely during homologous than HEs. Considering that HE is not a rare event, as reported in several allopolyploids and as estimated here as 0.1 HE event per meiosis in synthetic wheat, it might significantly contribute to gene and genome evolution.
Taken together, the data from wheat and other allopolyploids clearly show that HE events occur at the same hotspots and chromosomal regions as HR events, presumably through the same recombination machinery. However, unlike HR that is enriched in promoters and downstream of genes, HEs preferentially occurs within coding regions, in long stretches of homology, providing a mechanism for the generation of new genes and new proteins. While demonstrating new functions for such chimeric proteins is out of the scope of this study, we have identified a significant mechanism for neo- or subfunctionalization in allopolyploids.
Materials and Methods
Plant Materials and Karyotype Analysis.
The 11 individual plants of tetraploid wheat (AADD) from S6, S9, and S12 generations and their parent diploids were sampled for karyotype analysis and further sequencing analysis (SI Appendix, Fig. S1). FISH and genomic in situ hybridization (GISH) were conjoined for chromosome karyotyping. Details of the experimental procedures are described in SI Appendix, SI Materials and Methods.
Sequencing and Publicly Available Data.
For wheat individuals, young leaves were used for whole-genome resequencing and RNA-seq. Library construction and sequencing were performed by standard Illumina protocols. Clean data have been deposited the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/) with accession no. PRJNA608801. Other sequencing data (including Brassica, bsanana, Arabidopsis, rice, and peanut) were downloaded from the National Center for Biotechnology Information Sequence Read Archive collection. Detailed information of sequencing procedure and publicly available data are described in SI Appendix, SI Materials and Methods.
Bioinformatic Analysis.
CNVkit (72) and SNP-based methods were used to identify HE junctions. IGV was used to check chimeric-transcript–supported RNA-seq reads. MEME (49) and FIMO (73) were used to identify HE-enriched DNA motifs and their genome locations, respectively. The BAM files of each HE junction and 100-kb flanking region were deposited in the Sequence Read Archive database with accession no. PRJNA625880. Detailed analysis procedures, including HE junctions calling and chimeric transcript identification, are described in SI Appendix, SI Materials and Methods.
Validation of Chimeric Genes.
RT-PCR and Sanger sequencing were performed to obtain full-length chimeric genes. ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) was used to predict the ORF of chimeric transcripts (SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China NSFC #31830006 (to B.L.) and by a China Scholarship Council fellowship (to Z.Z.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession nos. PRJNA608801 and PRJNA625880).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003505117/-/DCSupplemental.
References
- 1.Wendel J. F., Genome evolution in polyploids. Plant Mol. Biol. 42, 225–249 (2000). [PubMed] [Google Scholar]
- 2.Levy A. A., Feldman M., The impact of polyploidy on grass genome evolution. Plant Physiol. 130, 1587–1593 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van de Peer Y., Maere S., Meyer A., The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Jiao Y. et al., Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Soltis P. S., Soltis D. E., Eds., Polyploidy and Genome Evolution, (Springer, 2013). [Google Scholar]
- 6.Comai L., The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Leitch A. R., Leitch I. J., Genomic plasticity and the diversity of polyploid plants. Science 320, 481–483 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Feldman M., Levy A. A., Genome evolution due to allopolyploidization in wheat. Genetics 192, 763–774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madlung A., Wendel J. F., Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet. Genome Res. 140, 270–285 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Wendel J. F., Jackson S. A., Meyers B. C., Wing R. A., Evolution of plant genome architecture. Genome Biol. 17, 37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding M., Chen Z. J., Epigenetic perspectives on the evolution and domestication of polyploid plant and crops. Curr. Opin. Plant Biol. 42, 37–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z. J., Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58, 377–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto S. P., The evolutionary consequences of polyploidy. Cell 131, 452–462 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Kashkush K., Feldman M., Levy A. A., Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33, 102–106 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Madlung A. et al., Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41, 221–230 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Feldman M. et al., Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 147, 1381–1387 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A. A., Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13, 1749–1759 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenan-Eichler M. et al., Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188, 263–272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kashkush K., Feldman M., Levy A. A., Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160, 1651–1659 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo M. J., Szadkowski E., Wendel J. F., Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110, 171–180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X. et al., Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA-sequencing. New Phytol. 209, 1264–1277 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Riley R., Chapman V., Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182, 713–715 (1958). [Google Scholar]
- 23.Jenczewski E. et al., PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164, 645–653 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lashermes P., Combes M. C., Hueber Y., Severac D., Dereeper A., Genome rearrangements derived from homoeologous recombination following allopolyploidy speciation in coffee. Plant J. 78, 674–685 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Chalhoub B. et al., Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Henry I. M. et al., The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of Arabidopsis. Plant Cell 26, 181–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chester M. et al., Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. U.S.A. 109, 1176–1181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Z. et al., Extensive homoeologous genome exchanges in allopolyploid crops revealed by mRNAseq-based visualization. Plant Biotechnol. J. 15, 594–604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurgobin B. et al., Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 16, 1265–1274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertioli D. J. et al., The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E., Osborn T. C., Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19, 3403–3417 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osborn T. C. et al., Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165, 1569–1577 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J. et al., Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor. Appl. Genet. 112, 509–516 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Lloyd A. et al., Homoeologous exchanges cause extensive dosage-dependent gene expression changes in an allopolyploid crop. New Phytol. 217, 367–377 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Li N. et al., DNA methylation repatterning accompanying hybridization, whole genome doubling and homoeolog exchange in nascent segmental rice allotetraploids. New Phytol. 223, 979–992 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Feldman M., Levy A. A., Allopolyploidy—A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109, 250–258 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Dubcovsky J., Dvorak J., Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldman M., Levy A. A., Genome evolution in allopolyploid wheat–A revolutionary reprogramming followed by gradual changes. J. Genet. Genomics 36, 511–518 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Gornicki P. et al., The chloroplast view of the evolution of polyploid wheat. New Phytol. 204, 704–714 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Marcussen T. et al., Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Middleton C. P. et al., Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe. PLoS One 9, e85761 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuoka Y., Evolution of polyploid triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 52, 750–764 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Sears E. R., Genetics society of canada award of excellence lecture an induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19, 585–593 (1977). [Google Scholar]
- 44.Zhang H. et al., Intrinsic karyotype stability and gene copy number variations may have laid the foundation for tetraploid wheat formation. Proc. Natl. Acad. Sci. U.S.A. 110, 19466–19471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gou X. et al., Transgenerationally precipitated meiotic chromosome instability fuels rapid karyotypic evolution and phenotypic diversity in an artificially constructed allotetraploid wheat (AADD). Mol. Biol. Evol. 35, 1078–1091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novikova P. Y. et al., Genome sequencing reveals the origin of the allotetraploid Arabidopsis suecica. Mol. Biol. Evol. 34, 957–968 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z. et al., Musa balbisiana genome reveals subgenome evolution and functional divergence. Nat. Plants 5, 810–821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling H.-Q. et al., Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557, 424–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey T. L. et al., MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37 (suppl. 2), W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shilo S., Melamed-Bessudo C., Dorone Y., Barkai N., Levy A. A., DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis. Plant Cell 27, 2427–2436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darrier B. et al., High-resolution mapping of crossover events in the hexaploid wheat genome suggests a universal recombination mechanism. Genetics 206, 1373–1388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zelkowski M., Olson M. A., Wang M., Pawlowski W., Diversity and determinants of meiotic recombination landscapes. Trends Genet. 35, 359–370 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Choi K. et al., Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45, 1327–1336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glémin S. et al., Pervasive hybridizations in the history of wheat relatives. Sci. Adv. 5, eaav9188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiong Z., Gaeta R. T., Pires J. C., Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. U.S.A. 108, 7908–7913 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharpe A. G., Parkin I. A. P., Keith D. J., Lydiate D. J., Frequent nonreciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome 38, 1112–1121 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Edger P. P. et al., Origin and evolution of the octoploid strawberry genome. Nat. Genet. 51, 541–547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Copenhaver G. P., Meiotic recombination: Mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Erik W. et al., The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2, e01426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi K., Henderson I. R., Meiotic recombination hotspots - a comparative view. Plant J. 83, 52–61 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Samans B., Chalhoub B., Snowdon R. J., Surviving a genome collision: Genomic signatures of allopolyploidization in the recent crop species Brassica napus. Plant Genome 10 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Dluzewska J., Szymanska M., Ziolkowski P. A., Where to cross over? Defining crossover sites in plants. Front. Genet. 9, 609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X., Li L., Yan J., Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat. Commun. 6, 6648 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kianian P. M. A. et al., High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat. Commun. 9, 2370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi K. et al., Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 28, 532–546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X., Bernatavichute Y. V., Cokus S., Pellegrini M., Jacobsen S. E., Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10, R62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gale M. D., Devos K. M., Comparative genetics in the grasses. Proc. Natl. Acad. Sci. U.S.A. 95, 1971–1974 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bennetzen J. L. et al., Grass genomes. Proc. Natl. Acad. Sci. U.S.A. 95, 1975–1978 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emmanuel E., Yehuda E., Melamed-Bessudo C., Avivi-Ragolsky N., Levy A. A., The role of AtMSH2 in homologous recombination in Arabidopsis thaliana. EMBO Rep. 7, 100–105 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma J., Bennetzen J. L., Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. U.S.A. 101, 12404–12410 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chalupska D. et al., Acc homoeoloci and the evolution of wheat genomes. Proc. Natl. Acad. Sci. U.S.A. 105, 9691–9696 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Talevich E., Shain A. H., Botton T., Bastian B. C., CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLOS Comput. Biol. 12, e1004873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.