Significance
Lutein is one of the most naturally abundant carotenoids and an essential nutrient for humans. The last reaction of lutein biosynthesis is the addition of a hydroxyl group to the β-ring and ε-ring located at the ends of the α-carotene linear polyene chain. The enzymes catalyzing such stereospecific reactions are two members of cytochrome P450 family 97, the most ancient plant P450s. Although their biological functions have been identified, the structures of these two enzymes have remained unsolved, and hence the molecular basis for the substrate specificity is unclear. We crystallized both enzymes and solved their structures. The structures, together with biochemical characterization, provide insights into the substrate binding and stereospecificity of these two P450 carotenoid hydroxylases.
Keywords: xanthophyll, P450, hydroxylation, chirality, stereospecificity
Abstract
Two cytochrome P450 enzymes, CYP97A3 and CYP97C1, catalyze hydroxylations of the β- and ε-rings of α-carotene to produce lutein. Chirality is introduced at the C-3 atom of both rings, and the reactions are both pro-3R–stereospecific. We determined the crystal structures of CYP97A3 in substrate-free and complex forms with a nonnatural substrate and the structure of CYP97C1 in a detergent-bound form. The structures of CYP97A3 in different states show the substrate channel and the structure of CYP97C1 bound with octylthioglucoside confirms the binding site for the carotenoid substrate. Biochemical assays confirm that the ferredoxin-NADP+ reductase (FNR)–ferredoxin pair is used as the redox partner. Details of the pro-3R stereospecificity are revealed in the retinal-bound CYP97A3 structure. Further analysis indicates that the CYP97B clan bears similarity to the β-ring–specific CYP97A clan. Overall, our research describes the molecular basis for the last steps of lutein biosynthesis.
Carotenoids, a family of tetraterpenoid pigments producing yellow-red colors, play diverse roles in plants (1, 2). Dietary carotenoids provide health benefits by their antioxidant properties and disease-preventing roles (3). The plant biosynthesis of carotenoids occurs within plastids, where enzymes catalyzing steps from phytoene to xanthophylls (oxygen-containing carotenoids) are localized in or associated with the membranes (4, 5). A better understanding of the carotenoid biosynthetic enzymes will help to harness the wealth of these versatile pigments by metabolic engineering of the pathway (6, 7).
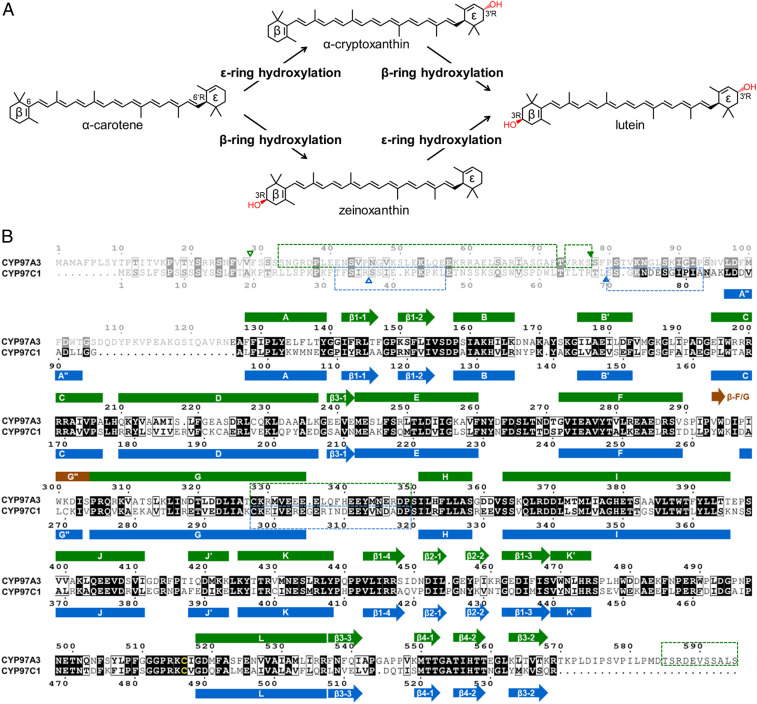
Lutein (3R,3′R,6′R-β,ε-carotene-3,3′-diol) is a dipolar xanthophyll derived from α-carotene (β,ε-carotene) (Fig. 1A). In humans, lutein accounts for more than half of the total carotenoids in the eye and brain, where it confers protection against light damage, oxidative damage, and inflammation (8). In chloroplasts, lutein comprises approximately half of the total carotenoids, accumulates in the thylakoid membranes, and participates in light harvesting and photoprotection of the antenna complexes (9–12). The transmembrane localization of lutein is ensured by the presence of the polar hydroxyl group at the β- and ε-rings, which differ in the position of the double bond within the cyclohexene moiety. The polyene chain is conjugated with the C-5,6 double bond in the β-ring but not with the C-4′,5′ double bond in the ε-ring, which makes a subtle structural difference in the absence or presence of a chiral center (6′R for the ε-ring). The hydroxylation reactions at the C-3 position of the β- and ε-rings are both pro-R–stereospecific. Two structurally unrelated enzymes catalyze the β-ring hydroxylation, namely the nonheme diiron hydrolase (HYD) and the heme-containing cytochrome P450 (CYP) monooxygenase. The HYD enzymes mainly participate in the dihydroxylation of β-carotene (β,β-carotene) to produce zeaxanthin (13, 14). The CYP monooxygenase hydroxylating the β-ring is CYP97A3 in the model plant Arabidopsis thaliana (15–17). The ε-ring hydroxylation is catalyzed by CYP97C1 (18–20). The CYP97A and CYP97C monooxygenases work in synergy to drive the dihydroxylation of α-carotene to produce lutein (14).
Fig. 1.
Stereospecific hydroxylation of α-carotene and alignment of CYP97A3 and CYP97C1. (A) Lutein formation from α-carotene via α-cryptoxanthin and zeinoxanthin. The hydroxyl group is in red. (B) Alignment of CYP97A3 and CYP97C1. Secondary-structure elements of CYP97A3 (green) are above the alignment, and those of CYP97C1 are below (blue). Elements in brown are only observed in the substrate-free CYP97A3 structure. The helix preceding the A helix or G helix is named A″ helix or G″ helix; the intermolecular β-strand is named β-F/G. Identical residues are in a black background and similar residues are boxed; the heme thiolate cysteine is in yellow. The dimmed residues are not observed in the structures. The blank triangle indicates the boundary of the predicted signal transit peptide; the filled triangle indicates the expression start site. The dashed box denotes the disordered region predicted by the DISOPRED server. The alignment figure was generated using the ESPript web server.
CYPs are a superfamily of heme-thiolate proteins and considered nature’s most versatile catalysts (21). The various spin states of iron within the heme prosthetic group bestow these enzymes with the ability to accomplish difficult oxidative reactions including oxygen insertion into a C–H bond (22). The overall structure of CYPs has a conserved triangular prism-like shape made by 12 or 13 α-helices and several β-strands, but their substrate-binding region is extremely diverse (23). Such structural diversity offers a wide range of biological activities besides the typical monooxygenase activity for this superfamily (24). Plants have evolved a large number of CYP genes including families from CYP71 to CYP99 and from CYP701 and above, but the majority of plant CYPs remain poorly characterized (25, 26). The CYP97 members lack a transmembrane helix and are peripherally bound to the membrane (14), resembling mitochondrial CYPs that use the ferredoxin-NADP+ reductase (FNR)–ferredoxin pair as redox partner (27, 28).
CYP97A3 and CYP97C1 stereospecifically add an oxygen atom to the pro-R hydrogen of C-3 in the cyclohexene ring. A structural elucidation of the pro-R stereospecificity and the recognition of β- and ε-rings is expected to provide molecular insights into the ultimate step of lutein biosynthesis. Here we describe the X-ray structures of CYP97A3 in ligand-free and retinal-bound forms and CYP97C1 in a detergent-bound form. The similarities between the CYP97 members and mitochondrial CYPs suggest that the enzymes share a similar electron transport mechanism.
Results
Characterization of Recombinant CYP97A3 and CYP97C1.
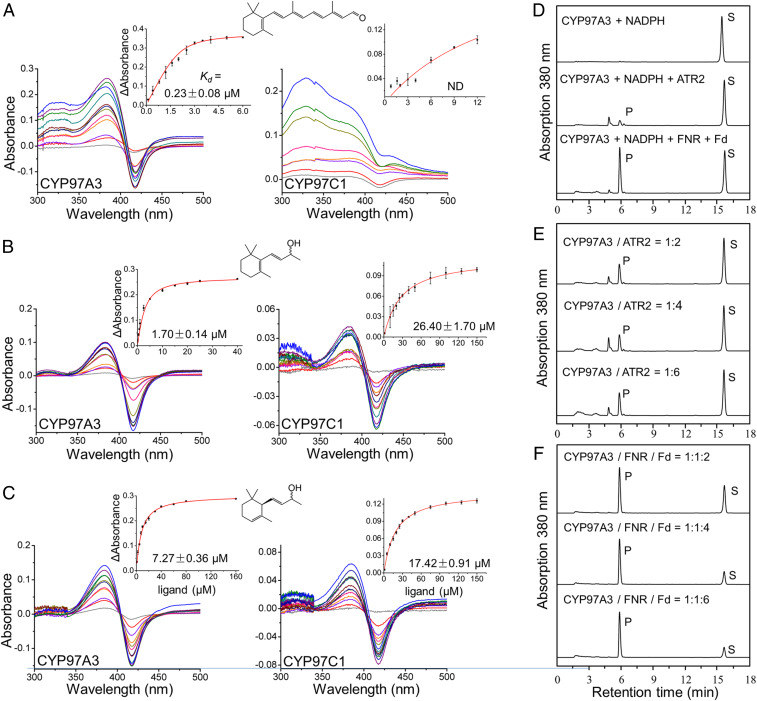
The A. thaliana genes LUTEIN DEFICIENT 5 (LUT5) and LUT1 encode CYP97A3 and CYP97C1, respectively. The CYP97A3 and CYP97C1 sequences share 233 identical residues (Fig. 1B). We successfully obtained the two recombinant enzymes (without the predicted transit peptide and disordered region) from heterogeneous expression in Escherichia coli (SI Appendix, Fig. S1). In an attempt to measure substrate binding to recombinant CYP97A3 and CYP97C1, we first used the natural substrates α-carotene and zeinozanthin. However, both compounds are extremely difficult to dissolve in aqueous buffer due to their hydrophobic long-chain hydrocarbon. Therefore, we used less hydrophobic substrate analogs, including the diterpenoid retinal, which mimics the β-ring–containing half-moiety of α-carotene and can act as a nonnatural substrate for CYP97A3, and the more hydrophilic β-ionol and α-ionol for the affinity assay. To measure the apparent binding constant (Kd) of retinal, β-ionol, and α-ionol to recombinant enzymes, a spectral titration method was performed (29). The Kd value of retinal to CYP97A3 was in the submicromolar range, while the Kd value to CYP97C1 was not detected (Fig. 2A). As for β-ionol, which also contains the β-ring, the affinity was 15-fold higher to CYP97A3 compared with that to CYP97C1 (Fig. 2B). These results suggest that the β-ring–containing ligands are less accessible to the heme iron of CYP97C1 than CYP97A3. However, for the ε-ring–containing α-ionol, it showed 2.4-fold higher affinity to CYP97A3 than CYP97C1 (Fig. 2C). It is possible that the four-carbon chain with a hydroxyl group prevents the ε-ring from traveling through the substrate channel to the buried active site, which is a common feature of CYP monooxygenases (30, 31). Alternatively, the interaction with CYP97A3 could be needed for facilitating substrate binding to CYP97C1 (14).
Fig. 2.
Characterization of recombinant CYP97A3 and CYP97C1. (A–C) Differential spectral titration of CYP97A3 and CYP97C1 with increasing concentrations of nonnatural ligand. (A–C) Sequential additions of (A) retinal, (B) β-ionol, and (C) α-ionol are individually colored. The structure of the ligand is shown above the spectra. (A–C, Insets) The Kd values are obtained from a nonlinear regression of the data. Error bar indicates the SD. ND, not determined. (D–F) HPLC analysis of CYP97A3-catalyzed retinal hydroxylation. (D) Incubation of retinal with CYP97A3 and NADPH in the absence (Top) and presence of the CPR ATR2 (Middle) or the FNR‒ferredoxin pair (Bottom). P and S stand for product and substrate. Arbitrary units were recorded. (E) Incubation of retinal, CYP97A3, and NADPH with increasing amounts of ATR2. (F) Incubation of retinal, CYP97A3, and NADPH with equal amounts of FNR coupled to increasing amounts of ferredoxin.
To test the retinal hydroxylation activity, we established an assay using recombinant CYP97A3. The product 3-hydroxy-retinal was measured by high-pressure liquid chromatography (HPLC) as described previously (32). Two redox partners, the FNR–ferredoxin pair and the NADPH-cytochrome P450 reductase (CPR) ATR2, were able to serve as electron donors for the hydroxylation reaction in the presence of NADPH (Fig. 2D). No reaction was detected with CYP97C1 (SI Appendix, Fig. S2). The FNR–ferredoxin pair was more efficient than CPR for 3-hydroxy-retinal production. Even when the amount of CPR was doubled, the product was still significantly lower than that by the ferredoxin-dependent system (Fig. 2 E and F). These results were consistent with previous work using a bacteria assay system (14), confirming that the CYP97 carotenoid hydroxylases are a mitochondria-type CYP monooxygenase that recruits the flavin adenine dinucleotide (FAD)-containing FNR and the iron-sulfur protein ferredoxin for electron transfer from NADPH to the heme center (33).
Substrate Channel.
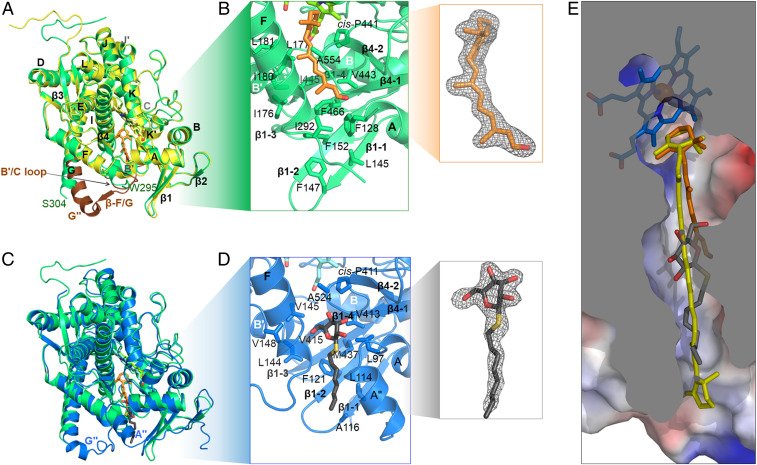
We crystallized and solved structures of CYP97A3 in substrate-free and retinal-bound forms at 2.4- and 2.0-Å resolution, respectively (SI Appendix, Table S1). The secondary-structure annotation is based on the traditional nomenclature (27, 34), in which α-helices and β-sheets are numbered as A to L and β1 to β4 (Fig. 1B). The substrate-free CYP97A3 structure differs from the retinal-bound structure in that it exists as a homodimer (SI Appendix, Fig. S1). The two monomers are nearly identical with an overall rmsd of 0.13 Å, and therefore one monomer (chain A) is used as the representative structure of substrate-free CYP97A3. The overall rmsd between the substrate-free and retinal-bound CYP97A3 is 0.40 Å, and obvious conformational differences exist in the F/G loop, the first half of the G helix, and the B′/C loop (Fig. 3A). This is consistent with the B′ and the F and G regions being the most structurally variable parts in CYPs (27). The retinal is positioned in the active site of CYP97A3 (Fig. 3B and SI Appendix, Fig. S3). Its polyene chain is held in a channel formed by hydrophobic residues, including Ile176, Leu177, Ile180, and Leu181 in the B′ helix, Ile292 in the F/G loop, Val443, Ile445, and Phe466 in the β1-sheet, and Ala554 from the β-hairpin in the β4-sheet. The aldehyde tail of retinal is covered by the surface-located Phe128, whose phenyl ring, together with the side chains of residues Leu145 and Phe147 in the β1-1 strand, Phe152 in the β1-2 strand, Val443 and Ile445 in the β1-4 strand, and Phe466 in the β1-3 strand, defines a hydrophobic patch along the extension of the polyene chain. However, our attempts to crystallize CYP97A3 in complex with α-carotene or β-carotene were unsuccessful.
Fig. 3.
Substrate channel in CYP97A3 and CYP97C1. (A) Superimposed structures of the substrate-free CYP97A3 (chain A, yellow) and retinal-bound CYP97A3 (green). The protein is shown as a ribbon with heme in stick representation. The retinal is in orange (stick representation). The B′/C loop, F/G loop, and N-terminal end of the G helix in the substrate-free CYP97A3 structure are in brown. Residues 296 to 305 are not observed in the retinal-bound structure. (B) Details of retinal-bound CYP97A3. The side chains of residues forming the hydrophobic channel are shown as sticks. The 2|Fo| − |Fc| map (gray mesh) of the retinal contoured at 1.0σ is shown (Inset). (C) Superimposed structures of retinal-bound CYP97A3 and OTG-bound CYP97C1 (blue). The carbons of the detergent are in black (stick representation). (D) Details of OTG-bound CYP97C1. The 2|Fo| − |Fc| map (gray mesh) of the detergent is shown (Inset). (E) Superimposition of the observed ligands (retinal and OTG) with a docked β-carotene molecule (yellow). The surface of CYP97A3 is colored by electrostatic potential (blue, positive; red, negative).
The 2.0-Å structure of CYP97C1 was determined in complex with the detergent octylthioglucoside (OTG). The OTG-bound CYP97C1 and retinal-bound CYP97A3 structures are similar, with <1.0 Å rmsd for 470 superimposed residues (Fig. 3C). Although their F/G loops differ significantly in conformation, the regions of the B′ helix, β1-sheet, and β-hairpin in the β-4 sheet have only slight changes, thus maintaining the shape of the channel holding the polyene chain. The channel hydrophobicity is kept by residues L144, Val145, Val148, Val413, Ile415, Met437, and Ala524 (Fig. 3D). The detergent OTG is situated in a patch composed by Leu114, Ala116, Phe121, Val413, Ile415, and Met437. The position of its glucose head is adjacent to the methyl group at the retinal tail as observed in CYP97A3 (Fig. 3E). Leu97, the counterpart of Phe128 in CYP97A3, allows entry of the sugar moiety into the substrate channel, while the hydrophobic residues within the channel prevent it from accessing the heme center. The alkene tail, resembling the entrance of the substrate channel, is in the vicinity of the β1-sheet. The superimposition of the ligands (retinal and OTG) in the crystal structures with a β-carotene molecule reveals that their positions are aligned along the alkene chain and compatible with a carotenoid substrate.
The Membrane-Binding Regions.
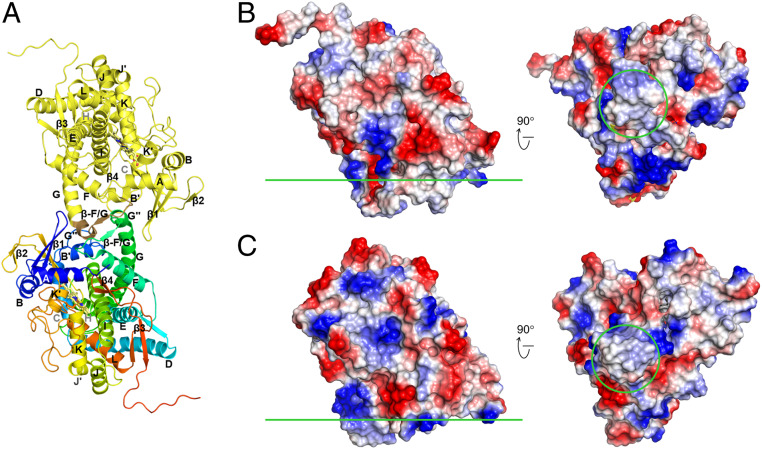
CYP97A3 and CYP97C1 share similar hydrophobic residues in the F/G loop (Fig. 1B), which is structurally near the β1-sheet. However, in the substrate-free dimeric CYP97A3 structure, the F/G loops interact with each other (Fig. 4A), which is similar to the observation in the rabbit CYP2B4 (35). The dimeric interactions suggested that dimerization might affect the catalytic activity. To test this possibility, we made a CYP97A3 mutant (CYP97A3m) by site-directed mutagenesis, in which Ser290, Trp300, and Ser304 were mutated to their respective counterparts in CYP97C1, Asp, Leu, and Val. The recombinant CYP97A3m was purified as a monomer (SI Appendix, Fig. S1). Furthermore, we determined the structures of CYP97A3m in the substrate-free and retinal-bound forms at the respective resolutions of 1.7 and 2.4 Å (SI Appendix, Fig. S4). Both structures are similar to that of retinal-bound CYP97A3 with rmsd values <0.18 Å. A retinal hydroxylation assay showed that CYP97A3m was enzymatically active (SI Appendix, Fig. S5), indicating that dimerization observed in the substrate-free CYP97A3 structure was dispensable for enzyme activity.
Fig. 4.
Membrane-binding regions. (A) The substrate-free dimeric CYP97A3 structure. Chain A is colored in yellow, and the F/G loop is colored in brown; chain B is colored in rainbow with the N terminus in blue and C terminus in red. (B) Electrostatic potential surface of the substrate-free CYP97A3 structure (chain A). The membrane interface is indicated by the green line (Left, side view; Right, bottom view). The F/G loop region is circled. (C) Electrostatic potential surface of the OTG-bound CYP97C1 structure.
The hydrophobic region within the F/G loop provides a membrane-binding site for mitochondrial CYPs (36). Similarly, the F/G loop in CYP97A3 and CYP97C1 shows up as a hydrophobic protrusion, which is surrounded by positively charged patches (Fig. 4 B and C). As the membrane surface is negatively charged (37), such a structure favors association of CYP97A3 and CYP97C1 with the membranes.
Structural Basis for Pro-3R Hydroxylation.
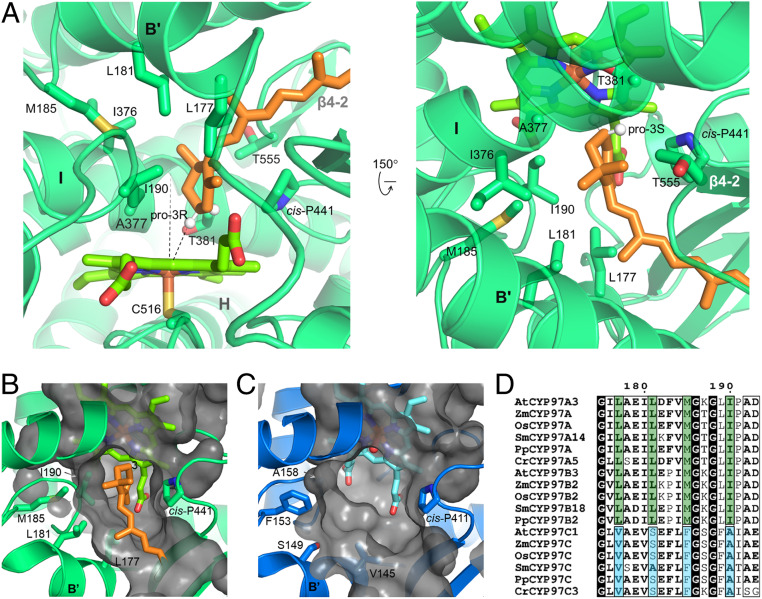
The retinal-bound CYP97A3 structure reveals how the pro-3R specificity is achieved (Fig. 5A). The C-3 atom of the β-ring is the closest nonbonded carbon to the heme iron. In the substrate-free CYP97A3 structure, a water molecule is coordinated as the distal axial ligand of the heme iron (SI Appendix, Fig. S6). Retinal displaces the bound water molecule, and thus the sulfur atom of Cys516 is the only axial ligand of the iron. The C-3 atom is tilting away ∼30° from the heme normal with its pro-3R hydrogen facing the iron, leaving a distance of ∼3.3 Å between the iron and the hydrogen. The conserved oxygen-activating threonine (34), Thr381, is the nearest residue to the C-3 atom. Ile190 in the B′/C loop, Ala377 and Thr381 in the I helix (distal helix), and Thr555 in the β4-sheet fix the β-ring through hydrophobic interactions. Such a configuration shows how the β-ring is precisely positioned to favor 3R hydroxylation.
Fig. 5.
Structural basis for pro-3R stereospecificity. (A) The active site in retinal-bound CYP97A3. The protein is shown in transparent ribbons and heme is in bright green. The C-3 hydrogens are positioned and shown as white spheres. The heme normal and the distance between the pro-3R hydrogen and iron are shown as dashes. (B) Surface representation of the CYP97A3 pocket. Side chains of Leu177, Leu181, Met185, and Ile190 are shown. (C) Surface representation of the CYP97C1 pocket. Side chains of Val145, Ser149, Phe153, and Ala158 are shown. (D) Alignment of the B′ helix and B′/C loop. Species include A. thaliana, Z. mays, Oryza sativa, Selaginella moellendorffii, Physcomitrella patens, and Chlamydomonas reinhardtii.
The configuration of the active site is structurally conserved between these two enzymes. A notable structural feature is the cis conformation of proline at position 441 in CYP97A3 and at position 411 in CYP97C1 (Fig. 5 B and C). This cis-proline lies at the pro-3S side of the cyclohexene ring. Other identical residues between CYP97A3 and CYP97C1 (with their corresponding positions in parentheses) include Ala377(347), Gly378(348), and Thr381(351) in the distal I helix, Leu444(414) in the β1-4 strand, and Ala554(524) and Thr555(525) in the turn of the β-4 sheet. On the proximal side of the heme, CYP97A3 and CYP97C1 share a conserved β-bulge referred to as the Cys pocket (27). All residues involved in heme binding are identical between the two enzymes (SI Appendix, Fig. S6).
Different residues (with those of CYP97C1 in parentheses) include Leu177(Val145), Leu181(Ser149), and Met185(Phe153) in the B′ helix, Ile190(Ala158) in the B′/C loop, and Ile376(Val346) in the I helix (Fig. 1B). These different residues are all located on the opposite side of the cis-proline. The Val/Ile variation in the I helix has almost no structural change, but variations in the B′ helix and B′/C loop result in notable differences with respect to the shape of the substrate channel. The side chains of Leu177–Leu181–Met185–Ile190 in CYP97A3 are longer than those of Val145–Ser149–Phe153–Ala158 in CYP97C1, forming a smaller pocket for accommodation of the more restrained β-ring. Furthermore, amino acid sequence alignment of the B′ helix and B′/C loop indicates that CYP97B3 has the same substrate specificity as CYP97A3 (Fig. 5D). This validates the speculation that the CYP97B family members are β-ring hydroxylases (14, 38).
Discussion
The genome of A. thaliana has 244 CYPs that account for nearly 1% of protein-coding genes, and about one-third have annotated biological functions which can be mapped to different biosynthetic or metabolic pathways (39). Currently, only CYP74A and CYP90B1 have been structurally investigated (40–42). CYP74A is a key enzyme in oxylipin biosynthesis and functions as an allene oxide synthase that does not need the reducing agent NADPH (36). CYP90B1 participates in brassinosteroid biosynthesis and relies on membrane-bound CPR for electron transfer (42, 43). Evolutionary analysis has shown that the CYP97 family members are the most ancient plant CYPs (44). Our activity assays and structural work confirm that the CYP97 family monooxygenases are mitochondrial CYPs and use the FNR–ferredoxin pair as redox partner (33).
Plant carotenoids play diverse roles, such as in regulation of light harvesting and protection against photooxidative stress and as precursors for developmental and environmental signals (2, 4, 5, 45). The common carotenoids include α-carotene, β-carotene, and their oxygenated derivatives lutein and zeaxanthin (3R,3′R-β,β-carotene-diol). The hydroxylation of α-carotene and β-carotene is catalyzed by two distinct classes of hydroxylases. The β-ring–specific HYDs are committed to zeaxanthin formation, and the CYP97 family members act synergically to form lutein (14, 15, 19, 20). The presented structures uncover the molecular details underlying the β-ring specificity of CYP97A3. Comparison of the substrate-binding sites offers clues about the structural determinants for the ε-ring by CYP97C1, and suggests that the CYP97B family members are β-ring–specific. The finding explains the observation of lutein production in the lut5(cyp97a3) b1(hyd-1) b2(hyd-2) triple mutant in A. thaliana (15, 17, 20) and supports the notion of a fourth β-ring–specific hydrolase (14, 16, 38).
Biologically, CYP97 family members are peripherally associated with membranes through the hydrophobic region (14). The membrane lipids play important roles in substrate accessibility for CYPs (36). In this study, the half-carotenoid molecule retinal was used to mimic the natural substrate, and was confirmed to be a nonnatural substrate for the β-ring–specific CYP97A3 in an in vitro activity assay. The retinal-bound CYP97A3 structures presented here show how the pro-3R stereospecificity is ensured. In the absence of retinal, CYP97A3 forms a homodimer through the F/G loop that is supposed to interact with the membrane lipids, and addition of retinal causes dissociation of the homodimer. The F/G loop appears to be involved in substrate binding besides its lipid-binding ability. No dimerization is observed for CYP97C1 or CYP97A3m that replaces three residues in the F/G loop region of CYP97A3 with those in CYP97C1. CYP97A3m retains retinal hydroxylation activity, which indicates that dimerization is not required for in vitro activity. However, it should be noted that the natural substrate α-carotene has a length that can cover the whole substrate channel and is extremely hydrophobic. It is possible that heterodimerization of CYP97A3 and CYP97C1 occurs in the membrane environment, which has been proposed to improve the efficiency of hydroxylation of α-carotene (14, 15).
The membrane-associated multienzyme complexes may facilitate substrate channeling between the carotenogenic enzymes (4, 5). The enzymes that produce α-carotene are lycopene β- and ε-cyclases, two flavoproteins that require FAD as cofactor. Lycopene β-cyclase has been found in the chloroplast envelope membrane, and lycopene ε-cyclase is suggested to be anchored to the envelope membrane by its transmembrane helix (12). Although the protein–protein interaction between lycopene cyclases and CYP97 family members still needs to be experimentally defined, their same localization on membranes suggests a multienzyme complex devoted to lutein biosynthesis. Further interesting work needs to be done to investigate such a complex.
Materials and Methods
Protein Expression and Purification.
The CYP97A3/LUT5 gene (Arabidopsis Genome Initiative [AGI] identifier At1g31800) without the 28-residue transit peptide and predicted disordered region (49 residues) was amplified by PCR. The amplified product was ligated between the EcoRI and SalI restriction sites of a modified pCW-MBP (maltose-binding protein) vector (46, 47). The expressed polypeptide contains an MBP-His6 tag followed by a tobacco etch virus (TEV) protease cleavage site and CYP97A3 (residues 78 to 595). The expression and purification procedure was adapted from ref. 47. The E. coli BL21 competent cells transformed with expression construct were cultured at 37 °C until the optical density at 600 nm reached ∼0.8. Then the culture was grown at 23 °C for 30 min before 5-aminolevulinic acid and isopropyl β-d-thiogalactoside were added, both to final concentrations of 0.5 mM. The cells were grown at 23 °C for 48 h before harvest by centrifugation, and the pellets were lysed in ice-cold buffer A (200 mM NaCl and 20 mM Tris⋅HCl, pH 7.5) with 5 mM imidazole. Cell debris was removed by centrifugation and the supernatant was subjected to affinity chromatography using an Ni2+-nitrilotriacetic acid (Ni-NTA; QIAGEN) resin, which was equilibrated with buffer A and washed with 5 mM imidazole in buffer A. The recombinant MBP-His6–tagged CYP97A3 was eluted with 200 mM imidazole in buffer A, and was further purified by size-exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare) eluted with buffer A. Peak fractions were pooled and incubated with His6-tagged TEV protease at 4 °C overnight to cut the affinity tag. The cleaved tag, uncleaved protein, and protease were removed by Ni-NTA resin. The cleaved protein was further purified by HiLoad 16/60 Superdex 200 column eluted with buffer A supplemented with 2 mM dithiothreitol. Fractions containing CYP97A3 were collected and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Expression and purification of CYP97A3m and CYP97C1 were essentially the same as described for CYP97A3. An exception was the length of the CYP97C1/LUT1 gene (AGI identifier At3g53130), whose coding product is 56 residues shorter than CYP97A3. Sequence corresponding to residues 70 to 539 of CYP97C1 was expressed. For more on the construction and purification of CYP97A3, CYP97A3m, and CYP97C1, see SI Appendix.
The fragment of A. thaliana ferredoxin-NADP+ oxidoreductase (At1g20020) lacking the N-terminal 55 residues and the fragment of Zea mays ferredoxin (FDX1) lacking the N-terminal 52 residues were PCR-amplified. Each fragment was cloned into the pET-22b(+) (Novagen) expression vector. Expression and purification of FNR and ferredoxin were performed as described previously (48). The fragment of A. thaliana NADPH-cytochrome P450 reductase 2 (ATR2; At4g30210) lacking the N-terminal 72 residues was expressed and purified as reported (47).
Ligand-Binding Assay.
The binding-induced spectral changes were monitored as a shift of the heme Soret peak. The dissociation constant of the enzyme–ligand complex was determined by differential spectral titration following a reported procedure (29). The binding of ligand to the purified recombinant protein was measured in 50 mM Tris⋅HCl (pH 7.5) at room temperature, with a final protein concentration of 1 μM. Freshly prepared aliquots of retinal, β-ionol, and α-ionol in ethanol were added to the sample cuvette during titration, and the total amount of ethanol added did not exceed 2% (volume [vol]/vol). The data were analyzed using Origin8 (OriginLab) and presented as means ± SEs.
Crystallization and Data Collection and Structure Determination.
Purified proteins were concentrated to 8 to 12 mg⋅mL‒1 for crystallization. Crystal trays were set up at 16 °C using the vapor diffusion method in a sitting drop consisting of 1 μL protein sample and 1 μL well solution. The substrate-free CYP97A3 was crystallized using 0.1 M 2-(N-morpholino)ethanesulfonic acid buffer (pH 6.5) and 12% (weight [wt]/vol) polyethylene glycol (PEG) 20,000. The retinal-bound CYP97A3 crystal was obtained using 0.2 M potassium sulfate, 20% (wt/vol) PEG 3,350, and 0.5 mM retinal. CYP97C1 bound with n-octyl-β-d-thioglucoside was crystallized using 0.18 M ammonium citrate, 20% (wt/vol) PEG 3,350, and 13.5 mM OTG. For data collection, the crystals were stepwise transferred into the reservoir solution containing increasing amounts of glycerol (5, 10, and 15% [vol/vol]) and then flash-cooled in liquid nitrogen. The data were collected at 100 K at beamline BL17U of the Shanghai Synchrotron Radiation Facility and at beamline BL19U of the National Center for Protein Science Shanghai. All diffraction data were processed and scaled with DENZO and SCALEPACK in the HKL2000 package (HKL Research).
Molecular replacement was performed with Phaser (49) in the CCP4 program suite (50). The substrate-free CYP97A3 structure was determined using the structure of the heme domain of Bacillus megaterium cytochrome P450 BM3 (Protein Data Bank [PDB] ID code 4KF2) (51) as search template. The structures of retinal-bound CYP97A3 and OTG-bound CYP97C1 were then solved using the substrate-free CYP97A3 structure as search template. The resulting model was rebuilt with PHENIX AutoBuild (52, 53). Manual correction was performed with Coot (54) according to |Fo| − |Fc| and 2|Fo| − |Fc| maps, and further refinement was carried out with the program phenix.refine in the PHENIX suite (55). The quality of the refined structures was evaluated by MolProbity (56). All structural figures were prepared using PyMOL (Schrödinger).
Enzyme Activity.
The assay was performed in a total volume of 200 μL of the following: 50 mM Tris⋅HCl (pH 7.5), 0.1% (vol/vol) Triton X-100, 1 mM NADPH, 10 mM glucose 6-phosphate, 5 units of glucose 6-phosphate dehydrogenase (Sigma-Aldrich), 0.5 mM substrate, 4 to 16 μM ATR2 (or 2 μM FNR and 2 to 16 μM FDX1), and 2 μM purified recombinant protein. Incubations were conducted at 26 °C for 12 h. The reaction was stopped by adding 1 μL 70% (wt/wt) perchloric acid. After the mixture was centrifuged at 15,000 × g for 10 min, 100 μL of supernatant containing the reaction products was subjected to HPLC analysis according to published procedures (32, 57) with minor modification. An Alltech Alltima HPLC reversed-phase C18 column (4.6 × 250 mm, 5 μm; Grace) was used. For assays with retinal, the following solvent system was used: A, ethyl acetate; B, acetonitrile:water:trimethylamine (8:2:0.01 [vol/vol/vol]). The column was developed at a flow rate of 1 mL⋅min−1, with a gradient from 100% B to 50% B within 22.5 min, and then to 100% A within 2.5 min, with the final conditions being maintained for another 5 min. For assays with β-ionol or α-ionol, the following solvent system was used: A, methanol:water:t-butylmethyl ether (5:1:5 [vol/vol/vol]); B, methanol:water:t-butylmethyl ether (5:5:1 [vol/vol/vol]). The column was developed at a flow rate of 1 mL⋅min−1, with a gradient from 100% B to 40% B within 20 min, and then to 100% A within 5 min, with the final conditions being maintained for another 5 min.
Data Availability.
Structural data have been deposited in the Protein Data Bank (https://www.wwpdb.org/) with PDB ID codes 6J94 for substrate-free CYP97A3, 6J95 for retinal-bound CYP97A3, 6L8I for substrate-free CYP97A3m, 6L8J for retinal-bound CYP97A3m, and 6L8H for OTG-bound CYP97C1 (details in SI Appendix, Table S1).
Supplementary Material
Acknowledgments
We are grateful to Dennis Stuehr at the Cleveland Clinic Lerner Research Institute for his kind provision of the pCW vector. We thank the staff at the Shanghai Synchrotron Radiation Facility and the National Center for Protein Science Shanghai for technical support, and Ming-Zhu Wang and Shi-Long Fan for assistance with data collection and processing. We also thank the Plant Science Facility of the Chinese Academy of Sciences Institute of Botany for the use of the HPLC system. This work was supported by the National Natural Science Foundation of China (Grants 31670794 and 31530006), National Key R&D Program of China (Grant 2017YFA0503703), Chinese Academy of Sciences (Grant XDB17030100), Chang Jiang Scholars Program (Award Q2017241), and Anhui Provincial Wanjiang Scholars Program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Structural data reported in this paper have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID codes 6J94, 6J95, 6L8I, 6L8J, and 6L8H).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001806117/-/DCSupplemental.
References
- 1.Frank H. A., Cogdell R. J., Carotenoids in photosynthesis. Photochem. Photobiol. 63, 257–264 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Shumskaya M., Wurtzel E. T., The carotenoid biosynthetic pathway: Thinking in all dimensions. Plant Sci. 208, 58–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao A. V., Rao L. G., Carotenoids and human health. Pharmacol. Res. 55, 207–216 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Moise A. R., Al-Babili S., Wurtzel E. T., Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 114, 164–193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nisar N., Li L., Lu S., Khin N. C., Pogson B. J., Carotenoid metabolism in plants. Mol. Plant 8, 68–82 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Wurtzel E. T., Kutchan T. M., Plant metabolism, the diverse chemistry set of the future. Science 353, 1232–1236 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Wurtzel E. T., Changing form and function through carotenoids and synthetic biology. Plant Physiol. 179, 830–843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mares J., Lutein and zeaxanthin isomers in eye health and disease. Annu. Rev. Nutr. 36, 571–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller P., Li X. P., Niyogi K. K., Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125, 1558–1566 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruban A. V., Johnson M. P., Xanthophylls as modulators of membrane protein function. Arch. Biochem. Biophys. 504, 78–85 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Jahns P., Holzwarth A. R., The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim. Biophys. Acta 1817, 182–193 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Sola M. Á., Rodríguez-Concepción M., Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arabidopsis Book 10, e0158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vallabhaneni R. et al., Metabolite sorting of a germplasm collection reveals the hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiol. 151, 1635–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinlan R. F. et al., Synergistic interactions between carotene ring hydroxylases drive lutein formation in plant carotenoid biosynthesis. Plant Physiol. 160, 204–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J., DellaPenna D., Defining the primary route for lutein synthesis in plants: The role of Arabidopsis carotenoid β-ring hydroxylase CYP97A3. Proc. Natl. Acad. Sci. U.S.A. 103, 3474–3479 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiore A., Dall’osto L., Fraser P. D., Bassi R., Giuliano G., Elucidation of the β-carotene hydroxylation pathway in Arabidopsis thaliana. FEBS Lett. 580, 4718–4722 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Kim J., Smith J. J., Tian L., DellaPenna D., The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol. 50, 463–479 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Pogson B., McDonald K. A., Truong M., Britton G., DellaPenna D., Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 8, 1627–1639 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian L., Magallanes-Lundback M., Musetti V., DellaPenna D., Functional analysis of β- and ε-ring carotenoid hydroxylases in Arabidopsis. Plant Cell 15, 1320–1332 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian L., Musetti V., Kim J., Magallanes-Lundback M., DellaPenna D., The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid ε-ring hydroxylation activity. Proc. Natl. Acad. Sci. U.S.A. 101, 402–407 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coon M. J., Cytochrome P450: Nature’s most versatile biological catalyst. Annu. Rev. Pharmacol. Toxicol. 45, 1–25 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Denisov I. G., Makris T. M., Sligar S. G., Schlichting I., Structure and chemistry of cytochrome P450. Chem. Rev. 105, 2253–2277 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Hasemann C. A., Kurumbail R. G., Boddupalli S. S., Peterson J. A., Deisenhofer J., Structure and function of cytochromes P450: A comparative analysis of three crystal structures. Structure 3, 41–62 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Podust L. M., Sherman D. H., Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 29, 1251–1266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapple C., Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 311–343 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Schuler M. A., Werck-Reichhart D., Functional genomics of P450s. Annu. Rev. Plant Biol. 54, 629–667 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Hannemann F., Bichet A., Ewen K. M., Bernhardt R., Cytochrome P450 systems—Biological variations of electron transport chains. Biochim. Biophys. Acta 1770, 330–344 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Iyanagi T., Xia C., Kim J. J., NADPH-cytochrome P450 oxidoreductase: Prototypic member of the diflavin reductase family. Arch. Biochem. Biophys. 528, 72–89 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeVore N. M., Smith B. D., Wang J. L., Lushington G. H., Scott E. E., Key residues controlling binding of diverse ligands to human cytochrome P450 2A enzymes. Drug Metab. Dispos. 37, 1319–1327 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cojocaru V., Winn P. J., Wade R. C., The ins and outs of cytochrome P450s. Biochim. Biophys. Acta 1770, 390–401 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Urban P., Lautier T., Pompon D., Truan G., Ligand access channels in cytochrome P450 enzymes: A review. Int. J. Mol. Sci. 19, e1617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alder A., Bigler P., Werck-Reichhart D., Al-Babili S., In vitro characterization of Synechocystis CYP120A1 revealed the first nonanimal retinoic acid hydroxylase. FEBS J. 276, 5416–5431 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Pylypenko O., Schlichting I., Structural aspects of ligand binding to and electron transfer in bacterial and fungal P450s. Annu. Rev. Biochem. 73, 991–1018 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Poulos T. L., Finzel B. C., Gunsalus I. C., Wagner G. C., Kraut J., The 2.6-Å crystal structure of Pseudomonas putida cytochrome P-450. J. Biol. Chem. 260, 16122–16130 (1985). [PubMed] [Google Scholar]
- 35.Scott E. E. et al., An open conformation of mammalian cytochrome P450 2B4 at 1.6-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 100, 13196–13201 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson E. F., Stout C. D., Structural diversity of eukaryotic membrane cytochrome P450s. J. Biol. Chem. 288, 17082–17090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dörmann P., Benning C., Galactolipids rule in seed plants. Trends Plant Sci. 7, 112–118 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Kim J. E., Cheng K. M., Craft N. E., Hamberger B., Douglas C. J., Over-expression of Arabidopsis thaliana carotenoid hydroxylases individually and in combination with a β-carotene ketolase provides insight into in vivo functions. Phytochemistry 71, 168–178 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Bak S. et al., Cytochromes p450. Arabidopsis Book 9, e0144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee D. S., Nioche P., Hamberg M., Raman C. S., Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 455, 363–368 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Li L., Chang Z., Pan Z., Fu Z. Q., Wang X., Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 105, 13883–13888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujiyama K. et al., Structural insights into a key step of brassinosteroid biosynthesis and its inhibition. Nat. Plants 5, 589–594 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Mizutani M., Ohta D., Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 116, 357–367 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson D., Werck-Reichhart D., A P450-centric view of plant evolution. Plant J. 66, 194–211 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Wurtzel E. T., Cuttriss A., Vallabhaneni R., Maize provitamin A carotenoids, current resources, and future metabolic engineering challenges. Front. Plant Sci. 3, 29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh D. K. et al., Characterization of the inducible nitric oxide synthase oxygenase domain identifies a 49 amino acid segment required for subunit dimerization and tetrahydrobiopterin interaction. Biochemistry 36, 10609–10619 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Niu G. et al., Structure of the Arabidopsis thaliana NADPH-cytochrome P450 reductase 2 (ATR2) provides insight into its function. FEBS J. 284, 754–765 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Liu L., Crystal structure and catalytic mechanism of 7-hydroxymethyl chlorophyll a reductase. J. Biol. Chem. 291, 13349–13359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy A. J. et al., Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winn M. D. et al., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler C. F. et al., Key mutations alter the cytochrome P450 BM3 conformational landscape and remove inherent substrate bias. J. Biol. Chem. 288, 25387–25399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terwilliger T. C. et al., Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams P. D. et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afonine P. V. et al., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen V. B. et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Norris S. R., Barrette T. R., DellaPenna D., Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7, 2139–2149 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural data have been deposited in the Protein Data Bank (https://www.wwpdb.org/) with PDB ID codes 6J94 for substrate-free CYP97A3, 6J95 for retinal-bound CYP97A3, 6L8I for substrate-free CYP97A3m, 6L8J for retinal-bound CYP97A3m, and 6L8H for OTG-bound CYP97C1 (details in SI Appendix, Table S1).