Significance
Acute myeloid leukemia (AML) is the most common acute leukemia in adults, with a 5-y survival of 29%. Immunotherapy is based on the premise that tumors suppress the immune system. We investigated the status of T cell immunity in AML at the time of diagnosis. We found a significant association between T cell percentage in the bone marrow and overall survival in newly diagnosed AML patients. When we evaluated the T cells from the bone marrow of patients with AML, one-third displayed profound functional impairment. Most of these compromised T cells, however, could be rescued using checkpoint inhibitors. Our results support the development of immune checkpoint therapy to combat this deadly disease.
Keywords: AML, T cell, checkpoint blockade, immune microenvironment, leukemia
Abstract
Acute myeloid leukemia (AML) is the most common acute leukemia in adults, with approximately four new cases per 100,000 persons per year. Standard treatment for AML consists of induction chemotherapy with remission achieved in 50 to 75% of cases. Unfortunately, most patients will relapse and die from their disease, as 5-y survival is roughly 29%. Therefore, other treatment options are urgently needed. In recent years, immune-based therapies have led to unprecedented rates of survival among patients with some advanced cancers. Suppression of T cell function in the tumor microenvironment is commonly observed and may play a role in AML. We found that there is a significant association between T cell infiltration in the bone marrow microenvironment of newly diagnosed patients with AML and increased overall survival. Functional studies aimed at establishing the degree of T cell suppression in patients with AML revealed impaired T cell function in many patients. In most cases, T cell proliferation could be restored by blocking the immune checkpoint molecules PD-1, CTLA-4, or TIM3. Our data demonstrate that AML establishes an immune suppressive environment in the bone marrow, in part through T cell checkpoint function.
Acute myeloid leukemia (AML) is a clinically and molecularly heterogeneous disorder. The treatment for AML remains largely unchanged over the last several decades, with high-dose chemotherapy remaining the mainstay of therapy (1). Although a majority of patients achieve remission following induction chemotherapy, most will relapse, leading to a 5-y survival rate of 29% (2). This has created an impetus to explore novel therapeutic approaches. Immunotherapies, such as immune checkpoint inhibitors (CPI), are treatments used to provoke the immune system to attack tumor cells (3–5). To date, this interest in testing CPI therapy in AML has resulted in over 30 clinical trials (6).
The rationale for using CPIs is based on the hypothesis that T cells in the tumor microenvironment (TME) are exhausted or suppressed by the presence of immune suppressive factors (7). A link between tumor-infiltrating T cells and outcome has been noted in several types of cancer and has been interpreted as an indication of immune recognition of the tumor (8–14). Currently, the functional status of T cells in the bone marrow of patients with AML remains unclear and there is little consensus on whether there is a role for T cells in the AML TME during disease development. A previous report described an association between high (i.e., ≥78.5% of lymphocytes) T cell percentages in the bone marrow and improved overall and leukemia-free survival in newly diagnosed AML patients (15). There is evidence indicating some degree of T cell dysfunction in AML, including reduced ability of T cells to proliferate or produce cytokines, but studies are limited (16–24). The mechanism behind T cell impairment remains elusive but may be the result of secreted factors from AML blasts, with some restoration in function if lymphocytes are removed from the suppressive environment (16, 18, 20, 23, 25). Additionally, gene-expression profiling has demonstrated global differences in T cell transcription profiles within patients with AML (18, 23). At the phenotypic level, CD8+ T cells from patients with AML have been reported to have an increased expression of exhaustion and senescence molecules (e.g., CD57, TIGIT, TIM-3, and PD-1) compared to healthy donors (23, 26–28). This activated phenotype is increased in relapsed disease and p53-mutant AML, suggesting that specific subsets of the disease may be more responsive to therapies aimed at restoring T cell function (23, 29).
To further investigate the impact of AML disease on immune function, with a particular focus on the unique bone marrow microenvironment where AML originates and therapy-resistant cells may persist, we developed a multifaceted study employing phenotypic and functional approaches to comprehensively examine the role of T cells in AML. We characterized the bone marrow T cells in a set of 49 patients with AML at initial diagnosis. T cells from AML patients varied widely in their ability to respond to stimulation, including roughly one-third that showed profound impairment. In the majority of those suppressed samples, however, functional responsiveness could be restored by interfering with checkpoint pathways, with PD-1 blockade being the most potent. We further associated T cell dysfunction with changes in the distribution of naïve and effector T cells. Finally, with clinical data on 80 newly diagnosed AML patients with a diagnostic bone marrow sample that was analyzed by flow cytometry, we found that a higher T cell percentage (of lymphocytes) in the TME correlated with longer patient survival, suggesting that T cells make important contributions to the status and outcome of disease.
Results
Functional Assessment of T Cells in Newly Diagnosed AML.
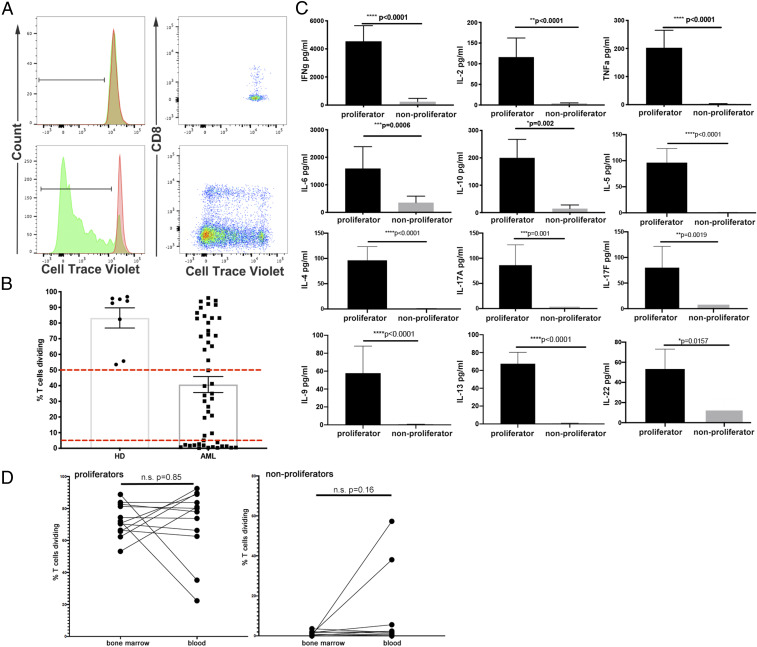
One of our primary goals for these studies was to determine the functional status of T cells in the bone marrow of patients with AML. To assess the impact of the immune microenvironment on T cell function, we assayed AML samples to measure the ability of T cells to proliferate and produce cytokines. Forty-nine bone marrow samples from patients with newly diagnosed AML were evaluated (patient characteristics listed in SI Appendix, Table S1). For functional comparisons, eight bone marrow samples were obtained from healthy donors. We determined T cell proliferation after stimulation of total bone marrow mononuclear cells (BMMC) with anti-CD3 agonistic antibody, allowing costimulation or inhibition to come from the surrounding cells. After 5 d of culture, we used flow cytometry to identify T cells (defined as positive for CD3 and single-positive for either CD4 or CD8) and assess their proliferation as measured by the dilution of CellTrace Violet (CTV). We observed samples in which the T cells did not dilute CTV after 5 d of anti-CD3 treatment (Fig. 1 A, Upper, green histogram), which was similar to wells that contained an irrelevant control mouse IgG (mIgG) (Fig. 1 A, Upper, red histogram). In contrast, we found that many samples responded to anti-CD3 by diluting CTV, with varying numbers of divisions (Fig. 1 A, Lower histograms). In these samples, T cell division occurred in both CD4+ and CD8+ subpopulations (Fig. 1 A, Right).
Fig. 1.
Impaired T cell proliferation and cytokine production in response to TCR stimulation in a subset of patients with AML. (A) Proliferation of T cells from two representative AML samples. CTV dilution by T cells is shown as histograms (mIgG, red; anti-CD3, green) or dot plots (CTV vs. CD8) of a nonproliferator (Upper) or a proliferator (Lower). (B) Summary of T cell proliferation data for all AML (n = 49) and healthy donor (n = 8) samples from the functional assay cohort. Dashed horizontal lines indicate cutoffs for the proliferator group (proliferation value > 50%, n = 20) and the nonproliferator group (proliferation value < 5%, n = 18). (C) Cytokine measurements from supernatants of cultures incubated with anti-CD3 + mIgG, categorized as proliferators or nonproliferators (error bars = SEM). P values shown are from a Student’s t test between groups and have not been adjusted for multiple comparisons. (D) Comparison of T cell proliferation in matched blood and bone marrow samples. Data shown are percentage of T cells diluting CTV as in B. Samples are grouped into proliferators (Left, n = 12) and nonproliferators (Right, n = 10) as defined in B. Statistical test for D is paired Student’s t test.
From this assessment of T cell function, AML samples were classified as either “proliferators” or “nonproliferators.” We assumed that AML samples with no T cell proliferative defect would behave similarly to samples from healthy donor bone marrow. Thus, if a sample’s proliferation value (defined as the percentage of T cells diluting CTV after anti-CD3 stimulation minus the percentage after treatment with control mIgG) was above the minimum proliferation value among the healthy donors (i.e., 50%), the sample was classified as a proliferator. On the other hand, samples were classified as nonproliferators if their anti-CD3–induced proliferation was less than 5%. A summary of T cell proliferation for the 49 BMMC samples is displayed in Fig. 1B, with red dotted lines indicating cutoffs for proliferators (n = 20, 40.8%) and nonproliferators (n = 18, 36.7%). The group of samples with anti-CD3–stimulated proliferation between 5% and 50% (n = 11, 22.4%) was excluded from further analysis in order to focus on the most extreme phenotypes. In addition to cell proliferation, we measured the production of cytokines in the cell culture supernatants of each sample following anti-CD3 stimulation. We detected significantly increased concentrations of IFN-γ, IL-2, TNF-α, IL-6, IL-10, IL-5, IL-4, IL-17A, IL-17F, IL-9, IL-13, and IL-22 in proliferators compared to nonproliferators (Fig. 1C).
Whether T cells residing in the AML bone marrow microenvironment are functionally and phenotypically different from those circulating in the blood is of great interest. Furthermore, knowing whether T cells in the blood are similar to those in the bone marrow would facilitate immune monitoring in clinical trials. Therefore, in addition to bone marrow-derived T cells, we examined peripheral blood T cells from a subset of our patient pool for which we received a matched peripheral blood sample at the same time as bone marrow. We hypothesized that the T cell proliferation that we observed in the bone marrow would be evident in the peripheral blood. The matched blood samples (n = 22) represented 12 proliferators and 10 nonproliferators, as defined by assay results with their corresponding bone marrow. Ten of the 12 samples we identified as proliferators from the bone marrow still met that criteria in the blood, while 2 had decreased T cell proliferation (88.8 to 35.2% and 72.1 to 22.3% T cell division) (Fig. 1 D, Left). Of the 10 blood samples that we identified as bone marrow nonproliferators, 2 showed marked proliferation by blood-derived T cells (0.1 to 57.3% and 1.1 to 38.1% T cell division) (Fig. 1 D, Right). As a result, although we did observe differences in proliferation status in a few matched samples, we did not find significant or consistent functional differences between blood and bone marrow resident T cells.
T Cell Suppression Is Associated with an Increased Naïve to Effector Memory T Cell Ratio.
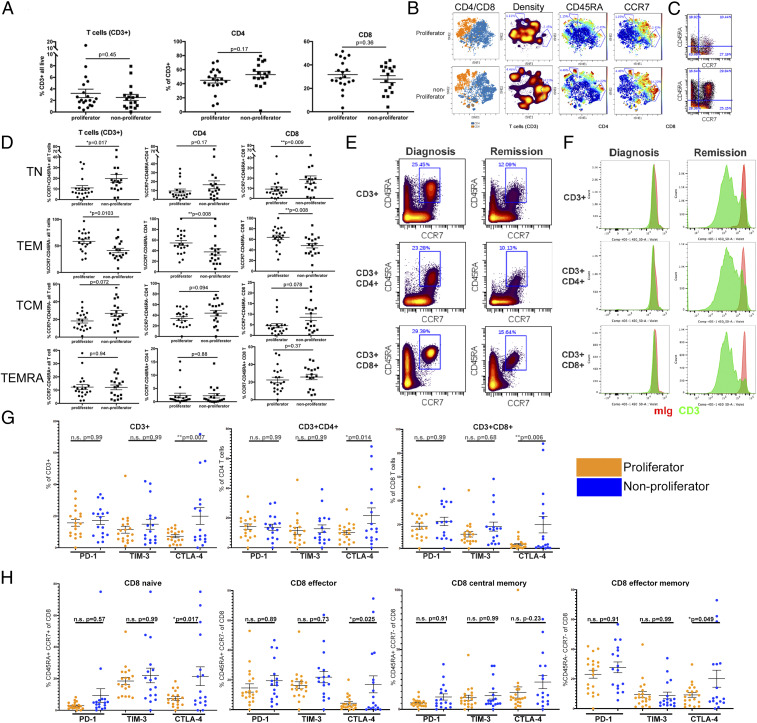
Along with determining the T cell functional status of our cohort, we performed mass spectrometry-based flow cytometry (CyTOF) to identify phenotypic parameters associated with each group. We quantified the major subsets of T cells for the samples in this cohort. We found no association between proliferation status and the percentages of total T cells (CD45+ CD3+) or CD4+ or CD8+ T cell subsets in AML bone marrow (Fig. 2A).
Fig. 2.
Impaired T cell proliferation is associated with an increase in naïve CCR7+ CD45RA+ T cell phenotype. (A) Frequency of T cells of all live cells (Left) and frequency of CD4+ (Center) or CD8+ (Right) of T cells. (B) Concatenated files representing all samples in the proliferator group (Upper) or nonproliferator group (Lower) projected in tSNE after gating on CD3+ cells. Distribution of CD4+ and CD8+ T cell subsets is shown in blue and orange, respectively (first column). Density plots show regions with reduced representation in the proliferator samples (second column). Intensity of CD45RA and CCR7 are mapped to the projections (third and fourth columns). (C) Representative plots of CCR7 vs. CD45RA staining from a sample in each group. (D) Percentages of T cell subpopulations gated on all T cells (Left column), CD4+ T cells (Center column), or CD8+ T cells (Right column). Differences in phenotypic distribution of TN (CCR7+ CD45RA+, Upper), TEM (CCR7− CD45RA−, second row), TCM (CCR7+ CD45RA−, third row), and TEMRA (CCR7− CD45RA+, Lower) T cell subsets. (E) T cell phenotype in an individual patient at diagnosis and after chemotherapy-induced remission. Frequency of CCR7+ CD45RA+ for indicated T cell subsets at the time of diagnosis (Left column) and remission (Right column). (F) CTV dilution from sample in E in response to incubation with mIgG + mIgG (red histogram) or anti-CD3 + mIgG (green histogram). (G) Frequency of T cells positive for PD-1, TIM-3, and CTLA-4 as a percentage of all CD3+ (Left), and CD4+ (Center) or CD8+ (Right) T cells, comparing proliferators (orange) and nonproliferators (blue). (H) Percentage of cells positive for PD-1, TIM-3, and CTLA-4 on CD8+ TN, TEM, TCM, and TEMRA subsets as defined in D. Bars represent mean ± SEM. Statistical tests are Student’s t test for A and D, one-way ANOVA with multiple comparisons of groups shown. Bonferroni correction for multiple testing was used.
In order to compare and identify differences in T cells classified as proliferators vs. nonproliferators, we concatenated the flow cytometry files and gated on the CD45+ CD3+ population. After down-sampling to 10,000 events per projection, we displayed the data using t-distributed stochastic neighbor-embedding (tSNE) based on all surface markers except gating parameters (Fig. 2B). We first color-coded the data based on CD4+ and CD8+ populations to confirm that our tSNE projections clearly resolved these major populations. When the T cells were projected based on event density, several populations emerged that were found to be greater in the nonproliferator group compared to the proliferator group (gated regions).
We then analyzed the distribution of CD45RA and CCR7 in order to identify the differentiation status of the T cells present in these AML samples (30, 31). We found that nonproliferator samples displayed an enrichment of T cells in the space corresponding to both CD45RA and CCR7 expression. We next analyzed the expression of CCR7 and CD45RA in each individual sample. We present a representative dot plot of CCR7 vs. CD45RA staining from the proliferator (Fig. 2 C, Upper) or nonproliferator Fig. 2 C, Lower) group. The T cell status of individual samples based on CCR7 and CD45RA expression is summarized in Fig. 2D. As expected from the tSNE analysis, we observed an enrichment in T cells expressing high levels of CCR7 and CD45RA, particularly in the CD8+ subset, in those samples that failed to proliferate compared to the samples that did. This increase in CCR7+ CD45RA+ “naïve” cells (TN) was at the expense of the CCR7− CD45RA− “effector memory” population (TEM). We did not observe statistically significant differences in the CCR7+ CD45RA− (TCM) or CCR7− CD45RA+ (TEMRA) populations residing in AML bone marrow.
In a single patient, we compared the disease status, phenotype and proliferative capacity of bone marrow T cells at the time of diagnosis and at remission following induction chemotherapy. The patient was diagnosed with normal karyotype AML containing mutations in FLT3 (non-ITD), DNMT3A, NPM1, NRAS, and TET2 according to targeted sequencing. At the time of diagnosis, the patient had a white blood cell (WBC) count of 116 (×103 cells/µL) with 51% blasts in the marrow. CCR7+ CD45RA+ cells accounted for 25% of total T cells, and 23% and 29% of CD4+ and CD8+ T cell subsets, respectively (Fig. 2E). After attaining remission following induction chemotherapy (∼3-wk postdiagnosis), the blast count was reduced to 1.5% and the patient tested negative for all of the previously detected mutations except DNMT3A. In this sample, the percentage of CCR7+ CD45RA+ cells was reduced to roughly half of the diagnostic sample. Functionally, T cell proliferation was impaired in response to anti-CD3 at the time of diagnosis (Fig. 2F, green histogram anti-CD3, red histogram mIgG control). At the time of remission, however, proliferative ability of both CD4+ and CD8+ T cell subsets was restored.
The impaired T cell responses that we observed in the nonproliferator group may be due to high levels of checkpoint molecule or ligand expession on the T cells or tumor. To test this possibility, we compared the positivity of PD-1, TIM-3, and CTLA-4 on T cells in the proliferator (Fig. 2G, orange) vs. the nonproliferator (Fig. 2G, blue) groups. We first examined these molecules on total T cells (CD3+), CD4+, and CD8+ subsets (Fig. 2G). We found that there was no difference in the percentage of T cells expressing PD-1 or TIM-3. Interestingly, we did observe an enrichment in the percentage of CTLA-4+ cells in all T cells, as well as CD4+ and CD8+ subsets. When we analyzed the CD8+ TN, TEM, TCM, and TEMRA subsets, we detected a similar result as in the total CD8+ T cells (Fig. 2H). We found the lowest percentages of T cells expressing PD-1, TIM-3, and CTLA-4 in the TN and TCM populations and higher percentages in the TEM and TEMRA, as expected (32, 33).
Association of T Cell Proliferative Capacity with Clinical Parameters.
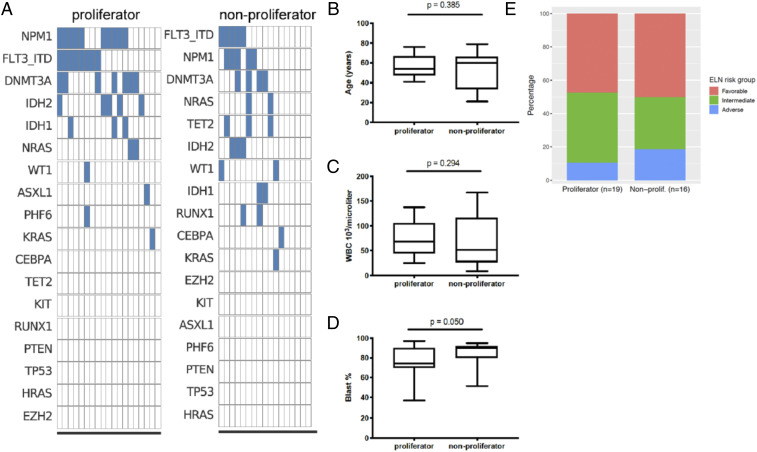
AML is a heterogeneous disease with a large diversity of mutations within each patient. We accessed whole-exome sequencing data from Vizome (http://www.vizome.org) to obtain the mutation data for the patient cohort that was analyzed for T cell function. A comparison of proliferators and nonproliferators (n = 19 and n = 17, respectively; data unavailable for one sample from each functional group) is displayed in Fig. 3A, with individual samples in each column and mutations ranked in order of prevalence. There was no observable difference in mutation profiles between the proliferators and nonproliferators. There was also no association between proliferation group and having a FLT3-ITD or NPM1 frameshift, the most common mutations in AML (SI Appendix, Table S1).
Fig. 3.
Comparison of clinical characteristics of proliferator and nonproliferator samples. (A) Mutations from whole exome sequencing, ranked in order of prevalence. (B–D) Box-and-whisker plots denoting medians and interquartile ranges of (B) age of patient at time of sample collection, (C) WBC count in peripheral blood and (D) blast percentage in the bone marrow. P values are from the Kruskal–Wallis test. (E) ELN risk group distribution.
To determine if other clinical or genetic parameters correlated with functional differences of bone marrow T cells from AML patients, we examined patient age (Fig. 3B), WBC count in peripheral blood (Fig. 3C), blast percentage in the bone marrow (Fig. 3D), European LeukemiaNet (ELN) prognostic risk group (Fig. 3E), and other features listed in SI Appendix, Table S1. We found that nonproliferators had a higher blast percentage (P = 0.050). However, “adverse” ELN risk (a group that includes complex karyotypes) did not correlate with anti-CD3 T cell proliferation status. Thus, despite prior findings that p53 mutations and complex karyotypes correlate with increased immune checkpoint expression, the T cell impairment we observed in our nonproliferator group could not be linked to abnormal cytogenetics (29).
Suppression of T Cell Proliferation through Immune Checkpoint Molecules.
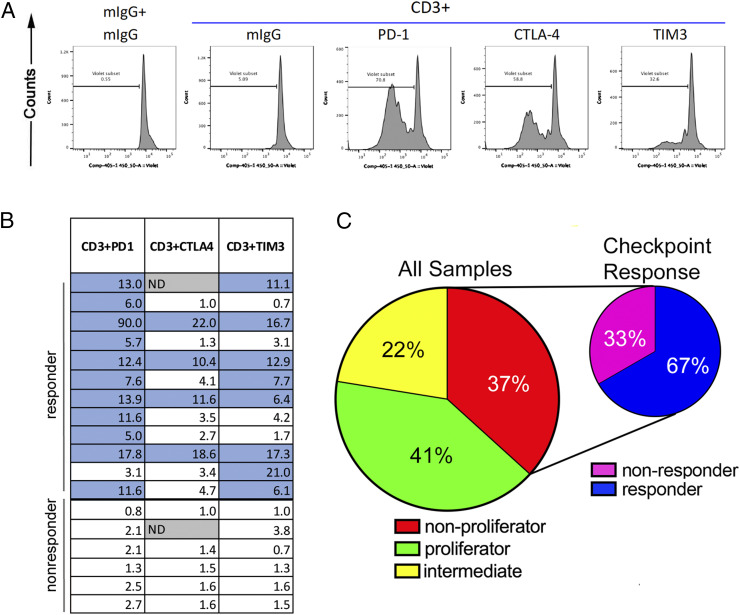
In an effort to understand the mechanisms underlying T cell suppression in AML samples, we added blocking antibodies against the checkpoint molecules PD-1, CTLA-4, and TIM3 to the proliferation assays. Results from these experiments revealed that in a majority of samples where anti-CD3 + mIgG did not induce T cell proliferation, anti-CD3 in combination with checkpoint blockade did effect some T cell division. Fig. 4A shows one sample in which T cells were nonresponsive to anti-CD3 + mIgG treatment but proliferated, to varying degrees, with the addition of a CPI. The response of T cells to CPI for all 18 nonproliferator samples is summarized in Fig. 4B. Numbers within boxes (Fig. 4B) represent the fold-change in proliferation, as determined by the proliferation measured with anti-CD3 and PD-1, CTLA-4, or TIM3 antibody treatment divided by the proliferation with anti-CD3 + mIgG (see SI Appendix, Table S2 for absolute proliferation data for all samples). Nonproliferators were considered “checkpoint responders” if proliferation increased five-fold over anti-CD3 + mIgG with the addition of a specific CPI (12 of 18, 67%, indicated in blue in SI Appendix, Table S2). We found no association between CPI response and age, blast percentage in the marrow, or WBC count (SI Appendix, Fig. S1).
Fig. 4.
Impaired T cell proliferation in response to TCR ligation can be reversed by immune checkpoint blockade in a subset of patients. (A) A representative “checkpoint responder” showing CTV dilution with the addition of antibodies against PD-1, CTLA-4, and TIM3. (B) A summary of the responses of all nonproliferators to checkpoint molecule blockade. Numbers within boxes represent the fold-change in proliferation, as determined by the proliferation measured with anti-CD3 and PD-1, CTLA-4, or TIM3 antibody treatment divided by the proliferation with anti-CD3 + mIgG. Blue-shaded boxes indicate checkpoint responders, defined as samples with at least a five-fold increase in proliferation value when comparing anti-CD3 + checkpoint antibody to anti-CD3 + mIgG. (C) Summary of results from the the patient cohort assayed for T cell function (n = 49).
To summarize the results from our studies on the T cell proliferative response to TCR stimulation in AML bone marrow (Fig. 4C), we found that 41% of samples exhibited T cell proliferation similar to healthy donors, while 37% showed nearly complete inhibition of T cell proliferation (i.e., division in <5% of T cells). The remaining samples (22%) displayed intermediate proliferation. Importantly, within the group of 18 nonproliferators, checkpoint blockade was able to induce T cell proliferation in 12 (67%) samples, with PD-1 blockade being the most effective.
Proliferation and Activation of T Cells Results in Secondary Immune Checkpoint Induction.
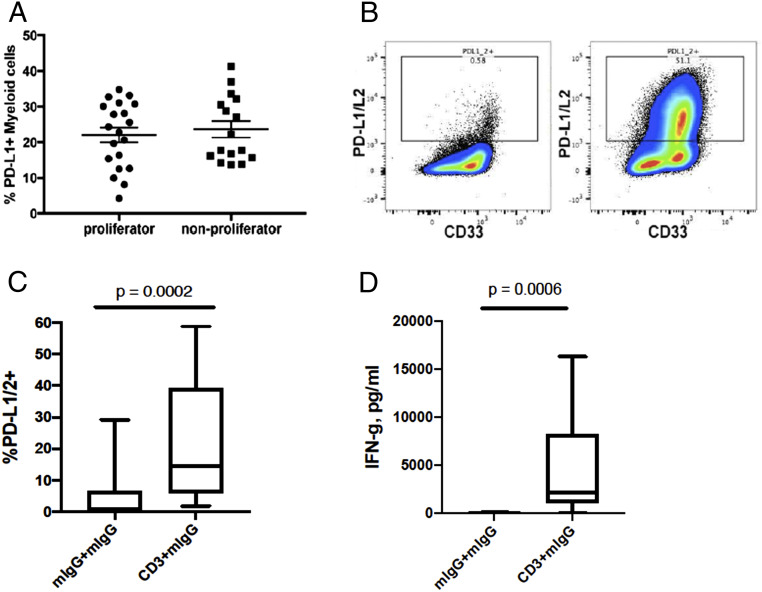
The presence of ligands that engage immune checkpoint molecules has been associated with clinical response to CPIs in some, but not all, tumor types (34). We measured the expression of PD-L1 by CyTOF and compared the levels in proliferator vs. nonproliferator AML samples. We found no association between PD-L1 expression on CD33+ cells and T cell response to anti-CD3 stimulation (Fig. 5A). We also examined expression of CD80, CD86, and VISTA on myeloid cells and found no statistically significant association between the percentage of positive myeloid cells and proliferation status (SI Appendix, Fig. S2). One possible explanation for how patients are able to respond to CPI treatment without detectable staining of checkpoint ligands is that those ligands are up-regulated secondary to immune activation. Both PD-L1 and PD-L2 are induced on cells upon exposure to type 1 and type 2 interferons (35). Therefore, these ligands may be expressed at very low levels if tumor-induced T cell suppression is present. We measured the expression levels of PD-1 ligands in cells from the proliferation assay to assess the ability of non-T cells to up-regulate these ligands in response to T cell proliferation and cytokine production. In general, expression levels of PD-1 ligands were low in cultures with control mIgG, which does not induce T cells to proliferate (Fig. 5 B and C). In contrast, PD-1 ligand expression was up-regulated in the majority of cells in samples whose T cells proliferated after CD3 ligation. We found a strong association between the induction of PD-1 ligands and T cell receptor (TCR) stimulation in our 20 proliferator samples (Fig. 5C). As expected, there was also a highly significant correlation between IFN-γ levels and TCR stimulation in these same samples (Fig. 5D).
Fig. 5.
Induction of checkpoint molecules in response to T cell proliferation and IFN-γ production. (A) PD-L1 expression on myeloid cells (CD33+) from the bone marrow of patients with AML. (B) Representative PD-1 ligand staining of a proliferator sample after 5 d of culture with either mIgG + mIgG (Left) or anti-CD3 + mIgG (Right). (C and D) Box-and-whisker plots denoting medians and interquartile ranges of (C) frequency of PD-1 ligand expressing cells and (D) IFN-γ from the culture supernatants of proliferator samples, with or without TCR stimulation. P values for C and D are from two-tailed paired t test.
We observed that CTLA-4, but not PD1 or TIM3, expression appeared to be increased on T cells from the nonproliferator group compared to proliferator (Fig. 2G). However, changes in expression of immune checkpoint molecules on the T cells during the proliferation assay culture period could contribute to impaired T cell proliferation. Therefore, we assessed expression levels of CTLA-4, PD1, and TIM3 on T cells following the proliferation assay culture. We found that after 5 d in culture with anti-CD3 (but not control mIgG) T cells in the proliferator group expressed PD-1, TIM-3, and CTLA-4, in both the divided (CTV diluted) and undivided (parental, CTV bright) populations (SI Appendix, Fig. S3). In contrast, we did not observe expression of PD-1, TIM-3, or CTLA-4 in the mIgG- or CD3-treated samples in the nonproliferator group after 5 d of culture. It is likely that the induced expression of checkpoint molecules on T cells in the proliferator group is due to the culture environment since these can be up-regulated by both TCR engagement and cytokine exposure (36–39).
AML Overall Survival Is Associated with Bone Marrow T Cell Percentage at the Time of Diagnosis.
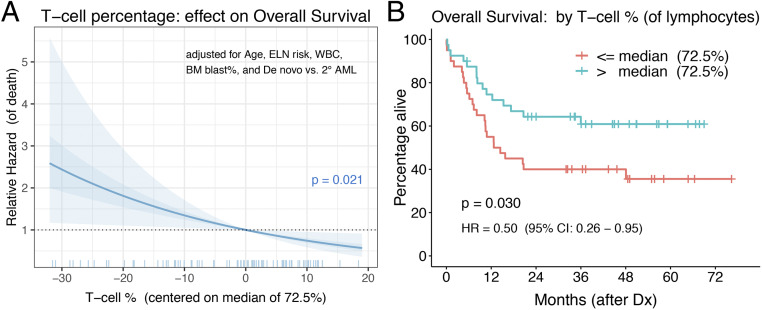
Strong associations between T cell infiltration and clinical outcome have been observed in various tumor types (8–13). A study from Egypt involving 66 de novo AML patients reported a survival benefit for those with a higher T cell percentage (defined as ≥78.5% [the sample mean] of lymphocytes) in their bone marrow (15). We sought to validate the prognostic impact of T cell abundance by analyzing a larger North American cohort of 80 adults diagnosed with AML (76 de novo, 4 secondary AML) at Oregon Health & Science University (OHSU) between 2010 and 2016 (See SI Appendix, Table S3 for cohort details). This is a separate patient cohort from the “functional cohort” analyzed above for T cell proliferation, but includes five overlapping patient samples. After adjusting for prognostic factors (i.e., patient age, ELN risk group, WBC count, and blast percentage in the marrow, and type of AML [de novo vs. secondary]), we found a statistically significant association between bone marrow T cell percentage (of lymphocytes) at the time of AML diagnosis and subsequent overall survival (P = 0.021, hazard ratio [HR] = 0.86 [95% CI: 0.76 to 0.98] for each five-point increase in T cell percentage) (Fig. 6A). This significant correlation between a higher T cell percentage and longer survival was maintained when removing the three acute promyelocytic leukemia patients (n = 77, P = 0.016, HR = 0.86 [95% CI: 0.75 to 0.97] for each five-point increase) and when limiting analysis to the 69 patients who received standard induction chemotherapy (P = 0.028, HR = 0.85 [95% CI: 0.74 to 0.98] for each five-point increase). When dichotomizing T cell percentage using the sample median of 72.5% (range was 41.0 to 90.9%), the “high” T cell group had a significant survival advantage over the “low” T cell group (P = 0.030, HR = 0.50 [95% CI: 0.26 to 0.95]) (Fig. 6B and SI Appendix, Table S3). Upon controlling for the five prognostic factors mentioned above, the high T cell group had a 41% reduced risk of death compared to the low group that did not reach statistical significance (P = 0.128, HR = 0.59 [95% CI: 0.30 to 1.17]).
Fig. 6.
Association between T cell relative abundance and overall survival. The T cell percentage of bone marrow lymphocytes at the time of AML diagnosis was evaluated for its association with overall survival. (A) Relative hazard of death (i.e., HR of T cell percentages compared to the median value of 72.5%, indicated as 0 on the x axis) across the range of observed T cell percent, as estimated from a Cox regression model that includes patient age, ELN risk group, bone marrow WBC count, bone marrow blast percent, and AML type as covariates. The T cell percent (as a continuous measure) P value of 0.021 is from a Wald test. The shaded area denotes the simulation-derived 95% CI for the relative hazard (40). (B) Kaplan–Meier survival curves for “low” (red) and “high” (blue) T cell percentage groups, defined by dichotomizing on the sample median of 72.5%. The P value of 0.030 is from the log-rank (or score) test and HR of 0.50 is estimated from a univariable Cox model.
Discussion
In this study, our goal was to investigate the status and impact of bone marrow-resident T cells in AML. From previously published literature, it is clear that T cells are critical for both achieving and maintaining remissions in leukemia (41–44). T cells from AML bone marrow have global differences in transcription profiles compared to T cells from healthy donors (24). Changes have been identified in genes associated with cytoskeleton formation, resulting in impaired immune synapse formation and reduced T cell activation (18). Patients in complete remission from AML have been reported to have reduced CD4+ T cells, while proliferative function remained normal (19). Immune checkpoint-mediated failure of T cells to function, T cell exhaustion, and loss of MHC expression have been identified in patients relapsing after transplant (45–47). T cells have also been shown to be critical for the function of FLT3 inhibitors after relapse in both animal models and patients (48). Because the TME postchemotherapy, at relapse, or following allogeneic transplant is different from the TME prior to transplant, we sought to better understand whether immune evasion is present in the leukemic bone marrow niche prior to clinical treatment and if so, whether immune function and proliferation could be rescued through checkpoint blockade as in other tumor types. We identified a range of responses, from undetectable to extensive T cell proliferation and cytokine production in response to stimulation with 1 µg/mL anti-CD3. Schnorfeil et al. (21) concluded that T cells are functionally unimpaired at diagnosis or relapse. There are several possible explanations for these conflicting results. First, we chose to direct our studies on fresh noncryopreserved, whole bone marrow samples to preserve the complex TME and more closely represent in vivo conditions, while Schnorfeil et al. isolated T cells for their functional assays. Their functional assessments used stimulation of peripheral blood mononuclear cells (PBMCs) with PMA and ionomycin, which circumvents the TCR complex, or anti-CD3 and anti-CD28, both stronger stimuli than what was used in our study. Even with the stronger stimulation, which might overcome mild immune suppression, it was reported that the T cells produced less IFN-γ, which is consistent with our results. In addition, differences in the function of T cells from the bone marrow and blood have been reported and may contribute to some of the differences observed in our results (27). A study using a defined 10:1 blast/T cell ratio and stimulation with both anti-CD3 and anti-CD28 also found impaired T cell proliferation when compared to T cells from healthy donors. Blasts were able to suppress healthy donor T cells to a lesser extent than AML patient T cells, which suggests that T cells in the marrow of patients with AML might be functionally different from those in the marrow of healthy individuals. This suppression was reversible in six of nine samples by the addition of PD-1–blocking antibodies (23). We believe our data support what has been shown by these studies.
Fifty percent of T cells dividing with anti-CD3 stimulation was our cutoff for a sample to be categorized as a proliferator. This was based on the lowest proliferation rate that we observed in our eight healthy donor samples. Our source for control healthy samples was bone marrow cells that had been collected from donors for allogeneic transplants. As such, the mean age of the healthy donors is lower in comparison to the AML samples. Nonetheless, we do not believe that age alone is the responsible factor of T cell impairment in patients with AML for several reasons: 1) Of the seven youngest patients (in their 20s and 30s) in our cohort, six were nonproliferators, one was intermediate, and none were proliferator; 2) the oldest healthy donor (58 y of age) was highly proliferative (88% T cell proliferation); and 3) there was no association between age of patient and proliferation status, with the median age of proliferators and nonproliferators being 54 and 60 y of age, respectively (SI Appendix, Table. S1).
We observed an association of the CCR7+ CD45RA+ T cell phenotype with samples showing suppressed T cell proliferation. This phenotype of T cells is described as naïve in the blood of healthy individuals (30). In AML bone marrow, this naïve cell population increased at the expense of the effector memory population (CCR7− CD45RA−) in the nonproliferator group (Fig. 2D). In some samples that displayed suppressed proliferation, the CCR7+ CD45RA+ population was greater than 80% of all T cells. It was surprising to find such high percentages of naïve T cells in the marrow of patients with AML. In healthy people, naïve T cell subsets decline with age; the population of CD4+ naïve T cells decreases from comprising ∼50% of the T cell population in people in their 20s to below 40% in people in their early 70s. There is a more dramatic drop for CD8+ naïve T cells across this same age span, falling from 50% to below 20% (49). As a consequence, it is possible that the cells displaying this phenotype in AML samples are not typical naïve T cell subsets. In support of this hypothesis, one study has described a CCR7+ CD45RA+ autoreactive Melan-A-specific CD8+ T cell population in healthy people that were suppressed, via regulatory T cells, in their ability to both proliferate and produce cytokines in response to TCR ligation (50). Other studies have also associated the naïve marker of CR45RA with anergic T cells (51). Further studies are needed to better understand the dynamics of T cell populations in the marrow of patients with AML.
Interestingly, we identified a patient whose T cells were unable to proliferate in response to TCR stimulation at the time of diagnosis but whose immunosuppression was reversed upon chemotherapy-induced tumor regression, demonstrating that normal immune function can be restored after the tumor burden is removed (Fig. 2F). This recovery corresponded with a decrease in the CCR7+ CD45RA+ population from diagnosis to remission (CD4+: 23.3 to 10.1%, CD8+: 29.3 to 15.6%) (Fig. 2E). These observations support the notion that the T cell suppression observed in some AML patients can be attributed to the presence of the tumor.
As with AML in general, the samples in our two cohorts were genetically heterogeneous (Fig. 3 and SI Appendix, Table S1) (52, 53). Available mutational information of the proliferator and nonproliferator groups did not reveal a clear association between the degree of immune suppression observed in our assays and common driver mutations, such as FLT3-ITD or NPM1. Our findings corroborate recent publications that describe an association, in AML samples, between CD8+ T cell function and the expression of genes involved in myeloid cell and leukocyte activation, immune regulation, and response to IFN-γ (23). Further studies are needed to better understand the functional differences in these pathways between subjects with highly proliferative vs. nonproliferative T cells. For example, hypoxia signaling promotes a protumorigenic microenvironment for many cancers, including AML, and leads to therapeutic resistance (54, 55). Changes in the expression of genes involved in this pathway suggest that patients with AML may have dysregulation in the tumor immune environment in addition to variation in T cell proliferation and function. This highlights the need to develop therapeutic strategies, such as CPIs, that target the immune environment to enhance and restimulate immune activation in patients with AML.
One important consideration beyond the scope of these studies is the question of antigen specificity. Our assay was designed to measure the effect of the immune environment on T cell responses downstream of TCR ligation. Response to CPI has been associated with diseases bearing a high mutational burden, including tumors with microsatellite instability (56–58). AML is a disease of relatively low mutational burden compared to other cancer types (59). Nonetheless, there are common mutations, such as the NPM1 frameshifts, that form functional CD8 epitopes (60). We (and others) are currently interrogating the immunogenic epitope repertoire in large cohorts of AML patients to identify other potential T cell targets. In addition to mutated antigens, T cell responses with specificity against tumor-associated antigens, such as WT1, RAGE, PRAME, and others have been detected (61). Despite the high percentage of patients that display expression and recognition of tumor-associated antigens, the occurrence of disease progression suggests the presence of antigen-specific immune suppression. Our results demonstrate that T cells are nonfunctional in a subset of patients and that, in some cases, one mechanism is through immune checkpoint molecules, many of which have been shown to be expressed by AML T cells (23, 27–29, 62–65). These findings may explain both the progression of disease in the presence of tumor-associated antigen responses and the poor clinical responses to AML vaccines to date (66). Recently, LILRB4, expressed on AML blasts of the monocytic (M4 and M5) subtypes, was found to regulate T cell proliferation (67). The inhibition caused by LILRB4+ AML cells was shown to be due to increased arginase function. Such findings indicate that the T cell suppressive effect of AML is complex and likely involves multiple mechanisms that include checkpoint molecules and other inhibitory enzymes, such as arginase and IDO/TDO (6).
There is currently great clinical interest in immune-based therapies for AML, with over 30 clinical trials that are testing CPIs for AML and myelodysplastic syndrome. The majority of these trials target PD-1, but CTLA-4 and TIM-3 blockade are also being evaluated (6). In the largest trial to date, Daver et al. (68, 69) showed encouraging results for patients with relapsed or refractory AML that were treated with combined anti-PD1 blockade (nivolumab) and a hypomethylating agent. Of the 70 patients that were treated, there was an overall response rate of 33% (n = 23), with 15 of these patients achieving either a complete remission or a complete remission with insufficient recovery of counts. Potential biomarkers of response were increased percentages of CD3+ and CD8+ T cells in the pretherapy bone marrow. Conversely, increased expression of CTLA-4 on effector CD4+ and CD8+ T cells was a predictor of a lack of response (69). Interestingly, in our studies we found an association between lack of T cell function and higher percentages of CTLA-4+ T cells (Fig. 2 G and H). While a consistently predictive measure has not been identified, these data suggest that there may a subpopulation of patients who may respond to CPIs and that pretherapy bone marrow could be a window into identifying these patients.
The most appropriate timing of administering CPIs in the clinical setting is unclear. Immune escape has been hypothesized as a contributor to the development of de novo disease and as a mechanism for relapse (70). Based on this assumption, there may be a role for CPIs during multiple phases of treatment. This is highlighted by the case discussed above (Fig. 2 E and F), where the patient had different degrees of immune dysfunction during different treatment timepoints. For CPIs to be effective, it is likely that a patient must have an adequate lymphocyte count, which may not be the case following a round of high-dose chemotherapy. Alternatively, the use of CPIs in the posttransplant setting is very enticing given their potential for enhancing the graft-vs.-leukemia effect. This potential for increased efficacy must be balanced with the risk of exacerbating graft-vs.-host disease, which can be fatal (71). Davids et al. (72) reported on 12 patients with AML who were treated with ipilimumab following an allogeneic stem cell transplant. Five of these patients achieved a complete response, with four having extramedullary disease, including three with leukemia cutis and one with myeloid sarcoma. Predictors of response were associated with in situ infiltration of CD8+ T cells, as well as enrichment of effector T cell subsets. This again suggests that, while only a minority of patients to date has shown benefit from CPIs, microenvironment features may help to identify those that may respond and allow clinicians to tailor immune-based therapies to the patient.
In summary, we have phenotypically and functionally characterized the bone marrow-resident T cells in patients with newly diagnosed AML. In a cohort of 80 patients, we found increased overall survival in those who displayed a higher percentage of T cells in their bone marrow, as has been described in other cancers (8–12, 14, 73). Furthermore, we have identified T cell suppression in a significant subset of patients with AML. This functional impairment appears to associate with blast count in the bone marrow but not with age, WBC count, ELN risk, or with the most common mutations found in AML. Impaired T cell proliferation was associated with an increase in T cell populations bearing a naïve phenotype. Remarkably, in the majority of samples that exhibited immunosuppression, some proliferative capacity was restored by blockade of one or more immune checkpoints. While expression of PD-1 ligands was low on cells analyzed prior to culturing to measure functional capacity, these were highly up-regulated in samples that displayed T cell proliferation. The findings from our study contribute to the growing body of literature regarding the use of CPIs in patients with hematologic malignancies and predictive models for efficacy (74, 75).
Methods
Specimens for T Cell Function Studies.
Bone marrow and peripheral blood samples from patients with newly diagnosed AML were obtained as part of the Beat AML study, approved by the Institutional Review Board (IRB) at OHSU (Protocol #4422) (76). Informed consent was obtained from all subjects in this study. Clinical annotations for individual samples (age, gender, WBC counts from corresponding peripheral blood, blast percent of bone marrow cells, mutation data) were retrieved from the BeatAML (lls scor) database from our AML cohort [as previously reported (76) and available at http://www.vizome.org] and through electronic medical records of the patient. Healthy donor bone marrow cells were de-identified and obtained by collecting residual cells from clinical transplant bags, considered medical waste.
T Cell Proliferation Assays and Cytokine Measurements.
Bone marrow aspirates and peripheral blood samples were subjected to Ficoll density gradient centrifugation to isolate BMMC or PBMC. All experiments were performed using freshly isolated cells. Total BMMC or PBMC were labeled with CTV (ThermoFisher) and cultured in 96-well plates coated with 1 µg/mL anti-CD3 (UCHT1) or control mIgG (MOPC-21). (All antibodies were purchased from BioLegend, unless noted otherwise.) Groups of wells also contained one of the following soluble antibodies at 10 μg/mL: Anti-CTLA-4 (L3D10), anti-PD-1 (EH12.2H7), anti-TIM3 (344823, R&D Systems), or control mIgG (MOPC-21). After 5 d, cells were stained with antibodies specific to CD45 (HI30), CD3 (HIT3a), CD4 (RPA-T4), CD8 (RPA-T8), CD33 (WM53), CD38 (HIT2), PD-L1 (M1H1, Becton Dickinson), PD-L2 (24F.10C12), PD-1 (MIH4, BD), TIM3 (7D3, BD), and CTLA-4 (BNI3, BD). Viability was determined by Zombie Aqua (BioLegend) staining and doublets were gated out of analysis by FSC-A vs. FSC-H. T cell proliferation was quantified based on the percentage of T cells (defined as CD3+ and single-positive for CD4 and CD8) that underwent at least one division from the parent population: Proliferation value (percent divided) = the percentage of T cells diluting CTV with anti-CD3 + mIgG minus the percentage of T cells diluting CTV with control mIgG + mIgG. Flow cytometry data were acquired on a BD LSRFortessa and analyzed using FlowJo v10 software.
Supernatants from the proliferation assay cultures were retained for cytokine analysis. Levels of IL-2, IL-4, IL-5, IL-6, IL-9, IL-10, IL-13, IL-17A, IL-17F, IL-21, IL-22, IFN-γ, and TNF-α were measured by multiplex bead-based assay (LEGENDplex, BioLegend).
Immunophenotyping by CyTOF.
Samples were stained for surface and intracellular molecules following protocols from Fluidigm, as detailed in SI Appendix, Supplemental Materials and Methods and as published in Lamble et al. (77), and analyzed on a CyTOF 1 instrument. Two separate panels of antibodies were used for phenotyping (SI Appendix, Table S4). All analyses were carried out using Cytobank.
Clinical Outcome Association with Bone Marrow T Cell Abundance.
Adult newly diagnosed AML patients (including three patients with antecedent hematological disorder and one with therapy-related AML) at OHSU, diagnosed between September 2010 and March 2016, with a bone marrow biopsy examined by the clinical flow cytometry laboratory at OHSU were analyzed (IRB Protocol #2305). The bone marrow samples of all included patients were immunophenotyped with multicolor flow cytometry and run on a targeted next-generation sequencing panel. Bone marrow cell estimates included WBC counts, blast percentages, and lymphocyte percentages. Subgroups of CD3+ T cells were defined by CD4 and CD8 expression to identify helper and cytotoxic T cells, respectively. T cell numbers (cells per microliter) were estimated using each group’s percentage of total lymphocytes and the size of the lymphocyte population (defined by CD45 expression and minimal side scatter). Besides marrow cell percentages and absolute numbers (per microliter), patient and sample characteristics that were considered as potential prognostic factors included AML type (de novo vs. secondary), gender, age, mutation calls (and allele frequencies) from the next-generation sequencing panel, and cytogenetic abnormalities as detected from standard karyotyping and FISH. The genomic and cytogenetic data were used to classify patients into risk groups (i.e., favorable, intermediate, adverse) according to the 2017 ELN recommendations (78). Overall survival was measured from the AML diagnosis date to the date of death or last contact and was estimated using the Kaplan–Meier method. Transplant status was considered as a time-varying covariate but was found to be unrelated to the risk of death. The association between patient characteristics and overall survival was evaluated with nonparametric log-rank tests (for categorical variables) and semiparametric Cox regression after checking the proportional hazards assumption by inspecting scaled Schoenfeld residuals. Univariable Cox regression produced unadjusted HR while multivariable models were built for each immune cell population by considering as covariates those patient and disease features that 1) correlated with survival time in the univariable setting or 2) are known to impact AML prognosis. Thus, HRs from these multivariable models quantify the relative risk of death for specific immune cell (e.g., CD3+ T cell) levels in the marrow when controlling for other prognostic factors.
Data Availability.
The data and protocols supporting the findings of this study are in the main text and SI Appendix. Other associated data are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
The authors thank all the patients whose generous donation of their bone marrow and blood made these studies possible, and the clinical and laboratory personnel who procured and processed the specimens used in our studies. This work was funded in part by generous support from the Leukemia and Lymphoma Society of America Beat AML project (Principal Investigators B.J.D. and J.W.T.). A.M. was supported by the Department of Pediatrics Biostatistics Pilot Grant, Oregon Health & Science University. J.W.T. received grants from the V Foundation for Cancer Research, the Gabrielle’s Angel Foundation for Cancer Research, and the National Cancer Institute (1R01CA183947, 1U01CA217862, 1U54CA224019). E.F.L. is supported by Grants U54CA224019 and U01CA217862 from the National Cancer Institute (Co-investigator). A.K. and M.M. are supported by NIH/National Cancer Institute Cancer Center Support Grant P30CA069533.
Footnotes
Competing interest statement: L.K.B., F.H., D.S., and H.A. are employees of Janssen Pharmaceuticals R&D, LLC. D.S. is currently an employee and holds stock in Genmab. C.V.L is an employee of Lab Connect LLC. J.N.S. receives research support form Kyn Therapeutics. B.J.D. has the following disclosures: Scientific Advisory Board for Aileron Therapeutics, ALLCRON, Cepheid, Vivid Biosciences, Celgene, RUNX1 Research Program, EnLiven Therapeutics, Gilead Sciences (inactive), Baxalta (inactive), Monojul (inactive); Scientific Advisory Board and Stock: Aptose Biosciences, Blueprint Medicines, Beta Cat, Iterion Therapeutics, Third Coast Therapeutics, GRAIL (inactive), CTI BioPharma (inactive); Scientific Founder: MolecularMD (inactive, acquired by ICON); Board of Directors and Stock: Amgen; Board of Directors: Burroughs Wellcome Fund, CureOne; Joint Steering Committee: Beat AML LLS; Founder: VB Therapeutics; Clinical Trial Funding: Novartis, Bristol-Myers Squibb, Pfizer; royalties from Patent 6958335 (Novartis exclusive license) and Oregon Health & Science University and Dana-Farber Cancer Institute (one Merck exclusive license). J.W.T. receives research support from Aptose, Array, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen, Petra, Seattle Genetics, Syros, and Takeda. J.W.T. is a cofounder of Leap Oncology. E.F.L. receives research support from Janssen Pharmaceuticals, Celgene Amgen, and Kyn Therapeutics.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916206117/-/DCSupplemental.
References
- 1.Deschler B., Lübbert M., Acute myeloid leukemia: Epidemiology and etiology. Cancer 107, 2099–2107 (2006). [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Institute Surveillance , Epidemiology, and End Results Program, Cancer Stat Facts: Leukemia — Acute Myeloid Leukemia (AML). https://seer.cancer.gov/statfacts/html/amyl.html. Accessed 26 May 2020.
- 3.Topalian S. L. et al., Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodi F. S. et al., Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Postow M. A., Callahan M. K., Wolchok J. D., Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 33, 1974–1982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamble A. J., Lind E. F., Targeting the immune microenvironment in acute myeloid leukemia: A focus on T cell immunity. Front. Oncol. 8, 213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D., Coussens L. M., Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Zhang L. et al., Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 348, 203–213 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Schumacher K., Haensch W., Röefzaad C., Schlag P. M., Prognostic significance of activated CD8(+) T cell infiltrations within esophageal carcinomas. Cancer Res. 61, 3932–3936 (2001). [PubMed] [Google Scholar]
- 10.Vesalainen S., Lipponen P., Talja M., Syrjänen K., Histological grade, perineural infiltration, tumour-infiltrating lymphocytes and apoptosis as determinants of long-term prognosis in prostatic adenocarcinoma. Eur. J. Cancer 30A, 1797–1803 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Roxburgh C. S. et al., The in situ local immune response, tumour senescence and proliferation in colorectal cancer. Br. J. Cancer 109, 2207–2216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakano O. et al., Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: Clinicopathologic demonstration of antitumor immunity. Cancer Res. 61, 5132–5136 (2001). [PubMed] [Google Scholar]
- 13.Governa V. et al., The interplay between neutrophils and CD8+ T cells improves survival in human colorectal cancer. Clin. Cancer Res. 23, 3847–3858 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Barnes T. A., Amir E., HYPE or HOPE: The prognostic value of infiltrating immune cells in cancer. Br. J. Cancer 118, e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ismail M. M., Abdulateef N. A. B., Bone marrow T-cell percentage: A novel prognostic indicator in acute myeloid leukemia. Int. J. Hematol. 105, 453–464 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Buggins A. G. et al., Microenvironment produced by acute myeloid leukemia cells prevents T cell activation and proliferation by inhibition of NF-kappaB, c-Myc, and pRb pathways. J. Immunol. 167, 6021–6030 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Kornblau S. M. et al., Recurrent expression signatures of cytokines and chemokines are present and are independently prognostic in acute myelogenous leukemia and myelodysplasia. Blood 116, 4251–4261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Dieu R. et al., Peripheral blood T cells in acute myeloid leukemia (AML) patients at diagnosis have abnormal phenotype and genotype and form defective immune synapses with AML blasts. Blood 114, 3909–3916 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lichtenegger F. S. et al., Impaired NK cells and increased T regulatory cell numbers during cytotoxic maintenance therapy in AML. Leuk. Res. 38, 964–969 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Orleans-Lindsay J. K., Barber L. D., Prentice H. G., Lowdell M. W., Acute myeloid leukaemia cells secrete a soluble factor that inhibits T and NK cell proliferation but not cytolytic function—Implications for the adoptive immunotherapy of leukaemia. Clin. Exp. Immunol. 126, 403–411 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnorfeil F. M. et al., T cells are functionally not impaired in AML: Increased PD-1 expression is only seen at time of relapse and correlates with a shift towards the memory T cell compartment. J. Hematol. Oncol. 8, 93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wendelbo Ø. et al., Functional characterization of T lymphocytes derived from patients with acute myelogenous leukemia and chemotherapy-induced leukopenia. Cancer Immunol. Immunother. 53, 740–747 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knaus H. A. et al., Signatures of CD8+ T cell dysfunction in AML patients and their reversibility with response to chemotherapy. JCI Insight 3, 120974 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Galen P. et al., Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mussai F. et al., Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood 122, 749–758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia B. et al., Eomes+T-betlow CD8+ T cells are functionally impaired and are associated with poor clinical outcome in patients with acute myeloid leukemia. Cancer Res. 79, 1635–1645 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Jia B. et al., Bone marrow CD8 T cells express high frequency of PD-1 and exhibit reduced anti-leukemia response in newly diagnosed AML patients. Blood Cancer J. 8, 34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C. et al., Tim-3 is highly expressed in T cells in acute myeloid leukemia and associated with clinicopathological prognostic stratification. Int. J. Clin. Exp. Pathol. 7, 6880–6888 (2014). [PMC free article] [PubMed] [Google Scholar]
- 29.Williams P. et al., The distribution of T-cell subsets and the expression of immune checkpoint receptors and ligands in patients with newly diagnosed and relapsed acute myeloid leukemia. Cancer 125, 1470–1481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sallusto F., Lenig D., Förster R., Lipp M., Lanzavecchia A., Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Sallusto F., Geginat J., Lanzavecchia A., Central memory and effector memory T cell subsets: Function, generation, and maintenance. Annu. Rev. Immunol. 22, 745–763 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Duraiswamy J. et al., Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults. J. Immunol. 186, 4200–4212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baitsch L. et al., Extended co-expression of inhibitory receptors by human CD8 T-cells depending on differentiation, antigen-specificity and anatomical localization. PLoS One 7, e30852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribas A., Hu-Lieskovan S., What does PD-L1 positive or negative mean? J. Exp. Med. 213, 2835–2840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Diaz A. et al., Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 19, 1189–1201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinter A. L. et al., The common gamma-chain cytokines IL-2, IL-7, IL-15, and IL-21 induce the expression of programmed death-1 and its ligands. J. Immunol. 181, 6738–6746 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Freeman G. J. et al., Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 192, 1027–1034 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latchman Y. et al., PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2, 261–268 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Chemnitz J. M., Parry R. V., Nichols K. E., June C. H., Riley J. L., SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol. 173, 945–954 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Gandrud C., simPH: An R Package for Illustrating Estimates from Cox Proportional Hazard Models Including for Interactive and Nonlinear Effects. Journal of Statistical Software 65 (3), 1–20 (2015). [Google Scholar]
- 41.Marmont A. M. et al., T-cell depletion of HLA-identical transplants in leukemia. Blood 78, 2120–2130 (1991). [PubMed] [Google Scholar]
- 42.Horowitz M. M. et al., Graft-versus-leukemia reactions after bone marrow transplantation. Blood 75, 555–562 (1990). [PubMed] [Google Scholar]
- 43.Ruggeri L. et al., Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 295, 2097–2100 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Ruggeri L. et al., Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood 94, 333–339 (1999). [PubMed] [Google Scholar]
- 45.Toffalori C. et al., Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat. Med. 25, 603–611 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Noviello M. et al., Bone marrow central memory and memory stem T-cell exhaustion in AML patients relapsing after HSCT. Nat. Commun. 10, 1065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jan M. et al., Recurrent genetic HLA loss in AML relapsed after matched unrelated allogeneic hematopoietic cell transplantation. Blood Adv. 3, 2199–2204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathew N. R. et al., Sorafenib promotes graft-versus-leukemia activity in mice and humans through IL-15 production in FLT3-ITD-mutant leukemia cells. Nat. Med. 24, 282–291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czesnikiewicz-Guzik M. et al., T cell subset-specific susceptibility to aging. Clin. Immunol. 127, 107–118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeda Y. et al., Detection of self-reactive CD8+ T cells with an anergic phenotype in healthy individuals. Science 346, 1536–1540 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Rochman Y., Yukawa M., Kartashov A. V., Barski A., Functional characterization of human T cell hyporesponsiveness induced by CTLA4-Ig. PLoS One 10, e0122198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel J. P. et al., Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 366, 1079–1089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ley T. J. et al.; Cancer Genome Atlas Research Network , Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdul-Aziz A. M. et al., HIF1α drives chemokine factor pro-tumoral signaling pathways in acute myeloid leukemia. Oncogene 37, 2676–2686 (2018). [DOI] [PubMed] [Google Scholar]
- 55.van Oosterwijk J. G. et al., Hypoxia-induced upregulation of BMX kinase mediates therapeutic resistance in acute myeloid leukemia. J. Clin. Invest. 128, 369–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Allen E. M. et al., Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le D. T. et al., Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawrence M. S. et al., Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexandrov L. B. et al.; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain , Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). Correction in: Nature 502, 258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Lee D. I. et al., Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J. Clin. Invest. 129, 774–785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greiner J. et al., Expression of tumor-associated antigens in acute myeloid leukemia: Implications for specific immunotherapeutic approaches. Blood 108, 4109–4117 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Zhu L. et al., Blimp-1 impairs T cell function via upregulation of TIGIT and PD-1 in patients with acute myeloid leukemia. J. Hematol. Oncol. 10, 124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan J. et al., Higher PD-1 expression concurrent with exhausted CD8+ T cells in patients with de novo acute myeloid leukemia. Chin. J. Cancer Res. 29, 463–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kong Y. et al., T-cell immunoglobulin and ITIM domain (TIGIT) associates with CD8+ T-cell exhaustion and poor clinical outcome in AML patients. Clin. Cancer Res. 22, 3057–3066 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Kong Y. et al., PD-1(hi)TIM-3(+) T cells associate with and predict leukemia relapse in AML patients post allogeneic stem cell transplantation. Blood Cancer J. 5, e330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Stasi A., Jimenez A. M., Minagawa K., Al-Obaidi M., Rezvani K., Review of the results of WT1 peptide vaccination strategies for myelodysplastic syndromes and acute myeloid leukemia from nine different studies. Front. Immunol. 6, 36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng M. et al., LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 562, 605–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daver N. et al., Hypomethylating agents in combination with immune checkpoint inhibitors in acute myeloid leukemia and myelodysplastic syndromes. Leukemia 32, 1094–1105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daver N. et al., Efficacy, safety, and biomarkers of response to azacitidine and nivolumab in relapsed/refractory acute myeloid leukemia: A nonrandomized, open-label, phase II study. Cancer Discov. 9, 370–383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Ijaz A. et al., Significant risk of graft-versus-host disease with exposure to checkpoint inhibitors before and after allogeneic transplantation. Biol. Blood Marrow Transplant. 25, 94–99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davids M. S. et al.; Leukemia and Lymphoma Society Blood Cancer Research Partnership , Ipilimumab for patients with relapse after allogeneic transplantation. N. Engl. J. Med. 375, 143–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y. Q. et al., Tumor secretion of CCL22 activates intratumoral Treg infiltration and is independent prognostic predictor of breast cancer. PLoS One 8, e76379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Masarova L. et al., Harnessing the immune system against leukemia: Monoclonal antibodies and checkpoint strategies for AML. Adv. Exp. Med. Biol. 995, 73–95 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Boddu P. et al., The emerging role of immune checkpoint based approaches in AML and MDS. Leuk. Lymphoma 59, 790–802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tyner J. W. et al., Functional genomic landscape of acute myeloid leukaemia. Nature 562, 526–531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamble A. J., Dietz M., Laderas T., McWeeney S., Lind E. F., Integrated functional and mass spectrometry-based flow cytometric phenotyping to describe the immune microenvironment in acute myeloid leukemia. J. Immunol. Methods 453, 44–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Döhner H. et al., Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129, 424–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and protocols supporting the findings of this study are in the main text and SI Appendix. Other associated data are available from the corresponding author upon request.