Significance
NER removes helix-destabilizing bulky adducts including UV lesions and crosslinks generated by formaldehyde. NER protects skin cells from carcinogenesis. XP patients deficient in NER develop progressive neurological dysfunction. We discovered that collaboration between TOP1 and the BER pathway can substitute for the lack of NER. This previously unappreciated repair pathway is initiated by TOP1-dependent sensing topological alterations near UV lesions and subsequent SSB formation followed by BER. Considering that the SSB formation by TOP1 is the rate-limiting step, and TOP1 expression is very low in the skin and neuronal tissues, our finding may help understand the molecular mechanisms and genetic determinants underlying the neurological dysfunction of XP patients, although TOP1-relevant endogenous lesions are currently unknown.
Keywords: 6–4PPs, UV damage, base excision repair, topoisomerase I
Abstract
Nucleotide excision repair (NER) removes helix-destabilizing adducts including ultraviolet (UV) lesions, cyclobutane pyrimidine dimers (CPDs), and pyrimidine (6–4) pyrimidone photoproducts (6–4PPs). In comparison with CPDs, 6–4PPs have greater cytotoxicity and more strongly destabilizing properties of the DNA helix. It is generally believed that NER is the only DNA repair pathway that removes the UV lesions as evidenced by the previous data since no repair of UV lesions was detected in NER-deficient skin fibroblasts. Topoisomerase I (TOP1) constantly creates transient single-strand breaks (SSBs) releasing the torsional stress in genomic duplex DNA. Stalled TOP1-SSB complexes can form near DNA lesions including abasic sites and ribonucleotides embedded in chromosomal DNA. Here we show that base excision repair (BER) increases cellular tolerance to UV independently of NER in cancer cells. UV lesions irreversibly trap stable TOP1-SSB complexes near the UV damage in NER-deficient cells, and the resulting SSBs activate BER. Biochemical experiments show that 6–4PPs efficiently induce stable TOP1-SSB complexes, and the long-patch repair synthesis of BER removes 6–4PPs downstream of the SSB. Furthermore, NER-deficient cancer cell lines remove 6–4PPs within 24 h, but not CPDs, and the removal correlates with TOP1 expression. NER-deficient skin fibroblasts weakly express TOP1 and show no detectable repair of 6–4PPs. Remarkably, the ectopic expression of TOP1 in these fibroblasts led them to completely repair 6–4PPs within 24 h. In conclusion, we reveal a DNA repair pathway initiated by TOP1, which significantly contributes to cellular tolerance to UV-induced lesions particularly in malignant cancer cells overexpressing TOP1.
Ultraviolet (UV) light induces two types of pyrimidine dimers, cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts (6–4PPs). The UV-induced lesions, helix-destabilizing bulky adducts, are highly mutagenic by causing base substitutions during DNA replication. CPDs are three to four times more frequent in comparison with 6–4PPs, while 6–4PPs distort duplex DNA to a greater extent and are more cytotoxic than CPDs (1–4). The DNA helix destabilizing activity of 6–4PPs is more stronger than that of CPDs, and the nucleotide excision repair (NER) machinery recognizes 6–4PPs more quickly than CPDs (5, 6). It is generally believed that NER is the only DNA repair pathway that removes these bulky adducts because of a lack of detectable removal of CPDs or 6–4PPs in NER-deficient primary fibroblasts even at 24 h after UV irradiation (7–11). Accordingly, X-ray repair cross-complementing group 1 (XRCC1) and DNA ligase III (Lig3), key base excision repair (BER) factors, have been invoked for their potential role in NER (12–15).
BER repairs lesions caused by alkylation, hydrolysis, and oxidation of nucleotides, so that lesions do not lead to strong structural alterations. No canonical BER factors recognize helix-destabilizing bulky adducts in mammalian cells (16). Typical BER is initiated by enzymatic removal of damaged nucleotides by DNA glycosylases, leading to the formation of apurinic/apyrimidinic (AP) sites (16). The AP-endonuclease 1 (APE1/APEX1) incises DNA 5′ to the AP sites, generating single-strand breaks (SSBs) with 3′-hydroxyl and 5′-2-deoxyribose–5′-phosphate (dRP) termini (17–19). SSB repair (SSBR) is a subpathway of BER, and the two subpathways share several repair effector molecules, including XRCC1, polynucleotide kinase phosphatase (PNKP), DNA polymerase β (POLβ), flap endonuclease 1 (FEN1), DNA ligase I (Lig1), and DNA ligase III (Lig3) (13, 20–22). The AP site is replaced by two types of the repair synthesis. Short-patch repair synthesis involves a single-nucleotide incorporation primarily by POLβ, whereas long-patch repair synthesis by a DNA polymerase (β, δ, or ε) incorporates two or more nucleotides by strand-displacement synthesis, creating a flap DNA intermediate that is removed by FEN1 (19, 23–25).
Topoisomerase I (TOP1) plays an essential role in DNA replication and transcription by relaxing supercoiled DNA (reviewed in ref. 26). During this process, TOP1 constantly creates transient TOP1-SSB complex, called a TOP1 cleavage complex (TOP1cc), where TOP1 is covalently bound to the 3′ end of SSB forming 3′-TOP1 DNA adduct. This transient TOP1cc permits rotation of the nicked strand around an intact strand of duplex genomic DNA, releasing its torsional stress. The transient TOP1-SSB complex formation occurs in the whole genome irrespective of the topological status of genomic DNA (26). Camptothecin, a chemotherapeutic TOP1 poison, binds at the interface of the DNA and TOP1 in the TOP1cc and prevents the religation of SSB, thereby trapping TOP1ccs (27). The resulting trapped TOP1ccs kill cycling cells by generating cytotoxic one-ended double-strand breaks during DNA replication (26, 28). Importantly, stable TOP1ccs are formed near a variety of DNA lesions, such as abasic sites, strand breaks, mismatches, and UV lesions, which can displace the 5′-OH end of the cleaved DNA within TOP1ccs and irreversibly prevent their religation (26, 29, 30). The removal of the TOP1-DNA adducts from such pathological TOP1ccs is carried out by both tyrosyl-DNA phosphodiesterase 1 and 2 (TDP1 and TDP2), which hydrolyze the 3′ tyrosyl blocking lesions (31, 32) (reviewed in ref. 26).
We here reveal that TOP1ccs trapped near 6–4PPs activate BER and remove 6–4PPs in NER-deficient cells. We also show that although NER-deficient primary fibroblasts did not remove 6–4PPs as shown previously (7–11), the ectopic expression of TOP1 allowed these cells to repair 6–4PPs. Biochemical studies provide evidence that purified TOP1 can be trapped in the vicinity of the 6–4PP generating SSBs and that long-patch repair synthesis primed from an SSB efficiently removes a 6–4PP localized eight nucleotides (8 nts) downstream of the SSB in vitro. Defects in both BER and NER caused a synergistic increase in UV sensitivity of cancer cells, which abundantly express TOP1. In conclusion, the current study uncovers the previously unappreciated TOP1-dependent excision repair that removes 6–4PPs independent of NER.
Results
BER Factors, POLβ and XRCC1, Contribute to Cellular Tolerance to UV Independently of NER.
To explore a functional overlap between BER and NER, we disrupted the POLβ and XRCC1 genes in NER-deficient XPA−/− cells derived from the human TK6 B cell line (SI Appendix, Fig. S1 A and B). The XPA protein is essential for NER and recruits the nucleases that make 5′ and 3′ incisions at UV damage (reviewed in ref. 33). We also generated XPA−/−, XRCC1−/−, and XPA−/−/XRCC1−/− cells from the human MCF-7 breast cancer cell line (SI Appendix, Fig. S1C). SI Appendix, Table S1 shows the list of gene-disrupted cells analyzed in this study. XRCC1−/− TK6 cells, but not XPA−/− TK6 cells, were sensitive to H2O2 and methyl methane sulfonate (SI Appendix, Fig. S1 D and E), which agrees with the role of XRCC1 in BER. XPA−/− and XRCC1−/− MCF-7 cells showed the same phenotype pattern as did the TK6 mutants (SI Appendix, Fig. S1 F and G).
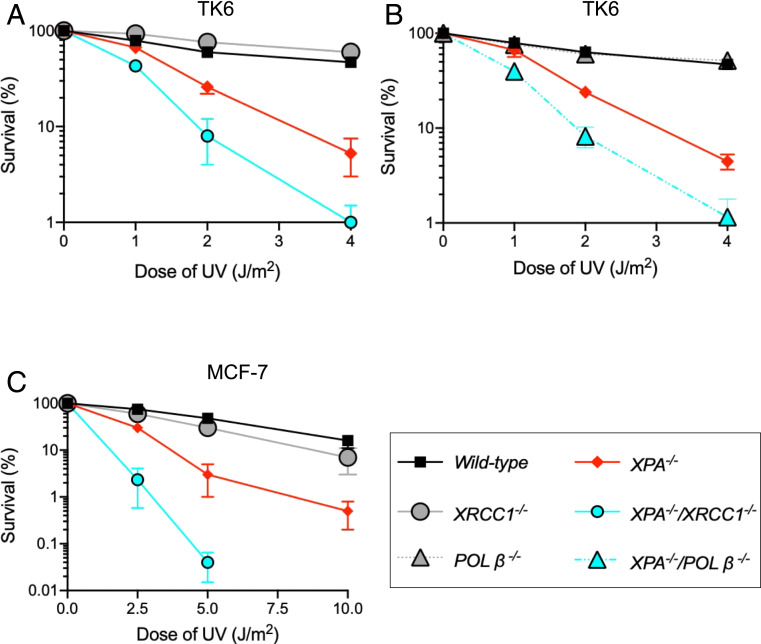
We first evaluated the capability of cells to repair UV-induced DNA damage by colony survival assays. As expected, XPA−/− cells, but not POLβ −/− plus XRCC1−/− cells, were sensitive to UV exposure (Fig. 1 A–C). Remarkably, in both double-null TK6 and MCF-7 cells, the XPA−/−/POLβ −/− and XPA−/−/XRCC1−/− cells displayed higher sensitivity to UV in comparison with XPA1−/− cells. Ectopic expression of XPA and XRCC1 in XPA1−/− and XPA−/−/XRCC1−/− MCF-7 cells rescued the UV sensitivity to a similar level as Wild-type and XPA1−/−cells, respectively (SI Appendix, Fig. S2 A–C). However, knockdown of APE1 in XPA1−/− MCF-7 cells had no impact on UV sensitivity of XPA1−/− MCF-7 cells (SI Appendix, Fig. S2 D and E). Therefore, we conclude that both POLβ and XRCC1 contribute to cellular tolerance to UV damage when NER is defective.
Fig. 1.
UV sensitivity of TK6 and MCF-7 cells as a function of loss of BER factors in the absence of XPA. (A–C) Colony survival of human TK6 (A and B) and MCF-7 cells (C) carrying the indicated genotypes following exposure to UV. The dose of DNA-damaging agents is displayed on the x axis on a linear scale, while the percent fraction of surviving cells is displayed on the y axis on a logarithmic scale. Error bars show the SD of mean for three independent experiments. Percent survival was calculated as the percentage of surviving cells treated with DNA-damaging agents relative to the untreated surviving cells.
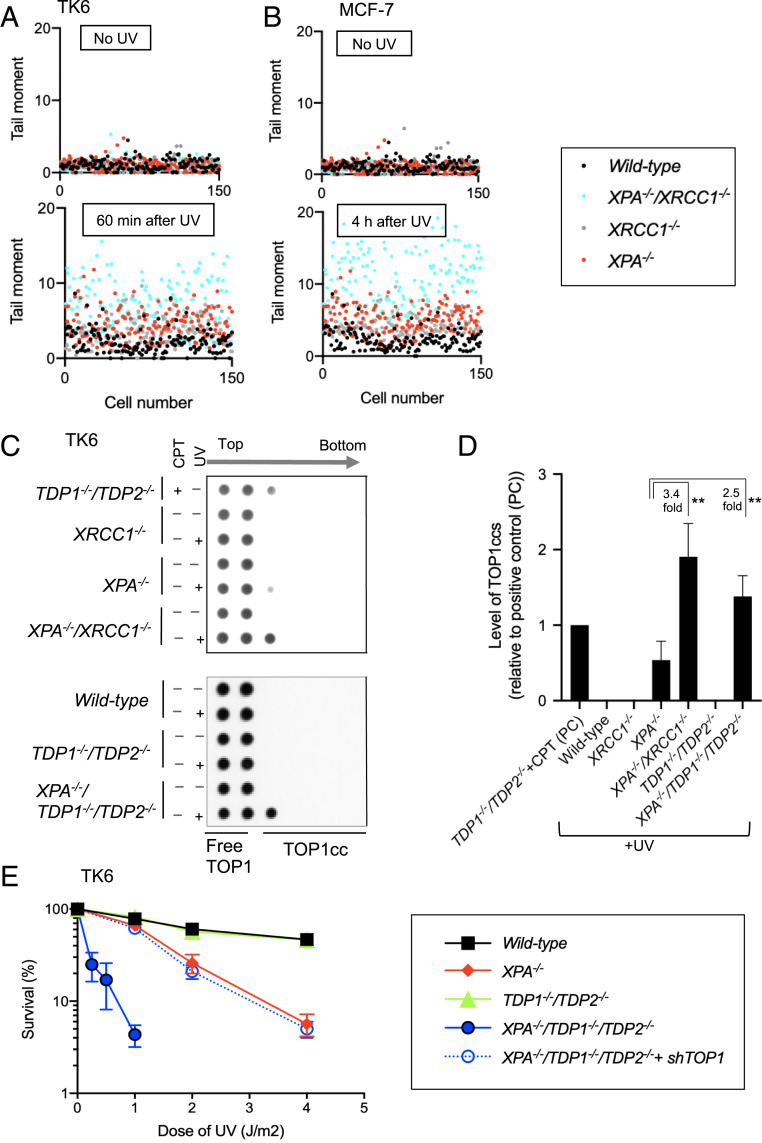
The involvement of POLβ and XRCC1 in cellular tolerance to UV in the absence of XPA suggests that BER is responsible for repair of UV-induced damage in NER-deficient cells. To address this notion, we measured the number of UV-induced SSBs, BER intermediates, by performing the alkaline comet assay (34, 35). UV-induced comet tails in abundance greater than that of wild-type controls in both XPA−/− TK6 and MCF-7 mutants (Fig. 2 A and B), indicating UV-dependent induction of SSBs when NER is deficient. Loss of XRCC1 further increased the UV-induced comet tails in XPA−/− TK6 and MCF-7 cells. These data suggest that XRCC1-dependent BER may remove UV lesions in the absence of NER.
Fig. 2.
Exposure of cells to UV increases SSBs as well as the amount of stable TOP1ccs in the absence of NER. (A and B) The number of SSBs, BER intermediates, were quantified in the indicated genotypes of TK6 (A) and MCF-7 cells (B) by the alkaline comet assay before and at the indicated time after treatments with UV. A scatter plot of the raw data from three of the experiments to show the level of variation in SSBs within single populations of cells. Each dot represents the tail moment of an individual cell, and 50 cells were scored per sample (in total 150 cells). (C) Representative Western blot quantitating the number of stable TOP1ccs in TK6 cells carrying the indicated genotypes having been treated with UV or camptothecin (CPT), a TOP1 poison. Quantification is shown in D, and experimental methods are shown in SI Appendix, Fig. S2C. (D) The amount of TOP1ccs in the indicated genotypes relative to the amount of TOP1ccs in CPT-treated TDP1−/−/TDP2−/− TK6 cells. CPT-treated TDP1−/−/TDP2−/− TK6 cells were analyzed as a positive control in every experiment. The y axis shows the number of UV-induced TOP1ccs in the indicated genotypes relative to that of the positive control. Every experiment was done independently at least three times, and the error bars represent SD. Statistical analyses (Student t test) are indicated (**P < 0.05). (E) Colony survival analysis following exposure to UV. Experiments were done as in Fig. 1A.
Exposure of NER-Deficient Cells to UV Induces Formation of Stable TOP1ccs.
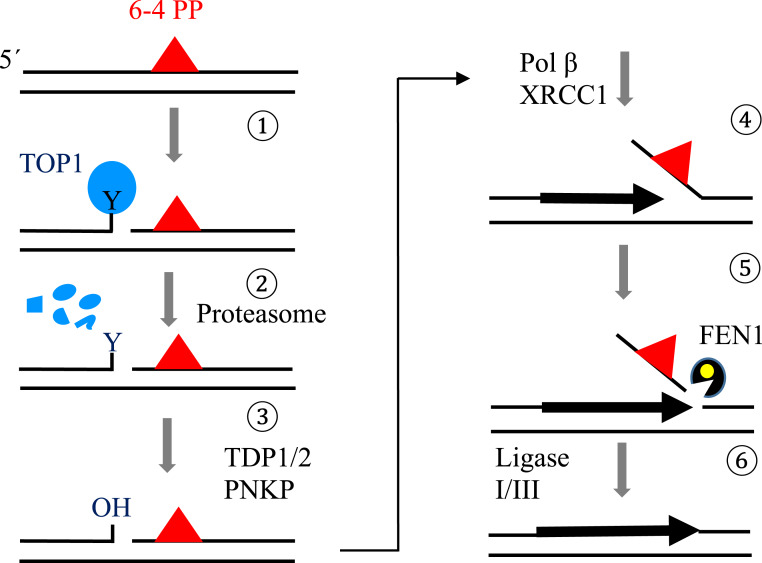
Canonical BER factors do not recognize the UV lesions (16), and an important question is the molecular mechanism underlying the induction of SSBs by UV. Since UV lesions have been suggested to interfere with the religation step of catalysis by TOP1 (26, 29, 30), stable TOP1ccs may represent UV-induced SSBs seen in NER-deficient cells (Fig. 2 A and B). To address this possibility, we analyzed the effect of camptothecin, a TOP1 poison, on cellular sensitivity to UV. If camptothecin and UV irradiation damage genomic DNA independently of each other, the two agents may have an additive cytotoxic effect. The two DNA-damaging agents indeed had an additive effect on the colony survival in wild-type cells (SI Appendix, Fig. S3A). By contrast, the two agents synergistically reduced the colony survival in XPA−/− cells (SI Appendix, Fig. S3B), indicating that camptothecin becomes more cytotoxic upon UV irradiation in NER-deficient cells. One possible scenario is that UV lesions might trap TOP1ccs in their vicinity, with camptothecin further increasing the number of trapped TOP1ccs. Based on the results of the genetic experiments described above, the BER system may be responsible for repairing both the TOP1 crosslink and the nearby UV lesion (Scheme 1).
Scheme 1.
Proposed model for TOP1-driven base excision repair of UV-induced 6–4PP damage (1). In the absence of NER, TOP1 recognizes the 6–4PP lesion and incises the lesion-containing strand 5′ upstream of the lesion. During this process, TOP1 becomes crosslinked to the DNA, forming a stable TOP1cc (2). Irreversible TOP1ccs are likely digested by the proteasome system, leaving the 3′-OH end at the TOP1 incision blocked by a phosphotyrosine group (3). TDP1/2 removes the Y (tyrosine) group, and PNKP removes the phosphate from the 3′ end of the SSB (4). POLβ and XRCC1 mediate strand-displacement DNA synthesis past the 6–4PP lesion (5). FEN1 incises the flap containing the lesion (6). DNA ligases seal the FEN1 product completing repair.
To test this possibility, we measured the amount of stable TOP1ccs following exposure to UV. To this end, we lysed cells and separated stable TOP1ccs from free TOP1 in the cellular lysates by subjecting cell extracts to cesium chloride (CsCl)-gradient ultracentrifugation-sedimentation under a denaturing condition (SI Appendix, Fig. S3C). We also measured the amount of genomic DNA in every cellular lysate to normalize the results (SI Appendix, Tables S2 and S3). Free TOP1 remains in the top two fractions, while TOP1ccs move to lower fractions of the CsCl-gradient that correspond to the migration of chromosomal DNA. TDP1, as well as TDP2, removes TOP1 adducts from SSBs, and deletion of both TDP enzymes generated TOP1ccs that were detected in the third fractions of the CsCl-gradient upon a CPT treatment (Fig. 2C, top row).
To detect UV-induced stalled TOP1ccs, we exposed cells to UV and measured the number of TOP1ccs at 30 min after UV. In UV-irradiated wild-type cells, only free TOP1 was observed, and no detectable TOP1 was seen in the lower CsCl-gradient fractions (Fig. 2C, the second row of the lower panel). In contrast, UV-irradiated XPA−/− cells displayed TOP1ccs in the third fraction (Fig. 2C). The number of UV-induced TOP1ccs was three times higher in XPA−/−/XRCC1−/− cells than XPA−/− cells (Fig. 2 C and D). Taken together, the results of these experiments indicate that UV induces stable TOP1ccs in the absence of NER and that those TOP1ccs may represent the UV-induced SSBs seen in Fig. 2 A and B.
TDP1 and TDP2 Contribute to the Repair of TOP1ccs and UV Damage in NER-Deficient Cells.
If stable TOP1ccs form 5′ to UV lesions, TDP1 and TDP2 should be involved in excising the TOP1 adducts to enable priming of repair synthesis (Scheme 1). To test this hypothesis, we created XPA−/−/TDP1−/−/TDP2−/− cells considering a substantial functional overlap between TDP1 and TDP2 in removal of TOP1-DNA adducts (22, 36, 37) (SI Appendix, Fig. S3D). While TDP1−/−/TDP2−/− cells exhibited no UV sensitivity, XPA−/−/TDP1−/−/TDP2−/− cells displayed significantly higher UV sensitivity than XPA−/− cells (Fig. 2E). The depletion of TOP1 in XPA−/−/TDP1−/−/TDP2−/− cells (SI Appendix, Fig. S3F) could reverse their UV sensitivity back to that of XPA−/− cells (Fig. 2E). Consistent with this result, the number of stable TOP1ccs was undetectable in TDP1−/−/TDP2−/− cells, but was higher in XPA−/−/TDP1−/−/TDP2−/− cells in comparison with XPA−/− and TDP1−/−/TDP2−/− cells (Fig. 2 C and D), and overexpression of TDP1 and TDP2 both in XPA−/−/TDP1−/−/TDP2−/− cells resulted in a decrease in number of TOP1cc similar to that of XPA−/− level (SI Appendix, Fig. S4 H and I). We conclude that the TDP enzymes contribute to UV damage repair when NER is not initiated. The data support the following scenario about the TOP1-dependent BER of UV lesions in NER-deficient cells. UV lesions trap TOP1ccs; the resulting TOP1cc adducts are removed by the TDP enzymes. Long-patch repair DNA synthesis initiated from the SSBs may remove the UV damage if stable TOP1ccs form 5′ to UV lesions (Scheme 1).
Repair Kinetics of 6–4PPs Correlates with the Amount of TOP1 in XPA−/− Cells.
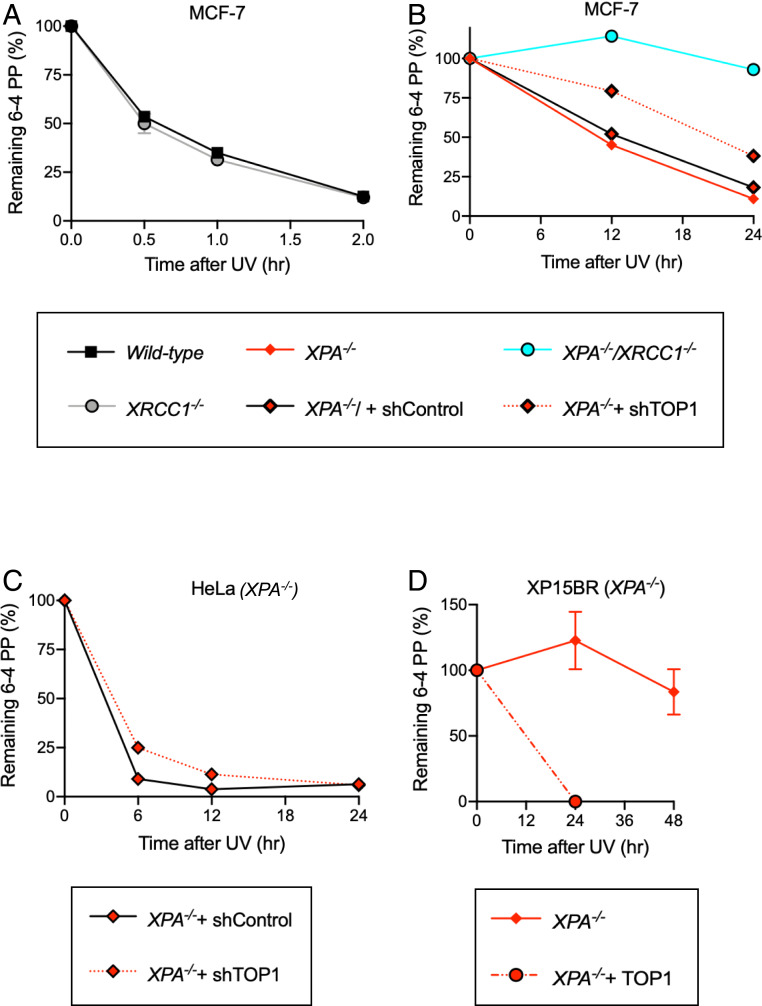
To verify the removal of UV-induced lesions independent of NER, we monitored repair kinetics of UV lesions, CPDs and 6–4PPs, in MCF-7 cells using specific antibodies (38). To prevent the dilution of UV lesions by DNA replication, we stopped the cell cycle by conducting serum-starvation and adding a CDK inhibitor. Four J/m2 UV irradiation completely inhibited DNA replication even at 1 d after the irradiation in XPA−/− MCF-7 cells (SI Appendix, Fig. S4A). We detected the removal of CPDs only in wild-type but not in XPA−/− MCF-7 cells (SI Appendix, Fig. S4B). Wild-type MCF-7 cells removed 6–4PPs within 2 h after UV irradiation (Fig. 3A). Remarkably, XPA−/− MCF-7 cells removed ∼90% of the UV-induced 6–4PPs by 24 h after UV irradiation (Fig. 3B). These data suggest that the higher UV sensitivity of XPA−/−/XRCC1−/− cells than XPA−/− cells (Fig. 1 A–C) is due to the slower removal of 6–4PPs in XPA−/−/XRCC1−/− cells in comparison with XPA−/− cells (Fig. 3B).
Fig. 3.
Repair kinetics of 6–4PPs correlates with the amount of TOP1 in NER-deficient cells. (A and B) The removal of 6–4PPs in G1-arrested MCF-7 cells carrying wild-type and XRCC1−/− genotypes (A) and XPA−/− and XPA−/−/XRCC1−/− genotypes (B). Cells were exposed to UV (4 J/m2) at time 0, and genomic DNA was isolated at the indicated time points shown on the x axis. The amount of 6–4PPs relative to the amount of 6–4PPs immediately after UV irradiation are shown on the y axis. The amount of TOP1 in short hairpin TOP1 (shTOP1)-treated cells is shown in SI Appendix, Fig. S4E. All experiments were done at least three times. Error bars show the SD of the mean value. (C) The removal of 6–4PPs in G1-arrested XPA−/− HeLa cells and those treated with shTOP1 (SI Appendix, Fig. S4F). Experiments were done as in A and B, and data are presented as in B. Data of wild-type HeLa cells are shown in SI Appendix, Fig. S4G. (D) The removal of 6–4PPs in XP patient-derived primary fibroblasts deficient in XPA (XP15BR). “+TOP1” indicates the overexpression of TOP1 (SI Appendix, Fig. S4E). The data are shown as in A and B.
Previous studies had shown that there is no repair of 6–4PPs in human primary skin fibroblasts deficient in XPA and XPC (7, 9, 10, 39). The apparent discrepancy between these previous studies and ours (Fig. 3B) might be due to the different cell types studied as well as the UV dose. We exposed MCF-7 cells to 4 J/m2 while the previous studies irradiated with 10 J/m2 or higher doses of UV. The UV dose, indeed, affected the repair kinetics of 6–4PPs as evidenced by the data that only ∼35% of the 6–4PPs induced by 10 J/m2 UV irradiation were removed by 24 h post-UV, which is in marked contrast with the removal of ∼90% of the 6–4PPs induced by 4 J/m2 UV irradiation by 24 h in MCF-7 cells (SI Appendix, Fig. S4C). This observation implies the presence of a rate-limiting factor that determines the kinetics of 6–4PP repair, which is more abundantly expressed in cancer cell lines than in primary skin fibroblasts. We hypothesized that the high amount of TOP1 in cancer cells might determine the repair kinetics because the TOP1cc trap and resulting SSB formation in the vicinity of 6–4PPs is likely to be the rate-limiting step, considering the removal of SSBs by BER is very quick, within a few hours (35). Primary skin fibroblasts may express TOP1 very poorly as evidenced by the data that a TOP1 protein expression profile among normal tissues shows an extremely low TOP1 expression level in skin cells (SI Appendix, Fig. S4D) (https://www.genecards.org/cgi-bin/carddisp.pl?gene=TOP1). Furthermore, actively cycling cancer cells may abundantly express TOP1 because it is essential for DNA replication (26). Thus, abundant expression of TOP1 in the MCF-7 cancer cell line might allow them to repair 6–4PPs in the absence of NER.
To study a correlation between TOP1 expression and repair kinetics of 6–4PPs, we depleted TOP1 in XPA−/− MCF-7 cells by expressing shRNA against TOP1 (SI Appendix, Fig. S4E) and examined 6–4PP repair kinetics. The removal of 6–4PPs was slower in TOP1-depleted XPA−/− MCF-7 cells than in control XPA−/− cells (Fig. 3B). We also examined serum-starved wild-type and XPA−/− HeLa cells arrested at G1 phase by adding a CDK inhibitor (Fig. 3C and SI Appendix, Fig. S4G). XPA−/− HeLa cells removed ∼90% of 6–4PPs by 6 h, and depletion of TOP1 (SI Appendix, Fig. S4E) delayed this removal (Fig. 3C). We next examined the effect of TOP1 overexpression on the repair of 6–4PPs in XPA-deficient human primary skin fibroblasts (XP15BR) (40). They poorly expressed TOP1, comparable to normal human primary skin fibroblasts (HDFa) (SI Appendix, Fig. S4 E and K) and displayed no detectable repair of 6–4PPs (Fig. 3D). The increased expression of TOP1 (SI Appendix, Fig. S4E), indeed, led these XPA-deficient fibroblasts to repair 6–4PPs (Fig. 3D) without changing their proliferation rate (SI Appendix, Fig. S4J). In conclusion, the abundant expression of TOP1 in MCF-7 and HeLa cancer cells allows them to repair 6–4PPs in the absence of NER.
UV Damage and Stabilizing TOP1ccs In Vitro.
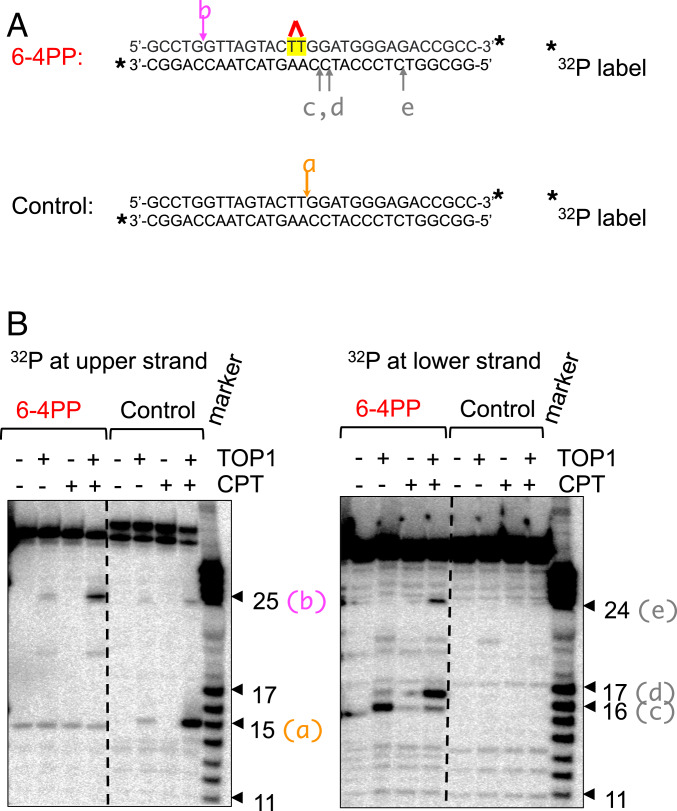
Next we investigated whether 6–4PPs caused the formation of stable TOP1ccs in vitro. To this end, we prepared two different duplex oligonucleotide DNAs containing a single 6–4PP lesion (Fig. 4A) and performed a DNA cleavage assay (41, 42). We incubated 32P end-labeled duplex oligonucleotides carrying the single UV lesion together with TOP1 and camptothecin and analyzed products using a denaturing polyacrylamide gel electrophoresis and autoradiography. Prominent TOP1-dependent DNA cleavage was detectable in an oligonucleotide carrying a 6–4PP, but not in the intact one in the presence of camptothecin (Fig. 4, site a). We prepared another set of duplex oligonucleotide DNAs carrying either a single CPD or 6–4PP lesion. We detected a strong TOP1cc signal upstream of the 6–4PP (SI Appendix, Fig. S5, site e). TOP1cc was not detectable in the oligonucleotide carrying the CPD or in the unmodified oligonucleotide (SI Appendix, Fig. S5 A and B). These data indicate that a 6–4PP can generate stable TOP1ccs near the DNA lesion in vitro. These results corroborate the stable TOP1cc formation in UV-irradiated XPA−/− cells (Fig. 2 C and D).
Fig. 4.
In vitro formation of TOP1ccs in a duplex oligonucleotide DNA containing 6–4PPs. (A) The sequence of a duplex DNA containing a 6–4PP is shown with its respective control. The 6–4PP site is highlighted by red. Star indicates 32P label at the 3′ end of either upper strand or lower strand. The arrows marked a–e show the positions of TOP1cc sites detected in B. (B) The representative TOP1 cleavage assay using oligonucleotides shown in A. (Left) Analysis of the upper strand and (Right) analysis of the lower strand shown in A. Following an incubation of the oligonucleotides with TOP1, they were subjected to denaturing polyacrylamide gel electrophoresis followed by autoradiography. The arrowheads marked (a–e) correspond to the TOP1 cleavage sites marked with a–e in A. The numbers shown as individual arrowheads represent the length of nucleotides from the 3′-32P label. The most prominent TOP1 cleavage site is indicated by the pink arrow with annotation b in A, Left.
Purified BER Factors Remove UV Damage In Vitro.
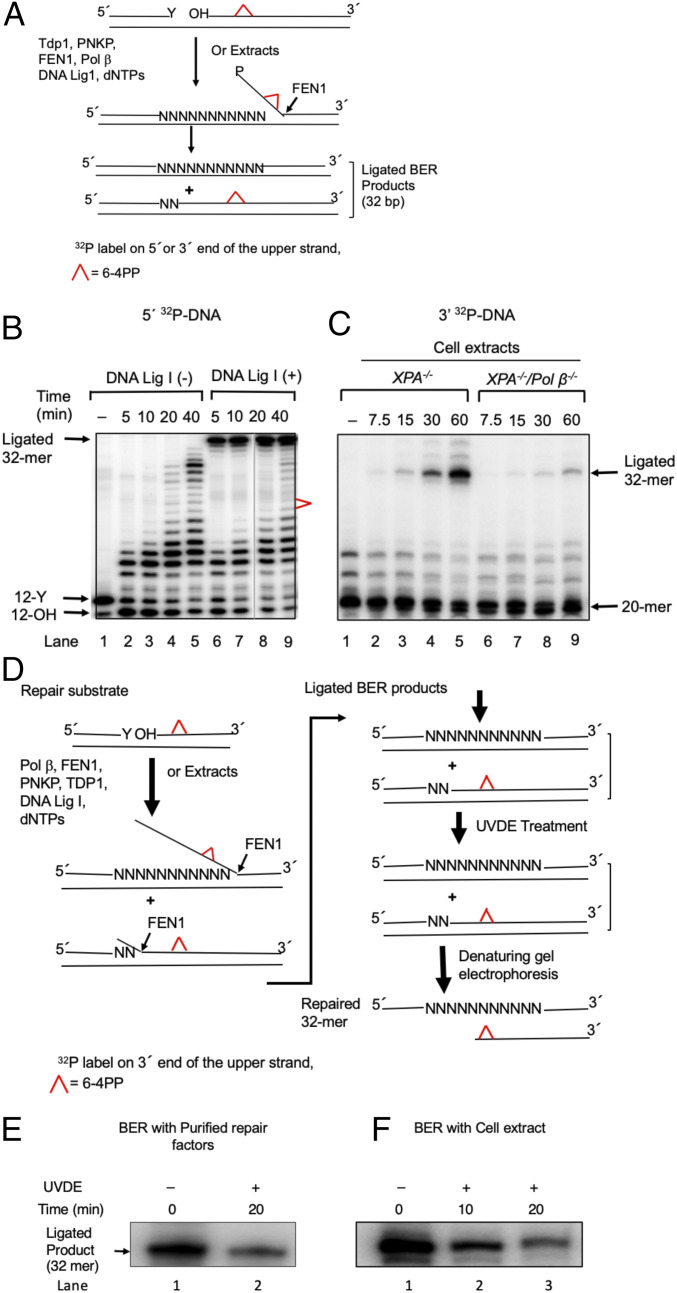
We investigated whether TOP1-initiated SSBs stimulated long-patch repair synthesis and also whether this synthesis could remove a UV lesion in vitro. We used 5′ 32P-labeled duplex DNA as substrate (32 bp) with an SSB 8 nts upstream of the 6–4PP lesion at positions 9 and 10 of the substrate DNA (SI Appendix, Fig. S6A). We found that POLβ conducted strand-displacement DNA synthesis beyond the 6–4PP lesion (SI Appendix, Fig. S6A), as expected from earlier works (43, 44). Thus, under these incubation conditions, POLβ performed strand-displacement DNA synthesis and generated flap structures that were cleaved by purified human FEN1 as shown in SI Appendix, Fig. S6A. We then verified whether FEN1 could process the model substrate that represents the strand-displacement product with a 10-nt flap structure attached to the downstream oligonucleotide (SI Appendix, Fig. S6B). The results confirmed that FEN1 generated an 11-nt-long product (SI Appendix, Fig. S6C); this represents the expected 10-nt flap plus one additional nucleotide (45), indicating that long-patch repair is capable of removing the 6–4PP lesion. We conclude that POLβ and FEN1 are capable of in vitro processing of the 6–4PP lesion located 8 nts downstream of an SSB.
We next prepared a model substrate that mimicked postproteasomal digestion of TOP1cc nicked DNA, where a 3′-phosphotyrosine group was present at the nick (SI Appendix, Fig. S6 D, Top). We found that the 3′ end blocking tyrosine was incised by TDP1 to generate the 3′-phosphate containing intermediate (12-P, SI Appendix, Fig. S6D; lane 2), and this was further processed by PNKP (46) to generate the 3′-OH containing BER intermediate (12-OH, SI Appendix, Fig. S6D; lane 4) that is required for priming of long-patch BER. Addition of POLβ and FEN1, indeed, caused a strand displacement beyond the 6–4PP lesion (SI Appendix, Fig. S6D; lane 5). We then examined the kinetics of repair synthesis by POLβ under the same reaction conditions as in lane 5 of SI Appendix, Fig. S6D and found that repair synthesis products were extended beyond the 6–4PP lesion at 40 min (Fig. 5B, lane 5). As expected, BER intermediates were converted to full-length ligated products upon the addition of DNA ligase I (Fig. 5B, lanes 6–9). In summary, purified POLβ, FEN1, PNPK, and TDP1 efficiently remove the 6–4PP lesion even when the 3′ end of an SSB localized upstream of this lesion was blocked by the phosphotyrosine group. These results are consistent with the model in Scheme 1.
Fig. 5.
In vitro long-patch BER synthesis removes the 6–4PP UV lesion. (A) Schematic representation of in vitro BER of a 6–4PP. Top represents the repair substrate. A 32-bp duplex DNA with a nick was prepared by annealing 32-mer template with both a 12-mer upstream primer having 3′ Y (tyrosine) and a 20-mer downstream sequence containing a 6–4PP lesion at positions 9 and 10. The tyrosine residue at the nick mimics a postproteasomal removal of stable TOP1ccs. The 5′ end of the 20-mer had a hydroxy residue. TDP1 removes the tyrosine residue, and polynucleotide kinase phosphatase (PNKP) cleaves the 3′-phosphate group and also phosphorylates the 5′ end of the 20-mer. POLβ inserts nucleotides (N) creating flap structures that are incised by FEN1, and DNA ligase I seals the resulting nick, leading to the formation of ligated BER products. The upper strand was labeled either at the 5′ end by 32P[γATP] (B) or at the 3′ end by 32P[αddCTP] (C). (B) Repair of 6–4PP lesion-containing DNA substrate by purified BER factors in vitro. The BER reaction was conducted without DNA ligase I (−) (lanes 2–5) or with DNA ligase I (lanes 5–9), and reaction products were analyzed at the indicated times. The positions of 32P-labeled substrate (12-Y), the products of TDP1 and PNKP catalysis (12-OH), the ligated 32-mer product, and 6–4PP are indicated. The results shown are typical of three experiments. (C) Repair of 6–4PP lesion-containing DNA by cell extracts. The BER reaction was conducted either with an XPA−/− extract (lanes 2–5) or with an XPA−/−/POLβ −/− extract (lanes 5–9), and the reaction products were analyzed at the indicated time intervals. The results show a requirement of POLβ to repair the 6–4PP lesion in cell extracts. The positions of the substrate (20-mer), BER intermediates, and ligated 32-mer products are indicated. The results shown are typical of three experiments. (D) Schematic representation of the method of analyzing the removal of a 6–4PP from the repair substrate and ligated 32-mer products. BER reaction mixtures were treated with UVDE that selectively incises 5′ to the 6–4PP lesion. Therefore, if the 6–4PP lesion was not repaired, the ligated product (32 bp) would be sensitive to UVDE digestion. However, if the 6–4PP lesion was repaired, the ligated product would be insensitive to UVDE digestion, as illustrated. (E and F) Repair of the 6–4PP lesion in in vitro BER. The repair reaction was performed with purified enzymes as in B (lane 9) or with an XPA−/− cell extract as in C (lane 5), respectively. Analysis of the ligated products was shown. Reaction mixtures were subjected to a UVDE treatment. The repaired products after 20-min (E) or 10- and 20-min (F) incubations with UVDE were analyzed. The ‘0’ time represents untreated reaction mixture. UVDE-resistant ligated products indicate repair of the 6–4PP lesion. Results show a significant amount of the 6–4PP lesion repair in both BER systems, the reconstitution with purified BER factors (E) and extract-based BER (F), respectively. A representative phosphorimage of two experiments is shown.
POLβ-Dependent Removal of UV Damage by Cell Extracts.
We next tested whether cell extracts could repair the model DNA repair substrate shown in Fig. 5A and SI Appendix, Fig. S5D. We used whole-cell extracts from XPA−/− and XPA−/−/POLβ −/− TK6 cells (Fig. 1B). XPA−/− cell extracts generated the 32-mer ligated product with a similar efficiency as purified BER factors (compare lane 9 of Fig. 5B with lane 5 of Fig. 5C). In contrast, XPA−/−/POLβ −/− cell extract generated approximately four times less repair product (compare lanes 2–5 with 6–9 in Fig. 5C), indicating an important role for POLβ in the repair of 6–4PPs in cell extracts derived from NER-deficient cells. To verify that the 6–4PP lesion in the substrate was repaired, the reaction mixtures obtained from the reconstituted BER reaction (Fig. 5B, lane 9) or from the cell extract-based BER reaction (Fig. 5C, lane 5) were treated with UV-damage endonuclease (UVDE) of Schizosaccharomyces pombe; UVDE generates SSBs 5′ to UV damage in DNA (47). Therefore, if the 6–4PP lesion was not repaired, the ligated product (32 bp) would be sensitive to UVDE digestion. However, if the 6–4PPs lesion was repaired, the ligated product would be insensitive to UVDE digestion, as illustrated in Fig. 5D. Significant amounts of the repair product were resistant to UVDE treatment (Fig. 5 E and F), indicating the removal of the 6–4PPs lesion by the reconstituted BER system (Fig. 5B) and by the extract-based BER system (Fig. 5C). These results demonstrate that long-patch repair synthesis is capable of removing the 6–4PPs lesion in DNA. In conclusion, once stable TOP1ccs form upstream of the 6–4PPs lesion, the long-patch BER pathway can repair this UV lesion in the absence of NER.
Discussion
We here reveal that a pathway, TOP1-dependent BER, is able to efficiently remove 6–4PPs from genomic DNA in human cells. The efficiency of this repair correlates with the amount of TOP1 expression (Fig. 3 and SI Appendix, Fig. S4). Cellular and biochemical studies indicate that 6–4PPs cause the formation of stable TOP1ccs (Figs. 2 C and D and 4 and SI Appendix, Fig. S5), and the resulting SSBs activate long-patch repair synthesis and remove the 6–4PPs lesion in vivo (Fig. 3) and in vitro (Fig. 5).
Until now, it was believed that only NER, but not any other DNA repair pathway, removed the 6–4PPs lesion. UV damage activates PCNA and APE1 in quiescent fibroblasts in the absence of NER (48–50). Nonetheless, previous studies of primary fibroblasts derived from NER-deficient patients and mice (7–11) failed to detect the repair pathway described here. We hypothesize that poor expression of TOP1 in primary cells might account for the lack of detectable repair of 6–4PPs in the absence of NER because the formation of stable TOP1ccs 5′ to 6–4PPs is likely to be the rate-limiting step considering that completion of conventional BER following the formation of SSBs, BER intermediates, is relatively quick, within 2 h after base damage (35). This hypothesis is consistent with our experimental data showing that ectopic expression of TOP1 in NER-deficient fibroblasts allowed them to remove 6–4PPs (Fig. 3D). We also confirmed that depletion of TOP1 in MCF-7 and HeLa cells reduced the repair kinetics of 6–4PPs (Fig. 3 B and C). In conclusion, the amount of TOP1 closely correlates with the efficiency of the 6–4PP removal in NER-deficient cells.
The excision repair pathway described here is reminiscent of “alternative excision repair” defined as the excision repair that is initiated by SSBs cleaved by endonucleases near DNA-damage sites (44, 47, 51, 52) (reviewed in ref. 53). This pathway is present in fungi and bacteria, but not in mammalian cells. The UVDE generates SSBs in response to various types of helix-destabilizing adducts and initiates alternative excision repair to remove the adducts independent of NER. Ectopic expression of UVDE in human cells allows the removal of both CPDs and 6–4PPs with an efficiency very similar to that of NER (44, 54, 55). UVDE-mediated repair of UV lesions depends on XRCC1, and the repair patch size is ∼7 nts in vivo, indicating that this repair is carried out by long-patch BER. The efficiency of the TOP1-mediated UV damage repair is lower than that of NER probably due to the prominent role of NER, which precludes the less frequent TOP1-induced SSB formation as well as a longer distance between UV-induced TOP1ccs and UV lesions in comparison with the UVDE-mediated repair. The full restoration of UV tolerance in XPA-deficient human cells by a UVDE transgene suggests that TOP1-induced BER is capable of repairing UV damage very efficiently once TOP1 is trapped within several nucleotides 5′ to UV damage (54). This mechanism is reminiscent of the recently described formation of TOP1cc 5′ to misincorporated ribonucleotides (56–59) and the potential role of TOP1 as a backup pathway for the repair of ribonucleotides incorporated during replication and not eliminated by RNase H2 (26, 60). A recent study showed that the absence of RNase H2 sensitizes cells to PARP poisons (61). Nonetheless, UV irradiation may not synergistically increase the sensitivity of XPA−/− cells to PARP poisons for the following reason. When TOP1 is trapped at misincorporated ribonucleotides (56–59), the removal of TOP1 generates a noncanonical SSB having one side with a 2′,3′‐cyclic phosphate end, which interferes with the rejoining of the SSB and effectively traps PARP. In contrast, TOP1ccs formed near 6–4PPs are quickly removed by coordinate action of TDP1/TDP2 and BER factors.
A question is whether TOP1-induced BER of 6–4PPs contributes to the symptom of NER-deficient XP patients. The UV sensitivity of XP patients results from both transcriptional suppression (62, 63) and replication blockage by UV lesions. CPDs contribute to the transcriptional suppression to a higher extent than 6–4PPs due to several times larger number of CPDs than 6–4PPs (1–4). Thus, TOP1-induced BER has very limited relevance for the skin problem of XP patients. An important question is whether TOP1-induced BER affects neurological symptoms of NER-deficient patients (64). It remains unclear why the loss of NER causes abnormality specifically in the brain. One possible scenario is that formaldehyde-induced DNA lesions might cause the neurological symptoms. NER, Fanconi anemia (FA) pathway, homologous DNA recombination (HR), and the enzyme alcohol dehydrogenase 5 (ADH5/GSNOR) prevent endogenous accumulation of formaldehyde-induced DNA damage (65–67). Unlike FA or HR, NER plays a crucial role in postmitotic cells including the cells in the central nervous system. Considering TOP1-dependent BER partially substitutes for lack of NER, very poor expression of TOP1 in the brain (SI Appendix, Fig. S4D) might explain prominent neurological symptoms of NER-deficient patients (64).
Materials and Methods
All materials and cell lines used in the paper are described in SI Appendix, Tables S1 and S4–S6. MCF-7 and primary fibroblast (XP15BR) cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Cat# 0845964, Gibco) containing FBS (10%, Gibco), penicillin (100 U/mL), and streptomycin (100 μg/mL, Nacalai Tesque). For G1/G0 arrest by serum-starvation, MCF-7 cells were incubated prior to analysis in phenol red-free DMEM (Cat# 21063029, GIBCO) for 2 d and in serum-free medium for 24 h. More than 95% of the cells were arrested in the G0/G1 phases by serum-starvation. Human TK6 B cells were incubated in RPMI1640 medium (Cat# 3026456, Nacalai Tesque) supplemented with horse serum (5%, Gibco), penicillin (100 U/mL), streptomycin (100 μg/mL, Nacalai), and sodium pyruvate (200 mg/mL, ThermoFischer). TK6 and MCF-7 mutants were generated by CRISPR/Cas9 gene targeting with a guide RNA designed and cloned into pX330 or pX459 (SI Appendix, Table S5). Details of targeting, clone selection, and screening are given in SI Appendix, Materials and Methods. Details of colony formation assay, alkaline comet assay, in vivo nucleotide excision repair assay, lentivirus vectors, production of lentiviral particles and infection for TOP1 knockdown, lentivirus infection of the TOP1 expression vector into primary fibroblast cells, measurement of TOP1 trapped by genomic DNA, in vitro TOP1 cleavage assay, and in vitro BER assay are also described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the members of the Radiation Genetics laboratory for their helpful comments on the manuscript. For technical assistance with UV irradiation, we are grateful to the staff of the Radiation Biology Center, Kyoto University. This work was conducted through the Joint Research Program of the Radiation Biology Center, Kyoto University. This work was directly supported by a Grant-in-Aid from the Ministry of Education, Science, Sport and Culture (KAKENHI 25650006, 23221005, 19K22561, and 16H06306 [to S.T.] and KAKENHI 16H02953, 18H04900, and 19H04267 [to H.S.]). This work was also supported by grants from the Takeda Research and Mitsubishi Foundation (to H.S.) and the Japan Society for the Promotion of Science Core-to-Core Program, Advanced Research Networks (to S.T.). This work was supported in part by a grant from the Japan Chemical Industry Association (JCIA) Long-Range Research Initiative (LRI). This work was supported in part by Intramural Research Programs of the NIH, National Institute of Environmental Health Sciences Project Z01 ES050159 (to S.H.W.) and National Cancer Institute Project Z01 BC006150 (to Y.P.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920165117/-/DCSupplemental.
References
- 1.Mitchell D. L., The induction and repair of lesions produced by the photolysis of (6-4) photoproducts in normal and UV-hypersensitive human cells. Mutat. Res. 194, 227–237 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Mitchell D. L., The relative cytotoxicity of (6-4) photoproducts and cyclobutane dimers in mammalian cells. Photochem. Photobiol. 48, 51–57 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Lo H. L. et al., Differential biologic effects of CPD and 6-4PP UV-induced DNA damage on the induction of apoptosis and cell-cycle arrest. BMC Cancer 5, 135 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masutani C. et al., Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 18, 3491–3501 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa R. M., Chiganças V., Galhardo R. d S., Carvalho H., Menck C. F., The eukaryotic nucleotide excision repair pathway. Biochimie 85, 1083–1099 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Riou L. et al., The relative expression of mutated XPB genes results in xeroderma pigmentosum/Cockayne’s syndrome or trichothiodystrophy cellular phenotypes. Hum. Mol. Genet. 8, 1125–1133 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Emmert S., Kobayashi N., Khan S. G., Kraemer K. H., The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6-4 photoproducts. Proc. Natl. Acad. Sci. U.S.A. 97, 2151–2156 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harada Y. N. et al., Postnatal growth failure, short life span, and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene. Mol. Cell. Biol. 19, 2366–2372 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiramoto T. et al., Repair of 254 nm ultraviolet-induced (6-4) photoproducts: Monoclonal antibody recognition and differential defects in xeroderma pigmentosum complementation groups A, D, and variant. J. Invest. Dermatol. 93, 703–706 (1989). [DOI] [PubMed] [Google Scholar]
- 10.Köberle B., Roginskaya V., Wood R. D., XPA protein as a limiting factor for nucleotide excision repair and UV sensitivity in human cells. DNA Repair (Amst.) 5, 641–648 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Nakane H. et al., High incidence of ultraviolet-B-or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature 377, 165–168 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Shen M. R., Zdzienicka M. Z., Mohrenweiser H., Thompson L. H., Thelen M. P., Mutations in hamster single-strand break repair gene XRCC1 causing defective DNA repair. Nucleic Acids Res. 26, 1032–1037 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbotts R., Wilson D. M. 3rd, Coordination of DNA single strand break repair. Free Radic. Biol. Med. 107, 228–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moser J. et al., Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol. Cell 27, 311–323 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Paul-Konietzko K., Thomale J., Arakawa H., Iliakis G., DNA ligases I and III support nucleotide excision repair in DT40 cells with similar efficiency. Photochem. Photobiol. 91, 1173–1180 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Howard M. J., Wilson S. H., DNA scanning by base excision repair enzymes and implications for pathway coordination. DNA Repair (Amst.) 71, 101–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doetsch P. W., Cunningham R. P., The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res. 236, 173–201 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Doetsch P. W., Helland D. E., Haseltine W. A., Mechanism of action of a mammalian DNA repair endonuclease. Biochemistry 25, 2212–2220 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Lindahl T., DNA repair enzymes. Annu. Rev. Biochem. 51, 61–87 (1982). [DOI] [PubMed] [Google Scholar]
- 20.Caldecott K. W., Single-strand break repair and genetic disease. Nat. Rev. Genet. 9, 619–631 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Friedberg E. C., The eureka enzyme: The discovery of DNA polymerase. Nat. Rev. Mol. Cell Biol. 7, 143–147 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Prasad R., Williams J. G., Hou E. W., Wilson S. H., Pol β associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Res. 40, 11571–11582 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rydberg B., Lindahl T., Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1, 211–216 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad R., Dianov G. L., Bohr V. A., Wilson S. H., FEN1 stimulation of DNA polymerase β mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem. 275, 4460–4466 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Fortini P. et al., Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry 37, 3575–3580 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Pommier Y., Sun Y., Huang S. N., Nitiss J. L., Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pommier Y., Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Strumberg D. et al., Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20, 3977–3987 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanza A., Tornaletti S., Rodolfo C., Scanavini M. C., Pedrini A. M., Human DNA topoisomerase I-mediated cleavages stimulated by ultraviolet light-induced DNA damage. J. Biol. Chem. 271, 6978–6986 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Subramanian D., Rosenstein B. S., Muller M. T., Ultraviolet-induced DNA damage stimulates topoisomerase I-DNA complex formation in vivo: Possible relationship with DNA repair. Cancer Res. 58, 976–984 (1998). [PubMed] [Google Scholar]
- 31.Yang S.-W. et al., A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 93, 11534–11539 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cortes Ledesma F., El Khamisy S. F., Zuma M. C., Osborn K., Caldecott K. W., A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461, 674–678 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Marteijn J. A., Lans H., Vermeulen W., Hoeijmakers J. H., Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 (2014). [DOI] [PubMed] [Google Scholar]
- 34.El-Khamisy S. F. et al., Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Hoch N. C. et al.; Care4Rare Canada Consortium , XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 541, 87–91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldecott K. W., Protein ADP-ribosylation and the cellular response to DNA strand breaks. DNA Repair (Amst.) 19, 108–113 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Zeng Z. et al., TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic Acids Res. 40, 8371–8380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori T. et al., Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 54, 225–232 (1991). [DOI] [PubMed] [Google Scholar]
- 39.Wittschieben B. O., Iwai S., Wood R. D., DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J. Biol. Chem. 280, 39982–39989 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Arlett C. F. et al., Clinical and cellular ionizing radiation sensitivity in a patient with xeroderma pigmentosum. Br. J. Radiol. 79, 510–517 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Pourquier P., Pommier Y., Topoisomerase I-mediated DNA damage. Adv. Cancer Res. 80, 189–216 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Dexheimer T. S., Pommier Y., DNA cleavage assay for the identification of topoisomerase I inhibitors. Nat. Protoc. 3, 1736–1750 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou E. W., Prasad R., Asagoshi K., Masaoka A., Wilson S. H., Comparative assessment of plasmid and oligonucleotide DNA substrates in measurement of in vitro base excision repair activity. Nucleic Acids Res. 35, e112 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asagoshi K. et al., DNA polymerase beta-dependent long patch base excision repair in living cells. DNA Repair (Amst.) 9, 109–119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y. et al., DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J. Biol. Chem. 280, 3665–3674 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Plo I. et al., Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair (Amst.) 2, 1087–1100 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Meulenbroek E. M. et al., UV damage endonuclease employs a novel dual-dinucleotide flipping mechanism to recognize different DNA lesions. Nucleic Acids Res. 41, 1363–1371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miura M., Domon M., Sasaki T., Kondo S., Takasaki Y., Two types of proliferating cell nuclear antigen (PCNA) complex formation in quiescent normal and xeroderma pigmentosum group A fibroblasts following ultraviolet light (uv) irradiation. Exp. Cell Res. 201, 541–544 (1992). [DOI] [PubMed] [Google Scholar]
- 49.Vrouwe M. G., Pines A., Overmeer R. M., Hanada K., Mullenders L. H., UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J. Cell Sci. 124, 435–446 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Li R., Hannon G. J., Beach D., Stillman B., Subcellular distribution of p21 and PCNA in normal and repair-deficient cells following DNA damage. Curr. Biol. 6, 189–199 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Takao M., Yonemasu R., Yamamoto K., Yasui A., Characterization of a UV endonuclease gene from the fission yeast Schizosaccharomyces pombe and its bacterial homolog. Nucleic Acids Res. 24, 1267–1271 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowman K. K. et al., A new ATP-independent DNA endonuclease from Schizosaccharomyces pombe that recognizes cyclobutane pyrimidine dimers and 6-4 photoproducts. Nucleic Acids Res. 22, 3026–3032 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasui A., Alternative excision repair pathways. Cold Spring Harb. Perspect. Biol. 5, a012617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okano S., Kanno S., Nakajima S., Yasui A., Cellular responses and repair of single-strand breaks introduced by UV damage endonuclease in mammalian cells. J. Biol. Chem. 275, 32635–32641 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Yasuhira S., Yasui A., Alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe operates both in nucleus and in mitochondria. J. Biol. Chem. 275, 11824–11828 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Kim N. et al., Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332, 1561–1564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim N., Jinks-Robertson S., The Top1 paradox: Friend and foe of the eukaryotic genome. DNA Repair (Amst.) 56, 33–41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang S. Y., Ghosh S., Pommier Y., Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J. Biol. Chem. 290, 14068–14076 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang S. N., Williams J. S., Arana M. E., Kunkel T. A., Pommier Y., Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks. EMBO J. 36, 361–373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams J. S. et al., Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell 49, 1010–1015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zimmermann M. et al., CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559, 285–289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walmacq C. et al., Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol. Cell 46, 18–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanz-Murillo M. et al., Structural basis of RNA polymerase I stalling at UV light-induced DNA damage. Proc. Natl. Acad. Sci. U.S.A. 115, 8972–8977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abeti R. et al., Xeroderma pigmentosum: Overview of pharmacology and novel therapeutic strategies for neurological symptoms. Br. J. Pharmacol. 176, 4293–4301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pontel L. B. et al., Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol. Cell 60, 177–188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burgos-Barragan G. et al., Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism. Nature 548, 549–554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Juarez E. et al., An RNAi screen in human cell lines reveals conserved DNA damage repair pathways that mitigate formaldehyde sensitivity. DNA Repair (Amst.) 72, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.