Significance
The preferential partitioning of single-span membrane proteins for ordered phase domains in phase-separated membranes is now reasonably well understood, but little is known about this phase preference for multispan helical membrane proteins. Here, it is shown that the disease-linked tetraspan membrane protein, PMP22, displays a pronounced preference to partition into the ordered phase, a preference that is reversed by disease mutations. This phase preference may be related to the role of PMP22 in cholesterol homeostasis in myelinating Schwann cells, a role that is also known to be disrupted under conditions of Charcot–Marie–Tooth disease (CMTD) peripheral neuropathy caused by pmp22 mutations.
Keywords: peripheral myelin protein 22, membrane phase domain, ordered
Abstract
The ordered environment of cholesterol-rich membrane nanodomains is thought to exclude many transmembrane (TM) proteins. Nevertheless, some multispan helical transmembrane proteins have been proposed to partition into these environments. Here, giant plasma membrane vesicles (GPMVs) were employed to quantitatively show that the helical tetraspan peripheral myelin protein 22 (PMP22) exhibits a pronounced preference for, promotes the formation of, and stabilizes ordered membrane domains. Neither S-palmitoylation of PMP22 nor its putative cholesterol binding motifs are required for this preference. In contrast, Charcot–Marie–Tooth disease-causing mutations that disrupt the stability of PMP22 tertiary structure reduce or eliminate this preference in favor of the disordered phase. These studies demonstrate that the ordered phase preference of PMP22 derives from global structural features associated with the folded form of this protein, providing a glimpse at the structural factors that promote raft partitioning for multispan helical membrane proteins.
Our current understanding of biological membranes has been shaped over the past 30 y by studies of membrane phase separation into ordered and disordered domains. Early on, these studies yielded the lipid raft hypothesis (1, 2), which was hotly debated in subsequent years (3–8). Based on a wealth of data it is now generally believed that phase separation does sometimes occur in sphingomyelin and cholesterol-rich membranes, such as in eukaryotic plasma membranes (PMs). While it is thought that phase-separated ordered membrane domains are often small in size, transient, and similar to the adjacent disordered phase in lipid composition (8–10), there also appear to be certain native membranes that are so cholesterol and sphingolipid-rich that their physical properties are, to a significant degree, akin to those of ideal liquid-ordered (Lo) phase model membranes (9, 11–13).
Early studies of membrane protein association with ordered membrane nanodomains were based largely on results involving the isolation of “detergent-resistant membranes” (14–17). More recent biophysical studies conducted in intact phase-separated membranes have confirmed that a number of single-pass transmembrane proteins do indeed have a preference to partition into ordered phase domains relative to surrounding disordered bilayers. Particularly important in this regard are studies that have employed “giant plasma membrane vesicles” (GPMVs), which can be formed from a variety of mammalian cell types (12). When GPMVs are isolated and then cooled, separation of microscopically observable ordered and disordered membrane phases can occur, enabling quantitative studies of protein partitioning between the two phases (3, 12, 18–21). GPMVs therefore provide facile experimental access to conditions in which large and stable ordered phase domains coexist with disordered membranes (18, 21, 22). Groundbreaking studies of single span membrane proteins in this medium led to development of a convincing quantitative model describing the structural basis for why some proteins of this class preferentially partition into the ordered phase (12, 20, 23–26). Studies of the phase preferences of multispan membrane proteins remain at a much earlier stage of development, with the exception of an important body of work for the perfringolysin O toxin (PFO), which autoinserts into membranes to form an oligomeric beta barrel in ordered phase domains (27–29). Here, we present an example of a helical multispan membrane protein that exhibits a pronounced preference for the ordered phase in GPMVs—the tetraspan peripheral myelin protein 22 (PMP22).
The human PMP22 is a 160-residue protein containing four transmembrane helices and intracellular N and C termini (Fig. 1A). PMP22 appears to play multiple roles in myelinating Schwann cells and peripheral myelin (30–35) including cholesterol homeostasis (36). This is especially important in Schwann cells given their specialized function as the factory for myelin production in the peripheral nervous system (PNS). Myelin membranes are unusually rich in both cholesterol and sphingolipids (37, 38) and are therefore highly ordered, as well suits their roles in providing electrical insulation and mechanical support to PNS axons. Mutations in the pmp22 gene result in >70% of all cases of Charcot–Marie–Tooth disease (CMTD, prevalence: 1:2,500) and related peripheral neuropathies (31, 32, 39). These closely related disorders are characterized by defective myelin membranes that contain altered cholesterol levels relative to healthy myelin (40–42).
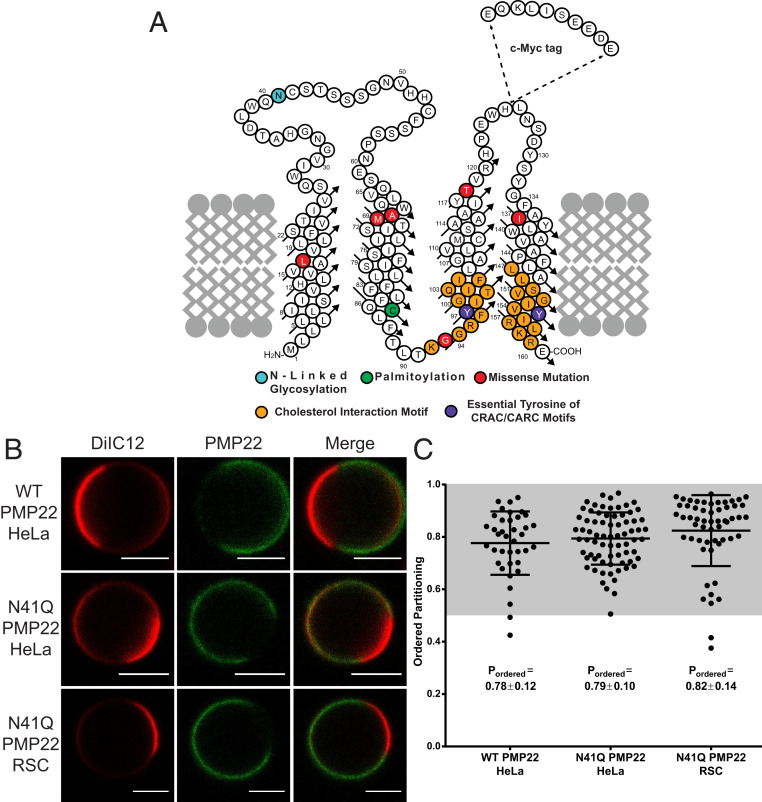
Fig. 1.
PMP22 partitions into ordered phase domains of GPMVs. (A) Cartoon topology map of PMP22 showing the locations of the myc-epitope tag, sites of posttranslational modifications (cyan, glycosylation; green, palmitoylation), cholesterol interaction motifs (orange) and their essential Tyr residues (purple), and the sites of the missense mutations examined in this study (L16P, A67T, M69K, G93R, I137V, and T118M; red). (B) Representative GPMVs containing WT or N41Q PMP22 derived either from HeLa cells or RSCs. (Scale bars, 5 µm.) The disordered phase marker is shown in red, and PMP22 is shown in green. (C) Quantification of PMP22 partitioning coefficients from three independent biological experiments for each condition with >10 GPMVs collected per replicate. Each point represents an individual GPMV. Mean ± SD is reported and plotted on graph.
The involvement of PMP22 in cholesterol trafficking as part of the process of myelin membrane formation suggests that this protein might have an intrinsic affinity for ordered membrane domains. Indeed, it has previously been reported that PMP22 is found in ordered membranes isolated from neurons following application of classical detergent-extraction methods (38, 43). However, some of these early methods of membrane domain isolation, including that used to identify PMP22 as an ordered-domain associated protein, are thought to be artifact-prone (6, 7, 14–16). Nevertheless, the likely role of PMP22 in Schwann cell cholesterol homeostasis combined with its residence in cholesterol and sphingolipid-rich myelin membranes suggest that the hypothesis that PMP22 may preferentially partition into ordered phase membranes has merit. This hypothesis is tested in this work.
Results
PMP22 Preferentially Partitions into Ordered Phase Membrane Domains of GPMVs.
To examine the preference of PMP22 for the ordered versus the disordered phase in plasma membranes (PM), we expressed human PMP22 (Fig. 1A) in HeLa cells and then prepared GPMVs using established protocols (21). To render PMP22 easily immunodetectable, the c-myc tag was inserted into the second extracellular loop, a modification that has no effect on PMP22 trafficking or function (44, 45). PMP22 in GPMVs was visualized using confocal fluorescence microscopy using an anti-myc antibody conjugated to AlexaFluor 647 (AF647, green). Disordered phase domains within GPMVs were identified using the fluorescent carbocyanine lipid DiIC12 (red), which partitions preferentially into disordered membrane domains (22). Fig. 1B shows representative PMP22-containing GPMVs. Within each phase PMP22 was uniformly distributed and showed no tendency to concentrate at the boundary between the ordered and disordered domains (Fig. 1B).
In our experiments we noticed an increase in the amount of PMP22-containing GPMVs when preparations were performed using cells overexpressing the N41Q N-glycosylation deficient variant of PMP22 (Fig. 1A, cyan) versus the WT protein. Eliminating glycosylation of PMP22 does not affect its function or turnover (46, 47). This observation is most likely due to an increased concentration of N41Q PMP22 at the PM compared to WT PMP22 (SI Appendix, Fig. S1). Representative N41Q PMP22-containing GPMVs derived from both HeLa cells and primary rat Schwann cells (RSCs) are also shown in Fig. 1B. Both glycosylated and nonglycosylated PMP22 show a clear preference for ordered phase membrane domains, as evidenced by the lack of colocalization of the red DiIC12 and green AF647 channels. PMP22 shows a distinct preference for ordered membrane domains in GPMVs prepared from both HeLa cells and RSCs.
AF647 fluorescence intensity was quantified in ordered and disordered GPMV membrane domains to determine the relative concentration of PMP22 in each membrane phase. Following image quantitation, the ordered domain partitioning fraction (Pordered) of PMP22 was calculated, where Pordered is [PMP22]ordered/([PMP22]ordered + [PMP22]disordered) and ranges from 0 to 1 with a value of 0.5, meaning the protein has equal affinity for both phases, while Pordered > 0.5 means that the protein prefers the ordered phase, and Pordered < 0.5 means that the protein prefers the disordered phase. For an example of GPMV analysis, see SI Appendix, Fig. S2 (25).
GPMVs from HeLa cells showed clear phase separation at 20 °C while those from RSCs exhibited phase separation at 15 °C. Quantification of Pordered for WT and N41Q PMP22 in GPMVs derived from HeLa cells as well as for N41Q PMP22 in GPMVs derived from RSCs is shown in Fig. 1C. Data were acquired from three independent biological experiments for each condition, and >10 GPMVs were analyzed per replicate. In HeLa GPMVs, WT PMP22 showed a Pordered of 0.78 ± 0.12 (mean ± SD) and N41Q PMP22 showed a Pordered of 0.79 ± 0.10. N41Q PMP22-containing RSC GPMVs displayed a Pordered of 0.82 ± 0.14. As a control, we measured the phase partitioning in both HeLa cells and RSC GPMVs of the well-studied mEGFP-labeled form of the single pass membrane protein, linker for activated T cells (tgLAT) (SI Appendix, Fig. S3). The measured Pordered values of 0.54 ± 0.11 and 0.55 ± 0.07 for tgLAT in GPMVs derived from HeLa and RSCs, respectively, are in line with those reported in the literature using the same GPMV preparation in rat basal leukemia cells (20, 24). Additionally, we measured Pordered for N41Q PMP22 in HeLa GPMVs using a different membrane phase marker, NBD-DSPE (SI Appendix, Fig. S4), which identifies ordered membrane phase domains (22). Using this marker, we calculated a Pordered of 0.82 ± 0.06. To ensure that potential antibody-induced dimerization of PMP22 was not impacting our partitioning results, we measured PMP22 phase partitioning using an AF647-labeled antigen-binding fragment (Fab; see SI Appendix, Fig. S4). While Pordered for PMP22 was slightly lower (Pordered = 0.71 ± 0.07) when the Fab was used for detection, PMP22 still strongly preferred ordered membrane domains, indicating that antibody-induced dimerization was not driving the observed phenomena. These results quantitatively demonstrate that PMP22 has a pronounced preference to partition into ordered membrane domains in GPMVs from both model mammalian cell lines and primary Schwann cells. Additionally, there were no significant differences in WT versus N41Q PMP22 phase partitioning indicating that this phase preference is not affected by N-linked glycosylation. In light of this result and because the level of surface expression for N41Q PMP22 made it easier to image, all subsequent experiments reported in this work utilized a N41Q PMP22 variant (hereto referred to as “PMP22” for the sake of simplicity). Moreover, because HeLa-derived GPMVs showed phase separation at a temperature closer to physiological levels and had higher transfection efficiencies compared to RSCs, all subsequent experiments utilized HeLa-derived GPMVs.
S-Palmitoylation of PMP22 Is Not a Significant Driver of Its Ordered Phase Preference.
It was recently shown that PMP22 is palmitoylated at Cys85 (Fig. 1A, green) (48). In that study, this posttranslational addition of a saturated fatty acid group on the cytosolic side of the protein was shown not to affect PMP22 processing/trafficking but was seen to be important for modulating epithelial cell shape and motility. For single-pass transmembrane proteins, it has repeatedly been shown that palmitoylation plays a significant role in mediating membrane phase partitioning; one study estimated that this modification contributes ∼0.5 kcal·mol−1 free energy per palmitoyl chain in favor or ordered phase partitioning (20, 24). The removal of palmitoylation from tgLAT disrupts the ordered phase preference of that protein and causes it to equally prefer both membrane phases (24). We therefore tested to see if palmitoylation affects PMP22 partitioning into ordered membrane phases.
To eliminate the palmitoylation of PMP22, we mutated Cys85 to an Ala residue. This mutation did not affect the localization of PMP22 in HeLa cells (SI Appendix, Fig. S1). We then measured the Pordered for C85A PMP22 in GPMVs. As seen in Fig. 2 A and B, palmitoylation did not dramatically affect the ordered phase preference of PMP22. We determined a Pordered for C85A PMP22 of 0.77 ± 0.10. This value is almost identical to that reported for WT and N41Q PMP22 in Fig. 1. Because the standard method for GPMV preparation requires the use of the reducing agent dithiothreitol (DTT;2 mM), we tested if PMP22 was still palmitoylated under these conditions. PMP22 transfected cells were incubated overnight with 100 mM 17-octadecynoic acid (17-ODYA), a palmitic acid analog containing an alkyne on the terminal carbon, which is known to be incorporated by thioesterases into palmitoylated proteins. Cells were then treated with or without 2 mM DTT for 90 min at 37 °C, lysed, and PMP22 was immunoprecipitated. We then added a biotin handle to palmitoylated proteins using biotin azide and classical “click” chemistry (48). Palmitoylated PMP22 was identified via Western blotting using an anti-biotin antibody (SI Appendix, Fig. S5). Palmitoylation was quantified as the intensity of the anti-biotin band over the anti-c-myc band and normalized to the amount of palmitoylation found in N41Q PMP22 samples without DTT treatment (Fig. 2C). As seen in Fig. 2C and SI Appendix, Fig. S5, treatment of HeLa cells for 90 min with 2 mM DTT did not affect PMP22 palmitoylation. As expected, C85A PMP22 was not palmitoylated in these experiments. These results show that S-palmitoylation of PMP22 is not a significant driver of ordered phase preference.
Fig. 2.
Palmitoylation is not required for PMP22 localization to the ordered phase. (A) Triple images are representative examples of PMP22-containing GPMVs from C85A/N41Q PMP22-transfected HeLa cells. (Scale bars, 5 µm.) (B) Quantification of C85A PMP22 partitioning coefficients from three independent biological experiments with >10 GPMVs collected per replicate. Mean ± SD is reported and plotted on the graph. (C) Quantification of the amount of palmitoylated PMP22 from three biological replicates based on Western blots shown in SI Appendix, Fig. S5. The amount of palmitoylated PMP22 from each sample is quantified by dividing the intensity from the biotin blot by the intensity of the myc blot and then normalized to the amount of palmitoylated PMP22 in the N41Q sample without DTT treatment.
Cholesterol Binding Motifs in PMP22 Do Not Mediate Its Ordered Phase Preference.
We next tested whether either or both of the predicted cholesterol binding sites in PMP22 play a role in its ordered phase preference. For the multispan beta barrel membrane protein perfringolysin O (PFO), it was shown that protein-associated sterols could alter the phase partitioning properties of the protein in phase-separated synthetic lipid vesicles (28). It has been shown that pmp22 −/− Schwann cells exhibit reduced plasma membrane levels and abnormal localization of cholesterol (33, 36). These cells also show reduced migration, adhesion, and lamellipodia extension, all of which can be restored through external supplementation of cholesterol in the culture media. PMP22 contains both a classical cholesterol-recognition amino acid consensus (CRAC) motif in TM4 (l-X1–5-Y-X1–5-K) and an inverted CRAC (CARC) motif in TM3 (K-X1–4-Y-X1–6-I), as illustrated in orange in Fig. 1A. While these motifs are loosely defined and are not always indicative of direct cholesterol interaction (49), there is substantial experimental and computational evidence supporting the notion that these motifs are sometimes directly involved in binding cholesterol (50, 51).
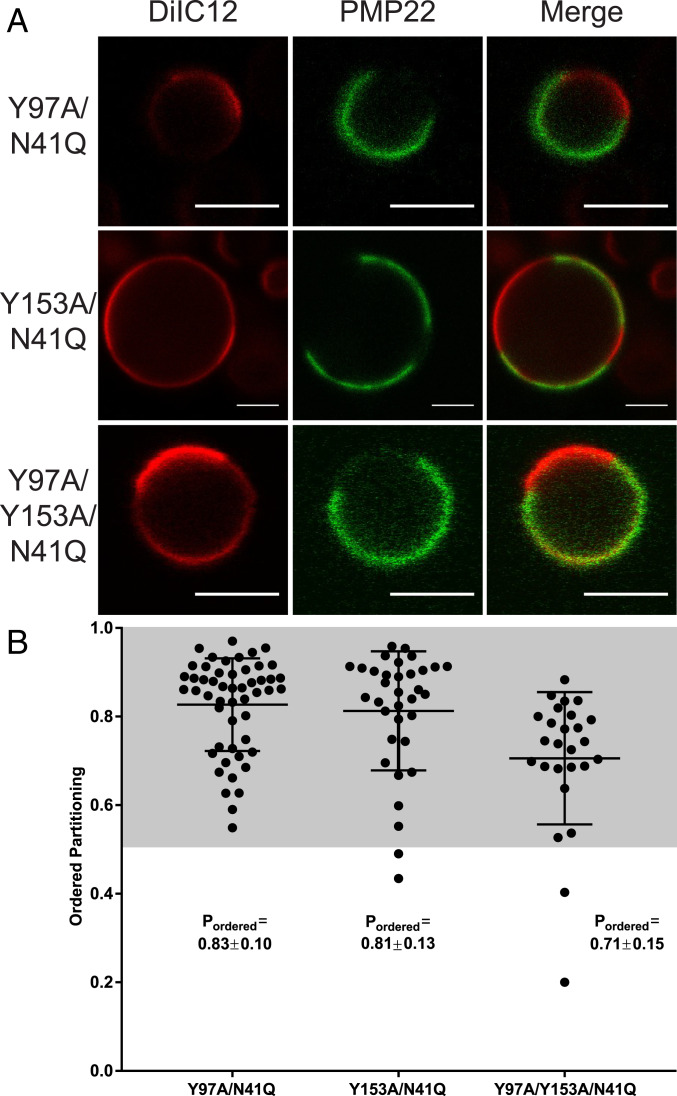
We mutated one or both of the essential Tyr residues in the CARC and CRAC motifs to Ala (Y97A, Y153A, and Y97A/Y153A mutants; Fig. 1A, purple). We then assessed the phase preference for each mutant in GPMVs. Mutation of Tyr97 had no effect on PM levels of PMP22, but mutation of Tyr153 or of both Tyr residues led to decreased PMP22 levels at the PM, suggesting lower expression and/or surface trafficking efficiency for these mutants (SI Appendix, Fig. S1). Nevertheless, the mutations did not significantly alter the ordered phase preference of PMP22 (Fig. 3 A and B). Y97A exhibited a Pordered of 0.83 ± 0.10, Y153A exhibited a Pordered of 0.81 ± 0.13, and the double mutant Y97A/Y153A yielded a Pordered of 0.71 ± 0.15. Pordered for the double mutant was only slightly reduced. We interpret these results to indicate that the presence of CRAC and/or CARC motifs are not significant drivers of the preference of PMP22 for ordered phase domains.
Fig. 3.
Cholesterol interaction motifs do not contribute to the ordered phase domain preference of PMP22. (A) Representative PMP22 containing GPMVs from Y97A/N41Q, Y153A/N41Q, and Y97A/Y153A/N41Q mutant forms of PMP22-transfected HeLa cells. (Scale bar, 5 µm.) (B) Quantification of PMP22 partitioning coefficients from three independent biological experiments with >10 GPMVs imaged per replicate. Mean ± SD is reported and plotted on the graph.
Phase Partitioning of Disease Mutant Forms of PMP22.
Since palmitoylation and cholesterol interaction motifs do not appear to play a major role in defining the phase preference of PMP22, we hypothesized that there is something intrinsic to the structure of the protein that drives its preferential association with the ordered phase. In order to test this hypothesis, we measured Pordered for a number of CMTD mutant forms of PMP22 (Fig. 1A). These different mutants were previously observed to demonstrate a range of in vitro conformational stabilities, plasma membrane trafficking efficiencies in cultured cells, and disease severity as quantitated by nerve conduction velocities in CMTD patients with these PMP22 variants (45).
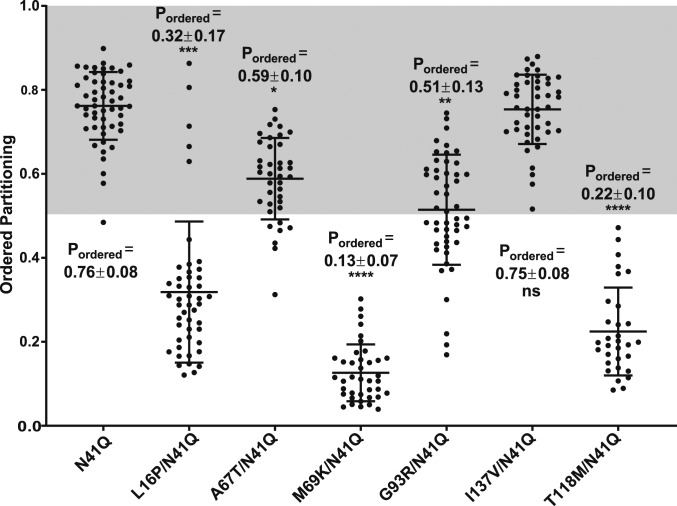
We first examined PMP22 containing a disease mutation in TM1: the L16P variant (Fig. 1A, red), which is also known as the “Trembler-J” mutation because of its mouse phenotype (31, 32, 45). Under WT/mutant heterozygous conditions, L16P PMP22 causes severe demyelination in both human and mice. Moreover, previous biophysical studies of L16P PMP22 showed that the L16P mutation introduces a flexible hinge in the TM1 helix, destabilizing the fully folded form of the protein and causing it to adopt an unfolded or folding-intermediate state in which TM1 is disassociated from TM2-4, which remain bundled, but only as a molten globule (45, 52, 53). Folding stability measurements in detergent micelles revealed L16P PMP22 to be destabilized compared to WT PMP22 with a ΔΔG of 3.3 ± 0.5 kcal·mol−1 (45). While introduction of the L16P mutation resulted in significantly reduced cell surface expression compared to PMP22 (SI Appendix, Fig. S1), we nevertheless were able to generate enough L16P PMP22-containing GPMVs to measure its phase preference—its Pordered was 0.32 ± 0.17 (Fig. 4; representative GPMV shown in SI Appendix, Fig. S6). The L16P mutation dramatically reverses the phase preference of PMP22 such that the protein now partitions preferentially into disordered membrane phase domains. This result led us to the hypothesize that the formation of stable tertiary structure in WT PMP22 is important for its preference to partition into ordered phase domains.
Fig. 4.
Phase domain partitioning of PMP22 missense mutants. Quantification of the PMP22 partitioning coefficients of WT-like glycosylation-deficient N41Q along with various missense mutants from three biological experiments with >10 GPMVs collected per replicate. Mean ± SD is reported and plotted on the graph. The nonparametric Mann–Whitney U test was used for all statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = not significant; all statistics compare missense mutations to N41Q PMP22. For representative GPMVs of each PMP22 mutant, see SI Appendix, Fig. S6.
To test this hypothesis, we measured Pordered for five additional PMP22 disease variants known to display a range of stabilities. The first three, M69K, G93R, and T118M PMP22 (mutation sites located in TM2, the intracellular loop, and TM4, respectively; Fig. 1A), are significantly destabilized compared to WT PMP22: M69K PMP22 exhibited a ΔΔG of 2.7 ± 0.5 kcal·mol−1, G93R PMP22 a ΔΔG of 2.9 ± 0.5 kcal·mol−1, and T118M a ΔΔG of 1.3 ± 0.6 kcal·mol−1 (45). The Pordered values for these variants showed significantly reduced ordered phase preferences relative to WT PMP22 (Fig. 4). M69K and T118M PMP22 were found to prefer disordered membrane phases with Pordered values of 0.13 ± 0.07 and 0.22 ± 0.10, respectively (Fig. 4; representative GPMV shown in SI Appendix, Fig. S6). G93R PMP22 was found to have no preference for either phase of the membrane, displaying a Pordered of 0.51 ± 0.13 (Fig. 4; representative GPMV shown in SI Appendix, Fig. S6). These results confirm that destabilized variants of PMP22 exhibit a reduced preference for ordered membrane phases.
We also examined the phase partitioning of two PMP22 variants that were only slightly destabilized, or not destabilized at all, compared to WT PMP22. In folding stability measurements, A67T and I137V PMP22 (located in TM2 and TM4, respectively; Fig. 1A) displayed ΔΔG values of −0.1 ± 0.9 kcal·mol−1 and 0.2 ± 0.7 kcal·mol−1 respectively—very similar to WT. In GPMVs, both of these PMP22 variants showed marked preference for ordered phase domains (Fig. 4). A67T PMP22 displayed a Pordered of 0.59 ± 0.10, a value only modestly reduced compared to that for WT PMP22 (Fig. 4; representative GPMV shown in SI Appendix, Fig. S6). The I137V variant exhibited a Pordered of 0.75 ± 0.08, which is almost identical to the Pordered measured for WT PMP22 in paired experiments, 0.79 ± 0.10 (Fig. 4; representative GPMV shown in SI Appendix, Fig. S6). These results show that variants of PMP22 that retain conformational stability still prefer ordered membrane domains over disordered domains. Taken together, these results indicate linkage between conformational stability and/or tertiary packing of PMP22, and the preference of the protein for ordered membrane domains.
PMP22 Alters the Biophysical Properties of GPMVs and Promotes Formation of Ordered Phase Domains.
Because it has been shown that PMP22 is critical for the formation of stable membrane domains in Schwann cells (33), we hypothesized that PMP22 may alter the stability of phase separation between ordered and disordered phase domains in GPMVs. To test this, we first determined the miscibility temperature (TMisc) of PMP22-containing GPMVs compared to GPMVs derived from cells transfected with an empty vector (“MOCK” conditions; Fig. 5A). TMisc is defined as the temperature at which 50% of the GPMVs exhibit phase separation (9, 12, 54). A higher TMisc suggests more stable phase-separated membrane domains. We collected images of >100 GPMVs at each temperature over temperatures ranging from 12.5 °C to 32.5 °C and calculated the fraction of phase-separated GPMVs at each temperature. Fitting this data to a sigmoidal curve allowed us to determine the TMisc of the GPMVs. Fig. 5B shows the TMisc seen for PMP22-containing GPMVs and for GPMVs from MOCK transfected cells. PMP22-containing GPMVs exhibited a TMisc of 20.2 ± 0.6 °C, whereas MOCK GPMVs showed a TMisc of 18.8 ± 0.4 °C. The TMisc seen for empty GPMVs was similar to what has previously been reported (9, 54). To validate that the increase in TMisc was not due to general overexpression of a TM protein at the plasma membrane, we measured the TMisc of tgLAT-containing GPMVs (SI Appendix, Fig. S7A). GPMVs containing tgLAT exhibited a TMisc of 18.4 ± 0.4 °C, similar to MOCK conditions (Fig. 5B), suggesting that the TMisc increase of PMP22-containing GPMVs was not due to generic membrane protein overexpression. Additionally, we measured the TMisc of L16P PMP22-containing GPMVs to assess if this increased domain stability was associated with PMP22 ordered domain partitioning. We found that L16P containing GPMVs had a TMisc of 19.2 ± 0.4 °C (Fig. 5B and SI Appendix, Fig. S7B). This value was not statistically different from GPMVs obtained from MOCK-transfected cells. These results indicate that WT PMP22, but not an unstable disease variant of the protein, stabilizes phase separation in GPMVs.
Fig. 5.
PMP22 alters the biophysical properties of GPMVs. (A) Percent phase separation of GPMVs containing WT-like N41Q PMP22 (green) or of GPMVs derived from cells transfected with an empty vector (MOCK; red) at various temperatures. Each point shows the average of three independent experiments, and the error bars represent the SEM. More than 100 GPMVs were measured at each temperature in each experiment. Plots are fit to a sigmoidal curve. (B) Calculation of the phase TMisc for GPMVs from MOCK-transfected cells, GPMVs containing glycosylation-deficient N41Q PMP22, L16P/N41Q PMP22, or tgLAT. TMisc is calculated from the fit of the sigmoidal curve for each independent experiment, and the SEM is calculated from the three replicates. The reported value represents the mean TMisc ± SEM. Significance was determined using a nonparametric Mann–Whitney U test comparing N41Q PMP22, L16P/N41Q PMP22, or tgLAT-containing GPMVs to MOCK GPMVs. (C) Ordered domain sizes (micrometers) from GPMVs obtained from cells transfected with either an empty vector (MOCK), GPMVs containing N41Q PMP22, or GPMVs containing L16P/N41Q PMP22. Data were obtained for three biological replicates for MOCK samples and L16P/N41Q PMP22 and six biological replicates for N14Q PMP22. The total number of vesicles measured is reported in the graph. Significance was determined using a nonparametric Mann–Whitney U test. *P ≤ 0.05; ns, not significant.
We also examined whether the presence of PMP22 increased the size of ordered phase domains in GPMVs. To do this, we imaged a large number of GPMVs derived from cells transfected with either an empty vector or one encoding WT or L16P PMP22 (three independent biological replicates with >30 GPMVs measured per replicate). For these images, we measured the radii of individual GPMVs as well as the fraction of each GPMV that contained the disordered phase maker DiIC12. From this information, we were able to calculate the relative size of ordered domains in GPMVs, as indicated in Fig. 5C (see SI Appendix, Fig. S8 for raw values). We found that GPMVs derived from cells transfected with an empty vector contained ordered domains with an average circumference of 11.8 ± 0.9 μm. GPMVs with PMP22 contained ordered domains with a circumference of 16.6 ± 0.1 μm, while GPMVs with L16P PMP22 contained ordered domains with a circumference of 11.9 ± 2.5 μm. These results show that PMP22, but not unstable L16P PMP22 causes a significant increase in ordered domain size and suggests that PMP22 is able to stabilize the ordered membrane domains of GPMVs. Combined with the fact that PMP22 increases the TMisc of GPMVs, we conclude that folded PMP22 is able to alter the biophysical properties of GPMVs to promote formation and stabilization of ordered phase domains.
Discussion
While there are a number of biochemical reports that identify multispan membrane proteins found to colocalize with biochemically isolated “detergent-resistant” cell fractions (55–57), this work quantitively demonstrates a preference to partition into ordered phase membrane domains by a multispan membrane protein in cell-derived GPMVs.
Factors Contributing to the Ordered Phase Domain Preference of PMP22.
Much effort has been devoted to understanding the driving forces of membrane phase preference for transmembrane proteins. In work conducted by the London laboratory, phase partitioning for the multispan beta barrel membrane protein PFO in synthetic lipid vesicles was found to be dependent on lipid composition, protein-associated sterols, pH, and hydrophobic matching between the protein TM segments and bilayer width (27–29). In the Levental group, previous work culminated in an elegant paper that provided a quantitative model describing the biochemical and biophysical features promoting ordered phase domain partitioning for single-pass transmembrane proteins (20). Ordered phase domain partitioning of single-pass proteins is promoted by increasing length for the transmembrane helix, by the presence of one or more palmitoyl chains, and by the presence of small amino acid side chains in the fully membrane-exposed TM segment, which reduces the exposed surface area in the plane of the membrane, especially for the half that occupies the exoplasmic bilayer leaflet. Whether components of this model can be extrapolated to ordered phase-preferring multispan membrane proteins is not yet clear. However, this study of PMP22 represents an important step in exploring this question.
The results demonstrated that the tetraspan integral membrane protein PMP22 has a distinct preference to partition into ordered phase membrane domains of GPMVs derived from both HeLa and primary Schwann cells. Unlike ordered phase-preferring single-pass transmembrane proteins (20, 23, 24), partitioning of PMP22 is not driven by palmitoylation. Although native PMP22 is palmitoylated, it retains its strong preference for the ordered phase even under conditions in which its palmitoylation site is mutated away. Our results demonstrate that this modification is not required for the ordered membrane phase preference of PMP22, as it seems to be for single-pass membrane proteins. Ordered phase partitioning of PMP22 was also found not to be associated with the putative CARC and CRAC cholesterol binding motifs present in PMP22. This suggests that unlike PFO, sterol binding does not drive PMP22 phase partitioning although we cannot rule out the possibility that PMP22 associates with cholesterol through mechanisms independent of the CRAC/CARC motifs.
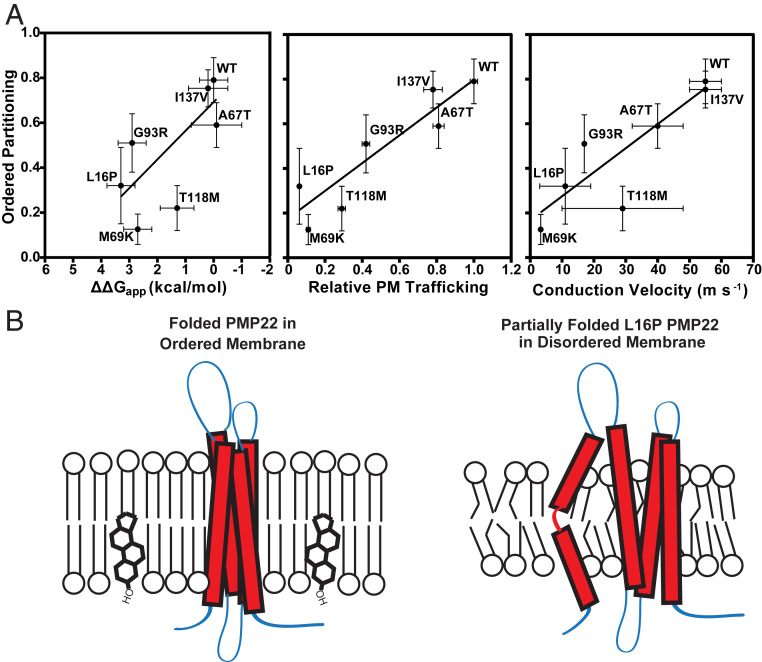
We observed that the conformational stability of the folded form of PMP22 plays a major role in mediating the phase preference of PMP22. Introduction of the destabilizing L16P mutation in the middle of TM1 reversed the phase preference of PMP22 so that it now favors the disordered phase. We have previously shown that the L16P mutant converts the straight and uninterrupted WT TM1 helical segment into a pair of helices linked by a flexible hinge (52, 53). This causes TM1 to dissociate from the other TM helices to favor a destabilized form of the protein in which TM2, TM3, and TM4 remain in contact as a molten globular bundle while TM1 is dissociated in the membrane, tethered to the rest of the protein by the TM1-TM2 loop. While we do not have the same in-depth structural information on M69K, G93R, and T118M PMP22, it has previously been shown that these mutations also destabilize the conformational stability of PMP22. Here, we find these mutations also decrease the affinity of PMP22 for ordered membrane domains, causing the disease variant forms of the protein to have either no preference for either phase (G93R) or to adopt a preference for disordered membrane phases (M69K and T118M). The A67T and I137V mutations, which either marginally destabilize or have no effect on the conformational stability of PMP22, retained a WT-like preference to partition into ordered phase membrane domains, further supporting our hypothesis about the role of conformational stability in the phase preference of PMP22.
In Fig. 6A, we plot Pordered for the seven forms of PMP22 examined in this work versus conformational stability, plasma membrane trafficking, and CMTD severity (as measured by nerve conduction velocity in patients with these single nucleotide polymorphisms) (45). We observed a strong positive correlation between stability, plasma membrane trafficking, and nerve conduction velocity with ordered phase partitioning of PMP22. Protein stability reflects the equilibrium constant between the folded conformation of a protein and the unfolded form. As illustrated in Fig. 6B, stable forms of PMP22, such as wild type, preferentially partition into ordered membrane domains, whereas unstable forms of PMP22, such as the L16P disease mutant, prefer the disordered phase. It is now clear that the protein folding quality control system of the endoplasmic reticulum (ER) has mechanisms for recognizing and retaining unstable forms of PMP22, eventually leading to ERAD pathway degradation of the protein. However, how quality control recognizes unstable PMP22 is not yet well understood (58). It is noteworthy that while the ER is the site for cholesterol biosynthesis in the cell, its membrane has only modest quantities of cholesterol. Most cholesterol is exported on to the Golgi apparatus (which has roughly 2× higher cholesterol in its membranes) and from there on to the plasma membrane (roughly 6× higher) (59). Given that ordered membrane domains in cells are cholesterol-rich, the results of this work lead us to speculate that one of the mechanisms that promotes “escape” of stable (predominately folded) forms of PMP22 from ER quality control may be its partitioning into cholesterol-rich ordered domains that are then likely to traffic on to the Golgi and thence to the plasma membrane.
Fig. 6.
Ordered phase domain partitioning correlates with PMP22 stability, trafficking, and CMTD severity. (A) Pordered determined for the PMP22 mutants examined in this study is plotted against tertiary structure stability (ΔΔG, relative to WT), relative plasma membrane trafficking efficiency (compared to WT), and CMTD severity (as reported by the nerve conduction velocity measured in patients harboring these PMP22 variants). Stabilities (ΔΔG), plasma membrane trafficking efficiencies, and nerve conduction velocities were obtained from ref. 45. Linear regression analysis in GraphPad was used to show correlations. (B) Model for preferential partitioning of folded and unfolded forms of PMP22 into distinct membrane domains. PMP22 that forms stable folded structure within the membrane partitions preferentially into ordered domains. PMP22 that is unstable, such as the L16P variant, and fails to adopt a stable tertiary fold partitions preferentially into disordered membrane domains.
That folded PMP22 favors ordered phase domains makes sense in light of the Levental model for single-span membrane proteins and with results from the London group on PFO. In the Levental model, one factor mediating membrane phase preference is the exposed protein surface area in the plane of the membrane. Minimizing this feature in a protein promotes ordered phase partitioning. Multispan helical membrane proteins that adopt a stable tertiary fold should have less exposed surface in the plane of the membrane compared to unfolded forms, which may promote ordered phase partitioning. Of course, not all folded multispan membrane proteins preferentially partition into ordered phase domains. This indicates that the folded structure of PMP22 has distinctive traits that favor ordered phase partitioning. For PFO, the London group discovered that hydrophobic matching of the length of its TM beta strands with bilayer width played a large role in phase partitioning (29). Therefore, forms of PFO with longer hydrophobic beta-strands more strongly prefer ordered membrane phases, which are slightly thicker than the surrounding disordered membrane phase. Examination of a homology/Rosetta model for the structure of PMP22 (52) suggest that two of its transmembrane helices may be longer than average (at least 26 residues each) and that the transmembrane domain has a fairly featureless surface. While there does not seem to the general preponderance of residues with small side chains in the exoplasmic half of the PMP22 transmembrane domain as appears to be a feature of ordered phase-preferring single-pass membrane proteins (20, 60), the presence of a Ser-Ala-Ala-Ala segment at the exoplasmic end of TM3 is intriguing. The intracellular “domain” of PMP22 is another distinctive feature, being comprised only of the N-terminal amino group of Met1, a four-residue loop connecting TM2 and TM3, and four charged residues that follow TM4. Testing whether any of these features are contributing factors to PMP22’s ordered phase preference will require many additional experiments, which we hope will be motivated by the results of this paper.
The Preference of PMP22 for Ordered Phase Membrane Domains in Cell-Derived GPMVs Does Not Extend to Lo Phase Domains in Synthetic Lipid Vesicles.
In previous work, we showed that purified recombinant PMP22 can be reconstituted into giant unilamellar vesicles (GUVs) containing synthetic lipids (61). Under GUV conditions in which the synthetic lipids separated into ideal liquid-disordered (LD) and Lo lipid phases, it was observed that PMP22 partitioned exclusively to the disordered LD phase. Why are the results for PMP22 partitioning in GPMVs at odds with what was observed in GUVs? Our current results show that neither of the known posttranslational modifications of PMP22, N-glycosylation at N41 or S-palmitoylation at C85, are required for the ordered phase preference of PMP22 in GPMVs. This allows us to rule out the possibility that the lack of posttranslational modifications of the recombinant PMP22 used in the earlier GUV studies is the basis for its LD phase preference. We suggest instead that the variance between the results from the GPMV and GUV studies point to the fact that the difference in order between phase-separated domains in cell-derived GPMVs is much reduced relative to GUVs comprised of a well-defined ternary mixture of synthetic lipids. This phenomenon has previously been documented for single-span membrane proteins (8, 18, 62). Given that the Pordered for PMP22 in GPMVs was seen in this work to be nearly 0.8 means, the energy by which PMP22 favors the ordered phase over the disordered phase in GPMVs is on the order of –RTln(4) = −0.8 kcal·mol−1. One can easily imagine that the highly ordered packing that occurs in ideal Lo phases (but only to a much lesser degree in GPMVs) would need to be disrupted to accommodate partitioning of a membrane protein and that this unfavorable energy contribution could easily reverse the overall energetics of partitioning in GUVs to favor the LD phase.
PMP22 Stabilizes Ordered Phase Domains and Promotes Their Formation.
While some proteins are thought to passively associate with raft-like ordered domains, others can actively promote their formation by clustering raft components and stabilizing ordered domains (25, 63). Proteins that modulate membrane order or fluidity also are capable of regulating phase separation (64, 65). Here, we identify PMP22 as an example of a protein that can directly stabilize ordered phase membrane domains.
PMP22-containing GPMVs exhibited a higher TMisc than GPMVs containing unstable L16P PMP22, tgLAT, or cells transfected with an empty vector (Fig. 5 A and B and SI Appendix, Fig. S7). That phase separation persists at higher temperatures in GPMVs containing PMP22 suggests that this protein can directly stabilize ordered phase membrane domains (9). This is consistent with previous results from studies of pmp22 −/− mice showing that the distribution of molecules typically associated with ordered phase membrane domains (such as cholesterol and GM1 ganglioside) are decreased at the plasma membrane (33). Moreover, Schwann cells isolated from these mice showed elongation and migration defects that could be corrected by external supplementation of the culture medium with cholesterol. Additionally, our results suggest that PMP22 is able to promote ordered domain formation. We showed that GPMVs containing PMP22 had ordered membrane domain circumferences on average ∼5 μm larger than those in GPMVs without PMP22 or unstable L16P PMP22 (Fig. 5C). This may be due to an increased concentration in cholesterol in PMP22-containing GPMVs since it has recently been shown that PMP22 regulates cholesterol PM trafficking (36). It seems likely that the mechanisms underpinning this regulatory function of PMP22, as well as its ability to promote ordered phase formation, is closely related to its preference to partition into ordered membrane phase domains.
Conclusions
We have documented PMP22 as a multispan helical membrane protein to exhibit a preference to partition into the ordered phase of cell-derived GPMVs. This phase preference appears to be closely linked to the formation of correct tertiary structure of the protein. Additional experiments will be required to determine exactly what features of its folded structure confer its preference for the ordered phase. Moreover, it remains unclear just how many other multispan helical membrane proteins will share the phase domain preference of PMP22 and whether they will resemble PMP22 in terms of driving traits. It is hoped that the results of this work will inspire future studies to address these issues.
Materials and Methods
Materials.
DiIC12 was purchased from Life Technology. NBD-PE was purchased from Avanti. Anti-myc AlexaFluora-647, anti-biotin, and anti-myc were purchased from Cell Signaling Technologies. Anti-myc AlexaFluora-647 labeled antigen binding fragment (Fab) was purchased from Promega. DTT, Tris(2-carboxyethyl)phosphine (TCEP), and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) were purchased from Research Products International; DTT was prepared fresh for every use. Paraformaldehyde (PFA) was purchased as a 16% stock solution from Electron Microscopy Sciences. CaCl2, NaCl, CuCl2, Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA), anti-myc magnetic beads, and propidium iodide were purchased from Fisher Scientific. 17-ODYA and biotin-azide were purchased from Cayman Chemicals. HeLa cell lines were acquired from American Type Culture Collection. Primary RSCs were a generous gift from the laboratory of Bruce Carter at Vanderbilt University (Nashville, TN).
Cloning.
Human cDNA for PMP22 was subcloned into a pCDNA3.1 mammalian expression vector. To make PMP22 immunologically detectable, we used QuikChange mutagenesis to insert a myc epitope into the second extracellular loop of PMP22 within the pCDNA3.1 vector (45). QuikChange mutagenesis was also used to make the various point mutations used in this study. Plasmids were purified using a GenElute HP Plasmid MidiPrep Kit (Sigma-Aldrich). The tgLAT construct (20) used in these studies was a generous gift from the laboratory of Dr. Ilya Levental at the McGovern Medical School, University of Texas (Houston, TX).
Cell Culture and Transfections.
HeLa and RSCs were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% pen/strep at 37 °C and 5% CO2. Culture medium for RSCs was supplemented with 2 µM forskolin (Sigma-Aldrich). Approximately 24 h prior to transfection, cells were plated so as to be 40–50% confluent at the time of transfection. Cells were transfected using FuGene Transfection Reagent (Promega) with a FuGene:DNA ratio of 3:1 in OptiMEM. Plates (6 cm2) were transfected with 1.5 µg of DNA. The transfection medium was removed from cells ∼12–15 h posttransfection, and cells were washed with Dulbecco's phosphate-buffered saline and fresh culture media was added to each plate.
GPMV Preparation.
Thirty-six hours after transfection, the medium was removed from cells and cells were washed three times with inactive GPMV buffer (10 mM Hepes, 150 mM NaCl, 2 mM CaCl2 pH 7.4). Cells were consistently 70–80% confluent at the time of GPMV prep. Active GPMV buffer (GPMV buffer plus 2 mM DTT and 25 mM formaldehyde) was then added to the plates, and cells were incubated at 37 °C with gentle shaking (70 rpm) for 90 min. DiIC12 or NBD-PE was then added to the plates from a stock solution of 0.5 mg/mL in EtOH to a final concentration of 0.5 µg/mL, and cells were gently rocked at room temperature for 15 min. The GPMV-containing supernatant was then decanted into 1.5-mL Eppendorf tubes, and an anti-myc AF647 mAb was added to the solution (1:750 μL dilution) and gently agitated in the dark at room temperature for at least 3 h. GPMVs were then allowed to settle in the dark to the bottom of the tube at 4 °C for 2–24 h (we observed no difference in GPMV quality whether we imaged immediately or at 24-h post-GPMV prep). Thirty minutes prior to imaging, 270 µL of GPMV solution was pipetted from the bottom of the Eppendorf tube and sandwiched between two coverslips coated with 0.1% bovine serum albumin and separated by a 0.5-mm-thick silicone isolator (Electron Microscopy Sciences).
GPMV Imaging.
GPMVs were imaged using a Zeiss LSM 510 confocal microscope using a 1.2 N.A. Zeiss Plan-Neofluor 40× objective. The confocal pinhole was set to 150 nm for all experiments. The fluorophores were excited using the 488-nm line of a 40 mW argon laser (NBD-PE, mEGFP, and AF-488), the 543-nm line of a HeNe laser (DiIC-12, propidium iodide), or the 633-nm line of a HeNe laser (AF-647). Images were collected at a 1× digital zoom for the case of miscibility temperature measurements and at 8–10× digital zoom for quantifying phase partitioning with a 512 × 512-pixel resolution. The stage was cooled using a Linkan Peltier Cooling system.
Quantifying GPMV Phase Partitioning.
SI Appendix, Fig. S2 shows a representative example of how Pordered was calculated. Briefly, GPMVs were labeled with either a disordered membrane phase marker (DiIC-12) or an ordered membrane phase marker (NBD-PE). PMP22-containing GPMVs were then labeled with anti-myc AF647-labeled antibodies. To determine the phase partitioning of PMP22, GPMVs were imaged in the green or red (NBD-PE or DiIC-12, respectively) and far-red channels, sequentially. To determine the phase partitioning of tg-LAT, GPMVs were imaged in the green and red channels sequentially. Line scans across a single GPMV were performed in all channels using the ImageJ software to determine the fluorescent intensity at every pixel. The position of the line was set so that it intersected with both an ordered and disordered region of the GPMV using the DiIC12 or NBD-PE channels as the references. This same line was used to measure the intensity in the protein (PMP22 or tgLAT) channel. The line scans were smoothened using a moving average (10 pixels) in Microsoft Excel. Ordered phase domain partitioning, Pordered was then calculated as previously described (25) as
| [1] |
where Iordered and Idisordered are the fluorescence intensity of the protein channels in the ordered and disordered phases, respectively. Three independent lines were chosen for each GPMV, and the mean Pordered was calculated and reported for individual GPMVs.
Quantification of Palmitoylation Using “Click” Chemistry.
Approximately 24 h after cells were transfected, cells were incubated overnight in media containing 100 μM 17-ODYA and a 1% final concentration of DMSO (or just DMSO for no 17-ODYA control). Cells were then incubated for 90 min with or without 2 mM DTT. Following incubation, cells were lysed for 1 h at 4 °C in 150 μL of lysis buffer (50 mM Tris, 150 mM NaCl, 0.3% CHAPS, 0.1% SDS, pH 7.4). Lysates were cleared via centrifugation for 15 min at 14,000 × g. Protein concentrations were determined via Bradford assay, and 75 μg of total protein was added to 10 μL of anti-myc conjugated magnetic beads for each lysate. Volumes for each lysate were brought up to 150 μL total in lysis buffer, and beads and lysates were incubated with end-over-end rotation at 4 °C overnight. The following day, beads were washed three times with lysis buffer and bound proteins were eluted with 25 μL of elution buffer (50 mM Hepes, 150 mM NaCl, 2% SDS pH 7.0) and transferred to fresh tubes. The following was added to the eluents: biotin azide to a final concentration of 20 μM, TCEP to a final concentration of 1 μM, 20 μM TBTA, and 2 μM CuSO4. Reactions were mixed at room temperature with end-over-end rotation for 2 h before being quenched via the addition of ethylenediaminetetraacetic acid to a final concentration of 1 mM. Samples were then split in two and analyzed via Western blotting for PMP22 (1:8,000 dilution of c-myc antibody) and biotin (1:1,000 dilution of biotin antibody).
Measurement of TMisc.
Tile scans (5 × 5) of GPMV samples were imaged at 1× digital zoom at temperatures ranging from 12.5 °C to 32.5 °C. Images were then randomized and GPMVs were blindly and manually classified as being either phase-separated or containing a single uniform phase. The fraction of vesicles that were phase separated at each temperature was then calculated. Plotting %-phase separated versus temperature yielded a curve that was fit to a sigmoidal function, and the TMisc was defined as the temperature at which 50% of GPMVs were phase separated. Three independent biological experiments were performed, and >100 GPMVs were imaged and classified for each temperature of each repeat. Classifications were performed blindly to the temperature at which the images were collected.
Fixed-Cell Imaging.
Cells were cultured on glass coverslips and transfected as described above. Thirty-six hours posttransfection, cells were fixed with 4% PFA and permeabilized with 0.25% Triton X-100. Cells were treated with 1 μg/mL RNase (Sigma-Aldrich). PMP22 was then detected with mouse anti-myc (1:500) and visualized with donkey anti-mouse AF-488 (Invitrogen; 1:500). Nuclei were stained with propidium iodide (Sigma-Aldrich; 0.33 µg/mL). Coverslips were mounted to a slide using ProLong Gold (Invitrogen) and allowed to dry for 24 h. Cells were then imaged using the 40× objective on the Zeiss LSM 510 confocal microscope at an optical zoom of 3–4×. The optical slice was set to <1 µM in each image.
Calculation of GPMV Ordered Domain Size.
Images of GPMVs derived from cells transfected either with an empty or N41Q PMP22 pCDNA3.1 vector were collected in a high-throughput manner on an ImageXpress Micro XL (Molecular Devices) using a 40× objective. MATLAB (MathWorks) was used to calculate the radius of each GPMV in pixels, and the percentage of GPMVs in the ordered phase using the DiIC12 dye to mark the disordered phase. The radius was converted from pixel to micrometer using the conversion factor of 0.34 μm:1 pixel for the 40× objective. The circumference of each GPMV was then calculated using the equation: circumference = 2π*radius, and ordered domain size was calculated by multiplying the fraction of each GPMV that was in the ordered phase by its circumference.
Statistical Analysis.
The nonparametric Mann–Whitney U test was used to compare pairs of data. Significance is reported when determined. GraphPad Prism was used to perform all statistical analysis.
Data Availability.
Raw data for all GPMV plots shown are available in Dataset S1: “Raw Values for GPMV Partitioning Plots.” Raw data used to plot and calculate TMisc can be found in Dataset S2: “Values for TMisc Plots.”
Supplementary Material
Acknowledgments
We thank Dr. Krishnan Raghunathan and Dr. Ajit Tiwari for introduction to the GPMV technique, Arina Hadziselimovic for assistance with cloning, the Vanderbilt Cell Imaging Shared Resource (CISR) for access to the confocal microscope, the Vanderbilt Institute of Chemical Biology and High Throughput Screening facility, Dr. Geoffrey Li for helpful discussion and comments on the mansuscript, Dr. Hui Huang for access and assistance with the ImageExpress, the laboratory of Bruce Carter at Vanderbilt for harvesting and help culturing RSCs, and Dr. Melanie Ohi (University of Michigan) for critical early revisions to the manuscript. This work was supported by NIH Grant R01 NS095989 (to C.R.S.) and a Vanderbilt Stanley Cohen Award (to C.R.S. and A.K.K.). J.T.M. was supported by NIH Fellowship F31 NS113494 and NIH Training Grant T32 NS00749. The CISR is supported by NIH Grants CA68485, DK20593, DK58404, DK59637, and EY08126.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000508117/-/DCSupplemental.
References
- 1.Simons K., Ikonen E., Functional rafts in cell membranes. Nature 387, 569–572 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Brown D. A., London E., Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14, 111–136 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Kraft M. L., Plasma membrane organization and function: Moving past lipid rafts. Mol. Biol. Cell 24, 2765–2768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veatch S. L., Keller S. L., Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 85, 3074–3083 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ackerman D. G., Feigenson G. W., Lipid bilayers: Clusters, domains and phases. Essays Biochem. 57, 33–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng X., Smith J. C., Biological membrane organization and cellular signaling. Chem. Rev. 119, 5849–5880 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Sezgin E., Levental I., Mayor S., Eggeling C., The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levental I., Veatch S., The continuing mystery of lipid rafts. J. Mol. Biol. 428, 4749–4764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veatch S. L. et al., Critical fluctuations in plasma membrane vesicles. ACS Chem. Biol. 3, 287–293 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Honerkamp-Smith A. R., Veatch S. L., Keller S. L., An introduction to critical points for biophysicists; observations of compositional heterogeneity in lipid membranes. Biochim. Biophys. Acta 1788, 53–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gielen E. et al., Rafts in oligodendrocytes: Evidence and structure-function relationship. Glia 54, 499–512 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Baumgart T. et al., Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 104, 3165–3170 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aureli M., Grassi S., Prioni S., Sonnino S., Prinetti A., Lipid membrane domains in the brain. Biochim. Biophys. Acta 1851, 1006–1016 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Brown D. A., Preparation of detergent-resistant membranes (DRMs) from cultured mammalian cells. Methods Mol. Biol. 1232, 55–64 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Brown D. A., Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 21, 430–439 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Heerklotz H., Triton promotes domain formation in lipid raft mixtures. Biophys. J. 83, 2693–2701 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K., Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. U.S.A. 100, 5795–5800 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levental K. R., Levental I., Giant plasma membrane vesicles: Models for understanding membrane organization. Curr. Top. Membr. 75, 25–57 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Lorent J. H., Levental I., Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem. Phys. Lipids 192, 23–32 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Lorent J. H. et al., Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 8, 1219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sezgin E. et al., Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 7, 1042–1051 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Klymchenko A. S., Kreder R., Fluorescent probes for lipid rafts: From model membranes to living cells. Chem. Biol. 21, 97–113 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Diaz-Rohrer B. B., Levental K. R., Simons K., Levental I., Membrane raft association is a determinant of plasma membrane localization. Proc. Natl. Acad. Sci. U.S.A. 111, 8500–8505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levental I., Lingwood D., Grzybek M., Coskun U., Simons K., Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 22050–22054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raghunathan K. et al., Glycolipid crosslinking is required for cholera toxin to partition into and stabilize ordered domains. Biophys. J. 111, 2547–2550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengupta P., Hammond A., Holowka D., Baird B., Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochim. Biophys. Acta 1778, 20–32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson L. D., Johnson A. E., London E., How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: Insights into the origin of perfringolysin O-lipid raft interaction. J. Biol. Chem. 283, 4632–4642 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Lin Q., London E., Transmembrane protein (perfringolysin o) association with ordered membrane domains (rafts) depends upon the raft-associating properties of protein-bound sterol. Biophys. J. 105, 2733–2742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Q., London E., Altering hydrophobic sequence lengths shows that hydrophobic mismatch controls affinity for ordered lipid domains (rafts) in the multitransmembrane strand protein perfringolysin O. J. Biol. Chem. 288, 1340–1352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snipes G. J., Suter U., Welcher A. A., Shooter E. M., Characterization of a novel peripheral nervous system myelin protein (PMP-22/SR13). J. Cell Biol. 117, 225–238 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Parker B., Martyn C., Natarajan C., Guo J., The PMP22 gene and its related diseases. Mol. Neurobiol. 47, 673–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Paassen B. W. et al., PMP22 related neuropathies: Charcot-Marie-Tooth disease type 1A and hereditary neuropathy with liability to pressure palsies. Orphanet J. Rare Dis. 9, 38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S. et al., PMP22 is critical for actin-mediated cellular functions and for establishing lipid rafts. J. Neurosci. 34, 16140–16152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jetten A. M., Suter U., The peripheral myelin protein 22 and epithelial membrane protein family. Prog. Nucleic Acid Res. Mol. Biol. 64, 97–129 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Mittendorf K. F. et al., Peripheral myelin protein 22 alters membrane architecture. Sci. Adv. 3, e1700220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y. et al., PMP22 regulates cholesterol trafficking and ABCA1-mediated cholesterol efflux. J. Neurosci. 39, 5404–5418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saher G. et al., High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 8, 468–475 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Gopalakrishnan G. et al., Lipidome and proteome map of myelin membranes. J. Neurosci. Res. 91, 321–334 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Suter U., Scherer S. S., Disease mechanisms in inherited neuropathies. Nat. Rev. Neurosci. 4, 714–726 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Larrouquère-Régnier S., Boiron F., Darriet D., Cassagne C., Bourre J. M., Lipid composition of sciatic nerve from dysmyelinating trembler mouse. Neurosci. Lett. 15, 135–139 (1979). [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y. et al., A neutral lipid-enriched diet improves myelination and alleviates peripheral nerve pathology in neuropathic mice. Exp. Neurol. 321, 113031 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Fledrich R. et al., Targeting myelin lipid metabolism as a potential therapeutic strategy in a model of CMT1A neuropathy. Nat. Commun. 9, 3025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasse B., Bosse F., Müller H. W., Proteins of peripheral myelin are associated with glycosphingolipid/cholesterol-enriched membranes. J. Neurosci. Res. 69, 227–232 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Liu N., Yamauchi J., Shooter E. M., Recessive, but not dominant, mutations in peripheral myelin protein 22 gene show unique patterns of aggregation and intracellular trafficking. Neurobiol. Dis. 17, 300–309 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Schlebach J. P. et al., Conformational stability and pathogenic misfolding of the integral membrane protein PMP22. J. Am. Chem. Soc. 137, 8758–8768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan M. C., Notterpek L., Tobler A. R., Liu N., Shooter E. M., Role of the peripheral myelin protein 22 N-linked glycan in oligomer stability. J. Neurochem. 75, 1465–1474 (2000). [DOI] [PubMed] [Google Scholar]
- 47.D’Urso D., Ehrhardt P., Müller H. W., Peripheral myelin protein 22 and protein zero: A novel association in peripheral nervous system myelin. J. Neurosci. 19, 3396–3403 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoltewicz S. J. et al., The palmitoylation state of PMP22 modulates epithelial cell morphology and migration. ASN Neuro 4, 409–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song Y., Kenworthy A. K., Sanders C. R., Cholesterol as a co-solvent and a ligand for membrane proteins. Protein Sci. 23, 1–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fantini J., Di Scala C., Baier C. J., Barrantes F. J., Molecular mechanisms of protein-cholesterol interactions in plasma membranes: Functional distinction between topological (tilted) and consensus (CARC/CRAC) domains. Chem. Phys. Lipids 199, 52–60 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Di Scala C. et al., Relevance of CARC and CRAC cholesterol-recognition motifs in the nicotinic acetylcholine receptor and other membrane-bound receptors. Curr. Top. Membr. 80, 3–23 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Mittendorf K. F., Kroncke B. M., Meiler J., Sanders C. R., The homology model of PMP22 suggests mutations resulting in peripheral neuropathy disrupt transmembrane helix packing. Biochemistry 53, 6139–6141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakakura M., Hadziselimovic A., Wang Z., Schey K. L., Sanders C. R., Structural basis for the Trembler-J phenotype of Charcot-Marie-Tooth disease. Structure 19, 1160–1169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levental I., Grzybek M., Simons K., Raft domains of variable properties and compositions in plasma membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 108, 11411–11416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohamed A., Robinson H., Erramouspe P. J., Hill M. M., Advances and challenges in understanding the role of the lipid raft proteome in human health. Expert Rev. Proteomics 15, 1053–1063 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Minogue S., Waugh M. G., Lipid rafts, microdomain heterogeneity and inter-organelle contacts: Impacts on membrane preparation for proteomic studies. Biol. Cell 104, 618–627 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Zheng Y. Z., Foster L. J., Contributions of quantitative proteomics to understanding membrane microdomains. J. Lipid Res. 50, 1976–1985 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marinko J. T. et al., Folding and misfolding of human membrane proteins in health and disease: From single molecules to cellular proteostasis. Chem. Rev. 119, 5537–5606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Meer G., Voelker D. R., Feigenson G. W., Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharpe H. J., Stevens T. J., Munro S., A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142, 158–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schlebach J. P. et al., Topologically diverse human membrane proteins partition to liquid-disordered domains in phase-separated lipid vesicles. Biochemistry 55, 985–988 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shogomori H. et al., Palmitoylation and intracellular domain interactions both contribute to raft targeting of linker for activation of T cells. J. Biol. Chem. 280, 18931–18942 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Johnson S. A. et al., Temperature-dependent phase behavior and protein partitioning in giant plasma membrane vesicles. Biochim. Biophys. Acta 1798, 1427–1435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Podkalicka J., Biernatowska A., Majkowski M., Grzybek M., Sikorski A. F., MPP1 as a factor regulating phase separation in giant plasma membrane-derived vesicles. Biophys. J. 108, 2201–2211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raggi C. et al., Caveolin-1 endows order in cholesterol-rich detergent resistant membranes. Biomolecules 9, 287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for all GPMV plots shown are available in Dataset S1: “Raw Values for GPMV Partitioning Plots.” Raw data used to plot and calculate TMisc can be found in Dataset S2: “Values for TMisc Plots.”