There is an emerging realization that plant genomes contain biosynthetic gene clusters (BGCs) for more specialized metabolism in certain cases (1). However, while horizontal gene transfer seems to drive the assembly of self-sufficient BGCs in microbes, the limitations of strict vertical gene transmission necessitate a distinct driving force for the assembly of BGCs in plants. The evidence for horizontal gene transfer of BGCs in microbes is derived from the appearance of homologous such loci, leading to biosynthesis of the same (or very closely related) natural products, in phylogenetically distinct species. By contrast, there has been much less investigation of such occurrences in plants. This is a result of many fewer examples of BGCs, as well as instances of natural products that skip across phylogenetic distances in this kingdom.
Fortunately, the momilactones provide a strikingly informative exception. These diterpenoids were originally found in rice (Oryza sativa), where the associated BGC was among the first few identified in plants (2), and where they serve as allelochemicals and/or phytoalexins (3, 4). Notably, momilactones are produced by not only other wild rice species within the Oryza genus but also barnyard grass (Echinochloa crus-galli), which falls within a separate clade of the Poaceae family. Intriguingly, beyond the expected orthologous momilactone BGCs in Oryza (5), a homologous BGC also was found in the phylogenetically more distant E. crus-galli (6). Even more interesting is the production of momilactones by the bryophyte Calohypnum plumiforme (formerly Hypnum plumaeforme), representing a very early diverging lineage of land plants (7). In PNAS, Mao et al. (8) report that the C. plumiforme genome contains a BGC for momilactone biosynthesis. Strikingly, this not only represents identification of a BGC in a bryophyte but, further, is clearly phylogenetically distinct from the BGCs associated with momilactone biosynthesis in the Poaceae. Nevertheless, despite their clearly independent evolutionary origins, comparison of these momilactone BGCs reveals intriguing hints about the distinct evolutionary pressures leading to not only their assembly but also that of BGCs in plants more generally.
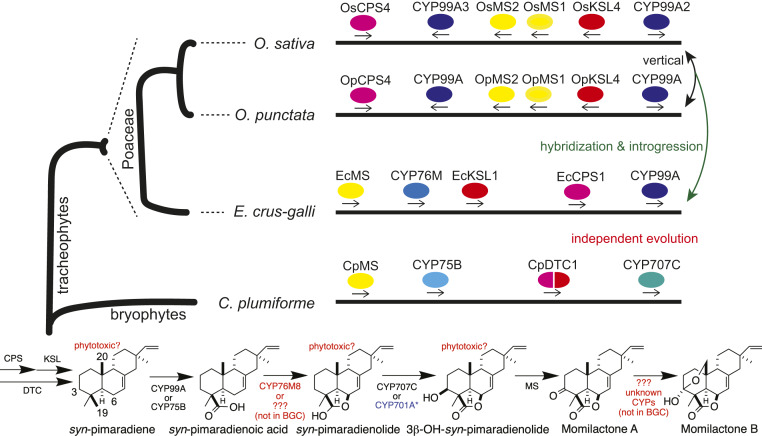
As labdane-related diterpenoids, the momilactones require dual cyclization reactions (9). Notably, identification of the two relevant cyclases revealed that the genes for these consecutively acting enzymes are close together in the rice genome (2). Moreover, the only other genes in the region encode two closely related cytochrome P450 (CYP) monooxygenases (CYP99A2 and CYP99A3) and two short-chain alcohol dehydrogenases/reductases (SDRs) (10). The two SDRs also are closely related, and both can catalyze oxidation of the penultimate intermediate 3β-hydroxy-syn-pimaradien-19,6β-olide to form the characteristic carbon-3 (C3) keto group of momilactone A (11). Accordingly, these have been termed momilactone synthases (MS1 and MS2) (12). However, while the diterpene cyclases together produce the olefin syn-pimaradiene, with CYP99A2 and CYP99A3 both functioning as subsequently acting C19 oxidases (13, 14), the resulting syn-pimaradien-19-oic acid requires further elaboration. In particular, formation of the eponymous lactone ring clearly requires oxidation at the C6β position, while hydroxylation at C3β also is required. Moreover, formation of the hemiketal ring that characterizes momilactone B requires hydroxylation at C20. Two other rice CYPs have been shown to react with syn-pimaradiene at two of these positions, with CYP701A8 acting as a C3β-hydroxylase (14), while CYP76M8 acts as a C6β-hydroxylase (15). Yet neither CYP76M8 or CYP701A8, nor any other CYP genes, are located in the rice momilactone BGC, which is then not self-sufficient (Fig. 1).
Fig. 1.
Momilactone BGC in various plants with corresponding pathway. Includes approximate phylogenetic tree for relevant plant species and corresponding momilactone BGC, with genes colored to indicate distinct enzymatic (sub)families (split coloring of CpDTC indicates bifunctional fusion of copalyl diphosphate synthase [CPS] and kaurene synthase-like [KSL] cyclases). Arrows and/or accompanying notes on the right side indicate hypothesized evolutionary relationships among the depicted momilactone BGCs. The biosynthetic pathway has been annotated to include numbering of the relevant carbons in syn-pimaradiene and the relevant enzymes from momilactone BGCs, with steps mediated by CYPs not contained within these noted, along with hypothesized phytotoxicity of certain intermediates (i.e., substrates for enzymes found within the momilactone BGCs, as discussed), in red text (CYP701A subfamily member from tandem gene array that includes the requisite ent-kaurene oxidase for gibberellin phytohormone biosynthesis).
A somewhat similar situation is observed in the momilactone BGCs in other species from the Poaceae. Consistent with their close phylogenetic relationship, Oryza punctata produces momilactones and contains a momilactone BGC exactly orthologous to that in O. sativa. However, only CYP99A subfamily members are present in this region in the more phylogenetically distant Oryza brachyantha, with no relevant genes found at all in the even more distant Leersia perrieri, neither of which makes momilactones. This indicates that such metabolism and the associated BGC arose within the Oryza lineage (5). Nevertheless, the much more phylogenetically distant E. crus-galli also produces momilactones and contains an apparently orthologous BGC. In particular, although the E. crus-galli momilactone BGC contains only single copies of CYP99A and MS (unlike in Oryza), and has undergone some change in gene order, phylogenetic analysis of the genes found in both indicates nearest homology to those from the Oryza BGC rather than with members of the relevant enzymatic families from more closely related plant species (6). Interestingly, it has been hypothesized that this orthology may reflect an ancient hybridization event and subsequent natural introgression of the momilactone BGC into the E. crus-galli lineage, which implies strong selective pressure for such metabolism (16). Regardless, the analogous composition of all of the momilactone BGCs from the Poaceae demonstrates that none is self-sufficient.
Intriguingly, the work of Mao et al. (8) reveals similar insufficiency in the independently evolved momilactone BGC from C. plumiforme. This BGC was found by sequencing the C. plumiforme genome and locating the previously identified bifunctional diterpene cyclase CpDTC (17), revealing that the surrounding region contains two distinct CYPs, members of the CYP75B and CYP707C subfamilies, as well as an SDR. Notably, the CYP75B subfamily member was shown to exhibit the same C19 oxidase activity as the Poaceae CYP99A subfamily members of their momilactone BGCs. Similarly, the SDR was shown to catalyze the same oxidation as the MS found in the Poaceae momilactone BGCs. On the other hand, the CYP707C subfamily member was found to act as a C3β-hydroxylase. Accordingly, the two independently evolved momilactone BGCs contain exactly the same enzymatic activities (i.e., diterpene cyclase, C19 oxidase, and MS), with the bryophyte BGC also additionally including the C3β-hydroxylase. This raises questions about these similarities, as well as difference, in composition.
The difference in composition of these momilactone BGCs seems to stem from more fundamental distinctions between bryophytes and tracheophytes, in particular, the gibberellin phytohormones that are required in the latter but not the former (18). The C3β-hydroxylase for momilactone biosynthesis in the Poaceae is a member of the CYP701A subfamily, which is otherwise closely associated with gibberellin biosynthesis. In rice, the relevant CYP701A8 is located in a tandem gene array that includes the paralogous ent-kaurene oxidase required for gibberellin phytohormone metabolism, CYP701A6, which then is an essential gene (10). Moreover, such CYP701A tandem gene duplication and diversion of a paralog to more specialized metabolism (i.e., neofunctionalization) seems to be widespread in the Poaceae (19). Hence, such linkage to an essential gene removes the need to “recruit” the C3β-hydroxylase to the momilactone BGC—in particular, as coinheritance is then accomplished regardless. This illustrates how vertical gene transmission impacts BGC assembly; that is, inheritance can be ensured even in the absence of self-sufficient BGCs. In addition, this argues against a role for coregulation in driving BGC assembly.
By contrast, the similarities in composition, despite the incomplete natures of both independently evolved momilactone BGCs, indicate that complete pathway inheritance is not sufficient to explain BGC assembly in plants. Indeed, it has been hypothesized that such assembly requires not only positive selection pressure provided by the bioactivity of the resulting natural product but also negative selection against partial pathway inheritance (20). The latter effect can be more specifically hypothesized to arise from phytotoxicity of certain intermediates, either directly or after alternative further metabolism. This hypothesis is supported by the observed similarities in momilactone BGCs, in particular, as this implies that the enzymatic substrates for the assembled genes exert deleterious effects. Evidence supporting this hypothesis has already been reported for the rice momilactone BGC, as loss of the second acting of the two diterpene cyclases leads to significantly decreased seed germination rates (3). Accordingly, it can be speculated that not only the syn-copalyl diphosphate substrate of this second cyclase but also the syn-pimaradiene substrate of the C19 oxidase and 3β-hydroxy-syn-pimaradien-19,6β-olide substrate of the MS, which are present in all momilactone BGCs, leads to phytotoxic effects. In addition, given the ensured coinheritance of the C3β-hydroxylase described above, its presumed syn-pimaradien-19,6β-olide substrate also may lead to a deleterious effect (Fig. 1). On the other hand, the syn-pimaradien-19-oic acid substrate of the C6β-hydroxylase that does not seem to be found in any of the momilactone BGCs would be expected to lack phytotoxicity. Similarly, consistent with its known accumulation and bioactivity as a phytoalexin, the momilactone A substrate of the C20 hydroxylase does not seem to exert any deleterious effect. Future examination of these predictions then can be expected to provide further insight into the distinct selective pressure driving assembly of BGCs in plants.
Acknowledgments
Research in R.J.P.’s laboratory is funded by NIH Grant GM131885.
Footnotes
The authors declare no competing interest.
See companion article, “Genomic evidence for convergent evolution of gene clusters for momilactone biosynthesis in land plants,” 10.1073/pnas.1914373117.
References
- 1.Nützmann H. W., Scazzocchio C., Osbourn A., Metabolic gene clusters in eukaryotes. Annu. Rev. Genet. 52, 159–183 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Wilderman P. R., Xu M., Jin Y., Coates R. M., Peters R. J., Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135, 2098–2105 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu M., et al. , Genetic evidence for natural product-mediated plant-plant allelopathy in rice (Oryza sativa). New Phytol. 193, 570–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu X., et al. , Inferring roles in defense from metabolic allocation of rice diterpenoids. Plant Cell 30, 1119–1131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyamoto K., et al. , Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. Plant J. 87, 293–304 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Guo L., et al. , Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 8, 1031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nozaki H., et al. , Momilactone A and B as allelochemicals from moss Hypnum plumaeforme: First occurrence in bryophytes. Biosci. Biotechnol. Biochem. 71, 3127–3130 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Mao L., et al. , Genomic evidence for convergent evolution of gene clusters for momilactone biosynthesis in land plants. Proc. Natl. Acad. Sci. U.S.A. 117, 12472–12480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters R. J., Two rings in them all: The labdane-related diterpenoids. Nat. Prod. Rep. 27, 1521–1530 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakamoto T., et al. , An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134, 1642–1653 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitaoka N., Wu Y., Zi J., Peters R. J., Investigating inducible short-chain alcohol dehydrogenases/reductases clarifies rice oryzalexin biosynthesis. Plant J. 88, 271–279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimura K., et al. , Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282, 34013–34018 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Wang Q., Hillwig M. L., Peters R. J., CYP99A3: Functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J. 65, 87–95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitaoka N., Wu Y., Xu M., Peters R. J., Optimization of recombinant expression enables discovery of novel cytochrome P450 activity in rice diterpenoid biosynthesis. Appl. Microbiol. Biotechnol. 99, 7549–7558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q., et al. , Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J. Biol. Chem. 287, 6159–6168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters R. J., Doing the gene shuffle to close synteny: Dynamic assembly of biosynthetic gene clusters. New Phytol., 10.1111/nph.16631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada K., et al. , HpDTC1, a stress-inducible bifunctional diterpene cyclase involved in momilactone biosynthesis, functions in chemical defence in the moss Hypnum plumaeforme. Sci. Rep. 6, 25316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zi J., Mafu S., Peters R. J., To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu. Rev. Plant Biol. 65, 259–286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Y., et al. , Multiple genes recruited from hormone pathways partition maize diterpenoid defences. Nat. Plants 5, 1043–1056 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Takos A. M., Rook F., Why biosynthetic genes for chemical defense compounds cluster. Trends Plant Sci. 17, 383–388 (2012). [DOI] [PubMed] [Google Scholar]