Significance
In order to establish conditions favorable for their intracellular survival, microbial pathogens modulate a wide range of host cell processes. The eukaryotic Hippo pathway is best known for its role in controlling cell proliferation and differentiation, yet we recently discovered that it also plays an important role during infection with Legionella pneumophila, the causative agent of Legionnaires’ pneumonia. We show that the Legionella kinase LegK7 mimics kinases from the host cell Hippo pathway in order to exploit the scaffold protein MOB1A for downstream target recruitment and phosphorylation. By employing this strategy of molecular mimicry, LegK7 can fulfill its function in a wide range of host cells, including amoeba in the environment and alveolar macrophages during infection in humans.
Keywords: microbial pathogen, type IV secretion system, translocated effector, crystal structure, MST1/2
Abstract
During infection, the bacterial pathogen Legionella pneumophila manipulates a variety of host cell signaling pathways, including the Hippo pathway which controls cell proliferation and differentiation in eukaryotes. Our previous studies revealed that L. pneumophila encodes the effector kinase LegK7 which phosphorylates MOB1A, a highly conserved scaffold protein of the Hippo pathway. Here, we show that MOB1A, in addition to being a substrate of LegK7, also functions as an allosteric activator of its kinase activity. A crystallographic analysis of the LegK7–MOB1A complex revealed that the N-terminal half of LegK7 is structurally similar to eukaryotic protein kinases, and that MOB1A directly binds to the LegK7 kinase domain. Substitution of interface residues critical for complex formation abrogated allosteric activation of LegK7 both in vitro and within cells and diminished MOB1A phosphorylation. Importantly, the N-terminal extension (NTE) of MOB1A not only regulated complex formation with LegK7 but also served as a docking site for downstream substrates such as the transcriptional coregulator YAP1. Deletion of the NTE from MOB1A or addition of NTE peptides as binding competitors attenuated YAP1 recruitment to and phosphorylation by LegK7. By providing mechanistic insight into the formation and regulation of the LegK7–MOB1A complex, our study unravels a sophisticated molecular mimicry strategy that is used by L. pneumophila to take control of the host cell Hippo pathway.
Microbial pathogens manipulate host cell signaling pathways by encoding molecular mimics of protein kinases. Kinases catalyze target phosphorylation by transferring the gamma phosphate group of adenosine 5′-triphosphate (ATP) onto side chains of substrate proteins, preferentially serine, threonine, or tyrosine residues, thereby altering the activity, localization, or stability of their substrates. Protein phosphorylation is one of the most abundant and most important posttranslational modifications in living cells, and mammals encode hundreds of kinases that target thousands of substrate proteins (1, 2). To promote infection, many pathogens, including bacteria and eukaryotic parasites, encode effector kinases that are translocated into the host cytosol where they alter signaling cascades, suggesting that kinase mimicry is a commonly used strategy to exploit host cell function (3, 4).
The Hippo pathway is an ancient kinase signaling cascade conserved in eukaryotes. It is best known for its role in controlling developmental processes such as cell-cycle progression, cell proliferation, differentiation, and apoptosis (5, 6). Mammalian Ste20-like kinases 1/2 (MST1/2), orthologs of the Drosophila Hippo kinase, phosphorylate MOB1A/B proteins (Mps one binder kinase activator 1A/B) (7), which in turn form a complex with LATS1/2 (large tumor suppressor kinases 1/2) or NDR1/2 (nuclear Dbf2-related kinase 1/2) and assist in their allosteric activation (8, 9). Active LATS1/2 phosphorylate the cotranscriptional regulator YAP1 (yes-associated protein 1) and its homolog TAZ (transcriptional coactivator with PDZ-binding motif). Phosphorylated YAP1 and TAZ are prevented from entering the nucleus by being either sequestered in the cytosol through binding to 14-3-3 proteins or targeted for proteolytic degradation (6, 8). Consequently, the main outcome of signal transduction along the Hippo pathway is changes in gene expression (6).
MOB1A and MOB1B share 96% sequence identity over 216 amino acid (a.a.) residues and are considered to be functionally redundant. They play a key role in the Hippo pathway by acting as scaffold proteins, coordinating a sequential, multistep phosphorylation cascade that runs from MST1/2 to the downstream components YAP1/TAZ (10, 11). When not phosphorylated, the flexible N-terminal extension (NTE) of MOB1A occupies the binding surface for LATS/NDR and interferes with binding of these cellular ligands to MOB1A. MST1/2 catalyzes the phosphorylation of two threonine residues, T12 and T35, within the NTE, thus destabilizing interaction with the remainder of MOB1A and exposing the LATS/NDR-binding epitope (8, 12). Only upon complex formation with MOB1A can allosterically activated LATS/NDR phosphorylate their downstream targets (5), though the exact molecular mechanism underlying YAP1/TAZ phosphorylation, in particular the role of MOB1A in this process, has yet to be determined.
We recently showed that the intracellular pathogen Legionella pneumophila alters the host Hippo signaling pathway during infection of macrophages (13). This gram-negative bacterium thrives within freshwater amoebae in the environment but, upon inhalation by humans, can also infect and replicate within alveolar macrophages, a scenario that in immunocompromised individuals often results in a potentially fatal pneumonia called Legionnaires’ disease (14, 15). L. pneumophila encodes the effector LegK7, which functions as a molecular mimic of MST1/2 (13). Upon translocation into host cells by the L. pneumophila Dot/Icm type IV secretion system (T4SS), LegK7 phosphorylates MOB1A on T12 and T35, thereby triggering a signaling cascade that results in the degradation of the downstream substrates YAP1/TAZ (13). Consequently, the expression of host genes controlled by several transcription factors is altered in ways that render macrophages more susceptible to L. pneumophila infection (13).
The finding that LegK7 targets MOB1A and manipulates its function points toward an unexpected role for the Hippo pathway in microbial pathogenesis (13). Since LegK7 appears to functionally mimic MST1/2, it became important to more thoroughly investigate this host–pathogen interaction and to determine, at a molecular level, how LegK7 exploits a signaling pathway that is engrained within the developmental program of eukaryotic cells. For example, it has remained unclear if LegK7 phosphorylates MOB1A to simply promote complex formation between MOB1A and cellular ligands such as LATS/NDR kinases. Or, does the bacterial kinase itself bind to phosphorylated MOB1A to possibly modify downstream signaling? And, if so, why does LegK7 not phosphorylate downstream substrates directly but instead target MOB1A, the scaffold component of the Hippo pathway? Here, we provide much-needed answers to these questions and, in the process, discovered an unexpected role for the N-terminal region of MOB1A during L. pneumophila infection.
Results
MOB1A Allosterically Activates the Kinase Activity of LegK7.
To monitor protein phosphorylation in our experiments, we adapted a previously reported in vitro kinase assay (16) that uses ATPγS (adenosine 5′-O-3-thiotriphosphate) as a phosphate donor for protein thiophosphorylation (see Materials and Methods for details). Upon incubation of ATPγS with glutathione-S-transferase (GST)-tagged MOB1A and maltose-binding protein (MBP)-tagged LegK7, we detected robust thiophosphorylation of GST-MOB1A (Fig. 1A), consistent with our earlier studies (13). Strikingly, the level of MBP-LegK7 autothiophosphorylation was also significantly enhanced when the bacterial kinase was coincubated with GST-MOB1A but not upon exposure to GST (Fig. 1A). No phosphorylation signal was detectable upon incubation of GST-MOB1A with the catalytically inactive variant MBP-LegK7(D307A). These results suggest that MOB1A not only served as a substrate of LegK7 but also as an allosteric activator for its kinase activity, a phenomenon reminiscent of what has been described for mammalian NDR/LATS kinases (9, 17). To determine whether the stimulatory effect of MOB1A was concentration-dependent, we added increasing amounts of GST-MOB1A to the kinase reconstitution assay and monitored MBP-LegK7 autothiophosphorylation (Fig. 1B). We found that the increase in thiophosphorylation of MBP-LegK7 was proportional to the concentration of GST-MOB1A added and that it reached saturation in the presence of a molar excess of GST-MOB1A (Fig. 1B), suggesting that MOB1A stimulates the kinase activity of LegK7 in a dose-dependent manner, most likely by binding to LegK7.
Fig. 1.
Human MOB1A binds to and allosterically activates LegK7. (A) MOB1A stimulates autophosphorylation of LegK7 in vitro. Thiophosphorylation (ps) of MBP-LegK7 and GST-MOB1A in a thiophosphate kinase assay was detected by immunoblot using a thiophosphate ester-specific antibody. The expected position of GST if it were thiophosphorylated is indicated by an arrow. (B) LegK7 autophosphorylation is proportional to the amount of MOB1A present. Autothiophosphorylation of MBP-LegK7 (0.2 μM) was detected by thiophosphate kinase assay in the presence of increasing concentrations of GST-MOB1A (as indicated). The relative level of thiophosphorylated/total MBP-LegK7 as a function of the concentration of GST-MOB1A is shown in the graph (Bottom). (C) LegK7(11–530) and MOB1A elute as a complex during size-exclusion chromatography (SEC). (C, Left) The absorption profiles of purified LegK7(11–530) (gray), MOB1A (blue), or a mixture of LegK7(11–530) and MOB1A (brown, dashed) are shown. Absorbance signals at 280 nm (mAU, milliabsorbance unit) are plotted versus the elution volume. (C, Right) SEC with multiangle light scattering profiles of the same proteins with calculated apparent molecular masses. (D) Binding constant for the formation of a complex between MOB1A or MOB1A33 proteins and LegK7(11–530) determined by ITC. See Materials and Methods for details. The Kd values were calculated with TA Instruments software. WT, wild type.
LegK7 is composed of 930 a.a. residues. Since the predicted kinase domain (amino acids 183 to 462) is located within the N-terminal half (13), we investigated whether the C-terminal half of LegK7 (∼400 a.a. residues in size) plays a role in allosteric activation of LegK7 by MOB1A. We purified several C-terminally truncated LegK7 variants, each encompassing the predicted kinase domain. The best-behaved construct was LegK7(11–530), which, in addition to the kinase domain, contained the flanking amino- and carboxyl-terminal regions of unknown function. Importantly, LegK7(11–530) exhibited kinase activity and robustly phosphorylated MOB1A and itself, indicating that, similar to the full-length protein, this truncated variant was allosterically activated by MOB1A (SI Appendix, Fig. S1A). Hence, in the following, we focused our investigation on the interaction between LegK7(11–530) and MOB1A.
We examined a possible complex formation between MOB1A and LegK7(11–530) in vitro. While LegK7(11–530) (61 kDa) and MOB1A (25 kDa) eluted according to their expected molecular masses during gel-permeation chromatography (Fig. 1C), an equimolar mixture of LegK7(11–530) and MOB1A formed a peak with an elution volume equivalent to that of a globular 81-kDa protein, matching the predicted size of a 1:1 complex between LegK7(11–530) and MOB1A (Fig. 1C). We further evaluated the strength of the interaction between LegK7(11–530) and MOB1A by isothermal titration calorimetry (ITC), which indicated a dissociation constant (Kd) of 1.26 ± 0.4 μM between the two proteins and confirmed that the complex follows a 1:1 stoichiometry (Fig. 1D). These data validated that LegK7(11–530) is sufficient for interaction with MOB1A, and that the C terminus (531–930) of LegK7 is not required for allosteric activation.
Crystallization of the LegK7(11–530)–MOB1A Complex.
To obtain in-depth insight into the interaction between LegK7 and MOB1A, we turned to protein crystallography. Initial attempts to crystallize a 1:1 ratio of LegK7(11–530) and full-length MOB1A yielded no diffraction-quality crystals despite extensive screening. Similarly, no crystals could be obtained for LegK7(11–530), either alone or in the presence of either ATP or its nonhydrolyzable analog adenosine-5′-(β,γ-imido) triphosphate (AMP-PNP). Since the NTE of MOB1A shows a high degree of flexibility (18, 19), which might interfere with crystal formation of the LegK7–MOB1A complex, we produced a truncated MOB1A variant lacking the first 32 residues (called MOB1A33 henceforth). When coincubated with LegK7, MOB1A33 showed reduced thiophosphorylation compared with full-length MOB1A (SI Appendix, Fig. S1B), likely due to the absence of T12 as one of the two preferred sites of phosphorylation (13). Despite the truncation, MOB1A33 retained the ability to directly interact with LegK7(11–530) (Fig. 1D). Crystallization trials with LegK7(11–530) and MOB1A33 yielded crystals that diffracted to 2.57-Å resolution (Table 1). Since LegK7(11–530) exhibited kinase activity toward itself and toward MOB1A (SI Appendix, Fig. S1A), we also carried out phosphorylation reactions to saturation (incubation with ATP overnight) and, after removal of free nucleotides (adenosine diphosphate and ATP) by size-exclusion chromatography, used the peak fractions containing the phosphorylated LegK7(11–530)–MOB1A33 complex for crystallization experiments with or without AMP-PNP. These experiments yielded crystals of the complex with AMP-PNP that diffracted to 2.5-Å resolution.
Table 1.
Summary of data collection and refinement statistics
| LegK7(11–530)–MOB1A33 (SeMet)* | LegK7(11–530)–MOB1A33 (native) | pLegK7(11–530)– pMOB1A33–AMP-PNP | |
| Wavelength, Å | 0.9787 | 0.9795 | 0.9795 |
| Resolution range, Å | 48.00–2.90 (2.98–2.90) | 48.00–2.57 (2.64–2.57) | 48.0–2.50 (2.56–2.50) |
| Space group | C2 | C2 | C2 |
| Cell parameters | 107.6, 112.6, 155.3, 90, 105.4, 90 | 107.4, 113.1, 157.9, 90, 106.0, 90 | 106.9, 112.4, 155.2, 90, 105.6, 90 |
| Total reflections | 301,848 (22,544) | 292,177 (20,942) | 310,866 (22,317) |
| Unique reflections | 77,669 (5,746) | 57,771 (4,248) | 61,056 (4,456) |
| Multiplicity | 3.9 (3.9) | 5.1 (4.9) | 5.1 (5.0) |
| Completeness, % | 99.8 (100.0) | 99.8 (99.8) | 99.8 (100.0) |
| Mean I/sigma(I) | 7.5 (1.8) | 9.7 (1.8) | 11.3 (1.9) |
| Wilson B factor, Å2 | 70.1 | 58.4 | 57.1 |
| Rmerge | 0.125 (0.903) | 0.103 (0.874) | 0.091 (0.973) |
| Rmeas | 0.136 (1.000) | 0.115 (0.980) | 0.102 (1.087) |
| CC1/2 | 99.6 (78.9) | 99.8 (74.8) | 99.8 (69.0) |
| Rwork/Rfree | 0.1941/0.2448 | 0.1910/0.2331 | |
| Ramachandran plot | |||
| Favored, % | 96.4 | 97.7 | |
| Allowed, % | 3.5 | 2.3 | |
| Outlier, % | 0.1 | 0 | |
| Clashscore | 4.8 | 5.7 | |
| Rmsd | |||
| Bonds, Å | 0.006 | 0.006 | |
| Angles, ° | 0.78 | 0.79 | |
| PDB ID code | 6MCQ | 6MCP |
Information for the highest-resolution shell is given in parentheses.
Friedel pairs were counted as separate reflections.
The structure of the unphosphorylated LegK7(11–530)–MOB1A33 complex was first solved by single-wavelength anomalous dispersion using selenomethionine (SeMet) derivatives of both proteins. The structure of the phosphorylated complex with AMP-PNP was solved by molecular replacement using the initial model of the unphosphorylated complex.
Structure of the LegK7–MOB1A Complex.
Since the complex of phosphorylated LegK7(11–530)–MOB1A33 diffracted to higher resolution, the following descriptions of structural features are based on this crystal form. The crystals belonged to the C2 space group and contained two copies of the complex in the asymmetric unit. The structure of the complex is shown in Fig. 2A. MOB1A33 was well-ordered in the crystal and the two copies were nearly identical (rmsd of 0.37 Å for all Cα atoms). The two LegK7(11–530) molecules superimposed with a somewhat larger rmsd of 0.70 Å for 483 out of 520 Cα atoms present in the model. Notably, the loop Ser180 to Asn191 was disordered in one molecule.
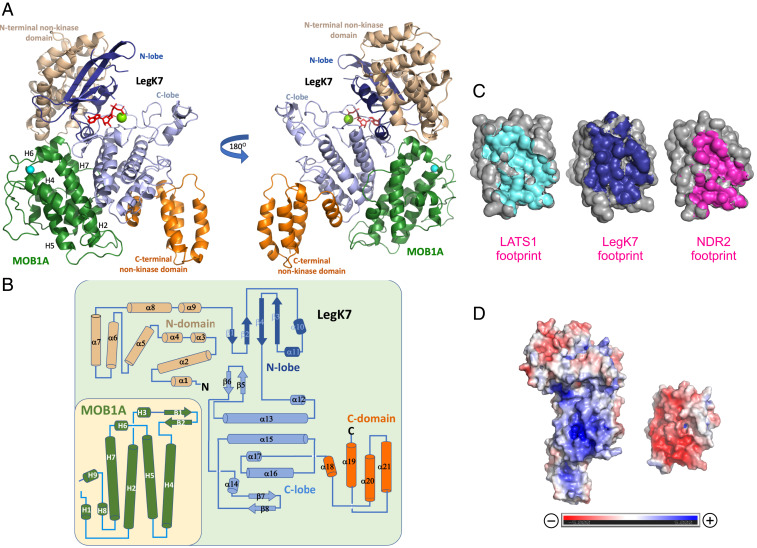
Fig. 2.
Crystal structure of the LegK7–MOB1A complex. (A) Cartoon representation of the LegK7(11–530)–MOB1A33 complex. The flanking N- and C-terminal domains of LegK7 are colored wheat and orange, respectively, the N lobe of the kinase domain is in blue, and the C lobe is in slate. AMP-PNP (red) is shown in stick representation and Mn2+ is shown as a green sphere. MOB1A33 is colored green and a bound Zn2+ ion is shown as a cyan sphere. MOB1A33 contacts predominantly the kinase domain of LegK7 with additional contacts to the N-terminal domain. (B) Topology diagram of MOB1A33 and LegK7(11–530). Color coding is as in A. The N and C termini are marked with black letters. (C) Interface between MOB1A and LATS1 (Left; PDB ID code 5BRK), LegK7 (Middle), or NDR2 (Right; PDB ID code 5XQZ). The solvent-accessible surface of MOB1A is shown in gray, while surfaces covered by LATS1 (cyan), LegK7 (blue), and NDR2 (magenta) are highlighted. The MOB1A molecules from each complex are shown in similar orientations, with the contact area of each facing the reader. (D) Electrostatic surface potential of LegK7 (Left) and MOB1A33 (Right). The molecules are separated along the interface and are shown with the binding sites facing the reader. Charges ranging from positive (dark blue) to negative (dark red) are indicated.
LegK7(11–530) consists of three domains: a eukaryotic-like kinase domain (amino acids 174 to 456) that is flanked by an N-terminal all–α-helical domain (amino acids 11 to 173) containing nine helices, α1 to α9, and a C-terminal domain containing four helices, α18 to α21 (amino acids 457 to 529), which form a tight bundle (Fig. 2A). The fold of the LegK7 kinase domain is composed of two lobes with the ATP binding site located in a deep groove between them (Fig. 2A). LegK7 contains all 12 subdomains characteristic of eukaryotic kinases (20), but significant alterations in some of the conserved motifs suggest that it is regulated differently from canonical eukaryotic kinases (21). Notably, the activation loop of LegK7 is well-ordered in both the phosphorylated and nonphosphorylated complex and appears to be stabilized by intrinsic hydrogen bonding. The fold of the kinase domain is similar to many eukaryotic protein kinases in their activated state as identified by the DALI server (22), including PIM1 [Protein Data Bank (PDB) ID code 1XR1 (23)], LIM domain kinase 1 [PDB ID code 5HVK (24)], or interleukin-1 receptor-associated kinase 4 [PDB ID code 4ZTL (25)]. Approximately 220 residues out of ∼280 can be superimposed on other kinases found in the database, with an rmsd of 2.5 to 2.9 Å (SI Appendix, Fig. S2A). The C lobe (amino acids 262 to 456) superimposes in almost its entirety, while the N lobe (amino acids 174 to 261) deviates more from homologous kinases despite its similar fold. The structural overlap with MST1 was somewhat poorer than with the above-mentioned homologous structures, which is not entirely surprising given the aforementioned lack of primary sequence homology between the proteins (13). A detailed comparison of the LegK7 kinase domain with eukaryotic kinases is provided in SI Appendix.
The flanking N-terminal α-helical domain (amino acids 11 to 173) of LegK7 contains seven helices arranged in three layers with helices in each layer inclined by ∼60° relative to the neighboring layer(s). Two additional C-terminal helices (α8 and α9) extend on the side of the domain, leading to the kinase domain. Searches with the DALI server (22) and PDBeFold (26) showed no notable similarity to known proteins, thus arguing that the region of amino acids 11 to 172 represents a fold of as-yet unknown function. The flanking C-terminal domain (amino acids 457 to 530) of LegK7 displays a tetratricopeptide repeat (TPR)-like fold. The three C-terminal helices α19 to α21 superimpose well on the N-terminal helices of the TPR of apoptosis signal-regulating kinase 1 [PDB ID code 5ULM, amino acids 272 to 323 (27), Z score 7.3, rmsd 1.7 Å for 54 Cα atoms] (SI Appendix, Fig. S2B), a module typically involved in protein–protein interactions.
MOB1A33 is a small globular α-helical protein composed of four up-and-down helices (H2, H4, H5, and H7; after ref. 18; Fig. 2 A and B), each five turns long, that are flanked by H1 at the N terminus and H8 and H9 at the C terminus. Two short, one-turn helices (H3 and H6) are also present, with the latter being located in the loop connecting helices H5 and H7 (Fig. 2 A and B). A Zn2+ ion is liganded by the side chains of Cys79 and Cys84 from helix H3, and His161 and His166 from the C terminus of helix H5 (Fig. 2A). Overall, MOB1A33 in complex with LegK7(11–530) is highly similar to previously described structures of MOB1A (rmsd of 0.9 Å), both alone (18) and in complex with a fragment of human LATS1 [PDB ID code 5BRK (8)] or NDR2 [PDB ID code 5XQZ (9)] (Fig. 2C). A small difference can be noticed in the conformation of residues 33 to 37. The preceding 32 residues, comprising the flexible NTE and not present in our structure, were found in different conformations and locations in the previously determined structures due to their flexibility (18, 19). The lack of success in obtaining well-diffracting crystals of LegK7(11–530) bound to full-length MOB1A suggests that the flexible NTE may have interfered with the formation of suitable crystal contacts. Notably, the electron density map of the partially phosphorylated complex, when compared with the unphosphorylated complex, showed extra density around T35 of MOB1A. However, this density was too weak to confidently place a phosphate group in that position, suggesting a low occupancy (low level of phosphorylation).
Direct Interaction with MOB1A Is Required for Allosteric Activation of LegK7 Kinase Activity.
LegK7 makes extensive contacts with MOB1A33 over an area of ∼1,500 Å2. Importantly, the LegK7 interface on MOB1A overlaps significantly with that for NDR/LATS (Fig. 2C). Despite being a direct substrate of LegK7, MOB1A33 binds LegK7 on the side opposite the ATP- and substrate-binding groove (Fig. 2A). The two proteins contact each other through relatively flat surfaces that contain only small incursions (Fig. 2D). Calculation of the electrostatic potential of each molecule revealed that the interface on LegK7 has a net-positive charge while that on MOB1A displays a net-negative charge (Fig. 2D). Therefore, the initial driving force toward binding appears to be electrostatic attraction. Interaction with MOB1A33 predominantly occurred through helices α13 and α17 (αE and αI following ref. 20) as well as the tip of the β-hairpin (N317 to R319) of the kinase domain of LegK7(11–530). The contacts are a mixture of H bonds, salt bridges (R420LegK7–E51MOB1A and K453LegK7–D63MOB1A), and hydrophobic contacts (Fig. 3A). Helix α8 of the N-terminal domain of LegK7 also participated in the interaction. This mode of binding differed significantly from that of NDR2 and LATS1 where a two-helix bundle stretches across the MOB1A interface almost perpendicular to helices α13 and α17 of LegK7 (SI Appendix, Fig. S2 C and D).
Fig. 3.
Complex formation with MOB1A is required for allosteric activation of LegK7. (A, Left) Close-up stereoview of the interface residues of the MOB1A33–LegK7 complex. Polypeptides are shown in stick representation with carbons colored slate (LegK7) and green (MOB1A). The MOB1A33 residues are more proximal to the reader. Residues mentioned in the text are highlighted as bold sticks and are labeled. (A, Right) Residues from each molecule located within a 3.3-Å radius from the other interface. Residues are shown in red (acidic), blue (basic), orange (polar), and black (hydrophobic). Residues opposing each other are connected by lines. (B) Substitution of interface residues affects allosteric activation of LegK7 by MOB1A. The levels of LegK7 autothiophosphorylation and MOB1A thiophosphorylation were determined by thiophosphate kinase assay using the indicated MBP-LegK7 and GST-MOB1A variants.
Surprisingly, the MOB1A-binding surface of LegK7 is only moderately conserved among LegK7 homologs (SI Appendix, Fig. S2E), thwarting identification of key contributors to MOB1A binding. Therefore, based on our structure of the complex (Fig. 3A), we substituted several LegK7 residues (P282E, R319A, R283A, Y286A, R420A, K453E, or N454E) that are involved in significant electrostatic or hydrophobic interactions with MOB1A and investigated by in vitro thiophosphorylation assay whether the resulting LegK7 mutant proteins are allosterically activated by MOB1A. LegK7(R319A) underwent degradation during production in Escherichia coli, suggesting that substituting this residue compromises the structural integrity of the protein. Of the remaining LegK7 mutants tested, only LegK7(P282E) and LegK7(Y286A) showed a significant reduction in thiophosphorylation of both MOB1A and themselves upon incubation with MOB1A, while LegK7(N454E) efficiently autothiophosphorylated but showed a mild reduction in MOB1A thiophosphorylation (Fig. 3B and SI Appendix, Fig. S3A). The remaining mutant proteins tested here in which the charges were either removed (R420A, R283A, or R319A) or reversed (K453E) displayed kinase activity comparable to that of wild-type LegK7, indicating that the individual arginine side chains contribute little to MOB1A binding (Fig. 3A). Thus, substitution of individual residues within the MOB1A-binding interface in LegK7 was, with few exceptions, tolerated.
We also substituted several MOB1A residues within the binding site for LegK7, namely L36D/G37D, A44L, or D63R, and tested how these changes affect allosteric activation of LegK7. Only substitution of D63 with arginine dramatically reduced the ability of MOB1A to stimulate LegK7 kinase activity, with autothiophosphorylation levels comparable to those of nonactivated MBP-LegK7 and significantly reduced levels of MOB1A thiophosphorylation (Fig. 3B). To validate that loss of allosteric activation was caused by loss of binding, we directly measured affinity of the complexes formed by the various MOB1A mutants and LegK7 using ITC. In line with the kinase assay results (Fig. 3B), we found that MOB1A(L36D/G37D) and MOB1A(A44L) bound LegK7 with similar or even higher affinity than wild-type MOB1A (Kd values of 0.3 ± 0.1 and 0.6 ± 0.2 μM, respectively), whereas interaction between MOB1A(D63R) and LegK7 was undetectable (SI Appendix, Fig. S3B). L36, G37, and A44 are located at the periphery of the LegK7-binding interface where their substitution might be accommodated by minor changes in orientation of MOB1A. In contrast, D63 is positioned at the center of the interface and contacts P282 of LegK7 (Fig. 3A) such that mutation of the latter to glutamate also abrogated binding, likely explaining why substitutions at this position in MOB1A were also not tolerated. Since all MOB1A mutant proteins showed melting temperatures similar to that of wild-type MOB1A (SI Appendix, Fig. S3C), it is unlikely that the inability of MOB1A(D63R) to form a stable complex with LegK7 resulted from a failure of the protein to fold correctly. Together, these findings confirmed that P282 and Y286 of LegK7 and D63 of MOB1 are critical for complex formation, and that direct interaction with MOB1A is required for allosteric activation of the kinase activity of LegK7.
The molecular mechanism underlying allosteric activation of LegK7 by MOB1A binding was not immediately apparent from the LegK7–MOB1A complex structure and would require its comparison with the structure of LegK7 alone. Unfortunately, all attempts to crystallize LegK7(11–530) were unsuccessful, which suggested significant flexibility of this protein in solution and/or conformational heterogeneity. We therefore resorted to molecular dynamics (MD) simulations and compared the dynamic behavior of this protein with that of LegK7 bound to MOB1A. The MD simulations revealed that several regions of LegK7 showed a prominent decrease in mobility when the protein was in a complex with MOB1A, most notably the regions directly interacting with MOB1A and regions of the kinase domain distal from the interacting surface (light blue and dark blue regions in SI Appendix, Fig. S4A). Although some regions of LegK7 had increased mobility upon MOB1A binding (pink regions in SI Appendix, Fig. S4A), these increases were far less prominent than the overall rms fluctuation decrease in the aforementioned regions that underwent stabilization. Consistent with this, we found that complex formation with either MOB1A or phosphorylated MOB1A increased the melting temperature of LegK7 to 38.7 or 41.7 °C, respectively (SI Appendix, Fig. S4B), further suggesting that MOB1A binding stabilizes LegK7. While the limited length of the simulation cannot rule out the possibility of large structural rearrangements in LegK7 upon binding to MOB1A, our MD simulations and melting temperature measurements allowed us to postulate that MOB1A binding decreased the conformational flexibility and stabilized the kinase domain of LegK7.
LegK7–MOB1A Complex Formation Promotes Target Phosphorylation in Mammalian Cells.
We had already demonstrated that LegK7 phosphorylates MOB1A on T12 and T35, both in vitro and within mammalian cells (13). Using ATP as a phosphate donor in an in vitro kinase reconstitution assay, we observed that phosphorylation of T12 and T35 was severely decreased in reactions containing LegK7(P282E) or MOB1A(D63R) (SI Appendix, Fig. S5A), suggesting that the MOB1A–LegK7 interaction is required for efficient phosphorylation of MOB1A. To evaluate the importance of complex formation between MOB1A and LegK7 for phosphorylation of endogenous MOB1, we produced green fluorescent protein (GFP)-tagged LegK7 wild type or D307A (catalytic mutant) or P282E (MOB1A-binding mutant) variants in transiently transfected human embryonic kidney (HEK293T) cells and probed their lysate for phosphorylation of endogenous MOB1 using an antibody directed against phospho-T35 (Fig. 4A). Compared with transiently transfected control HEK293T cells producing GFP, cells that produced GFP-LegK7 showed a robust increase in the relative level of T35 phosphorylation on MOB1 (Fig. 4A). In contrast, production of GFP-LegK7(D307A) or GFP-LegK7(P282E) caused only low levels of T35 phosphorylation. The increase in MOB1 phosphorylation in cells producing GFP-LegK7 did not occur via activation of cellular MST1 since no change in phosphorylation of Thr180, a key residue within the activation loop of MST1 (28), was observed in the presence of GFP-LegK7 or any of the LegK7 mutant proteins (SI Appendix, Fig. S5B). Moreover, treatment of HEK293T cells with XMU-MP-1, an MST1/2-specific inhibitor (29), had no effect on MOB1 phosphorylation by LegK7 but dramatically reduced MOB1 phosphorylation in response to okadaic acid-mediated stimulation of cellular MST1 (SI Appendix, Fig. S5C). Together, these studies show that allosteric activation of the kinase domain of LegK7 was required for efficient and direct phosphorylation of MOB1 within the context of living cells.
Fig. 4.
Complex formation promotes phosphorylation of MOB1 by LegK7 within mammalian cells. (A) Phosphorylation of endogenous MOB1 by GFP-LegK7. The indicated GFP-LegK7 proteins were produced in transiently transfected HEK293T cells, and the cell lysate was probed by immunoblot for phosphorylation of MOB1 on T35. (B) Complex formation with LegK7 is required for phosphorylation of MOB1 during L. pneumophila infection. Mouse RAW264.7 macrophages were challenged with an L. pneumophila Lp02ΔlegK7 strain complemented with expression plasmids encoding the indicated LegK7 variants. Relative phosphorylation levels were calculated as the ratio of pT35 MOB1 over total MOB1 with the vector control set as 1. Data are shown as mean ± SD (n = 3; *P < 0.05; two-tailed, unpaired t test).
Next, we determined whether interference with LegK7–MOB1A complex formation also affects phosphorylation of MOB1 during L. pneumophila infection. Mouse RAW264.7 macrophages were challenged with the L. pneumophila mutant strain Lp02(ΔlegK7) that was complemented with plasmids encoding either wild-type LegK7 (pflag-legK7) or the LegK7 variants D307A or P282E, and the relative level of T35-phosphorylated MOB1 within cell lysate was determined by immunoblot (Fig. 4B). While macrophages infected with Lp02ΔlegK7/pflag-legK7 showed elevated T35 phosphorylation levels compared with cells infected with a strain containing the empty vector control, complemented strains producing LegK7 variants with substitutions in either D307 or P282 showed near-baseline levels of T35 phosphorylation. The apparent fluctuation in total MOB1 levels is likely a consequence of proteasomal degradation (13) which is accelerated by phosphorylation and counteracted by complex formation with the kinase. These results confirm that efficient MOB1 phosphorylation during L. pneumophila infection requires that LegK7 forms a stable complex with MOB1.
Phosphorylation of the NTE Promotes MOB1A Binding to LegK7.
In the canonical Hippo pathway, MST1/2-mediated phosphorylation of MOB1A on T12 and T35 causes the flexible NTE to dissociate from the binding site for LATS/NDR, allowing MOB1A to form a complex with those kinases and to promote their activation (8, 9). Given that LegK7, like LATS/NDR, interacts with MOB1A (Figs. 1 and 2), we investigated how the phosphorylation state of MOB1A affects complex formation and allosteric activation of the LegK7 kinase domain. We phosphorylated full-length MOB1A to saturation by incubating it for 18 h with ATP and a small amount of LegK7(11–530), purified phosphorylated MOB1A using affinity and ion-exchange chromatography, and measured its interaction with LegK7(11–530) by ITC (Fig. 5A). While nonphosphorylated MOB1A bound LegK7(11–530) with a Kd of 1.26 μM (Fig. 1D), complex formation with phosphorylated MOB1A was ∼10-fold stronger (0.12 ± 0.09 μM) (Fig. 5A), suggesting that phosphorylation of MOB1A promotes the LegK7–MOB1A interaction.
Fig. 5.
MOB1A phosphorylation promotes binding and activation of LegK7. (A) Phosphorylation of MOB1A increases its affinity for LegK7. Dissociation constants between LegK7(11–530) and phosphorylated MOB1A or its phosphomimetic mutants T12D, T35D, and T74D were measured by ITC. For binding of LegK7(11–530) to unphosphorylated MOB1A, see Fig. 1D. (B) Contribution of MOB1A phosphorylation sites to allosteric activation of LegK7. MBP-LegK7 was incubated with the indicated GST-MOB1A variants, and autothiophosphorylation of MBP-LegK7 was determined by thiophosphate kinase assay. Thr residues in MOB1A that were replaced with either Asp or Ala are shown in red or black, respectively. Cartoon illustrates LegK7 conversion from low (blue line) to high (red line) kinase activity upon MOB1A binding.
In addition to T12 and T35, which are the primary target residues of phosphorylation by MST1/2, two other residues of MOB1A, T74 and T181, seem to play a role in NDR1 activation (30). To more precisely measure the contribution of each of the four known phosphorylation sites (T12, T35, T74, and T181) to MOB1A binding to LegK7, we first generated a GST-MOB1A variant in which all four threonine residues were replaced with alanines (4TA). We then introduced aspartate residues in place of the alanines to create phosphomimetic mutants (Fig. 5B). Upon testing the ability of these mutant proteins to allosterically activate the kinase activity of MBP-LegK7, we found that, unlike GST-MOB1A, which triggered strong autothiophosphorylation of MBP-LegK7, the MOB1A(4TA) variant failed to trigger kinase activity and caused low levels of autothiophosphorylation, as did GST-MOB1A(D63A), which does not bind LegK7 (Fig. 5B). Introduction of T12D or T35D into GST-MOB1A(4TA), either individually or together, resulted in a substantial increase in autothiophosphorylation of MBP-LegK7, though not quite to levels caused by GST-MOB1A (Fig. 5B). Notably, the stimulatory effect of T12D and T35D on LegK7 kinase activation was completely negated by the introduction of a T74D substitution in MOB1A. A T181D mutation, although in close proximity to T35, had no noticeable effect on the kinase activity of MBP-LegK7 (Fig. 5B).
To exclude the possibility that the stimulatory effect of T12D/T35D substitution on allosteric activation of LegK7 was present only when combined with the T74A/T181A mutations, we inserted the same phosphomimetic mutations into wild-type MOB1A (Fig. 5B). In contrast to MOB1A(T12A+T35A), which primarily exists in the closed conformation that cannot efficiently bind LegK7, MOB1A(T12D+T35D) robustly stimulated LegK7 autophosphorylation. Together, these findings indicate that phosphorylation of T12 and/or T35 enhances the affinity of MOB1A for LegK7, while phosphorylation of T74, which is located at the center of the interface, reduces binding, possibly by weakening MOB1A–LegK7 association. Consistent with this interpretation, we detected an increase in affinity between LegK7(11–530) and MOB1A(T12D) or MOB1A(T35D), with Kd values of 0.65 ± 0.13 and 0.96 ± 0.35 μM, respectively, and a substantial decrease in affinity with MOB1A(T74D) (Kd of 6.9 ± 1.2 μΜ) by ITC (Fig. 5A). Moreover, deletion of the NTE domain from MOB1A resulted in a lower Kd of the MOB1A33–LegK7(11–530) complex (0.55 ± 0.11 µM) that was close to the Kd of the complex with prephosphorylated MOB1A (Figs. 1D and 5A). Taken together, these studies demonstrate that LegK7, like MST1/2, phosphorylates MOB1A on T12 and T35 to promote NTE release and MOB1A–LegK7 complex formation, and that binding of MOB1A to LegK7 allosterically activates its kinase activity similar to what has been described for LATS/NDR (9, 17).
The NTE of MOB1A Creates an Epitope for Recruitment and Phosphorylation of Downstream Targets.
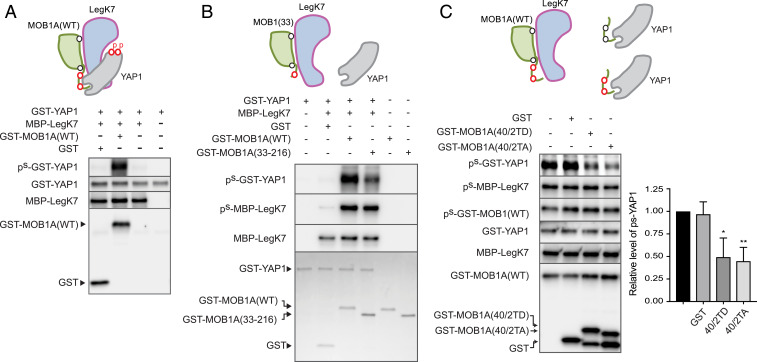
The finding that cellular MOB1A activates the LegK7 kinase domain, though intriguing, prompted us to question why LegK7 has not evolved into an autonomous kinase that functions independent of MOB1A. We reasoned that the presence of MOB1A may be required in order for LegK7 to fulfill its biological function, possibly by directing the kinase activity of LegK7 toward a downstream cellular target(s). We therefore tested whether LegK7, upon MOB1A binding, can directly phosphorylate the pseudosubstrate YAP1. While purified MBP-LegK7 coincubated with GST did not catalyze any detectable thiophosphorylation of YAP1, addition of GST-MOB1A resulted in a robust thiophosphorylation signal for YAP1 (Fig. 6A) but not for the GST control (SI Appendix, Fig. S6), demonstrating that LegK7, like mammalian LATS/NDR, phosphorylates YAP1 in a MOB1A-dependent manner in vitro.
Fig. 6.
NTE of MOB1A promotes YAP1 phosphorylation by LegK7. (A) LegK7 phosphorylates YAP1 in a MOB1A-dependent manner. Purified GST-YAP1 and MBP-LegK7 were incubated with either GST-MOB1A(WT) or GST in the presence of ATPγS, and thiophosphorylation levels were determined by immunoblot. (B) The NTE of MOB1A promotes phosphorylation of YAP1 by LegK7. Purified GST-YAP1 and MBP-LegK7 were incubated with either GST-MOB1A(WT) or GST-MOB1A33 in the presence of ATPγS, and thiophosphorylation levels were determined as in A. Total GST-YAP1, GST-MOB1A, GST-MOB1A33, and GST were detected by Coomassie staining. (C) NTE peptides interfere with efficient YAP1 phosphorylation by LegK7. A mixture of GST-YAP1, GST-MOB1A(WT), and MBP-LegK7 was incubated with either GST or GST-MOB1A NTE variants (40/2TD or 40/2TA), and thiophosphorylation of the indicated proteins was determined by immunoblot. Relative phosphorylation levels were calculated as the level of ps-YAP1 normalized to no GST control (set as 1). Data are shown as mean ± SD (n = 3; *P < 0.05, **P < 0.01; two-tailed, unpaired t test).
The structure of LegK7 in complex with MOB1A (Fig. 2A) yielded little insight into the molecular mechanism underlying YAP1 recruitment prior to its phosphorylation by LegK7. Given that MOB1A is phosphorylated on T12 and T35 both by MST1/2 during canonical Hippo signaling and by LegK7 during L. pneumophila infection and MOB1A activation precedes phosphorylation of YAP1, we hypothesized that the NTE, in addition to controlling complex formation with kinases, plays an additional role as a docking site for downstream substrates that are destined to be phosphorylated in response to MOB1A activation. If so, then deletion of the NTE from MOB1A should prevent recruitment and subsequent phosphorylation of YAP1 by LegK7. Upon comparing full-length MOB1A with MOB1A(33–216) in kinase reconstitution assays, we found that LegK7 incubated with MOB1A efficiently thiophosphorylated both YAP1 and itself, whereas MOB1A(33–216) promoted less efficient thiophosphorylation of YAP1 while still allowing allosteric activation and autothiophosphorylation of LegK7 (Fig. 6B). Thus, removal of the NTE from MOB1A, while maintaining a stimulatory effect on the LegK7 kinase activity, interfered with phosphorylation of YAP1, probably by eliminating the docking site for this downstream substrate of LegK7.
To determine whether the NTE of MOB1A suffices as a docking site for YAP1, we repeated the kinase reconstitution assays with full-length MOB1A but added, as a competitive inhibitor, purified MOB1A peptides composed of residues 1 to 40 in which T12 and T35 were substituted with either aspartates (40/2TD) or alanines (40/2TA). While the presence of GST had no competitive effect on YAP1 thiophosphorylation by the LegK7–MOB1A complex, addition of GST-MOB1A(40/2TD) strongly decreased thiophosphorylation of YAP1 (Fig. 6C) without impacting thiophosphorylation of either MBP-LegK7 or GST-MOB1A. Surprisingly, addition of GST-MOB1A(40/2TA) also reduced YAP1 thiophosphorylation to a degree similar to that of GST-MOB1A(40/2TD), showing that phosphorylation of T12 and T35, while important for alleviating MOB1A autoinhibition, was of little relevance for YAP1 binding. Together, these data demonstrate that the N terminus of MOB1A, in addition to its well-characterized function as a regulatory module for controlling accessibility to the kinase-interacting region, also plays a previously unappreciated role as a binding platform for the recruitment of YAP1 and possibly other downstream substrates by LegK7.
Discussion
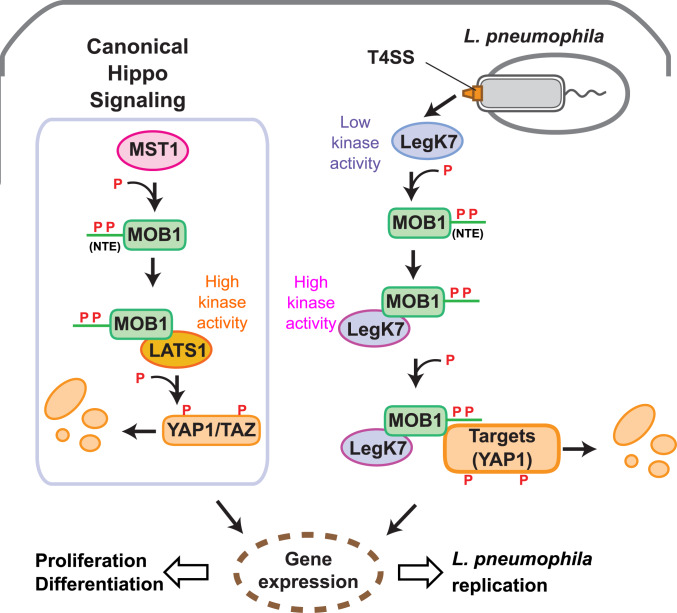
In this study, we provide detailed insights into the interplay between L. pneumophila LegK7 and mammalian MOB1A. We show that LegK7, like MST1/2, phosphorylates MOB1A on T12 and T35, thereby releasing the NTE from the globular domain of MOB1A and exposing the interface for LegK7 binding. Moreover, we demonstrate that this binding event triggers kinase activity, indicating that LegK7, like LATS1/2 and NDR1/2, utilizes MOB1A as an allosteric activator. Lastly, we provide evidence that LegK7 forms a complex with MOB1A in order to use its NTE as a binding platform for the recruitment of downstream substrates, possibly even YAP1, so that their phosphorylation can occur more efficiently (Fig. 7). Together, these findings suggest that LegK7 has evolved into a functional chimera of MST1/2 and NDR/LATS, and that it combines features from both of these core kinases of the Hippo pathway in order to exploit the downstream signaling cascade.
Fig. 7.
Cartoon comparing canonical Hippo signaling with signaling by Legionella LegK7. In the canonical Hippo pathway, MST1 phosphorylates the NTE (T12 and T35) of MOB1A to promote formation of the MOB1A–LATS1 kinase complex. The activated MOB1A–LATS1 kinase complex phosphorylates the transcriptional regulators YAP1/TAZ, which results in degradation or cytoplasmic retention of YAP1/TAZ and, consequently, expression of genes involved in cell proliferation or differentiation. During infection, L. pneumophila translocates the effector kinase LegK7 into host cells where it directly phosphorylates T12 and T35 on the NTE of MOB1A. Similar to LATS1, LegK7 interacts with phosphorylated MOB1A and becomes highly active. LegK7 exploits the NTE of MOB1A to recruit downstream substrates, possibly YAP1, for further phosphorylation and manipulates the conserved Hippo pathway to promote infection.
First clues for the formation of a stable complex between LegK7 and MOB1A came from in vitro kinase reconstitution assays in which we detected a robust increase in autophosphorylation of LegK7 upon incubation with GST-MOB1A but not GST (Fig. 1A). We subsequently confirmed by gel-permeation chromatography (Fig. 1C) and ITC (Fig. 1D) that the two proteins form a complex with a 1:1 stoichiometry. The crystal structure of the complex between MOB1A33 and LegK7(11–530) further revealed that the LegK7 kinase domain shows notable fold similarity to eukaryotic protein kinases (SI Appendix, Fig. S2A), suggesting a common evolutionary kinase ancestor. Notably, LegK7 binds MOB1A predominantly through its kinase domain, with MOB1A33 making extensive contacts with both the C- and N-terminal lobes of the kinase domain as well as the N-terminal flanking domain (amino acids 11 to 172) of unknown function (Fig. 2A). This binding mode markedly differed from that described for LATS1/2 and NDR1/2 which use a domain that precedes their kinase domain to bind MOB1A (SI Appendix, Fig. S2 C and D) (8, 9). Given that MOB1A33 binds to the back side of LegK7(11–530) opposite the ATP- and substrate-binding pocket (Fig. 2A), it is unlikely that MOB1A stimulates LegK7 kinase activity by directly participating in the catalytic reaction mechanism. Instead, our MD simulations suggest that MOB1A binding stabilizes several key regions within the LegK7 kinase domain, including the activation loop and the catalytic loop (SI Appendix, Fig. S4A), which might shift the conformation of LegK7 to a more active state. Consistent with this model, we found that substitution of interface residues in either LegK7 or MOB1A disturbed complex formation between both proteins in vitro (SI Appendix, Fig. S3B) which reduced allosteric activation of LegK7 (Figs. 3B and 4). The flexibility of the LegK7 molecule in the absence of MOB1A would also explain why LegK7 could not be crystallized on its own despite low polydispersity in solution. We previously reported a similar mechanism of allosteric activation binding of UbcH7∼ubiquitin to the back side of the Shigella flexneri effector kinase OspG (31, 32).
The structures of mammalian MOB1A both alone (18) and in complex with short fragments from cellular kinases such as LATS1 (8, 12), MST2 (8), and NDR2 (9) have been determined. The structure presented here shows mammalian MOB1A bound directly to an entire kinase domain to stimulate the phosphotransfer reaction, thus providing insight into the molecular details of this host–pathogen interaction. The LegK7 binding site on MOB1A partially overlaps with that of the MOB1A-binding domain of LATS1 (amino acids 602 to 704) and NDR2 (amino acids 25 to 88) (Fig. 2C) (8, 9, 12). In each case, about 30 residues of MOB1A make contacts with the kinase fragment (Figs. 2C and 3A), most notably residue D63 of MOB1A which is located at the center of the interface and that, when substituted with arginine or valine, resulted in a failure of mutant MOB1A to bind to either LegK7 or LATS1 (SI Appendix, Fig. S3B) (9). We found that substitution of P282E or Y286A on the surface of LegK7 resulted in loss of MOB1A binding, suggesting that hydrophobic interactions within the center of the interface are a main contributor to complex formation (Fig. 3B and SI Appendix, Fig. S3A). Notably, based on ITC measurements, the complex between phosphorylated MOB1A and LegK7 has a Kd value that is comparable to that of phosphorylated MOB1A and NDR2 and ∼20-fold lower than that of MOB1A with LATS1 (Figs. 1D and 5A) (9), suggesting that LegK7 can compete with cellular NDR/LATS for the same binding site on MOB1A.
In its nonphosphorylated form, MOB1A is believed to adopt an autoinhibited conformation, with the NTE occupying parts of the NDR/LATS kinase-binding surface (12). Phosphorylation of T12 and T35 by cellular kinases such as MST1/2 creates an electrostatic repulsion that destabilizes the interaction with the kinase-binding epitope and causes the NTE of MOB1A to dissociate, thereby contributing to NDR/LATS binding (8). We found a similar effect of MOB1A phosphorylation on LegK7 binding and allosteric activation, where introduction of phosphomimetic mutations in position 12 and/or 35 (T12D and T35D) of MOB1A or deletion of the NTE domain resulted in a robust increase in the affinity of MOB1A for LegK7 (Fig. 5A) as well as a substantial enhancement of the allosteric activation of the LegK7 kinase activity (Fig. 5B). Since MOB1A33, which lacks the NTE, stimulated the kinase activity of LegK7 to a similar degree as phosphorylated MOB1A (SI Appendix, Fig. S1B), it is likely that LegK7 promotes MOB1A binding mainly by relieving autoinhibition through phosphorylation of residues T12 and T35.
It may seem contradictory that activation of the LegK7 kinase domain requires binding of phosphorylated MOB1A when at the same time phosphorylation of MOB1A requires the LegK7 kinase domain to be active. The most likely explanation of this apparent paradox is that the NTE domain in its nonphosphorylated form undergoes continuous cycles of association with and dissociation from the central domain of MOB1A, thus allowing association with LegK7. Its phosphorylation on T12 and T35 then shifts the equilibrium between these two states toward the active, derepressed form of MOB1A, which favors LegK7 binding. Indeed, nonphosphorylated MOB1A has only a 10-fold weaker affinity for LegK7 than does phosphorylated MOB1A (Kd ∼1.26 μM vs. Kd ∼0.12 μM) (Figs. 1D and 5A), a difference that is similar to that observed for complex formation between MOB1A and the NDR2 fragment (phosphorylated MOB1A Kd ∼0.578 μM vs. nonphosphorylated MOB1A Kd ∼12.3 μM) (9). Given that both T12 and T35 are located far away from the LegK7 catalytic center (SI Appendix, Fig. S7), it is unlikely that they are phosphorylated in cis, meaning by the same LegK7 molecule MOB1A is bound to. Instead, we favor a model in which these residues are phosphorylated in trans, meaning by a neighboring LegK7–MOB1A assembly.
Importantly, we provide evidence that the NTE, besides its well-established function in autoinhibiting MOB1A, plays an unexpected role in recruiting downstream cellular targets into the complex with its associated kinase. Using purified YAP1 as a model substrate in the kinase reconstitution assay, deletion of residues 1 to 32 from MOB1A or addition of a molar excess of GST-MOB1A(1–40) as a competitive inhibitor resulted in a dramatic reduction in YAP1 phosphorylation by LegK7 (Fig. 6), consistent with the idea that LegK7 requires MOB1A with an intact NTE to recruit downstream targets into the protein complex so that phosphorylation can occur efficiently. We hypothesize that the NTE of MOB1A has evolved to serve as a binding platform for various cellular ligands, and that LegK7 relies on MOB1A to compensate for the diversification of downstream components of the Hippo signaling pathway in its various hosts. Unlike the core components of the Hippo pathway, factors such as YAP1 and TAZ appear to be more diverse among different eukaryotic species (33). For example, proteins from Homo sapiens and Acanthamoeba castellanii share 72% (MOB1A), 47% (MST1), and 46% (LATS1) sequence identity, while A. castellanii homologs to human YAP or TAZ are absent, suggesting that other transcriptional coactivators function in their place. Thus, in order for L. pneumophila to alter Hippo signaling in a diverse set of host species, it may be beneficial for LegK7 to engage the most conserved host factor, MOB1A, within this pathway and to use the intrinsic ligand recruitment ability of its NTE in order to target the more diverse downstream components of the Hippo signaling pathway. This hypothesis is supported by the fact that the first 30 amino acids of MOB1 are the least conserved segment among MOB1A homologs from various eukaryotes, with some MOB1A homologs (from Caenorhabditis elegans and Saccharomyces cerevisiae) even containing extended NTEs that may assist in the recruitment of a diverse set of ligands (SI Appendix, Fig. S8).
In summary, our data show that LegK7, by combining features from both MST1/2 and LATS1/2, cannot only activate MOB1 but also exploit its intrinsic ligand recruitment capability for the modification of downstream ligands. We propose that L. pneumophila, by using this strategy, has developed the ability to survive within a wide range of diverse host species, including humans.
Materials and Methods
Bacterial Strains.
E. coli strains and L. pneumophila strains used in this study are listed in SI Appendix, Table S1. E. coli strains were cultured in LB (Luria-Bertani) media at 37 °C. L. pneumophila transformed with pflag plasmids were cultured for 2 d at 37 °C on CAYET agar plates containing 2 g/L activated charcoal, 10 g/L ACES (N-(2-acetamido)-2-aminoethanesulfonic acid), 10 g/L yeast extract, 400 μg/mL cysteine, 100 μg/mL thymidine, 135 μg/mL ferric nitrate, and 5 μg/mL chloramphenicol. L. pneumophila patches were scraped from the plates and cultured overnight at 37 °C in liquid AYE media containing 10 g/L ACES, 10 g/L yeast extract, 400 μg/mL cysteine, 100 μg/mL thymidine, 135 μg/mL ferric nitrate, 5 μg/mL chloramphenicol, and 250 μM IPTG (isopropyl β-d-1-thiogalactopyranoside, an inducer for expression of flag-tagged LegK7).
Murine and Human Cell Lines.
RAW264.7 macrophages and human embryonic kidney 293T cells were cultured at 37 °C in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 2 mM l-glutamine under 5% CO2 in humidified incubators.
Plasmids, Reagents, and Antibodies.
Plasmids, oligonucleotides, reagents, and antibodies used in this study are listed in SI Appendix, Table S1.
Recombinant Protein Purification.
For purification of maltose-binding protein-tagged LegK7 (MBP-LegK7) proteins, E. coli BL21(DE3) strains carrying plasmids encoding MBP-LegK7 were cultured in 2xYT media at 37 °C for 3 h before production of MBP-LegK7 proteins was induced by addition of 500 μM IPTG. After 2 h of induction at 37 °C, the bacteria were pelleted and resuspended in MBP lysis buffer (20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM ethylenediaminetetraacetate, 1 mM β-mercaptoethanol, and 1× protease inhibitor mixture) for MBP-LegK7. Resuspended bacteria were lysed using a Microfluidics (M110P) cell breaker and then centrifuged to remove cell debris. The cleared lysate was incubated with amylose resins (New England BioLabs) to bind MBP-LegK7 proteins according to the manufacturer’s guidelines. For GST, GST-flag-YAP1, and GST-MOB1A proteins, E. coli BL21(DE3) strains carrying plasmids encoding GST, GST-flag-YAP1, or GST-MOB1A proteins were cultured in LB media at 37 °C for 3 h before production of the GST recombinant proteins was induced with 500 μM IPTG. Following further incubation overnight at 20 °C, the bacterial cells were pelleted and resuspended in GST lysis buffer (1× phosphate-buffered saline, pH 7.4, 1 mM β-mercaptoethanol, and 1× protease inhibitor mixture). The resuspended bacteria were then lysed using a Microfluidics (M110P) cell breaker and centrifuged to remove cell debris. The cleared lysate was incubated with glutathione Sepharose resins (GE Healthcare) to bind GST recombinant proteins according to the manufacturer’s manual. After affinity purification, the purified recombinant proteins were dialyzed in buffer containing 10 mM Hepes (pH 7.3) and 150 mM NaCl using Snakeskin dialysis tubing membranes (3.5-kDa pore size; Thermo Fisher Scientific) prior to use in the in vitro kinase assay.
Site-directed mutagenesis was performed with a Q5 Site-Directed Mutagenesis Kit (New England BioLabs) according to the manufacturer’s instructions. All mutant proteins were purified in the same manner as were the corresponding wild-type proteins.
In Vitro Kinase Assays.
Purified recombinant proteins were coincubated in 30 μL of 10 mM Hepes (pH 7.3), 150 mM NaCl, 10 mM MgCl2, and 1 mM ATPγS at 30 °C for 45 min. After incubation, thiophosphorylated proteins were alkylated for 2 h in the dark at room temperature following addition of 1.5 μL of 50 mM p-nitrobenzyl-mesylate. Alkylation was terminated by addition of 31.5 μL of 2× sodium dodecyl sulfate (SDS) protein sample buffer (100 mM Tris⋅HCl, pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol, and 200 mM β-mercaptoethanol) and incubation for 10 min at 95 °C. Alternatively, 1 mM regular ATP was used to replace ATPγS as a phosphate donor in the in vitro kinase assay. For kinase assays with ATP, the reaction was performed at 30 °C for 45 min and terminated by addition of 30 μL of 2× SDS protein sample buffer and incubation for 10 min at 95 °C. Protein samples from the kinase assays were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Thiophosphorylation of proteins was detected using a thiophosphate ester-specific primary antibody followed by a horseradish peroxidase (HRP)-conjugated secondary antibody. Phosphorylation of T12 and T35 of MOB1A proteins was detected using anti–phospho-T12 and anti–phospho-T35 primary antibodies followed by an HRP-conjugated secondary antibody. Blots were developed using an enhanced chemiluminescence (ECL) assay, and images of the blots were acquired using a ChemiDoc-MP Imaging System (Bio-Rad). All blots are representatives of at least three independent experiments.
Detailed materials and methods for this study are described in SI Appendix.
Data Availability Statement.
The coordinates and structure factors of the nonphosphorylated LegK7(11–530)–MOB1A(33–216) apo complex and phosphorylated LegK7(11–530)–MOB1A(33–216) complex with ATP analog were deposited in the Protein Data Bank with ID codes 6MCQ and 6MCP, respectively. The data that support the findings of this study are included within the main manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the M.P.M. and M.C. laboratories for comments and discussion. This work was funded by the Intramural Research Program of the NIH (Project no. 1ZIAHD008893-09) (to M.P.M.), Wayne State University startup funds (to P.-C.L.), and Canadian Institutes of Health Research (CIHR; Grant MOP-48370) (to M.C.). We thank the staff of beamline 08ID at the Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, National Research Council Canada, CIHR, Province of Saskatchewan, Western Economic Diversification Canada, and University of Saskatchewan. We gratefully acknowledge the use of instruments at the Protein Characterization and Crystallization Facility, College of Medicine, University of Saskatchewan funded by the Canadian Foundation for Innovation. We thank the members of Information and Communications Technology, University of Saskatchewan for access to the Research Computing Cluster resources, Mr. Denis Spasyuk for help with visualization of MD simulations, and Drs. Guanghui Wang and Marjan Gucek (both of the National Heart, Lung, and Blood Institute proteomics core) for technical support and sample processing.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors of the nonphosphorylated LegK7(11–530)–MOB1A(33–216) apo complex and phosphorylated LegK7(11–530)–MOB1A(33–216) complex with ATP analog have been deposited in the Protein Data Bank, https://www.pdb.org (PDB ID codes 6MCQ and 6MCP, respectively).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000497117/-/DCSupplemental.
References
- 1.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S., The protein kinase complement of the human genome. Science 298, 1912–1934 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Safaei J., Maňuch J., Gupta A., Stacho L., Pelech S., Prediction of 492 human protein kinase substrate specificities. Proteome Sci. 9 (suppl. 1), S6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grishin A. M., Beyrakhova K. A., Cygler M., Structural insight into effector proteins of gram-negative bacterial pathogens that modulate the phosphoproteome of their host. Protein Sci. 24, 604–620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park B. C., Reese M., Tagliabracci V. S., Thinking outside of the cell: Secreted protein kinases in bacteria, parasites, and mammals. IUBMB Life 71, 749–759 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hergovich A., MOB control: Reviewing a conserved family of kinase regulators. Cell. Signal. 23, 1433–1440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng Z., Moroishi T., Guan K. L., Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Praskova M., Xia F., Avruch J., MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18, 311–321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni L., Zheng Y., Hara M., Pan D., Luo X., Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 29, 1416–1431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulaberoglu Y. et al., Stable MOB1 interaction with Hippo/MST is not essential for development and tissue growth control. Nat. Commun. 8, 695 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hergovich A., Regulation and functions of mammalian LATS/NDR kinases: Looking beyond canonical Hippo signalling. Cell Biosci. 3, 32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishio M. et al., Cancer susceptibility and embryonic lethality in Mob1a/1b double-mutant mice. J. Clin. Invest. 122, 4505–4518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S.-Y., Tachioka Y., Mori T., Hakoshima T., Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway. Sci. Rep. 6, 28488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee P.-C., Machner M. P., The Legionella effector kinase LegK7 hijacks the host Hippo pathway to promote infection. Cell Host Microbe 24, 429–438.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser D. W. et al., Legionnaires’ disease: Description of an epidemic of pneumonia. N. Engl. J. Med. 297, 1189–1197 (1977). [DOI] [PubMed] [Google Scholar]
- 15.McDade J. E. et al., Legionnaires’ disease: Isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297, 1197–1203 (1977). [DOI] [PubMed] [Google Scholar]
- 16.Allen J. J. et al., A semisynthetic epitope for kinase substrates. Nat. Methods 4, 511–516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hergovich A., Schmitz D., Hemmings B. A., The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem. Biophys. Res. Commun. 345, 50–58 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Stavridi E. S. et al., Crystal structure of a human Mob1 protein: Toward understanding Mob-regulated cell cycle pathways. Structure 11, 1163–1170 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Couzens A. L. et al., MOB1 mediated phospho-recognition in the core mammalian Hippo pathway. Mol. Cell. Proteomics 16, 1098–1110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanks S. K., Hunter T., Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596 (1995). [PubMed] [Google Scholar]
- 21.Johnson L. N., Lewis R. J., Structural basis for control by phosphorylation. Chem. Rev. 101, 2209–2242 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Holm L., Kääriäinen S., Rosenström P., Schenkel A., Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian K. C. et al., Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J. Biol. Chem. 280, 6130–6137 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Hamill S., Lou H. J., Turk B. E., Boggon T. J., Structural basis for noncanonical substrate recognition of cofilin/ADF proteins by LIM kinases. Mol. Cell 62, 397–408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seganish W. M. et al., Discovery and structure enabled synthesis of 2,6-diaminopyrimidin-4-one IRAK4 inhibitors. ACS Med. Chem. Lett. 6, 942–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krissinel E., Henrick K., Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Weijman J. F. et al., Structural basis of autoregulatory scaffolding by apoptosis signal-regulating kinase 1. Proc. Natl. Acad. Sci. U.S.A. 114, E2096–E2105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Praskova M., Khoklatchev A., Ortiz-Vega S., Avruch J., Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 381, 453–462 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan F. et al., Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration. Sci. Transl. Med. 8, 352ra108 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Hirabayashi S. et al., Threonine 74 of MOB1 is a putative key phosphorylation site by MST2 to form the scaffold to activate nuclear Dbf2-related kinase 1. Oncogene 27, 4281–4292 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Grishin A. M. et al., Regulation of Shigella effector kinase OspG through modulation of its dynamic properties. J. Mol. Biol. 430, 2096–2112 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Grishin A. M. et al., Structural basis for the inhibition of host protein ubiquitination by Shigella effector kinase OspG. Structure 22, 878–888 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Sebé-Pedrós A., Zheng Y., Ruiz-Trillo I., Pan D., Premetazoan origin of the Hippo signaling pathway. Cell Rep. 1, 13–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors of the nonphosphorylated LegK7(11–530)–MOB1A(33–216) apo complex and phosphorylated LegK7(11–530)–MOB1A(33–216) complex with ATP analog were deposited in the Protein Data Bank with ID codes 6MCQ and 6MCP, respectively. The data that support the findings of this study are included within the main manuscript and SI Appendix.