Significance
Eukaryotic genomes harbor a vast number of “selfish” DNA elements, including transposable elements and repetitive sequences. They constitute nearly 50% of the human genome and need to be silenced to maintain the integrity of the genome. Aberrant expression of such sequences, possibly due to failure of silencing mechanisms, is associated with human diseases, including cancer. Silencing of these “selfish” DNAs involves methylation of specific lysine residues in the nucleosome core particles that help package these DNA elements into chromatin in the cell nucleus. Here we demonstrate that H1 linker histones, the most abundant chromatin-binding proteins, are critical for silencing of these sequences, by promoting repressive lysine methylation and further compacting these elements into more condensed chromatin structures.
Keywords: epigenetics, linker histones, repetitive elements, chromatin
Abstract
Nearly 50% of mouse and human genomes are composed of repetitive sequences. Transcription of these sequences is tightly controlled during development to prevent genomic instability, inappropriate gene activation and other maladaptive processes. Here, we demonstrate an integral role for H1 linker histones in silencing repetitive elements in mouse embryonic stem cells. Strong H1 depletion causes a profound de-repression of several classes of repetitive sequences, including major satellite, LINE-1, and ERV. Activation of repetitive sequence transcription is accompanied by decreased H3K9 trimethylation of repetitive sequence chromatin. H1 linker histones interact directly with Suv39h1, Suv39h2, and SETDB1, the histone methyltransferases responsible for H3K9 trimethylation of chromatin within these regions, and stimulate their activity toward chromatin in vitro. However, we also implicate chromatin compaction mediated by H1 as an additional, dominant repressive mechanism for silencing of repetitive major satellite sequences. Our findings elucidate two distinct, H1-mediated pathways for silencing heterochromatin.
Eukaryotic genomes are packaged into chromatin, a complex polymer of DNA, RNA, and protein that facilitates compaction of the long strands of linear DNA into the nucleus. The repeating unit of chromatin, the nucleosome, consists of a core histone octamer—two each of the core histones H2A, H2B, H3, and H4—wrapped around 147 bp of DNA. A fifth histone, the linker histone H1, dynamically associates (1, 2) with the nucleosome core particle at the dyad axis and facilitates the formation of more compact chromatin structures (3). In addition to controlling access to the underlying DNA via steric hinderance, the histone proteins carry a multitude of posttranslational modifications that influence many nuclear processes, including transcription, DNA repair, and replication (4).
Although linker histones are some of the most abundant nuclear proteins, our understanding of their functions of remains very incomplete. Mammals express multiple linker histone subtypes or variants, including seven somatic subtypes and four germ cell-specific subtypes (5). We previously generated mice inactivated for one or two H1 subtypes and discovered that cells can maintain their total H1 content through compensatory up-regulation of their remaining H1 genes (6). However, mice genetically ablated for three highly expressed H1 subtypes exhibit embryonic lethality, demonstrating that linker histones are essential for mammalian development (7). Embryonic stem (ES) cells derived from these triple-knockout (TKO) embryos have a reduced (∼50%) total H1 content and display specific changes in gene expression due to locus-specific alterations in DNA methylation and histone H3 lysine 4 methylation (8–10). These results, as well as data from numerous other studies (11, 12), have increasingly supported the view that in addition to their well-established structural roles in chromatin, H1 linker histones are intimately involved in epigenetic regulation of chromatin function. However, the extent, genomic contexts, and mechanisms of this regulation have not been fully explored. To further investigate the roles of H1 in epigenetic regulation, we have used CRISPR-Cas9–mediated genome editing to inactivate additional H1 genes in the H1 TKO ES cells and thereby deplete the H1 content to even lower levels.
Nearly 50% of the mouse and human genomes consist of repetitive sequences, including tandem repeats, such as satellite sequences, as well as interspersed elements, such as long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and endogenous retroviruses (ERVs) (13). Transcription of these sequences is tightly controlled during development (14). Whereas transcription of certain repetitive sequences has been reported to be required for normal cellular processes, such as heterochromatin formation and embryonic development (15–17), aberrant transcription of some repetitive sequences has been implicated in disease states, including cancer (18, 19).
The establishment and maintenance of a repressive chromatin environment is an important mechanism for the silencing of these repetitive sequences. Silencing of these sequences in mouse ES cells involves trimethylation of histone 3 lysine 9 (H3K9me3) catalyzed by three histone methyltransferases (HMTs): Suv39h1, Suv39h2, and SETDB1 (20–23). In mESCs null for both Suv39h1 and Suv39h2, H3K9me3 is lost from pericentric heterochromatin, and major satellite and LINE elements are de-repressed (20, 24). On the other hand, SETDB1 appears to be responsible for H3K9 methylation and silencing of class I and II ERVs (22, 23). However, H3K9 methylation alone is unlikely to be the sole determinant of the transcriptional status of the underlying repetitive sequence, because loss of H3K9 methylation does not always lead to their de-repression (23). Furthermore, DNA methylation extensively decorates repetitive elements in both ES cells and somatic tissues and in some cases is lost concomitantly with H3K9 methylation (20, 24). However, genetic ablation of the three DNA methyltransferases does not lead to widespread de-repression of repetitive element transcription (22); thus, the complex interplay between epigenetic marks and other nuclear factors in governing the transcription of these repetitive sequences has yet to be fully elucidated.
Here we describe an integral role for H1 linker histones in silencing repetitive element transcription in mouse ES cells. Using computational approaches, we uncover a set of H1 enriched domains and show that they significantly overlap with constitutive heterochromatin occupied by Suv39h1, Suv39h2, and SETDB1. Next, using ES cells with only one functional H1 allele, we show that severe H1 depletion leads to a profound de-repression of major satellite transcripts, to much higher levels than is seen in Suv39h1/2 double-null cells, as well as de-repression of LINE-1 and ERV transcripts. This de-repression is accompanied by a reduction in H3K9me3 at the affected loci. Mechanistically, H1 interacts directly with Suv39h1, Suv39h2, and SETDB1 and stimulates their methyltransferase activities toward chromatin in vitro. Finally, we provide evidence that in addition to promoting H3K9 methylation, H1-mediated chromatin compaction is a critical mechanism for fully silencing major satellites in pericentromeric heterochromatin.
Results
H1 Is Enriched in Constitutive Heterochromatin Silenced by Suv39h1/2 and SETDB1.
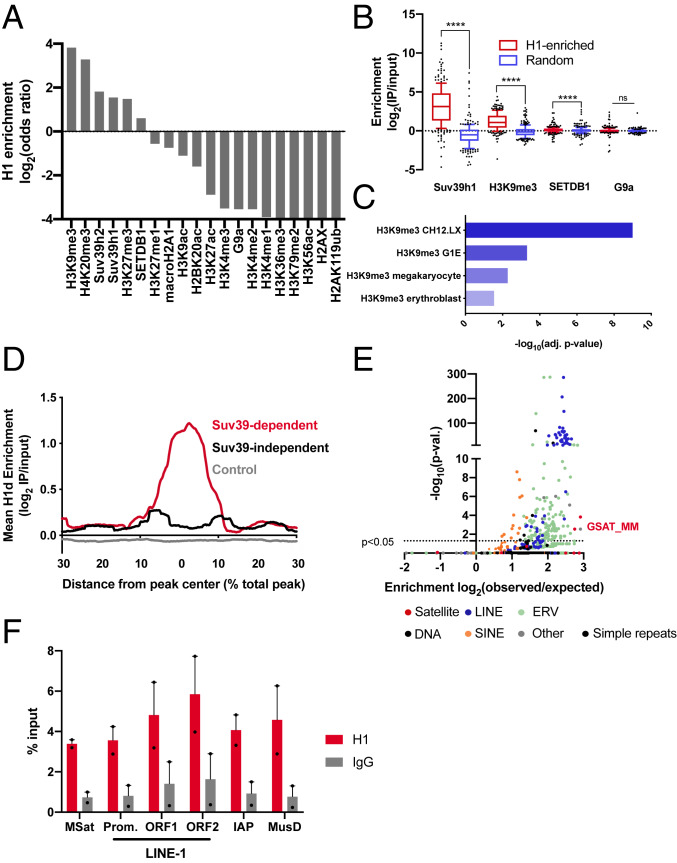
H1 linker histones are abundant nuclear proteins that are reported to display widespread genomic binding except at promoters of active genes, where they are depleted (25–27). To discover regions of H1 enrichment, we used the recently developed ISOR algorithm (28), and publicly available H1 ChIP-seq data in mouse embryonic stem cells (mESCs) (26) to analyze the genome-wide profile of H1 binding in these cells. ISOR iteratively segments the genome and compares ChIP and input reads to detect regions of variable length that are enriched or depleted for a given factor. We applied ISOR to ChIP-seq data of H1d, the most abundant H1 subtype in mESCs (8). Although mESCs express primarily five different H1 subtypes, nearly all studies of genome-wide occupancy of H1 have found very strong correlations among these H1 subtypes (25, 26); therefore, we used H1d occupancy as a surrogate for H1 occupancy generally. We compared H1-enriched regions (P < 0.00001; enrichment score [ES] >0.2) to published ChIP-seq data for several factors and histone posttranslational modifications in mESCs and identified chromatin features significantly enriched or depleted for H1 (Fig. 1A). Notably, this analysis indicates that H1 is significantly enriched (odds ratio >8) in trimethylated lysine 9 of histone H3 (H3K9me3)-marked chromatin. In addition, H1 is enriched in chromatin occupied by Suv39h1, Suv39h2, and SETDB1, H3K9 methyltransferases that are critical for the silencing of mammalian constitutive heterochromatin (22, 29). In contrast, the ISOR analysis showed that H1 is highly depleted from genomic regions occupied by G9a, an H3K9 di-methyltransferase associated primarily with euchromatin (30), as well as from regions of “active” chromatin containing several types of H3 acetylated residues (H3K27ac, H3K9ac, and H3K56ac) and H3K4 methylation. The latter result is consistent with previous studies reporting a depletion of H1 at active promoters, which are marked by H3K4me3 (25).
Fig. 1.
H1 is enriched within constitutive heterochromatin silenced by Suv39h1/2. (A) Overlap of H1d and histone posttranslational modifications or chromatin proteins in mESCs. ChIP-seq data for a number of histone PTMs and chromatin factors in mESCs was analyzed by ISOR to detect enriched regions. The overlap of H1d-enriched domains with each factor or modification is expressed as a log2(odds ratio). (B) The enrichment of Suv39h1, H3K9me3, SETDB1, and G9a within H1-enriched domains was compared to matched control genomic regions of equal size and number. Enrichment is expressed as log2(IP reads/input reads). (C) Significantly enriched histone posttranslational modifications in H1-enriched domains. H1-enriched ISOR regions (P < 0.01; ES >0.5) were analyzed by Enrichr (31) to detect significant overlaps with histone posttranslational modification ChIP-seq datasets. (D) H1 enrichment in Suv39-dependent, Suv39-independent, and control domains. Suv39-dependent and -independent H3K9me3 domains were determined (SI Appendix, Fig. S1 A and B), and the enrichment of H1d within these domains was analyzed using HOMER. Due to variable domain sizes, the x-axis is displayed as a percent of total peak length from the center. (E) Overlap of H1-enriched domains and repetitive elements genome-wide. H1 ISOR domains were analyzed by HOMER, and enrichment is expressed as log2(observed/expected) overlap. (F) Enrichment of H1a at repetitive elements. ChIP-qPCR was performed on fixed chromatin from ES cells using H1a and control IgG antisera. Enrichment is expressed a percentage of input DNA fragment recovered. ns, not statistically significant; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. The Wilcoxon signed-rank test was used to test for significance in B.
We further investigated the occupancy of some of the chromatin marks and protein factors identified in the ISOR analysis within H1-enriched domains. Suv39h1, H3K9me3, and, to a lesser extent, SETDB1 are significantly enriched over input in H1-enriched regions compared with random genomic regions of equal size and number (Fig. 1B). In contrast, we observed no difference in enrichment of G9a ChIP-seq reads within H1-enriched and control domains, supporting the ISOR analysis, which showed a negative odds ratio for the overlap of regions enriched for both G9a and H1.
To further explore genomic features associated with H1 enrichment, we used Enrichr, a web-based genomic enrichment tool (31). This analysis showed that several H3K9me3 ChIP-seq datasets from several cell types and tissues are enriched within H1 ISOR domains (Fig. 1C), suggesting that this relationship may be conserved in differentiated somatic tissues.
Given the significant enrichment of both H3K9me3 and Suv39h1 in H1 domains, we next asked whether H1 occupancy is increased specifically within genomic regions in which Suv39h1/2 is required for H3K9me3 deposition. Using published H3K9me3 ChIP-seq data in wild-type (WT) and Suv39h1/h2 double-null (Suv39-dn) mESCs (24), we defined a set of domains in which H3K9 trimethylation is decreased on deletion of these two factors (SI Appendix, Fig. S1 A and B). Within these Suv39-dependent H3K9me3 domains, we observe a marked enrichment of H1d ChIP-seq reads that is not present at matched control genomic regions (Fig. 1D). Similar results were obtained using ChIP-seq data of H1c, another highly expressed linker histone subtype in mESCs (SI Appendix, Fig. S1D).
The Suv39h1/h2 and SETDB1 methyltransferases have been implicated in repressing several different types of repetitive elements in mESCs (22–24). Therefore, we explored whether H1-enriched domains identified by ISOR are enriched in repetitive sequences genome-wide. Notably, H1-enriched domains significantly overlapped with several hundred annotated repeat regions, including LINEs, SINEs, and long-terminal repeat-containing ERVs (Fig. 1E). Importantly, none of the annotated repeats was significantly depleted within H1-enriched domains. In contrast, matched control genomic regions did not significantly overlap with repeat regions (SI Appendix, Fig. S1E). Of note, pericentromeric satellite sequences, a major target of Suv39-mediated silencing (20), are among the most highly enriched repeat sequences within the H1-enriched domains. ChIP-qPCR of H1 in mESCs confirmed the observed enrichment within major satellite, LINE, and ERV repeat sequences (Fig. 1F). Taken together, this analysis indicates that, whereas the genome-wide average stoichiometry of H1 to nucleosomes is 1:2 in mESCs (8), H1 is significantly enriched in repeat-rich regions of heterochromatin that are repressed by Suv39h1/2- and SETDB1-mediated H3K9 methylation in these cells.
H1 Is Required for Transcriptional Repression of Several Classes of Repetitive Sequences.
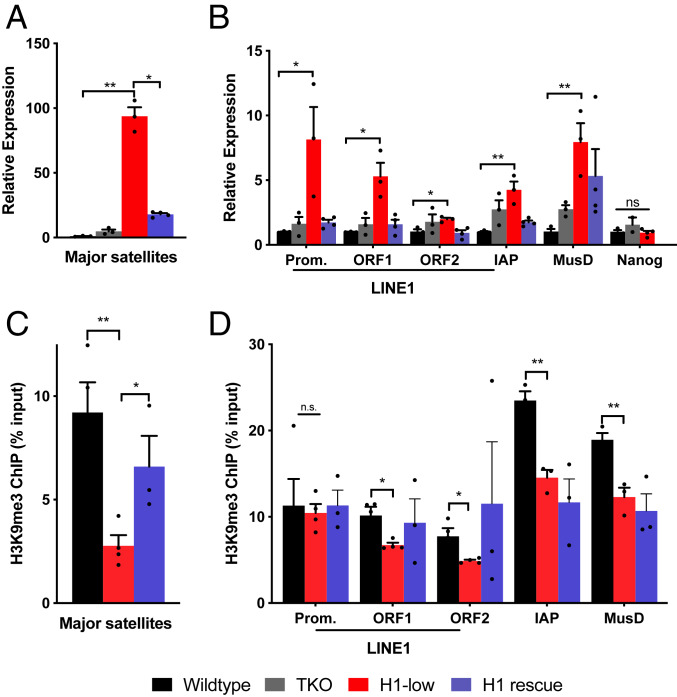
The strong enrichment of H1 in regions of the genome that harbor repetitive sequences and are repressed via Suv39h1/2- and SETDB1-mediated H3K9 methylation suggests that H1 may be required for maintaining silencing of these regions. To investigate this possibility, we measured the transcript levels of several repetitive elements within the satellite, LINE, and LTR families, specifically major satellites and LINE-1 elements that are repressed by Suv39h1/2 (20, 24) and ERV family members intracisternal A particle (IAP) and early transposon Mus musculus group D (ETn_MusD) that are repressed by SETDB1 (23). Transcript levels were compared in three clones of our previously described H1c/d/e TKO mESCs (8) and in WT control cells. For each of the repetitive elements assayed, we observed a mild increase in transcript level in some clones (Fig. 2 A and B).
Fig. 2.
H1 is required for transcriptional repression of several classes repetitive elements. (A and B) Transcript levels of the indicated repetitive sequences were measured by RT-qPCR in WT, TKO, H1-low, and H1 rescue lines stably expressing H1d. Values were normalized to Gapdh mRNA using the ΔΔCt method. Prom., promoter. Error bars represent SEM of three or four independently derived clones per condition. (C and D) H3K9me3 ChIP-qPCR in WT, H1-low, and H1 rescue lines. Cross-linked chromatin was immunoprecipitated with an H3K9me3 antibody, and recovered DNA was quantified by PCR using primers for the indicated repetitive sequences. Data are shown as a percentage of input DNA fragments. Error bars indicate SEM of three or four independent clones per condition. ns, not statistically significant; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
One possible explanation for the relatively modest effects of H1 depletion on repetitive sequence transcripts in TKO mESCs is that the total H1 content in these cells is reduced by only 50% compared with WT, owing to compensatory expression of the remaining H1 subtypes, H1a and H1b (8). Therefore, we sought to further reduce H1 levels in the TKO mESCs by targeted deletion of Hist1h1a and Hist1h1b with CRISPR-Cas9. We generated several mESC lines in which both alleles of Hist1h1b and one of the two alleles of Hist1h1a are deleted. These cells possess just 20% of the normal H1 level in mESCs (SI Appendix, Fig. S2 A and B). Strikingly, we observed a profound, 100-fold de-repression of major satellite transcripts in these highly H1-depleted (H1-low) mESCs (Fig. 2A). Similar, robust de-repression was also observed for the transcripts of several LINE-1 elements (promoter, ORF1, and ORF2) and ERVs (IAP and MusD) (Fig. 2B). In contrast, we did not observe significant differences in expression of the pluripotency gene Nanog among WT, TKO, and H1-low mESCs (Fig. 2B). These results indicate that H1 linker histones make a strong contribution to maintaining silencing of several different types of repetitive sequences in mESCs.
To determine whether H1 is simply required for maintaining silencing of these elements or whether it can initiate silencing, we used CRISPR-Cas9–mediated homology-directed repair to reintroduce an H1d gene into the H1-low mESCs by targeted integration of the gene into one of the previously inactivated H1d loci. Screening of clones by quantitative high-performance liquid chromatography allowed us to obtain several lines with total H1 levels similar to those observed in TKO mESCs (SI Appendix, Fig. S2 A and B). We observed that expression of the exogenous H1d protein in these cells restored repression of each type of repetitive element (Fig. 2 A and B). These results suggest that H1 is capable of initiating silencing of these repetitive sequences.
An important mechanism for silencing these repetitive sequences in mESCs is trimethylation of lysine 9 in nucleosomal histone H3, catalyzed by Suv39h1/2 and SETDB1. To determine whether H1-mediated silencing of these repetitive sequences involves H3K9 methylation, we performed ChIP-qPCR for H3K9me3 at these loci in H1-low, H1d restored, and WT mESC lines. We found that H3K9me3 was reduced at all the repetitive sequences except the LINE-1 promoter in H1-low mESCs (Fig. 2 C and D). Moreover, within major satellite sequences, H3K9 methylation was nearly fully restored in the cells in which exogenous H1d was expressed (Fig. 2 C and D). However, H3K9 methylation was not significantly restored at LINE-1 sequences, or at the IAP and MusD elements, despite the clear restoration of repression of these sequences by exogenous H1. These latter results suggest the operation of alternative, possibly redundant H1-mediated silencing pathways at these loci (see below). Nevertheless, the observations at major satellite sequences suggest that H1 linker histones cooperate with Suv39h1 and Suv39h2 to promote deposition of the repressive H3K9me3 mark and repress major satellites in mESCs.
Linker Histones Interact Directly with Suv39h1/h2 and SETDB1 and Promote H3K9 Methylation In Vitro.
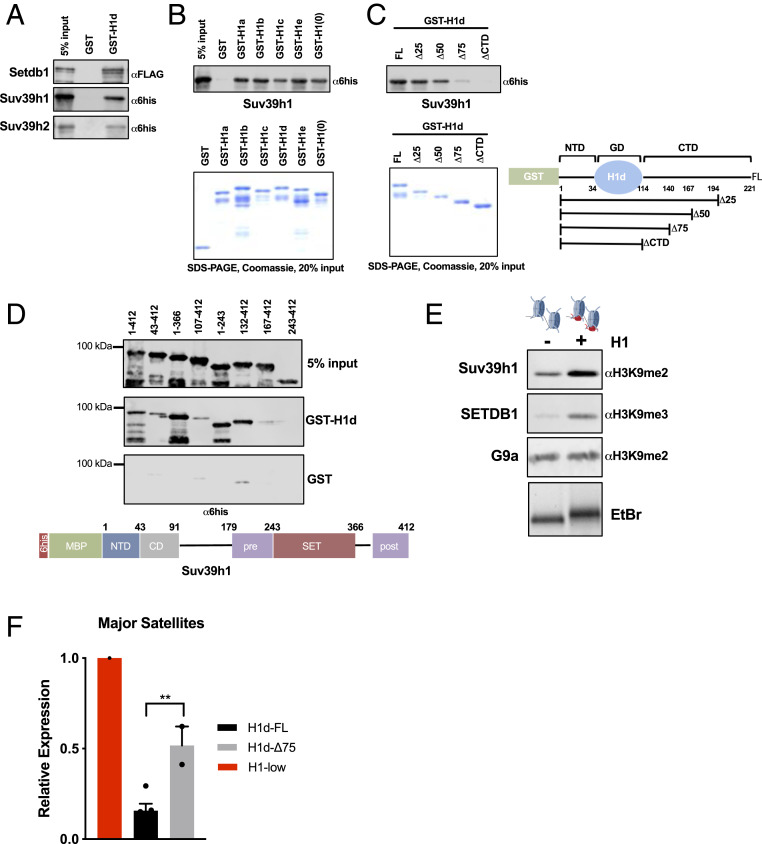
The preceding results suggest that H1 may cooperate with Suv39h1/2 and SETDB1 to promote silencing of several types of repetitive elements. Previous results from our laboratory showed that Drosophila H1 physically and functionally interacts with Su(var)3-9, the fly ortholog of Suv39h1/h2, to promote H3K9 methylation and repression of repetitive sequences in Drosophila (32). To begin to explore possible functional relationships between H1 and the three mammalian enzymes, we carried out in vitro protein–protein interaction studies and found that a GST-fusion protein of H1d interacts with recombinant Suv39h1, Suv39h2, and SETDB1 (Fig. 3A). This interaction is unlikely to be due to bridging by contaminating nucleic acids, because we found that interaction between GST-H1d and Suv39h1 was unaffected by inclusion of ethidium bromide in the pulldown assay (SI Appendix, Fig. S3A). Furthermore, we observed that five other H1 subtypes expressed in somatic cells interacted with both Suv39h1 and Suv39h2 (Fig. 3B and SI Appendix, Fig. S3B).
Fig. 3.
Linker histones interact with Suv39h1/h2 and SETDB1 and promote H3K9 methylation in vitro. (A) Interaction studies between H1 and H3K9 methyltransferases. Linker histone GST fusion proteins were purified, bound to glutathione Sepharose beads, and incubated with the indicated recombinant proteins tagged with either hexahistidine-maltose binding protein (6his-MBP) or Flag peptides. Bound proteins and 5% input control samples were analyzed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) followed by immunoblotting for the indicated peptides. (B and C) GST pulldown assays as in A with the indicated proteins. Coomassie blue-stained SDS/PAGE gel of purified proteins showing equal loading (Lower). (C, Right) Schematic representation of GST-H1d polypeptides used in interaction assays. Numbers indicate amino acid residues. (D) GST pulldown assays as in A with the indicated polypeptide fragments of Suv39h1. After incubation with GST or GST-H1d, pulldown products and 5% input controls were analyzed with a hexahistidine(6his)-specific antibody. A schematic illustration of Suv39h1 domain structure is shown below. (E) In vitro HMT assays with reconstituted chromatin in the presence or absence of H1. Chromatin was reconstituted in vitro using a DNA template bearing two repeats of the synthetic “601” nucleosome positioning sequence and recombinant histone octamers in either the presence or absence of H1. H1 incorporation was verified by nondenaturing agarose gel electrophoresis followed by ethidium bromide (EtBr) staining, demonstrating slower migration of H1-containing dinucleosomes (Lower). Dinucleosomes were incubated with the indicated enzymes (Suv39h1, 100 nM; SETDB1, 50 nM; G9a, 20 nM) under conditions described in Materials and Methods. Enzymatic activity was detected by immunoblotting for the indicated histone modifications. (F) RT-qPCR of major satellite transcripts. Expression of major satellites was quantified in H1-low lines in which either full-length H1d (H1d-FL) or H1d lacking 75% of the CTD (H1d-Δ75) bearing an N-terminal 3xFlag tag were reintroduced via stable transfection. Expression (relative to the parental line) was calculated using the ΔΔCt method normalized to Gapdh mRNA. n = 2 or 3 independent clones per condition.
Linker histones possess a tripartite domain structure consisting of a short, unstructured N-terminal domain (NTD), a central globular domain (GD), and a long, intrinsically disordered C-terminal domain (CTD) (33). We previously reported that the CTD of H1d is required for its physical and functional interactions with two DNA methyltransferases (9). To determine whether the CTD is also involved in the interactions of H1d with Suv39h1/2 and SETDB1, we prepared a series of GST fusion proteins in which portions of the CTD are deleted. We observed somewhat diminished binding of Suv39h1 to H1d terminated at residue 167, lacking 50% of the CTD, greatly diminished binding to H1d terminated at residue 140, and a complete loss of binding to H1d lacking its CTD (Fig. 3C). Similar results were observed with Suv39h2 (SI Appendix, Fig. S3B), and we determined that the H1d CTD is also required for interaction with SETDB1 (SI Appendix, Fig. S3C).
To further explore the H1–Suv39h1 interaction, we asked which domains of Suv39h1 interact with H1d. To do so, we prepared a series of N- and C-terminally deleted fragments of Suv39h1, as well as domains representing the NTD (residues 1 to 43), the chromodomain (CD; residues 43 to 91), the pre-SET domain (residues 179 to 243), the SET domain (residues 243 to 366) and a C-terminal fragment including the post-SET domain (residues 366 to 412) (Fig. 3D). Loss of the NTD fragment greatly diminished binding of Suv39h1 to H1d, although the NTD fragment alone did not bind H1d (Fig. 3D and SI Appendix, Fig. S3D). On the other hand, deletion of the SET and post-SET domains had no effect on H1d binding (Fig. 3D). However, binding experiments with individual domains showed that the CD, pre-SET, and SET domains each bound H1d (SI Appendix, Fig. S3D). Therefore, several regions of Suv39h1 may be involved in binding H1, and the binding appears to be influenced by the presence of the Suv39h1 NTD.
The direct interaction between H1 and Suv39h1/2 and SETDB1, coupled with the observation that H3K9me3 is reduced at the genomic targets of these enzymes in H1-depleted mESCs, suggests that H1 may promote the HMT activities of these enzymes toward chromatin. To test this possibility, we reconstituted chromatin in vitro with and without H1 (SI Appendix, Fig. S3 F and G) and measured the activity of Suv39h1 and SETDB1 toward these substrates. The presence of H1 in the chromatin strongly stimulated the activities of both enzymes to methylate H3 in the dinucleosome substrate (Fig. 3E). In contrast, the incorporation of H1 into chromatin had no effect on the activity of G9a, an HMT primarily responsible for H3K9 dimethylation in euchromatin (30) (Fig. 3E). In addition, the activity of MLL1, a major H3K4 methyltransferase, was actually decreased on nucleosome substrates containing H1 (SI Appendix, Fig. S3E), consistent with our previous results demonstrating inhibition of SET7/9 by H1d (9). These results are consistent with the computational analyses indicating significant overlap between H1-enriched domains and regions occupied by Suv39h1/2 and SETDB1, as well as depletion of G9a in such domains (Fig. 1 A and B).
As mentioned above, Suv39h1 and Suv39h2 together are required for the repression of major satellite transcription (20). The results presented here implicate H1 in the silencing of these loci, and protein–protein interaction experiments indicate that the CTD of H1d is required for its interaction with Suv39h1 and Suv39h2 in vitro. To test whether the H1 CTD is required for silencing of major satellite transcription in vivo, we compared the ability of exogenous full-length H1d and H1d lacking 75% of the CTD (H1d-Δ75) to restore repression of satellite transcription in H1-low mESCs. We used this construct because, like H1d lacking the CTD entirely (H1d-Δ100), H1d-Δ75 does not interact with Suv39h1/h2, and, unlike H1d-Δ100 (34), H1d-Δ75 retains substantial chromatin binding (SI Appendix, Fig. S3H). While full-length H1d was able to restore repression, H1d-Δ75 exhibited substantially impaired silencing activity (Fig. 3F). These results support the view that interaction of H1 with Suv39h1/h2 contributes to H1-mediated repression of major satellite transcription.
H1-Mediated Chromatin Compaction Is the Dominant Mechanism for Silencing Pericentric Heterochromatin.
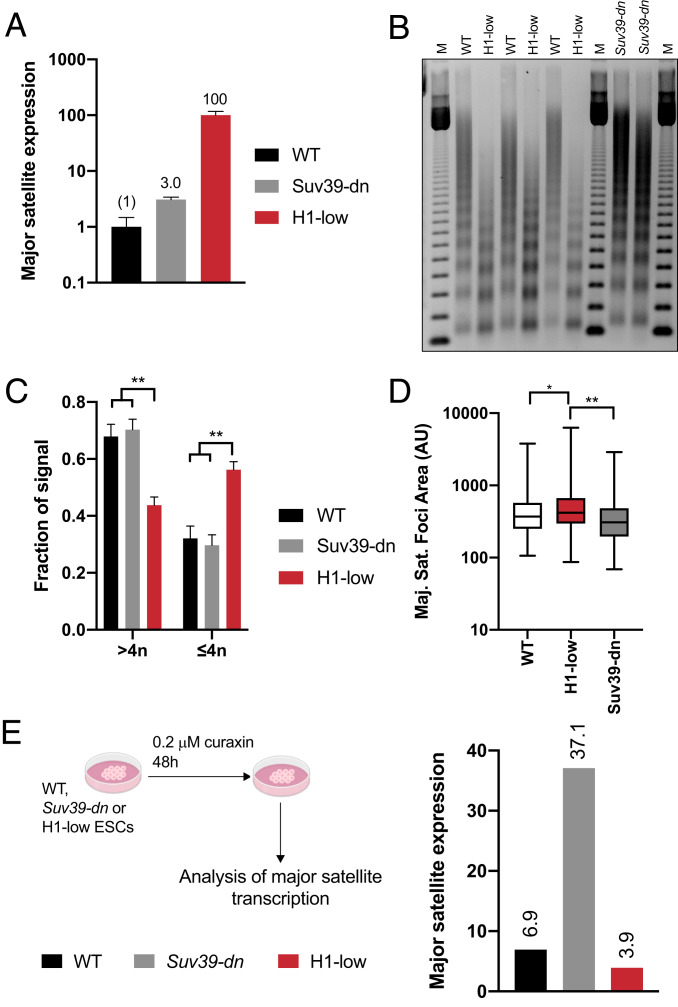
The preceding results indicate that H1 is a key player in silencing transcription of satellite DNA. However, we noted that the magnitude of satellite transcript de-repression in H1-depleted mESCs far exceeds the relatively mild (approximately threefold) de-repression in mESCs lacking both Suv39h1 and Suv39h2 (Fig. 4A), suggesting that H1 may use additional mechanisms to silence pericentric heterochromatin in these cells. Indeed, polycomb repressive complex 2 (PRC2)-mediated H3K27 methylation and DNA methylation have been reported to contribute to silencing pericentric heterochromatin (35–37). In addition, H1 has been reported to interact with components of PRC2 and to stimulate PRC2 activity in vitro (38), and we have shown that H1 interacts with DNA methyltransferases DNMT1 and DNMT3b and promotes DNA methylation in vitro and at certain loci in mESCs (8–10).
Fig. 4.
H1-mediated chromatin compaction is the dominant mechanism of major satellite repression. (A) Expression of major satellite transcripts in WT, Suv39-dn, and H1-low ES cells. Major satellite transcripts in WT, Suv39-dn, and H1-low ES cells were measured by RT-qPCR and normalized to Gapdh mRNA using the ΔΔCt method. Numbers above indicate expression relative to WT. Data are from three technical replicates of the indicated cell line. Similar results were obtained with several lines of the same genotype (see Fig. 2A). (B) Nucleosome spacing in WT, Suv39-dn, and H1-low ES cells. Nuclei were subjected to limited digestion with micrococcal nuclease and DNA was purified and analyzed by nondenaturing agarose gel electrophoresis, followed by ethidium bromide staining. (C) Analysis of chromatin condensation in WT, Suv39-dn, and H1-low ES cells. The fraction of signal arising from DNA fragments in B corresponding to tetranucleosomes and below (≤4n) and pentanucleosomes and above (>4n) was quantified and normalized to the total signal in each lane using ImageJ. n = 2 or 3 clones per genotype; error bars represent SD. (D) Area of major satellite foci in WT, Suv39-dn, and H1-low ESCs. At 24 h after transfection with TALEN specific to major satellite DNA fused to mClover, cells were sorted, applied to coverslips, and fixed. The area of major satellite foci was determined using ImageJ. WT, n = 180; Suv39-dn, n = 191; H1-low, n = 279. *P ≤ 0.05; **P < 0.01. Unpaired t test with Welch’s correction was used to test statistical significance. (E) Expression of major satellites in dimethyl sulfoxide (DMSO)- and curaxin-treated cells. Major satellite transcripts in WT, Suv39-dn, and H1-low ES cells treated with 0.2 µM curaxin or DMSO for 48 h were measured by RT-qPCR as in A. Data are shown as relative to DMSO control at 24 h.
To determine whether H1 promotes H3K27 methylation in pericentric heterochromatin in mESCs, we performed ChIP-qPCR for H3K27me3 in satellite chromatin in the two types of cells. The level of H3K27me3 in satellite chromatin was quite low in both WT and H1-low cells (SI Appendix, Fig. S4A), comparable to that within the promoter of a highly expressed negative control locus (Rps19). We also found that treatment of WT mESCs with GSK126, a potent PRC2 inhibitor, did not lead to de-repression of satellite transcription (SI Appendix, Fig. S4B), despite a strong reduction in H3K27me3 levels in bulk chromatin (SI Appendix, Fig. S4C). In a previous study from our laboratory, we did not detect significant differences in methylation of major satellite DNA in TKO mESCs (8), and DNMT TKO mESCs do not show elevation of satellite transcripts (39). Thus, neither H3K27 methylation nor DNA methylation appears to play a major role in H1-mediated repression of satellite transcription.
Another mechanism by which H1 could repress transcription of major satellite sequences is through a direct effect on chromatin compaction within pericentromeric chromatin. The ability of H1 to affect both nucleosome spacing and folding of chromatin into more compact structures is supported by numerous studies in vitro and in cells (8, 12, 40, 41). We showed previously that H1 TKO mESCs, which have 50% the WT level of H1, exhibit a 15-bp reduction in nucleosome repeat length (NRL) and a higher proportion of decondensed chromatin structures in electron micrographs of polynucleosome fractions isolated from these cells (8). Limited micrococcal nuclease (MNase) digestion of nuclei from H1-low mESCs revealed that they have a remarkably low NRL (166 bp) compared with WT mESCs (192 bp) and Suv39h1/2 double-null mESCs (194 bp) (Fig. 4B and SI Appendix, Fig. S4D). The NRL in H1-low mESCs is comparable to that reported in Saccharomyces cerevisiae, which has extremely low levels of a linker histone-like proteins (3). In addition, we determined that bulk chromatin isolated from H1-low mESCs was significantly less compact, as indicated by increased sensitivity to micrococcal nuclease digestion (Fig. 4C).
We also compared the accessibility of pericentric heterochromatin in WT, Suv39h1/2 double-null, and H1-low mESCs by treating nuclei with a minimal amount of MNase and measuring the amount of soluble major satellite chromatin released. We observed a slightly increased amount of satellite chromatin released from Suv39h1/h2 double-null mESCs compared with WT mESCs, but a much greater amount released from H1-low mESCs (SI Appendix, Fig. S4E).
To further characterize major satellite chromatin in WT, Suv39h1/2 double-null, and H1-low mESCs, we transfected these cells with transcription activator-like effector nucleases (TALENs) specific for major satellite DNA, fused to the mClover fluorescent protein (mClover-MajSat-TAL), and measured the area of major satellite foci (Fig. 4D and SI Appendix, Fig. S4F). We found that H1-low cells had larger major satellite foci than both WT and Suv39h1/2 double-null cells (Fig. 4D), indicating a less-condensed state of pericentric heterochromatin. Both measurements support the view that a marked reduction in satellite chromatin compaction may be responsible for the much larger effect of H1 depletion on satellite de-repression compared with that observed in Suv39h1/2 double-null mESCs.
To provide further support for this conclusion, we measured the level of satellite transcripts in WT, Suv39h1/h2 double-null, and H1-low mESCs following treatment with curaxin, a small molecule reported to decondense chromatin by inducing formation of alternative DNA structures that stimulate binding of the nucleosome chaperone FACT complex, leading to eviction of nucleosomal histones (42, 43). Treatment of WT and Suv39h1/h2 double-null mESCs led to a robust induction of major satellite transcripts but caused a smaller increase in H1-low mESCs (Fig. 4E). Taken together, studies of pericentric chromatin structure and the effects of curaxin on satellite transcription support the view that the much larger de-repression of major satellite transcription in H1-low compared with Suv39h1/2 double-null mESCs is due to the combined effects of reduced H1-mediated chromatin compaction and H1-stimulated Suv39h1/2-mediated deposition of H3K9 methylation at these loci.
Discussion
Silencing of repetitive elements in mESCs is thought to be mediated principally by trimethylation of histone H3 lysine 9 catalyzed by three HMTs: Suv39h1, Suv39h2, and SETDB1 (22–24). The results reported here indicate that H1 linker histones also play a central role in silencing the repetitive sequences targeted by these enzymes. Computational analyses show that H1 is enriched in constitutive heterochromatin, a fraction that contains H3K9me3 and is silenced by Suv39h1/h2 or SETDB1 (Fig. 1A). Depleting H1 to low levels in mESCs leads to a very robust activation of transcription of the repetitive sequences that are silenced by these enzymes, concomitant with the loss of H3K9 methylation at the affected sequences (Fig. 2 A–D), and reintroduction of H1 into the depleted cells restores repression (Fig. 2 A and B). Further evidence for an important role for linker histones in the silencing mechanism mediated by these enzymes comes from our findings that H1 linker histones interact directly with each of the enzymes and stimulate their activities toward chromatin in vitro (Fig. 3E). Importantly, our results are entirely consistent with an earlier report from our laboratory demonstrating that Drosophila H1 is a key regulator of heterochromatin silencing in flies through direct interaction with the H3K9 methyltransferase Su(var)3-9 (32). Thus, mechanistically, the role of H1 in silencing repetitive sequences appears to have been conserved in flies and mammals.
It also is important note that the role of H1 in these silencing processes appears to be specific for heterochromatin. Computational analyses indicate that H1 is depleted from chromatin containing G9a (Fig. 1A), a H3K9 methyltransferase associated with euchromatin (30). Furthermore, in contrast to the ability of H1 to stimulate the in vitro activities of Suv39h1, Suv39h2, and SETDB1 toward chromatin, H1 does not stimulate the HMT activity of G9a (Fig. 3E). A heterochromatin-specific role for H1 is further supported by our findings in Drosophila, where H1 was found to be required for silencing the heterochromatic copies of the stellate transposon located in heterochromatin, but not the transposon copies located in euchromatin (32).
Given that H1 binds to nucleosomes in both heterochromatin and euchromatin, how does H1 mediate these heterochromatin-specific effects? Recent studies on the regulation of Suv39h1 activity suggest that the enzyme exhibits both fast sampling of chromatin and more stable binding dependent on its N terminus (44). Our in vitro interaction data suggest that this region is also important for Suv39h1 binding to H1 (Fig. 3D). Thus, H1-containing chromatin may provide a preferred substrate for the enzyme by helping to anchor it to chromatin. In addition, heterochromatin-specific effects may be achieved through variations in H1 stoichiometry in different chromatin compartments. Our computational analyses indicate that H1 is enriched in chromatin domains marked by H3K9me3 and occupied by Suv39h1/h2 and SETDB1 (Fig. 1A), and H1 is enriched in large, heterochromatic “clutches” of nucleosomes by microscopy (45). Even subtle changes in H1 stoichiometry can significantly alter chromatin structure in vitro (41), and, given the findings of differential enrichment or depletion of H1 shown here, it is likely that this such alterations occur in vivo. Such H1-mediated changes in chromatin structure might influence the multivalent engagement of silencing factors, such as Suv39h1/2 and SETDB1, by affecting the binding of other factors that help recruit these enzymes. Indeed, several silencing factors demonstrate preferential binding to condensed chromatin structures through simultaneous binding of neighboring nucleosomes (46–48); thus, H1 may represent a critical component in the establishment of a chromatin state that facilitates binding of repressive factors or maintains their association with certain heterochromatic regions. Interestingly, the computational analysis indicates significant enrichment of KAP1 and ZFP57, two well-established repressive cofactors, in H1-enriched domains (SI Appendix, Fig. S1C). Indeed, KAP1 binds SETDB1 (49) and is reported to mediate its targeting (22), while ZFP57 itself has been shown to interact with KAP1 in ESCs (50).
While the results reported here clearly support a role for H1 in the silencing mechanism mediated by Suv39h1/h2 at major satellite sequences, we also found evidence for an additional, even more potent H1-dependent silencing mechanism that operates independently of the HMTs. We observed that strong H1 depletion results in a level of de-repression of major satellite transcription that far exceeds that in Suv39h1/h2 double-null ES cells (Fig. 4A). This additional silencing mechanism very likely involves H1-mediated alteration of the heterochromatin structure. Indeed, we found that major satellite chromatin is more accessible, and major satellite foci are larger, in the H1-low ES cells than in the Suv39h1/2-dn and WT ES cells (Fig. 4D and SI Appendix, Fig. S4 E and F). Furthermore, treatment of the Suv39h1/h2-dn cells with curaxin, a chromatin decompaction agent, led to a much greater activation of satellite transcripts in Suv39h1/2-dn ES cells than in the H1-low ES cells (Fig. 4E). These measurements support the view that H1 restricts access of the transcriptional machinery to major satellite sequences, not only by promoting H3K9 methylation of pericentric heterochromatin, but also through its effects on the higher-order structure of heterochromatin. A recent report found that contrary to expectation, the number and compaction of dense heterochromatic foci in murine fibroblasts was independent of HP1 binding or H3K9 trimethylation (51), consistent with the hypothesis that another factor is responsible for pericentric heterochromatin structure. We propose that this factor is H1. Indeed, this proposed H1-mediated compaction mechanism appears to be more robust than the effect of H1 on Suv39h1/2- and SETDB1-mediated H3K9 methylation, since restoration of H1 to the H1-depleted cells caused strong repression at all repetitive sequences examined but not a concomitant restoration of H3K9 methylation at LINE-1, IAP, and MusD elements. The properties of altered chromatin structures adopted on H1 binding in vivo merit further investigation.
The role of H1 as an essential repressive factor for several classes of repetitive sequences may also be important for disease mechanisms. Recent reports implicate repetitive sequence transcripts in cancer (19, 52, 53). Restricting the transcription of such elements may represent an important tumor suppression mechanism. A recent study demonstrated that transposable elements are often subjected to exaptation to drive oncogene activation in many human cancers (54). In this context, it is important to note that linker histone genes themselves are often mutated in certain human cancers (55–57), with especially high frequencies observed in diffuse large B-cell lymphomas (58). Many of these mutations occur within the H1 CTD which, in addition to mediating chromatin binding and compaction, is required for the physical interactions of H1 with the HMTs reported here and with DNA methyltransferases reported previously by our laboratory (9). The multifunctionality of the H1 CTD represents an exciting and expanding aspect of the field, and our results contribute to this emerging concept. The importance of the H1 CTD in disease etiology is underscored by the clustering of CTD mutations in HIST1H1E observed in a human overgrowth and intellectual disability syndrome (59). Therefore, it will be of interest to determine whether transcription of repetitive sequences is altered in disease conditions associated with mutations in H1 genes.
Materials and Methods
mESCs were cultured under standard conditions on gelatinized plates with 100 U/mL LIF (ESGRO; Millipore Sigma). Generation of H1-low ES cell lines is described in SI Appendix, Materials and Methods. Publicly available ChIP-seq data were accessed from the Gene Expression Omnibus database, aligned to the mouse genome (mm9) using STAR (60) and analyzed using the ISOR algorithm as described previously (28). More details on the computational analyses are provided in SI Appendix, Materials and Methods. qRT-PCR analyses of total RNA was performed as described previously (24). ChIP was carried out as described previously (9) with specific modifications described in SI Appendix, Materials and Methods. Procedures for recombinant protein purification, protein interaction studies, nucleosome reconstitution, and in vitro HMT are described in SI Appendix, Materials and Methods. Isolation and analysis of histones was carried out exactly as described previously (61). Analysis of chromatin compaction was performed as follows: 1 × 106 cells were resuspended in EZ Nuclei Lysis Buffer (Sigma-Aldrich) and digested with micrococcal nuclease (New England BioLabs) for 1 min at 37 °C. Purified DNA fragments were separated by 1.2% nondenaturing agarose gel electrophoresis and analyzed using ImageJ (NIH). sgRNA sequences are listed in SI Appendix, Table S1, and PCR primers are listed in SI Appendix, Tables S2 and S3.
Data Availability.
All unique reagents generated in this study are available by request from corresponding author A.I.S. (arthur.skoultchi@einsteinmed.org).
Supplementary Material
Acknowledgments
We thank Alexander Emelyanov and Dmitry Fyodorov for assistance with H1 purification; members of the Skoultchi laboratory, Cary Weiss, Charles Query, Robert Coleman and David Shechter for stimulating scientific discussion; Emmanuel Burgos for helpful advice regarding the HMT assays; Boris Bartholdy for guidance in computational approaches; Shirley Lee for providing the MLL1 complex; and the Einstein Flow Cytometry core facility (supported by National Cancer Institute [NCI] Grant P30-CA013330). This work was supported in part by funds from the National Institute of General Medical Sciences (NIGMS) (Grant R01-GM116143, to A.I.S.), the NCI (Grant F30-CA210539 to S.E.H.), and an NIH NIGMS Medical Scientist Training Program training grant (T32GM007288, to S.E.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920725117/-/DCSupplemental.
References
- 1.Misteli T., Gunjan A., Hock R., Bustin M., Brown D. T., Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Bednar J. et al., Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell 66, 384–397.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodcock C. L., Skoultchi A. I., Fan Y., Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14, 17–25 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T., Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Happel N., Doenecke D., Histone H1 and its isoforms: Contribution to chromatin structure and function. Gene 431, 1–12 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Fan Y., Sirotkin A., Russell R. G., Ayala J., Skoultchi A. I., Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol. Cell. Biol. 21, 7933–7943 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan Y. et al., H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 23, 4559–4572 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan Y. et al., Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Yang S.-M., Kim B. J., Norwood Toro L., Skoultchi A. I., H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc. Natl. Acad. Sci. U.S.A. 110, 1708–1713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maclean J. A. et al., The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone h1 and DNA methylation. Mol. Cell. Biol. 31, 1275–1287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hergeth S. P., Schneider R., The H1 linker histones: Multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 16, 1439–1453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fyodorov D. V., Zhou B. R., Skoultchi A. I., Bai Y., Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 19, 192–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterston R. H. et al.; Mouse Genome Sequencing Consortium , Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Friedli M., Trono D., The developmental control of transposable elements and the evolution of higher species. Annu. Rev. Cell Dev. Biol. 31, 429–451 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Macfarlan T. S. et al., Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Probst A. V. et al., A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev. Cell 19, 625–638 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Jachowicz J. W. et al., LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 49, 1502–1510 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Zhu Q. et al., Heterochromatin-encoded satellite RNAs induce breast cancer. Mol. Cell 70, 842–853.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee E. et al., Landscape of somatic retrotransposition in human cancers. Science 337, 967–971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martens J. H. A. et al., The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24, 800–812 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters A. H. F. M. et al., Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Matsui T. et al., Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Karimi M. M. et al., DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8, 676–687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulut-Karslioglu A. et al., Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell 55, 277–290 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Izzo A. et al., The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 3, 2142–2154 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Cao K. et al., High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet. 9, e1003417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millán-Ariño L. et al., Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 42, 4474–4493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H. et al., MacroH2A1.1 and PARP-1 cooperate to regulate transcription by promoting CBP-mediated H2B acetylation. Nat. Struct. Mol. Biol. 21, 981–989 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters A. H. F. M. et al., Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Tachibana M. et al., Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19, 815–826 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuleshov M. V. et al., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X. et al., Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science 340, 78–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X., The structure of histone H1 and its location in chromatin. Nature 288, 675–679 (1980). [DOI] [PubMed] [Google Scholar]
- 34.Kavi H., Emelyanov A. V., Fyodorov D. V., Skoultchi A. I., Independent biological and biochemical functions for individual structural domains of Drosophila linker histone H1. J. Biol. Chem. 291, 15143–15155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter M., Teissandier A., Pérez-Palacios R., Bourc’his D., An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. eLife 5, 1–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saksouk N. et al., Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol. Cell 56, 580–594 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Déjardin J., Switching between epigenetic states at pericentromeric heterochromatin. Trends Genet. 31, 661–672 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Martin C., Cao R., Zhang Y., Substrate preferences of the EZH2 histone methyltransferase complex. J. Biol. Chem. 281, 8365–8370 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Lehnertz B. et al., Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Carruthers L. M., Bednar J., Woodcock C. L., Hansen J. C., Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: Mechanistic ramifications for higher-order chromatin folding. Biochemistry 37, 14776–14787 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Routh A., Sandin S., Rhodes D., Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. U.S.A. 105, 8872–8877 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leonova K. et al., TRAIN (Transcription of Repeats Activates INterferon) in response to chromatin destabilization induced by small molecules in mammalian cells. Elife 7, 10.7554/eLife.30842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safina A. et al., FACT is a sensor of DNA torsional stress in eukaryotic cells. Nucleic Acids Res. 45, 1925–1945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller M. M., Fierz B., Bittova L., Liszczak G., Muir T. W., A two-state activation mechanism controls the histone methyltransferase Suv39h1. Nat. Chem. Biol. 12, 188–193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ricci M. A., Manzo C., García-Parajo M. F., Lakadamyali M., Cosma M. P., Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160, 1145–1158 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Machida S. et al., Structural basis of heterochromatin formation by human HP1. Mol. Cell 69, 385–397.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Poepsel S., Kasinath V., Nogales E., Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 25, 154–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan W. et al., Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337, 971–975 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J. 3rd, SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quenneville S. et al., In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 44, 361–372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erdel F. et al., Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell 78, 236–249.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bersani F. et al., Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. U.S.A. 112, 15148–15153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ting D. T. et al., Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jang H. S. et al., Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 51, 611–617 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasqualucci L. et al., Genetics of follicular lymphoma transformation. Cell Rep. 6, 130–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okosun J. et al., Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46, 176–181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landau D. A. et al., Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reddy A. et al., Genetic and functional drivers of diffuse large B Cell lymphoma. Cell 171, 481–494.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tatton-Brown K. et al.; Childhood Overgrowth Collaboration , Mutations in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am. J. Hum. Genet. 100, 725–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin Q. et al., Reductions in linker histone levels are tolerated in developing spermatocytes but cause changes in specific gene expression. J. Biol. Chem. 279, 23525–23535 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All unique reagents generated in this study are available by request from corresponding author A.I.S. (arthur.skoultchi@einsteinmed.org).