Significance
Proper resolution of inflammation is essential for restoration of homeostasis. Macrophages are particularly important for this process and dynamically change their functional phenotypes during the transition from inflammatory activation to resolution. we identify a long noncoding RNA, lncFAO, that promotes inflammation resolution. Deletion of lncFAO perturbed late suppression of inflammatory gene expression after LPS treatment and exacerbated endotoxic shock and skin wounds in vivo, which demonstrates that lncFAO is integral to inflammation resolution in macrophages. Mechanistically, lncFAO directly interacts with the HADHB subunit of mitochondrial trifunctional protein and activates fatty acid oxidation. Our data indicate that lncFAO is a mediator of metabolic reprogramming for resolution of inflammation in macrophages.
Keywords: Long noncoding RNA, macrophage, inflammation
Abstract
Proper resolution of inflammation is vital for repair and restoration of homeostasis after tissue damage, and its dysregulation underlies various noncommunicable diseases, such as cardiovascular and metabolic diseases. Macrophages play diverse roles throughout initial inflammation, its resolution, and tissue repair. Differential metabolic reprogramming is reportedly required for induction and support of the various macrophage activation states. Here we show that a long noncoding RNA (lncRNA), lncFAO, contributes to inflammation resolution and tissue repair in mice by promoting fatty acid oxidation (FAO) in macrophages. lncFAO is induced late after lipopolysaccharide (LPS) stimulation of cultured macrophages and in Ly6Chi monocyte-derived macrophages in damaged tissue during the resolution and reparative phases. We found that lncFAO directly interacts with the HADHB subunit of mitochondrial trifunctional protein and activates FAO. lncFAO deletion impairs resolution of inflammation related to endotoxic shock and delays resolution of inflammation and tissue repair in a skin wound. These results demonstrate that by tuning mitochondrial metabolism, lncFAO acts as a node of immunometabolic control in macrophages during the resolution and repair phases of inflammation.
Macrophages recognize molecules associated with pathogens and cell damage via pattern recognition receptors, such as Toll-like receptors (TLRs), which activate stimulus-regulated transcription factors, including NF-κB (1, 2). For example, TLR4 recognizes lipopolysaccharide (LPS) and activates an NF-κB–driven program of proinflammatory gene expression. In this way, macrophages play a key role as a first-line defense against pathogens and tissue damage. However, the actions of macrophages are not limited to this early response. They also play diverse and crucial roles in the resolution of inflammation, repair of tissue, and restoration of homeostasis. For example, early after tissue damage, proinflammatory activated macrophages promote acute inflammation, which is essential for elimination of cell debris and pathogens (3). After this early phase, inflammation subsides in coordination with activation of the repair phase. During this phase transition, macrophages change their phenotypes and play critical roles in inflammation resolution and tissue repair. Dysregulation of this phenotypic transition in macrophages prolongs inflammation and impedes tissue repair, which may underlie various chronic inflammatory diseases, including cardiovascular disease (4, 5). Accordingly, the activation states of macrophages must be tightly regulated (3).
Recent studies have identified cellular metabolism as a novel regulator of macrophage activation states (3, 6). For example, M1 proinflammatory activation is characterized by high glycolytic metabolism, whereas M2 alternative activation induced by IL-4 stimulates oxidative phosphorylation (OXPHOS) (6, 7). Importantly, interfering with that metabolic change impairs macrophage activation and function, demonstrating that metabolic reprogramming is a crucial part of the regulatory programs governing macrophage function and phenotype. Indeed, metabolic pathways and metabolites appear to be intricately linked to the cellular machinery that controls and executes macrophage activities (8).
Long noncoding RNAs (lncRNAs) are defined as RNAs over 200 nucleotides in length without protein-coding potential. It is becoming increasingly clear that lncRNAs are key regulatory mediators in a variety of biological processes. For example, lncRNAs are essential for controlling stemness and cell differentiation (9–12). A number of lncRNAs are also associated with various diseases, including cancer, cardiovascular diseases, and autoimmune diseases (13). Consequently, lncRNAs have attracted broad interest as novel biomarkers and potential therapeutic targets. So far, however, the functions of only a limited number of lncRNAs have been elucidated, although the results highlight the diverse modes of action of lncRNAs, including epigenetic regulation and various posttranscriptional and posttranslational mechanisms (14). Several lncRNAs are reportedly involved in regulating the functions of monocytes and macrophages (15). For example, lincRNA-Cox2, PACER, AS-IL1α, and FIREE are all acutely up-regulated by TLR activation and enhance inflammatory gene expression via transcriptional and posttranscriptional mechanisms (16–20). In contrast, lnc-IL7R, lincRNA-EPS, and Mirt2 suppress the inflammatory function of macrophages (21–23). lncRNAs are thus important regulators of macrophage activation. However, expression of the aforementioned lncRNAs is acutely (<6 h) controlled by LPS; little is known about the function of lncRNAs that respond later (e.g., during the resolution phase).
In the present study, we identified a lncRNA, designated lncFAO, that represses proinflammatory activation of macrophages during the late phase of inflammatory responses. We found that lncFAO exerts its effects by activating the β-subunit of mitochondrial trifunctional protein (hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase β-subunit; HADHB), a key enzyme in fatty acid β-oxidation (FAO), which is required for late suppression of proinflammatory cytokines. Within injured tissues, lncFAO is expressed in Ly6Chi macrophages during the inflammation resolution and tissue-repair phases. lncFAO deletion impairs resolution of inflammation and wound healing. These findings indicate that by controlling metabolic reprogramming, lncFAO mediates proper resolution of inflammation and is required for expression of proresolution/reparative phenotypes in macrophages.
Results
lncFAO, a Late-Response Macrophage lncRNA Was Identified.
In the present study we aimed to identify novel lncRNAs that regulate the inflammatory response of macrophages, particularly during the resolution phase (24). Because lncRNA expression is known to be highly diverse among different cell types (25, 26), we sought lncRNAs expressed in bone marrow-derived macrophages (BMDMs) by sequencing polyA+ RNAs. Upon assembling the transcripts (27), we found that LPS-treated BMDMs expressed >1,000 potential lncRNAs. Among them, we focused on lncRNAs whose expression was increased at late times after LPS stimulation (e.g., 24 and 48 h).
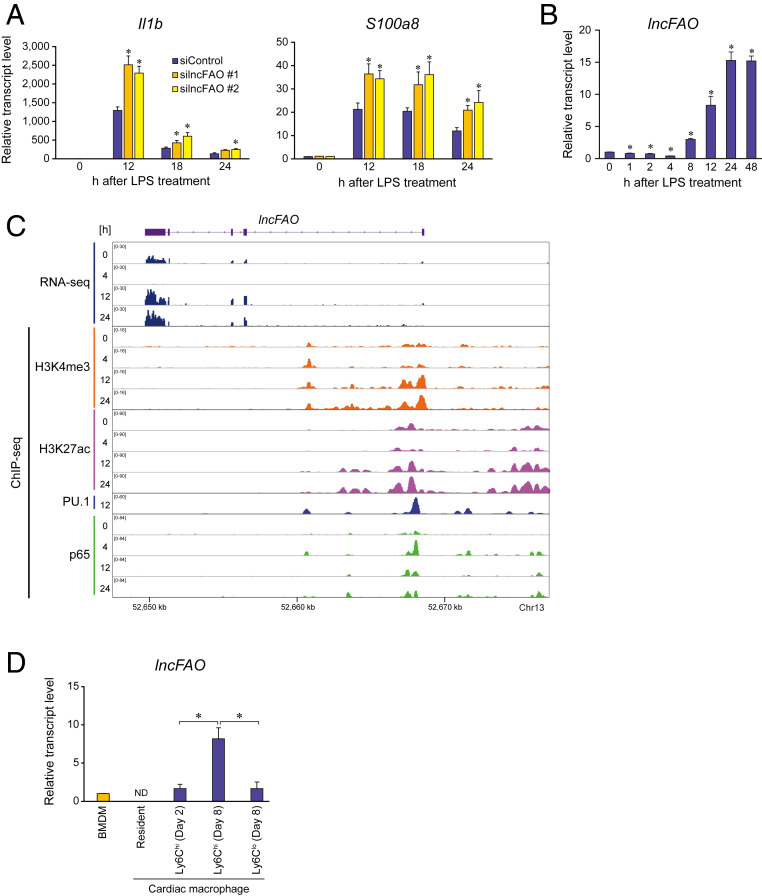
We chose 11 of the late-response lncRNAs identified because their expression levels were higher and were sustained for 24 or 48 h after LPS treatment in BMDMs (SI Appendix, Table S1A) and then assessed the effects of their knockdown on inflammatory gene expression (SI Appendix, Fig. S1 and Table S1B). Among the candidate lncRNAs, knockdown of a lncRNA designated lncFAO significantly increased expression of the proinflammatory genes Il1b and S100a8 in LPS-treated BMDMs (Fig. 1A and SI Appendix, Fig. S1 A and B). lncFAO was transiently down-regulated 4 h after LPS treatment but was markedly up-regulated 8 h after treatment (Fig. 1B). Gene set enrichment analysis (GSEA) of RNA-sequencing (RNA-seq) results showed that the Molecular Signatures Database (MSigDB) hallmark gene sets (28, 29) related to inflammatory responses and NF-κB signaling were up-regulated in lncFAO knockdown cells 24 h after LPS treatment (SI Appendix, Table S2). These results suggest that lncFAO may be important for suppressing LPS-induced inflammatory genes at late times (e.g., 24 h) and may contribute to the resolution of inflammation.
Fig. 1.
lncFAO is induced late after LPS treatment and affects proinflammatory cytokine expression. (A) Quantitative PCR analysis of Il1b and S100a8 expression in BMDMs transfected with control (siControl) or two types of siRNA targeting lncFAO (silncFAO). Analyses were performed at the indicated times after LPS treatment. mRNA levels were first normalized to those of 18s rRNA and then to the level in untreated control siRNA-transfected BMDMs. n = 3. *P < 0.05 vs. control siRNA at the same time point. Tukey–Kramer’s post hoc test. (B) Levels of lncFAO RNA expression in BMDMs treated with LPS. mRNA levels were first normalized to those of 18s rRNA and then to the level in untreated control BMDMs. n = 3. *P < 0.05 vs. 0 h in a two-tailed unpaired Student’s t test. (C) Gene locus of lncFAO. An IGV Genome Browser shot shows the RNA expression and ChIP-seq results of H3K4me3, H3K27ac, PU.1, and NF-κB p65 in BMDMs at the indicated times after TLR4 activation. (D) Levels of lncFAO expression in macrophages. Cardiac-resident macrophages (CD45+D11b+F4/80+Ly6G−) were isolated from mice in a steady state. MI was induced by ligating the left anterior descending artery, after which Ly6Chi macrophages (CD11b+F4/80+Ly6ChiLy6G−) and Ly6Clo macrophages (CD11b+F4/80+Ly6CloLy6G−) were isolated from the infarcted lesions at the indicated times post-MI. *P < 0.05. Tukey–Kramer’s post hoc test. ND, not determined.
lncFAO gene has a multiexonic structure (Fig. 1C). According to the widely used coding-potential assessment tools Annocript and CPAT (30, 31), the coding potential of lncFAO is low (Annocript noncoding potential score: 0.9653; CPAT coding probability: 0.0839). The transcription start site for lncFAO exhibited high H3K4me3 deposition (Fig. 1C), which is a hallmark of active promoters of mRNA-coding genes (32). We also found that the promoter, 5′-upstream regions, and an intronic region were all bound by PU.1, which is the macrophage lineage-determining transcription factor, and by NF-κB p65, which is the major signal-dependent transcription factor responding to TLR4 activation (33). Interestingly, after TLR4 activation, p65 binding associated with decreased H3K4me3 and H3K27ac deposition at 4 h (Fig. 1C), which reflects promoter and enhancer activity, respectively (34), and suggests NF-κB binding is involved in the transient repression of lncFAO transcription. Once expressed, the majority (>70%) of lncFAO transcripts were detected in the cytosol (SI Appendix, Fig. S1C).
Expression of lncFAO exhibited a tissue-selective pattern and was undetectable or very low in many tissues in mice under steady-state conditions (SI Appendix, Fig. S1D). But whereas lncFAO RNA was undetectable in normal hearts, its level was very high in the infarcted area 7 d after myocardial infarction (MI), which suggests its strong up-regulation by inflammation (SI Appendix, Fig. S1D). Whereas Ly6Chi monocyte-derived macrophages initially dominate during the inflammatory phase of MI, during the repair phase (days 3 to 7 post-MI) Ly6Clo macrophages become predominant (35). lncFAO was highly expressed in proinflammatory Ly6Chi macrophages in injured hearts, but was undetectable in cardiac-resident macrophages in the steady state (Fig. 1D and SI Appendix, Fig. S1E). As compared to Ly6Chi macrophages during the active inflammatory phase on day 2 after MI, expression of lncFAO was increased in Ly6Chi macrophages during the resolution phase on day 8 (36). Moreover, lncFAO expression was much higher in Ly6Chi macrophages than in Ly6Clo macrophages on day 8, suggesting that lncFAO is up-regulated in Ly6Chi macrophages during the resolution phase after MI.
To further delineate expression of lncFAO in MI, we analyzed its expression in a publicly available single-cell RNA-seq dataset (37). Farbehi et al. analyzed single-cell expression profiles of the total cardiac interstitial cells and Pdgfra+ fibroblasts in the ventricles on days 3 and 7 post-MI and on day 7 postsham. We found that lncFAO expression was confined to the populations of Ptprc+ leukocytes (SI Appendix, Fig. S2). The majority of lncFAO-expressing cells were also Cd68+Itgam+, indicating that they were monocytes/macrophages.
Analysis of Cd68+ macrophage populations showed that lncFAO is expressed in Ly6c2hiCcr2hiAdgre1int macrophages (cluster 1 in SI Appendix, Fig. S2C), which appear to correspond to CD11b+F4/80+Ly6Chi macrophages identified with flow cytometry (Fig. 1D) in the sham and day 7 post-MI hearts (SI Appendix, Fig. S3A), and a modest level lncFAO expression was also observed in a population of Ly6c2intMsr1hiArg1+ macrophages (cluster 2) on day 7 post-MI. lncFAO expression was transiently down-regulated on day 3 in cluster 1 cells. On day 7, cluster 1 cells were characterized by relatively high expression of inflammatory cytokines, such as Il6 and Il1b, as well as high expression of Tgfb1 (SI Appendix, Fig. S3A). Cluster 1 cells were also marked by higher expression of Nr4a1, which has been shown to limit inflammation during the repair phase after MI (SI Appendix, Fig. S2C) (35). Gene ontology enrichment analyses also showed that the transcriptome of cluster 1 cells was enriched in gene sets related to both inflammation and resolution, including “negative regulation of immune system process” and “wound healing” in Cd68+ cells (SI Appendix, Fig. S3B). GSEA also revealed that OXPHOS is enriched in cluster 1 cells (SI Appendix, Fig. S3C). The transcriptome of cluster 2 cells was characterized by genes related to cell migration, angiogenesis and wound healing. Cluster 2 cells also expressed relatively high levels of Il10 (SI Appendix, Fig. S3A). Collectively, these results show that lncFAO is expressed in monocyte-derived Ly6c2hi-int macrophages that appear to contribute to inflammation resolution and tissue repair after MI.
lncFAO Negatively Regulates Inflammatory Activation of Macrophages.
To analyze the function of lncFAO, we used CRISPR/Cas9 technology to generate several lncFAO knockout (lncFAO−/−) mouse lines. One of those lines had a 201-bp deletion with no additional mutations at predicted off-target sites (SI Appendix, Fig. S4 A–C). These mice were born at a normal Mendelian ratio and grew normally (SI Appendix, Fig. S4D), with no apparent gross abnormalities. Using qPCR detecting a region spanning exons 3 to 5, we confirmed that lncFAO transcripts were largely eliminated in BMDMs from lncFAO−/− mice (KO1 in SI Appendix, Fig. S4E). lncFAO RNA expression was also nearly eliminated in another line of lncFAO knockout mice that had a 711-bp deletion (KO2 in SI Appendix, Fig. S4 B and E). Mice from this second line also grew normally, and BMDMs derived from that line exhibited a cytokine gene-expression profile that was similar to that in cells from the KO1 line (SI Appendix, Fig. S4F). In the following experiments we used the KO1 line.
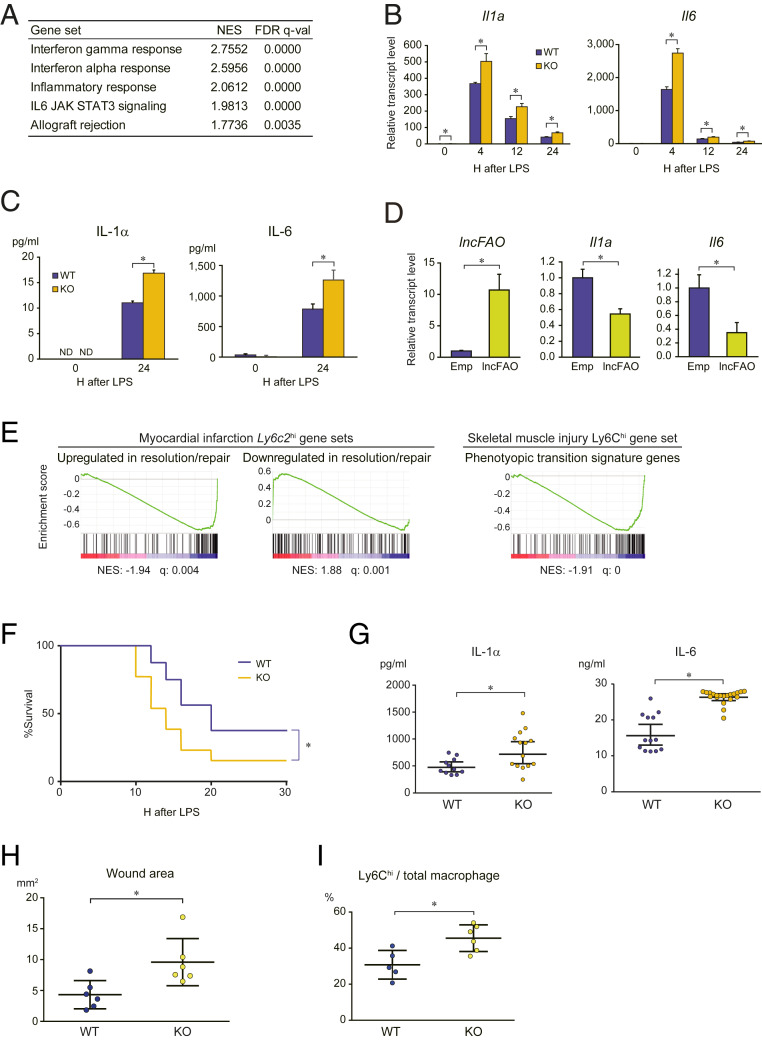
Transcriptome analysis of lncFAO−/− BMDMs 12 h after LPS treatment indicated increased expression of genes belonging to gene sets related to inflammatory responses (Fig. 2A). qPCR analysis showed that lncFAO deletion increased levels of Il1a and Il6 expression after LPS treatment (Fig. 2B). Secretion of IL-1α and IL-6 was also increased in lncFAO−/− BMDMs (Fig. 2C). Conversely, lncFAO overexpression decreased IL-1α and IL-6 levels in BMDMs 24 h post-LPS (Fig. 2D). Taken together, these results demonstrate that lncFAO negatively regulates expression of proinflammatory cytokines, such as IL-1α and IL-6 in response to LPS.
Fig. 2.
lncFAO deletion augments inflammatory responses. (A) Transcriptomes of BMDMs from WT and lncFAO−/− mice were analyzed using RNA-seq 12 h after LPS treatment. Gene set enrichment was analyzed using GSEA (28). Shown are false-discovery rate < 0.05 MSigDB hallmark gene sets up-regulated in lncFAO−/− cells as compared to WT cells. NES, normalized enrichment scores. (B) qPCR analysis of Il1a and Il6 in BMDMs from WT and lncFAO−/− mice. mRNA levels were first normalized to those of 18s rRNA and then to the level in untreated (0 h) WT BMDMs. n = 3 for each group. *P < 0.05, two-tailed unpaired Student’s t test. (C) IL-1α and IL-6 levels in medium collected from BMDM cultures 24 h after LPS treatment. n = 3. *P < 0.05, two-tailed unpaired Student’s t test. (D) BMDMs were transfected with empty vector (E) or an lncFAO expression vector; 24 h after transfection, the cells were treated with LPS, and mRNA levels were assessed after an additional 24 h. mRNA levels were first normalized to those of 18s rRNA and then to the level in WT BMDMs. n = 3. *P < 0.05, two-tailed unpaired Student’s t test. (E) Dysregulation of the signature genes representing the transition of Ly6Chi macrophages from the inflammation to resolution/repair phase after MI or skeletal muscle injury in lncFAO−/- BMDMs. Shown are GSEA enrichment scores in untreated lncFAO−/− BMDMs as compared to WT BMDMs (28). The MI gene sets consist of the genes up-regulated (107 genes) or down-regulated (144 genes) from day 3 to day 7 in Ly6c2hi macrophages in the single-cell RNA-seq dataset for MI (SI Appendix, Figs. S2 and S3) (37), and the skeletal muscle injury gene set consists of the genes enriched in Ly6Chi macrophages on day 4 in injured skeletal muscle (127 genes) (38). (F) Kaplan–Meier survival curves for WT and lncFAO−/− mice intraperitoneally injected with a high-dose of LPS (50 μg/mg). n = 10 for each group. *P < 0.05, log-rank test. (G) Mice were intraperitoneally injected with a low-dose of LPS (20 μg/mg), and blood levels of IL-1α and IL-6 were analyzed 12 h after the injection. Shown are concentrations of each mouse and the means ± SD n = 10 (WT) or 12 (KO) mice. *P < 0.05, two-tailed unpaired Student’s t test. (H) Quantification of the wound area 8 d after skin excision in mice transplanted with WT or lncFAO−/− bone marrow. *P < 0.05, two-tailed unpaired Student’s t test. (I) Flow cytometric analysis of the phenotypes of the macrophages (CD45.2+CD11b+F4/80+Ly6G−) infiltrating the wound area. The fraction of proinflammatory Ly6Chi macrophages is shown. *P < 0.05, two-tailed unpaired Student’s t test.
To gain additional insight into the effects of lncFAO deletion on BMDM function, we analyzed the transcriptome over the course of LPS stimulation. GSEA showed that the hallmark gene sets “inflammatory response” and “IFN-αi response” were up-regulated in lncFAO−/− cells at all timepoints (SI Appendix, Table S3). In untreated cells, gene sets related to “Myc signaling,” “angiogenesis,” “cell growth,” and “metabolism” were up-regulated. These results further confirm that lncFAO restricts inflammatory activation of BMDMs. In line with the transcriptome changes, immunostaining showed that 24 h after LPS treatment nuclear NF-κB levels remained higher in lncFAO−/− BMDMs than WT BMDMs (SI Appendix, Fig. S5), which supports the notion that lncFAO is important for resolution of LPS-induced inflammatory activation.
To assess the function of lncFAO within injured tissue, we analyzed the effects of lncFAO deletion on the signature genes representing the transcriptomic change from the inflammation to resolution/repair phase in Ly6Chi macrophages (cluster 1 cells) after MI (SI Appendix, Fig. S2). We generated gene sets consisting of the genes up-regulated or down-regulated in Ly6Chi macrophages from day 3 to day 7 post-MI. GSEA showed that lncFAO deletion dysregulated expression of these genes in untreated BMDMs (Fig. 2E and SI Appendix, Table S4). In another well-characterized mouse model, cardiotoxin-mediated skeletal muscle injury, Ly6Chi macrophages markedly alter the phenotype from proinflammation toward proresolution/repair on day 4 after injury (38). We found that the gene signature of Ly6Chi macrophages at the transition period was down-regulated in lncFAO−/− BMDMs (Fig. 2E). Taken together, these results suggest that lncFAO contributes to the acquisition of the phenotypic characteristic to Ly6Chi macrophages during the inflammation resolution and repair phases after tissue injury.
lncFAO Is Required for Proper Inflammatory Resolution and Healing.
To gain further insight into the function of lncFAO in inflammatory responses in vivo, we used an endotoxic shock model induced by intraperitoneal injection of LPS. In lethal-dose LPS experiments, lncFAO knockout reduced survival (Fig. 2F). Twelve hours after administration of a sublethal dose of LPS, serum IL-1α and IL-6 levels were significantly higher in lncFAO−/− than WT mice (Fig. 2G). It appears that lncFAO suppresses proinflammatory activation of macrophages, particularly during the late-resolution phase of the LPS response.
We also assessed the contribution of lncFAO in hematopoietic cells to inflammatory resolution using a skin wound excision model in mice that had received a bone marrow transplant (BMT) from WT or lncFAO−/− mice (WT-BMT and lncFAO−/−-BMT mice). The wound repair was slower in lncFAO−/−-BMT than WT-BMT mice (Fig. 2H and SI Appendix, Fig. S6A). In skin excision models, transition from predominantly Ly6Chi proinflammatory monocytes/macrophages to predominantly Ly6Clo macrophages has been shown to occur 2 to 3 d after the wounding (39). In lncFAO−/−-BMT mice, however, the Ly6Chi cell fraction remained high on day 8 postinjury (Fig. 2I). Moreover, infiltration of larger numbers of leukocytes was apparent in the wounds of lncFAO−/−-BMT mice (SI Appendix, Fig. S6B). These observations indicate that inflammation persists 8 d after skin excision in lncFAO−/−-BMT mice and that lncFAO in macrophages are important for proper resolution of inflammation and progression to healing.
lncFAO Associates with the HADHB Subunit of Mitochondrial Trifunctional Protein.
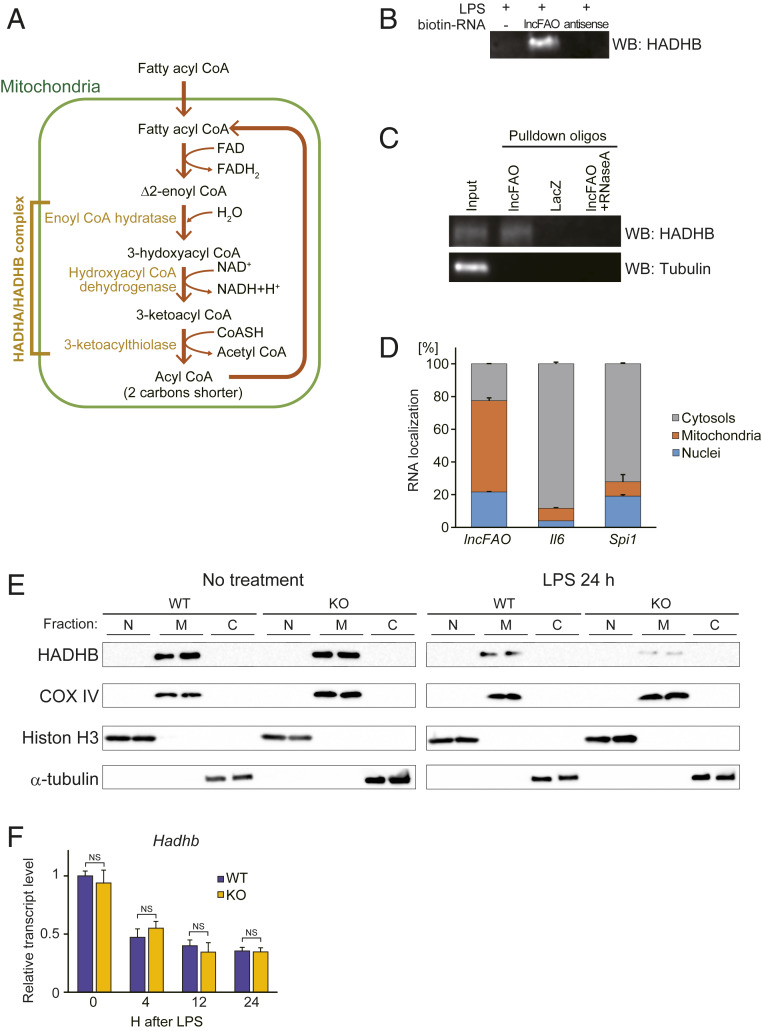
Although lncRNAs within the nucleus can control gene transcription through a variety of mechanisms (40–42), the major cytosolic localization of lncFAO prompted us to hypothesize that it may affect macrophage activation by interacting with cytosolic proteins. To test this idea, we constructed a biotinylated lncFAO RNA probe and used it to pull down protein complexes from cytosolic extracts from BMDMs by modifying the chromatin isolation by RNA purification (ChIRP) method to obtain cytosolic RNA–protein complexes (43, 44). Electrophoresis of the pulled down samples revealed a band that showed a higher intensity in samples from LPS-treated cells than from untreated cells or cells treated with an antisense probe (SI Appendix, Fig. S7A). Mass-spectrometric analysis of the gel band identified HADHB and several other proteins with high sequence coverages (SI Appendix, Fig. S7B). Because an earlier study showed that HADHB interacts with Ren mRNA, indicating its RNA binding capacity (45), we focused on HADHB as a potential interacting partner of lncFAO.
With its thiolase activity, HADHB catalyzes the last three steps of FAO to yield an acyl-CoA that is two carbons shorter than the original substrate plus acetyl-CoA (Fig. 3A). As expected, HADHB was detected in the precipitates of BMDM lysates using a biotinylated lncFAO RNA probe (Fig. 3B). That HADHB associates with lncFAO was further confirmed when HADHB was detected after using anti-lncFAO oligos to pull down endogenous complexes containing lncFAO from LPS-treated BMDM lysates (SI Appendix, Fig. S8). No HADHB was detected after pull-down using anti-LacZ oligos or in samples treated with RNase (Fig. 3C), which indicates that the ribonucleoprotein complexes were specifically retrieved by lncFAO, not as a result of nonspecific interactions between probe DNAs and HADHB. These data confirm that lncFAO associates with HADHB.
Fig. 3.
lncFAO binds to HADHB and enhances its enzymatic activity. (A) Schematic of the FAO pathway. (B) Western analysis of HADHB in samples pulled down from BMDM lysates 24 h after LPS treatment. The lysates were pulled down with biotinylated RNA probes for lncFAO or its antisense RNA. (C) Twenty-four hours after LPS treatment, BMDM lysates were incubated with biotin-conjugated antisense oligonucleotides targeting lncFAO and pulled down with streptavidin beads. Oligos against LacZ were used as a control. A RNaseA-treated sample was used an additional control. HADHB and tubulin were analyzed in the pulled down samples using Western blotting. (D) Distribution of RNA in the cellular compartments of BMDMs 24 h after LPS treatment. RNA abundances in the three cell subfractions were analyzed using qPCR. n = 3. (E) Western analysis of the nuclear (N), mitochondrial (M), and nonmitochondrial cytosolic (C) fractions of WT and lncFAO−/− BMDMs treated with or without LPS, as indicated. COX IV, Histone H3, and α-tubulin and were detected as internal controls for the mitochondrial, nuclear, and cytosolic fractions, respectively. (F) Hadhb mRNA levels in WT and lncFAO−/− BMDMs at the indicated times after LPS treatment. mRNA levels were first normalized to those of 18s rRNA and then to the level in untreated WT BMDMs. n = 3 for each group. NS, not significant in a two-tailed unpaired Student’s t test.
lncFAO Activates HADHB and FAO.
Because HADHB is localized in mitochondria, we further analyzed the localization of lncFAO by separating the mitochondrial fraction from the cytosolic fraction. We found that approximately half of lncFAO (∼56%) was present in the mitochondrial fraction of BMDMs 24 h after LPS treatment (Fig. 3D). FISH of lncFAO RNA also showed that the majority of lncFAO colocalizes with mitochondrial protein COXIV in BMDMs (SI Appendix, Fig. S9).
Mitochondrial levels of both HADHB protein (Fig. 3E) and Hadhb mRNA (Fig. 3F) were decreased by LPS treatment. Interestingly, the LPS-induced decrease in HADHB protein was greater in lncFAO−/− BMDMs than in WT cells, although the levels were comparable in untreated cells (Fig. 3E). In contrast, the lack of lncFAO did not affect Hadhb mRNA levels (Fig. 3F), which suggests lncFAO acts posttranscriptionally to affect mitochondrial HADHB levels after LPS treatment.
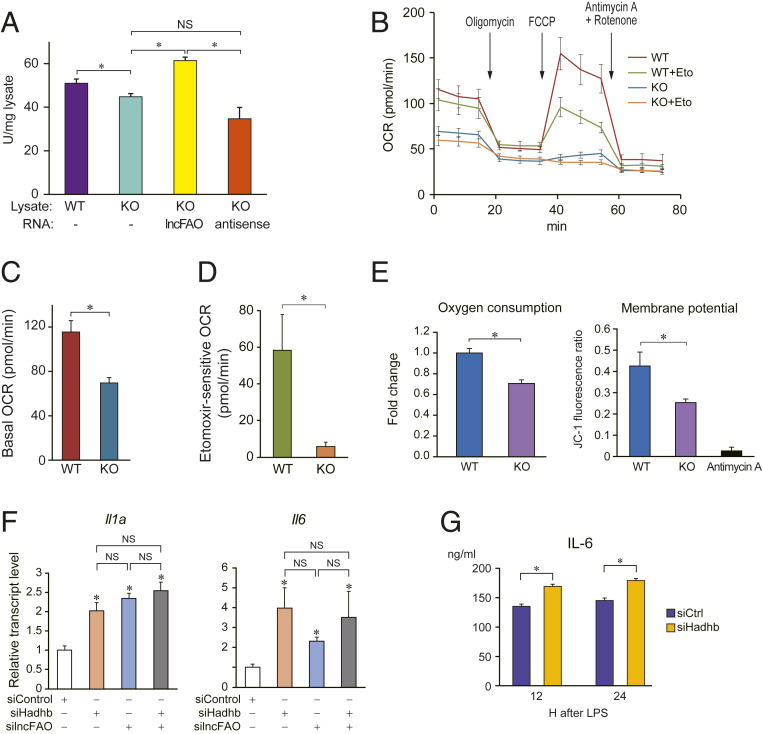
Because lncFAO in mitochondria binds HADHB, we hypothesized that lncFAO may affect HADHB enzymatic activity. To test that idea, we measured the thiolase-catalyzed conversion of acetoacetyl-CoA to acetyl-CoA in BMDM lysates. lncFAO−/− lysates exhibited less thiolase activity than WT lysates (Fig. 4A). Moreover, addition of lncFAO RNA to lncFAO−/− lysates enhanced the thiolase activity, while the antisense RNA for lncFAO failed to do so. This suggests lncFAO activates HADHB enzyme.
Fig. 4.
lncFAO modulates cellular metabolism. (A) Comparison of thiolase activity in lysates of untreated WT and lncFAO−/− (KO) BMDMs. In vitro transcribed lncFAO or its antisense RNA were added to lncFAO−/− lysates, as indicated. The y axis indicates the enzyme activity in the cell lysate that converts acetoacetyl-CoA to acetyl-CoA. One unit of activity is defined as the amount of enzyme that converts 1 μmol of acetoacetyl-CoA per minute. *P < 0.05, Tukey–Kramer’s post hoc test. (B) Changes in the OCR in WT and lncFAO−/− BMDMs treated sequentially with oligomycin (1 μM), FCCP (1.5 μM), and rotenone (0.5 μM) plus antimycin A (0.5 μM) in the presence of BSA and palmitate, with or without pretreatment with 40 μM etomoxir (Eto), an FAO inhibitor, as indicated. (C) OCR in untreated BMDMs. n = 6. *P < 0.05, two-tailed unpaired Student’s t test. (D) Etomoxir-sensitive fractions of OCR in FCCP-treated cells. n = 6. *P < 0.05, two-tailed unpaired Student’s t test. (E) The extracellular OCR and mitochondrial membrane potential were measured in WT and lncFAO−/− BMDMs 24 h after LPS treatment. OCR was measured using a phosphorescent oxygen probe (MitoXpress) (46). As an index of mitochondrial membrane potential, the ratio of the fluorescence intensity of JC-1 aggregates (595 nm) to that of JC-1 monomers (535 nm) is shown (47). The cells were also treated with the OXPHOS inhibitor antimycin A (10 μM) just before the measurement. n = 3. *P < 0.05, two-tailed unpaired Student’s t test. (F) BMDMs were transfected with siRNA targeting Hadhb, lncFAO, or control siRNA. Twelve hours after transfection, the cells were treated with LPS. Cellular mRNA levels were analyzed using qPCR 12 after LPS treatment. mRNA levels were first normalized to those of 18s rRNA and then to the level in siCtrl-transfected BMDMs. n = 3. Hadhb levels are shown in SI Appendix, Fig. S10. *P < 0.05, two-tailed unpaired Student’s t test. (G) BMDMs were transfected with siRNA targeting Hadhb or control siRNA. Twenty-four hours after transfection, the cells were treated with LPS. IL-6 levels in the medium were analyzed 12 and 24 h after LPS treatment. n = 3. *P < 0.05, two-tailed unpaired Student’s t test.
Because HADHB is essential for FAO, we used a flux analyzer to test whether lncFAO deletion affects cellular metabolism. The basal oxygen consumption rate (OCR) was much lower in lncFAO−/− BMDMs than in WT cells (Fig. 4 B and C). The fraction of the OCR sensitive to etomoxir (48), a carnitine palmitoyltransferase-1 (CPT-1) inhibitor, after addition of carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), a mitochondrial OXPHOS uncoupler that induces maximal respiration, was much smaller in lncFAO−/− cells than in WT cells (Fig. 4D). CPT-1 is the rate-limiting step in the transport of long-chain FAs into mitochondria for oxidation (49). The decrease in etomoxir-sensitive OCR in lncFAO−/− cells is thus indicative of reduced FAO capacity.
We also analyzed oxygen consumption in LPS-treated cells using a phosphorescent oxygen probe. Again, oxygen consumption was lower in LPS-treated lncFAO−/− cells than WT cells (Fig. 4E). In addition, mitochondrial membrane potential was reduced in lncFAO−/− cells, suggesting the lack of lncFAO impairs mitochondrial function. Taken together, these results show that lncFAO is important for mitochondrial FAO and health, acting at least in part through regulation of HADHB.
Finally, we tested whether HADHB dysfunction is responsible for the heightened proinflammatory activation of lncFAO−/− BMDMs. When Hadhb was knocked down in BMDMs using small-interfering RNA (siRNA), Il1a and Il6 expression was increased in LPS-treated BMDMs (Fig. 4F and SI Appendix, Fig. S10). Similarly, Hadhb knockdown led to increased IL-6 levels in conditioned medium (Fig. 4G). These results show that HADHB dysfunction enhances inflammatory activation of macrophages. In contrast, in the Hadhb knocked down cells an additional knockdown of lncFAO did not alter Il6 or Il1a levels (Fig. 4F), supporting the notion that Hadhb is required for the regulatory action of lncFAO. Moreover, Hadhb knockdown did not increase IL-6 levels in conditioned medium of lncFAO−/− BMDMs (SI Appendix, Fig. S11). These results suggest that lncFAO and HADHB work together in the same pathway to regulate the inflammatory activation of macrophages. Taking these data together, we find that lncFAO appears to be important for suppression of TLR4-induced proinflammatory cytokine expression during the resolution phase, in part by activating FAO through binding to HADHB.
Discussion
In the present study, we demonstrate that lncFAO is expressed in macrophages at late times after LPS treatment and that it suppresses proinflammatory cytokine expression. In LPS-treated BMDMs and Ly6Chi macrophages within injured tissues, lncFAO is induced at the resolution phase, which is consistent with the notion that lncFAO contributes to the resolution of inflammation. lncFAO acts by binding to HADHB and enhancing FAO, which is important for the late suppression of cytokine expression. These findings demonstrate that lncFAO is a mediator of late metabolic reprogramming of inflammatory macrophages that promotes the resolution of inflammation.
Functional transition of macrophage phenotypes from proinflammatory to proresolution/healing is vital for healing and restoration of homeostasis after tissue injury (3). Interestingly, down-regulation of genes associated with glycolysis and up-regulation of those associated with OXPHOS and FAO precedes the phenotypic transition of macrophages in a muscle injury model (38). In addition, IL-10, which contributes to this phenotypic transition after muscle injury (50), inhibits LPS-induced activation of glycolysis and promotes OXPHOS in BMDMs (51). These reports support the notion that activation of OXPHOS is integral to the macrophage functional transition in inflammation. Our present results demonstrate that lncFAO is an important regulator of the late changes in macrophage function and FAO during inflammatory responses in BMDMs. Deletion of lncFAO down-regulated the genes reflecting the change of Ly6Chi macrophages from the inflammatory to the reparative phase after MI or skeletal muscle injury in BMDMs, which suggests lncFAO is important for those changes (Fig. 2E). Consistent with those findings, lncFAO expression was increased in Ly6Chi macrophages during the repair phase after MI (e.g., ∼7 d post-MI), a time when the inflammation seen earlier (e.g., on day 2) was being resolved and reparative processes were progressing (Fig. 1D and SI Appendix, Figs. S2 and S3). On day 7 post-MI, lncFAO-expressing cells also expressed genes related to inflammatory resolution and wound healing, such as Tgfb1 (SI Appendix, Fig. S3). These findings suggest that lncFAO is a mediator of the metabolic reprograming required for induction of proresolution functionality in macrophages during inflammation.
We found that lncFAO interacts with HADHB. An earlier study reported that HADHB also binds to the 3′UTR of human REN mRNA, which decreases the stability of the mRNA (45). Although to our knowledge this report of an RNA affecting HADHB activity is unique, several proteins have been shown to interact with HADHB and modulate its activity (52, 53). For example, estrogen receptor α (ERα) interacts with HADHB within mitochondria and modulates its thiolase activity (52). This interaction may mediate estrogen-induced alterations in lipid metabolism. HADHB may also interact with other proteins and RNAs, enabling it to act as a hub enzyme that controls metabolism in response to multiple inputs.
Recent studies have identified RNA-binding activities in many metabolic enzymes, some of which affect the fate of the bound RNAs (54). For exampl, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) reduces translation of TNF mRNA in monocytes by binding to its 3′UTR (55). Moreover, the glycolysis rate is a key determinant of GAPDH binding to TNF mRNA, suggesting that GAPDH links the cellular metabolic state to cytokine production. Similarly, GAPDH binds to the 3′UTR of Ifng mRNA and suppresses its translation in effector T cells (56). Those findings and our present results highlight the notion that RNA-binding metabolic enzymes comprise the critical machinery that connects cellular metabolism to immune cell activation.
But while it appears that enzymes can affect the fate of RNAs through binding, the effects of RNA binding on enzymatic activity is less clear. We show here that interaction with lncFAO activates HADHB enzymatic activity (Fig. 4A) and positively regulates mitochondrial HADHB levels (Fig. 3E). Our results further demonstrate that RNA-binding enzymes can be regulated by specific RNAs. lncRNAs may also control cellular metabolism via multiple modes of action. For example, Tug1 controls mitochondrial bioenergetics by activating proliferator-activated receptor γ coactivator α (PGC-1α) in podocytes (57). SAMMSON interacts with p32, a critical regulator of tumor mitochondrial metabolism, and enhances its mitochondrial localization and function in melanoma cells (58). Because expression of lncRNAs is more highly cell-type–selective than expression of mRNAs (25, 59), lncRNAs may have important context-dependent regulatory functions in cellular metabolism. Future studies will need to further address this key biological function of lncRNAs.
Methods
Mice.
C57BL/6J mice were purchased from Japan Clea. lncFAO knockout mice were created using CRISPR-Cas9 technology (60). A mixture of in vitro-transcribed Cas9 mRNA and two types of single-guide RNA (sgRNA) were injected into fertilized eggs of C57BL/6J mice. To construct plasmids encoding each sgRNA, annealed oligonucleotides were inserted into a pDR274 vector digested with BsaI (61). The genomic target DNA sites and the sequences of the annealed oligonucleotides were as follows: sgRNA#1, 5′-AGCCTAACACAATGGGTGAGGGG-3′; sgRNA#2, 5′-GTAGCAGAGAGTTGTACGTCTGG-3′. These sequences were designed by CRISPR Design (https://zlab.bio/guide-design-resources).
To analyze deletion of the target region, genomic PCR was conducted using the primers 5′-GGGGAGAACCTGCTAGAAGC-3′ and 5′-GAGGTCTTATTCTTTCCAGAACACTG-3′. We generated five lines of mice, each with different types of deletions. By sequencing the genomic DNA of the vicinity of the guide RNA target sites, we found that two lines had an allele with a >200-bp deletion at the first exon of lncFAO and further characterized those lines.
To assess possible off-target deletions, off target candidate sites were identified using CRISPR Design tools and amplified from genomic DNA extracted from the tails of mice in the two lines. The target site was also amplified as a positive control. PCR products from WT and knockout mice were mixed, and DNA hybridization was performed as follows: Initial heating at 95 °C for 5 min, followed by gradually cooling to 4 °C at 2 °C/min. After hybridization, the mixture was treated with Guide-it Resolvase (Takara), which cuts DNA at mismatches. The primer sequences used for detection of off-target mutations were as follows: Off-target #1, GGCTGCACTCTGGGCATTAT and TCCGTGCAGACATGAACAGT; off-target #2, GGTAGTCTGGAAAGGGCAGTC and CCTCGTGACACGGATGGTTC; off-target #3, CTTCCACTCAACCACAAGCG and ACCACAGGATCGTTCCCTAA.
We generated mice homozygous for the targeted alleles in the two lines. Mice in the two lines did not show gross anatomical abnormalities and grew normally (SI Appendix, Fig. S2D). BMDMs prepared from the two lines showed similarly enhanced proinflammatory cytokine gene expression after LPS treatment (SI Appendix, Fig. S4F). We mainly used the first line (KO1) carrying a 201-bp deletion in the lncFAO gene for characterization of lncFAO (SI Appendix, Fig. S2A).
All experiments were approved by the University of Tokyo Ethics Committee for Animal Experiments and strictly adhered to the guidelines for animal experiments of the University of Tokyo.
Reagents and Antibodies.
For chromatin immunoprecipitation sequencing (ChIP-seq), anti-H3K4me3 (39159; Active motif), anti-PU.1 (sc-352; Santa Cruz), and anti-p65 (sc-372; Santa Cruz) antibodies were used. For Western blotting, anti-HADHB (sc-134922, Santa Cruz; 1:200), anti–α-tubulin (T6199, Sigma Aldrich; 1:2,000), anti-histone H3 (4620, Cell Signaling Technology; 1:2,000), and anti-COX IV (11967, Cell Signaling Technology; 1:1,000) antibodies were used. As secondary antibodies, horseradish peroxidase (HRP)-linked anti-rabbit IgG and anti-mouse IgG (Cell Signaling Technology; 1:2,000) were used. For immunofluorescent staining, anti-COX IV (11967, Cell Signaling Technology; 1:100), anti-HADHB (sc271495, Santa Cruz; 1:50), and anti-p65 (sc-8008, Santa Cruz; 1:50) antibodies were used as primary antibodies. As secondary antibodies, anti-mouse IgG, Alexa Fluor 647 (A-21235, Thermo Fisher; 1:200), and anti-rabbit IgG, Alexa Fluor 488 (A-11034, Thermo Fisher; 1:100) antibodies were used. For flow cytometry, anti-CD11b BV421 (101251, BioLegend), anti–F4/80-PE (123110, BioLegend), anti–Ly6c-PE-Cy7 (128018, BioLegend) and anti–Ly6g-PerCP/Cy5.5 (127615, BioLegend) antibodies were used.
BMDMs.
All cultures were maintained at 37 °C in a 5% CO2 water-jacketed incubator. Bone marrow cells were flushed from femurs and tibias of male mice between 6 and 9 wk of age, and were grown in DMEM/F12 (Thermo Fisher) supplemented with 10% FBS (HyClone), 1% penicillin–streptomycin (Thermo Fisher), 1% l-glutamine (Thermo Fisher), and 40 ng/mL of recombinant mouse M-CSF (576406; BioLegend) for 7 to 8 d. The medium was changed every 3 d. Differentiated BMDMs were detached from plates using StemPro Accutase Cell Dissociation Reagent (Thermo Fisher) and replated into 12-well tissue culture dishes at a density of 5 × 105 cells per well prior to cell stimulation. For LPS and Kdo2 lipid A (KLA) treatment, 100 ng/mL of LPS (Sigma-Aldrich) or KLA (Avanti polar lipids) was added to the medium. We found that LPS and KLA increased lncFAO transcription and induced histone modifications similarly (Fig. 1C). For Fig. 1C, data from LPS-treated cells (RNA-seq and ChIP-seq for H3K4me3 and PU.1) and KLA-treated cells (ChIP-seq for H3K27ac and p65) were assembled.
Wound-Healing Models.
Wound-healing assays were conducted 8 wk after bone marrow transplantation. Under anesthesia, a 6-mm disposable biopsy punch (Kai Medical) was used to generate two wounds in the dorsal skin of each animal (62). Close-up photos of the wounds were taken, and the sizes of the wounds were analyzed using Adobe Photoshop and normalized to a neighboring 6-mm circle seal (SI Appendix, Fig. S6A). After macroscopic analyses, the more rostral wounds were dissected for flow cytometric analyses, while the more caudal ones were used for pathological assessment.
Flow Cytometry.
Samples for flow cytometry were prepared as previously described (63). Briefly, mice were anesthetized and the heart was exposed and perfused with 10 mL PBS from the left ventricle. Thereafter, the whole heart tissue was excised, and the right and left atria and atrioventricular valves were removed. The excised biventricles were mechanically minced using a scalpel. The tissue was then incubated in DMEM containing 1% elastase (Worthington Biochemical) for 120 min at 37 °C. During the incubation, the cells in the suspension were dissociated by sequentially passing the extract through 20-, 21-, and 23-gauge needles at 30-min intervals. The cells were further dissociated by passing them through a 23-gauge needle three times and filtered through a 40-μm cell strainer (BD). The cells were then centrifuged at 300 × g for 5 min, washed with PBS, and resuspended in FACS buffer (PBS supplemented with 1% FBS). After removing erythrocytes by using BD PharmLyse solution (BD), isolated cells were stained with fluorochrome-conjugated antibody.
Skin macrophages within the wound were isolated as described previously (64). The excised skin, without the excess fat, was immersed for 30 min at 37 °C in HBSS, which lacked calcium and magnesium and contained 5 mM EDTA, 10 mM Hepes, and 10% FBS. The skin fragments were then cut into small pieces with a scissors and immersed for 30 min at 37 °C in the HBSS. After washing, the skin pieces collected using a 70-μm strainer (BD) were incubated for 30 min at 37 °C in the HBSS supplemented with 0.7 mg/mL collagenase D. The isolated cells from the digested skins were filtered through a 40-μm cell strainer and resuspended in FACS buffer. After removing erythrocytes with BD PharmLyse solution (BD), cells were stained with fluorochrome-conjugated antibody as above.
Segmentation of Cellular Compartments.
Cytosolic and nuclear compartments were isolated using a PARIS Kit (Thermo Fisher). RNA from each fraction was eluted in an equal amount of water, and equal volumes of RNA lysate were used for RT-PCR. The nuclear, mitochondrial, and cytosolic fractions were isolated using a Cell Fractionation Kit (ab109719, Abcam).
Purification of RNA and Real-Time PCR.
Total RNA was purified from cells and tissues by using RNeasy and RNeasy plus micro RNA Purification kits (Qiagen), respectively, according to the manufacturer’s instructions. Prior to RNA sequencing, the cell lysates were passed through a gDNA Eliminator spin column to eliminate potential contamination by genomic DNA. Total RNA was converted to cDNA using SuperScript III (Thermo Fisher). Quantitative real-time PCR analyses were carried out using a Lightcycler 480 system (Roche), with 18S rRNA serving as an internal control. The sequences of the primers used are as follows: 18S, 5′-AAACGGCTACCACATCCAAG-3′ and 5′-CGCTCCCAAGATCCAACTAC-3′; lncFAO, 5′-TGCTACCTCCTCGGTGCTAC-3′ and 5′-TGTTGCTAGGCACTGGAAAA-3′; Il1a, 5′-TTGGTTAAATGACCTGCAACA-3′ and 5′-GAGCGCTCACGAACAGTTG-3′; Il1b, 5′-TGTAATGAAAGACGGCACACC-3′ and 5′-TCTTCTTTGGGTATTGCTTGG-3′; Il6, 5′-GCTACCAAACTGGATATAATCAGGA-3′ and 5′-CCAGGTAGCTATGGTACTCCAGAA-3′; S100a8, 5′-TCCTTGCGATGGTGATAAAA-3′ and 5′-GGCCAGAAGCTCTGCTACTC-3′; Hadhb, 5′-GATGGAGGCCAGTATGCTTT-3′ and 5′-AGTCGGTCGCCTCCTTCTA-3′.
lncFAO and Hadhb Knockdown.
Cells were transfected with siRNAs using Lipofectamine RNAiMax (Thermo Fisher) according to the manufacturer’s instructions. Reverse transfection was performed by adding medium and detached BMDMs to prepared siRNA and lipid complexes inside wells. Stealth RNAi siRNA Negative Control Lo GC Duplex #2 (Thermo Fisher) served as a negative control. Predesigned Stealth RNAi siRNAs for Hadhb knockdown were purchased from Thermo Fisher Scientific. The sequences of two types of silncFAO were designed using BLOCK-iT RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/). The sequences of the siRNAs were as follows: silncFAO#1, 5′-CAAGAGAGAAAGUACUGUUUGGGUA-3′ (sense) and 5′-UACCCAAACAGUACUUUCUCUCUUG-3′ antisense; silncFAO#2, 5′-AGGGAUCAGUUUAGCAGCCUAGAUA-3′ (sense) and 5′-UAUCUAGGCUGCUAAACUGAUCCCU-3′ (antisense); siHADHB, 5′-GGAUCACCUCCUCUGGAGAAGUUUA-3′ (sense) and 5′-UAAACUUCUCCAGAGGAGGUGAUCC-3′ (antisense).
lncFAO Overexpression.
A full-length lncFAO cDNA was amplified from cDNA from LPS-treated BMDMs. The primer sequences used with PCR for cloning were 5′-gtcgacGCCATCCAGACGTACAACTCTCT-3′ and 5′-gcggccgcAGGAAAGAGTTTATTTGGAGCTTACATTTTC-3′. The cDNA was cloned into pGEM-T Easy Vector (Promega) and then the lncFAO insert was sublconed into pcDNA3.1(+) vector (Thermo Fisher). A mixture of 1 μg of the expression vector and 3 μL of lipofectamine 2000 (Thermo Fisher) was added to the medium in each well of a 12-well culture plate, after which 5 × 105 cells were added to each well. As a control the empty vector was transfected. Twenty-four hours after transfection the cell were treated with LPS.
Western Blotting.
Samples of protein lysate supplemented with 1× cOmplete proteinase inhibitor mixture (Sigma-Aldrich) and loading buffer were boiled, after which the proteins were separated by SDS/PAGE, transferred to a nitrocellulose blotting membrane (GE Healthcare), blocked with 5% skim milk, and incubated with primary antibodies at 4 °C overnight. Proteins were visualized using HRP-linked secondary antibody and ECL Prime (GE Healthcare) and then captured using ImageQuant LAS 4000 (GE Healthcare). The same membrane was then stripped using Stripping solution (Wako) and sequentially incubated with other primary antibodies, as multiple proteins were targeted.
FISH and Immunofluorescent Staining.
BMDMs were seeded into eight-well chamber slides at a density of 105 cells per well, fixed with 10% neutral buffered formalin at room temperature for 30 min, dehydrated with ethanol, and stored at −20 °C. RNA FISH was performed using a RNAscope 2.5 HD Reagent Kit-RED (Advance Cell Diagnostics) according to the manufacturer’s protocol. Briefly, the fixed cells were rehydrated followed by treatment with hydrogen peroxide and protease III. The sample was hybridized with probes designed for lncFAO by the manufacturer for 2 h at 40 °C, after which the remainder of the assay protocol was implemented.
For immunofluorescent staining, the samples were washed with PBS, blocked by 2% BSA for 30 min, and incubated with a primary antibody against COX-IV and then the secondary antibody for 1 h each at room temperature. After washing the samples with PBS, nuclei were counterstained with DAPI Fluoromount-G (SouthernBiotech). The fluorescent signals were visualized and captured using a confocal laser scanning microscope (LSM510 Meta, Zeiss)
Analysis of lncFAO-Binding Proteins.
To identify lncFAO binding proteins, RNA pull-down assays were performed using biotinylated lncFAO RNA probe as described previously (41, 65). The plasmid carrying the lncFAO cDNA was linearized using NotI and used as the template of in vitro transcription. Biotinylated lncFAO RNA was generated using T7 RNA polymerase (Promega) and Biotin RNA labeling mix (Sigma Aldrich). As a control, an antisense lncFAO RNA probe was also generated. The in vitro transcribed RNA was treated with RNase-free DNase I and purified using a RNeasy mini kit (Qiagen). RNA was denatured at 60 °C for 10 min and the cooled down to 4 °C before adding it to cell lysates for RNA pull-down.
After treating 108 BMDMs with or without LPS for 24 h, the cytosolic fraction was purified from the cells using a PARIS Kit (Thermo Fisher) and then were diluted to 1 mL with buffer A (150 mM KCl, 25 mM Tris⋅HCl pH 7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% Nonidet P-40) supplemented with 1× cOmplete protease inhibitor mixture (Sigma Aldrich) and 100 U/mL SUPERase In (Thermo Fisher). The resultant solution was divided into three samples and incubated for 1 h at 4 °C with 20 μg of biotinylated RNAs, followed by incubation for 45 min with 60 μL of Dynabeads M280 and T1 (Thermo Fisher). A mixture of different size beads was used to increase sedimentation efficacy (66). After incubation, the beads were washed five times with buffer A and boiled with SDS loading buffer. The eluted samples were loaded into 4 to 15% Mini-PROTEAN Precast Gels (Bio-Rad), which was followed by silver staining using a Silver Stain kit (APRO science). The extracted gel band was submitted to APRO science for LC-MS/MS analysis.
Pull-Down of Endogenous lncFAO-Containing Complexes.
To pull down proteins associated with lncFAO, we modified the procedures used for the ChIRP method (43, 44). The original ChIRP protocol was developed to isolate genomic regions bound by an RNA of interest. We modified that protocol to obtain proteins that associate with lncFAO RNA within cells. Endogenous RNA–protein complexes were retrieved using 10 biotinylated oligonucleotides corresponding to the complementary sequence of lncFAO. Briefly, 24 h after LPS treatment, BMDMs were fixed with 3% formaldehyde for 30 min at room temperature, quenched in 0.125 M glycine for 5 min, and snap frozen. For experimentation, the cell pellets were thawed in a 10× volume of lysis buffer (50 mM Tris⋅HCl pH 7.0, 10 mM EDTA, 1% SDS, supplemented with cOmplete and 0.2 U/mL SUPERase-In) and sonicated for 8 min using a Covaris S220. The resultant lysates were diluted with two volumes of hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris⋅HCl 7.0, 1 mM EDTA, 15% Formamide, cOmplete and SUPERase-In) and divided into three equal aliquots. The aliquots were incubated first with 50 pmol/mL biotinylated anti-lncFAO oligos or anti-LacZ oligos, or with 10 μg/mL RNase A (Sigma Aldrich) at 37 °C overnight. This was followed by incubation for an additional 30 min with 100 μL/mL Dynabeads MyOne Streptavidin C1 (Thermo Fisher). After the incubations, the beads were washed five times with wash buffer (1× SSC, 0.5% SDS, and cOmplete) and incubated in biotin elution buffer (12.5 mM biotin, 7.5 mM Hepes pH 7.5, 75 mM NaCl, 1.5 mM EDTA, 0.15% SDS, 0.075% sarkosyl, and 0.02% Na-Deoxycholate) for 20 min at room temperature and then at 65 °C for 10 min. For protein elution, the eluent was mixed with a 25% total volume of trichloroacetic acid, and the proteins were precipitated at 4 °C overnight. Ten types of anti-lncFAO and four types of anti-LacZ 20-mer oligos were designed using online tools available at (https://www.biosearchtech.com/stellaris). Oligos modified with 3′ biotin-TEG were purchased from Eurofins Genomics. For pull-down, a mixture of oligos were used. The sequences of oligos against lncFAO were as follows: AGTATCACCATAGGAGCTTT, CCAGGTAAGGTGATACATCT, ATGTCTGAGTGTATCTAGGC, CTGGCTGGAATCTATAGTCT, CATTCCTCTTTACCTTTCAA, GCTGTGTGTTTATTCTCTTT, GTGAGCTCAGACTGTAGATT, TCAAGATGCATTATCTGCGC, GGACTCATGACTCTTCTTTC, CTTGACTGATGTGATGTTCC. The sequences of oligos against LacZ were described previously (43).
Cytokine ELISA.
Culture media from BMDMs in 12-well plates were harvested and centrifuged at 700 × g to remove debris. ELISAs were then performed following the instructions provided with BioLegend’s ELISA MAX kits (mouse IL-1α: 433401, IL-1β: 432601, and IL-6: 431301). Cytokine concentrations were calculated based on the subtraction of absorbance at 570 nm from that at 450 nm. Absorbances were measured using an EnSpire plate reader (PerkinElmer).
RNA Sequencing.
Poly-A mRNA was extracted from total RNA by using an Oligo-dT beads of TruSeq RNA Sample Prep Kit v2 (Illumina) or Oligo-dT beads of NEBNext Poly(A) RNA Magneic Isolation Module (New England Biolabs), after which RNA-seq libraries were prepared using a ScriptSeq v2 RNA-Seq Library Preparation Kit (Epicentre) or NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturers’ protocols. Libraries were single-end–sequenced or paired-end–sequenced on GAIIx or HiSEq. 1500 sequencer (Illumina). Reads were aligned to the mm9 or mm10 mouse genome using Bowtie (66) and TopHat (67) or STAR (68). Aligned read files were analyzed using Cufflinks (69) and HOMER (70). Cufflinks identified gene loci of novel lncRNAs, including lncFAO, from the reads of RNA-seq, and the 5′ end of lncFAO was confirmed with reference to CAGE data from FANTOM (https://fantom.gsc.riken.jp/zenbu/). Expression analysis of the RNA-seq data were performed using HOMER and Genomatix. Gene set enrichment was performed using GSEA (28) with rank files generated as previously described (71) from expression data analyzed using DESeq2 (72, 73).
ChIP Sequencing.
ChIP assays were performed as described previously (74). After BMDMs were cross-linked with 1% formaldehyde for 10 min at room temperature and quenched with 0.25 M glycine, cell pellets were lysed in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris⋅HCl pH 8.1, supplemented with cOmplete). Chromatin was sheared to 250-bp to 400-bp fragments using a Covaris S220 μLtrasonicator, diluted 10× in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16 mM Tris⋅HCl pH 8.1, 167 mM NaCl), and incubated with 1 μg of primary antibodies at 4 °C overnight. The protein–DNA complexes immunoprecipitated with Dynabeads Protein A or G (Thermo Fisher) were washed twice with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris⋅HCl pH 8.1, 150 mM NaCl), twice with high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris⋅HCl pH 8.1, 500 mM NaCl), twice with LiCl buffer (0.25 M LiCl, 0.5% Nonidet P-40, 0.5% Na Deoxycholate, 1 mM EDTA, 10 mM Tris⋅HCl pH 8.1), and once with TE (10 mM Tris⋅HCl pH 7.6, 1 mM EDTA). Bound protein–DNA complexes were eluted using elution buffer (1% SDS, 100 mM Na2HCO3) and reversed by addition of 5 M NaCl overnight at 65 °C. ChIP-seq libraries were prepared using a TruSeq ChIP Library Preparation kit (Illumina) according to the manufacturer’s protocol. Libraries were single-end-sequenced on GAIIx or HiSEq. 1500 sequencers (Illumina).
Single-Cell Transcriptome Analysis.
FASTQ data downloaded from ArrayExpress (E-MTAB-7376) (37) were processed using 10X Genomics Cell Ranger count software. The sequence of lncFAO was appended to the mm10 reference. The standard procedures of filtering, log-normalization, and variable gene selection were performed using Seurat v3 (75). We filtered cells that expressed fewer than 200 unique molecular identifiers, and the percentage of counts mapped to the mitochondrial genome was ≥5%. To exclude possible doublets, cells with high unique molecular identifier counts were also removed. Six datasets (sham, day 3 and 7 post-MI of total interstitial cells and Pdgfra+ cells) (37) were aggregated using Seurat’s standard data integration procedure with 2,000 highly variable genes. The expression matrix was dimensionally reduced using principal component analysis of the corrected integrated gene matrix. Clusters were identified using a graph-based approach with the Louvian modularity optimization algorithm. We employed t-distributed stochastic neighbor embedding for dimensionality reduction and visualization of our datasets. For visualization of gene expression, the MAGIC algorism was used to impute drop-out values (76).
We first analyzed expression of lncFAO in the total dataset (SI Appendix, Fig. S2A). Most of the cells expressing lncFAO were found in the clusters expressing Ptprc, Itgam, and Cd68. We then analyzed a subset of clusters expressing Ptprc and Cd68, after which the cells were clustered again to further identify subpopulations (SI Appendix, Fig. S2 B and C).
To identify genes differentially expressed between day 3 and day 5 post-MI in a cluster, MAGIC-imputed data were analyzed using Seurat’s FindMarkers function with the MAST algorithm (77). Highly expressed genes that had an adjusted P < 0.001 and log2 fold-change > 0.25 were selected. The gene sets were used for the GSEA shown in Fig. 2E. To identify the signature genes of a cluster on day 7, gene expression was compared with that of the remaining clusters and the more highly expressed genes that had adjusted P < 0.05 and log2 fold-change > 0.2 were selected. The gene set enrichment was analyzed using Metascape (78) with the default parameters for SI Appendix, Fig. S3B. For GSEA (SI Appendix, Fig. S3C), the results of the MAST test of the genes with log2 fold-change > 0.1 were used to generate rank files.
Skeletal Muscle Signature Gene Set.
Microarray data analyzed in supplemental table 1 in Varga et al. (38) were used to identify genes enriched in Ly6Chi macrophages on day 4 postinjury in cardiotoxin-injured skeletal muscle. The genes whose expression was significantly higher in Ly6Chi macrophages on day 4 than on day 2 or day 8 were selected from the data and used as the signature genes of Ly6Chi macrophages on day 4 postinjury in Fig. 2E.
Thiolase Activity.
HADHB activity was assessed by monitoring thiolytic cleavage of acetoacetyl-CoA (52, 53). In brief, BMDMs were lysed by sonication in 1 mL of lysis buffer (100 mM Tris⋅HCl, pH 8.3, 200 mM NaCl, 0.1% hexamethylphosphoric triamide, 2 mM β-mercaptoethanol, 0.5 mM EDTA, 0.5% Tween-20 and protease inhibitor cOmplete) per 100 mg of cell pellets. Cell lysates diluted 10 times in reaction buffer (100 mM Tris⋅HCl, pH 8.3, 25 mM MgCl2, 100 μM CoA, 40 μM acetoacetyl-CoA) were incubated for 5 min at 30 °C. After the homogenate was centrifuged at 18,000 × g for 5 min, the supernatant was used for enzyme assays. Thiolase activity was spectrophotometrically measured by monitoring the absorbance at 303 nm (79). Mixtures of acetoacetyl CoA and CoA at several concentrations in reaction buffer were measured at the same time to construct a standard curve. One unit of activity was defined as the amount of enzyme that converted 1 μmol of acetoacetyl-CoA in 1-min. For each treatment, the enzymatic activity was measured three times.
Measurement of Energy Metabolism.
The OCR of BMDMs was analyzed using a XF-96 Extracellular Flux Analyzer (Seahorse Bioscience). BMDMs were plated in a 96-well Seahorse plate (40,000 cells per well) and incubated overnight in a complete medium (DMEM/F12 containing l-glutamine, 10% FBS and 40 ng/mL M-CSF). The next day, the medium was changed to FAO assay medium (111 mM NaCl, 4.7 mM KCl, 1.25 mM CaCl2, 2.0 mM MgSO4, 1.2 mM NaH2PO4, 2.5 mM glucose, 0.5 mM carnitine and 5 mM Hepes) and incubated for 30 min. Then, after a pretreatment with etomoxir (40 μM) for 15 min, palmitate-BSA (200 μM palmitate conjugated with 34 μM BSA) or BSA (34 μM) (Seahorse Bioscience) was added, and the assays were initiated. OCR was obtained as pmol of O2 consumed per minute. At the same time, extracellular acidification rate was measured as a change in pH. The wells were sequentially treated with the ATP synthase inhibitor oligomycin (1.0 μM), the chemical uncoupler FCCP (1.5 μM), and the electron transport inhibitor antimycin A/rotenone (0.5 μM each).
OCR and mitochondrial membrane potential were also measured using MitoXpress Xtra (80) (600880; Cayman) according to the manufacturer’s protocol. BMDMs were plated in a 96-well plate at 60,000 cells per well and incubated for 6 h, after which they were treated with 100 ng/mL LPS for 24 h as described above in the section on culture of BMDMs. The following day, the cells were incubated with JC-1 staining solution for 30 min and, after refreshing the medium, MitoXpress Xtra solution was added. The time-resolved fluorescence (excitation 380 nm/emission 650 nm) was measured using an EnSpire multiplate reader at 37 °C for 60 min. Subsequently, fluorescence intensities for JC-1 monomers (excitation 485 nm/emission 535 nm) and JC-1 aggregates (excitation 535 nm/emission 590 nm) were measured.
Data Availability.
All RNA-seq and ChIP-seq data are available in the Gene Expression Omnibus under accession number GSE130056. Our previous ChIP-seq results used in this study are also available (accession number GSE95712) (81).
Supplementary Material
Acknowledgments
We thank Drs. Atsushi Okabe, Masaki Fukuyo, and Bahityar Rahmutulla for next-generation sequencing; and M. Hayashi, X. Yingda, N. Yamanaka, M. Hatase, and K. Ogasawara for excellent technical assistance. This study was supported in part by the Grant-in-Aid for Scientific Research 18K08061 (to Y.N.), 17H04171 and 19K22615 (to K.F.), 20H03679 (to Y.O.), and 17KT0047 and 19H03648 (to I.M.); Grant-in-Aid for Scientific Research on Innovative Areas Preventive Medicine through Inflammation Cellular Sociology 20H04956 (to Y.O.), 20H04938 (to I.M.) from the MEXT Japan; JP19ek0310013, JP19jm0210052, and JP19gm6110006 (to K.F.), JP19gm5910021h9903 (to Y.O.), and JP20gm5010002 (to I.M.) from the Japan Agency for Medical Research and Development; Precursory Research for Embryonic Science and Technology JPMJPR13M7 (to K.F.) from Japan Science and Technology Agency; and grants from MSD Life Science Foundation International and the Japan Foundation for Applied Enzymology (to Y.N.), Takeda Science Foundation (to Y.O. and I.M.), and Uehara Memorial Foundation (to I.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE130056 and GSE129963).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005924117/-/DCSupplemental.
References
- 1.Ghisletti S. et al., Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Glass C. K., Natoli G., Molecular control of activation and priming in macrophages. Nat. Immunol. 17, 26–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oishi Y., Manabe I., Macrophages in inflammation, repair and regeneration. Int. Immunol. 30, 511–528 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Novak M. L., Koh T. J., Phenotypic transitions of macrophages orchestrate tissue repair. Am. J. Pathol. 183, 1352–1363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oishi Y., Manabe I., Macrophages in age-related chronic inflammatory diseases. NPJ Aging Mech. Dis. 2, 16018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Bossche J., O’Neill L. A., Menon D., Macrophage immunometabolism: Where are we (Going)? Trends Immunol. 38, 395–406 (2017). [DOI] [PubMed] [Google Scholar]
- 7.O’Neill L. A. J., Kishton R. J., Rathmell J., A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck M. D., Sowell R. T., Kaech S. M., Pearce E. L., Metabolic instruction of immunity. Cell 169, 570–586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttman M. et al., lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loewer S. et al., Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 42, 1113–1117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu W., Yuan B., Flygare J., Lodish H. F., Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 25, 2573–2578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klattenhoff C. A. et al., Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu F., Jin L., Jin Y., Nie Z., Zheng H., Long noncoding RNAs in autoimmune diseases. J. Biomed. Mater. Res. A 107, 468–475 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Bhat S. A. et al., Long non-coding RNAs: Mechanism of action and functional utility. Noncoding RNA Res. 1, 43–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Zheng Y., lncRNAs regulate innate immune responses and their roles in macrophage polarization. Mediators Inflamm. 2018, 8050956 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter S. et al., A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krawczyk M., Emerson B. M., p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. eLife 3, e01776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z. et al., The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. U.S.A. 111, 1002–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan J. et al., Cutting edge: A natural antisense transcript, AS-IL1α, controls inducible transcription of the proinflammatory cytokine IL-1α. J. Immunol. 195, 1359–1363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y. et al., The NF-κB-responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J. Immunol. 199, 3571–3582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui H. et al., The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 44, 2085–2095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atianand M. K. et al., A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du M. et al., The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat. Commun. 8, 2049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oishi Y. et al., SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab. 25, 412–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derrien T. et al., The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hon C. C. et al., An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C. et al., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A. et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberzon A. et al., The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musacchia F., Basu S., Petrosino G., Salvemini M., Sanges R., Annocript: A flexible pipeline for the annotation of transcriptomes able to identify putative long noncoding RNAs. Bioinformatics 31, 2199–2201 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Wang L. et al., CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barski A. et al., High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Heinz S., Romanoski C. E., Benner C., Glass C. K., The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nord A. S. et al., Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155, 1521–1531 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilgendorf I. et al., Ly-6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ. Res. 114, 1611–1622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindsey M. L., Saucerman J. J., DeLeon-Pennell K. Y., Knowledge gaps to understanding cardiac macrophage polarization following myocardial infarction. Biochim. Biophys. Acta 1862, 2288–2292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farbehi N. et al., Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife 8, e43882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varga T. et al., Highly dynamic transcriptional signature of distinct macrophage subsets during sterile inflammation, resolution, and tissue repair. J. Immunol. 196, 4771–4782 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Kimball A. et al., Ly6CHi blood monocyte/macrophage drive chronic inflammation and impair wound healing in diabetes mellitus. Arterioscler. Thromb. Vasc. Biol. 38, 1102–1114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J., Sun B. K., Erwin J. A., Song J. J., Lee J. T., Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinn J. L. et al., Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yap K. L. et al., Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu C., Qu K., Zhong F. L., Artandi S. E., Chang H. Y., Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu C. et al., Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams D. J. et al., HADHB, HuR, and CP1 bind to the distal 3′-untranslated region of human renin mRNA and differentially modulate renin expression. J. Biol. Chem. 278, 44894–44903 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Buler M., Aatsinki S.-M., Izzi V., Uusimaa J., Hakkola J., SIRT5 is under the control of PGC-1α and AMPK and is involved in regulation of mitochondrial energy metabolism. FASEB J. 28, 3225–3237 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Kumar B., Koul S., Khandrika L., Meacham R. B., Koul H. K., Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 68, 1777–1785 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Liu P.-S. et al., α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Pike L. S., Smift A. L., Croteau N. J., Ferrick D. A., Wu M., Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim. Biophys. Acta 1807, 726–734 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Deng B., Wehling-Henricks M., Villalta S. A., Wang Y., Tidball J. G., IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J. Immunol. 189, 3669–3680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ip W. K. E., Hoshi N., Shouval D. S., Snapper S., Medzhitov R., Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z., Zhou J., Du Y., Estrogen receptor alpha interacts with mitochondrial protein HADHB and affects beta-oxidation activity. Mol. Cell Proteomics 11, M111.011056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kao Y.-T. et al., Japanese encephalitis virus nonstructural protein NS5 interacts with mitochondrial trifunctional protein and impairs fatty acid β-oxidation. PLoS Pathog. 11, e1004750 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castello A. et al., Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Millet P., Vachharajani V., McPhail L., Yoza B., McCall C. E., GAPDH binding to TNF-α mRNA contributes to posttranscriptional repression in monocytes: A novel mechanism of communication between inflammation and metabolism. J. Immunol. 196, 2541–2551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang C.-H. et al., Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long J. et al., Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Invest. 126, 4205–4218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leucci E. et al., Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531, 518–522 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Cabili M. N. et al., Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang H. et al., One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hwang W. Y. et al., Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eichenfield D. Z. et al., Tissue damage drives co-localization of NF-κB, Smad3, and Nrf2 to direct Rev-erb sensitive wound repair in mouse macrophages. eLife 5, e13024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujiu K. et al., A heart-brain-kidney network controls adaptation to cardiac stress through tissue macrophage activation. Nat. Med. 23, 611–622 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Benck C. J., Martinov T., Fife B. T., Chatterjea D., Isolation of infiltrating leukocytes from mouse skin using enzymatic digest and gradient separation. J. Vis. Exp., e53638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao J. et al., Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40, 939–953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.So B. R. et al., A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat. Struct. Mol. Biol. 23, 225–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C. et al., Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heinz S. et al., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ziemann M., How to generate a rank file from gene expression data Genome Spot (2015). http://genomespot.blogspot.com/2015/01/how-to-generate-rank-file-from-gene.html. Accessed 2 June 2020.
- 72.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mootha V. K. et al., PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Lai F. et al., Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stuart T. et al., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Dijk D. et al., Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finak G. et al., MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou Y. et al., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Middleton B., 3-Ketoacyl-CoA thiolases of mammalian tissues. Methods Enzymol. 35, 128–136 (1975). [DOI] [PubMed] [Google Scholar]
- 80.Salvioli S., Ardizzoni A., Franceschi C., Cossarizza A., JC-1, but not DiOC6(3) or rhodamine 123, is a reliable fluorescent probe to assess delta psi changes in intact cells: Implications for studies on mitochondrial functionality during apoptosis. FEBS Lett. 411, 77–82 (1997). [DOI] [PubMed] [Google Scholar]
- 81.Oishi Y. et al., Bmal1 regulates inflammatory responses in macrophages by modulating enhancer RNA transcription. Sci. Rep. 7, 7086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq and ChIP-seq data are available in the Gene Expression Omnibus under accession number GSE130056. Our previous ChIP-seq results used in this study are also available (accession number GSE95712) (81).