Significance
CD5 is a surface receptor that negatively regulates T cell activation, but T cells expressing more CD5 have been shown to survive better in the effector/memory phase after pathogenic insult. Our data suggest a mechanistic framework for this duality, by identifying the NF-κB pathway as a target of CD5 signaling. CD5 controls the amounts of lymphocyte IκBα independently of the canonical binding partner SHP-1 and in addition to T cell receptor signals. The increased basal levels of NF-κB transcription factors in CD5hi cells also conferred an NF-κB–dependent survival advantage.
Keywords: T cell activation, TCR signaling, CD5
Abstract
Immature T cells undergo a process of positive selection in the thymus when their new T cell receptor (TCR) engages and signals in response to self-peptides. As the T cell matures, a slew of negative regulatory molecules, including the inhibitory surface glycoprotein CD5, are up-regulated in proportion to the strength of the self-peptide signal. Together these regulators dampen TCR-proximal signaling and help avoid any subsequent peripheral activation of T cells by self-peptides. Paradoxically, antigen-specific T cells initially expressing more CD5 (CD5hi) have been found to better persist as effector/memory cells after a peripheral challenge. The molecular mechanisms underlying such a duality in CD5 function is not clear. We found that CD5 alters the basal activity of the NF-κB signaling in resting peripheral T cells. When CD5 was conditionally ablated, T cells were unable to maintain higher expression of the cytoplasmic NF-κB inhibitor IκBα. Consistent with this, resting CD5hi T cells expressed more of the NF-κB p65 protein than CD5lo cells, without significant increases in transcript levels, in the absence of TCR signals. This posttranslationally stabilized cellular NF-κB depot potentially confers a survival advantage to CD5hi T cells over CD5lo ones. Taken together, these data suggest a two-step model whereby the strength of self-peptide–induced TCR signal lead to the up-regulation of CD5, which subsequently maintains a proportional reserve of NF-κB in peripheral T cells poised for responding to agonistic antigen-driven T cell activation.
The ability of the adaptive immune system to defend against a broad range of environmental pathogens while avoiding autoimmune responses against self-antigens is critically dependent on generating and managing the repertoire of antigen-specific receptors expressed by T and B cells (1). T cell antigen receptors (TCR) are generated by semirandom recombination as T cells develop in the thymus. Afterward, those that bind with high affinity to self-antigens are eliminated by negative selection or diverted into other cell fates, such as regulatory T cells (Tregs) or CD8αα intraepithelial cells (2). These processes still appear to allow some TCRs with lower affinity for self-antigens to complete maturation and migrate to the periphery, necessitating multiple mechanisms of peripheral tolerance in order to prevent autoimmune disease (3, 4). However, in the thymus, prior to the negative selection step, newly acquired TCRs on developing T cells are all forced to undergo a quality-control step known as positive selection (5, 6). This step ensures that every new TCR can react specifically with at least one self-antigen peptide-MHC (pMHC) complex in the thymus; and cells whose TCRs cannot do so, undergo apoptosis (referred to as nonselection or death by neglect) (7–10). Paradoxically, this key step in thymic development also ensures that all T cells that reach the periphery are nominally self-reactive, at least with a self-antigen that is presented in the thymus (11, 12).
Several dominant or cell-extrinsic mechanisms (such as control of costimulation, activity of Tregs, and so forth) as well as T cell-intrinsic alterations cooperatively limit the unwarranted activation of T cells to these and other self-antigens in the periphery. The latter include a “tuning” of each T cell’s activation threshold by the developmental modulation of the biochemical signal-transduction capability of the TCR (13, 14). During tuning, alterations in the expression levels of many key regulators of signaling result in lowering (or also possibly raising) of the antigen-sensitivity of that T cell, ensuring that the levels of self-antigens that initially positively selected that TCR can no longer trigger a strong enough intracellular signal and drive pathological T cell activation. Such alterations may be in the expression of specialized regulators, including the cell surface glycoprotein CD5 (13, 15), the microRNA miR-181a (16), voltage-gated sodium channels (17), the adapter Themis (18, 19), or in molecules such as LCK, CD4, CD8, and others. Importantly, despite the process of thymic tuning, it is increasingly evident that peripheral T cells not only continue to be aware of their specific self-ligands but that these tonic TCR–self-ligand interactions can play important roles in supporting survival, activation, differentiation, and memory formation to pathogen-derived antigens (12, 20–24). Understanding the mechanisms by which differing TCR-affinities for self-peptides during thymic selection events continue to influence peripheral function is likely to have a significant translational relevance (25).
The surface level of CD5 is one the earliest characterized markers of positive selection. CD5 is up-regulated in thymocytes proportional to the strength of TCR signals generated from interaction with self-pMHC ligands (15). CD5 has also proven to be a surrogate marker for TCR-affinity to self-peptides in mature, peripheral T cells to the extent that many investigators use CD5 expression to parse out T cells that bind to self-ligands strongly or weakly (24, 26, 27). In recent years, such approaches have led to an appreciation of differences in the survival or function of peripheral CD5hi or CD5lo T cells (22, 23, 28). The precise affinities of TCRs (e.g., in the CD5hi versus CD5lo fraction) for their self-ligands have been difficult to measure since the identity of natural self-peptides, which positively select or deliver tonic signals, are known for only a very few TCRs. The central tenets uncovered by these approaches have, however, been supported by indirect evidence from examining differences in TCR signaling as well as the consequences of genetically manipulating signal transduction pathways in T cells. A sticking point that remains is the functional contribution of CD5 itself.
CD5 is a scavenger receptor cysteine-rich (SRCR) family transmembrane glycoprotein expressed on all T and some B cells (29). Despite the presence of three canonical SRCR domains in its extracellular region, the potential ligands for CD5 or even the extracellular domain itself does not seem to be critical for CD5’s ability to negatively regulate TCR signaling (30). The intracellular domain of CD5 has multiple tyrosines that are phosphorylated at various stages of T cell activation (31). The activation induced recruitment of molecules, such as the phosphatase SHP-1, the ubiquitin ligase cbl-b, the kinase casein kinase II, and others. to these residues and thereby to the TCR signalosome is largely thought to mediate the negative regulatory impact of CD5 in lymphocytes (32–42). The consequences of CD5 signaling has been measured by its impact on TCR proximal adapters, cytokine secretion, proliferation of T cells, and so forth (34, 40, 43–47). This close association with the TCR has also made it difficult to parse out any distinct consequences of signaling directly mediated by CD5 itself. This is especially relevant in experiments describing different peripheral functions of T cells expressing varying levels of CD5. Intriguingly, many such experiments attribute a net gain in pathogen responsiveness from CD5hi T cells, which is inconsistent with the known negative regulatory activities of CD5. While these data have been interpreted with the understanding that CD5hi T cells have higher self-reactive TCRs, which could potentiate their pathogen responses, the role of CD5 itself would need to be understood to develop a complete picture of the logical signaling networks in T cells.
Toward this end, herein we examined the biochemical signaling intermediates that are differentially expressed between resting T cells displaying high and low levels of CD5 in mouse models. We found that intracellular levels of the regulator of NF-κB nuclear translocation, IκBα, strongly tracks with surface CD5 expression. The genetic ablation of CD5 resulted in a reduction of IκBα expression in T cells, revealing a critical role for CD5 in posttranscriptionally regulating the NF-κB signaling pathway. Importantly, inducible ablation of CD5 showed that rather than a secondary consequence of the strength of TCR signaling, the tonic levels of IκBα in T cells was dynamically maintained by continued expression of CD5. Finally, overexpressing CD5 in cell lines, without additional TCR signaling, led to the stabilization of IκBα as well. The consequence of this regulation was the maintenance of a larger reservoir of NF-κB in T cells that had a higher expression of CD5. This basal difference in the signaling capacity of CD5hi and CD5lo T cells also translated with greater thymocyte survival of the former, in an NF-κB–dependent fashion. Taken together, these data suggest a model by which levels of CD5 directly control the basal state of NF-κB signaling in thymocytes and resting peripheral T cells.
Results
The Amount of Intracellular IκBα in Peripheral T Cells Correlates with the Surface Expression Levels of CD5.
The functional differences in peripheral T cells with varying levels of CD5 from thymic development signals or from tonic signals in the periphery have been described (15, 22, 23, 26, 27), but the underlying molecular mechanisms are not fully understood. This is especially true in the context of reports that CD5hi T cells respond better to foreign antigens (22, 23, 48), although CD5 is reported to be a negative regulator of TCR signaling. Previous studies have examined gene-expression changes between CD5hi and CD5lo cells, as well as direct consequences on TCR signaling (22). We reasoned that any basal differences in the signaling capacity of these cells might also be discerned by analyzing the protein expression of key mediators of signaling in T cells, in relation to the levels of CD5 (Fig. 1).
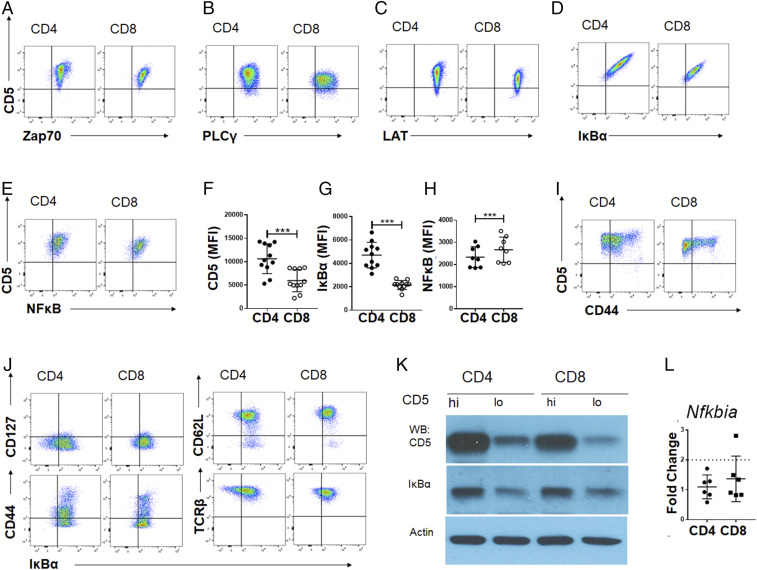
Fig. 1.
Components of NF-κB signaling are selectively altered in CD5hi compared to CD5lo peripheral T cells. The surface levels of CD5 and intracellular levels of Zap70 (A), PLCγ (B), LAT (C), IκBα (D), or NF-κB (E), are shown for either CD4 (Left) or CD8 (Right) T cells. The gMFI of CD5 (F), IκBα (G), or NFκB (H) expressed by CD4 or CD8 T cells from lymph node or spleen cells, gated on live, TCR-β+ cells (n = 11 from 3 independent experiments). Statistical significance was calculated using a paired t test. ***P < 0.001; anything not marked is not statistically significant. (I) The surface levels of CD5 and CD44 are shown for CD4 (Left) or CD8 (Right) T cells. (J) The distribution of IκBα expression with other cell surface markers (CD127, CD44, CD62L, TCR-β) on polyclonal peripheral T cells from lymph node or spleen gated on live, TCR-β+ cells are shown for CD4 (Left) or CD8 (Right). CD5 CV of 66.83 ± 2.91 in CD4 CD44hi; CD5 CV of 52.98 ± 0.69 in CD4 CD44lo; CD44 CV of 65.65 ± 1.24 in CD4 CD44hi; CD44 CV of 60.80 ± 2.35 in CD4 CD44lo. (K) Western blot analysis from B6 lymphocytes sorted on 20% CD5hi and CD5lo within CD4+ and CD8+ T cell (TCR-β+) populations. Representative Western blot is shown (n = 3 from 3 independent experiments). Sorting strategy is shown in SI Appendix, Fig. S1. (L) Peripheral T cells from WT animals were sorted on 20% CD5hi and CD5lo expression as indicated in SI Appendix, Fig. S1. Cells were lysed and RNA was harvested for reverse transcription. cDNA was analyzed for expression of Actb and Nfkbia between groups. Fold-change (2ΔΔCT) of CD5hi cells compared to CD5lo cells of either CD4 or CD8 populations normalized to expression of Actb (n = 6 from 2 independent experiments).
TCR signal transduction is initiated by the activity of the CD4/CD8 coreceptor-associated kinase LCK, which phosphorylates immunoreceptor tyrosine-based activating motifs on the CD3ζ chain, resulting in recruitment of the kinase ZAP70 and subsequent signaling through LAT, ERK, PLCγ, p38, and others. Since peripheral T cells express a range of CD5 (15) (see y axis in Fig. 1 A–E and I) we first examined whether higher CD5 expression correlated with higher or lower expression of any of these TCR signaling intermediates. There was no apparent correlation between CD5 levels to the total intracellular levels of Zap70, PLCγ, or LAT (Fig. 1 A–C). Typically, the TCR proximal signaling machinery converges on key transcription factor (TF) complexes that activate gene expression. Of these, the NF-κB pathway involves a tightly regulated cytoplasm to nucleus transit of the TF complex. Typically, the dimeric TFs of NF-κB p50 or p65 are complexed with the inhibitor, IκB, which masks their nuclear localization signals and retains them in the cytoplasm. Upon TCR signaling, proximal kinases phosphorylate IκB and mark it for ubiquitin tagging and degradation, allowing the cytoplasmic pool of NF-κB TF to translocate to the nucleus and drive gene expression. We found that the expression levels of IκBα in T cells consistently showed a broad distribution similar to the distribution of CD5 in CD4 and CD8 T cells (Fig. 1D). Furthermore, as shown in Fig. 1F, peripheral CD4 T cells typically express a higher level of CD5 (mean fluorescence intensity [MFI] of 10,631 ± 3,025) than CD8s (geometric MFI [gMFI] of 5,953 ± 2,252) and the IκBα levels showed a similar trend (gMFI of 4,709 ± 1,041 in CD4s and gMFI of 2,155 ± 364 in CD8s), as seen in Fig. 1G. We did find a subtle, yet significant, increase in total NF-κB p65 levels in CD8 compared to CD4 T cells, but there was not a strong correlation between CD5 and the TF subunit NF-κB p65 (Fig. 1 E and H). This striking correlation between CD5 and IκBα expression was not shared with other surface receptors, such as CD127, CD44, CD62L, or TCR-β (Fig. 1 I and J) or signaling molecules, as discussed above (Fig. 1 A–C). As we discuss later in Fig. 2, the overall levels of CD5 are higher in the discrete memory T cell population identified as CD44hi T cells; but the distribution of CD44 itself did not follow a correlation with either CD5 or IκBα (Fig. 1 I and J, Left, Lower).
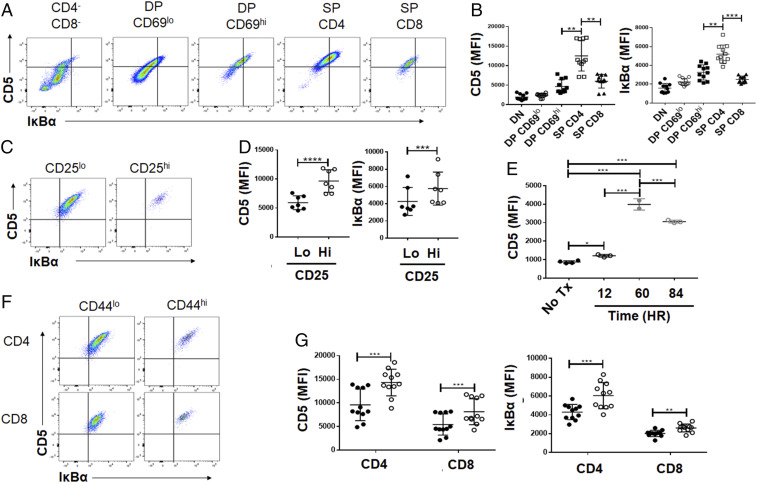
Fig. 2.
Concomitant up-regulation of IκBα with developmental and peripheral modulation of CD5 levels. (A) Representative plots showing the levels of CD5 versus IκBα progressive stages of T cell development in the thymus. Thymocytes were stained for CD4, CD8, and CD69 and gated on markers listed on top of each panel (gating strategy shown in SI Appendix, Fig. S2A). DP = CD4+CD8+ and SP = single positive for either CD4 or CD8 expression as designated. (B) The CD5 and IκBα gMFI on thymocyte subsets as identified in A (n = 11 from 3 independent experiments; statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparisons). (C) Representative peripheral expression levels of CD5 and IκBα in T cells gated by CD4+, TCR-β+, and CD25 expression, to enrich for regulatory T cells in pooled lymph node (the CD25hi population). (D) The CD5 and IκBα gMFI on CD25hi and CD25lo T cells (n = 7 from 3 independent experiments; statistical significance was calculated using paired t tests). (E) 5C.C7 TCR-tg CD4+ T cells were adoptively transferred to congenically distinct B10.A mice and challenged with MCC peptide and LPS challenge in B10.A hosts. Expression of CD5 was analyzed at varying time points after challenge on adoptively transferred TCR-tg CD4+ T cells (n = 2 to 3 from 1 independent experiment; statistical significance was calculated using Tukey’s multiple comparisons). (F) Representative flow plots gated on live, TCR-β+ and CD4+ or CD8+ cells, further subdivided by CD44 expression (gating strategy is shown in SI Appendix, Fig. S2). (G) CD5 and IκBα gMFI on CD4 or CD8 cells gated as indicated in F from lymph node or spleen (n = 11 from 3 independent experiments; statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparisons). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Anything not marked is not statistically significant.
The correlation between CD5 and IκBα was further validated with Western blot analysis on CD4 and CD8 peripheral lymphocytes that were FACS-sorted based on their expression of CD5 (Fig. 1K; gating shown in SI Appendix, Fig. S1A). We then examined if CD5hi cells expressed more Nfkbia mRNA, the gene that encodes the IκBα protein (Fig. 1L). Cells with high and low CD5 expression did not show significant differences in Nfkbia transcript by qRT-PCR (Fig. 1L). Furthermore, additional analysis of transcripts for NF-κB subunits by qRT-PCR did not show significant differences between CD5hi and CD5lo T cells (SI Appendix, Fig. S1B). Consistent with this, querying an RNA-sequencing (RNA-seq) dataset that analyzed FACS-sorted CD5lo and CD5hi T cells (both CD4 and CD8) also did not show differences between multiple NF-κB–related genes (SI Appendix, Fig. S1C). These data suggest that the regulation of IκBα protein levels in correspondence to CD5 levels involves posttranscriptional mechanisms. Taken together, this identifies IκBα as a key signaling molecule whose expression levels strictly covary with surface levels of CD5 in steady-state (resting) peripheral CD4 and CD8 T cells.
CD5 and IκBα Expression Are Dynamically Regulated during T Cell Development and Peripheral Activation.
The increasing levels of CD5 on T cells typically reflects a developmental continuum in the thymus, which continues to change as the T cells undergo antigen activation and differentiation into effector and memory fates in the periphery. As previously reported, early thymic precursors to T cells (CD4−CD8− double-negative [DN] thymocytes) express very little surface CD5 (15). During the positive selection step (transition of the CD4+CD8+ double-positive [DP] cells to express CD69), there is a proportional up-regulation of CD5 to the strength of the positively selecting TCR signal (15). We therefore examined if the levels of IκBα are also dynamically regulated with CD5 during T cell development and transition to the periphery (Fig. 2; gating shown in SI Appendix, Fig. S2A).
As previously described, CD4−CD8− thymocytes have lower expression of CD5 (gMFI 1917.55 ± 729.76) compared to developing T cells that are DP CD69hi cells (gMFI 4647.27 ± 1644.39) (Fig. 2 A and B). In concert with CD5 up-regulation, IκBα expression also increases through these developmental stages (Fig. 2 A and B). This suggested that not only does IκBα expression correlate with CD5 in the periphery, as demonstrated in Fig. 1, but may be a novel quantitative marker for positive selection and TCR self-reactivity as previously reported for CD5 and Nur77 (49).
Typically, T cells that complete thymic development and emigrate to the periphery maintain the levels of CD5 they acquired in the thymus. Tregs, which also have highly self-reactive TCRs, have been shown to express high levels of CD5 (50, 51). We also found a higher IκBα expression in the peripheral CD25hi population of CD4+ T cells compared to the CD25lo population (Fig. 2 C and D; gating shown in SI Appendix, Fig. S2B). Finally, CD5 is further up-regulated when peripheral T cells undergo antigen-dependent activation (52) (Fig. 2E). We examined this during an in vivo activation time course, by adoptively transferring a model antigen- (pigeon cytochrome C) specific 5C.C7 TCR-tg CD4+ T cells with a subsequent peptide and adjuvant (LPS) challenge, before recovering them at various time points for analysis of surface CD5 expression (Fig. 2E). Within 12 h after challenge, there was a significant up-regulation of CD5 on the adoptively transferred 5C.C7 population that continues to increase up to 60 h after challenge (Fig. 2E). As discussed later (see Fig. 4H), IκBα is also proportionally up-regulated after TCR stimulation in peripheral T cells. Additionally, peripheral “memory” phenotype T cells in a polyclonal B6 repertoire (identified as CD44hi as indicated in SI Appendix, Fig. S2B) also maintain higher CD5 expression (Fig. 2 F and G, Left). Similarly, the basal IκBα expression in peripheral CD44hi CD4 and CD8 T cells increased correspondingly (Fig. 2 F and G, Right; gating shown in SI Appendix, Fig. S2B). Taking these data together, this suggests that antigen exposure up-regulates both CD5 and IκBα expression. The continuum of CD5 expression during a T cell’s life is also reflected in IκBα expression, which covaries with the TCR stimulus or antigen exposure history of the cell, whether the relevant stimulus is encountered during thymic selection or peripheral activation.
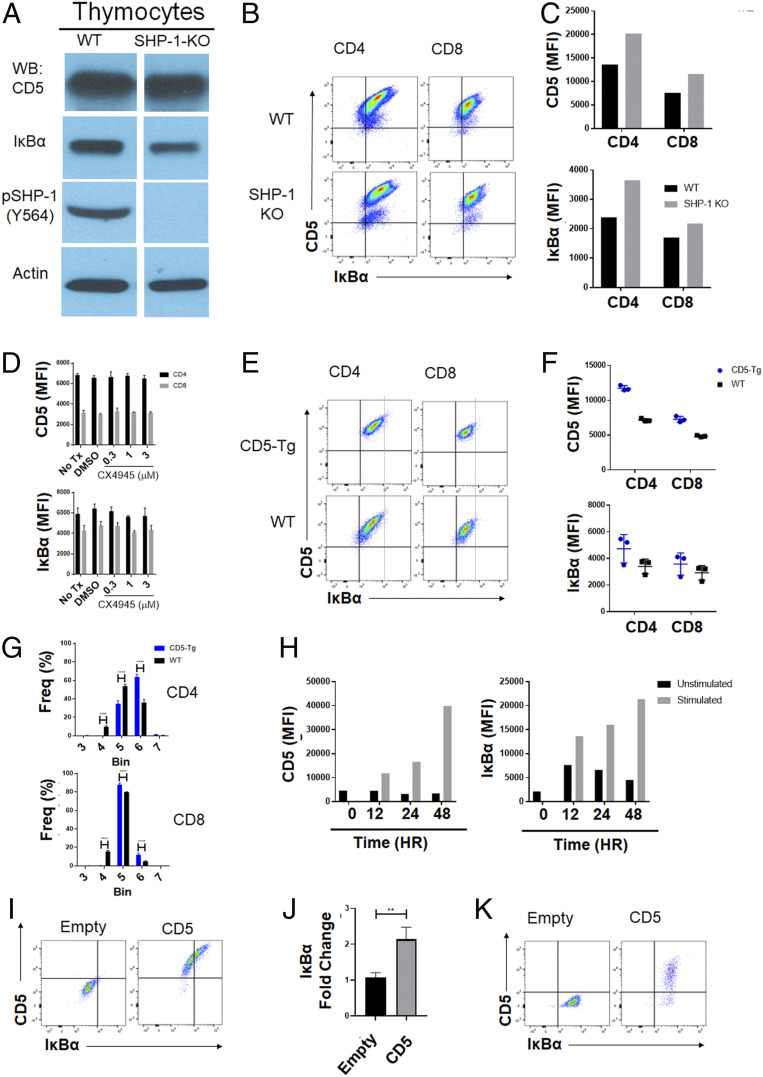
Fig. 4.
CD5-IκBα correlation is independent of SHP-1 and has a saturable level of IκBα up-regulation. (A) Western blot analysis of thymocytes from WT and SHP-1–KO animals (n = 1 from 1 independent experiment). (B) Flow cytometry analysis of WT and SHP-1–KO splenocytes (n = 1 from 1 independent experiment). (C) CD5 and IκBα gMFI in WT and SHP-1–KO splenocytes (n = 1 from 1 independent experiment). (D) CD5 and IκBα gMFI of peripheral T cells after 24 h ex vivo culture in the presence or absence of varying concentrations CX4945 (n = 3 from 1 independent experiment; statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparisons test). (E) WT and CD5 transgenic (CD5-Tg) thymocytes were harvested and analyzed for expression of CD5 and IκBα. Representative flow plots are shown. (F) CD5 and IκBα gMFI on peripheral CD4 and CD8 T cells from WT and CD5-Tg animals (n = 3 from 1 independent experiment; statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparisons test). (G) WT and CD5-Tg lymphocytes were harvested from pooled lymph nodes and grouped into bins by CD5 expression as detailed in SI Appendix, Fig. S5. Frequency of population in each bin is displayed for CD4 and CD8 peripheral T cells (n = 3 from 1 independent experiment; statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparisons test). (H) Lymphocytes from SMARTA CD4+ TCR-Tg mice were isolated and stimulated ex vivo with TCR-αβ KO antigen presenting cells and cognate peptide for the indicated time points. Cells were harvested and analyzed by flow cytometry. CD5 and IκBα gMFI of SMARTA CD4+ TCR-Tg T cells (n = 1 from 1 independent experiment). (I) BW-5147 cells were transduced with empty or CD5 retroviral vector. Cells were rested for 5 d after transduction and then analyzed by flow cytometry. Representative flow plots are shown. (J) CD5 and IκBα gMFI on retrovirally transduced BW-5147 cells (n = 4 from 3 independent experiments; statistical significance was calculated using one-way ANOVA with Tukey’s multiple comparisons test). **P < 0.01. (K) 3T3 cells were transduced with empty retroviral vector or with CD5 retroviral vector. Cells were rested for 48 h after transduction and then analyzed by flow cytometry. (n = 1 from 1 independent experiment).
The Setting of Basal IκBα Levels in T Cells Requires Continued Expression of CD5.
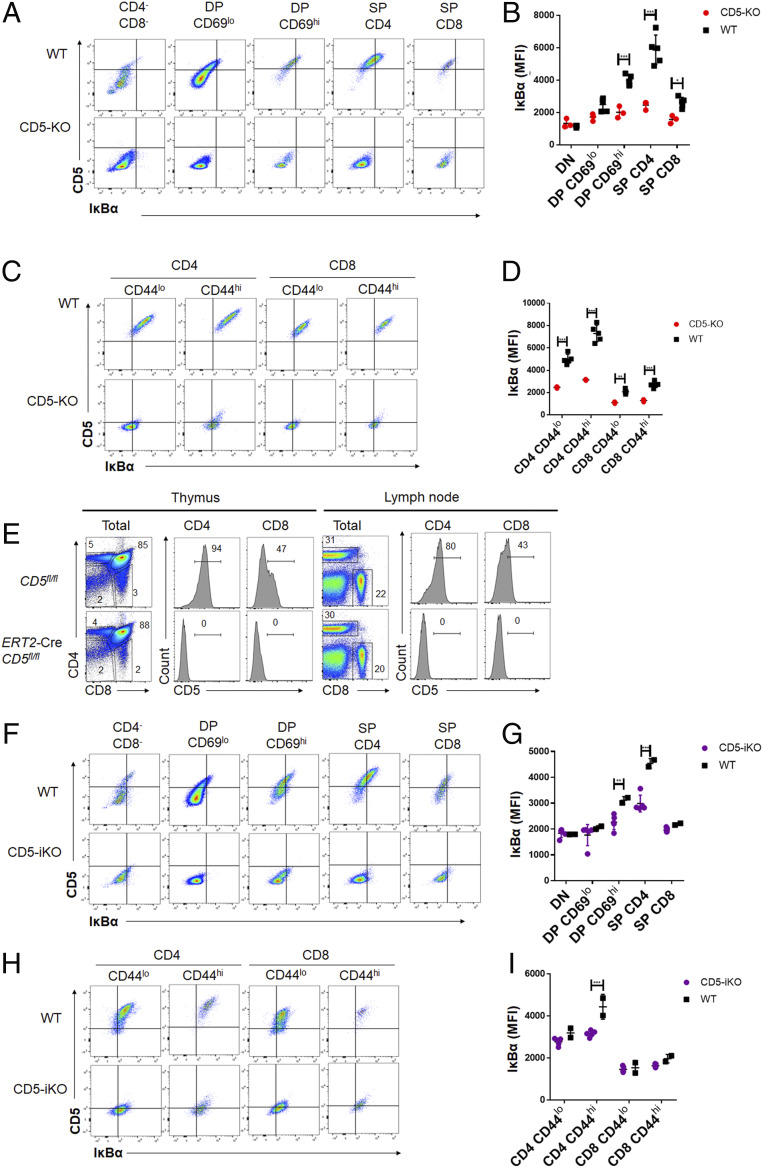
Given our current understanding of how TCR affinity to self-ligands, and by extension TCR signal intensity, influences CD5 expression, it was formerly possible that the concordance we observed between CD5 and IκBα expression were both independently regulated by TCR, where higher affinity interactions with self-peptides resulted in up-regulation of IκBα as well as CD5. If this were true, we would expect the IκBα levels in a CD5 knockout (CD5-KO) T cell to be either unchanged relative to a WT T cell or potentially slightly higher due to stronger TCR signals from a loss of CD5 inhibition. In the absence of CD5, to correlate with the levels of IκBα, we examined the gradation of IκBα levels as the CD5-KO T cells progress from DN through positive selection and then peripheral differentiation from naïve through memory (as discussed in Fig. 2).
CD5-KO mice had significantly lower expression of IκBα in DP CD69hi, single-positive (SP) CD4, and SP CD8 thymocytes compared to WT (CD5+/+) animals (Fig. 3 A and B) but not within the CD4−CD8− and DP CD69lo. This suggests a failure to up-regulate IκBα in CD5-KO (CD5−/−) animals as thymocyte development progresses through TCR-dependent selection stages. Similarly, IκBα expression was decreased in both the peripheral CD4 and CD8 T cells of CD5-KO mice, regardless of the expression of CD44 (Fig. 3 C and D). This suggested that IκBα up-regulation during antigen encounter required the presence of CD5.
Fig. 3.
The surface expression of CD5 is necessary for maintaining heterogenous levels of IκBα in T cells. (A) A similar gating strategy as in Fig. 2A was used to examine different thymocyte subsets in WT (Upper) or CD5-KO (Lower) mice and representative flow plots are shown. (B) The gMFI of CD5 and IκBα on thymocyte populations as gated in Fig. 2A from WT or CD5-KO mice (n = 3 to 5 from 1 experiment; statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparisons test). (C) Peripheral live, TCR-β+ CD4, or CD8 T cells from the pooled lymph nodes of WT or CD5-KO mice were analyzed for CD5 and IκBα expression and a representative set of flow plots are shown. (D) The gMFI of CD5 and IκBα gMFI in peripheral CD4 or CD8 T cells gated as indicated above from lymph nodes, respectively (n = 3 to 5 from 1 experiment; statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparisons test). (E) Representative flow plots of thymocytes and peripheral T cells isolated from lymph nodes of CD5-floxed mice with CAG-CreERT2 transgene (CD5 inducible knockouts; CD5-iKO) after treatment with tamoxifen for 5 d. Histograms of CD5 expression are shown. (F) The expression of CD5 and IκBα in thymocytes of either wild-type (WT, Upper) or CD5-floxed mice with CAG-CreERT2 transgene (CD5-iKO, Lower) after treatment with tamoxifen for 5 d. Representative flow plots are shown. (G) CD5 and IκBα gMFI on thymocytes as gated in Fig. 2A from WT or CD5-iKO mice (n = 2 to 5 from 1 experiment; statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparisons test). (H) The expression of CD5 and IκBα in peripheral T cells in either WT or CD5-iKO mice after treatment with tamoxifen for 5 d. Representative flow plots are shown. (I) CD5 and IκBα of peripheral CD4 or CD8 T cells gated as indicated above from lymph nodes of WT or CD5-iKO mice (n = 2 to 5 from 1 experiment; statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparisons test).
While these data reveal an unexpected role for CD5 in directly modulating IκBα levels, the possibility still existed that our observations resulted from the selection of different TCRs in the WT versus CD5-KO mice (34, 43). Since CD5 regulates TCR signals in developing thymocytes, the absence of CD5 could lead to the selection of a lower self-affinity TCR repertoire resulting in basal differences of IκBα expression. To evaluate this, we generated an inducible KO mouse model in which CD5 was ablated only after administration of Tamoxifen (referred to as CD5-iKO), as shown in SI Appendix, Fig. S3. Characterization of this model is shown in Fig. 3E. Tamoxifen administration resulted in the ablation of CD5 in both the thymus and periphery after 5 d of treatment (Fig. 3E).
Using this model of inducible ablation of CD5, we found that there is a decreased expression of IκBα in T cells (Fig. 3 F–I). We first examined the thymocyte populations within the CD5-iKO. While the ablation of CD5 will impact the repertoire that was selected, we wanted to evaluate whether the loss of CD5 during the selection process impacted IκBα expression accordingly. Within the thymocyte populations, loss of IκBα expression is most significant within the DP CD69hi and SP CD4 cells, unlike the CD5-KO where all populations after positive selection had significant loss of IκBα expression (Fig. 3 F and G). To address the impact of T cell repertoire, we next examined the peripheral population of T cells in the CD5-iKO mice. Although the ablation of CD5 will impact the recent thymic emigrants within the periphery, this comprises less than 25% of the total peripheral population within the window of analysis and therefore should not significantly impact the repertoire selection and the analysis of IκBα expression (53). We found that the loss of IκBα expression is significant in the CD44hi CD4 T cells, but not within the CD44lo CD4 T cells (Fig. 3 H and I). Although CD8 T cells also trended similarly, the levels of IκBα in the WT versus CD5-iKO in these cells were not statistically significant at the time frame of CD5 ablation or with the number of mice used here. Taken together, these data suggest that CD5 necessarily and continuously up-regulates IκBα expression both during TCR-dependent developmental steps and after antigen encounter in the periphery.
Regulation of IκBα by CD5 Involves a Saturable Signaling Mechanism Independent of SHP-1.
We next investigated the molecular mechanisms that maintain the regulatory nexus between CD5 and IκBα. Although recent reports hint at “noncanonical” roles for CD5 (45), it is typically thought to act by negatively regulating the TCR-proximal signaling pathway. The intracellular tail of CD5 recruits negative regulators, most prominently the phosphatase SHP-1 to TCR signaling complexes (34). We therefore examined the levels of CD5 and IκBα in SHP-1 knockout (SHP-1–KO) mice (Fig. 4 A–C). Intriguingly, both by Western blot analysis and by intracellular staining, the proportional relationship between CD5 and IκBα expression was maintained despite an increase in the overall expression of both CD5 and IκBα in SHP-1–KO T cells (Fig. 4C). A second molecule involved in CD5 signaling is casein kinase II (CKII). CKII binds to the cytoplasmic tail of CD5 influences T cell function and differentiation and has also been shown to modulate IκBα activation (40, 54, 55). Treatment with a well-characterized CKII inhibitor CX4945 (56, 57) also did not affect the levels of CD5 or IκBα in peripheral T cells in a dose-dependent manner (Fig. 4D and SI Appendix, Fig. S4). To address whether further synthetic increases in CD5 levels can also increase IκBα expression, we utilized a CD5 transgenic mouse model (13). In these mice CD5 levels in peripheral T cells increase by less than twofold (1.52-fold increase in CD4 T cells; 1.39-fold increase in CD8 T cells) (Fig. 4 E and F, Upper). Accordingly, a subtle shift in IκBα (gMFI of 4,734 ± 867 in CD5-Tg and 3,412 ± 452 in WT) in CD4 T cells, was also observed but did not approach significance (Fig. 4 E and F, Lower). Importantly, although CD5 overexpression in the transgenic mice did increase, this was relatively small compared to the normal expression range. For example, the gMFI of CD5 in peripheral T cells range from 1,060 to 32,150 in CD4 T cells and 1,211 to 14,873 in CD8 T cells, while in CD5-Tg mice this was 4,193 to 37,951 and 3,202 to 19,213, respectively. Intriguingly, on further examination of the data, we observed that the maximum value of IκBα expression between WT and CD5-Tg mice was similar (Fig. 4E, dotted line) but, the average of IκBα expression trended higher in CD5-Tg cells compared to WT cells (Fig. 4F).
To further elaborate this, we examined the frequency of the population of T cells in seven bins based on equivalent CD5 levels in both WT and CD5-Tg mice (SI Appendix, Fig. S5). Plotting of the frequency of T cells in each bin clearly demonstrates slight skewing, with significantly less of the population of CD5-Tg T cells in the lower bins (Fig. 4G, bin 4) and increased populations in the higher bins (Fig. 4G, bin 6). This suggests that within resting (steady-state, nonantigen-activated) T cells, the levels of IκBα are limited to a point of saturation despite the enforced, marginal increase in CD5 expression. Importantly, this saturation of IκBα expression is overcome during T cell activation by antigen; that is, the physiological increase of CD5 in such cases leads to a corresponding and further increase in IκBα (Fig. 4H) beyond the saturating limit found in resting T cells. This reveals another level of regulation for CD5-mediated control of IκBα in peripheral T cells.
Given the difficulty in effectively separating the roles of tonic TCR signals and CD5 toward the regulation of IκBα levels, we used a cell culture model, using the T cell (lymphoma) line BW-5147 that lacks TCR expression and expresses relatively low levels of CD5 natively. Retroviral transduction for overexpression of CD5 in these cells also resulted in a proportional up-regulation of IκBα (Fig. 4 I and J). Interestingly, in a control group using the fibroblastic cell line NIH 3T3, we did not find any consequence to IκBα levels even though high levels of CD5 is expressed (Fig. 4K). Apart from demonstrating a necessary and sufficient role for CD5 in the control of lymphocytic IκBα, this also suggests the involvement of lineage-specific interactors to mediate the proportional relationship between CD5 and IκBα.
CD5hi T Cells Retain a Larger Reserve of the NF-κB TF.
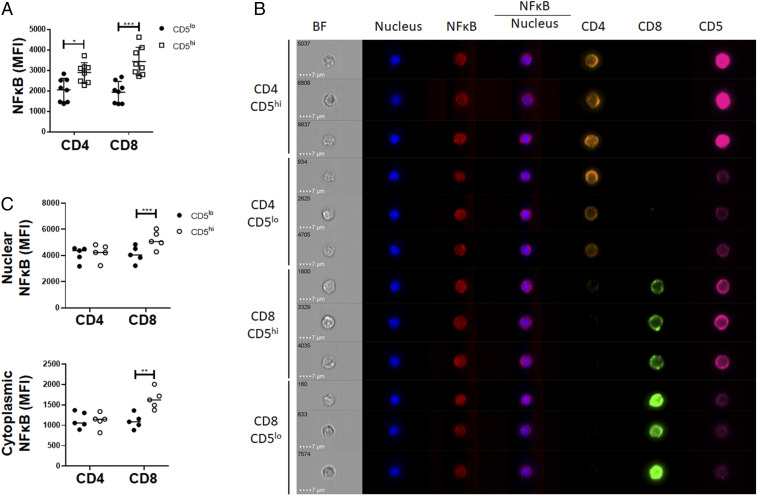
Although IκBα is well characterized as a cytoplasmic protein that binds components of the NF-κB TF that prevents them from triggering gene expression in the nucleus, the expression of IκBα is also regulated by NF-κB itself (58, 59). Therefore, it is not trivial to discern if the enhanced IκBα expression would lead to lower expression of NF-κB target genes or conversely it simply reflects a greater amount of NF-κB activity within the cell. We found that CD5hi cells have an increase in expression of p65 compared to CD5lo cells in both CD4 and CD8 T cells and, as discussed previously, CD8 T cells have higher expression of NF-κB p65 than CD4 T cells (Figs. 1H and 5A). To resolve whether this implies higher NF-κB activity, we used localization of NF-κB p65 as a surrogate marker, since active NF-κB is translocated to the nucleus. Given the low amounts of cytoplasm relative to the nucleus in resting T cells, analysis of this distribution from primary cells ex vivo tended to demonstrate greater NF-κB in CD5hi T cells but analysis was not statistically significant over multiple experiments (SI Appendix, Fig. S6A).
Fig. 5.
The CD5hi and CD5lo fractions of peripheral T cells show differential pool of cytoplasmic NF-κB. (A) gMFI of NF-κB in peripheral CD4 and CD8 T cells isolated from lymph nodes (n = 8 from 3 independent experiments). (B) Purified WT T cells were stained and analyzed by imaging flow cytometry. Screenshot of dashboard from IDEAS analysis from data collected on Amnis ImageStream. (C) Mean NF-κB expressed as a ratio of CD5hi to CD5lo within CD4 and CD8 peripheral resting T cells (n = 5 from 1 independent experiment). *P < 0.05; **P < 0.01; ***P < 0.001; anything not marked is not statistically significant.
We then quantitated the amounts of NF-κB in steady-state T cells using imaging flow cytometry (Fig. 5). This allows us to measure, albeit with the challenges of quantitating a small cytoplasmic volume to a larger nuclear one, the distribution of NF-κB p65, in both cellular components in individual CD5hi versus CD5lo T cells (Fig. 5B). As expected from the data discussed so far, overall CD5hi T cells had more NF-κB in both compartments relative to CD5lo T cells (SI Appendix, Fig. S6B). Yet, most clearly in the CD8 T cell population, we observed a larger fraction of the NF-κB was retained in the cytoplasm relative to the nucleus of the CD5hi T cell (Fig. 5C).
Combining with the previous literature on the dynamic nature of the pool of NF-κB factors in T cells (58), we suggest that the stabilization of IκBα by CD5 allows a greater cellular reserve of NF-κB to be built up in CD5hi T cells. This is maintained cytoplasmically, (at least in CD8 T cells) providing an advantageous depot of NF-κB TFs, ready to be translocated to the nucleus upon activation.
The NF-κB Reserve in CD5hi T Cells Promotes Enhanced Steady-State Survival.
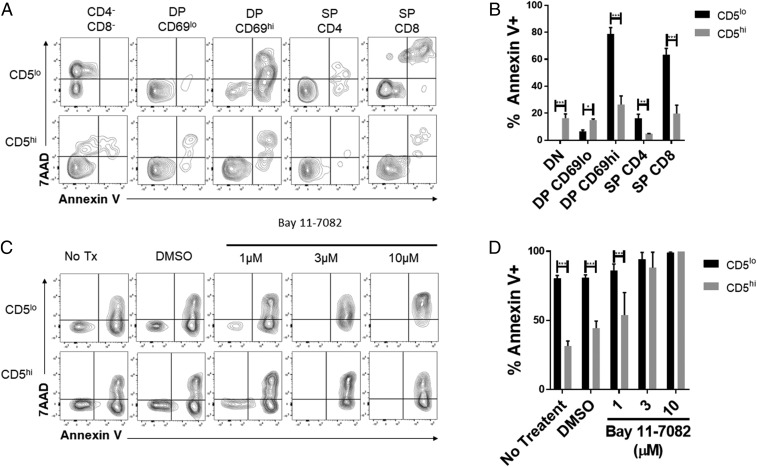
We then evaluated if this poising of NF-κB proteins in the cells has functional consequences for CD5hi versus CD5lo T cells. NF-κB transcription factors are involved in multiple cellular functions, including the survival of thymocytes and T cells both under homeostatic conditions or after activation by antigen receptors. We examined the survival of CD5hi and CD5lo thymocytes using a simple ex vivo culture assay (mimicking trophic factor withdrawal-induced apoptosis). After 24 h of culture without additional cytokines, we found that the expression of CD5 had a significant impact on cell death (Fig. 6 A and B). CD5hi populations have lower proportions of Annexin V+ cells in the developmental stages after positive selection, including DP CD69hi, SP CD4, and SP CD8. This suggests that the up-regulation of CD5 after TCR stimulation, and by extension IκBα, during positive selection provides a survival advantage, potentially through NF-κB signaling.
Fig. 6.
CD5 expression confers thymocyte survival advantage. (A) Thymocytes from B6 mice were cultured ex vivo for 24 h. Cells were stained and gated as in SI Appendix, Fig. S2A, together with Annexin V and 7AAD to assess cell death. The populations were grouped based on top and bottom 20% of CD5 expression within each population. Representative flow plots are shown. (B) The frequencies of Annexin V+ cells within the top and bottom 20% of CD5 expressers are shown (n = 4 from 1 experiment is shown and n = 3 from a second independent experiment). (C) Thymocytes from B6 mice were cultured ex vivo for 15 h with or without varying Bay 11-7082 doses. Cells were stained and gated as in SI Appendix, Fig. S2, together with Annexin V and 7AAD to assess cell death. Representative flow plots are shown for the double-positive CD69hi population. (D) The frequencies of Annexin V+ cells within the top and bottom 20% of CD5 expressers of the double-positive CD69hi population are shown (n = 3 from 1 independent experiment). *P < 0.05; **P < 0.01; ***P < 0.001; anything not marked is not statistically significant.
We then investigated if NF-κB signaling was required for this survival effect. We cultured thymocytes ex vivo in the presence or absence of an NF-κB inhibitor, Bay 11-7082, at varying concentrations. After 15 h, cell death was analyzed. We found that with increasing doses of Bay 11-7082 the significant differences in death between the CD5hi and CD5lo in postselection DP CD69hi, SP CD4, and SP CD8 populations is abrogated (Fig. 6 C and D and SI Appendix, Fig. S7). This suggests that the survival advantage of CD5hi cells is mediated through NF-κB signaling.
Discussion
CD5 plays a critical role as a rheostat for TCR signaling in T cells, but the molecular mechanisms by which it regulates multiple aspects of T cell biology are not completely understood. This is especially true in the context of responses to pathogens, where unexpectedly, T cells with higher levels of CD5 (a negative regulator) survive better and form a larger memory compartment than their CD5lo companions. In this report, we identify a molecular mechanism that can help further clarify CD5 function. We found that CD5 regulates the levels IκBα, starting in the thymus and throughout various stages of T cell development and maturation. The increased expression of IκBα allows for an accumulation of NF-κB p65, potentially helping to “jumpstart” the NF-κB pathways during T cell activation by antigen. The ease of triggering robust NF-κB–driven gene expression in these cells can also help explain the enhanced survival of the cells previously observed during peripheral immune responses. Indeed, in our experiments, inhibiting NF-κB abrogated any survival differences between CD5hi and CD5lo thymocytes (Fig. 6). Overall, these data suggest an intriguing model for the duality of CD5 function in peripheral T cells (i.e., the role as a negative regulator, which also imbues T cells with better reactivity to agonist stimulation). The enhanced expression of IκBα confers a basal inhibitory component to the CD5hi T cells. However, these T cells also express a higher level of NF-κB p65, potentially restrained in the cytoplasm (at least in the case of CD8 T cells). This “poised” state of the NF-κB pathway in CD5hi T cells may also confer a faster activation of NF-κB–dependent gene activation when it receives strong agonistic TCR signals to facilitate IκBα degradation. Thus, CD5 dependent differences in depots of NF-κB may account for heterogeneity and intrinsic biases in subsequent antigen-responses by peripheral T cells.
The identification of CD5 as a rheostat for NF-κB function can also help resolve many biological paradoxes in T cell development and activation. CD5 has been implicated in multiple different signaling activities in peripheral T cells. CD5hi cells have been shown to have reduced calcium fluxes after stimulation via CD3 cross-linking (consistent with its role as an inhibitory molecule) yet generate faster IL-2 secretion after anti-CD3/CD28 stimulation (24). Interestingly, there is less IFN-γ secretion in CD5hi cells after 5 d of TCR stimulation, but the early kinetic differences have yet to be explored (60). While biochemical data support a TCR proximal effect of CD5 (e.g., on TCRζ phosphorylation), it has also been shown to affect phosphorylation of ERK after phorbol 12-myristate 13-acetate (PMA) stimulation (28), which does not typically require TCRζ phosphorylation. As discussed above, the duality inherent in the enhanced survival of CD5hi T cells in the context of an infectious challenge is also not intuitive, given its negative regulatory role. One explanation, consistent with current literature, could be that CD5 dampens TCR signals (via negative regulators, such as SHP-1) to limit the terminal effector differentiation of CD5hi T cells (61, 62). While this mechanism has also not been experimentally evaluated, it could reasonably explain the duality of a negative regulator promoting memory responses. Indeed, in a recent work from Sood et al. (60) steady-state CD5lo T cells seem better poised to produce more effector cytokines. Nevertheless, the basic idea of a weaker antigenic signal driving this bias is not consistent with other reports: For example, of SHP-1–KO T cells generating better memory T cells (63). In fact, the latter observation is also consistent with the propensity of CD5hi T cells to better contribute to the memory pool, since we found that SHP-1–KO mice have a higher level of CD5 (Fig. 4C). The causal chain between SHP-1 and CD5, in this case, will have to be explored in future experiments with double-KO mice.
Indeed, our data suggest that independent of SHP-1, some of these disparate phenotypes stem from the differential signaling potential of the NF-κB pathway in CD5hi compared to CD5lo cells. Since NF-κB is involved in regulating the expression of multiple genes in T cells, some of the other altered functions in CD5hi T cells discussed above could very well stem from the gene products of NF-κB signaling that are triggered either at basal state or more rapidly following T cell activation. The NF-κB pathway can directly account for some of the responses, such as the increased early IL-2 production and increased survival in CD5hi T cells. Future studies will need to parse out the direct role of CD5 signaling versus indirect effects due to differential expression of NF-κB target genes. Taking the data together, we find that the involvement of an NF-κB reserve (expected to augment T cell activation) using an inhibitor (IκBα) can be a helpful concept in resolving these questions.
In resting or steady-state T cells, although IκBα follows a striking correlation with CD5 (Fig. 1) in WT mice, artificially increasing CD5 through transgenic overexpression in the CD5-Tg mice cannot increase IκBα beyond the maximum value that WT T cells normally have (Fig. 4E). The simplest explanation for this is that CD5 primarily regulates the posttranslational levels of IκBα, whereas antigen activation by TCR signals allows a transcriptional up-regulation of IκBα (64). Indeed, this is validated by the further increase in IκBα in antigen-activated T cells (Fig. 4H). Yet, it is clear from experiments on BW-5147 cells (Fig. 4J) that the ability of CD5 to enhance IκBα levels does not require parallel TCR signaling. In other words, if a lymphocyte expresses sufficient mRNA for IκBα then CD5 is necessary (based on the knockouts in Fig. 3) and sufficient (based on overexpression in Fig. 4) to posttranslationally increase the intracellular levels of the protein, but it cannot increase the message levels by itself. Indeed, these data can also be used to discriminate between another potential mechanism. Since NF-κB directly regulates IκBα expression [as a feedback process (59)], it was quite possible that CD5 is increasing IκBα via the regulation of NF-κB activity. However, this would have resulted in an increase in IκBα transcripts in the CD5hi T cells, which we did not observe (Fig. 1L).
It is not yet clear if additional increases in IκBα mediated by a further transcriptional up-regulation during TCR signaling would serve a positive or negative role in T cell activation. While the basal levels of IκBα can help maintain an NF-κB depot, the further increase (driven by TCR signals rather than CD5) may in fact serve a feedback inhibitory role as it has been commonly studied. Furthermore, antigen-activated T cells, which may rest down from the acute TCR exposure, set IκBα levels higher than naïve T cells (Fig. 2) in a CD5-dependent fashion (Fig. 3). This is critical because the protein levels of NF-κB members are known to require posttranslational control of their stoichiometry (58, 59). CD5 could be the mechanism to fix the gain in IκBα expression by the TCR signals; stabilizing a larger NF-κB pool in resting-memory phenotype T cells. Whether this increased NF-κB depot in memory T cells could drive some of the rapid responses associated with memory function is a significant question that remains to be studied. It is also important to note that NF-κB does not necessarily mean positive or negative responses, but rather relies on the unique chromatin landscape in a cell. Therefore, in cells like Tregs the effect of an NF-κB depot could be quite contrary to that in memory T cells, depending on the genes transcribed, given the varied accessibility of genes between these two populations. Linking CD5 to the regulation of the dynamics of NF-κB in resting T cells therefore allows us to develop a more complete picture of the intracellular signaling networks controlling T cell fate.
Materials and Methods
Cell Culture.
BW-5147 (α−β−) cells originally generated by White et al. (65) were a gift from Philippa Marrack, National Jewish Health, Denver, CO, to B. J. Fowlkes, National Institute of Allergy and Infectious Diseases (NIAID), NIH, Bethesda, MD, and maintained as described by White et al. NIH 3T3 cells were obtained from ATCC. Cells were maintained in RPMI 1640 (Gibco) supplemented with 10% FCS (Gemini), 2% glutamine (Gibco), 1% sodium pyruvate (Gibco), and 1% antibiotic/antimycotic (Gibco). Phoenix-GP cells were obtained from ATCC and maintained in DMEM (Gibco) supplemented with 10% FCS (Gemini), 2% glutamine (Gibco), 1% sodium pyruvate (Gibco), and 1% antibiotic/antimycotic (Gibco). Primary murine thymocytes and T cells were maintained in RPMI-1640 (Gibco) supplemented with 10% FCS (Gemini), 1% glutamine (Gibco), 1% antibiotic-antimycotic (Gibco), and 0.000014% β-mercaptoethanol.
Mice.
WT mice were obtained from both Jackson Laboratories (C57BL/6J and B6.SJL-PtprcaPepcb/BoyJ) and Taconic Biosciences (B6NTac and B6.SJL-Ptprca/BoyAiTac). B6 mice were bred for dual congenic expression using a dam or sire from opposing place of purchase to eliminate any strain drift between the two WT strains. WT B10.Q/Ai mice were originally obtained from Ron Schwartz, Institute of Allergy and Infectious Diseases, NIH, Bethesda, MD. 5C.C7 TCR transgenic mice were obtained from Taconic Biosciences [B10.A-Rag2tm1FwaH2-T18aTg (Tcra5CC7,Tcrb5CC7)lwep]. SMARTA TCR transgenic mice were obtained from the Jackson Laboratories [B6.Cg-Ptprca Pepcb Tg(TcrLCMV)1Aox/PpmJ]. TCR-αβ KO mice were obtained from the Jackson Laboratories (B6.129S2-Tcratm1Mom/J). THEMIS knock out animals were generated as described previously (18). SHP-1–KO, CD5 KO, and CD5-Tg animals were generated as described previously (13, 66) and maintained at the Animal facilities at the National Institute of Child Health and Human Development, NIH. CD5 inducible KO animals were generated by microinjection of constructs shown in SI Appendix, Fig. S3 into C57SBL/6 embryonic stem cells and implantation into superovulated females. Animals were bred and maintained in modified specific pathogen-free facilities at the University of Maryland Baltimore or at the National Institute of Child Health and Human Development, NIH. Experiments were performed with animals at least 6 wk of age and approved by University of Maryland Baltimore Institutional Animal Care and Use Committee or by the Animal Care and Use Committee of the National Institute of Child Health and Human Development.
T Cell Purification.
Lymph node and spleen were isolated from mice and mechanically processed into single-cell suspensions by mashing tissues through a 100-μM nylon mesh. The cell suspensions were then processed through a Ficoll-HiPaque Density Gradient to isolate lymphocytes. Magnetic depletion was performed with MyOne Streptavidin T1 DynaBeads with biotinylated anti-CD45R/B220 (RA3-6B2, Biolegend #103204), biotinylated anti-NK1.1 (PK136, Biolegend #108704), biotinylated anti-I-A/I-E (M5/114, Biolegend #107604), and biotinylated anti-CD11b (M1/70, Biolegend #101204) for isolation of CD4 and CD8 T cells according to the manufacturer’s instructions.
Flow Cytometry Staining.
Fc-receptor blocking was performed for 15 min at 4 °C in 1× PBS supplemented with 2% FCS, 0.01% azide, 1% mouse serum, 1% hamster serum, and 1% rat serum (FcBlock). Surface staining was performed for 30 min at 4 °C in PBS supplemented with 2% FCS, 0.01% azide, and 0.02% EDTA (FACS Buffer) using the antibodies provided in SI Appendix, Table S1. Cells were washed once with FACS Buffer. Intracellular staining was performed with eBioscience Fixation/Permeabilization Kit. Cells were resuspended in 1× eBioscience Fixation/Permeabilization buffer and incubated overnight at 4 °C. Intracellular staining was performed overnight at 4 °C in 1× eBioscience Permeabilization Buffer using the antibodies listed in SI Appendix, Table S1. Stained cells were washed twice with FACS buffer. Cells were analyzed on an LSR-Fortessa flow cytometer (BD). All data were analyzed using FlowJo (Tree Star).
Flow-Assisted Cell Sorting.
Magnetically depleted T cells were labeled with the antibodies listed in SI Appendix, Table S1 and 7AAD in PBS supplemented with 2% FCS and 1% antibiotic-antimycotic (Gibco). Cells were sorted on a BD FACSAria II cytometer into the following groups: 1) 7AAD−CD44loCD4+CD5hi, 2) 7AAD−CD44loCD4+CD5lo, 3) 7AAD−CD44loCD8+CD5hi, or 4) 7AAD−CD44loCD8+CD5lo.
Adoptive Transfer and Antigen Challenge.
Lymph nodes were isolated from 5C.C7 TCR-tg and mechanically dissociated by mashing tissues through a 100-μM nylon mesh. Next, 100,000 cells were transferred into B10.A mice and 24 h later were challenged with moth cytochrome C (MCC) and LPS or PBS as a control. Spleen and lymph node tissue were isolated at varying time points (12, 60, and 84 h postchallenge) and analyzed by flow cytometry.
Imaging Flow Cytometry Analysis.
Cells were stained with antibodies to CD4 (BV605), CD8 (BB515), CD5 (PE-Cy7), and NF-κB (AF647). The nucleus was stained with DAPI. Cells were acquired on an ImageStream MarkII imaging flow analyzer (Luminex) equipped with two CCD cameras, at 60× magnification and low speed (7-mm core stream). Analysis was performed using the IDEAS software package (Luminex). After spectral compensation, in-focus, single cells were identified on the first camera bright-field images and were further subdivided using 2D plots of CD4 and CD8 followed by CD5 histogram plots. The upper and lower 20% of CD5-expressing subsets of CD4+ and CD8+ cells were analyzed for NF-κB nuclear translocation and for nuclear NF-κB fluorescence intensity measurements. Nuclear translocation (Fig. 5C and SI Appendix, Fig. S6A) was determined by masking the nucleus based on DAPI staining and expressed as the log-transformed Pearson’s correlation coefficient between the nuclear (DAPI)- and N-FκB (AF647) -masked images. The amount of nuclear NF-κB (Fig. 5C) was expressed as the gMFI of AF647 staining signal within the DAPI-masked nuclear image.
Western Blot Analysis.
Sorted cells as described above were lysed in 1% Nonidet P-40, 10 mM Tris⋅HCl (pH 7.2), 140 mM NaCl, 2 mM EDTA, 5 mM iodoacetamide, 1 mM Na3VO4, and complete protease inhibitor mixture (Roche) and incubated for 30 min on ice. Lysates were spun at 14,000 rpm at 4 °C and clarified lysate was removed. Samples were run on an SDS/PAGE gel and transferred onto nitrocellulose membranes. Membranes were blocked with 5% skim milk at room temperature for 1 h. Membranes were incubated with primary antibodies of anti-CD5 (SC6986, SantaCruz Biotechnology), anti-IκBα (9242, Cell Signaling Technology), antiphosphor-SHP-1(Y564) (8849, Cell Signaling Technology), antiactin (A5441, Sigma Aldrich), anti-PLCγ1 (SC81, SantaCruz Biotechnology), anti-p65 NF-κB (SC372, SantaCruz Biotechnology), or anti-SP1 (SC59, SantaCruz Biotechnology), for 4 h followed by HRP-conjugated anti-mouse IgG, anti-rabbit IgG or anti-goat IgG. Membranes were developed and visualized using the ECL technique (Thermo Scientific). Band densitometry was analyzed with MultiGauge software (Fujifilm).
Cellular Fractionation.
Nuclear and cytoplasmic extracts were prepared by using Subcellular Protein Fractionation Kit (78840, Thermo Scientific).
Thymocyte Ex Vivo Culture.
Thymus tissue was isolated and mechanically dissociated by mashing tissue through a 100-μM nylon mesh. Next, 200,000 thymocytes were plated in T cell media and incubated for 24 h at 37 °C. Cells were harvested and surface-stained as described above. Cells were washed twice with 1× Annexin Binding Buffer (BD). Cells were resuspended in 1× Annexin Binding Buffer with Annexin V PE (BD) and 7AAD (BD) and incubated at 27 °C for 10 min and then analyzed by flow cytometry.
CX4945 and Bay 11-7082 Ex Vivo Culture.
Thymocytes or peripheral T cells (lymph node and spleen) were plated at a density of 100,000 cells per well in T cell media. CX4945 (Enzo) or Bay 11-7082 (Sigma-Aldrich) was resuspended in DMSO, diluted in T cell media, and then plated with thymocytes. Samples were incubated at 37 °C and harvested for flow cytometric analysis at varying time points (12, 15, 24, and 48 h).
Real-Time Quantitative PCR Analysis.
Sorted cells were resuspended in TRIzol and frozen at −80 °C. TRIzol suspension was filtered through a QIAShredder Mini Column (Qiagen). Chloroform was applied to filtrate and RNA layer was removed. RNA was further isolated using RNEasy MinElute Kit (Qiagen) according to the manufacturer’s instructions. Total RNA was transcribed using SuperScript III reverse transcriptase and oligo(dT)20 primers (Invitrogen). The expression of the mRNA for Nfkbia, Nfkb1, Nfkb2, and Actb were determined by real-time PCR using the following primers. Gene-specific primer sets used in the real-time PCR assays were: Nfkbia 5′-GGAGACTCGTTCCTGCACTT-3′ and 5′-TCTCGGAGCTCAGGATCACA-3′; Nfkb1 5′-GAAATTCCTGATCCAGACAAAAAC-3′ and 5′-ATCACTTCAATGGCCTCTGTGTAG-3′; Nfkb2 5′-CTGGTGGACACATACAGGAAGAC-3′ and 5′-ATAGGCACTGTCTTCTTTCACCTC-3′; Actb 5′-GGAGCACCCTGTGCTGCTCACCGAGG-3′ and 5′-ATCTACGAGGGCTATGCTCTCCC-3′. The expression of genes was normalized to Actb.
In Vitro Overexpression of CD5.
CD5 cDNA (amplified from Origene, Catalog #MR207937) was cloned into MIGR1 retroviral expression vector (submitted to Addgene, Catalog #27490) after PCR amplification (primers 5′-CGCCGGAATTAGATCCCATTATCCATGGACTCCCACGAAGT-3′ and 5′-GGGGGGGGCGGAATTTTACAGTCTCTGAGCCACTTGCAG-3′) and in-fusion reaction. CD5 retroviral constructs and empty vectors were used to transfect Phoenix-GP cells, retroviral supernatants collected, concentrated, and used to transduce BW-5147 TCR− cells by spinoculation.
RNA-Seq and Data Analysis.
Total lymph node cells from C57BL/6 mice were stained with fluorochrome-conjugated antibodies against CD4, CD8, CD5, CD25, CD62L, and CD44. Sorted populations consisted of the one-third of CD25−CD62Lhi CD44lo CD4+ or CD8+ T cells expressing the highest surface levels of CD5 (CD5hi) or the one-third of CD25−CD62Lhi CD44lo CD4+ or CD8+ T cells expressing the lowest surface levels of CD5(CD5lo). RNA was extracted from FACS-sorted T cells using TRIzol reagent (Invitrogen) followed by purification and DNase treatment with RNeasy columns (Qiagen) and submitted for library construction by the TruSeq RNA sample preparation kit v2 or TruSeq stranded mRNA sample preparation kit (Illumina). Sequencing was done on an Illumina HiSEq. 2500 sequencer, yielding 30 to 70 million reads of 101 bp. The raw reads were aligned using STAR v2.5.2a (67) and reads quantified with featureCounts v1.4.6-p3 (68). Differential expression analysis was performed using DESeq2 (69). We used a false-discovery rate cutoff of <1% to define statistical significance. RNA-seq data are will be made publicly available at the time of publication.
Data Availability.
All protocols and reagents are detailed in the main text. Plasmid constructs for CD5 overexpression are available via Addgene (https://www.addgene.org/Nevil_Singh/). RNA-seq data are available in the Gene Expression Omnibus (accession GSE151395).
Supplementary Material
Acknowledgments
We thank the members of the N.J.S. laboratory, Allison Gerber, Kenneth Rosenberg, and Gideon Wolf, for extended discussions and critiques during the development of this work; and the University of Maryland Greenebaum Comprehensive Cancer Center Flow Cytometry Core and Dr. Xiaoxuan Fan for FACS Sorting and support. This work was supported by Grant R01 AI110719 (to N.J.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Plasmid constructs for CD5 overexpression are available via Addgene (https://www.addgene.org/Nevil_Singh/). RNA-seq data are available in the Gene Expression Omnibus (accession GSE151395).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922525117/-/DCSupplemental.
References
- 1.Singh N. J., Schwartz R. H., Primer: Mechanisms of immunologic tolerance. Nat. Clin. Pract. Rheumatol. 2, 44–52 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Pobezinsky L. A. et al., Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nat. Immunol. 13, 569–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu W. et al., Clonal deletion prunes but does not eliminate self-specific αβ CD8(+) T lymphocytes. Immunity 42, 929–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano M., Toda M., Sakaguchi N., Sakaguchi S., Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J. Exp. Med. 184, 387–396 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Love P. E., Bhandoola A., Signal integration and crosstalk during thymocyte migration and emigration. Nat. Rev. Immunol. 11, 469–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein L., Kyewski B., Allen P. M., Hogquist K. A., Positive and negative selection of the T cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 14, 377–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogquist K. A., Gavin M. A., Bevan M. J., Positive selection of CD8+ T cells induced by major histocompatibility complex binding peptides in fetal thymic organ culture. J. Exp. Med. 177, 1469–1473 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebert P. J., Jiang S., Xie J., Li Q. J., Davis M. M., An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat. Immunol. 10, 1162–1169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo W.-L. et al., An endogenous peptide positively selects and augments the activation and survival of peripheral CD4+ T cells. Nat. Immunol. 10, 1155–1161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juang J. et al., Peptide-MHC heterodimers show that thymic positive selection requires a more restricted set of self-peptides than negative selection. J. Exp. Med. 207, 1223–1234 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris G. P., Allen P. M., How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat. Immunol. 13, 121–128 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Davis M. M. et al., T cells as a self-referential, sensory organ. Annu. Rev. Immunol. 25, 681–695 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Azzam H. S. et al., Fine tuning of TCR signaling by CD5. J. Immunol. 166, 5464–5472 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Grossman Z., Paul W. E., Autoreactivity, dynamic tuning and selectivity. Curr. Opin. Immunol. 13, 687–698 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Azzam H. S. et al., CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188, 2301–2311 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q. J. et al., miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129, 147–161 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Lo W. L., Donermeyer D. L., Allen P. M., A voltage-gated sodium channel is essential for the positive selection of CD4(+) T cells. Nat. Immunol. 13, 880–887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lesourne R. et al., Themis, a T cell-specific protein important for late thymocyte development. Nat. Immunol. 10, 840–847 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu G. et al., Themis sets the signal threshold for positive and negative selection in T-cell development. Nature 504, 441–445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wülfing C. et al., Costimulation and endogenous MHC ligands contribute to T cell recognition. Nat. Immunol. 3, 42–47 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Chu H. H. et al., Positive selection optimizes the number and function of MHCII-restricted CD4+ T cell clones in the naive polyclonal repertoire. Proc. Natl. Acad. Sci. U.S.A. 106, 11241–11245 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulton R. B. et al., The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nat. Immunol. 16, 107–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandl J. N., Monteiro J. P., Vrisekoop N., Germain R. N., T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity 38, 263–274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinzow-Kramer W. M., Weiss A., Au-Yeung B. B., Adaptation by naïve CD4+ T cells to self-antigen-dependent TCR signaling induces functional heterogeneity and tolerance. Proc. Natl. Acad. Sci. U.S.A. 116, 15160–15169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogquist K. A., Jameson S. C., The self-obsession of T cells: How TCR signaling thresholds affect fate “decisions” and effector function. Nat. Immunol. 15, 815–823 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquez M. E. et al., CD8 T cell sensory adaptation dependent on TCR avidity for self-antigens. J. Immunol. 175, 7388–7397 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Stamou P. et al., Chronic exposure to low levels of antigen in the periphery causes reversible functional impairment correlating with changes in CD5 levels in monoclonal CD8 T cells. J. Immunol. 171, 1278–1284 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Persaud S. P., Parker C. R., Lo W. L., Weber K. S., Allen P. M., Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat. Immunol. 15, 266–274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raman C., CD5, an important regulator of lymphocyte selection and immune tolerance. Immunol. Res. 26, 255–263 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Bhandoola A. et al., CD5-mediated inhibition of TCR signaling during intrathymic selection and development does not require the CD5 extracellular domain. Eur. J. Immunol. 32, 1811–1817 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Voisinne G., Gonzalez de Peredo A., Roncagalli R., CD5, an undercover regulator of TCR signaling. Front. Immunol. 9, 2900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raman C., Kimberly R. P., Differential CD5-dependent regulation of CD5-associated CK2 activity in mature and immature T cells: Implication on TCR/CD3-mediated activation. J. Immunol. 161, 5817–5820 (1998). [PubMed] [Google Scholar]
- 33.Raman C., Kuo A., Deshane J., Litchfield D. W., Kimberly R. P., Regulation of casein kinase 2 by direct interaction with cell surface receptor CD5. J. Biol. Chem. 273, 19183–19189 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Perez-Villar J. J. et al., CD5 negatively regulates the T-cell antigen receptor signal transduction pathway: Involvement of SH2-containing phosphotyrosine phosphatase SHP-1. Mol. Cell. Biol. 19, 2903–2912 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gary-Gouy H. et al., The pseudo-immunoreceptor tyrosine-based activation motif of CD5 mediates its inhibitory action on B-cell receptor signaling. J. Biol. Chem. 275, 548–556 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Ochi H., Watanabe T., Negative regulation of B cell receptor-mediated signaling in B-1 cells through CD5 and Ly49 co-receptors via Lyn kinase activity. Int. Immunol. 12, 1417–1423 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Thien C. B. et al., Loss of c-Cbl RING finger function results in high-intensity TCR signaling and thymic deletion. EMBO J. 24, 3807–3819 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demydenko D., c-Cbl mediated ubiquitylation and regulation of cell surface exposure of CD5. Biochem. Biophys. Res. Commun. 392, 500–504 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Bamberger M. et al., A new pathway of CD5 glycoprotein-mediated T cell inhibition dependent on inhibitory phosphorylation of Fyn kinase. J. Biol. Chem. 286, 30324–30336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sestero C. M. et al., CD5-dependent CK2 activation pathway regulates threshold for T cell anergy. J. Immunol. 189, 2918–2930 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong B. et al., CD5-mediated inhibition of TCR signaling proceeds normally in the absence of SHP-1. Int. J. Mol. Med. 38, 45–56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voisinne G. et al., Co-recruitment analysis of the CBL and CBLB signalosomes in primary T cells identifies CD5 as a key regulator of TCR-induced ubiquitylation. Mol. Syst. Biol. 12, 876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peña-Rossi C. et al., Negative regulation of CD4 lineage development and responses by CD5. J. Immunol. 163, 6494–6501 (1999). [PubMed] [Google Scholar]
- 44.Chan S., Waltzinger C., Tarakhovsky A., Benoist C., Mathis D., An influence of CD5 on the selection of CD4-lineage T cells. Eur. J. Immunol. 29, 2916–2922 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Zhang C. et al., CD5 binds to interleukin-6 and induces a feed-forward loop with the transcription factor STAT3 in B cells to promote cancer. Immunity 44, 913–923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henderson J. G., Opejin A., Jones A., Gross C., Hawiger D., CD5 instructs extrathymic regulatory T cell development in response to self and tolerizing antigens. Immunity 42, 471–483 (2015). [DOI] [PubMed] [Google Scholar]
- 47.McGuire D. J. et al., CD5 enhances Th17-cell differentiation by regulating IFN-γ response and RORγt localization. Eur. J. Immunol. 44, 1137–1142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fulton R. B. et al., The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nat. Immunol. 16, 107–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashouri J. F., Weiss A., Endogenous Nur77 is a specific indicator of antigen receptor signaling in human T and B cells. J. Immunol. 198, 657–668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delpoux A. et al., TCR signaling events are required for maintaining CD4 regulatory T cell numbers and suppressive capacities in the periphery. J. Immunol. 193, 5914–5923 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Sprouse M. L. et al., High self-reactivity drives T-bet and potentiates Treg function in tissue-specific autoimmunity. JCI Insight 3, 97322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freitas C. M. T., Hamblin G. J., Raymond C. M., Weber K. S., Naïve helper T cells with high CD5 expression have increased calcium signaling. PLoS One 12, e0178799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Houston E. G. Jr., Higdon L. E., Fink P. J., Recent thymic emigrants are preferentially incorporated only into the depleted T-cell pool. Proc. Natl. Acad. Sci. U.S.A. 108, 5366–5371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mier-Aguilar C. A., Cashman K. S., Raman C., Soldevila G., CD5-CK2 signaling modulates Erk activation and thymocyte survival. PLoS One 11, e0168155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chu Z. L., McKinsey T. A., Liu L., Qi X., Ballard D. W., Basal phosphorylation of the PEST domain in the I(kappa)B(beta) regulates its functional interaction with the c-rel proto-oncogene product. Mol. Cell. Biol. 16, 5974–5984 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibson S. A. et al., Protein kinase CK2 controls the fate between Th17 cell and regulatory T cell differentiation. J. Immunol. 198, 4244–4254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jang S. W. et al., Casein kinase 2 is a critical determinant of the balance of Th17 and Treg cell differentiation. Exp. Mol. Med. 49, e375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scott M. L., Fujita T., Liou H. C., Nolan G. P., Baltimore D., The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 7, 1266–1276 (1993). [DOI] [PubMed] [Google Scholar]
- 59.Sun S. C., Ganchi P. A., Ballard D. W., Greene W. C., NF-kappa B controls expression of inhibitor I kappa B alpha: Evidence for an inducible autoregulatory pathway. Science 259, 1912–1915 (1993). [DOI] [PubMed] [Google Scholar]
- 60.Sood A. et al., Differential interferon-gamma production potential among naïve CD4+ T cells exists prior to antigen encounter. Immunol. Cell Biol. 97, 931–940 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Chandok M. R., Farber D. L., Signaling control of memory T cell generation and function. Semin. Immunol. 16, 285–293 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Kaech S. M., Cui W., Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snook J. P., Kim C., Williams M. A., TCR signal strength controls the differentiation of CD4+ effector and memory T cells. Sci. Immunol. 3, eaas9103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oh H., Ghosh S., NF-κB: Roles and regulation in different CD4(+) T-cell subsets. Immunol. Rev. 252, 41–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White J. et al., Two better cell lines for making hybridomas expressing specific T cell receptors. J. Immunol. 143, 1822–1825 (1989). [PubMed] [Google Scholar]
- 66.Tarakhovsky A., Müller W., Rajewsky K., Lymphocyte populations and immune responses in CD5-deficient mice. Eur. J. Immunol. 24, 1678–1684 (1994). [DOI] [PubMed] [Google Scholar]
- 67.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao Y., Smyth G. K., Shi W., featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All protocols and reagents are detailed in the main text. Plasmid constructs for CD5 overexpression are available via Addgene (https://www.addgene.org/Nevil_Singh/). RNA-seq data are available in the Gene Expression Omnibus (accession GSE151395).