Abstract
Functional inhibitory peptides of human dipeptidyl peptidase 4 (hDPP4) have been highly anticipated as the active ingredient of functional food for type II diabetes; however, the molecular mechanism of hDPP4 inhibition remains unclear. In this study, we focused on dipeptides and tripeptides, which display structure-function correlations that are relatively easy to analyze, and examined their interactions with hDPP4 on an atomic level using a combination of docking studies and an hDPP4 inhibition assay. First, we performed comprehensive binding mode analysis of the dipeptide library and demonstrated that the formation of a tight interaction with the S1 subsite composing part of the substrate pocket is essential for dipeptides to compete with the substrate and strongly inhibit hDPP4. Next, we synthesized tripeptides by adding various amino acids to the C-terminus of Ile-Pro and Val-Pro, which have especially high inhibitory activity among compounds in the dipeptide library, and measured the hDPP4 inhibitory activity of the tripeptides. When hydrophobic amino acids (Ile, Met, Val, Trp) were added, the inhibitory activity increased several-fold. This phenomenon could be explained as follows: the C-terminal amino acid of the tripeptide formed hydrophobic interactions with Tyr547 and Trp629, which compose the S1′ subsite located relatively outside the substrate pocket, thereby stabilizing the hDPP4-peptide binding. The structural information on the interaction between hDPP4 and peptide inhibitors attained in this study is anticipated to be useful in the development of a more potent hDPP4 competitive inhibitor.
Keywords: Biochemistry, Bioinformatics, Biophysics, Structural biology, Computer simulation, Biophysical chemistry, Pharmaceutical chemistry, Material science of foods, Biochemical characterization of food, Computer-aided drug design, Peptides, Drug binding, Structure activity relationship, Human dipeptidyl peptidase 4, Molecular docking, Protein-compound binding mode, Type II diabetes, Dipeptide inhibitor, Tripeptide inhibitor
Biochemistry; Bioinformatics; Biophysics; Structural biology; Computer simulation; Biophysical chemistry; Pharmaceutical chemistry; Material science of foods; Biochemical characterization of food; Computer-aided drug design; Peptides; Drug binding; Structure activity relationship; Human dipeptidyl peptidase 4; Molecular docking; Protein-compound binding mode; Type II diabetes; Dipeptide inhibitor; Tripeptide inhibitor.
1. Introduction
Human dipeptidyl peptidase 4/CD26 (hDPP4) is a serine protease with high specificity and is widely expressed in various tissues such as lung, brain, pancreas, and kidney and by many different cell types. It recognizes peptides with proline or alanine as the second amino acid from the N-terminus [1]. The most significant substrates of hDPP4 are incretins such as glucagon-like peptide-1 and glucose-dependent insulinotropic polypeptide [1]. These incretins increase postprandial insulin secretion (insulin secretion activity); thus, functional inhibition of DPP4 is expected to improve blood glucose control in type II diabetic patients. DPP4 inhibitors such as sitagliptin, vildagliptin, saxagliptin, and alogliptin have been developed previously and have been used clinically since 2007 [2]. From the perspective of functional food, Atlantic salmon skin gelatin-derived DPP4-inhibiting peptide has been reported to improve blood glucose levels [3]. These findings indicate the potential of DPP4-inhibiting substances as active ingredients in medications and functional food. In order to develop a more potent DPP4-inhibiting component, it is useful to thoroughly understand the mechanism of inhibition.
With X-ray crystallography analysis, the interactions between DPP4 and small molecule inhibitors have been well studied at the atomic level. DPP4 encompasses a relatively large substrate binding pocket composed of four subsites (S1, S1′, S2, S2 extensive), and most existing inhibitors stably bind to DPP4 by fitting into multiple subsites [4]. In particular, the S1 and S2 pockets are at the core of the binding, and central scaffolds of all Class 1, 2, and 3 inhibitors bind to these pockets [4]. Moreover, additional interaction at the S2 extensive subsite contributes to improved affinity. For example, when hydrophobic interactions in this region were strengthened in teneligliptin-related compounds, the inhibitory activity increased approximately 1000-fold [5, 6].
In contrast to DPP4-small molecule inhibitor interaction, the hDPP4-peptide co-crystal structure has only been described in three reports [7, 8, 9]. Although all of these structures display salt bridges between the N-terminal amino group of the peptide and Glu205/Glu206, the binding pattern to DPP4 differs for each peptide. For example, when Pro is the second amino acid from the N-terminus, the Pro side chain binds to the S1 pocket [7, 8]; when Pro is the third amino acid from the N-terminus, the amino acid side chain of the N-terminus binds to the S1 pocket [9]. Thus, the binding pattern to DPP4 differs depending on the amino acid sequence of the peptide. Furthermore, the structural binding mechanism with regards to the interaction at the peptide C-terminus is still not well understood.
Multiple DPP4-inhibiting peptides have been found in various food protein hydrolysates by FitzGerald et al., Li-Chan et al., and other research groups [10, 11, 12, 13, 14]. The current consensus is that substrate-like peptides with Pro or Ala as the second amino acid from the N-terminus have high inhibitory effects. Our research group previously conducted a comprehensive analysis of a dipeptide library to elucidate a full picture of DPP4 inhibition [15]. We endeavored to identify the amino acid residue associated with the inhibitory activity based on the attained results. Although we observed a high inhibitory effect of dipeptides containing Trp at the N-terminus, we could not obtain an overall definitive rule. The mix of competitive inhibition and uncompetitive/non-competitive inhibition as the inhibitory mechanism of dipeptides signifies the presence of multiple dipeptide binding sites on DPP4. For this reason, we most likely could not find a definitive rule with direct interpretation of the comprehensive analysis results [16, 17].
To fully understand the DPP4-inhibiting mechanism of peptides, it is necessary to conduct a comprehensive analysis from a new perspective. In this study, we therefore focused on dipeptides and tripeptides in which the structure-function correlation can be relatively easily analyzed, and we investigated their interactions with DPP4 using in silico docking studies, which have contributed to the identification of new DPP4 inhibitors in the past [18, 19]. First, with the hypothesis that all dipeptides exhibit competitive inhibition, we constructed a calculation protocol that can reproduce the inhibitory activity of the dipeptide. A certain level of calculation precision could be achieved with this approach presumably because most of the potent DPP4-inhibiting peptides display competitive inhibition. From the DPP4-dipeptide docking poses obtained through the calculation protocol, we extracted characteristic interactions of dipeptides possessing a high inhibitory activity. Next, we analyzed tripeptides with various amino acids added to the C-terminus of dipeptides with high inhibitory activity (Ile-Pro and Val-Pro) with docking studies using the same protocol and hDPP4 inhibition assay. By integrating the results from these studies, we identified additional interactions at the S1′ subsite that contribute to the elevation in inhibitory activity. Dipeptides and tripeptides are abundant in food and are close in size as small molecule drugs. Thus, findings obtained in this study may be useful in designing molecules that are competitive inhibitors of DPP4.
2. Materials & methods
2.1. Molecular docking of DPP4 and dipeptides/tripeptides
DPP4-peptide docking studies were conducted with ensemble docking using the three-dimensional structures of DPP4 extracted from four DPP4-peptide co-crystal structures (PDBID: 2BGR, 1NU8, 2BGN, 1R9N) [20]. A series of water of crystallization was removed because its preservability at the inhibitor binding site was not observed among these co-crystal structures. Moreover, proteins, dipeptides, and tripeptides were protonated using the Protonate 3D module of Molecular Operating Environment [21] such that it was in a dominant ionization state at pH 7.0. GOLD 5.5 [22] was used as the docking program. The standard default settings for the genetic algorithm were used, and all protein atoms were fixed during molecular docking. The docking search area was the spherical area with a 10-Å radius with the center at the midpoint of E205Cδ and Y547OH. Molecular docking was performed 25 times for each compound, and binding poses with the top five scores were saved. Next, the binding stabilities of the five docking poses were calculated with Molecular Mechanics Generalized Born Surface Area (MM-GBSA) [23] to determine the docking pose with the highest binding stability. In the MM-GBSA calculation, 100-steps energy minimization in implicit solvent with the conjugate gradient algorithm was carried out for each complex structure model using the minab module of AMBER Tools [24], and then the DPP4-peptide binding energy was calculated using the MMPBSA.py module [25]. During MM-GBSA calculation process, the salt concentration was set at 150 mM.
The DPP4-peptide interaction fingerprint was calculated as follows. First, for each peptide, the docking pose on each of the four protein structures was extracted. Next, the 16 amino acids positioned 5 Å from the bound ligand of the DPP4-peptide co-crystal structure (PDBID: 1NU8) (Ile-Pro-Ile) were defined as pocket-composing residues; for each amino acid, the number of atoms in contact with the docked peptide within 5 Å was counted, and the mean between the four docking poses was calculated. The atomic contact was calculated for heavy and hydrogen atoms in DPP4 and the docked peptide.
2.2. DPP4 inhibition assay
A commercial DPP4 inhibitor screening assay kit (Item No. 700210, Cayman Chemical Company, Michigan, USA) was used to measure the ability of the designed tripeptides to inhibit DPP4 activity. Human recombinant DPP4 was incubated with 100 μM Gly-Pro-aminomethylcoumarin (AMC) at 37 °C for 30 min in the presence of each tripeptide in a buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM EDTA. Fluorescence was recorded using a microplate reader (Spectra Max i3, Molecular Devices, California, USA) at excitation and emission wavelengths of 350 nm and 450 nm, respectively. For tripeptides that exhibit high inhibition ratios of more than 50% in the presence of the test tripeptide (10 μg/ml), the IC50 was determined from measurements at three different concentrations of the tripeptide. In cases of IPN (the inhibition ratio of 31.9%), IPW (32.6%), IPY (32.6%), VPD (24.2%), VPL (31.0%), VPS (45.4%), and VPV (11.1%), the IC50 was hard to be determined because of their weak inhibitory activities.
3. Results & discussion
3.1. Docking study of DPP4-dipeptide
We previously reported the inhibitory activity of a dipeptide library in DPP4 protease activity as an inhibition ratio (%) relative to Glycyl-proline-4-methyl-coumaryl-7-amide (Gly-Pro-MCA) [15]. We therefore conducted docking studies to analyze the DPP4-dipeptide binding mode with the hypothesis that all dipeptides exhibit competitive inhibition, and searched for an intermolecular interaction that is important in inhibitory activity. First, we verified whether or not the molecular docking can accurately reproduce the inhibitory data of the dipeptide. From the dipeptide library, we extracted 17 compounds exhibiting a relatively high activity (inhibition ratio >50%) and 99 compounds with no activity (inhibition ratio <5%), and determined the discriminatory ability of highly active and inactive peptides based on the docking score (MM-GBSA score [23]) that reflects the dipeptide binding stability. Single docking using a DPP4 co-crystal structure (PDBID: 1NU8 [7]) resulted in a discriminatory ability (the area under the receiver operating characteristic curve, AUC) of 0.74 (Figure 1A); however, ensemble docking using four DPP4 co-crystal structures [20] resulted in an AUC of 0.84, indicating a higher discriminatory ability compared to single docking (Figure 1B). MM-GBSA scores were weakly correlated with inhibitory activities for 237 dipeptides exhibiting DPP4 inhibition (inhibition ratio >5%) (e.g. dipeptides with stronger DPP4 inhibitory activity tend to have a higher binding stability, Figure 1C). These results suggested that the DPP4 inhibitory activities of dipeptides/tripeptides may be ranked by ensemble docking. Although dipeptides with different inhibitory mechanisms are included in the dipeptide library, in reality [16, 17], this calculation result might indicate that most dipeptides exhibit competitive inhibition.
Figure 1.
Prediction precisions of (A) single docking using a DPP4 co-crystal structure (PDBID: 1NU8) and (B, C) ensemble docking using four types of co-crystal structures (PDBID: 1NU8, 1R9N, 2BGN, and 2BGR); (A, B) Receiver operating characteristic (ROC) curves for (A) single docking and (B) ensemble docking, which are calculated from the MM-GBSA scores of 17 active dipeptides (inhibition ratio >50%) and 99 inactive dipeptides (inhibition ratio <5%), using the R package ROCR [28]. (C) MM-GBSA scores are plotted against inhibitory activity for 237 dipeptides that exhibit DPP inhibition (inhibition ratio >5%), where a lower MM-GBSA score indicates a higher binding stability.
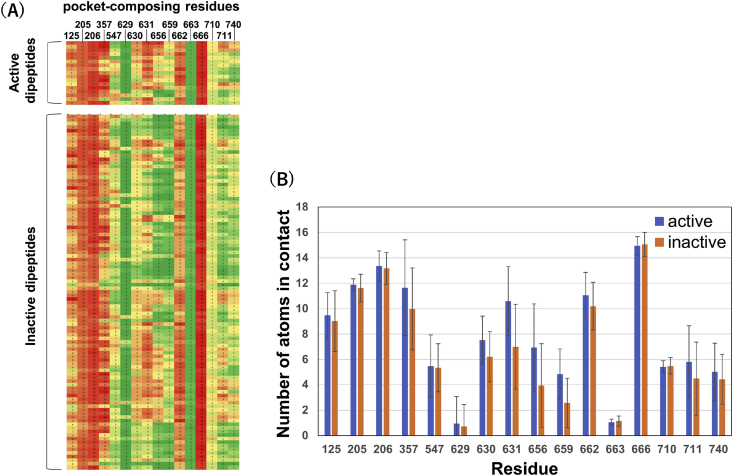
Next, we compared the DPP4-peptide interaction pattern between highly active and inactive dipeptides by calculating a fingerprint of the contact between a bound dipeptide and each pocket-composing residue based on the docking pose [26]. Although marked differences were not observed at most residues, we found that the highly active dipeptides tend to interact tightly with S630, Y631, V656, and W659 (Figure 2). Here, S630 is the active center, and Y631, V656, and W659 form the S1 pocket located in the deepest area of the substrate binding site [4] (Figure 3A). Superimposing the distribution of docking poses also showed that highly active dipeptides tend to interact closely with these four residues and bind slightly more deeply in the S1 pocket (Figure 3B). These results suggest that attaining a tight interaction with the S1 pocket is essential for dipeptides to compete with the substrate and strongly inhibit the protease activity.
Figure 2.
DPP4-dipeptide interaction fingerprint. (A) DPP4-dipeptide interaction fingerprint of 17 highly active dipeptides (inhibition ratio >50%) and 99 inactive dipeptides (inhibition ratio <5%) is shown. Pocket-composing residues are located 5 Å from the bound ligand, Ile-Pro-Ile, in the co-crystal structure (PDBID: 1NU8). The number of atoms in contact with the dipeptide was counted for each residue. Residues with a relatively high (low) count are shown in red (green). (B) Interaction patterns of DPP4-dipeptide for active and inactive dipeptides are shown. The number of DPP4 atoms in contact with each dipeptide was averaged across the docking poses observed for 17 active compounds and 99 inactive compounds. S630 (p = 8×10−3), Y631 (p = 3×10−5), V656 (p = 2×10−3), and W659 (p = 2×10−4) were identified as residues that exhibit a significant difference between the active and inactive dipeptides (p < 0.01).
Figure 3.
Docking pose distribution of highly active and inactive dipeptides. (A) DPP4 substrate pocket for PDBID: 1NU8 is shown. S630, the active center, and Y631, V656, and W659, which compose the S1 pocket, are highlighted. (B) Docking pose distribution of 17 active compounds (inhibition ratio >50%, left) and 99 inactive compounds (inhibition ratio <5%, right) is shown. S630, Y631, V656, and W659 in the co-crystal structure (PDBID: 1NU8) are visualized with sphere models. When the shortest atomic distance between V656 (gamma carbon, Cγ2) and the docked dipeptide is averaged across the docking poses [68 (=17×4) for active dipeptides and 396 (=99×4) for inactive dipeptides], the mean shortest distances for active and inactive dipeptides are 3.8 ± 1.5 and 4.9 ± 1.8 (Å), respectively (p = 6×10−7).
3.2. Additional interaction at the S1′ subsite that contributes to increased DPP4 inhibitory activity
The N-terminus of competitive DPP4-inhibiting peptides often contains hydrophobic/aromatic amino acids such as Ile, Leu, Val, Phe, Trp, or Tyr [14]; dipeptides Ile-Pro and Val-Pro especially exhibit high inhibitory activity [15]. We therefore added various kinds of amino acids to the C-terminus of these two dipeptides to create tripeptides (Ile-Pro-XXX, Val-Pro-XXX) and measured their DPP4 inhibitory activity to evaluate the contribution of amino acids on the C-terminal side of tripeptides (Figure 4). For both peptides, when a polar amino acid such as Thr or Arg was added to the C-terminus, the inhibitory activity either remained the same or tended to decrease slightly. In contrast, addition of hydrophobic amino acid (Ile, Met, Val, Trp) resulted in several-fold increases in the inhibitory activity.
Figure 4.
Inhibitory activity of tripeptides against DPP4. The 50% inhibitory concentrations (IC50) of (A) Ile-Pro-XXX and (B) Val-Pro-XXX against DPP4 are shown. In cases of IPN, IPW, IPY, VPD, VPL, VPS, and VPV, the IC50 was hard to be determined because of their weak inhibitory activities (See Materials & Methods). (C) MM-GBSA scores are plotted against IC50 for these tripeptides, where a lower MM-GBSA score indicates a higher binding stability, showing that tripeptides with higher binding stability tend to exhibit a stronger DPP4 inhibitory activity.
To identify the factors that determine the difference in inhibitory activities among these tripeptides, we analyzed the DPP4-tripeptide co-crystal structure and the complex structure models predicted by molecular docking. The characteristics of the co-crystal structure of DPP4 and Ile-Pro-Ile (PDBID: 1NU8) are that the Pro side chain in the tripeptide fits into the S1 pocket and that the N-terminal amino group forms salt bridges with Glu205 and Glu206 [7] (Figure 5A). The Ile-Pro-Ile binding mode predicted by molecular docking correctly reproduced these structural characteristics observed in the co-crystal structure (Figure 5A). This suggests that the docking protocol constructed for dipeptides can be applied in predicting the binding structures of tripeptides. The docking poses of six tripeptides that exhibited relatively high inhibitory activity (Ile-Pro-Ile, Val-Pro-Ile, Val-Pro-Trp, Val-Pro-Met, Ile-Pro-Met, Ile-Pro-Val) all showed that the C-terminal amino acid is located near the Tyr547 and Trp629 that compose the S1′ subsite of DPP4 (Figure 5A-F). Moreover, when the DPP4-peptide interaction pattern was compared between highly active and low-active tripeptides, we found that highly active tripeptides tended to interact more closely with Tyr547 and Trp629 (Figure 6). These results suggest that the tripeptides with high inhibitory activity form additional hydrophobic interactions at the S1′ subsite located somewhat outside the substrate pocket. This additional interaction appears to stabilize the DPP-peptide binding, thereby attaining a higher inhibitory activity that exceeds that of the corresponding dipeptide.
Figure 5.
Docking poses of tripeptides with ameliorated inhibitory activity. Docking poses (green) of (A) Ile-Pro-Ile, (B) Val-Pro-Ile, (C) Val-Pro-Trp, (D) Val-Pro-Met, (E) Ile-Pro-Met, and (F) Ile-Pro-Val in the protein structure (electrostatic surface model) of the DPP4-Ile-Pro-Ile co-crystal structure (PDBID: 1NU8) are shown. Ile-Pro-Ile binding pose from the co-crystal structure is superimposed (cyan).
Figure 6.
DPP4-tripeptide interaction fingerprint. (A) DPP4-tripeptide interaction fingerprint of six highly active tripeptides (IC50 < 0.01 mM) and six low-active tripeptides (IC50 > 0.03 mM). Pocket-composing residues are located 5 Å from the bound ligand, Ile-Pro-Ile, in the co-crystal structure (PDBID: 1NU8). The number of atoms in contact with the tripeptide was counted for each residue. Residues with a relatively high (low) count are shown in red (green). (B) Interaction patterns of DPP4-tripeptide for highly active and low-active tripeptides. The number of DPP4 atoms in contact with each tripeptide was averaged across the docking poses observed for six highly active tripeptides and six low-active tripeptides.
A previous report has demonstrated an improved affinity of small molecule inhibitors through additional interactions at the S2 extensive subsite [4]. In contrast, for tripeptides such as Ile (Val)-Pro-Xaa that exhibit competitive inhibition, an interaction with the S1′ subsite may contribute to increasing DPP4 inhibitory activity because the C-terminus of the peptide is positioned at this subsite when binding to DPP4. Although we have mainly discussed their role as DPP4 inhibitors, tripeptides with a penultimate proline residue appear to be slowly hydrolyzed by DPP4 because of their substrate-like nature [27], suggesting transient DPP4 binding of Ile-Pro-XXX and Val-Pro-XXX. However, the structural information on the temporarily-formed DPP4-tripeptide interactions is expected to be useful in the molecular design of novel DPP4 inhibitors.
Declarations
Author contribution statement
M. Araki and H. Iwata: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
K. Masuda: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Y. Okuno: Conceived and designed the experiments; Wrote the paper.
N. Kanegawa: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Y. Sagae: Analyzed and interpreted the data.
K. Ito: Analyzed and interpreted the data; Wrote the paper.
Funding statement
Y. Okuno was supported by Ministry of Education, Culture, Sports, Science and Technology of Japan as “Priority Issue on Post-K computer (Building Innovative Drug Discovery Infrastructure Through Functional Control of Biomolecular Systems)” (hp190154). K. Masuda was supported by Suntory Global Innovation Center Limited.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
This research used computational resources of the HPCI system provided by Cybermedia Center, Osaka University through the HPCI System Research Project (Project ID: hp190154).
Contributor Information
Katsuyoshi Masuda, Email: Katsuyoshi_Masuda@suntory.co.jp.
Yasushi Okuno, Email: okuno.yasushi.4c@kyoto-u.ac.jp.
References
- 1.Mentlein R. Dipeptidyl-peptidase IV (CD26)–role in the inactivation of regulatory peptides. Regul. Pept. 1999;85(1):9–24. doi: 10.1016/s0167-0115(99)00089-0. [DOI] [PubMed] [Google Scholar]
- 2.Cahn A., Cernea S., Raz I. An update on DPP-4 inhibitors in the management of type 2 diabetes. Expet Opin. Emerg. Drugs. 2016;21(4):409–419. doi: 10.1080/14728214.2016.1257608. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh C.H., Wang T.Y., Hung C.C., Chen M.C., Hsu K.C. Improvement of glycemic control in streptozotocin-induced diabetic rats by Atlantic salmon skin gelatin hydrolysate as the dipeptidyl-peptidase IV inhibitor. Food Funct. 2015;6(6):1887–1892. doi: 10.1039/c5fo00124b. [DOI] [PubMed] [Google Scholar]
- 4.Nabeno M., Akahoshi F., Kishida H., Miyaguchi I., Tanaka Y., Ishii S., Kadowaki T. A comparative study of the binding modes of recently launched dipeptidyl peptidase IV inhibitors in the active site. Biochem. Biophys. Res. Commun. 2013;434(2):191–196. doi: 10.1016/j.bbrc.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida T., Sakashita H., Akahoshi F., Hayashi Y. [(S)-gamma-(4-Aryl-1-piperazinyl)-l-prolyl]thiazolidines as a novel series of highly potent and long-lasting DPP-IV inhibitors. Bioorg. Med. Chem. Lett. 2007;17(9):2618–2621. doi: 10.1016/j.bmcl.2007.01.110. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida T., Akahoshi F., Sakashita H., Kitajima H., Nakamura M., Sonda S., Takeuchi M., Tanaka Y., Ueda N., Sekiguchi S., Ishige T., Shima K., Nabeno M., Abe Y., Anabuki J., Soejima A., Yoshida K., Takashina Y., Ishii S., Kiuchi S., Fukuda S., Tsutsumiuchi R., Kosaka K., Murozono T., Nakamaru Y., Utsumi H., Masutomi N., Kishida H., Miyaguchi I., Hayashi Y. Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-y lcarbonyl]thiazolidine): a highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Bioorg. Med. Chem. 2012;20(19):5705–5719. doi: 10.1016/j.bmc.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Thoma R., Loffler B., Stihle M., Huber W., Ruf A., Hennig M. Structural basis of proline-specific exopeptidase activity as observed in human dipeptidyl peptidase-IV. Structure (London, England: 1993) 2003;11(8):947–959. doi: 10.1016/s0969-2126(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 8.Aertgeerts K., Ye S., Tennant M.G., Kraus M.L., Rogers J., Sang B.C., Skene R.J., Webb D.R., Prasad G.S. Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation. Protein Sci.: Publ. Protein Soc. 2004;13(2):412–421. doi: 10.1110/ps.03460604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weihofen W.A., Liu J., Reutter W., Saenger W., Fan H. Crystal structures of HIV-1 Tat-derived nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) bound to the active site of dipeptidyl-peptidase IV (CD26) J. Biol. Chem. 2005;280(15):14911–14917. doi: 10.1074/jbc.M413400200. [DOI] [PubMed] [Google Scholar]
- 10.Lacroix I.M., Li-Chan E.C. Isolation and characterization of peptides with dipeptidyl peptidase-IV inhibitory activity from pepsin-treated bovine whey proteins. Peptides. 2014;54:39–48. doi: 10.1016/j.peptides.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Li-Chan E.C., Hunag S.L., Jao C.L., Ho K.P., Hsu K.C. Peptides derived from atlantic salmon skin gelatin as dipeptidyl-peptidase IV inhibitors. J. Agric. Food Chem. 2012;60(4):973–978. doi: 10.1021/jf204720q. [DOI] [PubMed] [Google Scholar]
- 12.Nongonierma A.B., FitzGerald R.J. Dipeptidyl peptidase IV inhibitory and antioxidative properties of milk protein-derived dipeptides and hydrolysates. Peptides. 2013;39:157–163. doi: 10.1016/j.peptides.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Nongonierma A.B., FitzGerald R.J. Susceptibility of milk protein-derived peptides to dipeptidyl peptidase IV (DPP-IV) hydrolysis. Food Chem. 2014;145:845–852. doi: 10.1016/j.foodchem.2013.08.097. [DOI] [PubMed] [Google Scholar]
- 14.Nongonierma A.B., Mooney C., Shields D.C., FitzGerald R.J. In silico approaches to predict the potential of milk protein-derived peptides as dipeptidyl peptidase IV (DPP-IV) inhibitors. Peptides. 2014;57:43–51. doi: 10.1016/j.peptides.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 15.Lan V.T., Ito K., Ohno M., Motoyama T., Ito S., Kawarasaki Y. Analyzing a dipeptide library to identify human dipeptidyl peptidase IV inhibitor. Food Chem. 2015;175:66–73. doi: 10.1016/j.foodchem.2014.11.131. [DOI] [PubMed] [Google Scholar]
- 16.Nongonierma A.B., Fitzgerald R.J. Inhibition of dipeptidyl peptidase IV (DPP-IV) by tryptophan containing dipeptides. Food Funct. 2013;4(12):1843–1849. doi: 10.1039/c3fo60262a. [DOI] [PubMed] [Google Scholar]
- 17.Lan V.T., Ito K., Ito S., Kawarasaki Y. Trp-Arg-Xaa tripeptides act as uncompetitive-type inhibitors of human dipeptidyl peptidase IV. Peptides. 2014;54:166–170. doi: 10.1016/j.peptides.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 18.Li C., Lu W., Lu C., Xiao W., Shen X., Huang J., Liu G., Tang Y. Identification of diverse dipeptidyl peptidase IV inhibitors via structure-based virtual screening. J. Mol. Model. 2012;18(9):4033–4042. doi: 10.1007/s00894-012-1394-3. [DOI] [PubMed] [Google Scholar]
- 19.Guasch L., Ojeda M.J., Gonzalez-Abuin N., Sala E., Cereto-Massague A., Mulero M., Valls C., Pinent M., Ardevol A., Garcia-Vallve S., Pujadas G. Identification of novel human dipeptidyl peptidase-IV inhibitors of natural origin (part I): virtual screening and activity assays. PloS One. 2012;7(9) doi: 10.1371/journal.pone.0044971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craig I.R., Essex J.W., Spiegel K. Ensemble docking into multiple crystallographically derived protein structures: an evaluation based on the statistical analysis of enrichments. J. Chem. Inf. Model. 2010;50(4):511–524. doi: 10.1021/ci900407c. [DOI] [PubMed] [Google Scholar]
- 21.Molecular Operating Environment (MOE) 2016. Chemical Computing Group Inc., 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7. [Google Scholar]
- 22.Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 23.Onufriev A., Bashford D., Case D.A. Modification of the generalized Born model suitable for macromolecules. J. Phys. Chem. B. 2000;104(15):3712–3720. [Google Scholar]
- 24.Case D.A., Darden T.A., Cheatham I.T.E., Simmerling C.L., Wang J., Duke R.E., Luo R., Walker R.C., Zhang W., Merz K.M., Roberts B., Hayik S., Roitberg A., Seabra G., Swails J., Götz A.W., Kolossváry I., Wong K.F., Paesani F., Vanicek J., Wolf R.M., Liu J., Wu X., Brozell S.R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D.R., Mathews D.H., Seetin M.G., Salomon-Ferrer R., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., Kollman P.A. University of California; San Francisco: 2012. AMBER 12. [Google Scholar]
- 25.Miller B.R., 3rd, McGee T.D., Jr., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theor. Comput. 2012;8(9):3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 26.Sato M., Hirokawa T. Extended template-based modeling and evaluation method using consensus of binding mode of GPCRs for virtual screening. J. Chem. Inf. Model. 2014;54(11):3153–3161. doi: 10.1021/ci500499j. [DOI] [PubMed] [Google Scholar]
- 27.Rahfeld J., Schierhorn M., Hartrodt B., Neubert K., Heins J. Are diprotin A (Ile-Pro-Ile) and diprotin B (Val-Pro-Leu) inhibitors or substrates of dipeptidyl peptidase IV? Biochim. Biophys. Acta. 1991;1076(2):314–316. doi: 10.1016/0167-4838(91)90284-7. [DOI] [PubMed] [Google Scholar]
- 28.Sing T., Sander O., Beerenwinkel N., Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics (Oxford, England) 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]