Significance
RIG-I, an up-regulated gene in ATRA-induced myeloid differentiation of APL cells, has been repeatedly described as a cytoplasmic RNA receptor that plays a vital role in the innate antiviral immunity. However, it is still unclear what function RIG-I exerts in hematopoiesis. Here, we aimed to decipher the mechanisms promoting myeloid differentiation by RIG-I. We showed that RIG-I could bind TRIM25 mRNA, resulting in enhanced RNA stability and up-regulated protein expression. Meanwhile, RIG-I could promote ISGylation by up-regulating the key genes in the ISGylation pathway through cooperation with STAT1/2 and IRF1. The integrity of ISGylation was essential in ATRA or RIG-I–induced differentiation. These findings reveal a unique molecular mechanism implicated in myeloid differentiation relevant to RIG-I-TRIM25-ISGylation axis.
Keywords: RIG-I, TRIM25, ISGylation, acute promyelocytic leukemia (APL), myeloid differentiation
Abstract
Retinoic acid-inducible gene I (RIG-I) is up-regulated during granulocytic differentiation of acute promyelocytic leukemia (APL) cells induced by all-trans retinoic acid (ATRA). It has been reported that RIG-I recognizes virus-specific 5′-ppp-double-stranded RNA (dsRNA) and activates the type I interferons signaling pathways in innate immunity. However, the functions of RIG-I in hematopoiesis remain unclear, especially regarding its possible interaction with endogenous RNAs and the associated pathways that could contribute to the cellular differentiation and maturation. Herein, we identified a number of RIG-I–binding endogenous RNAs in APL cells following ATRA treatment, including the tripartite motif-containing protein 25 (TRIM25) messenger RNA (mRNA). TRIM25 encodes the protein known as an E3 ligase for ubiquitin/interferon (IFN)-induced 15-kDa protein (ISG15) that is involved in RIG-I–mediated antiviral signaling. We show that RIG-I could bind TRIM25 mRNA via its helicase domain and C-terminal regulatory domain, enhancing the stability of TRIM25 transcripts. RIG-I could increase the transcriptional expression of TRIM25 by caspase recruitment domain (CARD) domain through an IFN-stimulated response element. In addition, RIG-I activated other key genes in the ISGylation pathway by activating signal transducer and activator of transcription 1 (STAT1), including the modifier ISG15 and several enzymes responsible for the conjugation of ISG15 to protein substrates. RIG-I cooperated with STAT1/2 and interferon regulatory factor 1 (IRF1) to promote the activation of the ISGylation pathway. The integrity of ISGylation in ATRA or RIG-I–induced cell differentiation was essential given that knockdown of TRIM25 or ISG15 resulted in significant inhibition of this process. Our results provide insight into the role of the RIG-I-TRIM25-ISGylation axis in myeloid differentiation.
Retinoic acid-inducible gene I (RIG-I) is a highly expressed gene during the differentiation and maturation of the acute promyelocytic leukemia (APL) cell line NB4 induced by all-trans retinoic acid (ATRA) (1). Over the past two decades, a large body of evidence has been accumulated that RIG-I can bind to virus-specific 5′-ppp-double-stranded RNA (dsRNA) and activate interferon regulatory factor 3 (IRF3) and nuclear factor NF-kappa-B (NFκB) to induce type I interferons, thereby activating the antiviral innate immune responses of the host (2, 3). Therefore, RIG-I is considered to be a cytoplasmic RNA receptor (4).
Notably, tripartite motif-containing protein 25 (TRIM25), an E3 ligase for K63-linked polyubiquitination of RIG-I, is shown to be principal in the RIG-I–mediated antiviral pathway (5). In addition, it has been demonstrated that TRIM25 plays an essential role in ISGylation, a ubiquitin-like modification (6). In this pathway, interferon (IFN)-induced 15-kDa protein (ISG15) serves as a ubiquitin-like modifier and is conjugated to protein substrates through a cascade of enzymatic reactions, which involve an activating enzyme, a conjugating enzyme, and ligases, in a process similar to ubiquitination (7). ISGylation is substantially involved in immune regulation, DNA repair, tumorigenesis, and hematopoiesis (7–9). It has been reported that ISGylation also plays an important and conservative role for normal tissue differentiation, especially in placental and fetal development, with ISG15 and other ISGylation-associated genes being specifically and highly expressed in the placenta of a wide range of species from mouse to human (10–15).
However, it remains unclear what function of RIG-I may exert in the ATRA-induced cell differentiation, a process unrelated to viral infection and requiring the reprogramming of cell activities at multiple layers. We and others have found that Rig-I-deficient mice were partially embryonic lethal and that postnatal mice exhibited enteritis, abnormal activation of T cells, and other abnormal immune functions (16, 17). In addition, abnormal hematopoietic phenotypes such as myeloproliferative neoplasms and hepatosplenomegaly were observed in Rig-I−/− mice (16, 18). These studies suggested that RIG-I could be involved in the intrinsic regulation of hematopoiesis and immunity. Indeed, our previous studies have shown that RIG-I can inhibit leukemia cell proliferation by activating signal transducer and activator of transcription 1 (STAT1) (19) and that RIG-I can restrain leukemic stemness by modulating Src-mediated AKT activation (20).
Apart from its classic way of binding to 5′-ppp-dsRNA (2, 21), several studies have shown that RIG-I can interact with dsRNAs with or without phosphate groups (22, 23) as well as the poly-U/UC tracts of hepatitis C virus RNAs or U/A-rich regions of the influenza virus NS1 gene, inducing type I interferons (24, 25). A previous study showed that RIG-I could bind to 3′-untranslated region (UTR) of Nfkb1 messenger RNA (mRNA) and regulate its expression (26). Recently, it has been found that RIG-I binds self-noncoding RNA lnc-Lsm3b at a late stage of viral infection to restrict the RIG-I–mediated innate immune response (27). These findings suggest that the ligands of RIG-I vary according to the physiological or pathological settings, which encouraged us to further explore the functions exerted by RIG-I as a cytoplasmic RNA receptor in hematopoietic cell differentiation and maturation.
Therefore, we hypothesize that RIG-I, as a highly expressed protein during APL cell differentiation, may serve as a hub regulator through the interaction between RNA and protein in hematopoiesis. We characterized the features of the interaction between RIG-I and the endogenous mRNAs in NB4 cell line using RNA-binding protein immunoprecipitation-sequencing (RIP-seq). Interestingly, we found that RIG-I could bind TRIM25 mRNA, which led to the enhanced stability of TRIM25 transcripts and up-regulation of TRIM25 protein level. The detailed mechanisms underlying the interaction between RIG-I and TRIM25 were further studied with regard to myeloid differentiation.
Results
RIG-I Binds Endogenous RNAs in NB4 Cells Undergoing ATRA-Induced Differentiation.
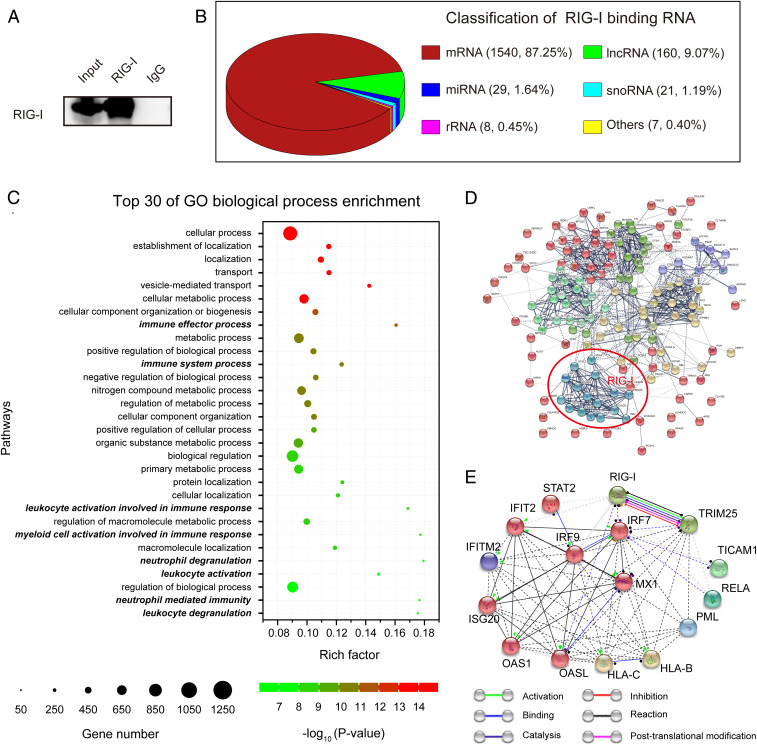
To explore whether RIG-I could bind endogenous RNAs during ATRA-induced differentiation, we performed high-throughput sequencing of the RNA immunoprecipitated by the anti-RIG-I antibody (Fig. 1A). The RIP-seq data showed that RIG-I bound 1,765 endogenous RNAs, including mRNA, long noncoding RNA (lncRNA), microRNA (miRNA), small nucleolar RNA (snoRNA), ribosomal RNA (rRNA), and other RNA molecules (Fig. 1B and Dataset S1). Gene ontology enrichment analysis results revealed that the transcripts of protein-coding genes bound by RIG-I were primarily involved in some basic biological processes such as metabolism, cellular/protein localization and vesicle-mediated transport, and some specific functional processes such as immune regulation and leukocyte/myeloid/neutrophil regulation (Fig. 1C and Dataset S2).
Fig. 1.
RIG-I binds endogenous RNAs in NB4 cells undergoing ATRA-induced differentiation. (A) RNA-binding protein immunoprecipitation (RIP) was performed with an anti-RIG-I N terminus antibody in NB4 cells treated with ATRA for 3 d, followed by immunoblotting to verify the efficiency of immunoprecipitation. (B) Classification of RIG-I–binding endogenous RNAs revealed by RIP-seq. (C) The top 30 most significant Gene Ontology biological process terms of the protein-coding RNAs bound by RIG-I. Rich factor refers to the ratio of observed gene count and background gene count. Immune, leukocyte, myeloid, and neutrophil regulation-related terms are marked in bold italics. (D) Prediction of the protein-protein association network between RIG-I and the 149 fuctional genes in the immune effect process. RIG-I is clustered with 15 functionally related genes, which are marked in red circle. (E) The interactions between RIG-I and the 15 genes clustered in the protein-protein interaction network.
In order to identify genes functionally associated with RIG-I, we further inputted RIG-I with the 149 genes enriched in the immune-related pathway with the lowest P value (GO: 0002252, immune effector process) for protein-protein association network analysis by using STRING with a confidence model of network edges (28). The interaction network was divided into six clusters by using k-means clustering algorithm and revealed a relatively independent gene cluster composed of 16 genes associated with RIG-I (Fig. 1D and Dataset S3). We then extracted this cluster for protein-protein interaction analysis using a molecular action model. The results showed that TRIM25 was most functionally related to RIG-I. There were mutual specific actions between RIG-I and TRIM25 at six levels, including activation, binding, catalysis, inhibition, reaction, and posttranslational modification (Fig. 1E and Dataset S4) with a combined interaction score of 0.995 (Dataset S5).
RIG-I Binds TRIM25 mRNA.
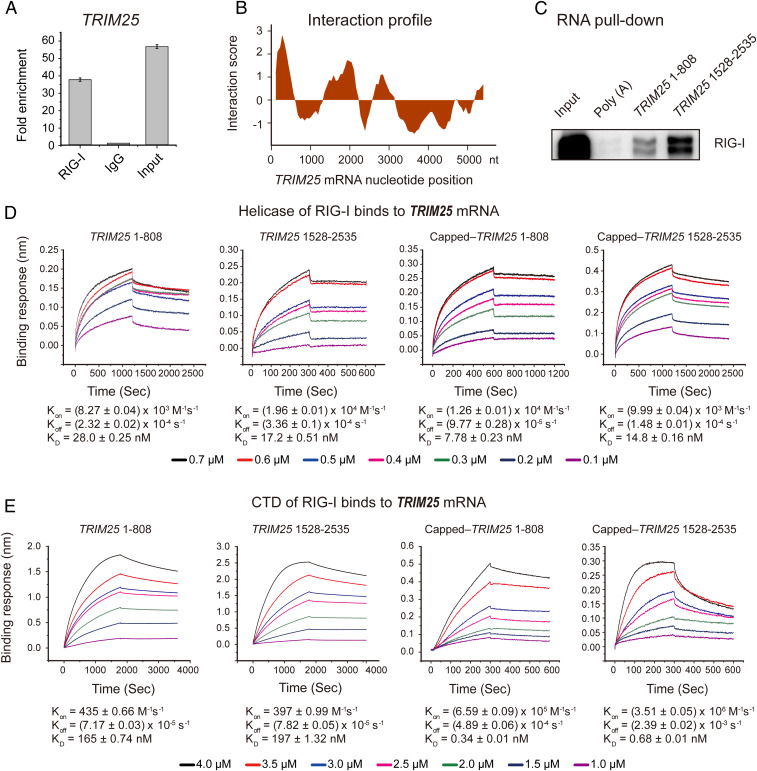
TRIM25 encodes a protein known to catalyze the K63-linked polyubiquitination of RIG-I as an indispensable step in RIG-I–mediated innate antiviral immune responses. Based on the RIP-seq data performed in a myeloid differentiation model and their cooperation in the antiviral immunity, we speculated TRIM25 could play an important role in RIG-I–mediated differentiation. First, we confirmed the interaction between RIG-I and TRIM25 mRNA by RIP-qPCR (Fig. 2A). Second, the catRAPID algorithm (29), an online analysis tool, was used to predict the possible fragment of TRIM25 responsible for the interaction between RIG-I and TRIM25 mRNA. The data showed that RIG-I could interact with two major fragments of TRIM25 mRNA (Fig. 2B). Third, we performed RNA pull-down assay using in vitro transcribed RNA of these two candidate fragments, which indicated that RIG-I did interact with TRIM25 mRNA (Fig. 2C).
Fig. 2.
The interaction between RIG-I and the TRIM25 gene cluster. (A) The interaction between RIG-I and TRIM25 mRNA verified by RIP-qPCR. (B) The TRIM25 mRNA fragments responsible for the interaction between RIG-I and TRIM25 mRNA predicted by the catRAPID algorithm. The predicted binding regions are those with interaction score > 0. (C) Analysis of the interaction between RIG-I and the various TRIM25 mRNA fragments using an RNA pull-down assay and immunoblotting. (D and E) Affinities of the interactions between TRIM25 mRNA and RIG-I helicase (D) or CTD (E) measured by a biolayer interferometry technique (Kon, on-rate constant; Koff, off-rate constant; KD, binding constant).
Several studies have shown that the C-terminal regulatory domain (CTD) of RIG-I can interact with viral specific 5′-ppp-dsRNA (30–32). The catRAPID algorithm prediction showed that the helicase domain and CTD of RIG-I could bind TRIM25 mRNA (SI Appendix, Fig. S2 A and B). To determine the dynamics of the interaction between RIG-I and TRIM25 mRNA, the helicase domain, CTD and caspase recruitment domain (CARD) of RIG-I were cloned and expressed as glutathione-S-transferase (GST)-fusion proteins, respectively (SI Appendix, Fig. S2B). Then, we measured the kinetic constants of the interaction between RIG-I domains and the TRIM25 mRNA fragments using the biolayer interferometry technique as previously described (33). The results showed that RIG-I could indeed interact with the TRIM25 mRNA fragments through the helicase domain (Fig. 2D) and CTD (Fig. 2E), but not the CARD domain (SI Appendix, Fig. S2C). In parallel, to mimic the “cap” of mature mRNAs, we modified the 5′ end of the transcribed TRIM25 mRNA with m7G analogs as previously described (34). Pull-down data showed that the capped TRIM25 mRNA interacted with RIG-I, particularly the fragment 1528 to 2535 (SI Appendix, Fig. S2D). On the other hand, kinetic data showed that the affinity between the capped TRIM25 mRNA and RIG-I was similar to that between 5′-ppp-dsRNA and RIG-I (Fig. 2 D and E and SI Appendix, Fig. S2E), while the capped fragments of TRIM25 had even stronger affinity with RIG-I than the triphosphorylated one (Fig. 2 D and E). These results suggested that RIG-I was able to bind TRIM25 mRNA through the helicase domain and CTD independently of the triphosphate groups, a property that allows RIG-I to bind endogenous RNAs in the context of nonviral infection.
RIG-I Up-Regulates TRIM25 Expression at mRNA and Protein Levels.
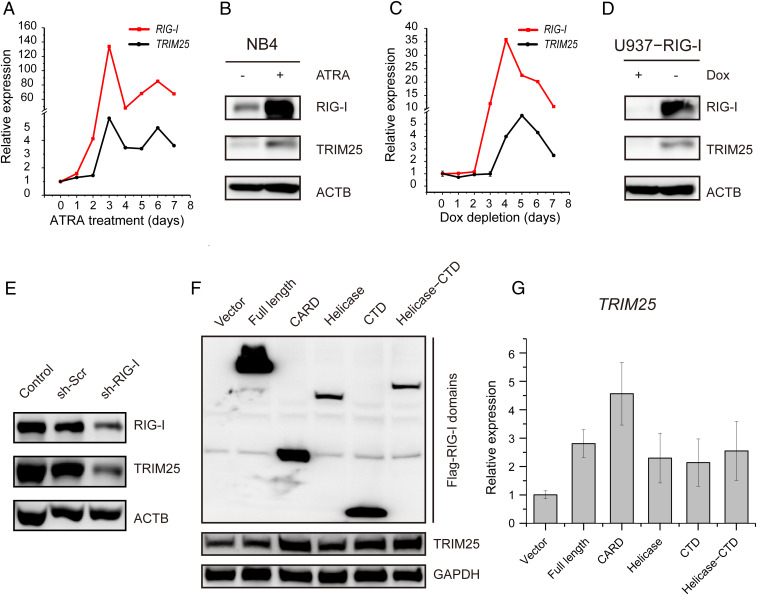
After confirming the interaction between RIG-I and TRIM25 mRNA, we investigated the biological significance of this interaction. It was found that both RIG-I and TRIM25 were significantly up-regulated at mRNA and protein levels with ATRA induction in NB4 cells (Fig. 3 A and B). To exclude the interference of other genes involved in the differentiation of NB4 cells induced by ATRA (1, 35), we further investigated the direct effect of RIG-I on up-regulating TRIM25 in a RIG-I-inducible cell line U937-RIG-I that was established in our previous study (19). The induction of RIG-I by doxycycline depletion increased the expression of TRIM25 (Fig. 3 C and D). Moreover, TRIM25 was down-regulated by the knockdown of RIG-I in NB4 cells (Fig. 3E). These results supported our hypothesis that RIG-I could up-regulate the expression of TRIM25.
Fig. 3.
The expression of TRIM25 regulated by RIG-I. (A and B) The mRNA (A) and protein (B) levels of TRIM25 in NB4 cells with ATRA treatment. (C and D) The mRNA (C) and protein (D) levels of TRIM25 in U937-RIG-I cells with RIG-I induction upon doxycycline depletion. (E) Down-regulation of TRIM25 expression upon the knockdown of RIG-I in NB4 cells. (F) Up-regulation of TRIM25 protein by overexpressing the CARD, CTD, and helicase domain of RIG-I in HEK 293T cells, respectively. (G) The transcriptional level of TRIM25 was up-regulated upon the overexpression of three distinct domains of RIG-I in HEK 293T cells.
To explore which domain of RIG-I played a key role in the up-regulation of TRIM25, we transduced the pFLAG-CMV4 plasmids carrying various domains of RIG-I into human embryonic kidney (HEK) 293T cells. It was found that both the RNA-binding domains (helicase domain and CTD) and CARD of RIG-I could obviously augment the protein level of TRIM25 (Fig. 3F). Of note, the CARD domain increased the mRNA level of TRIM25 more significantly than the helicase domain and CTD did (Fig. 3G), suggesting that RIG-I could modulate the expression of TRIM25 through distinct mechanisms by using different domains.
RIG-I Enhances RNA Stability and Up-Regulates the Transcription of TRIM25.
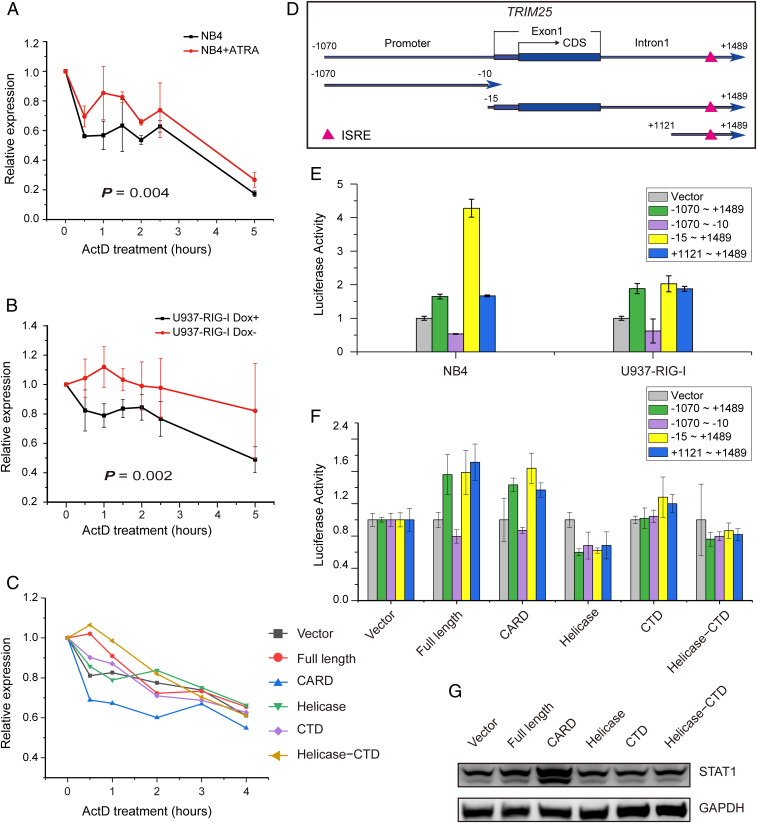
It has been reported that RNA-binding proteins usually play key roles in the regulation of mRNA stability (36). To investigate the mechanism underlying the up-regulation of TRIM25 mediated by RIG-I, we tested the stability of TRIM25 mRNA through an actinomycin D (ActD) chase experiment in NB4 cells before and after ATRA induction. Remarkably, the stability of TRIM25 mRNA was significantly increased by ATRA treatment (Fig. 4A). Similarly, the TRIM25 mRNA stability was also significantly increased in the U937-RIG-I cell line with RIG-I induction by doxycycline depletion (Fig. 4B). Furthermore, we found that RIG-I enhanced the stability of TRIM25 mRNA mainly through the helicase and CTD domain in HEK 293T cells (Fig. 4C).
Fig. 4.
Enhanced RNA stability and transcriptional activation of TRIM25 mediated by RIG-I contribute to the up-regulation of TRIM25. (A) Enhancement of stability of TRIM25 mRNA in NB4 cells with ATRA treatment. (B) RIG-I enhanced TRIM25 mRNA stability in U937-RIG-I cells with RIG-I induction upon doxycycline depletion. (C) The RNA-binding domains of RIG-I stabilized TRIM25 mRNA in HEK 293T cells. (D) Schematic diagram of TRIM25 promoter and DNA fragments near the transcription initiation site of the TRIM25 gene. (E) The Luciferase assay of the transcriptional activity of the sequence containing ISRE in the first intron of TRIM25 upon treatment with ATRA in NB4 cells and RIG-I induction in U937-RIG-I cells, respectively. (F) Activation of the ISRE in the first intron of TRIM25 by the CARD domain of RIG-I in HEK 293T cells. (G) Up-regulation of STAT1 mediated by the CARD domain of RIG-I, which was capable of binding to the ISRE in the first intron of TRIM25.
Next, we explored the role of the CARD in the up-regulation of TRIM25. It has been reported that RIG-I can activate STAT1 by inhibiting its dephosphorylation through the interaction of CARD with STAT1 (37). The first intron of the TRIM25 gene contains an IFN-stimulated response element (ISRE), which is the binding site for STAT1 (38). Therefore, we hypothesized that RIG-I could promote the transcriptional expression of TRIM25 by activating STAT1 through the CARD domain. To verify this hypothesis, luciferase reporter assays were performed in NB4 and U937-RIG-I cells, which were infected with lentivirus carrying promoter DNA fragments of TRIM25 (Fig. 4D) and then induced by ATRA treatment or doxycycline depletion, respectively. As expected, the transcriptional activity of TRIM25 was activated via the sequences containing ISRE in the first intron but not the canonical promoter sequence upstream of the transcription initiation site of TRIM25, following upon treatment with ATRA or RIG-I induction (Fig. 4E). Consistently, the full length and CARD of RIG-I activated the transcriptional activity of TRIM25 via the sequences containing ISRE in HEK 293T cells (Fig. 4F). Notably, it was the CARD, but not the RNA-binding domains of RIG-I, that up-regulated and activated STAT1 (Fig. 4G). Taken together, these data suggested that RIG-I could up-regulate the expression of TRIM25 via two distinct mechanisms: 1) the RNA-binding domains could enhance mRNA stability of TRIM25 through protein-RNA interaction and 2) the signal transduction domain could up-regulate the transcriptional level of TRIM25 via STAT1 activation.
RIG-I Promotes ISGylation in Cooperation with STAT and IRF Family Transcription Factors.
TRIM25 is widely recognized to be an ubiquitination E3 ligase that modifies RIG-I during RIG-I–mediated antiviral activity (5). Nevertheless, ubiquitination of RIG-I was not detectable during ATRA-induced NB4 differentiation (SI Appendix, Fig. S3A). Besides, although TRIM25 was significantly up-regulated (Fig. 3 A and B), the total ubiquitination of the lysate in NB4 cells was not significantly changed during ATRA-induced differentiation (SI Appendix, Fig. S3B). We therefore assumed that there might be another regulatory mechanism that was distinct from antiviral response underlying the interaction between RIG-I and TRIM25 during myeloid differentiation. Coincidentally, when we first cloned RIG-I, we identified another retinoic acid-induced gene B (RIG-B) (1), which encodes a ubiquitin/ISG15-conjugating enzyme (currently defined as ubiquitin/ISG15-conjugating enzyme E2L6, UBE2L6 or UBCH8) (35, 39, 40). These findings led us to speculate that ATRA might activate the ISGylation pathway.
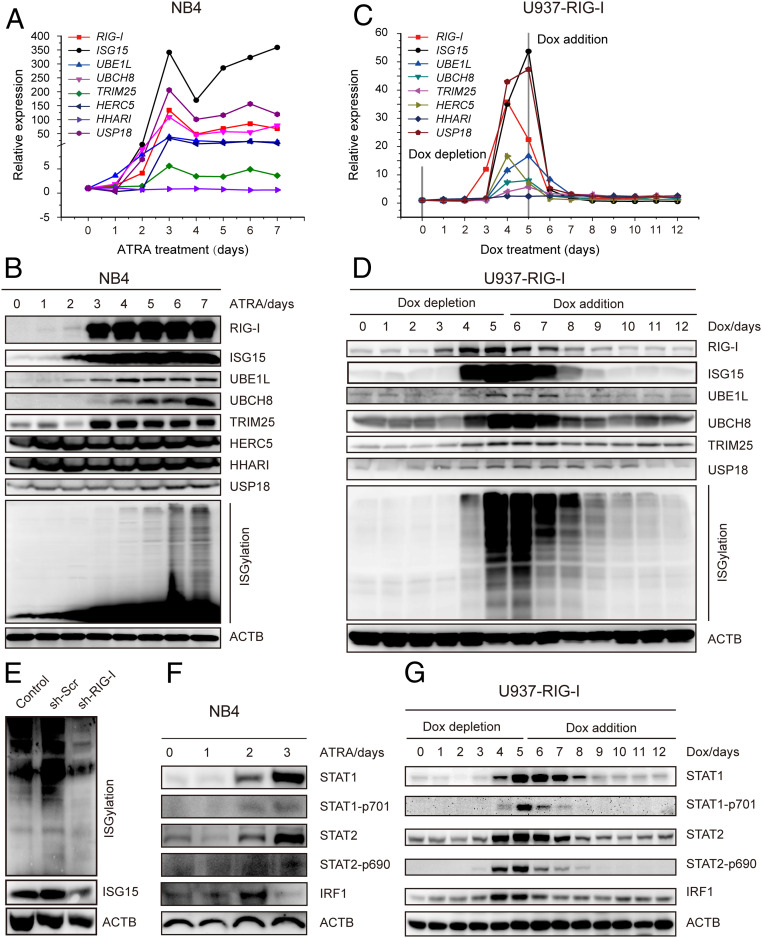
We subsequently investigated the expression status of all genes known to participate in the ISGylation modification pathway, including ISG15 (modifier) (41), ubiquitin-activating enzyme E1-like (UBE1L, as an activating enzyme) (42), UBCH8 (conjugase) (39, 40), TRIM25 (ligase) (6), HECT domain and RCC1-like domain-containing protein 5 (HERC5, as a ligase) (43), human homolog of Drosophila ariadne (HHARI, as a ligase) (44), and ubiquitin specific peptidase 18 (USP18 as a de-ISGylation proteinase) (45). Quantitative RT-qPCR assays showed that all of these ISGylation pathway genes except for HHARI were up-regulated following ATRA induction in NB4 cells (Fig. 5A). Furthermore, the protein levels of the ISGylation pathway except for HHARI and HERC5 increased in parallel (Fig. 5B). In addition, we discovered that the ISGylation pathway genes were also up-regulated in other ATRA responsive cell lines, such as HL-60 and U937, but not in the cell line that is insensitive to ATRA, such as K562 (SI Appendix, Fig. S4 A–F). These data implied that the modifier ISG15, together with the cascade of the ISGylation enzymes UBE1L, UBCH8, USP18, and TRIM25, would be involved in the process of ATRA-induced cell differentiation.
Fig. 5.
Activation of the ISGylation pathway by RIG-I. (A and B) ATRA induced the expression of ISGylation pathway genes. NB4 cells were collected at various time points after the treatment with ATRA. The expression levels of the mRNAs (A) and proteins (B) of the ISGylation pathway genes were evaluated respectively. (C and D) RIG-I promoted the expression of the ISGylation pathway genes. U937-RIG-I cells were collected at various time points after RIG-I induction, and the expression levels of the mRNAs (C) and proteins (D) of ISGylation pathway genes were evaluated respectively. (E) RIG-I knockdown by shRNA down-regulated ISG15 expression and total ISGylation in NB4 cells. (F) ATRA treatment up-regulated and activated STAT1, STAT2, and IRF1 in NB4 cells. (G) RIG-I up-regulated and activated STAT1, STAT2, and IRF1 in U937-RIG-I cells.
Following RIG-I induction in U937-RIG-I cells, the ISGylation pathway genes were significantly up-regulated at mRNA and protein levels, in a manner similar to that in NB4 cells, while the shut-down of RIG-I expression with doxycycline treatment resulted in the decreased expression of the ISGylation pathway genes (Fig. 5 C and D). Additionally, ISGylation, but not ubiquitination, was down-regulated after RIG-I knockdown by short hairpin RNA (shRNA) in NB4 cells (Fig. 5E and SI Appendix, Fig. S3C). These results provided evidence for a critical role played by RIG-I in activating the ISGylation pathway.
According to the Encyclopedia of DNA Elements (ENCODE) database, there are binding motifs of transcription factor STAT1, STAT2, or IRF1 in the promoter regions of the above-mentioned ISGylation pathway genes (SI Appendix, Fig. S5). These transcription factors usually cooperate in regulating the expression of target genes (46, 47). Our results showed that STAT1, STAT2, and IRF1 were all induced in NB4 by ATRA (Fig. 5F). In U937-RIG-I, these transcription factors were also modulated by RIG-I, with their protein levels increased upon RIG-I induction and declined following RIG-I depletion (Fig. 5G). Knockdown of STAT1 down-regulated the ISGylation pathway genes and reduced total ISGylation (SI Appendix, Fig. S6). Of note, the CARD domain of RIG-I induced an increased expression of the ISGylation pathway genes (SI Appendix, Fig. S7). We therefore confirmed a cooperation between RIG-I and STAT/IRF family transcription factors in promoting the ISGylation pathway.
ISGylation Affects Myeloid Differentiation and Maturation.
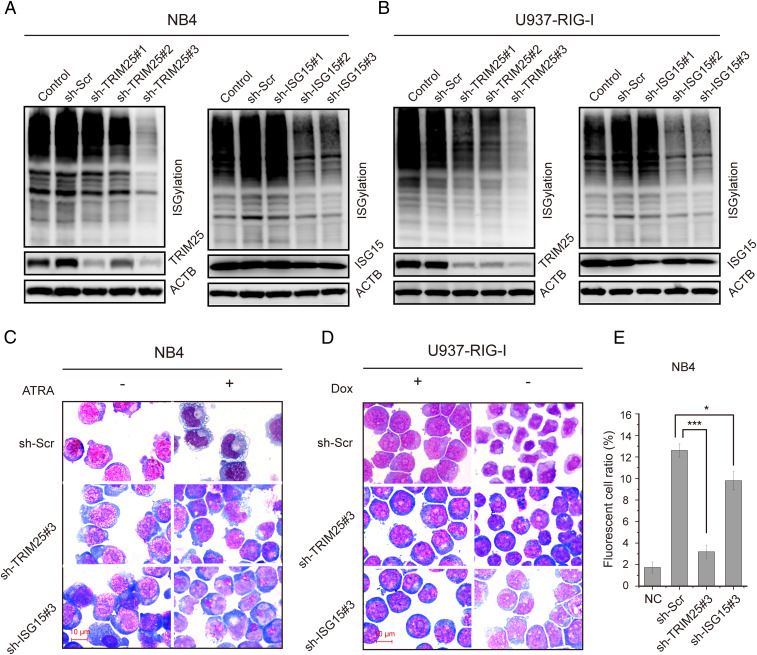
Since we have demonstrated that RIG-I activated the ISGylation pathway genes during ATRA-induced cell differentiation, we then addressed the next issue whether ISGylation could affect the differentiation. Our data showed that knockdown of TRIM25 and ISG15 reduced total ISGylation but not ubiquitination of the cell lysate, respectively (Fig. 6 A and B and SI Appendix, Fig. S8). Morphologically, NB4 cells were capable of differentiating into mature stages of granulocytes (metamyelocytes and band neutrophils) after 4 d of treatment with 1 μM ATRA, whereas NB4 cells with knockdown of TRIM25 or ISG15 were blocked at the promyelocytic stage (Fig. 6C). Knockdown of TRIM25 or ISG15 also affected the differentiation of U937-RIG-I cells following RIG-I induction (Fig. 6D).
Fig. 6.
The effect of ISGylation on myeloid differentiation and maturation. (A) Knockdown of TRIM25 or ISG15 in NB4 cells inhibited the ATRA-induced up-regulation of ISGylation. Immunoblotting was used to detect ISGylation expression in NB4 cells with TRIM25 or ISG15 knockdown after 3 d of treatment with 1 µM ATRA. (B) TRIM25 or ISG15 knockdown in U937-RIG-I cells inhibited ISGylation. Immunoblotting was used to detect ISGylation expression in U937-RIG-I cells with TRIM25 or ISG15 knockdown. (C) Knockdown of TRIM25 or ISG15 in NB4 cells inhibited granulocytic differentiation stained with Wright-Giemsa. (D) TRIM25 or ISG15 knockdown in U937-RIG-I cells inhibited cell differentiation as revealed by morphological examination with Wright-Giemsa staining. (E) Flow cytometric analysis of phagocytosis in NB4 cells with TRIM25 or ISG15 knockdown. NC: control group without ATRA treatment. The experimental groups were treated with 1 µM ATRA for 3 d. *P < 0.05; ***P < 0.001.
Meanwhile, granulocytic maturation surface marker CD11b was significantly reduced in the NB4 cells with knockdown of TRIM25 or ISG15 as compared to the control cells upon ATRA induction (SI Appendix, Fig. S9A). TRIM25 or ISG15 knockdown in U937-RIG-I cells also abrogated CD11b up-regulation with RIG-I induction (SI Appendix, Fig. S9B). Likewise, the knockdown of STAT1 reduced the CD11b expression in NB4 cells (SI Appendix, Fig. S9C). Moreover, the phagocytic capacity of NB4 cells with ATRA induction was also significantly decreased by TRIM25 and ISG15 knockdown, respectively (Fig. 6E). These results suggested that RIG-I contributed to myeloid differentiation and maturation through the modification of TRIM25-mediated protein ISGylation.
Discussion
The treatment of APL with ATRA is considered to be a successful example of cancer differentiation therapy (48). ATRA in combination with an arsenic compound can cure more than 90% of APL patients, and both drugs have been found to target the leukemogenic driver promyelocytic leukemia/retinoic acid receptor alpha (PML-RARα), leading to the differentiation, apoptosis, and senescence of APL cells (49, 50). In an early study, to elucidate the molecular and cellular mechanisms underlying ATRA-induced differentiation therapy, we cloned RIG-I as one of the downstream genes regulated by retinoic signaling in the APL cell line NB4 (1). Since then, the role of RIG-I in innate immunity has been extensively explored, and the protein has been recognized as a receptor for the viral dsRNA in host cells (2, 4). However, the exact mechanisms of RIG-I in cell differentiation and maturation remain obscure. The aim of the present study, therefore, was to elucidate the regulatory function of RIG-I in myeloid differentiation and maturation.
Using RIP-seq technology, we found that RIG-I has potential to bind endogenous RNAs in NB4 cells and identified a significant interaction between RIG-I and TRIM25 mRNA. It has been known that TRIM25 encodes a protein which may function as an E3 ligase for ubiquitin or ISGylation. As a ubiquitin E3 ligase, TRIM25 plays an essential role in RIG-I–associated innate immunity (5). In the present work, although TRIM25 was found significantly up-regulated during ATRA-triggered cell differentiation, no obvious ubiquitination of RIG-I was detectable. Instead, the up-regulated RIG-I protein showed a high ability to bind TRIM25 mRNA via its helicase and CTD domains, thereby enhancing TRIM25 transcript stability. In addition, we conformed that RIG-I could promote the transcriptional expression of TRIM25 through activating STAT1 via CARD (37). Hence, RIG-I might function as a driver of myeloid differentiation via TRIM25 protein expression, which subsequently might function as an E3 ligase of ISG15 for the relevant protein substrates. This possibility was also supported by a previous study showing that ATRA-triggered NB4 cell differentiation exhibited cross-talk with the activation of the IFN pathway (51). Coincidentally, RIG-B might also be involved in ATRA-induced granulocytic differentiation (1). Of note, several studies showed that the ISGylation pathway could play an important role in regulating the differentiation of monocytes (52), erythroid elements (53), and dendritic cells (54), respectively, upon the induction by physiological cytokines or exogenous interleukin-6, erythropoietin, or type I interferons. Intriguingly, a previous work reported that bexarotene, a second generation drug of retinoic acid approved by the US Food and Drug Administration (FDA) for the treatment of cutaneous T cell lymphoma, could inhibit the growth of lung cancer cells by inducing UBE1L and ISGylation (55). In addition, it has been shown that mutations of the ISGylation pathway genes ISG15, UBE1L, and USP18 could result in abnormal phenotypes not only associated to immunity but also related to hematopoiesis (8, 53, 54, 56, 57). Here, through a series of hypothesis-driven experiments, we found that RIG-I cooperated with TRIM25 to activate cellular ISGylation through up-regulating the expression of a whole set of ISGylation pathway genes in cell differentiation. Since the integrity of ISGylation enzymatic machinery was essential for myeloid differentiation and maturation, the RIG-I-TRIM25-ISGylation axis might represent a target for modulation of granulopoiesis.
It is widely recognized that the ligands of RIG-I are highly diverse and include not only virus-specific 5′-ppp-dsRNA but also the noncoding regions of viruses and bacteria. One of the findings of our study was that RIG-I could bind to endogenous RNA molecules without 5′-end triphosphate groups. RNA pull-down and kinetic assays further confirmed the interaction between RIG-I protein and TRIM25 mRNA. Now that RIG-I could bind to TRIM25 mRNA in a way different from that to 5′-ppp-dsRNA, it might also interact with other cellular RNA species. The diversity of RIG-I in binding different RNA ligands and its interaction with relevant protein partners may therefore allow this important regulatory protein to exert a wider range of physiological functions than initially expected.
Taken together, our study showed that RIG-I up-regulates TRIM25 to activate the ISGylation pathway in myeloid differentiation (SI Appendix, Fig. S10). Therefore, our findings may shed light on the mechanisms of RIG-I in regulating the process of myeloid differentiation and the therapeutic response.
Materials and Methods
Reagents.
The key reagents used in this study are shown in SI Appendix, Table S1.
RIP-Seq.
RNA-binding protein immunoprecipitation (RIP) was performed with an anti-RIG-I N-terminus antibody using cell lysate of NB4 cells after ATRA was treated for 3 d. The enriched RNAs and input RNA were analyzed by high-throughput sequencing. Functionally associated genes screened from RIG-I binding protein-coding genes were analyzed by using STRING version 11.0 as described in detail in SI Appendix.
RNA Pull-Down Assay.
The fragments of TRIM25 RNA or 5′ m7G analog capped RNA were transcribed and labeled with biotin. RNA pull-down assays were performed with a Pierce Magnetic RNA-Protein Pull-Down Kit as described previously (58) and detailed in SI Appendix.
Kinetic-Binding Experiments.
The affinity constants for the interaction between the domains of RIG-I and the TRIM25 RNA fragments were measured with Octet Red 96 (Pall ForteBio) equipped with a streptavidin sensor as described previously (33). Octet System Data Analysis 7.1 was used to fit the affinity constants for the interactions (Kon, Koff, KD) using a 1:1 model. The details are described in SI Appendix.
RNA Stability Assay.
Actinomycin D treatment assay was performed for detecting TRIM25 mRNA stability and is detailed in SI Appendix.
Luciferase Assay.
Metridia luciferase assay was performed in NB4 and U937-RIG-I cells. The dual luciferase reporter assay was performed in HEK 293T cells. The details are described in SI Appendix.
Flow Cytometry Analysis.
CD11b expression in NB4 and U937-RIG-I cells was determined with a BD FACS Calibur analyzer (BD Biosciences). Phagocytosis assay was performed as previously reported (59) and detailed in SI Appendix.
Data Availability.
More details and the other associated procedures, including cell culture, plasmids construction, transfection, lentiviral infection, RIP-seq data analysis, RT-qPCR, and immunoblot are described in SI Appendix. The raw data of RIP-seq were deposited in the National Genomics Data Center (https://bigd.big.ac.cn, accession number: PRJCA002547).
Supplementary Material
Acknowledgments
We thank our colleagues from Shanghai Institute of Hematology. This work was supported by Mega-projects of Scientific Research for the 12th Five-Year Plan (2013ZX09303302), National Natural Science Foundation of China (81770182), the National High-tech Research and Development [863] Program of China (2012AA02A505), the Overseas Expertise Introduction Project for Discipline Innovation (111 Project, B17029), Double First-Class Project (WF510162602) of Shanghai Jiao Tong University, Shanghai Collaborative Innovation Program on Regenerative Medicine and Stem Cell Research (2019CXJQ01), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant (20152507), Shanghai Jiao Tong University Tang Scholar Program (2017), SMC-Morningstar Young Scholars Program (2014), and Samuel Waxman Cancer Research Foundation Co-Principal Investigator Program.
Footnotes
The authors declare no competing interest.
Data deposition: The raw data of RIP-seq were deposited in the National Genomics Data Center (https://bigd.big.ac.cn, accession number: PRJCA002547).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918596117/-/DCSupplemental.
References
- 1.Liu T. X. et al., Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood 96, 1496–1504 (2000). [PubMed] [Google Scholar]
- 2.Hornung V. et al., 5′-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M. et al., The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Schlee M., Master sensors of pathogenic RNA–RIG-I like receptors. Immunobiology 218, 1322–1335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gack M. U. et al., TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Zou W., Zhang D. E., The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J. Biol. Chem. 281, 3989–3994 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Zhang D., Zhang D. E., Interferon-stimulated gene 15 and the protein ISGylation system. J. Interferon Cytokine Res. 31, 119–130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X. et al., Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature 517, 89–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han H. G., Moon H. W., Jeon Y. J., ISG15 in cancer: Beyond ubiquitin-like protein. Cancer Lett. 438, 52–62 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Bebington C., Bell S. C., Doherty F. J., Fazleabas A. T., Fleming S. D., Localization of ubiquitin and ubiquitin cross-reactive protein in human and baboon endometrium and decidua during the menstrual cycle and early pregnancy. Biol. Reprod. 60, 920–928 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Yang L. et al., Up-regulation of expression of interferon-stimulated gene 15 in the bovine corpus luteum during early pregnancy. J. Dairy Sci. 93, 1000–1011 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Joyce M. M. et al., Interferon stimulated gene 15 conjugates to endometrial cytosolic proteins and is expressed at the uterine-placental interface throughout pregnancy in sheep. Endocrinology 146, 675–684 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Johnson G. A. et al., Conceptus-uterus interactions in pigs: Endometrial gene expression in response to estrogens and interferons from conceptuses. Soc. Reprod. Fertil. Suppl. 66, 321–332 (2009). [PubMed] [Google Scholar]
- 14.Schanz A. et al., Interferon stimulated gene 15 expression at the human embryo-maternal interface. Arch. Gynecol. Obstet. 290, 783–789 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Rempel L. A. et al., Ubp43 gene expression is required for normal Isg15 expression and fetal development. Reprod. Biol. Endocrinol. 5, 13 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato H. et al., Cell type-specific involvement of RIG-I in antiviral response. Immunity 23, 19–28 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Wang Y. et al., Rig-I-/- mice develop colitis associated with downregulation of G alpha i2. Cell Res. 17, 858–868 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Zhang N. N. et al., RIG-I plays a critical role in negatively regulating granulocytic proliferation. Proc. Natl. Acad. Sci. U.S.A. 105, 10553–10558 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang L. J. et al., RA-inducible gene-I induction augments STAT1 activation to inhibit leukemia cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 108, 1897–1902 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X. Y. et al., RIG-I modulates Src-mediated AKT activation to restrain leukemic stemness. Mol. Cell 53, 407–419 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Schmidt A. et al., 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U.S.A. 106, 12067–12072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang S. Y. et al., 5′-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res. 40, 2724–2733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C., Ranjith-Kumar C. T., Hao L., Kao C. C., Li P., Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 39, 1565–1575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis W. G. et al., The 3′ untranslated regions of influenza genomic sequences are 5′PPP-independent ligands for RIG-I. PLoS One 7, e32661 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnell G., Loo Y. M., Marcotrigiano J., Gale M. Jr., Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog. 8, e1002839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H. X. et al., Rig-I regulates NF-κB activity through binding to Nf-κb1 3′-UTR mRNA. Proc. Natl. Acad. Sci. U.S.A. 110, 6459–6464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang M. et al., Self-Recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173, 906–919.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Szklarczyk D. et al., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellucci M., Agostini F., Masin M., Tartaglia G. G., Predicting protein associations with long noncoding RNAs. Nat. Methods 8, 444–445 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Wang Y. et al., Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 17, 781–787 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu C. et al., The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure 18, 1032–1043 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui S. et al., The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29, 169–179 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Mukherjee K., Korithoski B., Kolaczkowski B., Ancient origins of vertebrate-specific innate antiviral immunity. Mol. Biol. Evol. 31, 140–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yisraeli J. K., Melton D. A., Synthesis of long, capped transcripts in vitro by SP6 and T7 RNA polymerases. Methods Enzymol. 180, 42–50 (1989). [DOI] [PubMed] [Google Scholar]
- 35.Zheng P. Z. et al., Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation/apoptosis of promyelocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 102, 7653–7658 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdelmohsen K., “Modulation of gene expression by RNA binding proteins: mRNA stability and translation” in Binding Protein, Abdelmohsen K., Ed. (InTech, 2012), pp. 123–138. [Google Scholar]
- 37.Hou J. et al., Hepatic RIG-I predicts survival and interferon-α therapeutic response in hepatocellular carcinoma. Cancer Cell 25, 49–63 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Nakasato N. et al., A ubiquitin E3 ligase Efp is up-regulated by interferons and conjugated with ISG15. Biochem. Biophys. Res. Commun. 351, 540–546 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Zhao C. et al., The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-α/β-induced ubiquitin-like protein. Proc. Natl. Acad. Sci. U.S.A. 101, 7578–7582 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim K. I., Giannakopoulos N. V., Virgin H. W., Zhang D. E., Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 24, 9592–9600 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narasimhan J., Potter J. L., Haas A. L., Conjugation of the 15-kDa interferon-induced ubiquitin homolog is distinct from that of ubiquitin. J. Biol. Chem. 271, 324–330 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Pitha-Rowe I., Hassel B. A., Dmitrovsky E., Involvement of UBE1L in ISG15 conjugation during retinoid-induced differentiation of acute promyelocytic leukemia. J. Biol. Chem. 279, 18178–18187 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Dastur A., Beaudenon S., Kelley M., Krug R. M., Huibregtse J. M., Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 281, 4334–4338 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Okumura F., Zou W., Zhang D. E., ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 21, 255–260 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malakhov M. P., Malakhova O. A., Kim K. I., Ritchie K. J., Zhang D. E., UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277, 9976–9981 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Li X., Leung S., Qureshi S., Darnell J. E. Jr., Stark G. R., Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-alpha. J. Biol. Chem. 271, 5790–5794 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Matikainen S., Lehtonen A., Sareneva T., Julkunen I., Regulation of IRF and STAT gene expression by retinoic acid. Leuk. Lymphoma 30, 63–71 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Wang Z. Y., Chen Z., Acute promyelocytic leukemia: From highly fatal to highly curable. Blood 111, 2505–2515 (2008). [DOI] [PubMed] [Google Scholar]
- 49.de Thé H., Pandolfi P. P., Chen Z., Acute promyelocytic leukemia: A paradigm for oncoprotein-targeted cure. Cancer Cell 32, 552–560 (2017). [DOI] [PubMed] [Google Scholar]
- 50.de Thé H., Differentiation therapy revisited. Nat. Rev. Cancer 18, 117–127 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Xiao S. et al., RIG-G as a key mediator of the antiproliferative activity of interferon-related pathways through enhancing p21 and p27 proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 16448–16453 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L. Q. et al., A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol. Cell. Biol. 19, 3029–3038 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maragno A. L. et al., ISG15 modulates development of the erythroid lineage. PLoS One 6, e26068 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cong X. L. et al., Usp18 promotes conventional CD11b+ dendritic cell development. J. Immunol. 188, 4776–4781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng Q. et al., UBE1L causes lung cancer growth suppression by targeting cyclin D1. Mol. Cancer Ther. 7, 3780–3788 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cong X., Yan M., Yin X., Zhang D. E., Hematopoietic cells from Ube1L-deficient mice exhibit an impaired proliferation defect under the stress of bone marrow transplantation. Blood Cells Mol. Dis. 45, 103–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bogunovic D. et al., Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science 337, 1684–1688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang P. et al., The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Sun H. M. et al., PALLD regulates phagocytosis by enabling timely actin polymerization and depolymerization. J. Immunol. 199, 1817–1826 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
More details and the other associated procedures, including cell culture, plasmids construction, transfection, lentiviral infection, RIP-seq data analysis, RT-qPCR, and immunoblot are described in SI Appendix. The raw data of RIP-seq were deposited in the National Genomics Data Center (https://bigd.big.ac.cn, accession number: PRJCA002547).