Significance
TAK1 deficiency has been shown to promote liver cancers; however, the mechanism is unclear. Here we show that TAK1 deficiency promotes RIPK1-mediated inflammation to promote cancer development. Our study highlights the role of RIPK1 as an important driver of inflammatory cytokine production in livers, which includes CCL2 and CCR2+ macrophage infiltration that are prominent in human pathology from liver pathology, and implications for additional human diseases, such as inflammatory bowel disease and Alzheimer’s disease.
Keywords: RIPK1, TAK1, cancer, inflammation, cell death
Abstract
Transforming growth factor β-activated kinase1 (TAK1) encoded by the gene MAP3K7 regulates multiple important downstream effectors involved in immune response, cell death, and carcinogenesis. Hepatocyte-specific deletion of TAK1 in Tak1ΔHEP mice promotes liver fibrosis and hepatocellular carcinoma (HCC) formation. Here, we report that genetic inactivation of RIPK1 kinase using a kinase dead knockin D138N mutation in Tak1ΔHEP mice inhibits the expression of liver tumor biomarkers, liver fibrosis, and HCC formation. Inhibition of RIPK1, however, has no or minimum effect on hepatocyte loss and compensatory proliferation, which are the recognized factors important for liver fibrosis and HCC development. Using single-cell RNA sequencing, we discovered that inhibition of RIPK1 strongly suppresses inflammation induced by hepatocyte-specific loss of TAK1. Activation of RIPK1 promotes the transcription of key proinflammatory cytokines, such as CCL2, and CCR2+ macrophage infiltration. Our study demonstrates the role and mechanism of RIPK1 kinase in promoting inflammation, both cell-autonomously and cell-nonautonomously, in the development of liver fibrosis and HCC, independent of cell death, and compensatory proliferation. We suggest the possibility of inhibiting RIPK1 kinase as a therapeutic strategy for reducing liver fibrosis and HCC development by inhibiting inflammation.
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and a major leading cause for cancer-related deaths worldwide (1). HCC development has been attributed to chronic liver damage induced by hepatocyte death and inflammation, which in turn drives cirrhosis and fibrosis as well as compensatory proliferation (2). The relative contribution of different disregulated processes, including hepatocytic cell death, inflammation, and compensatory proliferation, to HCC development remains to be clarified. Furthermore, the mechanism that drives liver inflammation and methods to control such inflammation to block HCC development are still unclear.
Transforming growth factor β-activated kinase1 (TAK1) is an important regulator of cellular inflammatory pathways. In the signaling pathways of TNF-α and Toll-like receptor (TLR) ligands, TAK1—associated with TAB1, TAB2, and TAB3—is recruited by K63 ubiquitination chain-modified signaling complexes to promote its activation. Activated TAK1 phosphorylates the downstream components IKK complex or p38, JNK, and ERK to mediate NF-κB and MAPK pathway activation (3–6). TAK1 deficiency in hepatocytes and cholangiocytes in mice leads to liver cell death, inflammation, fibrosis, and HCC (7, 8). TAK1 liver deficiency in mice presents similarity to the gene-expression signature of human HCC (9, 10). Thus, TAK1 deficiency in liver provides a good model for studying the mechanism of human HCC.
RIPK1 (receptor interacting protein kinase 1) plays important roles in mediating cell death, including necroptosis and RIPK1-dependent apoptosis (RDA) (11, 12). Recent evidence suggests that RIPK1 kinase may mediate inflammation, independent of cell death (13, 14). While the mechanisms of RIPK1 in mediating necroptosis and RDA have been well-studied, we still know little about the mechanism and pathophysiological significance of RIPK1-mediated inflammation. TAK1 can directly suppress the activation of RIPK1 kinase by phosphorylation, as well as indirectly by promoting the activation of IKKs, which in turn perform inhibitory phosphorylation on RIPK1 (15, 16). Inhibition of TAK1 promotes RDA and necroptosis. Tak1ΔHEP mice (Tak1f/f;Alb-cre mice), where TAK1 is deleted exclusively in hepatocytes using Albumin Cre (Alb-cre), show chronic hepatocellular-specific damage, compensatory proliferation, hepatitis, fibrosis, and spontaneously develop HCC (8). However, since TAK1-deficient hepatocytes have been suggested to undergo Fas-associated death domain (FADD)-dependent apoptosis, independent of RIPK1 (17), if and how RIPK1 kinase might be involved in hepatocytes specific TAK1 deficiency induced HCC is unknown.
Inflammation is important in driving pathology of human diseases. In particular, CCL2, also known as monocyte chemoattractant protein-1 (MCP-1), is a master regulator of monocyte/macrophage function in response to tissue injury, and regulates the production of proinflammatory cytokines, including TNF-α, IL-6, and IL-1β. Elevated levels of CCL2 have been found in various human conditions, from liver pathology to inflammatory bowel disease and Alzheimer’s disease (18–20). Up-regulation of CCL2 in livers has been shown to promote inflammation, fibrosis, and steatosis in metabolic diseases, including diabetes and obesity-mediated insulin resistance. CCL2 knockout mice have been shown to be highly resistant to chronic alcoholic liver injury and steatosis, independent of the NF-κB pathway or its known receptor CCR2 (21). Pharmacological inhibition of CCL2 is being considered for blocking liver macrophage infiltration and steatohepatitis (22). However, the mechanism that regulates the transcriptional expression of CCL2 is still unclear.
In this study, we investigated the mechanism by which RIPK1 kinase mediated liver injury and HCC development using Tak1ΔHEP mice as a model. Our results demonstrate that while genetic inhibition of RIPK1 kinase using D138N knockin mutation has minimum effect on the death of hepatocytes or compensatory proliferation induced by TAK1 deficiency, inhibition of RIPK1 strongly reduces the development of tumor biomarkers and the incidence of HCC development in Tak1ΔHEP mice. We find that RIPK1 kinase primarily promotes the transcription of key inflammatory cytokines/chemokines, such as CCL2 (MCP-1), in TAK1-deficient hepatocytes and macrophage infiltration to modulate the tumor microenvironment, which in turn accelerates liver fibrosis and HCC development. Our study highlights an important but previously unappreciated role and mechanism of RIPK1-mediated inflammation in promoting the development of HCC in both a cell-autonomous and noncell-autonomous manner.
Results
RIPK1 Kinase-Dependent and -Independent Cell Death and Compensatory Proliferation in Tak1ΔHEP Mice.
To investigate the mechanism by which RIPK1 mediates hepatocellular damage, compensatory proliferation, fibrosis, and HCC development, we generated Tak1f/f;Alb-cre/+;Ripk1D138N/D138N (Tak1ΔHEP;Ripk1D138N/D138N) mice by crossing Ripk1D138N/D138N mice with Tak1f/f; Alb-cre/+ (Tak1ΔHEP) mice. As reported previously, Alb-cre drives the loss of TAK1 specifically in hepatocytes of Tak1f/f;Alb-cre mice (8). We found that TAK1 protein levels in hepatocytes of Tak1ΔHEP mice were markedly decreased, which was not affected by RIPK1 D138N mutation in Tak1ΔHEP;Ripk1D138N/D138N mice (SI Appendix, Fig. S1A).
We next characterized the time course of apoptosis using immunostaining of cleaved caspase-3 (CC3) as a biomarker in the livers of Tak1ΔHEP mice. Low levels of CC3 became detectable in the livers of Tak1ΔHEP mice at 4 wk of age, peaked at 6 wk, and remained elevated at 12 wk of age. The apoptotic CC3 levels were reduced and maintained at low levels in Tak1ΔHEP;Ripk1D138N/D138N mice at these ages (SI Appendix, Fig. S1 B and C). The result was further confirmed by immunohistochemistry of CC3 in the mice of 6 wk age (SI Appendix, Fig. S1D). Strong CC3+ signals were detected around the necrotic areas and weak CC3+ signals were detected in hepatocytes in the liver of Tak1ΔHEP mice (SI Appendix, Fig. S1D). These results suggest that TAK1 deficiency promotes hepatocytes to undergo RDA.
Hepatocytic damage can be directly measured by alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in serum (23, 24). ALT and AST levels in the serum of Tak1ΔHEP mice were elevated compared with that of Tak1f/f mice (WT) at 4, 6, and 12 wk. No difference in the levels of ALT and AST was detected between Tak1ΔHEP;Ripk1D138N/D138N mice and Tak1ΔHEP mice at 4 wk of age or that of ALT alone at 6 wk of age (SI Appendix, Fig. S1E). Inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138/D138N mice reduced the serum levels of ALT at 12 wk of age and that of AST at 6 and 12 wk of age (SI Appendix, Fig. S1E). H&E staining also confirmed that liver necrotic damage in Tak1ΔHEP mice peaked at 6 and 12 wk of age, which was reduced upon inhibition of RIPK1 (SI Appendix, Fig. S1 F and G). Thus, inhibition of RIPK1 can reduce necrotic liver damage.
Increased serum bilirubin level is an indication for bile duct damage (7). We detected increased serum bilirubin levels in Tak1ΔHEP mice at 12 wk of age (undetectable before 12 wk). Inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138N/D138N mice significantly reduced the serum levels of bilirubin (SI Appendix, Fig. S1H). Since TAK1 expression is maintained in cholangiocytes in Tak1ΔHEP mice (8), this result suggests that RIPK1 is involved noncell-autonomously in promoting the bile duct damage as a secondary response to chronic hepatocyte damage. Taken together, these results suggest that activation of RIPK1 can mediate apoptosis of TAK1-deficient hepatocytes cell-autonomously as well as noncell-autonomously to promote bile duct damage (SI Appendix, Fig. S1 B, C, and H); however, an additional RIPK1-independent mechanism was also activated to mediate hepatocytic injury in Tak1ΔHEP mice (SI Appendix, Fig. S1E).
We next investigated the effect of RIPK1 kinase on the liver compensatory proliferation. Compensatory proliferation after liver damage has been suggested to be involved in promoting hepatocarcinogenesis (8, 25, 26). Immunohistochemistry of Ki67 was used to measure the rate of compensatory proliferation in livers. The numbers of Ki67+ cells were at the high levels at age of 4 wk when the liver damage was first detected; and inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138N/D138N mice had no effect on the numbers of Ki67+ cells (SI Appendix, Fig. S2 A and B). The compensatory proliferative response in Tak1ΔHEP mice was highest at 4 wk of age; inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138N/D138N mice did not have a significant reducing effect on the numbers of Ki67+ cells in liver at any age analyzed, and even increased it modestly at 6 wk of age (SI Appendix, Fig. S2 A and B). The cellular morphology of Ki67+ cells in livers analyzed suggested that they were hepatocytes (SI Appendix, Fig. S2A). To validate this conclusion, we used a BrdU assay to label the proliferative cells and coimmunostain them with a hepatocyte marker HNF4α. The result demonstrated that BrdU+ cells were indeed hepatocytes that could be coimmunostained with HNF4α and furthermore, inhibition of RIPK1 kinase had no effect on the numbers of Ki67+ hepatocytes (SI Appendix, Fig. S2 C and D).
Compensatory liver regeneration may involve not only hepatocytes but also other cell types in livers (27). We found that the numbers of liver cells positive for biliary cytokeratin-19 (CK19+), a marker for cholangiocytes, were increased in the Tak1ΔHEP mice liver at 4 to 12 wk of age. Inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138N/D138N mice at 4 wk, but not at 6 or 12 wk of age, reduced the number of CK19+ cells (SI Appendix, Fig. S2 E and F), suggesting the function of RIPK1 in the cholangiocyte regeneration at early stage (4 wk) but not later stages.
Taken together, these results suggest that although RIPK1 kinase is partially involved in hepatocytic cell damage and subsequent secondary bile duct injury induced by hepatocytic-specific TAK1 deficiency, inhibition of RIPK1 does not have a strong overall effect on the compensatory proliferation of hepatocytes in Tak1ΔHEP mice.
RIPK1 Kinase Promotes Tumor Development in Tak1ΔHEP Mice.
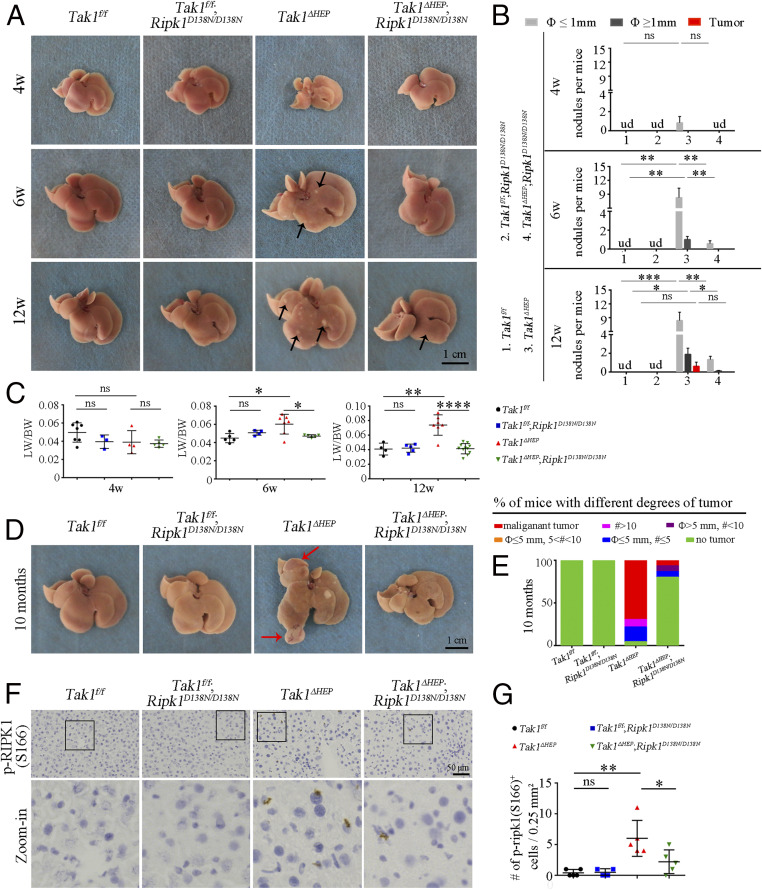
Next, we examined the hepatic phenotypes in the mice at 4, 6, and 12 wk of age. Small but macroscopically visible nodules (diameter smaller than 1 mm) were found in the livers of Tak1ΔHEP mice at age of 4 wk, which were increased in number and size at the age of 6 wk and developed into small visible tumors at 12 wk. Inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138N/D138N mice reduced the development of such nodules and tumors (Fig. 1 A and B), suggesting that RIPK1 D138N mutation can suppress the early-stage hepatocarcinogenesis caused by hepatocytic TAK1 deficiency. Consistently, we also observed the increased liver-to-body weight (LW/BW) ratio in Tak1ΔHEP mice from 6 wk to 12 wk, which is known to occur with liver hepatocarcinogenesis (28–31). RIPK1 D138N mutation also restored the LW/BW ratio to the normal level (Fig. 1 A and C).
Fig. 1.
Genetic inhibition of RIPK1 blocks tumor development in Tak1ΔHEP mice. (A and B) Liver images of indicated genotypes and ages, arrows indicates the nodules (A); the quantification is shown in B (Tak1f/f n = 7 to 9, Tak1f/f;Ripk1D138N/D138N n = 5 to 11, Tak1ΔHEP n = 6 to 12, Tak1ΔHEP;Ripk1D138N/D138N n = 7 to 15 for each age). (C) The ratio of LW/BW of indicated genotypes from different ages; each dot represents one mouse. (D and E) Images of liver tumors (D) (red arrows point to the tumors) and comparison of the percentages of mice with different amount of tumors (E) (the malignant tumor means the tumor with visible vessels) in indicated genotypes of 10-mo-old mice (Tak1f/f n = 21, Tak1f/f;Ripk1D138N/D138N n = 16, Tak1ΔHEP n = 23, Tak1ΔHEP;Ripk1D138N/D138N n = 15). (F and G) Immunohistochemistry (F) and quantification (G) of p-RIPK1 (S166) in liver of indicated genotypes at the age of 6 wk; each dot represents one mouse (magnification of zoom-in in F: 16×). The results are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant; ud, undetectable; Student’s t test was performed.
Tak1ΔHEP mice began to develop liver tumors as early as at 3-mo-old (Fig. 1B) and most Tak1ΔHEP mice displayed obvious liver tumors at 9 to 10 mo of age; in comparison, the tumor burden in the liver of Tak1ΔHEP;Ripk1D138N/D138N mice was considerably lower, while Tak1f/f and Tak1f/f;Ripk1D138N/D138N mice developed no tumor (Fig. 1 D and E). The liver tumor burden in Tak1ΔHEP mice was also demonstrated by the increased ratio of LW/BW, which was restored to normal levels by RIPK1 D138N mutation (SI Appendix, Fig. S3A). The serum ALT and AST levels in 10-mo-old Tak1ΔHEP mice remained elevated compared to that of Tak1f/f mice but lower compared to the Tak1ΔHEP mice at 1 to 3 mo of ages, and were also reduced upon inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138N/D138N mice at 10 mo of age (SI Appendix, Fig. S3B). At the histological level, the liver tumors in Tak1ΔHEP mice exhibited severe dysplasia with an increased nuclear-to-cytoplasmic index, enlarged and hyperchromatic nuclei, and loss of normal liver architecture (SI Appendix, Fig. S3C). The tumors in Tak1ΔHEP mice are highly Ki67+ (SI Appendix, Fig. S3D). RIPK1 activation was confirmed by p-RIPK1 (S166) immunohistochemistry in the liver of Tak1ΔHEP mice. The activation of RIPK1 was inhibited by RIPK1 D138N mutation (Fig. 1 F and G). These results suggest that inhibition of RIPK1 kinase using RIPK1 D138N mutation is able to mitigate the morphologic abnormality, early-stage tumor nodule formation and tumor development in the livers of Tak1ΔHEP;Ripk1D138N/D138N mice.
RIPK1 Kinase Inhibition Reduces the Development of Tumor Biomarkers in Tak1ΔHEP Mice.
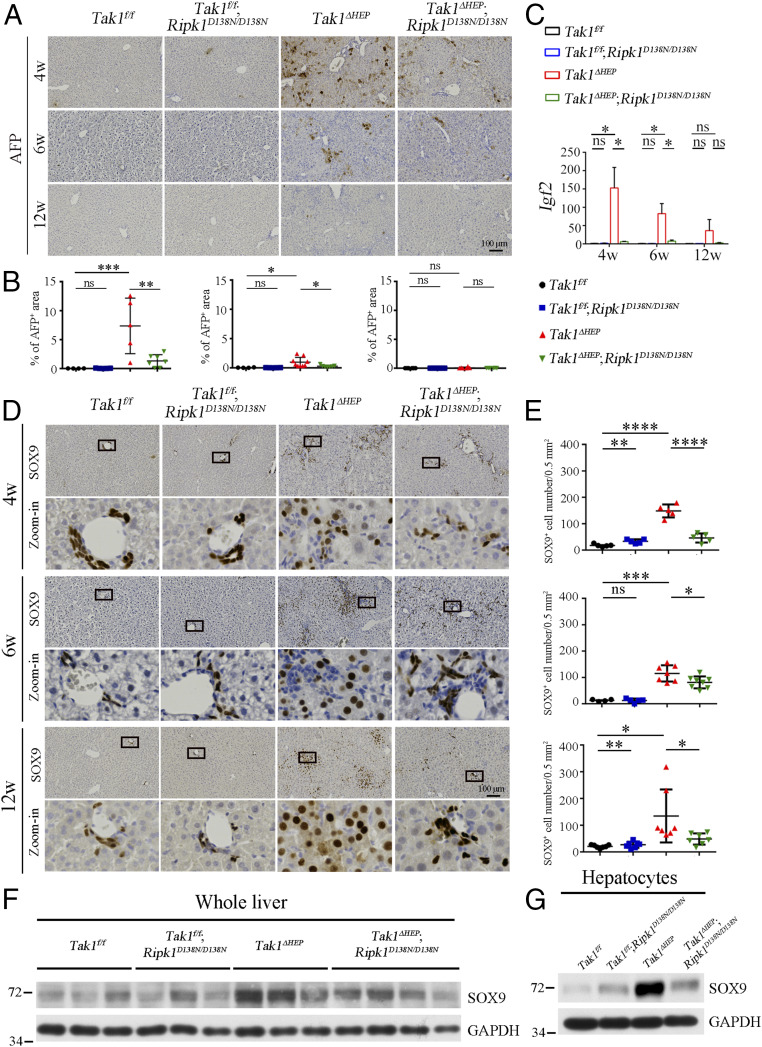
We next investigated the expression of liver tumor biomarkers in these mice. α-Fetoprotein (AFP) is a clinical serum marker for diagnosing HCC in humans (32–34). The levels of AFP were increased and at peak in the liver of Tak1ΔHEP mice at 4 wk of age, but reduced at 6 and 12 wk of age (Fig. 2 A and B). IGF2, another biomarker of liver cancer (35, 36), showed the same pattern (Fig. 2C). Interestingly, inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138N/D138N mice strongly suppressed the increases in levels of both AFP and IGF2 to that comparable with WT at same ages (Fig. 2 A–C).
Fig. 2.
RIPK1 kinase promotes the development of tumor biomarkers in Tak1ΔHEP mice. (A and B) Immunohistochemistry (A) and quantification (B) of AFP in liver of indicated genotypes and ages; each dot represents one mouse. (C) qRT-PCR analysis of mRNA levels of cancer stem cell marker Igf2 in whole liver of indicated genotypes and ages (Tak1f/f n = 5 to 8, Tak1f/f; Ripk1D138N/D138N n = 5 to 9, Tak1ΔHEP n = 8 to 16, Tak1ΔHEP;Ripk1D138N/D138N n = 7 to 11 for each age). (D and E) Immunohistochemistry of SOX9 (D) and quantification the numbers of SOX9+ cells (E) in the liver of indicated genotypes and ages; each dot represents one mouse (magnification of zoom-in in D: 40×). (F and G) Western blotting analysis of the protein level of SOX9 in the whole liver (F) or hepatocytes (G) of indicated genotypes of 12-wk-old mice; each group n = 3 in G. The results are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant; Student’s t test was performed.
The protein SRY [sex determining region Y-box 9 (SOX9)] is a transcription factor that plays a critical role in self-renewal and tumor propagation of liver cancer stem cells with elevated expression levels in solid tumors, such as HCC (37, 38). Interestingly, we found that the levels of SOX9 were up-regulated in the whole liver or hepatocytes of Tak1ΔHEP mice compared to that of Tak1f/f and Tak1f/f;Ripk1D138N/D138N mice, but strongly suppressed by genetic inhibition of RIPK1 kinase in 4-, 6-, and 12-wk-old Tak1ΔHEP;Ripk1D138N/D138N mice (Fig. 2 D–G). Furthermore, the tumors in Tak1ΔHEP mice showed elevated levels of AFP and IGF2 and are highly SOX9+ (SI Appendix, Fig. S3 E–H).
Taken together, these results suggest that the activation of RIPK1 in the hepatocytic TAK1 condition promotes hepatocarcinogenesis as indicated by the elevated expression of multiple liver tumor biomarkers, including AFP, IGF2, and SOX9. Furthermore, since AFP, IGF2, and SOX9 are the biomarkers for liver cancer stem/progenitor cells (37, 39, 40), these results also suggest that RIPK1 kinase is also involved in promoting the proliferation of cancer stem/progenitor cells in the livers of Tak1ΔHEP mice.
RIPK1 Kinase Promotes Liver Fibrosis in Tak1ΔHEP Mice.
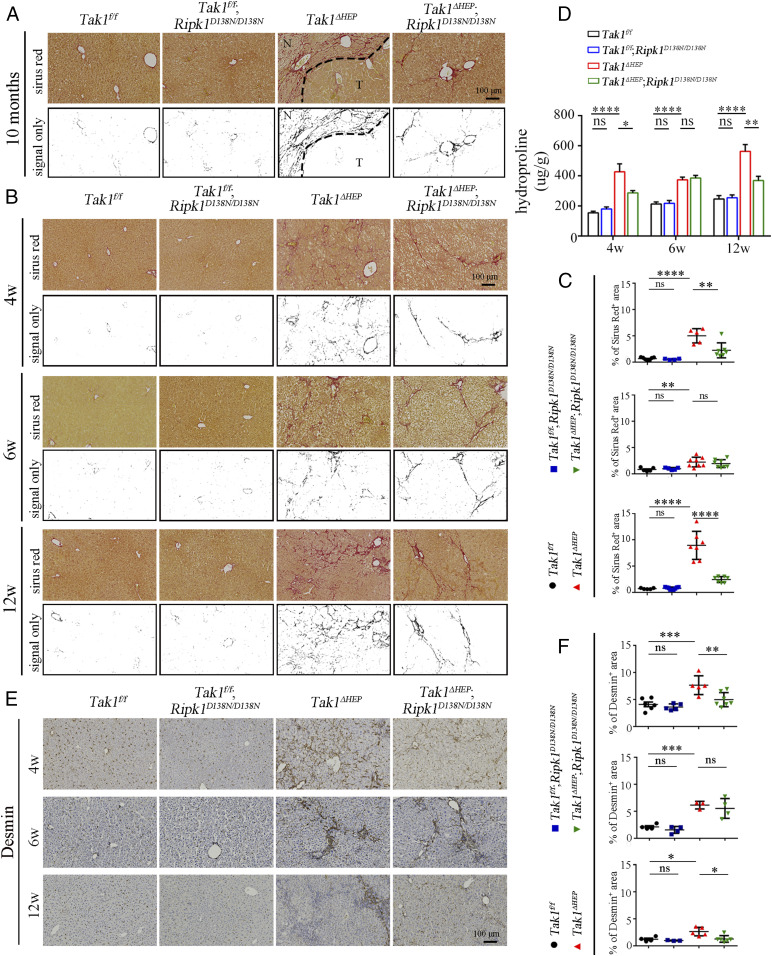
Hepatocytic damage is known to activate hepatic stellate cells (HSC), which then release extracellular matrix proteins, including collagen, to promote the formation of fibrotic scars (41, 42). We noted that in the livers of Tak1ΔHEP mice at 10 mo of age, as well as earlier at 4 and 12 wk of age, the fibrillar collagen deposition displayed chaotic patterns; in comparison, the extent of fibrosis was reduced and became ordered, often forming a “bridge” between the two blood vessels in the livers of Tak1ΔHEP;Ripk1D138N/D138N mice (Fig. 3 A–C). We next used an hydroxyproline assay (total collagen) to further evaluate fibrillar collagen deposition in younger mice. This analysis showed that RIPK1 D138N reduced liver fibrosis of Tak1ΔHEP mice at 4 wk and 12 wk but not for 6-wk-old mice (Fig. 3D). These results suggest that inhibition of RIPK1 was able to reduce liver fibrosis in Tak1ΔHEP mice.
Fig. 3.
RIPK1 kinase promotes liver fibrosis caused by TAK1 deficiency. (A) Sirius red staining evaluates the liver fibrosis at 10-mo-old; the signals only represent the morphology of the fibers (Tak1f/f n = 15, Tak1f/f;Ripk1D138N/D138N n = 11, Tak1ΔHEP n = 23, Tak1ΔHEP;Ripk1D138N/D138N n = 13). “N” means no tumor region and “T” means tumor region. (B and C) Sirius red staining of livers from mice with indicated genotypes at 4, 6, and 12 wk (B) and the percentage of Sirius red-positive area is shown in C; each dot represents one mouse. (D) Quantification of the collagen content in liver tissues from mice with indicated genotypes and ages by Hydroxyproline-assay (Tak1f/f n = 5 to 9, Tak1f/f;Ripk1D138N/D138N n = 5 to 11, Tak1ΔHEP n = 6 to 12, Tak1ΔHEP;Ripk1D138N/D138N n = 6 to 13 for each age). (E and F) Immunohistochemistry of HSC marker Desmin in the livers of indicated genotypes and ages (E). The quantification is shown in F; each dot represents one mouse. The results are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant; Student’s t test was performed.
To explore how RIPK1 kinase is involved in promoting Tak1ΔHEP-induced liver fibrosis, we next analyzed the expression of Desmin, the marker of HSC, in the livers of mice. As shown in Fig. 3 E and F, the numbers of HSC were increased in Tak1ΔHEP mice at the age of 4, 6, and 12 wk compared with that of Tak1f/f mice, which was reduced by RIPK1 D138N mutation at 4 and 12 wk but not at 6 wk. In addition, the mRNA levels of Col1a1, Tgfb1, and Timp1, which are biomarkers for liver fibrosis, were increased in the livers of Tak1ΔHEP mice at 4, 6, and 12 wk, and were reduced by RIPK1 D138N mutation at 6 and 12 wk (SI Appendix, Fig. S4). These results indicate that HSC activation induced by liver injury in Tak1ΔHEP mice was partially regulated by RIPK1 kinase activity.
Evaluating the Impact of TAK1 Hepatocyte Deficiency Using Single-Cell RNA Sequencing.
Single-cell RNA sequencing (scRNA-seq) provides a quantitative method to determine changes in the cell types in different genetic backgrounds (43). We developed a strategy to isolate individual liver cells from mice of four different genotypes (6-wk-old) for transcriptome analysis using droplet microfluidics (10X Genomics) (SI Appendix, Fig. S5A). Overall, on average 90% of reads aligned to genomes, among which 75% mapped to exons, 6.3% mapped to introns, and 5.5% mapped to intergenic regions. We finally retained 34,420 cells for further analysis (12,118 cells for Tak1f/f mice, 11,933 cells for Tak1f/f;Ripk1D138N/D138N mice, 6,377 cells for Tak1ΔHEP, and 3,992 cells for Tak1ΔHEP;Ripk1D138N/D138N mice).
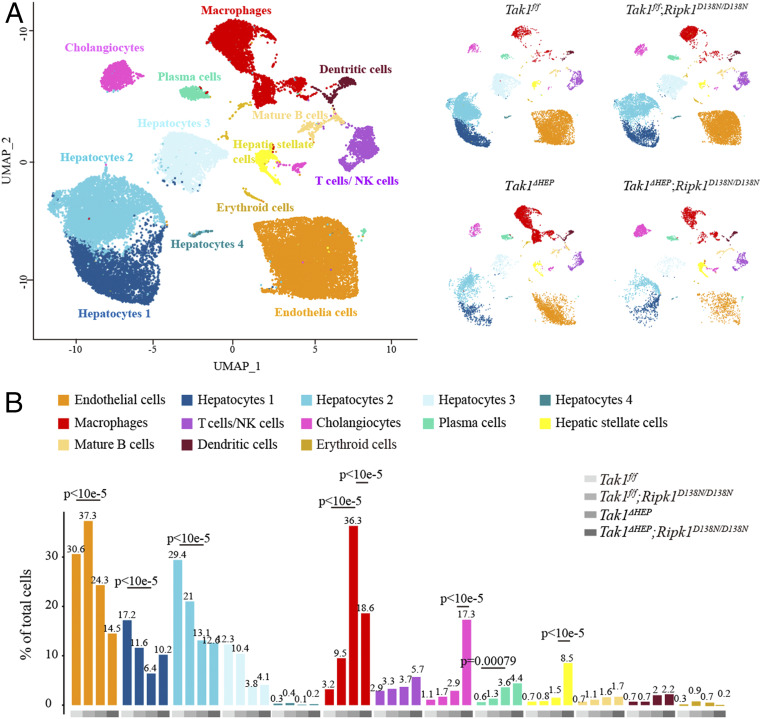
The 34,420 filtered cells were clustered using Seurat 3.0 (44) into 13 distinct clusters (Fig. 4A). We used principal component analysis (PCA) dimension reduction followed by graph-based clustering, which was visualized by uniform manifold approximation and projection (UMAP). Each cluster contained cells from all samples, indicating overall reproducible transcriptional identities among different genotypes. We performed differential expression analysis to identify marker genes that were significantly enriched in particular clusters. We subsequently defined cell clusters to known cell types based on outspoken signature genes identified in the clusters (SI Appendix, Fig. S5B). The enriched biological processes for each cluster are shown in SI Appendix, Fig. S5C.
Fig. 4.
scRNA-seq of livers with TAK1 deficiency and the effect of RIPK1 inhibition. (A) Unbiased clustering of scRNA-seq based on the transcriptional profiles of 34,420 liver cells isolate from mice with indicated genotypes (6-wk-old). (B) Differential proportion analysis of different cell types in indicated genotypes.
We observed major cellular responses between different genotypes. To determine whether changes in the percentage of cell population were greater than expected by chance, we used a permutation-based analysis, as previously described (45). This analysis confirmed the lack of effect of RIPK1 D138N mutation on the reduction in the numbers of hepatocytes in Tak1ΔHEP mice, as the numbers of hepatocytes in Tak1ΔHEP mice and Tak1ΔHEP;Ripk1D138N/D138N mice were both reduced compared to that of Tak1f/f mice and Tak1f/f;Ripk1D138N/D138N mice (Fig. 4 A and B).
The scRNA-seq data suggest that the numbers of cholangiocytes and stellate cells were increased in Tak1ΔHEP;Ripk1D138N/D138N mice compared to that of Tak1ΔHEP mice (Fig. 4 A and B). However, the quantitative in situ measurement of cholangiocytes using its marker CK19 and that of stellate cells using marker Desmin, demonstrated reduction or no change in the numbers of cholangiocytes (SI Appendix, Fig. S2 E and F) and stellate cells (Fig. 3 E and F) in the livers of Tak1ΔHEP;Ripk1D138N/D138N mice compared to that of Tak1ΔHEP mice. This result suggests the apparent increases in cholangiocytes and stellate cells in scRNA-seq data might have occurred during the single-cell isolation process as the inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138N/D138N mice might offer resistance to apoptosis, which made it easier to isolate single cells from the populations of cholangiocytes and stellate cells than that from the livers of Tak1ΔHEP mice.
Inhibition of RIPK1 Blocks Liver Inflammation Induced by TAK1 Deficiency.
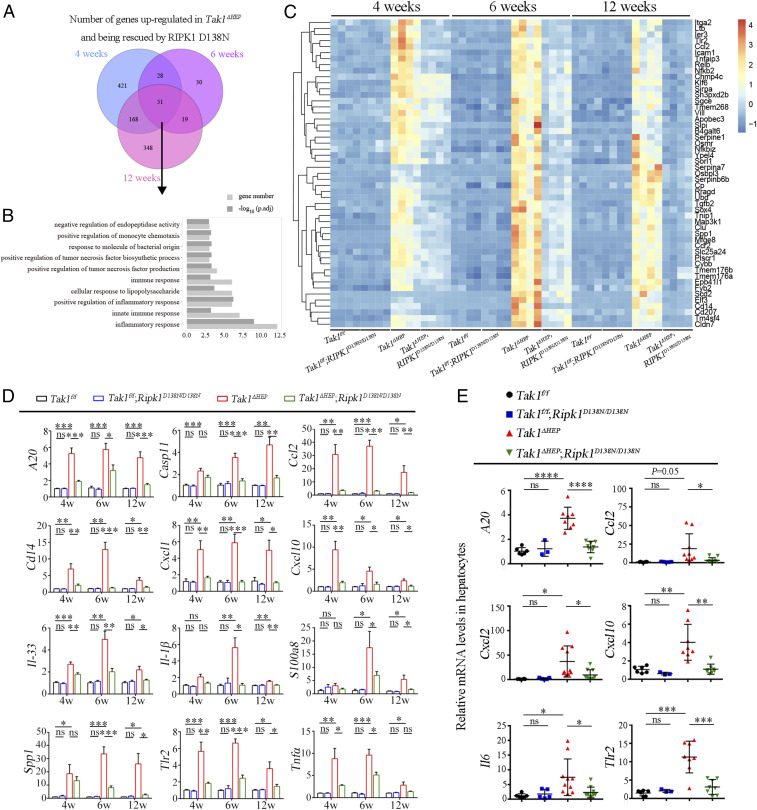
scRNA-seq analysis revealed that inhibition of RIPK1 had a strong effect in reducing the numbers of macrophages (Fig. 4). To control for the potential artifact introduced by single-cell isolation, we next conducted total liver RNA-seq to verify this result. Whole livers were isolated from mice of four genotypes (Tak1f/f, Tak1f/f;Ripk1D138N/D138N, Tak1ΔHEP, and Tak1ΔHEP;Ripk1D138N/D138N) at 4, 6, and 12 wk of age and analyzed by bulk-RNA sequencing (SI Appendix, Fig. S6A). The PCA and Volcano plots showed that transcriptional patterns of Tak1f/f and Tak1f/f;Ripk1D138N/D138N mice were similar, suggesting that RIPK1 D138N mutation in normal conditions does not alter the transcription pattern in the liver (SI Appendix, Fig. S6 B and C). The gene transcriptional patterns from the livers of Tak1ΔHEP mice showed dramatic changes compared to that of Tak1f/f mice; inhibition of RIPK1 with RIPK1 D138N mutation in Tak1ΔHEP;Ripk1D138N/D138N mice restored the expression of a subset of these genes to the normal levels (SI Appendix, Fig. S6 B–F). Gene ontology (GO) analysis showed that hepatocytic TAK1 deficiency lead to significant increase in the expression levels of those genes involved in mediating inflammatory and immune response, and indeed most of them were restored to normal levels by RIPK1 D138N mutation. In addition, the oxidation-reduction process and epoxygenase P450 pathway were significantly down-regulated in Tak1ΔHEP mice; RIPK1 D138N mutation restored them to that of Tak1f/f control levels (SI Appendix, Fig. S6 D–F). Venn diagraming showed that 51 genes were up-regulated in Tak1ΔHEP, which were rescued by RIPK1 D138N mutation at 4, 6, and 12 wk (Fig. 5A). GO analysis of these commonly changed gene expressions showed that most enriched pathways were indeed related to inflammatory response (Fig. 5 B and C).
Fig. 5.
RIPK1 kinase inhibition blocks liver inflammation induced by TAK1 deficiency. (A) Venn diagram of genes up-regulated in Tak1ΔHEP mice and being rescued by RIPK1 D138N mutation at 4, 6, and 12 wk. (B and C) GO analysis (B) and heat map analysis (C) of common genes at 4, 6, and 12 wk in A. (D) qRT-PCR analysis of the mRNA levels of inflammatory cytokines in the livers of mice with indicated genotypes (Tak1f/f n = 4 to 11, Tak1f/f;Ripk1D138N/D138N n = 4 to 12, Tak1ΔHEP n = 4 to 13, Tak1ΔHEP;Ripk1D138N/D138N n = 4 to 12 for each age). (E) The mRNA levels of inflammatory cytokines in hepatocytes of indicated genotypes at 6 wk were measured by qRT-PCR; each dot represents one mouse. The results are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant; Student’s t test was performed.
We further verified the increased RIPK1-dependent inflammatory response in Tak1ΔHEP livers by quantifying the expression levels of important genes involved in mediating inflammatory and immune response using qRT-PCR. This analysis confirmed that the levels of A20, Casp11, Ccl2, Cd14, Il-1β, Il33, Tlr2, Tnfα, Cxcl1, Cxcl10, S100a8, and Spp1 were increased in Tak1ΔHEP mice and restored toward to the normal levels in Tak1ΔHEP;Ripk1D138N/D38N mice (Fig. 5D). Since cytokines such as Il33 and Ccl2 are highly expressed in human HCC (46, 47), these results suggest that RIPK1-mediated inflammatory response may play an important role for HCC development. Thus, we were able to use unbiased total RNA-seq of whole livers, which does not involve isolation of single cells, to confirm the role of RIPK1 in promoting the inflammatory response at the transcriptional levels in Tak1ΔHEP mice.
We next isolated hepatocytes from the livers of Tak1f/f (WT) mice, Tak1f/f;Ripk1D138N/D138N mice, Tak1ΔHEP mice, and Tak1ΔHEP;Ripk1D138N/D138N mice and examined the expression of inflammatory cytokines in hepatocytes by qRT-PCR. Our results demonstrated a robust increase in the levels of proinflammatory cytokine mRNA in Tak1ΔHEP hepatocytes, such as CCL2, which were inhibited in Tak1ΔHEP;Ripk1D138N/D138N hepatocytes (Fig. 5E). This result suggests that the activation of RIPK1 in TAK1-deficient hepatocytes promotes the transcriptional expression of key proinflammatory cytokines, such as CCL2, in cell-autonomous manner.
Hepatocytic TAK1 Deficiency Promotes Liver Infiltration of Inflammatory Cells in RIPK1-Dependent Manner.
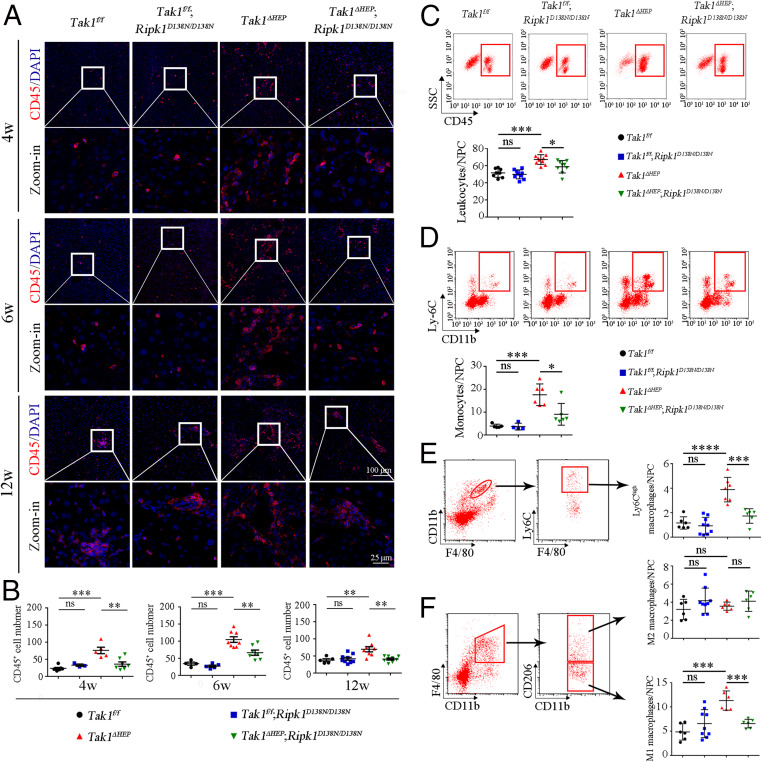
We next analyzed the mechanism by which RIPK1-mediated inflammation might promote HCC in Tak1ΔHEP mice. CD45 is an important marker for infiltrating lymphocytes in the tumor microenvironment (48, 49). We found that TAK1 deficiency in Tak1ΔHEP mice caused the prominent increases in the liver recruitment of CD45+ cells, which was suppressed by RIPK1 D138N mutation (Fig. 6 A and B). This result was further validated by sorting CD45+ cells using fluorescence activated cell sorting (FACS) (Fig. 6C). In addition, we analyzed the levels of CD11b+ myeloid cells, the monocyte-derived macrophages in liver, and Ly6Chigh monocyte-derived macrophages (Ly6Chigh macrophages), which are linked to promoting liver inflammation (50, 51). As shown in Fig. 6D, monocyte-derived macrophages were indeed increased in the liver of Tak1ΔHEP mice, which can be inhibited by RIPK1 D138N mutation. Inflammatory Ly6Chigh macrophages showed the similar pattern (Fig. 6E). We also detected infiltrated monocytes by Gr-1 (Ly6C, Ly6G) immunostaining: Liver-infiltrating Gr-1+ cells in Tak1ΔHEP mice were found at 4 wk of age and increased at 6 wk of age, which was blocked upon inhibition of RIPK1 kinase in Tak1ΔHEP;Ripk1D138N/D138N mice (SI Appendix, Fig. S7 A and B).
Fig. 6.
Activation of RIPK1 upon hepatocyte-specific deletion of TAK1 promotes infiltrating inflammatory macrophages. (A and B) Immunostaining of CD45 for infiltrating immune cells in liver of indicated genotypes and ages (A). The quantification of the number of CD45+ cells is shown in B; each dot represents one mouse. (C–F) FACS analysis of infiltrating leukocytes (CD45+) (C), monocytes-derived macrophages (CD11b+ Ly6C+) (D), Ly6Chigh monocyte-derived macrophages (CD11bhigh F4/80intermediate Ly6Chigh) (E), M1 macrophages (CD11b+ F4/80+ CD206−), and M2 macrophages (CD11b+ F4/80+ CD206+) (F) in the livers of mice with indicated genotypes and ages. Quantifications are shown on the right; each dot represents one mouse. The results are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant; Student’s t test was performed.
Liver macrophages can be divided into M1 (proinflammatory phenotype) and M2 macrophages (antiinflammatory phenotype) based on their cell surface markers (47, 51, 52). Using FACS we sorted M2 macrophages based on the cell surface marker CD11b+ F4/80+ CD206+ and M1 macrophages based on CD11b+ F4/80+ CD206−. Interestingly, while no significant difference was found among four different genotypes for M2 macrophages, the number of M1 macrophages was increased in the livers of Tak1ΔHEP mice, which was corrected by RIPK1 D138N (Fig. 6F). In addition, p65, a key regulator of NF-κB activation, was up-regulated in the livers of Tak1ΔHEP mice, which may be due to the recruitment of p65 high immune cells into the livers of Tak1ΔHEP mice. Up-regulated p65 was inhibited in Tak1ΔHEP;Ripk1D138N/D138N mice at 12 wk of age (SI Appendix, Fig. S7 C and D).
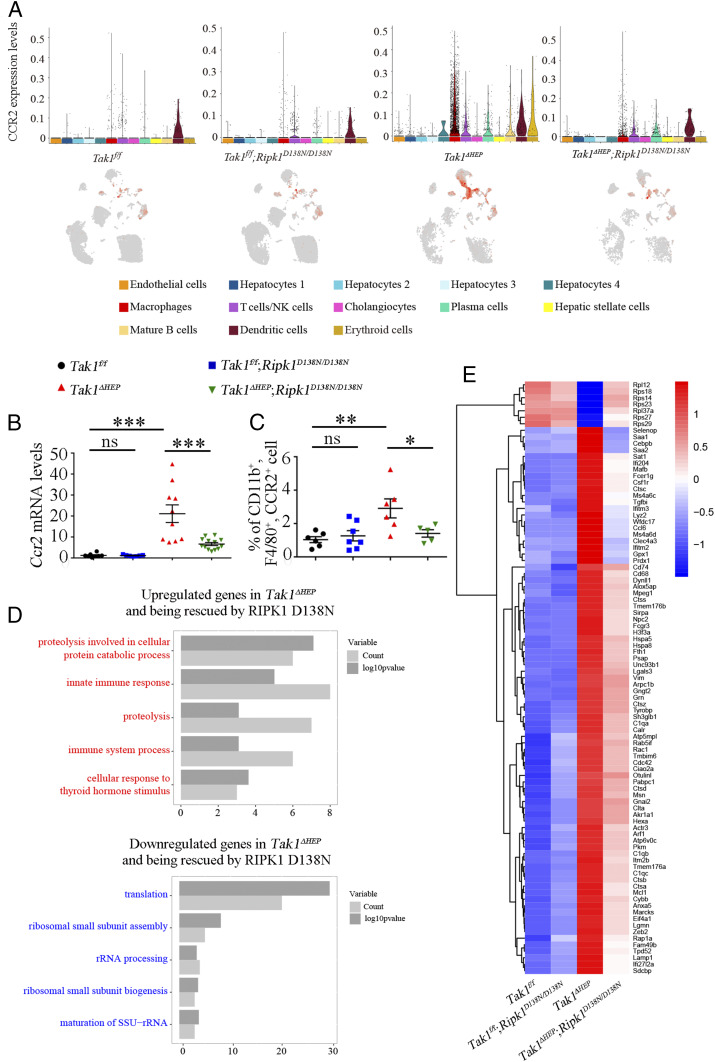
The CCL2/CCR2 axis has been investigated as a potential target to treat HCC by blocking remodeling of tumor microenvironment by inflammatory macrophages (47, 53). We next analyzed the expression of CCR2 across different liver cell types from scRNA-seq data. Interestingly, we found that CCR2 was highly expressed in macrophages in Tak1ΔHEP mice compared to that of Tak1f/f mice, which was blocked upon the inhibition of RIPK1 kinase in Tak1ΔHEP;Ripk1D138N/D138N mice (Fig. 7A). To further validate this result, we measured the mRNA levels of CCR2 in whole liver and also sorted CCR2+ macrophages from hepatic nonparenchymal cells. Both the expression of CCR2 and the number of CCR2+ macrophages were increased in the livers of Tak1ΔHEP mice and reduced upon inhibition of RIPK1 kinase in Tak1ΔHEP;Ripk1D138N/D138N mice (Fig. 7 B and C).
Fig. 7.
CCR2+ macrophages are important for hepatocytic TAK1 deficiency induced RIPK1 kinase-dependent inflammation. (A) Expression of CCR2 gene (inflammatory cell marker) across different liver cell types in mice with indicated genotypes as visualized by Violin plot (Upper) and UMAP plot (Lower). (B) qRT-PCR measures the mRNA levels of Ccr2 in whole livers of mice with indicated genotypes (6-wk-old); each dot represents one mouse. (C) FACS analysis of the percentages of CCR2+ microphages in hepatic nonparenchymal cells of 6-wk-old mice with indicated genotypes; each dot represents one mouse. (D) GO Biological Process reveals enriched pathways dys-regulated by hepatocytic Tak1 loss which was rescued by RIPK1 D138N in CCR2+ cells. (E) Heat map analysis of differentially expressed genes in CCR2+ cells. The results are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant; Student’s t test was performed.
To further explore the role of CCR2+ populations in response to hepatocytic TAK1 deficiency, we analyzed the gene expression of CCR2+ populations in scRNA-seq data among four different genotypes. Compared to that of Tak1f/f and Tak1f/f;Ripk1D138N/D138N mice, Tak1ΔHEP mice showed markedly elevated expression of genes involved in immune response, such as Cd74, Csf1r, Unc93b1, Lyz2, Sirpa, genes involved in positive regulation of phagocytosis (such as Cd68) and proteolysis; and down-regulated translation and ribosomal small subunit assembly pathways. Interestingly, these abnormal gene-expression patterns were corrected by RIPK1 D138N in Tak1ΔHEP;Ripk1D138N/D138N mice (Fig. 7 D and E). These results suggest that CCR2+ inflammatory cells contributed to RIPK1 kinase-dependent inflammation activated by hepatocyte-specific TAK1 deficiency.
These above results demonstrated the essential role of RIPK1 in mediating liver infiltration macrophages, such as CCR2+ macrophages, under a hepatocyte-specific TAK1-deficiency condition. Thus, RIPK1 kinase is critically involved in promoting liver fibrosis and HCC development by mediating inflammation in both a cell-autonomous manner from hepatocytes as well as a noncell-autonomous manner in macrophages, which are known to be closely related to liver fibrosis and HCC development in humans (47, 50, 53–55).
Discussion
In this study, we demonstrate that the activation of RIPK1 promotes RIPK1-dependent inflammation cell-autonomously as well as noncell-autonomously to mediate liver fibrosis and HCC development. Liver fibrosis and HCC development is a dynamic and multifactorial process, including cell death, compensatory proliferation of liver stem cells, and inflammation (56). Inhibition of RIPK1 reduces the liver inflammation and development of HCC induced by TAK1 deficiency with minimum effect on the compensatory proliferation and regeneration of hepatocytes, suggesting RIPK1-mediated inflammation plays a key role in HCC development. HCC development is often associated with liver fibrosis (57). Inhibition of RIPK1 also partially reduces liver fibrosis at 4 and 12 wk but not at 6 wk in Tak1ΔHEP mice. The contribution of additional factors is likely, such as the proliferation of cholangiocytes, which become prominent at 6 wk but reduced at 12 wk, since proliferative cholangiocytes play an important role in promoting liver fibrosis progression (58, 59). Inhibition of RIPK1 reduces the proliferation of cholangiocytes at 4 wk, but not at 6 wk or later (SI Appendix, Fig. S2 E and F). Since TAK1 expression in cholangiocytes is largely maintained in Tak1ΔHEP mice, inhibition of RIPK1 is likely to exert a partial effect cell-nonautonomously on cholangiocyte proliferation. In addition, since Ripk1D138N/D138N mice carry a global RIPK1 inactive mutation, inhibition of RIPK1 also likely inhibits inflammation of myeloid compartment (e.g., macrophages). M1 macrophages have been shown to modulate liver fibrosis by promoting HSC apoptosis (60) and Ly6Clow macrophages promote liver fibrosis resolution (50).
scRNA-seq was used in this study to decipher the functional role of RIPK1 in driving liver tumorigenesis. scRNA-seq has become a powerful tool in cell biology. However, our study highlights the importance of validating scRNA-seq data using additional methods to eliminate potential artifacts introduced during the single-cell isolation process. Our study demonstrates the role of RIPK1 in driving the expression of proinflammatory cytokines, such as CCL2, and the recruitment of CCR2+ macrophages induced by liver damage. Since the TAK1-deficient hepatocytes undergo cell death in both an RIPK1-dependent manner and -independent manner, inhibition of RIPK1 kinase had minimum effect on the survival of TAK1-deficient hepatocytes. Consistently, inhibition of RIPK1 has minimum effect on the compensatory proliferation and regeneration of hepatocytes, which is directly linked with hepatocytic loss. Inhibition of RIPK1 also has only little effect on the proliferation of cholangiocytes or stellate cells, both of which are known as the hallmarks of regenerating livers after hepatic damage. The most striking result of inhibition of RIPK1 in the livers of Tak1ΔHEP mice is the reduction in the numbers of proinflammatory infiltrating macrophages and the levels of proinflammatory cytokines, such as CCL2. Thus, inhibition of a proinflammatory mechanism mediated by RIPK1 kinase is able to reduce liver fibrosis and HCC formation in the presence of long-term continuing liver injury and compensatory proliferation induced by hepatic specific TAK1 deficiency. Our results demonstrate the role of RIPK1-mediated inflammatory response in driving HCC development.
CCL2, secreted by hepatocytes, stellate cells, and Kupffer cells in liver pathology, is known to be a key chemokine that activates monocytes and macrophages. Elevated levels of CCL2 have been implicated in mediating various liver pathology, including acute liver injury, cirrhosis, and tumorigenesis (61), as well as in other human inflammatory and degenerative diseases, such as inflammatory bowel disease and Alzheimer’s disease. Our study demonstrates the role of RIPK1 in driving the transcriptional expression of CCL2 and suggests that elevated levels of CCL2 may serve as a biomarker for the activation of RIPK1 in human clinical studies. The role of RIPK1 in driving the expression of CCL2 also suggests the possibility of developing RIPK1 inhibitor for the treatment of chronic liver injury and steatosis.
Chronic cycles of liver damage and regeneration have been proposed to promote HCC development by inducing chromosomal instability and accumulating cancer-promoting mutations (7). Inhibition of RIPK1 has been shown to reduce hepatocarcinogenesis in Tak1LPC-KO mice (17). Our results highlight the important role of proinflammatory mechanism mediated by RIPK1 in promoting liver fibrosis and HCC formation, largely independent of cell death and compensatory proliferation. In this regard, it is interesting to note that down-regulation of A20, an important suppressor of RIPK1 and the NF-κB pathway, has been associated with invasiveness of human HCCs and reduced disease-free survival (62). Mutant mice with A20 knockout specifically in liver parenchymal cells spontaneously develop chronic liver inflammation but no fibrosis or HCCs (63). In addition, inhibition of hepatocyte apoptosis induced by liver parenchymal cell-specific TAK1 by FADD knockout can also protect against HCC (17). Thus, chronic inflammation mediated by RIPK1 and continuing cycles of liver damage/regeneration may represent two “hits” that are critical for the development of HCC.
The activation of RIPK1 kinase downstream of TNFR1 upon TNF-α stimulation can promote both apoptosis and necroptosis (11, 64). Since liver injury and fibrosis of Tak1ΔHEP mice can be at least partially rescued upon TNFR1 knockout, the activation of RIPK1 in Tak1ΔHEP mice is most likely mediated by the stimulation of TNF-α with its receptor TNFR1. However, since the predominant effect of inhibiting RIPK1 in Tak1ΔHEP mice is the reduction of infiltrating macrophages, our study revealed a previously underappreciated role of RIPK1 in promoting liver inflammation. RIPK1-mediated inflammation might be involved in HSC activation by modulating Ly6Chigh monocyte-derived macrophages, which have been reported to play important role in liver fibrosis progression (50, 54, 55). Our study also suggests the possibility of inhibiting RIPK1 to reduce the development of M1 macrophages. In addition, we show that RIPK1 plays an important role in modulating CCR2+ macrophages, which has been recognized to be involved in HCC development by remodeling of the tumor microenvironment (47, 53). Inflammation plays a central role in modulating the HCC microenvironment and immunotherapy is emerging as a new frontier for HCC treatment (65–67). Our results suggest that the activation of RIPK1 plays a critical role in modulating inflammation and tumor microenvironment.
Our results demonstrate that inhibition of RIPK1 kinase activity leads to reduction in the levels of multiple liver progenitor/cancer stem cell biomarkers, including AFP, IGF2, and SOX9. These results suggest that inhibition of hepatocarcinogenesis upon inhibition of RIPK1 in Tak1ΔHEP;Ripk1D138N/D138N mice is at least in part mediated by the blockage of key transcriptional production of these cancer promoters. Specifically, SOX9 is a transcriptional factor known to be critical for promoting liver regeneration, HCC initiation, and propagation (68, 69). A high level of SOX9 is negatively correlated with the survival of HCC patients (37, 38). In our study, the loss of TAK1 in hepatocyte promotes the expression of SOX9 in hepatocytes in RIPK1 kinase-dependent manner. Thus, targeting RIPK1 might suppress the expression of multiple tumor promoters in hepatocytes to block the development of HCC.
Materials and Methods
Animals.
Tak1f/f mice were kindly provided by Shizuo Akira of Osaka University, Japan (6). Albcre/cre mice were from The Jackson Laboratory (Catalog No. 003574). Tak1f/f;AlbCre/+ mice were crossed with Ripk1D138N/D138N mice to generate Tak1f/f;AlbCre/+;Ripk1D138N/D138N mice. Ripk1D138N/D138N mice were generated via CRISPR/Cas9 system. Briefly, the mixture of single-guide RNA, donor DNA and Cas9 mRNA was microinjected into the zygotes of C57BL/6 mice, single-guide RNA (5′- tgacaaaggtgtgatacaca-3′) targeted exon 4 of Ripk1 to direct Cas9 endonuclease to specifically cut the Ripk1 gene, and induced a double-stranded break, then mutated the aspartate (GAC) to asparagine (AAC) at position 138 of RIPK1 through donor DNA-mediated homology-directed repair (SI Appendix, Fig. S8). The Ripk1D138N/D138N mice have been backcrossed to C57BL/6 background over 10 generations. All animals were maintained in a specific pathogen-free environment, and animal experiments were conducted according to the protocols approved by the Standing Animal Care Committee at Interdisciplinary Research Center of Biology and Chemistry, Shanghai Institute of Organic Chemistry.
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR.
Total RNA was extracted from liver tissues using RNAiso Plus (Takara, 9109) according to the manufacturer’s instructions. The following cDNA synthesis and quantitative real-time PCR were performed as described in SI Appendix, SI Materials and Methods.
Single-Cell Isolation from Liver and Sequencing.
Single cells from liver tissues were isolated by a modified two-step collagenase perfusion method (70). Liver was perfused in anesthetized mouse via the portal vein delivery of ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid tetrasodium salt buffer (37 °C) at 5 mL/min for 5 min, followed by collagenase (Sigma, C5138) buffer (37 °C) at 5 mL/min for 6 to 8 min. The liver was removed and dissociated in suspension buffer and filtered with 40-μM cell strainer (Falcon, 352340). After washing two times with ice-cold Dulbecco’s phosphate-buffered saline (DPBS), single cells were resuspended in DPBS (Gibico, 14190-144) with 1% BSA (Genebase, 9048-46-8). Then cells were stained with PI (stained dead cell) and Hoechst 33342 (Invitrogen, H1399, stained total cell). Cell numbers were counted using cell counting chamber (Germany) and cell viability was assessed by living cell number/total cell number.
Cells were loaded onto 10X Genomics with a targeted cell recovery estimate of 10,000 cells for each sample. A gene-expression library was constructed using Single Cell 3′ Reagent Kit V3 (10X Genomics) by following the manufacturer’s protocol.
For the analysis of 10× scRNA-seq data, see SI Appendix, SI Materials and Methods.
Histology and Immunochemistry.
Livers were harvested from mice with different genotypes and fixed with 4% paraformaldehyde. Fixed tissues were embedded with Milestone Cryoembeeding Compound (Milestone, 51420, for cryostat sections) or paraffin (for paraffin sections). Fluorescent images were collected by the Leica TSC SP8 confocal microscopy system using a 20× or 40× objective. The signals of immunochemistry were detected using SignalStain DAB Substrate Kit (CST, 8059).
The details for immunostaining and immunochemistry can be found in SI Appendix, SI Materials and Methods.
Data Availability.
The raw data files of sequencing experiments have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus. The accession number is GEO: GSE148859 (71).
Supplementary Material
Acknowledgments
The authors thank Professor Shizuo Akira of Osaka University for kindly providing Tak1f/f mice. This work was supported in part by grants from the National Key Research ad Development Program of China (2016YFA0501900) and the China National Natural Science Foundation (31530041, 91849204, 21837004, and 91849109) (to N.L.).
Footnotes
Competing interest statement: J.Y. is a consultant for Denali Therapeutics.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE148859).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005353117/-/DCSupplemental.
References
- 1.Forner A., Reig M., Bruix J., Hepatocellular carcinoma. Lancet 391, 1301–1314 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Weber A., Boege Y., Reisinger F., Heikenwälder M., Chronic liver inflammation and hepatocellular carcinoma: Persistence matters. Swiss Med. Wkly. 141, w13197 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Adhikari A., Xu M., Chen Z. J., Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26, 3214–3226 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Ajibade A. A., Wang H. Y., Wang R. F., Cell type-specific function of TAK1 in innate immune signaling. Trends Immunol. 34, 307–316 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Ninomiya-Tsuji J., et al. , The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398, 252–256 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Sato S., et al. , Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6, 1087–1095 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Bettermann K., et al. , TAK1 suppresses a NEMO-dependent but NF-kappaB-independent pathway to liver cancer. Cancer Cell 17, 481–496 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Inokuchi S., et al. , Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 844–849 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dow M., et al. , Integrative genomic analysis of mouse and human hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 115, E9879–E9888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friemel J., et al. , Characterization of HCC mouse models: Towards an etiology-oriented subtyping approach. Mol. Cancer Res. 17, 1493–1502 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Yuan J., Kroemer G., Alternative cell death mechanisms in development and beyond. Genes Dev. 24, 2592–2602 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christofferson D. E., Li Y., Yuan J., Control of life-or-death decisions by RIP1 kinase. Annu. Rev. Physiol. 76, 129–150 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Tao P., et al. , A dominant autoinflammatory disease caused by non-cleavable variants of RIPK1. Nature 577, 109–114 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Ofengeim D., et al. , RIPK1 mediates a disease-associated microglial response in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 114, E8788–E8797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dondelinger Y., et al. , Serine 25 phosphorylation inhibits RIPK1 kinase-dependent cell death in models of infection and inflammation. Nat. Commun. 10, 1729 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geng J., et al. , Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat. Commun. 8, 359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishna-Subramanian S., et al. , RIPK1 and death receptor signaling drive biliary damage and early liver tumorigenesis in mice with chronic hepatobiliary injury. Cell Death Differ. 26, 2710–2726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y., Beller D. I., Frendl G., Graves D. T., Monocyte chemoattractant protein-1 regulates adhesion molecule expression and cytokine production in human monocytes. J. Immunol. 148, 2423–2428 (1992). [PubMed] [Google Scholar]
- 19.Kmieć Z., Cytokines in inflammatory bowel disease. Arch. Immunol. Ther. Exp. (Warsz.) 46, 143–155 (1998). [PubMed] [Google Scholar]
- 20.Morgan A. R. et al.; NIMA Consortium; Annex: NIMA–Wellcome Trust Consortium for Neuroimmunology of Mood Disorders and Alzheimer’s Disease , Inflammatory biomarkers in Alzheimer’s disease plasma. Alzheimers Dement. 15, 776–787 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandrekar P., Ambade A., Lim A., Szabo G., Catalano D., An essential role for monocyte chemoattractant protein-1 in alcoholic liver injury: Regulation of proinflammatory cytokines and hepatic steatosis in mice. Hepatology 54, 2185–2197 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baeck C., et al. , Pharmacological inhibition of the chemokine CCL2 (MCP-1) diminishes liver macrophage infiltration and steatohepatitis in chronic hepatic injury. Gut 61, 416–426 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Kim H. C., et al. , Normal serum aminotransferase concentration and risk of mortality from liver diseases: Prospective cohort study. BMJ 328, 983 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee T. H., Kim W. R., Benson J. T., Therneau T. M., Melton L. J. 3rd, Serum aminotransferase activity and mortality risk in a United States community. Hepatology 47, 880–887 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Maeda S., Kamata H., Luo J. L., Leffert H., Karin M., IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 121, 977–990 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Yang L., et al. , Transforming growth factor-beta signaling in hepatocytes promotes hepatic fibrosis and carcinogenesis in mice with hepatocyte-specific deletion of TAK1. Gastroenterology 144, 1042–1054.e4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raven A., et al. , Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camargo F. D., et al. , YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Dong J., et al. , Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu L., et al. , Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. U.S.A. 107, 1437–1442 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao B., Lei Q. Y., Guan K. L., The hippo-YAP pathway: New connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 20, 638–646 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aoyagi Y., et al. , The fucosylation index of alpha-fetoprotein and its usefulness in the early diagnosis of hepatocellular carcinoma. Cancer 61, 769–774 (1988). [DOI] [PubMed] [Google Scholar]
- 33.Kelsten M. L., Chan D. W., Bruzek D. J., Rock R. C., Monitoring hepatocellular carcinoma by using a monoclonal immunoenzymometric assay for alpha-fetoprotein. Clin. Chem. 34, 76–81 (1988). [PubMed] [Google Scholar]
- 34.McMahon B. J., et al. , A comprehensive programme to reduce the incidence of hepatitis B virus infection and its sequelae in Alaskan natives. Lancet 2, 1134–1136 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Livingstone C., IGF2 and cancer. Endocr. Relat. Cancer 20, R321–R339 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Brouwer-Visser J., Huang G. S., IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 26, 371–377 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Liu C., et al. , Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. Hepatology 64, 117–129 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Guo X., et al. , Expression features of SOX9 associate with tumor progression and poor prognosis of hepatocellular carcinoma. Diagn. Pathol. 7, 44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Quetglas I., et al. , IGF2 is up-regulated by epigenetic mechanisms in hepatocellular carcinomas and is an actionable oncogene product in experimental models. Gastroenterology 151, 1192–1205 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Zhu M., et al. , HBx drives alpha fetoprotein expression to promote initiation of liver cancer stem cells through activating PI3K/AKT signal pathway. Int. J. Cancer 140, 1346–1355 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Bataller R., Brenner D. A., Liver fibrosis. J. Clin. Invest. 115, 209–218 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman S. L., Mechanisms of hepatic fibrogenesis. Gastroenterology 134, 1655–1669 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halpern K. B., et al. , Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farbehi N., et al. , Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife 8, e43882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang P., et al. , Detection of interleukin-33 in serum and carcinoma tissue from patients with hepatocellular carcinoma and its clinical implications. J. Int. Med. Res. 40, 1654–1661 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Li X., et al. , Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut 66, 157–167 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Hermiston M. L., Xu Z., Weiss A., CD45: A critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 21, 107–137 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Shahar E., Gorodetsky R., Aizenshtein E., Lalush L., Pitcovski J., Modulating the innate immune activity in murine tumor microenvironment by a combination of inducer molecules attached to microparticles. Cancer Immunol. Immunother. 64, 1137–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramachandran P., et al. , Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc. Natl. Acad. Sci. U.S.A. 109, E3186–E3195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tacke F., Zimmermann H. W., Macrophage heterogeneity in liver injury and fibrosis. J. Hepatol. 60, 1090–1096 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Tang P. M.-K., Nikolic-Paterson D. J., Lan H.-Y., Macrophages: Versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 15, 144–158 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Eggert T., et al. , Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell 30, 533–547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baeck C., et al. , Pharmacological inhibition of the chemokine C-C motif chemokine ligand 2 (monocyte chemoattractant protein 1) accelerates liver fibrosis regression by suppressing Ly-6C(+) macrophage infiltration in mice. Hepatology 59, 1060–1072 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Karlmark K. R., et al. , Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 50, 261–274 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Stauffer J. K., Scarzello A. J., Jiang Q., Wiltrout R. H., Chronic inflammation, immune escape, and oncogenesis in the liver: A unique neighborhood for novel intersections. Hepatology 56, 1567–1574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakurai T., Kudo M., Molecular link between liver fibrosis and hepatocellular carcinoma. Liver Cancer 2, 365–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X., et al. , Osteopontin induces ductular reaction contributing to liver fibrosis. Gut 63, 1805–1818 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Glaser S. S., Gaudio E., Miller T., Alvaro D., Alpini G., Cholangiocyte proliferation and liver fibrosis. Expert Rev. Mol. Med. 11, e7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma P. F., et al. , Cytotherapy with M1-polarized macrophages ameliorates liver fibrosis by modulating immune microenvironment in mice. J. Hepatol. 67, 770–779 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Sahin H., Trautwein C., Wasmuth H. E., Functional role of chemokines in liver disease models. Nat. Rev. Gastroenterol. Hepatol. 7, 682–690 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Yi P. S., et al. , Emerging role of zinc finger protein A20 as a suppressor of hepatocellular carcinoma. J. Cell. Physiol. 234, 21479–21484 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Catrysse L., et al. , A20 prevents chronic liver inflammation and cancer by protecting hepatocytes from death. Cell Death Dis. 7, e2250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanden Berghe T., Kaiser W. J., Bertrand M. J., Vandenabeele P., Molecular crosstalk between apoptosis, necroptosis, and survival signaling. Mol. Cell. Oncol. 2, e975093 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J., Gingold J. A., Su X., Immunomodulatory TGF-β signaling in hepatocellular carcinoma. Trends Mol. Med. 25, 1010–1023 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Hou J., Zhang H., Sun B., Karin M., The immunobiology of hepatocellular carcinoma in humans and mice: Basic concepts and therapeutic implications. J. Hepatol. (2019). [DOI] [PubMed] [Google Scholar]
- 67.Lu C., et al. , Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: Challenges and opportunities. Mol. Cancer 18, 130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang Y., et al. , β-Catenin deficiency in hepatocytes aggravates hepatocarcinogenesis driven by oncogenic β-catenin and MET. Hepatology 67, 1807–1822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song C. Q., et al. , Genome-wide CRISPR screen identifies regulators of mitogen-activated protein kinase as suppressors of liver tumors in mice. Gastroenterology 152, 1161–1173.e1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu J., et al. , Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science 343, 416–419 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Tan S., Zhao J., Liu N., Yuan J., Hepatocyte-specific TAK1 deficiency drives RIPK1 kinase-dependent inflammation to promote liver fibrosis and hepatocellular carcinoma. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148859. Deposited 27 April 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data files of sequencing experiments have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus. The accession number is GEO: GSE148859 (71).