Significance
The significance of this work lies in the application of innovative analytical, sequencing, and computational tools to discover a novel epigenetic regulatory mechanism. Here, we discovered that phosphorothioate (PT) DNA modifications are maintained at a constant density in a genome in part by DNA shape-driven target site selection. While structurally similar GAAC, GTTC, and GATC motifs are all modified by PT-catalyzing proteins, methylation of G6mATC by Dam methyltransferase blocks PT modification of GATC and shifts PTs to the other two sites in Salmonella enterica, maintaining a constant number of PTs in the genome.
Keywords: epigenetics, DNA modification, ChIP-seq, DNA target selection, restriction-modification
Abstract
Phosphorothioate (PT) DNA modifications—in which a nonbonding phosphate oxygen is replaced with sulfur—represent a widespread, horizontally transferred epigenetic system in prokaryotes and have a highly unusual property of occupying only a small fraction of available consensus sequences in a genome. Using Salmonella enterica as a model, we asked a question of fundamental importance: How do the PT-modifying DndA-E proteins select their GPSAAC/GPSTTC targets? Here, we applied innovative analytical, sequencing, and computational tools to discover a novel behavior for DNA-binding proteins: The Dnd proteins are “parked” at the G6mATC Dam methyltransferase consensus sequence instead of the expected GAAC/GTTC motif, with removal of the 6mA permitting extensive PT modification of GATC sites. This shift in modification sites further revealed a surprising constancy in the density of PT modifications across the genome. Computational analysis showed that GAAC, GTTC, and GATC share common features of DNA shape, which suggests that PT epigenetics are regulated in a density-dependent manner partly by DNA shape-driven target selection in the genome.
The discovery of DNA modifications and their functions in restriction-modification (R-M) paralleled the discovery of DNA function in gene expression (1), with a fast-forward to the more recent concept of the epigenetic regulation of gene expression by DNA modifications (2). Viewed from either perspective, we now recognize a diversity of microbial DNA modifications that do not change the genetic code but do regulate DNA physiology. Classical R-M systems pair a sequence-specific methyltransferase that establishes “self” and a cognate restriction endonuclease that cleaves unmodified “non-self” DNA (3). Included here are phosphorothioate (PT) (4) and 7-deazaguanine (5) modifications, with the latter (6) and likely others shared by DNA-based bacteriophage coevolving with the bacteria (7). This diversity of modification chemistry is accompanied by a diversity of functions. The bridge between R-M and epigenetics was crossed when the initial discovery of bacteriophage restriction by 2′-deoxyadenosine methyltransferase (Dam) (8), which N6-methylates A in GATC motifs, led to the realization that Dam lacked a partner restriction endonuclease as a so-called “orphan” methyltransferase. It is now known that Dam and G6mATC play important epigenetic roles in chromosome replication, DNA mismatch repair, and gene regulation in γ-proteobacteria, including Escherichia coli and Salmonella enterica (9–11).
Here we explore the impact of cooccurring Dam-mediated 6mA and PT modifications in bacteria. Originally developed by Eckstein and coworkers (12, 13) as synthetic nuclease-resistant oligodeoxynucleotide modifications, PTs were later rediscovered as natural DNA modifications in a wide range of bacteria and archaea (14–16). The PT modification genes dndABCDE often function as part of an R-M system, together with restriction genes dndFGHI in bacteria or pbeABCD in archaea (16–19). However, PTs also appear to perform other epigenetic functions, with about half of PT systems lacking obvious restriction genes (20) and PTs endowing cells with multiple characteristics, including regulation of gene expression (20, 21) and redox homeostasis (20).
PTs differ from classical methylation-based epigenetic and R-M systems in several ways. First, redox-sensitive PTs are subject to damage and subsequent turnover (22). More importantly, while PTs typically occur in 3- to 4-nucleotides (nt) consensus sequences, such as GPSAAC/GPSTTC in E. coli B7A and S. enterica serovar Cerro 87, only about 10% of the consensus sites are modified in an organism, and some bistranded PT modification consensus sites are modified on only one strand (23, 24). This partial modification behavior raises questions about how both the PT modification and restriction proteins select their genomic targets. With many bacteria possessing multiple DNA modification systems (25, 26), questions arise about the consequences of coexisting PTs and classical R-M machinery. For example, Eckstein and coworkers showed that PTs can substitute for methyl-based DNA modifications with resistance to cognate restriction enzymes in vitro (12, 13). Similarly, we previously analyzed the effects of coexpression of Dnd proteins from Hahella chejuensis KCTC2396, which lacks Dam, in an E. coli K12 strain possessing Dam. This artificial hybrid system revealed that PT and 6mA could coexist in GPS6mA motifs since the modification proteins shared the same GATC consensus sequence (27). Moreover, G6mATC was found to be resistant to cleavage by the H. chejuensis KCTC2396 DndFGH restriction system (27). While biochemically interesting, these results left unanswered the question of how PT modification proteins choose their DNA targets and why not all consensus sequences are not modified.
Here we addressed this problem by focusing on an S. enterica strain that was known to naturally possess Dnd proteins that inserted PTs at GPSAAC/GPSTTC motifs and Dam that synthesized G6mATC. Our studies of this epigenetic competition revealed an unexpected DNA target site plasticity for Dnd proteins and an unusual density-dependent regulation of PTs across bacterial genomes. These results point to critical factors other than DNA sequence in the target selection and restriction by the PT modification system.
Results
DndCDE Proteins Preferentially Bind to Dam GATC Sites, Not the Established GAAC/GTTC Consensus.
We initiated these studies with an analysis of Dnd protein target selection using chromatin immunoprecipitation sequencing (ChIP-seq). To this end, S. enterica 87 ∆dnd mutants were engineered with FLAG-tagged Dnd proteins as illustrated in SI Appendix, Fig. S1A; all strains are detailed in SI Appendix, Tables S1 and S2. In one case, a ∆dndC mutant was transfected with FLAG-tagged DndC. To ensure equal expression of all three essential PT modification proteins (DndCDE), six other constructs were engineered using a ∆dndBCDE mutant or ∆dndBCDEFGH mutant transfected with one of three expression vectors containing the FLAG-tagged protein of interest along with the other two native Dnd proteins (SI Appendix, Fig. S1A). Control strains were also constructed in which the FLAG tag was missing or the FLAG tag was not fused to the Dnd proteins (SI Appendix, Fig. S1A and Table S2). Two members of the five-gene Dnd cluster were not considered here: DndB is a transcriptional regulator not involved in PT biochemistry and DndA is a cysteine desulfurase replaced by an endogenous enzyme in S. enterica. Functional validation of the seven expression vectors and FLAG-tagged Dnd proteins was performed by mass spectrometric quantification of PT dinucleotides in DNA from these S. enterica strains, which revealed PT levels ranging from 225 to 555 per 106 nt of GPSA and GPST (SI Appendix, Fig. S1B). This compares favorably with PT levels in genomic DNA from the wild-type (WT) S. enterica strain at 362 ± 9 and 370 ± 11 per 106 nt, respectively (15).
These constructs were then used in ChIP-seq studies outlined in SI Appendix, Fig. S2. We performed ChIP experiments with three biological replicates of the YF11 FLAG-DndC strain of the S. enterica 87∆dndC mutant, single analyses for each of three different WXL1 strains with a FLAG tag fused to DndC, DndD, or DndE in the S. enterica 87∆dndBCDE mutant, and single analyses for each of three different XTG103 strains with a FLAG tag fused to DndC, DndD, or DndE in the S. enterica 87∆dndBCDEFGH mutant. We also included negative controls lacking anti-FLAG antibody (“mock”) and strains either lacking the FLAG tag or in which the FLAG tag was not fused to the Dnd protein (SI Appendix, Fig. S1A and Table S2).
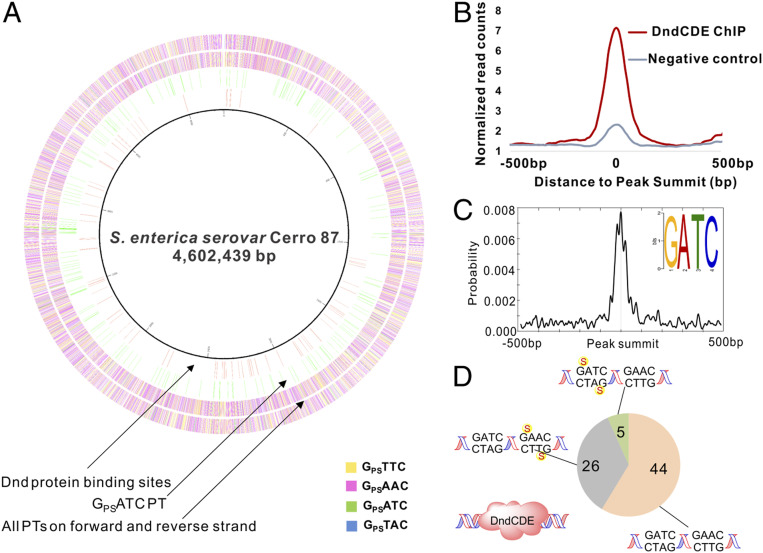
Analysis of the ChIP-seq data for these strains revealed multiple consistent Dnd protein binding sites in the nine strains analyzed (Dataset S1), with 75 detected in the same genomic regions in the nine FLAG-tagged Dnd strains, as depicted in the inner circle of the circos plot of the S. enterica genome in Fig. 1A. The significance of the read pile-ups in these 75 Dnd protein binding regions was based on peak detection criteria (window statistic ≥ 5, P ≤ e−6), with threefold higher coverage of the read counts at each region in ChIP samples compared to negative controls, as shown in Fig. 1B, and examples of the reproducible patterns apparent for the nine FLAG-tagged Dnd experiments in two genomic regions shown in SI Appendix, Fig. S3. Surprisingly, an alignment of the sequences in these 75 regions predicted a GATC consensus motif and not the expected GAAC/GTTC consensus for PT modifications previously established in S. enterica (Fig. 1C) (15, 23). SI Appendix, Fig. S4 shows the pronounced density of read counts at the GATC centers of the Dnd-bound regions. Additionally, there were 343 GATC sites located within 200 base pairs (bp) of the centers of the 75 Dnd-bound regions.
Fig. 1.
Sites of Dnd protein binding and PT modification in S. enterica. (A) Inner to outer: the Circos plot shows the genomic locations of DndC binding sites determined by ChIP-seq (circle 1, pink), PTs at GPSATC sites mapped by TdT-seq (circle 2, green), and all PT sites in the YF11 FLAG-DndC strain mapped by TdT-seq (circles 3 and 4 for forward and reverse strands, respectively). (B) The average normalized read coverage (Materials and Methods) in peaks for ChIP-seq samples is threefold higher than negative controls, validating the ChIP-seq results. (C) GATC was identified as the most frequent motif associated with the Dnd ChIP-seq read pileups and was strongly localized at peak summits. The motif search was performed using MEME, and motif enrichment analysis was performed with CentriMo using the 75 Dnd ChIP-seq peaks noted in A. (D) There are few PT modifications near Dnd protein binding sites. With the same cell samples used for ChIP-seq, TdT-seq showed that <1% of 14,168 PT modifications occurred within 200 bp of the 75 peak summits in the ChIP-seq Dnd binding regions. Of the 343 GATC sites identified within 200 bp of the 75 ChIP-seq peaks, five had PT at GPSATC and no other PTs, 26 had unmodified GATC but possessed PTs in GPSAAC/GPSTTC, and 44 lacked PTs entirely. The GATC sites were all presumably modified with 6mA by Dam.
The observation of a GATC consensus sequence associated with Dnd protein binding raised questions about the modification of this site in S. enterica. To address this question, we used a terminal transferase DNA sequencing method (terminal deoxynucleotidyl transferase sequencing [TdT-seq]) (28) to map sites of PT modification on each strand of the genomes of the S. enterica strains used for ChIP-seq analysis. The TdT-seq method (28) involves first blocking background strand breaks with dideoxyNTPs (ddATP, ddCTP, ddGTP, ddTTP) and DNA polymerase I, then converting PTs into single-strand breaks by selective oxidation with iodine (23, 28), and finally using terminal transferase to create poly(dT)-tails at the new 3′-ends of the PT break sites. Sequencing libraries were then constructed using Clontech’s DNA SMART (Switching Mechanism at the 5′ end of RNA Template) technology (29). After genomic alignment of the sequencing data, the sites of PT modification 1) were identified as the 5′-ends of poly(dT) tracts that did not align with known poly(dA/dT) runs in S. enterica (30), 2) had a coverage of more than five reads total, 3) had more than five times more reads than sites up- and downstream of the putative cleavage site, and 4) had more than five times more reads than the same sites in negative controls which were not treated by iodine (28). The sequencing results are detailed in Dataset S2. As summarized in Fig. 1A, the YF11 FLAG-DndC S. enterica strain had 14,168 PT modification sites, of which 41% were GPSTTC, 52% GPSAAC, 6% GPSTAC, and 1.4% GPSATC sites. This is consistent with our previous sequencing analysis of E. coli B7A, which possesses Dnd proteins nearly identical to those of S. enterica (23). The locations of these PT modification sites were then compared to ChIP-seq Dnd binding regions (Fig. 1A). Of the 14,168 PT sites, less than 1% occurred within 200 bp of the peak summits in the ChIP-seq Dnd binding regions. Of the 343 GATC sites identified within 200 bp of the 75 ChIP-seq peaks, only five were modified with PT (GPSATC), which suggests that Dnd proteins bind to GATC sites lacking PTs (Fig. 1D).
Dam Methylation Reshapes the PT Map, but PT Density Remains Constant.
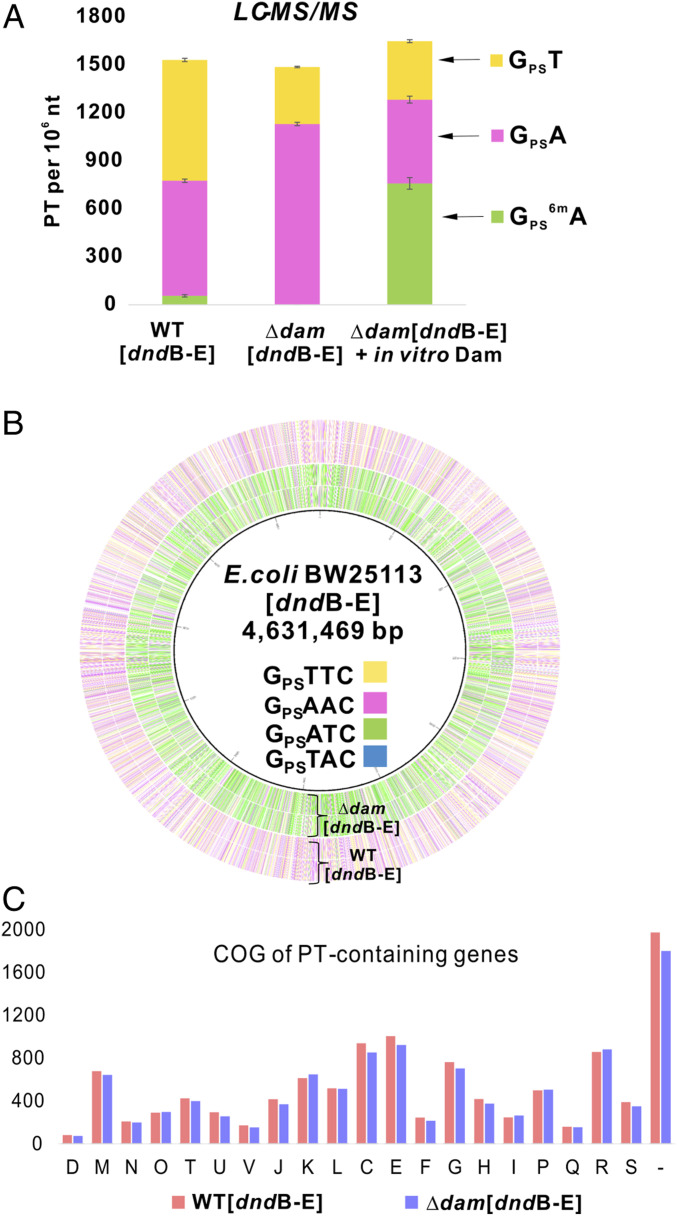
Dam methyltransferase catalyzes N6-dA methylation at GATC motifs in γ-proteobacteria (11). Given the ChIP-seq identification of GATC as the major Dnd binding site in S. enterica, we next set out to determine the role of 6mA in the limited (1.4%) PT modification of GATC sites. We were unable to knock out dam in S. enterica 87 so we inserted the S. enterica dnd genes DndBCDE into the dam-expressing E. coli BW25113 strain and in a BW25113 mutant lacking dam. These two strains were then assessed for levels of PT-linked dinucleotides by liquid chromatography-mass spectrometry (LC-MS) (15). Expression of S. enterica 87 Dnd proteins in WT E. coli BW25113 produced GPST at 756 per 106 nt and GPSA at 720 per 106 nt, which is similar to the levels of 225 to 555 per 106 nt in the S. enterica strains used here, as noted earlier. We also detected the doubly modified GPS6mA at 56 per 106 nt, which is again favorable to the GPS6mA levels of 3 to 54 per 106 nt detected in the S. enterica strains (SI Appendix, Fig. S1). These results validate the expression of S. enterica dnd genes in E. coli.
A comparison of the levels of the PT dinucleotides in the dam-expressing and Δdam E. coli strains revealed that the total number of PT dinucleotides was similar in both strains (1,532 vs. 1,487 per 106 nt, respectively) (Fig. 2A). However, the distribution shifted significantly, with an increase in GPSA to 1,131 per 106 nt and a decrease in GPST to 356 per 106 nt, and no detectable GPS6mA (Fig. 2A). The ability of Dam to methylate its PT-modified GPSATC consensus sequence was assessed by treating purified DNA from the ∆dam DNA with Dam and S-adenosylmethionine (SAM) in vitro and quantifying PT dinucleotides by LC-MS. As shown in Fig. 2A, the level of GPSA decreased from 1,131 to 524 per 106 nt and the level of GPS6mA increased from zero to 759 per 106 nt. This establishes Dam’s ability to methylate PT-modified GATC motifs. These results demonstrate that the Dnd proteins are flexible in their target selection, preferring to modify GATC rather than the previously observed GAAC/GTTC predominance when 6mA is present at GATC. The results also show that the presence of 6mA prevents PT modification of GATC.
Fig. 2.
Dam-dependent changes in quantities and genomic locations of PTs in E. coli possessing DndB-E from S. enterica. (A) Dam activity blocks PT at GATC in vivo, but Dam can methylate PT-modified GATC in vitro. PT dinucleotides were quantified (LC-MS) in DNA purified from E. coli expressing S. enterica dndB-E and possessing (WT[dndB-E]) or lacking (∆dam[dndB-E]) dam. DNA purified from ∆dam[dndB-E] was treated with Dam and SAM in vitro and analyzed by LC-MS for GPS6mA dinucleotide (∆dam[dndB-E] + in vitro Dam). (B) Circos plot of PT sites detected by TdT-seq in E. coli possessing (WT[dndB-E]) and lacking dam (∆dam[dndB-E]). Inner to outer: circles 1 and 2, (forward, reverse strands) are PT sites in ∆dam[dndB-E]; circles 3 and 4, (forward, reverse strands) are PT sites in WT[dndB-E]. (C) The presence of Dam and 6mA does not grossly alter the distribution of PTs in the E. coli genome. Genomic locations of PTs in the E. coli Dnd-expressing strains possessing and lacking dam were quantified by TdT-seq, and the distributions among gene families were analyzed with clusters of orthologous groups (COGs): C, energy production and conversion; D, cell cycle control, cell division and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation; K, transcription; L, replication; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, posttranslational modification, protein turnover, chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking and secretion; V, defense mechanisms; and –, not in COGs. The y axis represents the PT-containing gene numbers in each defined COG type. All results shown here are based on expressing dndBCDE from a high-copy number plasmid in E. coli BW25113. The similar results from a low-copy number plasmid are shown in SI Appendix, Fig. S5.
Given the relatively constant density of PTs at ∼1,500 per 106 nt despite 6mA-dependent shifts in the quantities of different PT dinucleotides, we sought to define 6mA-dependent changes in the types and locations of PT consensus sequences. Here, we used TdT-seq to map PTs across the genomes of the S. enterica DndBCDE-possessing E. coli BW25113 and its ∆dam mutant. The circos plot in Fig. 2B shows the overall shift from a predominance of GPSAAC and GPSTTC in BW25113 expressing Dam (outer circles) to a large increase in GPSATC when Dam is lost (inner circles). These changes are quantified in Table 1 and SI Appendix, Fig. S5. The results confirm our previous observation that only 10 to 15% of consensus sequences are modified with PTs (23). SI Appendix, Fig. S5C shows that, while the bulk of the PT modifications occur at the same sites in BW25113 and the ∆dam mutant, 5 to 35% of the sites differ between the two strains, which is consistent with previous studies (24). As shown in Fig. 2C and Dataset S3, these shifts in consensus sequence modification frequency were not accompanied by gross changes in the distribution of PTs in families of genes in the E. coli genome. Similar results were obtained with a low-copy number dndBCDE expression plasmid (SI Appendix, Fig. S5 and Dataset S4). Overall, the results reveal two interdependent features of this PT modification system: the plasticity of S. enterica Dnd protein target selection and the conservation of the density of PT modifications in bacterial genomes. Dam methylation of GATC forces Dnd proteins to modify other sites while maintaining the same density and general distribution of PTs. This raises questions about the mechanisms governing target selection by S. enterica Dnd proteins and, given the fact that DNA methylation can prevent PT-based restriction enzyme cleavage (27), the potential for PTs to substitute for 6mA function as an epigenetic mark at GATC sites.
Table 1.
Dam-dependent shifts in PT sites detected by TdT-seq in E. coli BW25113
| GPSAAC | GPSTTC | GPSATC | GPSTAC | Total | |
| [dndB-E] | 5,264* (15%) | 4,629 (13%) | 200 (0.5%) | 544 (2.3%) | 10,637 |
| [dndB-E] Δdam | 2,783 (8%) | 2,412 (7%) | 4,600 (12%) | 125 (0.5%) | 9,920 |
| Total no. of motifs | 35,600 | 35,600 | 37,772 | 23,800 | 132,772 |
Data represent the number of PT-modified sequence motifs in each E. coli genome, based on TdT-seq analyses of a high-copy number plasmid for expressing dnd genes in E. coli BW25113.
Partial PT Modification of GATC Does Not Substitute for 6mA Epigenetic Function.
Dam methylates the majority of GATC sites throughout the genomes of many γ-proteobacteria, with the 6mA functioning as an epigenetic mark for many bacterial cell processes (9). Based on the observation of PT modification of 12% (Table 1) of GATC sites in the E. coli BW25113 ∆dam mutant, we next asked if PT could substitute for 6mA in GATC sites and restore defects caused by loss of Dam. Here, we interrogated two of the epigenetic functions of Dam-dependent G6mATC methylation: synchronization of DNA replication initiation (31) and discrimination between newly synthesized DNA and the parental strand (10, 32, 33). As shown in SI Appendix, Fig. S6, there were no apparent differences between the E. coli BW25113 WT strain, the E. coli BW25113 [dndB-E] strain, and their ∆dam mutants: loss of dam disrupted replication synchrony and caused a growth defect due to mismatch repair activation whether dnd genes were present or not. Transcriptional profiling by RNA-seq with these strains was consistent with previous microarray and RNA-seq studies on ∆dam mutants (34–36), with data showing increased expression of DNA SOS response genes dinI, recN, and recX, and decreased expression of the Dam- and OxyR-dependent phase variation gene, agn43, in the ∆dam mutants and no apparent effect of dnd genes (Dataset S5). We conclude that PTs cannot substitute for 6mA modification at GATC either due to structural incompatibility or to the low frequency of PT modifications compared to nearly complete methylation of GATC sites by Dam.
DNA Shape-Based Target Selection by S. enterica Dnd Proteins.
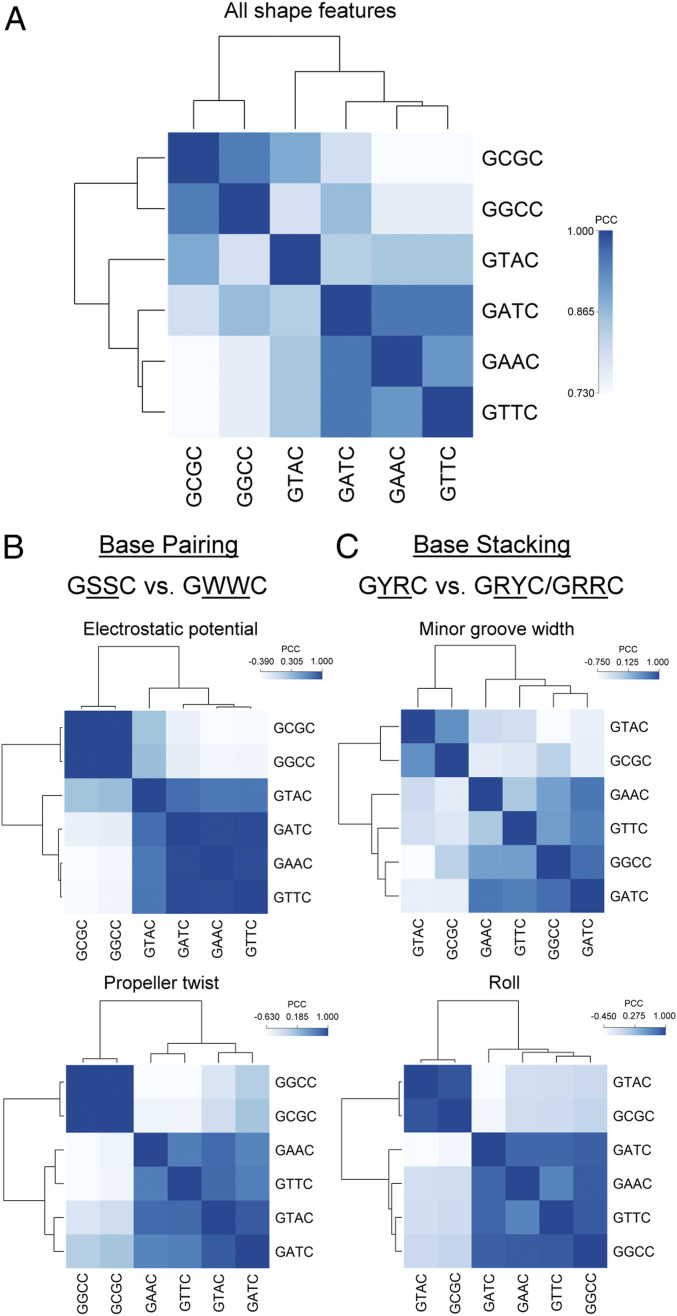
The Dam-dependent shifts in PT modification of consensus sequences raised the question of how Dnd proteins select their targets. We have previously observed low levels of PT-containing dinucleotides that differ from the main modification sites in nearly all bacteria studied (15), which suggests some degree of “sloppiness” in target selection by Dnd proteins. However, the present studies show that loss of Dam increases PT modification of GATC from 2 to 46% of all PT sites (Table 1). Here, we tested the role of intrinsic DNA shape in target selection by Dnd proteins. We have previously shown that many DNA-targeting proteins recognize sequence-dependent variations in DNA shape and electrostatic potential rather than simply reading a unique chemical signature of the DNA bases (37, 38). The ability of S. enterica Dnd proteins to variously read GAAC/GTTC, GATC, and GTAC suggests that DNA shape rather than nucleotide sequence defines the target site. There are six different motifs of the form GXXC (where X represents any of the four possible bases), most of which have been identified as PT modification sites in other bacteria, with the exception of GCGC. GPSGCC has been observed in Streptomyces lividans and Pseudomonas fluorescens Pf0-1 (15). We used the DNAshapeR (39) algorithm to predict 13 geometric DNA shape features (40, 41) and minor groove electrostatic potential (EP) (42) for each sequence motif set located within a constant 30-bp context. We averaged the predicted features for each motif set and calculated the Pearson correlation coefficient (PCC) and corresponding P value between motif sets with either all 14 or individual DNA features (Dataset S6). Based on the PCC correlation matrix, we performed hierarchical clustering and visualized the result with dendrograms (Fig. 3).
Fig. 3.
Heat map of the correlation matrix and hierarchical clustering of sequences with PT motifs based on all 14 DNA features (A) and individual DNA features (B and C). We scanned the E. coli genome using six PT modification motifs—GAAC, GTTC, GATC, GTAC, GGCC, and GCGC—and extracted the DNA sequences of length 30 bp centered around these motifs. We then quantified the DNA features of these sequences using DNAshapeR, a high-throughput method to predict 13 DNA shape features and minor groove EP. We averaged the predicted features for each motif set, normalized the resulting features using min-max normalization with the global minimum and maximum values retrieved from the DNAshape pentamer query table, and calculated the PCC and corresponding P value between motif sets using either all 14 or individual DNA features. Using the PCC matrix, we performed hierarchical clustering using the complete linkage algorithm and demonstrated the result with dendrograms. Differences between motifs based on individual DNA features were most predominant for features related to base pairing (B) when comparing GSSC and GWWC motifs (S: C or G; W: A or T) or base stacking (C) when comparing GYRC and GRYC/GRRC motifs (R: A or G; Y: C or T).
The two largest sequence groups, one containing motifs of the form GAAC/GTTC/GATC/GTAC (GWWC with W being either A or T) and the other containing motifs of the form GGCC/GCGC (GSSC with S being either G or C), which result from the clustering based on all DNA features demonstrate the structural similarity between the motifs within each of the groups (Fig. 3A). Within the GWWC group, GTAC shows the least structural similarity with GATC/GAAC/GTTC. This agrees with the PT modification frequency at each site in the E. coli BW25113 [dndB-E] ∆dam strain (Table 1) and with the idea that the Dnd proteins from S. enterica recognize GAAC, GTTC, and GATC motifs based on shape similarity, with GTAC serving as a minority site due to its distinct structure. The dissimilar GGCC motif recognized by P. fluorescens Dnd proteins is consistent with the significant sequence divergence of these proteins from those in S. enterica (15). This behavior generalizes in the striking phylogenic correlation between the PT sequence preference and Dnd protein sequence similarity, with bacteria possessing GPSG motifs clearly distinguished from bacteria with GPSA and GPST motifs (15).
Regarding the individual DNA features, EP and propeller twist (ProT) (Fig. 3B) show two even more distinct groups, GAAC/GTTC/GATC/GTAC and GGCC/GCGC, which can be categorized as International Union of Pure and Applied Chemistry (IUPAC) motifs GSSC and GWWC. These two IUPAC motifs have distinct characteristics that can be explained by DNA shape and EP. The A/T base pairs in the center of the GWWC motif can have a larger negative ProT due to the only two hydrogen bonds compared to the G/C base pair with three hydrogen bonds. The larger ProT narrows the minor groove at A/T base pairs compared to that at G/C base pairs (43, 44), which is associated with enhanced negative EP (38). In addition, while the G/C base pair has a guanine N2 amino group in the minor groove, the absence of such a group in the A/T base pair leads to a more negative EP (42). Thus, A/T base pairs carry more negative EP, which can attract positively charged amino acids such as arginine (38), lysine (45), and histidine (46). These structural characteristics suggest possible mechanisms for S. enterica Dnd proteins to recognize their binding sites and differentiate the GGCC motif recognized by P. fluorescens Dnd proteins.
Intriguingly, when the clustering is based on the DNA features minor groove width (MGW) or roll, the GGCC motif, which is recognized by P. fluorescens Dnd proteins, is clustered with the motifs GAAC, GTTC, and GATC, which are typically recognized by S. enterica Dnd proteins, as shown in Fig. 3C. Compared to the GCGC and GTAC motifs, which are minority PT sites, the majority PT sites GAAC, GTTC, GATC, and GGCC exhibit better geometric overlap of adjacent base pairs, which increases the strength of hydrophobic stacking interactions (37, 47, 48) that seem to be structurally selected by Dnd proteins.
Lack of Convergence of PT and Dam-Mediated 6mA in the Same Sequence Motifs.
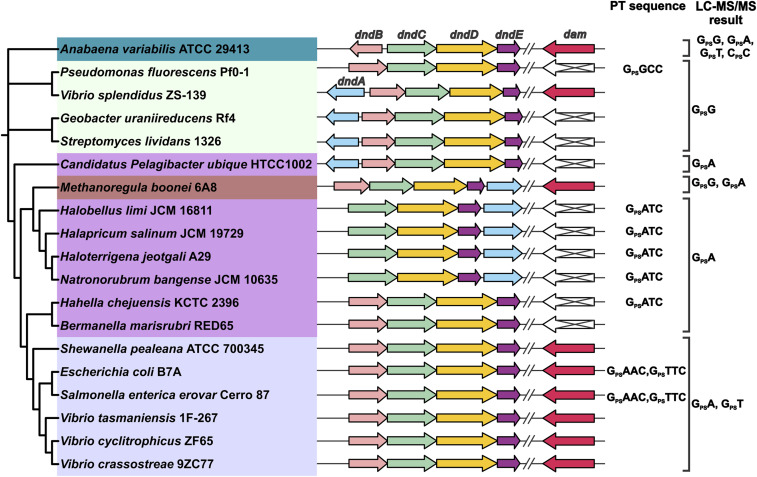
While we previously observed that 6mA and PT could coexist in GPS6mATC in an engineered bacterium (27), our observation of low levels of GPS6mATC in a naturally occurring S. enterica strain and the inability of Dnd proteins from this strain to modify G6mATC raised the question of the generality of PT and 6mA segregation. Here, we explored the relationships among Dnd and Dam genes and PT consensus sequences in diverse bacteria by building a phylogenetic tree using DndCDE protein sequences and correlating the tree with the presence of the dam gene and PT consensus sequences and dinucleotides derived from DNA sequencing and LC-MS analyses (15, 16, 20, 23, 24, 27). As shown in Fig. 4, the PT dinucleotides identified in each strain specified three major groups that coincided with Dnd protein sequence clusters and Dnd gene arrangements: the GPSA/GPST group, GPSG group, and GPSA. Interestingly, the dam gene was not present in strains containing only the GPSA dinucleotide or GPSATC motif without the complementary GPST or GPSTTC (Fig. 4). In parallel, strains possessing dam had PT consensus sequences other than GPSATC. Coupled with the biochemical incompatibility of G6mATC with S. enterica Dnd proteins (Fig. 2), this relatively small set of results raise the possibility of an evolutionary pressure to avoid the convergence of 6mA and PT. However, we previously observed GPS6mATC in dam-containing E. coli engineered with dndB-E from dam-naive H. chejuensis KCTC2396 (27). This forced combination reveals the existence of Dnd protein sequences that biochemically accommodate a neighboring 6mA. While the clustering of dnd genes on mobile genetic elements suggests facile transfer to dam-containing bacteria, the coexistence of 6mA-tolerating Dnd proteins with Dam in the same genome remains to be discovered.
Fig. 4.
Correlation between PT modification motifs and the presence of dam. Using MEGA, we constructed a phylogenetic tree from the protein sequences of 19 bacterial and archaeal DndCDE homologs. Different color shading was then applied to distinguish the PT sequence contexts present in the various strains, with the PT motifs noted as either GXXC or as LC-MS–detected PT dinucleotides on the right. The genomic organization of the dnd gene clusters and dam is also noted for each strain. All data are derived from our previous publications (15, 16, 23).
Discussion
The studies presented here were motivated by our observation that PT modifications occurred at only 10 to 15% of the 3- to 4-nt consensus sequences identified in diverse bacteria (23), which raised the question of how Dnd proteins select their DNA targets. Here, we used ChIP-seq to define DNA binding sites for the well-studied Dnd modification proteins from S. enterica serovar Cerro 87. ChIP-seq has been used in attempts to define the mechanisms of DNA target selection by DNA methylation enzymes, with no direct sequence-specific interactions identified (49–51). So it was not surprising that we did not observe specific binding of S. enterica Dnd proteins to the GAAC/GTTC motif identified from PT modification analyses (15, 23). This lack of detectable binding of DNA modification proteins to their genomic target sequences is reasonable given the potentially transient nature of the DNA–protein interactions at specific sites for processive enzymes such as Dam (52) and distributive enzymes involved in R-M systems (53, 54), with rapid release of the enzymes from modified sites yielding low steady-state levels of bound proteins.
What was surprising, however, was the observation of Dnd proteins stably bound to GATC sites not previously thought to be targets for PT modification in S. enterica. Coupled with the genomic mapping of PTs, this expanded the repertoire of PT modification sites selected by S. enterica Dnd proteins to include four of the six possible GXXC motifs. That readout of base sequence seems to play a smaller role than shape readout in the selection of DNA targets by S. enterica Dnd proteins is supported by the observed correlation between PT modification frequency in the absence of Dam and the similarity of DNA shape features: GATC and GAAC/GTTC show most significant structural similarity (Fig. 3) and are nearly equally modified (12% vs. 15%, respectively) (Table 1) while GTAC differs in shape from GATC and GAAC/GTTC (Fig. 3) and is modified only 0.5% of the time (Table 1). The difference between these motifs is that the TpA base pair step is a flexible hinge due to weak stacking interactions compared to the ApT and ApA (TpT) base pair steps. GGCC and GCGC motifs are totally different structurally from GAAC/GTTC motifs when clustered based on all DNA features so it was not surprising that we did not detect any PTs at these sites in TdT-seq mapping or GPSG or GPSC dinucleotides by LC-MS in S. enterica. In addition to these observations, 6mA modification of GATC sites by Dam may disrupt this apparent shape recognition by Dnd proteins, as the example that m5C modification alters the DNA structure due to the addition of a bulky methyl group (55–57), rendering the site resistant to PT modification and shifting the PT distribution almost completely to GAAC/GTTC (Table 1). This behavior could be common to bacteria possessing both Dam and Dnd proteins homologous to those from S. enterica serovar Cerro 87 and E. coli B7A (Fig. 4). Given the apparent avoidance of simultaneous GPSA and 6mA in GATC in S. enterica, we predict that bacteria possessing GATC as the sole PT modification site will not possess Dam and that the presence of Dam will shift PT modifications to sites other than GATC. If the behavior of Dam from E. coli BW25113 is generalizable, Dam appears to be the dominant enzyme here given its ability to methylate GPSATC and the inability of S. enterica Dnd proteins to modify G6mATC.
It was also surprising that, while the presence or absence of Dam caused large shifts in the PT modification landscape, there was a constant density of PT modifications at ∼1,500 per 106 nt by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (Fig. 1) (∼2,200 per 106 nt by TdT-seq analysis in Table 1). This constancy is not a matter of modifying all available sites since only 10 to 15% of all modifiable sequence motifs contain PTs under any circumstance, and PTs are relatively evenly distributed across the bacterial genome (23, 24) (Figs. 1 and 2). Previous studies have shown that expression of DndB, the transcriptional regulator of the dnd gene cluster, modulates the level of PT dinucleotides, with deletion of dndB in S. enterica causing a twofold increase in both GPSA and GPST (58). So it is possible that the “density” of PT modifications is regulated at the level of dnd gene expression and Dnd protein abundance. This model is supported by the observation of a uniform increase in the level of all PT modifications, both high- and low-frequency sites, with increasing expression of the full set of Dnd proteins (15). Two intriguing questions motivate ongoing research: Why the density is maintained at 10 to 15% of available modification sites and what keeps Dnd restriction enzymes in check in the face of this state of partial modification.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacteria strains and plasmids used in this study are listed in SI Appendix, Table S1 and depicted in SI Appendix, Fig. S1A. S. enterica 87 and its dnd knockout mutants were prepared as described previously (19), and the FLAG-tagged strains for ChIP-seq were prepared using a homologous recombination method described elsewhere (17). For YF11[pFLAG-DndC] (SI Appendix, Fig. S1A), the FLAG-tagged dndC gene was amplified by PCR from S. enterica 87 genomic DNA using primers containing the FLAG sequence and cloned into a p15 origin plasmid (https://www.addgene.org/44249/) using BglII and XhoI restriction enzymes. For the other six ChIP-seq strains (WXL1[FLAG-DndC], WXL1[FLAG-DndD], WXL1[FLAG-DndE] derived from the S. enterica 87∆dndBCDE mutant; and 103[FLAG-DndC], 103[FLAG-DndD], and 103[FLAG-DndE] derived from the S. enterica 87∆dndBCDEFGH mutant) (SI Appendix, Fig. S1A), the FLAG tag was introduced into a DndBCDE-containing plasmid pJTU1980 (low copy number) or pJTU1238 (high copy number) using the NEB Q5 Site-Directed Mutagenesis Kit. The genetics of the bacterial mutants and plasmids were confirmed by Sanger DNA sequencing. Strains of S. enterica 87 and E. coli K12 expressing Salmonella DndBCDE were grown in Luria–Bertani (LB) broth or M9 minimal medium (22) at 37 °C.
LC-MS/MS Analysis for PT Quantification.
PT modifications were quantified as PT-linked dinucleotides by LC-MS/MS analysis as described elsewhere (22). Briefly, DNA was hydrolyzed with nuclease P1 and alkaline phosphatase, and the resulting mixture of nucleosides and PT-linked dinucleotides was resolved by high-pressure liquid chromatography (HPLC) (Agilent 1290) equipped with a reversed-phase HPLC column (Synergy Fusion RP column, 2.5-μm particle size, 100-Å pore size, 100-mm length, 2-mm inner diameter) with an in-line diode array detector and coupled to an Agilent 6490 triple quadrupole mass spectrometer. 2′-Deoxynucleosides were quantified by ultraviolet absorbance, and PT-containing dinucleotides were identified and quantified by tandem quadrupole mass spectrometry. Enantiomerically pure, PT-containing dinucleotide standards were obtained from IBA BioTechnology (Germany) and used to prepare calibration curves for the dinucleotides.
ChIP-Seq and Data Analysis.
ChIP-seq using anti-FLAG antibodies was performed with the engineered bacterial strains listed in SI Appendix, Table S2 and Fig. S1A, using previously established methods (59). The workflow is depicted in SI Appendix, Fig. S2. Bacteria were fixed with formaldehyde (1%) for 30 min, and cross-linking was stopped by adding glycine (250 mM) for 15 min. The fixed bacteria were then sheared using a Covaris ME220 sonicator to produce DNA fragment sizes averaging 200 bp (Peak Incident Power 74 W, duty factor 24%, cycle/burst 1,000, 10 s on/10 s off). Anti-FLAG antibody (10 μL; Sigma) was added to each cell lysate followed by overnight incubation at 4 °C. Protein–antibody complexes were immunoprecipitated using protein G agarose (Thermo Fisher) by incubation at 4 °C for 1 h and ambient temperature for 2 h. The protein–antibody–agarose complex was sequentially washed five times with IPP150 buffer (10 mM Tris⋅HCl, pH 8, 150 mM NaCl, and 0.1% Nonidet P-40) and twice with TE buffer (10 mM Tris⋅HCl, 1 mM disodium ethylenediaminetetraacetic acid, pH 8.0) at ambient temperature. Reverse cross-linking was performed by addition of Proteinase K (1 mg/mL) and degraded by incubating at 65 °C overnight. ChIP DNA was purified using a Qiagen QIAquick PCR Purification Kit. Sequencing libraries were prepared using the NEB Next Ultra II DNA Library Prep Kit for Illumina. Single-end 75-bp reads were generated using an Illumina NextSeq 500 sequencer. Read quality control of raw reads was performed using FastQC. Adapter sequences were then removed from the reads using cutadapt (60), and the reads were aligned to the corresponding genome (CP008925.1) using Bowtie 2 (61) with the default option. Peak calling was performed using CisGenome (62), with parameters set at window statistic ≥5 and P value ≤ e−6 using the two-sample analysis mode to identify significantly enriched regions of ChIP reads relative to the negative control reads based on a conditional binomial model. BAM files (the alignment file generated by Bowtie2) were converted into .tdf format for visualization using IGVtools and viewed on the GenomeView (63) genome browser. The normalized (1×) coverage of each genome position was calculated by bamCoverage in deepTools2 (64) based on alignment files on the Galaxy server (65). The normalized coverage at the ChIP-seq binding regions was visualized and checked by plotHeatmap in deepTools2 (64). Dnd binding motifs were predicted using MEME Suite based on the 100-bp up- and downstream region of the peak summit, and a consensus motif was deduced and verified by CentriMo (66). In Fig. 2B, the average normalized read coverage in peaks for ChIP-seq samples and negative controls was calculated by 1) normalizing read counts to 1× coverage across the genome; 2) visually determining peak shift between forward and reverse reads as a criterion for evaluating peak calls; and 3) averaging the data from ChIP and negative control samples and then plotting read counts versus distance to peak summit. Data were averaged as follows: DndCDE ChIP—the three biological replicates of YF11[pFLAG-DndC] and single replicates of WXL1[FLAG-DndC], WXL1[FLAG-DndD], WXL1[FLAG-DndE, 103[FLAG-DndC], 103[FLAG-DndD], and 103[FLAG-DndE]; negative control—single replicates of each of the 15 control strains.
TdT-Seq for Genomic Mapping of PTs.
PT modifications were mapped in bacteria using a TdT sequencing method (TdT-seq) (28). Briefly, following blocking of existing DNA strand breaks with DNA polymerase I and dideoxyNTPs (ddATP, ddCTP, ddGTP, and ddTTP), PT modifications were converted to DNA strand breaks by mild treatment with iodine. The 3′-end of each break site was then extended as a poly(dT) tail by TdT and dTTP. This poly(dT) tail was then exploited to construct Illumina sequencing libraries using the Clontech SMART ChIP-seq kit (Clontech) (28). The purified libraries were sequenced using an Illumina NextSeq 500 instrument for 75-bp paired-end sequencing. The raw reads were processed using the Galaxy web platform (65). Initially, the paired-end reads were preprocessed with Trim Galore! to remove adapters, and the GGG sequence was added at the library preparation stage. All of the reads were aligned to the corresponding genome using Bowtie 2 (61). Peak calling with the aligned reads was performed with BamTools, BEDTools, and Rstudio, and the results were filtered based on R2 (67). The 5′ read coverage (experimental sample) or full read coverage (negative controls without iodine cleavage) at each position was calculated based on the filtered results by BEDTools (positive and negative strand separately) (68). A read pileup at a site was considered significant if the value was five times greater than pileups immediately upstream and downstream and in the negative control. Using this method, more than 95% of the detected PT sites involved consensus sequences previously identified using single-molecule real-time sequencing (23).
Flow Cytometry Analysis of Replication Synchrony.
Dam regulation of replication synchrony was quantified as described elsewhere (69). Overnight cultures were diluted 100-fold in growth medium and grown to an optical density at 600 nm (OD600) of ∼0.25. Cultures were then harvested or treated with rifampicin (300 μg/mL) and cephalexin (10 μg/mL) for 4 h, followed by harvesting of cells, washing in 1× phosphate-buffered saline (PBS) (pH 7.4), and fixation in 70% cold ethanol at −20 °C overnight. The bacteria were then washed in 1× PBS (pH 7.4) and resuspended in 1 mL of a buffer containing 1× PBS (pH 7.4), 0.1% Triton X-100 (Sigma), 0.1 μg/mL RNase A (DNase-free; Sigma), and 30 μM propidium iodide (Sigma), followed by incubation at 37 °C for 30 min. The samples were then analyzed using an Attune NxT Acoustic Focusing flow cytometer. The peak fluorescence intensity of an overnight culture in the same medium was regarded as one copy of a DNA chromosome.
2-Aminopurine Sensitivity Analysis.
To quantify Dam regulation of mismatch repair (MMR), we performed a 2-aminopurine (2AP) sensitivity assay, as described elsewhere (32). Overnight cultures were diluted 100-fold in LB without 2AP and grown to log phase (OD600 of ∼0.6). Log phase cultures were then subjected to 10-fold serial dilutions in LB and spotted on LB agar plates containing 2AP (350 mg/mL). Plates were incubated at 37 °C for 24 h, and colon size was quantified in photographic images.
RNA-Seq and Data Analysis.
For RNA-seq analysis, total RNA was extracted using TRIzol Reagent (Invitrogen), and ribosomal RNA was removed using a Ribo-Zero Magnetic kit (EpiCentre). An Illumina Truseq RNA sample prep kit was used to prepare the RNA-seq library, which was sequenced using an Illumina HiSeq sequencer for 2 × 150-bp paired-end sequencing. Raw reads were then processed using SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle), with the reads then aligned to the E.coli K12 BW25113 genome using Bowtie 2 (61). Quantification of gene expression was carried out by RSEM in the form of FPKM (fragments per kilobase million) (70). edgeR was used for analysis of differential gene expression (71). Genes with P < 0.01 and |log2(Fold-change)| >2 were regarded as differentially expressed genes.
DNA Shape Analysis.
The 13 DNA shape and EP features of the GAAC, GTTC, GATC, GTAC, GGCC, and GCGC motifs were determined using DNAshapeR (39) based on analysis of ∼1,500 30-bp sequences from the E. coli K12 genome, each containing a centrally located motif. DNAshapeR predicts DNA shape features in an ultrafast, high-throughput manner from genomic sequencing data, which uses a sliding pentamer window to derive the structural features from all-atom Monte Carlo simulations (40). The DNA features include six intrabase pair parameters (buckle, opening, ProT, shear, stagger, and stretch), six interbase pair parameters (helix twist, rise, roll, shift, slide, and tilt), MGW, and EP (41, 42). For each GXXC motif, the DNA shape features calculated for the ∼1,500 sequences were averaged, and the resulting DNA shape profiles are depicted in SI Appendix, Fig. S7. The averaged shape features were normalized between 0 and 1 using min-max normalization with the global minimum and maximum values retrieved from the DNAshape pentamer query table. For the all shape features analysis, the normalized shape features were concatenated as a vector shape. The PCCs and corresponding P values between motif sets with all or individual DNA features were calculated and are provided in SI Appendix, Table S1. With the PCC correlation matrix, a complete-linkage clustering was performed and visualized as a dendrogram.
Phylogenetic Tree and Evolution Analysis.
The protein sequences of DndC and DndD were used to generate the phylogenetic trees, and the methods for the tree construction were described previously (16). The LC-MS/MS and PacBio Single Molecule, Real-Time sequencing results of each strain were obtained from previous studies (15, 16, 20, 23, 24, 27). The National Center for Biotechnology Information genome database was used to determine the presence of dam in the various bacterial genomes (72).
Material and Data Availability.
ChIP-seq, TdT-seq, and RNA-seq data have been deposited in the Gene Expression Omnibus database under accession numbers GSE135768, GSE135910, and GSE135938. The DNAshapeR algorithm can be obtained at Bioconductor Open Source Sofware for Bioinformatics (https://www.bioconductor.org/packages/release/bioc/html/DNAshapeR.html). Sources of code for TdT-seq and ChIP-seq data processing are detailed in Materials and Methods. Bacterial strains can be obtained from the corresponding authors.
Supplementary Material
Acknowledgments
We thank Stuart Levine for discussions about the ChIP-seq work; and Michael S. DeMott, Liang Cui, Peiying Ho, and Hooi Linn Loo for technical support. We also acknowledge the Massachusetts Institute of Technology (MIT) BioMicro Center, Genome Institute of Singapore, and Singapore-MIT Alliance for Research and Technology (SMART) for the use of their facilities for LC-MS/MS, flow cytometry, and Illumina sequencing. This work was supported by grants from the National Natural Science Foundation of China (Grants 31720103906 and 31925002), the China National Key Research and Development Program (Grant 2019YFA0904300), the Fundamental Research Funds for the Central Universities of China, the SMART Antimicrobial Resistance Interdisciplinary Research Group sponsored by the National Research Foundation of Singapore, the US National Science Foundation (Grant CHE-1709364) (to P.C.D. and J.E.G.), and the NIH (Grant R35GM130376) (to R.R.). X.W. was supported by a fellowship from the China Scholarship Council (201606270163) and T.-P.C. by a University of Southern California–Taiwan Postdoctoral Fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.L. is a guest editor invited by the Editorial Board.
Data deposition: ChIP-seq, TdT-seq, and RNA-seq data have been deposited in the Gene Expression Omnibus database under accession numbers GSE135768, GSE135910, and GSE135938.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002933117/-/DCSupplemental.
References
- 1.Boyer H., Genetic control of restriction and modification in Escherichia coli. J. Bacteriol. 88, 1652–1660 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadesús J., Low D., Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70, 830–856 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasu K., Nagaraja V., Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77, 53–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L. et al., Phosphorothioation of DNA in bacteria by dnd genes. Nat. Chem. Biol. 3, 709–710 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Thiaville J. J. et al., Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc. Natl. Acad. Sci. U.S.A. 113, E1452–E1459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutinet G. et al., 7-Deazaguanine modifications protect phage DNA from host restriction systems. Nat. Commun. 10, 5442 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weigele P., Raleigh E. A., Biosynthesis and function of modified bases in bacteria and their viruses. Chem. Rev. 116, 12655–12687 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Marinus M. G., Morris N. R., Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J. Bacteriol. 114, 1143–1150 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wion D., Casadesús J., N6-methyl-adenine: An epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4, 183–192 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adhikari S., Curtis P. D., DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol. Rev. 40, 575–591 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Marinus M. G., Casadesus J., Roles of DNA adenine methylation in host-pathogen interactions: Mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33, 488–503 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen D. B., Kotzorek G., Eckstein F., Investigation of the inhibitory role of phosphorothioate internucleotidic linkages on the catalytic activity of the restriction endonuclease EcoRV. Biochemistry 29, 9546–9551 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Olsen D. B., Kotzorek G., Sayers J. R., Eckstein F., Inhibition of the restriction endonuclease BanII using modified DNA substrates. Determination of phosphate residues critical for the formation of an active enzyme-DNA complex. J. Biol. Chem. 265, 14389–14394 (1990). [PubMed] [Google Scholar]
- 14.Barski A. et al., High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Wang L. et al., DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc. Natl. Acad. Sci. U.S.A. 108, 2963–2968 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong L. et al., A new type of DNA phosphorothioation-based antiviral system in archaea. Nat. Commun. 10, 1688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu T., Yao F., Zhou X., Deng Z., You D., A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 38, 7133–7141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao B. et al., Pathological phenotypes and in vivo DNA cleavage by unrestrained activity of a phosphorothioate-based restriction system in Salmonella. Mol. Microbiol. 93, 776–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan R. et al., DNA phosphorothioate modifications influence the global transcriptional response and protect DNA from double-stranded breaks. Sci. Rep. 4, 6642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong T. et al., Occurrence, evolution, and functions of DNA phosphorothioate epigenetics in bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, E2988–E2996 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L., Jiang S., Deng Z., Dedon P. C., Chen S., DNA phosphorothioate modification-a new multi-functional epigenetic system in bacteria. FEMS Microbiol. Rev. 43, 109–122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellner S. et al., Oxidation of phosphorothioate DNA modifications leads to lethal genomic instability. Nat. Chem. Biol. 13, 888–894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao B. et al., Genomic mapping of phosphorothioates reveals partial modification of short consensus sequences. Nat. Commun. 5, 3951 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J. et al., Quantitative mapping of DNA phosphorothioatome reveals phosphorothioate heterogeneity of low modification frequency. PLoS Genet. 15, e1008026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blow M. J. et al., The epigenomic landscape of prokaryotes. PLoS Genet. 12, e1005854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krebes J. et al., The complex methylome of the human gastric pathogen Helicobacter pylori. Nucleic Acids Res. 42, 2415–2432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C. et al., Convergence of DNA methylation and phosphorothioation epigenetics in bacterial genomes. Proc. Natl. Acad. Sci. U.S.A. 114, 4501–4506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao B. et al., Nick-seq for single-nucleotide resolution genomic maps of DNA modifications and damage. bioRxiv, 10.1101/845768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y. Y., Machleder E. M., Chenchik A., Li R., Siebert P. D., Reverse transcriptase template switching: A SMART approach for full-length cDNA library construction. Biotechniques 30, 892–897 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Vardi O., Shamir I., Javasky E., Goren A., Simon I., Biases in the SMART-DNA library preparation method associated with genomic poly dA/dT sequences. PLoS One 12, e0172769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mott M. L., Berger J. M., DNA replication initiation: Mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5, 343–354 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Burdett V., Baitinger C., Viswanathan M., Lovett S. T., Modrich P., In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 98, 6765–6770 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guarné A., Charbonnier J. B., Insights from a decade of biophysical studies on MutL: Roles in strand discrimination and mismatch removal. Prog. Biophys. Mol. Biol. 117, 149–156 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Løbner-Olesen A., Marinus M. G., Hansen F. G., Role of SeqA and Dam in Escherichia coli gene expression: A global/microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 100, 4672–4677 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshima T. et al., Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45, 673–695 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Robbins-Manke J. L., Zdraveski Z. Z., Marinus M., Essigmann J. M., Analysis of global gene expression and double-strand-break formation in DNA adenine methyltransferase- and mismatch repair-deficient Escherichia coli. J. Bacteriol. 187, 7027–7037 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohs R. et al., Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 79, 233–269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohs R. et al., The role of DNA shape in protein-DNA recognition. Nature 461, 1248–1253 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiu T. P. et al., DNAshapeR: An R/Bioconductor package for DNA shape prediction and feature encoding. Bioinformatics 32, 1211–1213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou T. et al., DNAshape: A method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 41, W56–W62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J. et al., Expanding the repertoire of DNA shape features for genome-scale studies of transcription factor binding. Nucleic Acids Res. 45, 12877–12887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiu T. P., Rao S., Mann R. S., Honig B., Rohs R., Genome-wide prediction of minor-groove electrostatic potential enables biophysical modeling of protein-DNA binding. Nucleic Acids Res. 45, 12565–12576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haran T. E., Mohanty U., The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 42, 41–81 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Rohs R., Sklenar H., Shakked Z., Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2-DNA binding sites. Structure 13, 1499–1509 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Deng Z., Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 6, 7642 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang Y. P., et al. , Mechanism of origin DNA recognition and assembly of an initiator-helicase complex by SV40 large tumor antigen. Cell Rep. 3, 1117–1127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shakked Z., Rabinovich D., The effect of the base sequence on the fine structure of the DNA double helix. Prog. Biophys. Mol. Biol. 47, 159–195 (1986). [DOI] [PubMed] [Google Scholar]
- 48.Mack D. R., Chiu T. K., Dickerson R. E., Intrinsic bending and deformability at the T-A step of CCTTTAAAGG: A comparative analysis of T-A and A-T steps within A-tracts. J. Mol. Biol. 312, 1037–1049 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Jin B. et al., Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells. Cell Rep. 2, 1411–1424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verma N. et al., TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat. Genet. 50, 83–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki T. et al., RUNX1 regulates site specificity of DNA demethylation by recruitment of DNA demethylation machineries in hematopoietic cells. Blood Adv. 1, 1699–1711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urig S. et al., The Escherichia coli dam DNA methyltransferase modifies DNA in a highly processive reaction. J. Mol. Biol. 319, 1085–1096 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Surby M. A., Reich N. O., Contribution of facilitated diffusion and processive catalysis to enzyme efficiency: Implications for the EcoRI restriction-modification system. Biochemistry 35, 2201–2208 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Albu R. F., Jurkowski T. P., Jeltsch A., The Caulobacter crescentus DNA-(adenine-N6)-methyltransferase CcrM methylates DNA in a distributive manner. Nucleic Acids Res. 40, 1708–1716 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiu T. P., Xin B., Markarian N., Wang Y., Rohs R., TFBSshape: An expanded motif database for DNA shape features of transcription factor binding sites. Nucleic Acids Res. 48, D246–D255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dantas Machado A. C. et al., Evolving insights on how cytosine methylation affects protein-DNA binding. Brief. Funct. Genomics 14, 61–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao S. et al., Systematic prediction of DNA shape changes due to CpG methylation explains epigenetic effects on protein-DNA binding. Epigenet. Chromatin 11, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He W. et al., Regulation of DNA phosphorothioate modification in Salmonella enterica by DndB. Sci. Rep. 5, 12368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aquino P. et al., Coordinated regulation of acid resistance in Escherichia coli. BMC Syst. Biol. 11, 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J., 10.14806/ej.17.1.200 (2011). [DOI] [Google Scholar]
- 61.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji H. et al., An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26, 1293–1300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abeel T., Van Parys T., Saeys Y., Galagan J., Van de Peer Y., GenomeView: A next-generation genome browser. Nucleic Acids Res. 40, e12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramírez F. et al., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Afgan E. et al., The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bailey T. L. et al., MEME suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barnett D. W., Garrison E. K., Quinlan A. R., Strömberg M. P., Marth G. T., BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27, 1691–1692 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jha J. K., Chattoraj D. K., Inactivation of individual SeqA binding sites of the E. coli origin reveals robustness of replication initiation synchrony. PLoS One 11, e0166722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wheeler D. L., et al. , Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 36, D13–D21 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.