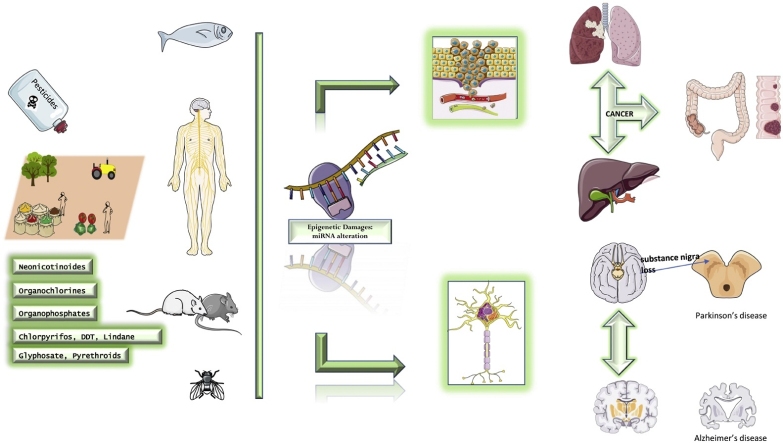
Graphical abstract
Keywords: Pesticides, miRNA, Cancer, Neurodegenerative diseases
Highlights
-
•
Current knowledge linking pesticide exposure, cancer and neuro-degenerative diseases to dysregulation of microRNA network was summarized.
-
•
Literature indicates differential miRNA expression targeting biomolecules and pathways involved in cancer and neurodegenerative diseases.
-
•
Evaluation of miRNA expression may be used to develop new non-invasive strategies for the prediction and prognosis of diseases including cancer.
-
•
The application of miRNAs as diagnostic and therapeutic biomarkers in the clinical field is extremely challenging.
Abstract
This review summarizes the current knowledge linking cancer and neuro-degenerative diseases to dysregulation of microRNA network following pesticide exposure. Most findings revealed differential miRNA expression targeting biomolecules and pathways involved in various neoplastic localizations and neurodegenerative diseases.
A growing body of evidence in recent literature indicates that alteration of specific miRNAs can represent an early biomarker of disease following exposure to chemical agents, including pesticides. Different miRNAs seem to regulate cell proliferation, apoptosis, migration, invasion, and metastasis via many biological pathways through modulation of the expression of target mRNAs.
The evaluation of miRNA expression levels may be used to develop new non-invasive strategies for the prediction and prognosis of many diseases, including cancer. However, the application of miRNAs as diagnostic and therapeutic biomarkers in the clinical field is extremely challenging.
1. Introduction
Literature data suggest that exposure to xenobiotic is strongly associated with human diseases. In particular, epidemiological and occupational studies indicate that carcinogenic substances such as metals, organic pollutants or pesticides reach the soil and aquifer persisting for a long time in the environment [1]. Hence, they join the food chain, accumulating in tissues and blood [2]. These compounds are also able to cross the placental barrier, leading to detrimental effects on the fetus. Effects can be both acute and chronic ranging from simple eye or skin irritation to systemic effects on the nervous system, immune system, reproductive system till cancer development [3,4]. Furthermore, exposure to xenobiotics even at low doses can also have effects on the endocrine system by acting on the hypothalamic / pituitary / endocrine gland axis with consequent onset of numerous pathologies [5]. For this reason the competent EU authorities have tried to monitor their toxic action through the introduction of toxicological reference values [6]. Many diseases potentially linked to pesticides exposure are thought to be caused by several mechanisms, including genetic damages and oxidative stress [7,8]. As regards oxidative stress, it derives from the cell's inability to neutralize an excess of oxidant agents induced by exposure to xenobiotics. This exposure can lead to molecular alterations in tissues, cells and biological macromolecules with potential mutagenic effects. Furthermore, some individuals appear to have a higher susceptibility due to the presence of genetic polymorphism that influences the metabolism of these xenobiotics [9,10]. Also the diet seems to influence the oxidative balance, since a diet rich in antioxidant substances such as polyphenols can have positive implications in terms of health [11]. A recent study has proposed the use of 8-Hydroxydeoxyguanosine (8−OHdG) as a biological marker of DNA damage mediated by oxidative stress in workers exposed to low-dose of benzene [12]; moreover, exposure to benzene causes oxidative stress with consequent production of ROS, which act by damaging the cells and forming new metabolites which are used as markers of oxidant / antioxidant imbalance [13]. Still, exposure to toxicants may affect the immune system by modulating cellular functionality [14]. In particular experimental and epidemiological studies suggest that pesticides can alter immune function and induce greater development of immunological diseases [15]. This modulation result in activation of proinflammatory processes responsible for genetic damage accumulation that DNA repair systems can not properly remove; macrophages alteration seem to be involved in the onset of tumorigenesis and autoimmune diseases [16]. A wealth of data demonstrates that epigenetic modifications may be one of the mechanisms by which pesticides can bring to harmful effects on human health and may have significance as biomarkers of carcinogen exposure even if the precise role of miRNA in chemical related carcinogenesis is not well cleared [[17], [18], [19], [20], [21], [22], [23]].
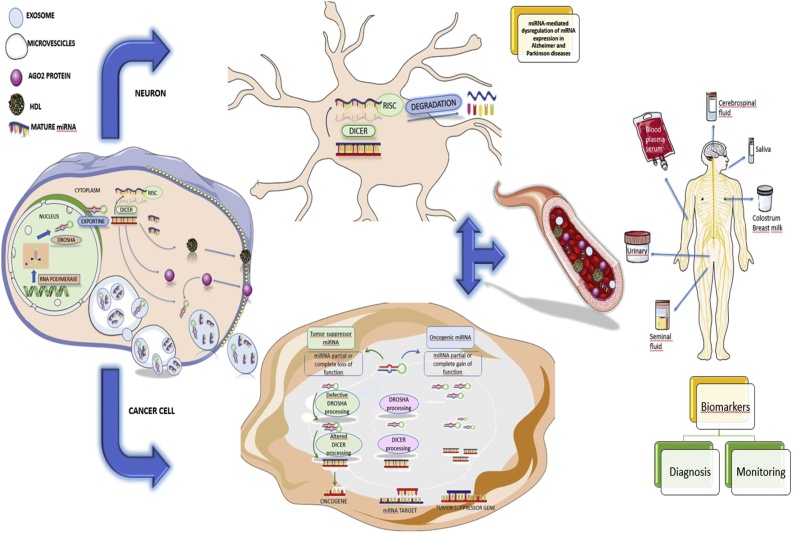
MiRNAs belong to a class of short single-stranded RNAs produced by genes present in all multicellular organisms. They are small non-coding RNA species that negatively regulate gene expression at post-transcriptional level by binding to the 3′UTR region of messenger RNA (mRNA) inducing degradation or preventing its translation into proteins. From a functional point of view it has been shown that a single miRNA can regulate the expression of different genes and that a particular gene can be regulated by different miRNAs that act in a synergistic way between them: the genes that produce the microRNAs are initially transcribed in RNA molecules plus l nails that are called primary microRNA (pri-miRNA), that folding in hairpins (pre-miRNA) are converted into mature miRNA. In particular, the pri-miRNA is processed by the nuclear enzyme Drosha and it is transformed into a double-strand molecule called pre-miRNA of 70 nucleotides. Pre-miRNA are then exported to the cytoplasm where the Dicer proteic complex cuts them into mature single-strand miRNAs of approximately 20–25 nucleotides [24].
In man, more than 2650 mature miRNA have been identified, present in all human tissues especially in body fluids, such as blood and urine [25]. Experimental studies show that miRNAs play a key role in many cellular processes such as proliferation, apoptosis and differentiation, and in many physiological processes such as metabolism, the nervous and immune system development [22]; still it has been shown that miRNAs are involved in different pathologies: infertility, cardiovascular diseases, neurological disorders and especially cancer [[26], [27], [28], [29], [30]]. Altered levels of miRNAs could be considered sensitive indicators of pesticide exposure and therefore used as biomarkers. A study showed that following exposure to paraquat in fish serum there was an overexpression of the circulating miR-122 with consequent damage to the liver, as this miRNA is specific for hepatocytes. For this reason, it could be considered a biomarker of liver damage. Finally, it was seen that the miR-122 and the mir-125b can also induce immunotoxicity. Moreover, miR-155overexpression following to paraquat exposure activates inflammatory response in fish [31]. The research on miRNA topic is rapidly growing, aiming to discover new mechanisms relevant to physiology and pathology. Furthermore, it is possible to use sophisticated research tools for the study of miRNAs: screening tools for mRNA, tools for forecasting bioanalytical targets, tools for targeting and manipulative tools for miRNA expression [32]. The experimental evidence collected until now show that miRNA could represent valid diagnostic and prognostic markers especially in human tumors. Despite this, the role of miRNA and responsive mechanisms leading to disease development following pesticides exposure are still not very clear. The purpose of this review is to evaluate the existing results on miRNAs associated with exposure to various classes of pesticides by focusing their role in the development of certain diseases.
2. Methods
A literature research was performed using PubMed database for detecting full text in vivo and in vitro studies, conducted during the last ten years (2010–2019) and using the following keywords: "miRNA pesticide cancer" [Pharmacological Action]OR "miRNA cancer pesticides" [MeSH Terms] OR "miRNA cancer pesticides" [All fields] OR "miRNA pesticides cancer" [All fields]; ["miRNA pesticides immunotoxicity" [MeSH terms] OR ["miRNA pesticides immunotoxicity" [All fields]; "miRNA pesticides immunotoxicity" [All fields] OR "miRNA pesticides immunotoxicity "[All Fields]; ["Parkinson's miRNA pesticides"[MeSH Terms] OR ["Parkinson's miRNA pesticides"[All fields]; "Parkinson's miRNA pesticides"[All Fields] OR"Parkinson's miRNA pesticides"[All Fields] OR "Parkinson's miRNA pesticides" [All fields]; ["Alzheimer's miRNA pesticides" [MeSH terms] OR ["Alzheimer's miRNA pesticides" [All fields] And "Alzheimer's miRNA pesticides" [All fields] OR "miRNA pesticides Alzheimer's"[All Fields] as search terms.
Articles published in languages other than English have not been included. The search produced a total of 50 results. The reference lists of the selected articles have been further screened for relevance, identifying additional records. A final number of 105 articles were selected for this review.
3. Cancer
Cancer is one of the leading causes of death in the world with a rising of the number of new cases [33]. Despite the evolution of cancer pharmacological treatment ameliorate the therapeutic strategies available for cancer patients, still today a significant fraction of cancer patients does not benefit from treatments mainly because of the lack of effective diagnostic biomarkers and the consecutive late diagnosis [34,35]. In order to identify novel diagnostic and prognostic biomarkers for cancer patients, many studies investigated the role of epigenetics modification in the development and progression, as well as in the diagnosis of cancer [[36], [37], [38], [39], [40], [41]]. The most important epigenetic alterations consist of DNA methylation, histone/chromatin structure, nucleosome placement and non-coding RNAs.
In cancer, miRNAs' expression is altered. In particular, miRNA classes involved in cancer initiation and metastatization could act with oncogenic function. Many studies have shown that miRNA aberrant expression could inhibit tumor suppressor genes or inappropriately activate oncogenes resulting in the onset of cancer and increased invasiveness and the appearance of metastases. Some authors have observed differential miRNA expression not only between normal and tumor tissue, but also between primary and metastatic tumor. These differences are tumor-specific and in some cases may be associated with prognosis. At the base of variations in miRNA expression levels there are three different mechanisms: co-localization of coding genes for miRNAs and cancer-associated genomic regions, epigenetic regulation and altered protein expression involved processing and biogenesis of microRNAs [42].
The availability of several bioinformatic prediction tools and a large amount of data relating to the expression levels of miRNAs and mRNA contained in public databases such as The Cancer Genome Atlas and GEO DataSets has favored the growing number of computational studies aimed at identifying miRNAs to be used as diagnostic and prognostic biomarkers for different tumors.
Such computational approaches allowed the identification of effective miRNA biomarkers for the early diagnosis and the prediction of prognosis of almost all cancers. By using multiple computational studies, diagnostic and prognostic miRNAs were identified for the most common and aggressive tumors, including colorectal cancer [43], breast cancer [44], lung cancer [45], oral cancer [46], bladder cancer [47,48], uveal melanoma [49].
Recent studies suggest that the expression of miRNA is altered by certain environmental chemicals, including metals, organic pollutants, cigarette smoke, pesticides and carcinogenic drugs [50]. In particular, experimental studies have shown that exposure to pesticides involves changes in the epigenome. The most studied mechanism is DNA methylation, however, recent research has investigated the effects on histone modifications and miRNAs. In agricultural workers urinary miRNAs were used for their ability to act as biomarkers of environmental exposure to pesticides representing an early biological response [18].
Among the pesticides responsible for the alteration of miRNAs are pyrethroids. In particular, cypermethrin is a type II pyrethroid, widely used as an insecticide for domestic and agricultural purposes, inducing genotoxicity, oxidative stress and cytotoxic and cytostatic effects both in mouse and human peripheral blood lymphocytes [51]. Acute and chronic exposure to this pesticide has been shown to cause neurodegenerative diseases and cancer. Some authors have showed that cypermethrin can promote tumor metastatization through inhibition of proinflammatory macrophages. This action is driven by repression of the expression of miR-155 [52].
Other authors have studied the side effects of niclosamide, used to control aquatic pests and as an antiparasitic agent both for humans and animals. In particular, they investigated its anticancer properties through inhibition of vasculogenic mimicry formation mediated by upregulating miR-124 and downregulating STAT3 [53]; another anticancer mechanism could be the upregulation of let-7a and downregulation of STAT3 [54].Wang et al. have shown that a new O-alkylamino-bound derivative of niclosamide, an inhibitor targeting STAT3 signaling and the miR-21/β-catenin axis, inhibits STAT3 with consequent antitumor effects against human head and neck squamous cell carcinoma [55]. Furthermore, Suliman et al. showed that treatment of colon cancer cells with niclosamide inhibits their growth and induces apoptosis, allowing to hypothesize that this treatment could have therapeutic implications for the management of colon cancer. Niclosamide also acts on the miR-200 family leading to an upregulation that resulted in inhibition of colon cancer progression [56].
Another study evaluated the alteration of miRNA expression following exposure to atrazine of zebrafish embryos. Atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine) is an herbicide used to prevent the growth of broadleaf and grassy weeds on crops such as corn, sorghum grass, cane from sugar, and wheat. In particular the main alteration involves miR-10 and suggests targeting of epigenetic regulators of cell cycle and cell signaling, possibly leading to cancer; miR-126 could also influence the cellular processes associated with angiogenesis [57].
Recent reviews reported exposure to pesticides, in particular organochlorine and organophosphates acting as endocrine disruptors, as a risk factor for the onset of breast cancer [58]. These compounds, due to their bio-persistency, have become widespread pollutants after decades of extensive use. Also, experimental studies suggest that exposure to organophosphates and carbamates could lead to the onset of non-Hodgkin lymphoma and increase the risk of developing cancer in various organs such as liver, kidney, thyroid, adrenal glands, bladder, uterus, bones and nervous system. Carbamates studied because possible carcinogens in humans are mancozeb, maneb, metiram, chlorpropham and diallate [59,60].
Exposure to chlordane and especially trans-nonachlor (TNC), one component of technical chlordane, is associated with an increased risk of cutaneous melanoma after correction for sun sensitivity and exposure. TNC is able to downregulate miR-141-3p in normal human melanocytes to levels commonly found in melanocytic cells, predisposing exposed individuals for the development of melanoma; remarkably, in a Drosophila model, TNC also decreased the level of miR-8,promoting an epigenetic multigenerational inheritance of the miR-141-3p/miR-8 defect [61].
Organophosphates such as chlorpyrifos, dichlorvos, monocotophos, malathion and parathion are widely used as pesticides in industrial and domestic environments [62]. A recent study showed that chlorpyrifos acts on circulating steroids and gonadotropins reducing their levels; for this reason it increases the number of alveolar structures in the mammary gland of rats and consequently increase the frequency of benign proliferative lesions, contributing to breast tumorigenesis [63]. Omethoateis another widely used highly toxic, broad-spectrum organophosphate which has been correlated with human tumorigenesis and genetic damage such as changes in telomere length by consistent data in literature. In particular, polymorphism of miRNA genes was closely related to the occurrence of multiple tumors. Wei Wang et all.in a study involving 180 long-term omethoate-exposed workers, found a correlation between encodingmiR-145rs353291gene polymorphic locus and telomere length [64].
Finally conazoles, agricultural fungicides with a tumorigenic effect, act by alteration of miRNA expression. In particular these chemicals induce liver tumors after prolonged administration. Other studies have highlighted the correlation between miRNA deregulation and the onset of liver cancer in workers exposed to fungicides. Specifically, exposure to tumorigenic doses of propiconazole induced alterations in the expression of 63 miRNAs in mouse liver, while triadimefon modulated the expression of 28 miRNAs [17,65].
4. Neurodegenerative diseases
Epidemiological evidence suggests that exposure to environmental toxicants, mainly pesticides, could increase the risk of developing neurodegenerative diseases. Several studies confirmed a role of chronic oxidative stress in the pathogenesis of age-related neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS) and Huntington’s disease [66]. Oxidative stress can induce mitochondrial DNA mutations, depression of the mitochondrial respiratory chain functions, alteration of membrane permeability and also impairment of the mitochondrial antioxidant system. Prolonged rise in the production of mitochondrial ROS increments calcium uptake by protein oxidation and promotes cell death, possibly leading to the pathogenesis of neurodegenerative diseases. Pesticides, by increasing oxidative stress and triggering inflammatory responses, seem to cause miRNA alterations [67].
Subjects exposed to high-dose pesticides show signs of neurotoxicity; in particular, organophosphates and carbamates act by specifically inhibiting the activity of the enzyme acetylcholine in different species including human [68].
37 miRNAs, mainly miR-29b miR-138 and miR-155, were significantly modified in the serum of OP poisoned patients. Target prediction analysis by TargetScan and miRDB databases, followed by analysis of the target genes with the KEGG pathway database also revealed that the identified miRNAs and relative target genes were associated with several pathophysiological pathways which may be involved in neuromuscular disorders: up-regulated miRNAs were involved in pathways targeting mainly axon guidance, neurotrophin, regulation of the actin cytoskeleton, dilated cardiomyopathy, endocytosis, intercellular junctions, as well as signalling pathways like calcium, ErbB, MAPK, Wnt and TGF-beta and ubiquitin mediated proteolysis; target genes of down-regulated miRNAs were involved in the mTOR signalling pathway and the ubiquitin mediated proteolysis. Down-regulation of miR-155 was also highlighted in patients affected by Guillain-Barre Syndrome, a polyneuropathic disorder affecting the peripheral nervous system, confirming the significantly stimulated production of Th1-type cytokines by miR-155 silencing in vitro [69].
Glyphosate, the most used herbicide world-wide, is a phosphonate compound with a controversial safety profile. Glyphosate neurotoxicity is known to be associated with glutamate excitotoxicity and oxidative stress; there is evidence of deleterious effects of gestational exposure on neurodevelopment, including an increased risk of attention deficit hyperactivity disorder, also in human studies. A recent study identified several potential miRNA target genes and their possible neurological pathways using GO term enrichment and KEGG databases; authors evaluated the expression pattern of these miRNA in the prefrontal cortex of postnatal day mouse offspring following glyphosate exposure during pregnancy and lactation. The results indicated 53 differentially expressed miRNAs, with 11 (in particular miR-34b-5p, miR-19b-3p, miR-324-5p, miR-320-3p and miR-322-5p) involved in neurogenesis regulation, neuron differentiation, brain development. Dysregulated expression of miRNAs may be thus involved in the mechanism of glyphosate-induced neurodevelopmental toxicity [70]. In adult rats exposed to glyphosate, changes in dopaminergic markers have been demonstrated; these alterations resulted in behavioral impairment, while chronic exposure is assumed to be responsible for the onset of depression-like behaviors [71].
miRNAs are able to modulate genes involved in the differentiation of neurons in the central nervous system and they seem to be involved in Parkinson's disease (PD) development.
PD is characterized by selective degeneration of dopaminergic neurons in the substantia nigra of pars compacta possibly induced by impaired mitochondrial activity, altered function of lysosomal and proteosomal system or alpha-synuclein (a-syn) protein aggregation. It has been shown that exposure to pesticides induces overexpression of alpha-synuclein. Exposure to atrazine can cause dopaminergic neurotoxicity as it reduces the amount of dopamine in the striatum. In a rat model of atrazine-induced PD, it resulted that a-syn level increased in the substantia nigra with increasing atrazine dose, while peripheral blood levels were not affected. Thus, blood a-syn concentration would not be a diagnostic biomarker of PD. However, atrazine exposure also altered miRNA levels in blood and substantia nigra including miR-7, the expression of which has been shown to be inversely related to a-syn protein level. In particular miR-7 represses the expression of a-syn and consequently cell death mediated by this protein [72].
Also chlorpyrifos, an organophosphorus insecticide and one of the most commonly employed pesticides, has been associated with increasedα-syn expression in neurons [73]. But no evidence of an association between exposure to chlorpyrifos, butyryl cholinesterase-chlorpyrifos adducts or cholinesterase inhibition and increased blood α-syn levels was found in agricultural workers. Slightly higher α-syn blood levels were found only with the PON1-108T (lower paraoxonase enzyme) allele, but with no clear dose-response relation [74]. 287 subjects with idiopathic PD and population controls, characterized by exposure to diazinon, chlorpyrifos and parathion in residential and workplace setting, were enrolled. Three well-known functional PON1 SNPs were genotyped: two exonic polymorphisms (PON1L55 M and PON1Q192R) and the promoter region variant (PON1C-108 T). The exposed carriers of the "faster" metabolizer genotypes, ML or LL, presented lower odds ratios (1.20–1.39) than carriers of the "slower" metabolizer genotype MM (1.78–2.45) relative to unexposed carriers of the faster genotypes. Similarly increased ORs were found for PON1Q192R genotypes, but not for PON1C-108 T genotypes [75]. However, the detailed molecular mechanisms remain unclear and no predictive biomarker has yet been identified. Chlorpyrifos acts through the activation of oxidative stress, which leads to the accumulation of lipid peroxidation products generating reactive oxygen species (ROS). It is known that organophosphate pesticides can induce oxidative stress in exposed subjects. This leads to an increase in free radicals with consequent damage to biological macromolecules and the formation of new compounds such as advanced glycation end products (AGE) and advanced oxidation protein products (AOPP) [76]. Chlorpyrifos causes oxidative stress and miR-19a-AMPK axis disorder while inducing apoptosis and autophagy in common carp kidney [77]. It also seems to inhibit cell proliferation, increase susceptibility to oxidative stress-induced toxicity by increasing miR-181 through down-regulation of the SIRT1 / PGC-1α / Nrf2 pathway in human SH-SY5Y neuroblastoma cells [78]. Subchronic exposure to chlorpyrifos is also implicated in cognitive dysfunctions such as learning and memory deficits. In chlorpyrifos-exposed rats, miR-132 and miR-212 are elevated in the hippocampus CA1 region, and this has been suggested to play a role in the disruption of neurotrophin-mediated cognitive processes. Dichlorvos, another organophosphate compound, can produce both neurotoxicity and non-neuronal toxicity. In porcine kidney epithelial cells, it produces aberrant expression of miRNAs, and this coincides with inhibition of cell proliferation in a dose- and time-dependent manner, which has been suggested to be a result of dichlorvos-induced apoptosis [17].
MPP+ (1-methyl-4-phenylpyridinium) is a neurotoxin acting by promoting the formation of reactive free radicals in mitochondria and causing parkinsonism in primates by suppression of dopamine-producing neurons in the substantia nigra. Since the discovery of its PD-inducing activity, MPP + has given a relevant contribution to PD research. The chloride salt of MPP+, namely cyperquat, has been used in the 1970s as an herbicide; the closely related structural analog paraquat is still used in agriculture in some countries, though raising some safety concerns because exposure has been significantly correlated with PD [79]. Paraquat is characterized by chemical affinity with 1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine (MPTP), a prodrug of MPP + and impurity of synthetic heroin. It has been shown that exposure to paraquat, as well as to MPTP, could alter the expression profile of miRNAs involved in the degeneration of dopaminergic neurons of the substantia nigra. Particularly, one of the toxic effects of paraquat is mediated by the downregulation of miR-17−5p. It is known that miR-17−5p negatively regulates the transcription factor E2F1, a target of c-myc; this mechanism may be at the basis of the increased brain cells apoptosis observed after paraquat treatment [80].
MPP + treatment of SH-SY5Y neuroblastoma cells represents an in vitro experimental model which allowed to observe modulation of several miRNAs related to PD; for this reason, some miRNAs have been suggested as early biomarkers for the diagnosis of this pathology. MPP + induces death in neuroblastoma cells through upregulation of miR-505, which is also highly expressed in the plasma of patients with PD [81]; miR-212 resulted downregulated in SH-SY5Y cells, and its overexpression reduced MPP+ -induced damage; the neuroprotective effect of this miRNA in SH-SY5Y cells could be mediated by the factor 4 (KLF4)/Notch pathway [82]; MPP + also induces upregulation of miR-210-3p with consequent reduction of brain-derived neurotrophic factor (BDNF) production and damage of dopaminergic neurons [83]. Moreover, Geng et al. further demonstrated that miR-494-3p inhibition could play a neuroprotective role in neuroblastoma cells treated with MPP + by negatively regulating SIRT3, suggesting that its suppression could be a potential therapeutic target [84]. Inhibition of miR-384-5p could suppress in vitro rotenone-induced neurotoxicity, while miR-384-5p overexpression worsen neurotoxicity in subjects exposed to rotenone, suggesting miR-384-5p as an important regulator of PD [85]. miR-7 seem to have a neuroprotective effect improving glycolysis and allowing neuronal cells to satisfy energy needs when oxidative phosphorylation is inhibited by MPP + . This miRNA is expressed in the brain and appears to regulate neuronal functions such as neuronal differentiation and neurite growth. Furthermore, it showed a protective function on neuronal cells in different PD models. The first neuroprotective mechanism of miR-7 consists in the downregulation of α-syn aiming at the 3′-UTR of its mRNA. Moreover, it was shown that miR-7 protects against MPP+ -induced cell death by downregulating the RelA subunit of NF-κB [86].
These results suggest that increased levels or miR-7 activity may represent a novel therapeutic strategy for PD [87].
Other authors have shown altered expression of miR-34a, miR-141 and miR-9 in PC12 pheochromocytoma cells; still, in this experimental setting MPP + downregulates the expression of SIRT1, BCL2 and BDNF [88]. Also miR-181b seems to play an important role in the same in vitro model; a recent study suggested that its downregulation inhibits autophagy and reduces cell death by acting on the PTEN/AKT/mTOR signaling pathway [89]. Finally, miR-133b has been supposed to promote neurite growth in dopaminergic neurons and improve axon degeneration following exposure to MPP+; it could exert these beneficial effects by attenuating MPP+ -induced upregulation of α-syn [90].
miRNAs can also act in the pathophysiology and progression of Alzheimer's disease (AD), a neurodegenerative disorder characterized by the formation of intracellular neurofibrillary tangles and extracellular deposition of amyloid-βwith consequent impairment or loss of language, memory, behavior and cognition.
Decremented expression of multiple miRNAs was observed in the brain of AD patients, suggesting a correlation with neuroprotection due to reduced amyloid-β (Aβ) and phosphorilated τ-protein, acting through target genes like APP, BACE1, sirt1, tau. Conversely, other miRNAs induced an increment of Aβ, phosphorilated τ-protein and inflammation, thus promoting neurodegeneration targeting Rb1, BDNF and other genes [91].
The expression of microRNA-153 (miR-153) is reduced in the brains of patients with advanced AD and in addition the same miRNA suppresses the Aβ precursor protein (APP) expression in human fetal brain cells in culture [92]. Other authors have shown that the expression of microRNA-26b (miR-26b) is upregulated in AD and that its overexpression facilitates τ-phosphorylation and consequently the onset of Alzheimer's disease, probably through upregulation of Rb1 gene [91,93]. Also, it was observed that miR-128 dissolves amyloid β-mediated neurotoxicity by targeting PPAR-gamma and inactivating intracellular NF-kB pathway [94]. The phenyl-pyrazole insecticide fipronil and the broad spectrum insecticide/miticide triazophos have been shown to alter miRNA expression in zebrafish and have been suggested to serve as biomarkers for toxicity. Paraquat produces lung toxicity through redox cycling and formation of superoxide anion and eventually hydroxyl radicals leading to lipid peroxidation. In human neural progenitor cells, 66 miRNAs have been found to be differentially regulated in proliferating cells upon paraquat treatment, and in silico analysis has shown that the targets of these miRNAs include genes involved in neural proliferation and differentiation, as well as cell cycle and apoptosis [17].
5. Discussion
This review summarizes the current knowledge linking cancer and neuro-degenerative diseases to dysregulation of microRNA network following pesticide exposure. As summarized in Table 1, most findings revealed differential miRNA expression targeting biomolecules and pathways involved in various neoplastic localizations, PD, AD and other and neurodegenerative diseases.
Table 1.
Literature overview of differential miRNA involvement in neoplastic transformation and neurodegeneration.
| Pesticides | miRNA | Up/down regulation | Study model | Effects | Authors |
|---|---|---|---|---|---|
| Paraquat | miR-122 | ⇧ | fish serum | Liver cancer | [31] |
| miR-155 | ⇧ | fish serum | Inflammatory response activation | [31] | |
| miR-17−5 | ⇩ | Neuro-2 A cells | Brain cells apoptosis Parkinson’s diseases | [80] | |
| Cypermethrin | miR-155 | ⇩ | in vitro | Tumor metastatization | [52] |
| Niclosamide | miR124 | ⇧ | in vitro | Anticancer properties through inhibition of vasculogenic mimicry | [53] |
| STAT 3 | ⇩ | in vitro | Anticancer properties through inhibition of vasculogenic mimicry | [53] | |
| Let-7 | ⇧ | in vitro | Anticancer properties | [53] | |
| miR-200 | ⇧ | in vitro | Inhibition of colon cancer progression | [56] | |
| Trans-nonachlor | miR-141−3p | ⇩ | human melanocytes | Melanoma | [61] |
| miR 8 | ⇩ | Drosophila m. | Epigenetic multigenerational inheritance of the miR-141-3p/miR-8 defect | [61] | |
| Organophosphates | miR 155 | ⇩ | human serum | Guillain-Barre Syndrome, polyneuropathic disorder affecting the peripheral nervous system | [69] |
| Atrazine | miR-7 | ⇩ | blood and substantia nigra | Parkinson diseases due to accumulation of α-syn | [72] |
| Chlorpyrifos | miR-181 | ⇧ | human SH-SY5Y neuroblastoma cells | Inhibit cell proliferation, increase susceptibility to oxidative stress-induced toxicity | [78] |
| miR-132 and miR-212 | ⇧ | rat | Disruption of neurotrophin-mediated cognitive processes | [17] | |
| MPP+ | miR-505 | ⇧ | in vitro | Death in neuroblastoma cells. Parkinson’s diseases | [81] |
| miR-212 | ⇩ | SH-SY5Y cells | Neuroprotective effect | [82] | |
| miR-210-3p | ⇧ | in vitro | Reduction of brain-derived neurotrophic factor (BDNF) production and damage of dopaminergic neurons | [83] | |
| miR-494-3p | ⇩ | in vitro | Neuroprotective role in neuroblastoma cells | [84] | |
| miR-181b | ⇩ | in vitro | Inhibits autophagy and reduces cell death | [89] | |
| miR-133b | ⇧ | rat dopaminergic neurons | [90] | ||
| Rotenone | miR-384-5p | ⇩ | in vitro | Neurotoxcity | [85] |
A growing body of evidence in recent literature indicates that alteration of specific miRNAs can represent an early biomarker of disease following exposure to chemical agents, including pesticides. Examination of current literature suggests that different miRNAs seem to regulate cell proliferation, apoptosis, migration, invasion, and metastasis via many biological pathways through modulation of the expression of target mRNAs: miR-155 following exposure to cypermethrin, miR-21 after niclosamide treatment, miR-10 and miR-126 following atrazine, miR-141-3b and miR-8 with chlordane and TNC. Also, the expression level of miR-145 was significantly down-regulated in many types of tumor following omethoate exposure while dozens of miRNAs including miR-135b have been identified as related to tumor formation after exposure to conazoles.
The evidence highlighted mainly shows that in PD patients or in PD in vivo and in vitro models exposure to chlorpyriphos is associated with dysregulation of miR-19a, miR-132, miR-212 and miR181, while paraquat is associated with modulation of miR-17−5 levels; MPP + is in relation with altered levels of miR-505, miR-210, miR-212, miR-494-3p,miR-384-5p,miR-34a, miR-141,miR-9 andmiR-181b. A possible therapeutic use of miRNAs is suggested by the neuroprotective effect of miR-7 andmiR-133b against experimental PD induced by MPP + and atrazine. Finally in fipronil, triazophos and paraquat -treated cell cultures, multiple miRNAs have been found to be differentially regulated.
However, the precise function of miRNAs in pesticide-induced tumorigenesis and neurotoxicity still necessitates further investigation. Current research focuses on different hypotheses: the disruption of appropriate biogenesis of miRNA, the identification of miRNAs targeting specific disease genes, the epigenetic alterations. These hypotheses have been investigated also using bioinformatics. In recent years, an attempt has been made to extrapolate from experimental studies data about the interaction between miRNAs and target genes in order to establish the function and the complex molecular network responsible for gene regulation. In this way, mathematical algorithms for the prediction of hypothetical miRNAs targeted mRNA were developed.
These computational techniques can be divided into two categories: algorithms based on sequence complementarity, possibly considering evolutionary conservation sites in homologous genes (miRanda, TargetScan, and PicTar); algorithms based on the prediction of the most energy-friendly secondary structure of dual-stranded RNA molecules (RNAhybrid and PITA).
Bioinformatics for the study of target mRNAs is certainly an important tool to better understand the functional role of miRNAs, but the predicted hypothetical targets must always be validated by specific experimental methodologies.
There are several collections of miRNA-mRNA defined as validated, as they are reproduced in vitro, through which it is possible to demonstrate the regulation of target genes by miRNAs and compare both the different prediction algorithms as well as other methodologies aiming to investigate the regulatory action of miRNAs. These include miRecords, TaRBase, mirWalk and mirTarBase [95,96].
miRTarBase provides information on experimentally validated miRNA-target (MTI) interactions [97]. This is an important biological database, continuously updated which gives relevant information related to miRNA. It could also provide a reliable database platform for a wide range of scientific services. The latest version of miRTarBase 8.0 (September 15, 2019) contains 479,340 target genes [98].
However, for both researchers who study miRNAs and those who develop next-generation programs for target prediction, these interaction databases have a strong limit due to the very small fraction of validated compared to the total of possible interactions; and laboratory testing is still very expensive [99].
Epigenetic modifications are stable over time and have been shown to be influenced by the environment. In particular, exposure to pesticides can induce epigenomic alterations. Experimental, clinical and epidemiological studies have made it possible to increase knowledge on the mechanisms of action that alter gene expression. As represented in Fig. 1, it was widely demonstrated that miRNAs de-regulation and other epigenetics modifications represent early events of neoplastic transformation and neurodegeneration [100,101]. Starting from the stability of such epigenetics biomarkers and their predictive value for the early recognition of precancerous lesions or early brain damages, nowadays novel methods based on the evaluation of circulating DNA and miRNAs represent effective strategies for the early diagnosis of chronic-degenerative diseases [[102], [103], [104], [105]].
Fig. 1.
Molecular link between miRNA biogenesis, secretion and neoplastic transformation and neurodegeneration.
In particular, most of the studies conducted so far have focused attention on DNA methylation, but recent research has also investigated the effects on histone modifications and miRNAs. However, further studies are needed to understand whether, for example, the effects observed may depend on exposure to a single pesticide or a complex mixture of different chemicals.
In conclusion it has been seen that following exposure to environmental chemicals and especially to pesticides, alterations in miRNA expression can be observed. In particular, pesticides exposure modulate the expression levels of both oncogenes and tumor suppressor miRNAs, representing early events of neoplastic transformation. Therefore, the evaluation of miRNA expression levels may be used to develop new non-invasive strategies for the prediction and prognosis of many diseases, including cancer. However, the application of miRNAs as diagnostic and therapeutic biomarkers in the clinical field is extremely challenging. Therefore this review aimed to highlight the challenges in the application of microRNA in guiding disease discrimination decisions and its future prospects as a non-invasive diagnostic biomarker.
The major weakness of comparing these studies is that unaccountability of the consistently increasing known and predicted human miRNAs over a period (www.mirbase.org).
Therefore, to compare the data between studies, re-performing of expression profile in the same samples is warranted. Though the miRNA detection technology is quickly evolving, there is a lack of consensus among scientists in using an optimal approach to analyse large-scale miRNA profile. Also, lack of databases providing information regarding temporal and inter-individual miRNA expression variations are limiting the identification of miRNA pattern.
If we overcome these barriers, the richness of information associated with miRNA profiles could partake eventual clinical translation. To design and evaluate more effective diagnostic and therapeutic interventions based on miRNA, ultimately requires appropriate interpretation of differentially expressed miRNAs and their related family members that underpin the PD development and progression. A signature pattern of a family of miRNA can considerably strengthen their diagnostic value over single candidate miRNA. The future investigations should also focus on normal variations of miRNAs associated with PD and related disorders within and between individuals, over time with age, gender, and other aspects of the disease condition. This might give fascinating results to interpret the levels of individual or family of miRNAs significantly varied between individuals without any pathological significance or discern donor-specific variations. Besides, this could help us to define and build a database to understand the human individuality and their association with the disease.
If a new technological platform provides an opportunity for faster miRNA extraction or direct analysis without an extraction step is established, that could significantly improve usability of miRNAs in clinical settings.
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- 1.Falzone L., Marconi A., Loreto C., Franco S., Spandidos D.A., Libra M. Occupational exposure to carcinogens: benzene, pesticides and fibers. Mol. Med. Rep. 2016;14(5):4467–4474. doi: 10.3892/mmr.2016.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsatsakis A.M., Kouretas D., Tzatzarakis M.N., Stivaktakis P., Tsarouhas K., Golokhvast K.S., Rakitskii V.N. Simulating real-life exposures to uncover possible risks to human health: a proposed consensus for a novel methodological approach. Hum. Exp. Toxicol. 2017;36(6):554–564. doi: 10.1177/0960327116681652. [DOI] [PubMed] [Google Scholar]
- 3.Gangemi S., Gofita E., Costa C., Teodoro M., Briguglio G., Nikitovic D., Tzanakakis G. Occupational and environmental exposure to pesticides and cytokine pathways in chronic diseases (review) Int. J. Mol. Med. 2016;38(4):1012–1020. doi: 10.3892/ijmm.2016.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa C., Tsatsakis A., Mamoulakis C., Teodoro M., Briguglio G., Caruso E., Tsoukalas D. Current evidence on the effect of dietary polyphenols intake on chronic diseases. Food Chem. Toxicol. 2017;110 doi: 10.1016/j.fct.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 5.Petrakis D., Vassilopoulou L., Mamoulakis C., Psycharakis C., Anifantaki A., Sifakis S., Docea A., Tsiaoussis J., Makrigiannakis A., Tsatsakis A. Endocrine disruptors leading to obesity and related diseases. Int. J. Environ. Res. Public Health. 2017;14(10):1282. doi: 10.3390/ijerph14101282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fountoucidou P., Veskoukis A.S., Kerasioti E., Docea A.O., Taitzoglou I.A., Liesivuori J., Tsatsakis A., Kouretas D. A mixture of routinely encountered xenobiotics induces both redox adaptations and perturbations in blood and tissues of rats after a long-term low-dose exposure regimen: the time and dose issue. Toxicol. Lett. 2019;317(December):24–44. doi: 10.1016/j.toxlet.2019.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Afolabi O., Kayode, Aderibigbe F.A., Folarin D.T., Arinola A., Wusu A.D. Oxidative stress and inflammation following sub-lethal oral exposure of cypermethrin in rats: mitigating potential of epicatechin. Heliyon. 2019;5(8):e02274. doi: 10.1016/j.heliyon.2019.e02274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rapisarda V., Ledda C., Matera S., Fago L., Arrabito G., Falzone L., Marconi A., Libra M., Loreto C. Absence of t(14;18) chromosome translocation in agricultural workers after short-term exposure to pesticides. Mol. Med. Rep. 2017;15(5):3379–3382. doi: 10.3892/mmr.2017.6385. [DOI] [PubMed] [Google Scholar]
- 9.Costa C., Miozzi E., Teodoro M., Briguglio G., Fenga C. Influence of genetic polymorphism on pesticide-induced oxidative stress. Curr. Opin. Toxicol. 2019;13(February):1–7. doi: 10.1016/j.cotox.2018.12.008. [DOI] [Google Scholar]
- 10.Teodoro M., Briguglio G., Fenga C., Costa C. Genetic polymorphisms as determinants of pesticide toxicity: recent advances. Toxicol. Rep. 2019;6:564–570. doi: 10.1016/j.toxrep.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa C., Miozzi E., Teodoro M., Briguglio G., Rapisarda V., Fenga C. New insights on ‘old’ toxicants in occupational toxicology. Mol. Med. Rep. 2017;15(5):3317–3322. doi: 10.3892/mmr.2017.6374. [DOI] [PubMed] [Google Scholar]
- 12.Fenga C., Gangemi S., Teodoro M., Rapisarda V., Golokhvast K., Docea A.O., Tsatsakis A.M., Costa C. 8-hydroxydeoxyguanosine as a biomarker of oxidative DNA damage in workers exposed to low-dose benzene. Toxicol. Rep. 2017;4 doi: 10.1016/j.toxrep.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa C., Ozcagli E., Gangemi S., Schembri F., Giambò F., Androutsopoulos V., Tsatsakis A., Fenga C. Molecular biomarkers of oxidative stress and role of dietary factors in gasoline station attendants. Food Chem. Toxicol. 2016;90(April):30–35. doi: 10.1016/j.fct.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Fenga C., Gangemi S., Di Salvatore V., Falzone L., Libra M. Immunological effects of occupational exposure to lead. Mol. Med. Rep. 2017;15(5):3355–3360. doi: 10.3892/mmr.2017.6381. [DOI] [PubMed] [Google Scholar]
- 15.Fenga C., Gangemi S., Catania S., De Luca A., Costa C. IL-17 and IL-22 serum levels in greenhouse workers exposed to pesticides. Inflamm. Res. 2014;63(11):895–897. doi: 10.1007/s00011-014-0769-6. [DOI] [PubMed] [Google Scholar]
- 16.Ostuni R., Kratochvill F., Murray P.J., Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36(4):229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Miguel V., Yue Cui J., Daimiel L., Espinosa-Díez C., Fernández-Hernando C., Kavanagh T.J., Lamas S. The role of MicroRNAs in environmental risk factors, noise-induced hearing loss, and mental stress. Antioxid. Redox Signal. 2018;28(9):773–796. doi: 10.1089/ars.2017.7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weldon B.A., Pacheco Shubin S., Smith M.N., Workman T., Artemenko A., Griffith W.C., Thompson B., Faustman E.M. Urinary MicroRNAs as potential biomarkers of pesticide exposure. Toxicol. Appl. Pharmacol. 2016;312(December):19–25. doi: 10.1016/j.taap.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q., Kappil M.A., Li An, Dassanayake P.S., Darrah T.H., Friedman A.E., Friedman M. Exploring the associations between MicroRNA expression profiles and environmental pollutants in human placenta from the national children’s study (NCS) Epigenetics. 2015;10(9):793–802. doi: 10.1080/15592294.2015.1066960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brieno-Enriquez M., Garcia-Lopez J., Cardenas D.B., Guibert S., Cleroux E., Ded L., de D. Hourcade Juan, Peknicova J., Weber M., Del Mazo J. Exposure to endocrine disruptor induces transgenerational epigenetic deregulation of microRNAs in primordial germ cells. PLoS One. 2015;10(4):e0124296. doi: 10.1371/journal.pone.0124296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaiserman Alexander. Early-life exposure to endocrine disrupting chemicals and later-life health outcomes: an epigenetic bridge? Aging Dis. 2014;5(6):419–429. doi: 10.14336/AD.2014.0500419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collotta M., Bertazzi P.A., Bollati V. Epigenetics and pesticides. Toxicology. 2013;307(May):35–41. doi: 10.1016/j.tox.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Ferrante M., Conti G.O. Environment and neurodegenerative diseases: an update on miRNA role. MicroRNA. 2017;6(3) doi: 10.2174/2211536606666170811151503. [DOI] [PubMed] [Google Scholar]
- 24.Banzhaf-Strathmann J., Benito E., May S., Arzberger T., Tahirovic S., Kretzschmar H., Fischer A., Edbauer D. MicroRNA 125-b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014;33(15):1667–1680. doi: 10.15252/embj.201387576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozomara A., Birgaoanu M., Griffiths-Jones S. MiRBase: from MicroRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hajibabaie F., Kouhpayeh S., Mirian M., Rahimmanesh I., Boshtam M., Sadeghian L., Gheibi A., Khanahmad H., Shariati L. MicroRNAs as the actors in the atherosclerosis scenario. J. Physiol. Biochem. 2019;(December) doi: 10.1007/s13105-019-00710-7. [DOI] [PubMed] [Google Scholar]
- 27.Candido S., Lupo G., Pennisi M., Basile M., Anfuso C., Petralia M., Gattuso G. The analysis of MiRNA expression profiling datasets reveals inverse MicroRNA patterns in glioblastoma and Alzheimer’s disease. Oncol. Rep. 2019;(June) doi: 10.3892/or.2019.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Battaglia R., Palini S., Vento M.E., La Ferlita A., Lo Faro M.J., Caroppo E., Borzì P. Identification of extracellular vesicles and characterization of MiRNA expression profiles in human blastocoel fluid. Sci. Rep. 2019;9(1):84. doi: 10.1038/s41598-018-36452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drusco A., Croce C.M. 2017. MicroRNAs and Cancer: A Long Story for Short RNAs; pp. 1–24. [DOI] [PubMed] [Google Scholar]
- 30.Hafsi S., Candido S., Maestro R., Falzone L., Soua Z., Bonavida B., Spandidos D.A., Libra M. Correlation between the overexpression of Yin Yang 1 and the expression levels of MiRNAs in Burkitt’s lymphoma: a computational study. Oncol. Lett. 2016;11(2):1021–1025. doi: 10.3892/ol.2015.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J., Li Y., Wu M., Zhang C., Che Y., Li W., Li X. Serum immune responses in common carp (Cyprinus carpio L.) to paraquat exposure: the traditional parameters and circulating MicroRNAs. Fish Shellfish Immunol. 2018;76(May):133–142. doi: 10.1016/j.fsi.2018.02.046. [DOI] [PubMed] [Google Scholar]
- 32.Dangwal S., Bang C., Thum T. Novel techniques and targets in cardiovascular microRNA research. Cardiovasc. Res. 2012;93(4):545–554. doi: 10.1093/cvr/cvr297. [DOI] [PubMed] [Google Scholar]
- 33.Galun D. Hepatocellular carcinoma: from clinical practice to evidence-based treatment protocols. World J. Hepatol. 2015;7(20):2274. doi: 10.4254/wjh.v7.i20.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falzone L., Salomone S., Libra M. Evolution of cancer pharmacological treatments at the turn of the third millennium. Front. Pharmacol. 2018;9(November) doi: 10.3389/fphar.2018.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neal R.D., Tharmanathan P., France B., Din N.U., Cotton S., Fallon-Ferguson J., Hamilton W. Is increased time to diagnosis and treatment in symptomatic cancer associated with poorer outcomes? Systematic review. Br. J. Cancer. 2015;112(S1):S92–S107. doi: 10.1038/bjc.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nebbioso A., Tambaro F.P., Dell’Aversana C., Altucci L. Cancer epigenetics: moving forward. PLoS Genet. 2018;14(6):e1007362. doi: 10.1371/journal.pgen.1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biswas S., Mallikarjuna Rao C. Epigenetics in cancer: fundamentals and beyond. Pharmacol. Ther. 2017;173(May):118–134. doi: 10.1016/j.pharmthera.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 38.Lin S., Gregory R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banaudha K.K., Verma M. Epigenetic biomarkers in liver cancer. Methods Mol. Biol. 2015;1238:65–76. doi: 10.1007/978-1-4939-1804-1_4. [DOI] [PubMed] [Google Scholar]
- 40.Shin V. Yvonne, Chu K.M. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J. Gastroenterol. 2014;20(30):10432–10439. doi: 10.3748/wjg.v20.i30.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuce D., Ersoy E.O. Lung cancer and epigenetic modifications. Tuberk. Toraks. 2016;64(2):163–170. doi: 10.5578/tt.10231. [DOI] [PubMed] [Google Scholar]
- 42.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M. Human MicroRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falzone L., Scola L., Zanghì A., Biondi A., Di Cataldo A., Libra M., Candido S. Integrated analysis of colorectal cancer MicroRNA datasets: identification of microRNAs associated with tumor development. Aging. 2018;10(5):1000–1014. doi: 10.18632/aging.101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He K., Li W.-X., Guan D., Gong M., Ye S., Fang Z., Huang J.-Fi, Lu A. Regulatory network reconstruction of five essential microRNAs for survival analysis in breast cancer by integrating MiRNA and MRNA expression datasets. Funct. Integr. Genomics. 2019;19(4):645–658. doi: 10.1007/s10142-019-00670-7. [DOI] [PubMed] [Google Scholar]
- 45.Liu X., Wang J., Chen M., Liu S., Yu X., Wen F. Combining data from TCGA and GEO databases and reverse transcription quantitative PCR validation to identify gene prognostic markers in lung cancer. Onco. Ther. 2019;12(January):709–720. doi: 10.2147/OTT.S183944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falzone L., Lupo R., Crimi A., Salemi R., Rapisarda V., Libra M., Candido S. Identification of novel MicroRNAs and their diagnostic and prognostic significance in oral cancer. Cancers. 2019;11(5):610. doi: 10.3390/cancers11050610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polo A., Crispo A., Cerino P., Falzone L., Candido S., Giudice A., De Petro G. Environment and bladder cancer: molecular analysis by interaction networks. Oncotarget. 2017;8(39) doi: 10.18632/oncotarget.18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falzone L., Candido S., Salemi R., Basile M.S., Scalisi A., McCubrey J.A., Torino F., Signorelli S.S., Montella M., Libra M. Computational identification of MicroRNAs associated to both epithelial to mesenchymal transition and NGAL/MMP-9 pathways in bladder cancer. Oncotarget. 2016;7(45) doi: 10.18632/oncotarget.11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falzone L., Romano G., Salemi R., Bucolo C., Tomasello B., Lupo G., Anfuso Carmelina, Spandidos D., Libra M., Candido S. Prognostic significance of deregulated MicroRNAs in uveal melanomas. Mol. Med. Rep. 2019;(February) doi: 10.3892/mmr.2019.9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li M., Huo X., Davuljigari C.B., Dai Q., Xu X. MicroRNAs and their role in environmental chemical carcinogenesis. Environ. Geochem. Health. 2019;41(1):225–247. doi: 10.1007/s10653-018-0179-8. [DOI] [PubMed] [Google Scholar]
- 51.Costa C., Rapisarda V., Catania S., Di Nola C., Ledda C., Fenga C. Cytokine patterns in greenhouse workers occupationally exposed to α-cypermethrin: an observational study. Environ. Toxicol. Pharmacol. 2013;36(3):796–800. doi: 10.1016/j.etap.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 52.Huang B., Warner M., Gustafsson J. Estrogen receptors in breast carcinogenesis and endocrine therapy. Mol. Cell. Endocrinol. 2015;418(December):240–244. doi: 10.1016/j.mce.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 53.Li X., Ding R., Han Z., Ma Z., Wang Y. Targeting of cell cycle and Let-7a/STAT3 pathway by niclosamide inhibits proliferation, migration and invasion in oral squamous cell carcinoma cells. Biomed. Pharmacother. 2017;96(December):434–442. doi: 10.1016/j.biopha.2017.09.149. [DOI] [PubMed] [Google Scholar]
- 54.Li X., Yang Z., Han Z., Wen Y., Ma Z., Wang Y. Niclosamide acts as a new inhibitor of vasculogenic mimicry in oral cancer through upregulation of MiR-124 and downregulation of STAT3. Oncol. Rep. 2017;(December) doi: 10.3892/or.2017.6146. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., Wang S., Wu Y., Ren Y., Li Z., Yao X., Zhang C. Suppression of the growth and invasion of human head and neck squamous cell carcinomas via regulating STAT3 signaling and the MiR-21/β-catenin axis with HJC0152. Mol. Cancer Ther. 2017;16(4):578–590. doi: 10.1158/1535-7163.MCT-16-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suliman M.A., Zhang Z., Na H., Ribeiro A.L.L., Zhang Y., Niang B., Hamid A.S., Zhang H., Xu L., Zuo Y. Niclosamide inhibits colon cancer progression through downregulation of the notch pathway and upregulation of the tumor suppressor MiR-200 family. Int. J. Mol. Med. 2016;38(3):776–784. doi: 10.3892/ijmm.2016.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wirbisky S.E., Weber G.J., Schlotman K.E., Sepúlveda M.S., Freeman J.L. Embryonic atrazine exposure alters zebrafish and human MiRNAs associated with angiogenesis, cancer, and neurodevelopment. Food Chem. Toxicol. 2016;98(December):25–33. doi: 10.1016/j.fct.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fenga C. Occupational exposure and risk of breast cancer. Biomed. Rep. 2016;4(3):282–292. doi: 10.3892/br.2016.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koutros S., Harris S.A., Spinelli J.J., Blair A., McLaughlin J.R., Zahm S.H., Kim S. Non-hodgkin lymphoma risk and organophosphate and carbamate insecticide use in the North American pooled project. Environ. Int. 2019;127(June):199–205. doi: 10.1016/j.envint.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piel C., Pouchieu C., Carles C., Béziat B., Boulanger M., Bureau M., Busson A. Agricultural exposures to carbamate herbicides and fungicides and central nervous system tumour incidence in the cohort AGRICAN. Environ. Int. 2019;130(September) doi: 10.1016/j.envint.2019.05.070. [DOI] [PubMed] [Google Scholar]
- 61.Verrando P., Capovilla M., Rahmani R. Trans-nonachlor decreases MiR-141-3p levels in human melanocytes in vitro promoting melanoma cell characteristics and shows a multigenerational impact on MiR-8 levels in Drosophila. Toxicology. 2016;368–369(August):129–141. doi: 10.1016/j.tox.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 62.Roman P., Cardona D., Sempere L., Carvajal F. Microbiota and organophosphates. NeuroToxicology. 2019;75(December):200–208. doi: 10.1016/j.neuro.2019.09.013. [DOI] [PubMed] [Google Scholar]
- 63.Ventura C., Zappia C.D., Lasagna M., Pavicic W., Richard S., Bolzan A.D., Monczor F., Núñez M., Cocca C. Effects of the pesticide chlorpyrifos on breast cancer disease. Implication of epigenetic mechanisms. J. Steroid Biochem. Mol. Biol. 2019;186(February):96–104. doi: 10.1016/j.jsbmb.2018.09.021. [DOI] [PubMed] [Google Scholar]
- 64.Wang W., Zhang H., Duan X., Feng X., Wang Ti, Wang P., Ding M. Association of genetic polymorphisms of MiR-145 gene with telomere length in omethoate-exposed workers. Ecotoxicol. Environ. Saf. 2019;172(May):82–88. doi: 10.1016/j.ecoenv.2019.01.023. [DOI] [PubMed] [Google Scholar]
- 65.Ross J.A., Blackman C.F., Thai S.F., Li Z., Kohan M., Jones C.P., Chen T. A potential MicroRNA signature for tumorigenic conazoles in mouse liver. Mol. Carcinog. 2010 doi: 10.1002/mc.20620. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 66.Tan L., Yu J.T., Tan L. Causes and consequences of MicroRNA dysregulation in neurodegenerative diseases. Mol. Neurobiol. 2015;51(3):1249–1262. doi: 10.1007/s12035-014-8803-9. [DOI] [PubMed] [Google Scholar]
- 67.Agnihotri A., Aruoma O.I. Alzheimer’s disease and Parkinson’s disease: a nutritional toxicology perspective of the impact of oxidative stress, mitochondrial dysfunction, nutrigenomics and environmental chemicals. J. Am. Coll. Nutr. 2020;39(1):16–27. doi: 10.1080/07315724.2019.1683379. [DOI] [PubMed] [Google Scholar]
- 68.Xie H.Q., Xu T., Chen Y., Li Y., Xia Y., Xu S.L., Wang L., Tsim K.W.K., Zhao B. New perspectives for multi-level regulations of neuronal acetylcholinesterase by dioxins. Chem. Biol. Interact. 2016;259(November):286–290. doi: 10.1016/j.cbi.2016.06.030. [DOI] [PubMed] [Google Scholar]
- 69.Yuan H., Yuan M., Tang Y., Wang B., Zhan X. MicroRNA expression profiling in human acute organophosphorus poisoning and functional analysis of dysregulated MiRNAs. Afr. Health Sci. 2018;18(2) doi: 10.4314/ahs.v18i2.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ji H., Xu L., Wang Z., Fan X., Wu L. Differential MicroRNA expression in the prefrontal cortex of mouse offspring induced by glyphosate exposure during pregnancy and lactation. Exp. Ther. Med. 2017;(December) doi: 10.3892/etm.2017.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu N., Tong Y., Zhang D., Zhao S., Fan X., Wu L., Ji H. Circular RNA expression profiles in hippocampus from mice with perinatal glyphosate exposure. Biochem. Biophys. Res. Commun. 2018;501(4):838–845. doi: 10.1016/j.bbrc.2018.04.200. [DOI] [PubMed] [Google Scholar]
- 72.Li B., Jiang Y., Xu Y., Li Y., Li B. Identification of MiRNA-7 as a regulator of brain-derived neurotrophic factor/α-synuclein Axis in atrazine-induced Parkinson’s disease by peripheral blood and brain MicroRNA profiling. Chemosphere. 2019;233(October):542–548. doi: 10.1016/j.chemosphere.2019.05.064. [DOI] [PubMed] [Google Scholar]
- 73.Slotkin T.A., Seidler F.J. Developmental exposure to organophosphates triggers transcriptional changes in genes associated with Parkinson’s disease in vitro and in vivo. Brain Res. Bull. 2011;86(5–6):340–347. doi: 10.1016/j.brainresbull.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nielsen S.S., Checkoway H., Zhang J., Hofmann J.N., Keifer M.C., Paulsen M., Farin F.M., Cook T.J., Simpson C.D. Blood α-Synuclein in agricultural pesticide handlers in central Washington State. Environ. Res. 2015;136(January):75–81. doi: 10.1016/j.envres.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee P.C., Rhodes S.L., Sinsheimer J.S., Bronstein J., Ritz B. Functional paraoxonase 1 variants modify the risk of Parkinson’s disease due to organophosphate exposure. Environ. Int. 2013;56(June):42–47. doi: 10.1016/j.envint.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Costa C., Gangemi S., Giambò F., Rapisarda V., Caccamo D., Fenga C. Oxidative stress biomarkers and paraoxonase 1 polymorphism frequency in farmers occupationally exposed to pesticides. Mol. Med. Rep. 2015;12(4):6353–6357. doi: 10.3892/mmr.2015.4196. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Q., Zheng S., Wang S., Wang W., Xing H., Xu S. Chlorpyrifos induced oxidative stress to promote apoptosis and autophagy through the regulation of MiR-19a-AMPK Axis in common carp. Fish Shellfish Immunol. 2019;93(October):1093–1099. doi: 10.1016/j.fsi.2019.07.022. [DOI] [PubMed] [Google Scholar]
- 78.Zhao M.W., Yang P., Zhao L.L. Chlorpyrifos activates cell pyroptosis and increases susceptibility on oxidative stress-induced toxicity by MiR-181/SIRT1/PGC-1α/Nrf2 signaling pathway in human neuroblastoma SH-SY5Y cells. Environ. Toxicol. 2019;34(6):699–707. doi: 10.1002/tox.22736. [DOI] [PubMed] [Google Scholar]
- 79.Tangamornsuksan W., Lohitnavy O., Sruamsiri R., Chaiyakunapruk N., Scholfield C.N., Reisfeld Brad, Lohitnavy M. Paraquat exposure and Parkinson’s disease: a systematic review and meta-analysis. Arch. Environ. Occup. Health. 2019;74(5):225–238. doi: 10.1080/19338244.2018.1492894. [DOI] [PubMed] [Google Scholar]
- 80.Wang Q., Zhan Y., Ren N., Wang Z., Zhang Q., Wu S., Li H. Paraquat and MPTP alter MicroRNA expression profiles, and downregulated expression of MiR-17-5p contributes to PQ-Induced dopaminergic neurodegeneration. J. Appl. Toxicol. 2018;38(5):665–677. doi: 10.1002/jat.3571. [DOI] [PubMed] [Google Scholar]
- 81.Zhu J., Wang S., Liang Y., Xu X. Inhibition of MicroRNA-505 suppressed MPP+ -induced cytotoxicity of SHSY5Y cells in an in vitro Parkinson’s disease model. Eur. J. Pharmacol. 2018;835(September):11–18. doi: 10.1016/j.ejphar.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 82.Song Y., Liu Y., Chen X. MiR-212 attenuates MPP + -induced neuronal damage by targeting KLF4 in SH-SY5Y cells. Yonsei Med. J. 2018;59(3):416. doi: 10.3349/ymj.2018.59.3.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang S., Chen S., Liu A., Wan J., Tang L., Zheng N., Xiong Y. Inhibition of BDNF production by MPP + through up-regulation of MiR-210-3p contributes to dopaminergic neuron damage in MPTP model. Neurosci. Lett. 2018;675(May):133–139. doi: 10.1016/j.neulet.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 84.Geng L., Zhang T., Liu W., Chen Y. MiR-494-3p modulates the progression of in vitro and in vivo parkinson’s disease models by targeting SIRT3. Neurosci. Lett. 2018;675(May):23–30. doi: 10.1016/j.neulet.2018.03.037. [DOI] [PubMed] [Google Scholar]
- 85.Jiang M., Yun Q., Shi F., Niu G., Gao Y., Xie S., Yu S. Downregulation of MiR-384-5p attenuates rotenone-induced neurotoxicity in dopaminergic SH-SY5Y cells through inhibiting endoplasmic reticulum stress. Am. J. Physiol.-Cell Physiol. 2016;310(9):C755–C763. doi: 10.1152/ajpcell.00226.2015. [DOI] [PubMed] [Google Scholar]
- 86.Chaudhuri A., Kabaria D.S., Choi D.C., Mouradian M.M., Junn E. MicroRNA-7 promotes glycolysis to protect against 1-methyl-4-phenylpyridinium-induced cell death. J. Biol. Chem. 2015;290(19):12425–12434. doi: 10.1074/jbc.M114.625962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi D.C., Chae Y.-J., Kabaria S., Chaudhuri A.D., Jain M.R., Li H., Mouradian M.M., Junn E. MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-Induced cell death by targeting RelA. J. Neurosci. 2014;34(38):12725–12737. doi: 10.1523/JNEUROSCI.0985-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delavar M.R., Baghi M., Safaeinejad Z., Kiani-Esfahani A., Ghaedi K., Hossein Nasr-Esfahani M. Differential expression of MiR-34a, MiR-141, and MiR-9 in MPP+-treated differentiated PC12 cells as a model of Parkinson’s disease. Gene. 2018;662(July):54–65. doi: 10.1016/j.gene.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 89.Li W., Jiang Y., Wang Y., Yang S., Bi X., Pan X., Ma A., Li W. MiR-181b regulates autophagy in a model of Parkinson’s disease by targeting the PTEN/Akt/MTOR signaling pathway. Neurosci. Lett. 2018;675(May):83–88. doi: 10.1016/j.neulet.2018.03.041. [DOI] [PubMed] [Google Scholar]
- 90.Niu M., Xu R., Wang J., Hou B., Xie A. MiR-133b ameliorates axon degeneration induced by MPP+ via targeting RhoA. Neuroscience. 2016;325(June):39–49. doi: 10.1016/j.neuroscience.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 91.Reddy P.H., Williams J., Smith F., Bhatti J.S., Kumar S., Vijayan M., Kandimalla R., Kuruva C.S., Wang R., Manczak M., Yin X., Reddy A.P. Prog. Mol. Biol. Transl. Sci. 2017;146:127–171. doi: 10.1016/bs.pmbts.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 92.Long Justin M., Ray Balmiki, Lahiri Debomoy K. MicroRNA-153 physiologically inhibits expression of amyloid-β precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J. Biol. Chem. 2012;287(37):31298–31310. doi: 10.1074/jbc.M112.366336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu H., Chu W., Gong L., Gao X., Wang W. MicroRNA-26b is upregulated in a double transgenic mouse model of Alzheimer’s disease and promotes the expression of Amyloid-β by targeting insulin-like growth factor 1. Mol. Med. Rep. 2016;13(3):2809–2814. doi: 10.3892/mmr.2016.4860. [DOI] [PubMed] [Google Scholar]
- 94.Geng L., Zhang T., Liu W., Chen Y. Inhibition of MiR-128 Abates Aβ-mediated cytotoxicity by targeting PPAR-γ via NF-KB inactivation in primary mouse cortical neurons and neuro2a cells. Yonsei Med. J. 2018;59(9):1096. doi: 10.3349/ymj.2018.59.9.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sethupathy P. TarBase: a comprehensive database of experimentally supported animal MicroRNA targets. RNA. 2005;12(2):192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dweep H., Sticht C., Pandey P., Gretz N. MiRWalk – database: prediction of possible MiRNA binding sites by ‘walking’ the genes of three genomes. J. Biomed. Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 97.Chou C.H., Shrestha S., Yang C.-D., Chang N.-W., Lin Y.L., Liao K.W., Chi Huang W. MiRTarBase update 2018: a resource for experimentally validated MicroRNA-Target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang H., Yuan, Lin Y.C.D., Li J., Huang K.Y., Shrestha S., Chin Hong H., Tang Y. MiRTarBase 2020: updates to the experimentally validated MicroRNA–target interaction database. Nucleic Acids Res. 2019;(October) doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., Tsai W.T. MiRTarBase: a database curates experimentally validated MicroRNA–target interactions. Nucleic Acids Res. 2011;39(suppl_1):D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sulas P., Di Tommaso L., Novello C., Rizzo F., Rinaldi A., Weisz A., Columbano A., Roncalli M. A large set of MiRNAs is dysregulated from the earliest steps of human hepatocellular carcinoma development. Am. J. Pathol. 2018;188(3):785–794. doi: 10.1016/j.ajpath.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 101.Arshad A.R., Sulaiman Siti A., Saperi A.A., Jamal R., Ibrahim N.M., Abdul Murad N.A. MicroRNAs and target genes As biomarkers for the diagnosis of early onset of Parkinson disease. Front. Mol. Neurosci. 2017;10(October) doi: 10.3389/fnmol.2017.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tuaeva, Falzone, Porozov, Nosyrev, Trukhan, Kovatsi, Spandidos Translational application of circulating DNA in oncology: review of the last decades achievements. Cells. 2019;8(10):1251. doi: 10.3390/cells8101251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fortunato O., Gasparini P., Boeri M., Sozzi G. Exo-MiRNAs as a new tool for liquid biopsy in lung cancer. Cancers. 2019;11(6):888. doi: 10.3390/cancers11060888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Salemi R., Falzone L., Madonna G., Polesel J., Cinà D., Mallardo D., Ascierto P.A., Libra M., Candido S. MMP-9 as a candidate marker of response to BRAF inhibitors in melanoma patients with BRAFV600E mutation detected in circulating-free DNA. Front. Pharmacol. 2018;9(August) doi: 10.3389/fphar.2018.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zheng H., Liu J.Y., Song F.J., Chen K.X. Advances in circulating MicroRNAs as diagnostic and prognostic markers for ovarian cancer. Cancer Biol. Med. 2013;10(3):123–130. doi: 10.7497/j.issn.2095-3941.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]