Abstract
Snakebite is a socio-economic problem in tropical countries and it is exacerbated by geographical venom variation of snakes. We investigated on venom variation in geographically distinct populations of Echis carinatus from three ecologically distinct regions: Tamil Nadu (ECVTN), Goa (ECVGO), and Rajasthan (ECVRAJ). Venom was fractionated by RP-HPLC, combined with SDS-PAGE, and subjected to tandem mass spectrometry. Toxins were identified, and their relative abundance was estimated. Using NCBI database of Echis genus, we queried the MS/MS spectra, and found 69, 38 and 38 proteins in ECVTN, ECVGO and ECVRAJ respectively, belonging to 8–10 different toxin families. The differences in the venom profiles were due to change in the relative composition of the toxin families. Snake venom metalloproteinase (svMP), Snaclecs and Phospholipase A2 (PLA2) were the major venom components in all the venoms. Heteromeric Disintegrins were found in ECVTN and absent in other venoms. ECVRAJ showed higher abundance of low-molecular-weight (>30 kDa) proteins than ECVTN and ECVGO. Cysteine-rich venom protein (CRISP) was highest in ECVRAJ (7.34%), followed by ECVTN (0.01%) and in ECVGO, it was not detected. These findings highlight the need for evaluating the efficacy of the polyvalent anti-venom to neutralize the toxins from geographically distinct venoms of E. carinatus.
Keywords: Venomics, Mass spectrometry, Snake venom, Envenomation, Toxins, De-complexation, RP-HPLC
Graphical abstract
Highlights
-
•
Relative abundance of toxin families in E. carinatus venom (ECV) varied in the three regions.
-
•
Heteromeric Disintegrins were found only in the ECV from Tamil Nadu.
-
•
Proteolytic processing of P-II and P-III svMP varied considerably in the venoms from different regions.
-
•
Efficacy of the polyvalent anti-venom on geographically distinct ECVs needs testing.
1. Introduction
Snakebites are a major health concern in India, and it claims at an average 46900 deaths annually, among them, a considerable number of victims sustain permanent physical disability (Mohapatra et al., 2011). This alarming mortality rate is mainly due to people resorting to faith healers, poor health infrastructure, limitations in the efficacy of antivenom, and poor economic condition of victims (Padma et al., 2018, Whitaker and Whitaker, 2012). In India, more than 90% of snakebites are from the “Big-four” snakes which include Spectacle Cobra (Naja naja), Common Krait (Bungarus caeruleus), Russell's viper (Daboia russelli), Saw-scaled viper (Echis carinatus) (Whitaker and Whitaker, 2012). A close relative of E. carinatus, E. ocellatus probably bite and kill more people than any other venomous snake in their geographic distribution (Warrell et al., 1977).
Clinical management for snake envenomation depends on the ability of Polyvalent Anti-venom (PAV) to neutralize the venom toxins. The venoms used for producing PAV in India are procured mainly from Irula Cooperative Society based in Tamil Nadu, South India (Whitaker and Whitaker, 2012). Since geographical venom variation in Russell's viper and Indian cobra has been reported, the effectiveness of PAV has been questioned (Jayanthi and Veerabasappa Gowda, 1988, Mukherjee and Maity, 1998, Sharma et al., 2015, Shashidharamurthy et al., 2002). Clinical symptoms of envenomation caused by E. carinatus from different geographical regions are known to vary considerably (Ali et al., 2004, Gillissen et al., 1994, Patra et al., 2017). It implies that the composition of the venom probably varies from one geographic region to another.
The venom proteome of E. carinatus has been elucidated from venom from a single source, the Irula Cooperative Society (Patra et al., 2017). Since extensive post-genomic venom variation has been reported in the Echis genus (Casewell et al., 2014), there is a need to examine the geographical variation in E. carinatus venom (ECV).
Our study investigated geographical venom variation in three distinct populations of Echis carinatus, from Tamil Nadu (ECVTN), Goa (n = 3, ECVGO), Rajasthan (n = 1, ECVRAJ). The three regions represent distinct ecological conditions and we hypothesize that venom composition in snakes might be different. To test this, we used the snake venomics protocol (Calvete et al., 2007, Eichberg et al., 2015) to identify the relative toxin composition from these regions. De-complexation of the venom was carried out using RP-HPLC and SDS-PAGE, and subjected to in-gel digestion, and tandem mass spectrometry, to identify toxins, and estimate relative abundance of toxins.
2. Materials and methods
2.1. Venom collection
Echis carinatus venom from Tamil Nadu was procured from Irula Cooperative Society, Tamil Nadu. Echis carinatus from Goa and Rajasthan were captured from their habitat (see Supplementary material S1, Table 1). Venom was milked by allowing snakes to bite the membrane fastened on the glass beaker (Mirtschin et al., 2006), and pooled for the region. It was transported in dry ice and lyophilized in ScanVac Freeze dryer (Labogene, Denmark).
2.2. Fractionation of venom components
2–3 mg of lyophilized ECV was dissolved in 200 μl of 5% ACN, 0.1% TFA solution and centrifuged at 13000 rpm for 10 min. The supernatant was fractionated by Reverse-phase chromatography using HPLC 1200 series (Agilent Technologies, CA, USA) and Zorbax 300SB C-18 (4.6 × 250 mm, 5 μm) column (Agilent Technologies, CA, USA). A linear gradient of 0.1% TFA (Solvent A) and 100% Acetonitrile (Solvent B) using following elution condition: 5% B for 5 min, 5–25% B for 25 min, 25–40% B for 120 min and 40–70% for 20 min at a flow rate of 1 ml/min. Protein was detected at 215 nm with a reference wavelength of 600 nm (Eichberg et al., 2015). Fractions were collected manually and vacuum dried using Savant™ SpeedVac (ThermoFisher Scientific, Waltham, MA, USA).
Dried fractions were reconstituted in double-distilled water and loaded on SDS-PAGE in reducing, and non-reducing conditions. The bands were cut from short SDS run, reduced with 10 mM DTT in 50 mM Ammonium bicarbonate at 56 °C for 45 min. It was later alkylated with 55 mM iodoacetamide in 50mM Ammonium bicarbonate at RT for 30 min. In-solution digestion was performed for the fractions not visible on SDS-PAGE using the standardized protocol (Wiśniewski et al., 2009). The samples were then digested with sequencing grade Trypsin from bovine Pancreas (Roche, Risch-Rotkreuz, Switzerland). Desalting of peptides were done using C18 Pierce® Zip tips (Thermo Fisher Scientific, Waltham, MA, USA) before loading in a Nanospray capillary column (PepMap™ RSLC C18, Thermo Fisher Scientific, Waltham, MA, USA), and subjected to sequencing by MS/MS in Q-Exactive HF (Thermo Fisher Scientific, Waltham, MA, USA). The spectra were analyzed using Proteome Discoverer (Version 2.2) against the NCBI protein database of Echis Genus (NCBI txid: 8699). MS/MS mass tolerance was set to 10 ppm. Carbamidomethyl cysteine was set as a fixed modification. Oxidation of methionine and deamidation of Arginine, and Glutamine were set as variable modification.
2.3. Calculating relative abundance of toxin fractions
Peak area at 215 nm was calculated using the tangent skim method (Haver T, n.d.) on Chemstation (Agilent technologies, CA, USA). The area under each peak was divided by the total area of all the peaks to obtain a relative percentage of total peptide bonds present in each peak. For all the peaks containing multiple bands in SDS PAGE, the proportion of each band in the lane was estimated using densitometry analysis by ImageJ software. The relative percentage of peptide bonds was converted to the relative percentage of molecules by dividing it by the average number of peptide bonds in the respective peaks (Eichberg et al., 2015). Relative abundances of toxins were estimated by compiling data from peak fractions of RP-HPLC and precursor abundance from MS spectra.
2.4. MS/MS data analysis
The MS/MS spectra were analyzed using proteome discoverer (Version 2.2) using the workflow described in Supplementary material S1, Fig. 1. Double and triple charge peptides were selected and matched proteins were filtered for two unique peptides. The abundance of each protein was estimated by summing up all the matched peptide intensity for that protein. The proteins were assigned to its toxin family and the relative percentage for each toxin family was estimated (Supplementary material S2, Tables 1–3). Unique accession from each toxin family were aligned using Clustal omega (https://www.ebi.ac.uk/Tools/msa/clustalo). Peptides sequenced by MS/MS were searched and highlighted on the aligned protein sequences using a macro code snippet (Chen. V, n.d.), see Supplementary S1, Fig. 5. Protein isoforms with at least one unique peptide was included in the list (Supplementary material S3, Figs. 1–3).
3. Results
3.1. SDS-PAGE analysis of venoms
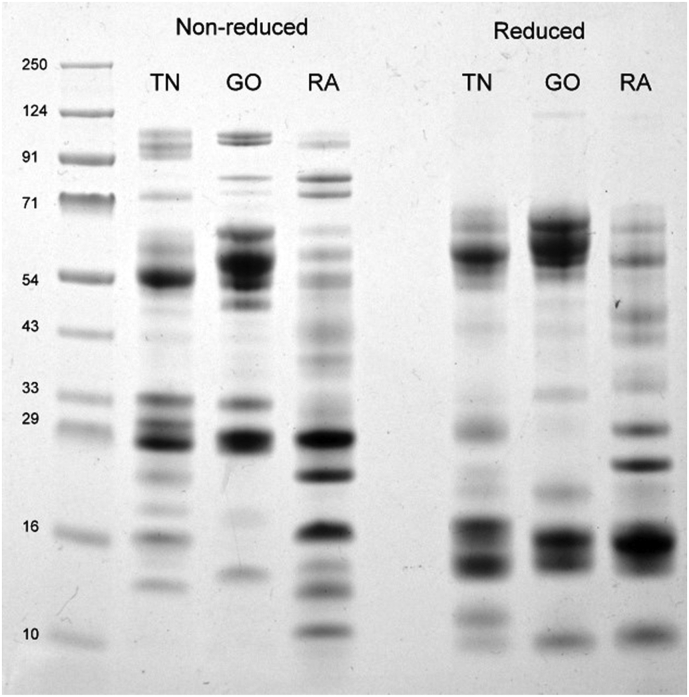
20μg of ECV from three different geographic regions were loaded on 12% polyacrylamide gels under reducing and non-reducing conditions (Fig. 1). Densitometry analysis using ImageJ software revealed that the majority of protein bands had molecular weight from 45 to 70 kDa and 18–35 kDa for ECVTN and ECVGO (Supplementary material S1, Table 2). ECVRAJ had a high abundance of low molecular weight proteins less than 30 kDa when compared with ECVGO and ECVTN. Protein bands above 70 kDa in the non-reducing lanes were not seen in the reducing lanes. Venom proteins at 27–33 kDa in non-reducing lanes of ECVTN and ECVGO were reduced to 12–16 kDa (Fig. 1).
Fig. 1.
20μg of Crude ECV from Tamil Nadu (TN), Goa (GO) and Rajasthan (RA) was loaded on 12% SDS-PAGE under reducing and non-reducing condition. Bands from each lane were cut, in-gel digested and subjected to MS/MS to identify toxin families and compared across lanes.
3.2. Relative abundance of venom components
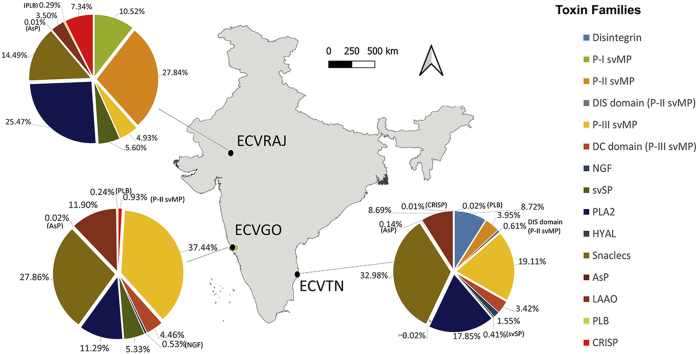
We were able to detect 69, 38, 38 unique proteins from ECVTN, ECVGO and ECVRAJ respectively (See Supplementary material S2, Table 4, 5 and 6). The identified proteins were assigned to 8–10 different toxin families with variation in composition seen across different venoms (Fig. 5). Estimation of relative abundance revealed three predominant class of toxin families - Snake venom metalloproteinase (svMP), Snaclecs and Phospholipase A2 (PLA2). Renin-like Aspartic Protease (AsP), Nerve growth factor (NGF), Hyaluronidase (HYAL), and Phospholipase B (PLB) were detected in trace amounts. Heteromeric disi ntegrins and Disintegrin domain of P-II svMP were only detected in ECVTN. DC-domain of PIII-svMP was detected in ECVTN and ECVGO. Low molecular weight toxins including P–I svMP, PLA2, and CRISP had higher abundance in ECVRAJ compared to others validating the densitometry result from SDS-PAGE (Supplementary material S1, Table 2).
Fig. 5.
Pie chart representing relative abundance of toxin families for Echis carinatus venom. svMP (snake venom Metalloproteinases), NGF (Neural growth factors), Phospholipase A2 (PLA2), Phospholipase B(PLB), renin-like aspartic protease (AsP), svSP (snake venom Serine proteinase), DIS domain (P-II svMP)- Disintegrin domain of P-II svMP, DC domain (Disintegrin-like and cysteine domain of P-III svMP), HYAL (Hyaluronidase), LAAO (L-amino acid oxidase), PLB (Phospholipase B), CRISP (Cysteine-rich secretory protein).
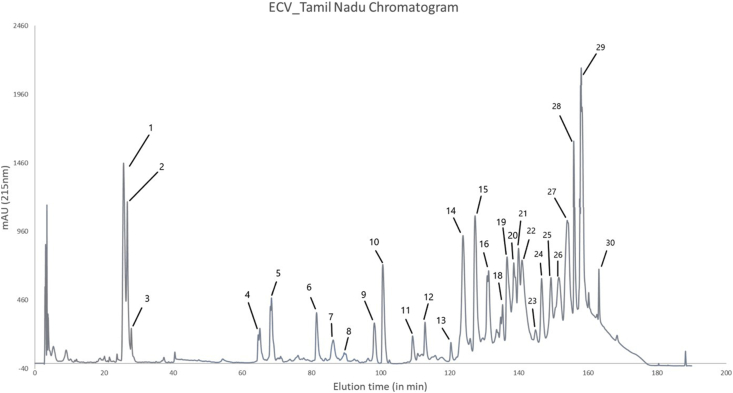
3.3. Venom profile of ECVTN
Peptides sequenced in HPLC peaks 1–3 were detected using in-solution digestion, and were matched to short disintegrins (5.4 kDa) (Fig. 2). Peaks 4 and 5 eluted out between 60th and 80th minute had heterodimeric disintegrins-16 kDa (non-reducing) and two bands at 12–14 kDa (reducing), and homodimeric disintegrins-22 kDa (non-reducing) and single band at 12 kDa (reducing) respectively (Supplementary material S1, Fig. 2). Disintegrin domain of P-II svMP was also detected in these peaks. Peaks 6 showed the band at 22 kDa in the non-reducing, and the reducing gel, and was matched to DC domain of P-III svMP. Peak 9–11 were at 14 kDa in both the gels, with peptides that matched with Phospholipase A2 (PLA2). NGF was detected in Peaks 7–10, but were not observed on the gels. Peaks 12–16 were identified as toxins from Snaclec family. They showed 3–4 bands at 25–30 kDa in the non-reducing gel, which were reduced to 12–16 kDa in the reducing gel. All the SDS-PAGE lanes from peaks 18–30 had bands in the range of 50–70 kDa corresponding to the presence of Snake venom metalloproteinase (P-II and P-III svMP) and L-amino acid oxidase (LAAO). Faint band profiles of Snaclec toxin family were seen from peaks 18–30. High-molecular-weight svMPs were observed in Peak 25, 28, and 29, but they were not observed in the reducing gels. Renin-like Aspartic protease (AsP), Hyaluronidase and Phospholipase B were detected in traces at Peaks 25–28 that eluted between 160th and 180th minute.
Fig. 2.
Reverse phase-HPLC of ECV from Tamil Nadu (ECVTN). 2–3 mg of venom was loaded and 30 fractions were collected manually, dried and reconstituted with double distilled water.
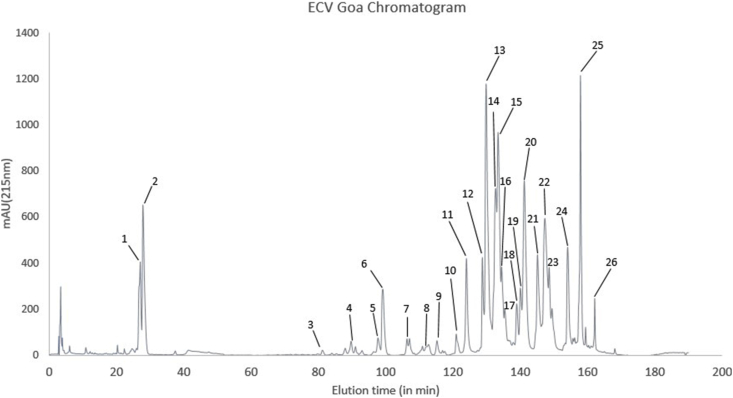
3.4. Venom profile of ECVGO
The peaks between 20 and 30th min of RP-HPLC (Peak 1 and 2) were short disintegrins (5.4 kDa) (Fig. 3). PLA2 eluted out between 90th and 100th minute, in peaks 4 to 6, corresponding to the band at 14 kDa, in reducing and non-reducing gels. DC domain of P-III svMP corresponding to 30 kDa band at the reducing, and the non-reducing gel was identified in peak 8. Toxins from Snaclec family were found in peaks 10–12 eluted between 120th and 10th minute. Peaks 13–25 showed the presence of svMP and LAAO corresponding to 50–70 kDa in the non-reducing and the reducing gel. Serine proteases were detected in trace amounts between peak 8 and peak 12. There were high molecular weight bands (~120kDa) seen at peak 16, 23 and 25, which were not seen in the reducing gel. Snaclecs co-eluted at peak 23 corresponded to the band at 25–30 kDa in the non-reducing and 12–16 kDa in the reducing gel (Supplementary material S1, Fig. 3). Traces of NGF and renin-like aspartic proteases were identified in peak 25, but were not seen on the SDS-PAGE.
Fig. 3.
Reverse phase-HPLC of Echis carinatus venom from Goa (ECVGO). 2–3 mg of venom was loaded and 26 fractions were collected manually, dried and reconstituted with double distilled water.
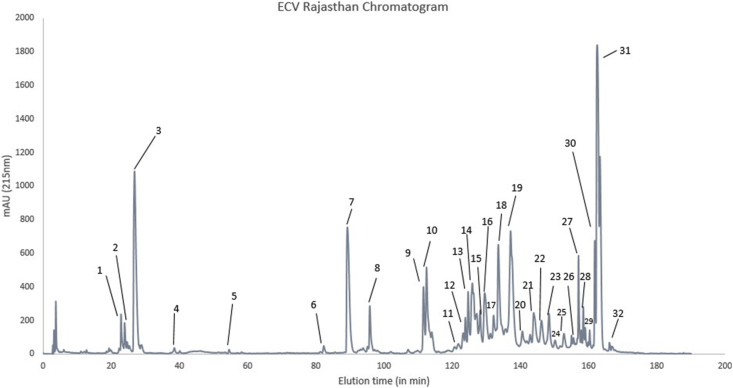
3.5. Venom profile of ECVRAJ
Peaks 1–3 eluted between 20 and 30 min, and it matched with short disintegrins (5.4 kDa) (Fig. 4). PLA2 was present in peak 7 and 8. It corresponded to the bands at 14 kDa, that eluted between 90th and 100th minutes. Peaks 9 and 10 eluted between 110th and 120th minutes, and they correspond to 30 kDa band. This band had Cysteine-rich venom secretory protein (CRISP), that co-eluted with serine protease. The bands from peaks from 12 to 18 were seen at 25–30 kDa in the non-reducing and at 12–16 kDa in the reducing gel. The peptides from these peaks matched with Snaclec toxin. The peaks from 20 to 30 had bands in SDS at 50–70kDa, with the presence of svMP and LAAO. Peaks 21–23 had distinct ladder-like bands in the reducing gel. High molecular weight svMP were identified in peaks 23, 27 and 28. In peak 31, P–I svMP and P-II svMP were detected at 22kDa (Supplementary material S1, Fig. 4).
Fig. 4.
Reverse phase-HPLC of Echis carinatus venom from Udaipur, Rajasthan (ECVRAJ). 2–3 mg of venom was loaded and 32 fractions were collected manually, dried and reconstituted with double distilled water.
4. Discussion
The SDS-PAGE analysis of ECVTN, ECVGO and ECVRAJ under reducing and non-reducing conditions provided us with the preliminary idea about differences in toxin profiles of venom collected from different regions (Fig. 1). Previous proteomics profiles of Echis carinatus from Tamil Nadu suggested toxins belonging to 6–15 toxin families (Casewell et al., 2014, Patra et al., 2017, Senji Laxme et al., 2019) namely, snake venom metalloproteinases and LAAO (~50–120 kDa); Snaclecs, CRISP, NGF and Serine proteases at 25–35 kDa; Disintegrins and PLA2 at 12–16 kDa. All venoms contained high-molecular-weight proteins (70–120 kDa) probably belonging to P-IV svMP or dimers of P-III svMP. The densitometry analysis of band patterns suggests svMP and Snaclecs as prominent in ECVGO and ECVTN whereas low molecular weight proteins, such as, PLA2 (~14 kDa) and CRISP (~30 kDa) were present in ECVRAJ (Fig. 1).
Venoms from three different regions showed distinct RP-HPLC profiles with toxins eluting out at similar time frames but with different peak areas suggesting differences in the composition of each toxin. Venom proteome of species belonging to Echis genus has high inter-specific variation in the relative composition of toxins (Casewell et al., 2014). Broadly, Echis group of snakes have venoms that contain high abundance of svMP, PLA2 and Snaclecs. Our data from ECVTN and ECVGO corroborated with this finding, but ECVRAJ deviated from it. Toxins belonging to Snaclecs, Serine proteases and svMPs appeared in multiple RP-HPLC peaks, could be because the toxins exist as multiple isoforms (Supplementary material S3, Figs. 1, 2, 3). Previously, toxin families and their relative compositions were identified in E. carinatus venom from Irula cooperative society, Tamil Nadu using size exclusion chromatography and SDS-PAGE (Patra et al., 2017, Senji Laxme et al., 2019). Their findings matched with our results from ECVTN broadly, but with differences in the relative abundance of few toxins.
Disintegrins are a strong inhibitor of platelet aggregation and thereby prevents blood coagulopathy (Calvete, 2013, Calvete, 2005). They are a group of non-enzymatic toxins released in the venom by proteolytic processing of svMPs. Short disintegrins were present in all the venoms in initial RP-HPLC peaks that are known to block the function of integrin receptors (Calvete et al., 2007). ECVTN had two peaks between 60th and 70th minute identified as heteromeric disintegrins (8.72%), which were absent in ECVGO and ECVRAJ (Fig. 5). These disintegrins could be result of proteolytic processing of svMP (Doley and Kini, 2009). This indicates variation in proteolytic processing mechanism of svMPs in the geographically distinct venoms of ECV.
Phospholipase A2 is known to be the most potent pharmacologically active venom component, having a wide range of effects including myotoxicity, hemolytic activity, haemorrhage and cardiotoxicity (Doley and Kini, 2009). It eluted out between 90th and 100th minute in RP-HPLC gradient, and showed variable peaks in different venoms. ECVRAJ had the highest proportion of PLA2 (25.5%), followed by ECVTN (18%) and ECVGO (11.3%) (Fig. 5). Differences in the relative abundance of PLA2 could influence the symptoms from E. carinatus bites in these regions.
Snake venom serine proteases (svSP) catalyze various reactions in the blood coagulation pathway (Matsui et al., 2000). They were detected in all the venoms between 110th and 140th minute in RP-HPLC. The proteomes of snake venoms have repeatedly revealed multiple isoforms of Serine proteases (Supplementary material S3, Figs. 1–3). Since we detected a five-fold variation in the relative proportion of svSP in ECV from three regions, we anticipate that concomitant changes in blood coagulation symptoms in victims could be manifested.
Snaclecs (Snake Lectins) are known to either inhibit or activate platelets thereby responsible for consumption coagulopathy reported in viper bites patients (Clemetson, 2010). It was the second most predominant toxin in ECVTN and ECVGO. Four different toxins namely: C-Type lectins, Echicetin, Coagulation factor X/IX, and EMS 16 were identified. We observed four distinct bands at 25–30 kDa in the non-reducing gel, that were reduced to 16-12 kDa bands in the reducing gels. ECVTN had the highest relative abundance of Snaclecs (32.2%), followed by ECVGO (27.9%) and ECVRAJ (14.5%). We propose that symptoms mediated by Snaclecs might be pronounced in the E. carinatus bites in Tamil Nadu and Goa.
Snake venom Metalloproteinase (svMP) toxins facilitate the entry of other toxins by breaking the extracellular fibrinogen, and basement membrane, thereby inducing haemorrhage (Doley and Kini, 2009, Matsui et al., 2000). Based on their domain structure, they are categorized into P–I, P-II, P-III and P-IV. It is the most abundant toxin family in ECV. P-II and P-III svMP were observed in all the venoms. P–I svMP was only detected in ECVRAJ, which eluted at peak 31 in RP-HPLC (Fig. 5). There were 37, 19, and 19 isoforms from svMP family retrieved in ECVTN, ECVRAJ and ECVGO respectively (Supplementary material S3, Figs. 1–3). The pooling of venom from different individuals at Irula cooperative society, TN might be the reason for more number of isoforms in ECVTN. In the HPLC run, time of elution for svMP is 140th to 160th minutes for all the venoms. High molecular weight svMPs (~120kDa) were present, which were reduced to 54 kDa protein. This signified the presence of either dimeric P-III svMP of P-IV svMP, with different subunits attached by cysteine bonds.
L-amino oxidases (LAAO) is a flavoprotein which is involved in the oxidative deamination of L-amino acid and releasing hydrogen peroxide resulting in various symptoms, including: edema formation, platelet aggregation, apoptosis and blood anticoagulation (Doley and Kini, 2009). In our study, it co-eluted with svMP toxins with a molecular weight of ~54 kDa. The peptides of LAAO were detected after 150th min of HPLC in all the venoms. ECVTN and ECVGO showed similar relative abundance (8.69% and 11.9% respectively). In ECVRAJ, the relative abundance of these peptides was low (3.5%).
5. Conclusion
Although research on snake venom has gained momentum in the last few years in India, incompatibility of methods used has eluded comparisons of proteomes. We suggest that de-complexation strategies are essential for venom proteome analyses (Eichberg et al., 2015, Tan et al., 2019). Since, only <10% of the peptides in E. carinatus venom could be matched to the NCBI database, we emphasize the need for whole-genome sequence and transcriptome data for venomous snakes. We speculate that the variation in the composition of toxins from geographically distinct venoms of E. carinatus, could be one of the reasons for the low efficacy of PAV, and an increase in the burden of treatment.
Credit author statement
Siddarth Bhatia: Conceptualization; Methodology; Investigation; Formal analysis; Validation; Data Curation; Data Curation; Writing - Review & Editing; Visualization. Karthikeyan Vasudevan: Conceptualization; Methodology; Investigation; Formal analysis; Validation; Resources; Supervision; Project administration; Funding acquisition; Writing - Review & Editing.
Ethical statement
The research was approved by the Centre for Cellular and Molecular Biology, Hyderabad Internal ethics committee. Appropriate permissions from the Forest Department of Goa and Rajasthan were obtained for venom extraction.
Declaration of competing interest
The authors have no conflict of interest.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank CSIR-CCMB providing support for the study. Council for Scientific and Industrial Research, India provided graduate student scholarship to SB for conducting his research. SERB-Department of Science and Technology provided support to KV for the study through grant - SERB/MenuPage" title = "https://www.serbonline.in/SERB/MenuPage">EMR/2017/005515. State Forest departments of Goa and Rajasthan provided permissions to venom collection from snakes. We thank Mr Nirmal Kulkarni and the Nature and Wildlife Rescue Society, Udaipur for helping with the collection of snakes. We thank the anonymous reviewers of the manuscript for their valuable inputs. Mrs V Krishna Kumari, Mrs Sivakama Sundari, Mrs Y Kameshwari and Mr B. Raman provided valuable assistance for use of RP-HPLC and Mass spectrometry.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.toxcx.2020.100048.
Appendix B. Supplementary data
The following are the supplementary data related to this article:
Supplementary material S1 contains Fig. 1 contains workflow for assignment of peptides to MS/MS spectra using Proteome discoverer (version 2.2); Figs. 2–4 contains SDS-PAGE for RP-HPLC fractions of ECVTN, ECVGO and ECVRAJ respectively; Fig. 5 contains macro code snippet; Table 1 shows collection of venom samples from different locations; Table 2 shows densitometry for venoms loaded on SDS-PAGE.
Supplementary material S2 contains Tables 1–3 that shows assignment of peptides to MS/MS spectra with estimation of relative abundance of each protein and Tables 4–6 that shows unique proteins matched in the data along with toxin families.
Supplementary material S3 contains; Figs. 1, 2 and 3 show alignment of identified peptides on matched proteins from Echis genus database.
References
- Ali Gazanfar M., Bagh Tulsi, Ali G., Kak M., Kumar M., Bali S.K., Tak Si, Hassan G., Wadhwa M.B. vol. 14. 2004. (Acute Renal Failure Following Echis Carinatus (Saw-Scaled Viper) Envenomation). [Google Scholar]
- Calvete Juan J. The continuing saga of snake venom disintegrins. Toxicon. 2013;62:40–49. doi: 10.1016/j.toxicon.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Calvete Juan J., Juárez Paula, Sanz Libia. vol. 42. 2007. “Snake venomics. Strategy and applications.” pp. 1405–14. (Journal of Mass Spectrometry). [DOI] [PubMed] [Google Scholar]
- Calvete, Juan J., Cezary marcinkiewicz, daniel monleón, vicent esteve, bernardo celda, paula Juárez, and libia Sanz. 2005. “Snake venom disintegrins: evolution of structure and function.” Toxicon 45(8):1063–1074. [DOI] [PubMed]
- Casewell Nicholas R., Simon C., Wagstaff Wolfgang Wuster, Darren A.N., Cook, Fiona M. S. Bolton, Sarah I. King, Davinia Pla, Sanz Libia, Calvete Juan J., Harrison Robert A. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl. Acad. Sci. U. S. A. 2014;111(25):9205–9210. doi: 10.1073/pnas.1405484111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Vera. n.d. “2 Quick Ways to Find Multiple Items in One Word Document at the Same Time.” Retrieved March 26, 2020 (https://www.datanumen.com/blogs/2-quick-ways-find-multiple-items-one-word-document-time/).
- Clemetson Kenneth J. Snaclecs (snake C-type lectins) that inhibit or activate platelets by binding to receptors. Toxicon. 2010;56(7):1236–1246. doi: 10.1016/j.toxicon.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Doley R., Kini R.M. Protein complexes in snake venom. Cell. Mol. Life Sci. 2009;66(17):2851–2871. doi: 10.1007/s00018-009-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichberg Susann, Sanz Libia, Calvete Juan J., Davinia Pla. Constructing comprehensive venom proteome reference maps for integrative venomics. Expet Rev. Proteonomics. 2015;12(5):557–573. doi: 10.1586/14789450.2015.1073590. [DOI] [PubMed] [Google Scholar]
- Gillissen Adrian, Theakston R. David G., Barth Jürgen, May Burkard, Krieg Michael, Warrell David A. “Neurotoxicity, haemostatic disturbances and haemolytic anaemia after a bite by a Tunisian saw-scaled or carpet viper (Echis ’pyramidum’-complex): failure of antivenom treatment. Toxicon. 1994;32(8):937–944. doi: 10.1016/0041-0101(94)90372-7. [DOI] [PubMed] [Google Scholar]
- Haver, Tom. n.d. “Intro. to Signal Processing:Integration and Peak Area Measurment.” Retrieved February 25, 2020 (https://terpconnect.umd.edu/~toh/spectrum/Integration.html).
- Jayanthi G.P., Veerabasappa Gowda T. “Geographical variation in India in the composition and lethal potency of Russell's viper (Vipera russelli) venom. Toxicon. 1988;26(3):257–264. doi: 10.1016/0041-0101(88)90216-4. [DOI] [PubMed] [Google Scholar]
- Matsui Taei, Fujimura Yoshihiro, Titani Koiti. Snake venom proteases affecting hemostasis and thrombosis. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000;1477(1–2):146–156. doi: 10.1016/s0167-4838(99)00268-x. [DOI] [PubMed] [Google Scholar]
- Mirtschin Peter J., Dunstan Nathan, Hough Ben, Hamilton Ewan, Klein Sharna, Lucas Jonathan, Millar David, Frank Madaras, Nias Timothy. Venom yields from Australian and some other species of snakes. Ecotoxicology. 2006;15(6):531–538. doi: 10.1007/s10646-006-0089-x. [DOI] [PubMed] [Google Scholar]
- Mohapatra Bijayeeni, Warrell David A., Wilson Suraweera, Bhatia Prakash, Dhingra Neeraj, Jotkar Raju M., Rodriguez Peter S., Mishra Kaushik, Whitaker Romulus, Jha Prabhat. Snakebite mortality in India: a nationally representative mortality survey. PLoS Neglected Trop. Dis. 2011;5(4) doi: 10.1371/journal.pntd.0001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A.K., Maity C.R. The composition of Naja Naja venom samples from three districts of West Bengal, India. Comparative Biochemistry and Physiology - A Molecular and Integrative Physiology. 1998;119(2):621–627. doi: 10.1016/s1095-6433(97)00475-3. [DOI] [PubMed] [Google Scholar]
- Padma Y., Sarojinidevi N., Ratnam K.V. Management of snake bite by Indian quackery: a medical mockery. J. Adv. Med. 2018;7(1):13–15. [Google Scholar]
- Patra Aparup, Kalita Bhargab, Chanda Abhishek, Ashis K., Mukherjee Proteomics and antivenomics of Echis carinatus carinatus venom: correlation with pharmacological properties and pathophysiology of envenomation. Sci. Rep. 2017;7(1):1–17. doi: 10.1038/s41598-017-17227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senji Laxme R.R., Khochare Suyog, Francisco de Souza Hugo, Ahuja Bharat, Suranse Vivek, Martin Gerard, Whitaker Romulus, Sunagar Kartik. “Beyond the ‘big four’: venom profiling of the medically important yet neglected Indian snakes reveals disturbing antivenom deficiencies. PLoS Neglected Trop. Dis. 2019;13(12) doi: 10.1371/journal.pntd.0007899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma Maitreyee, Das Diganta, Janaki Krishnamoorthy Iyer. Manjunatha Kini R., Doley Robin. Unveiling the complexities of Daboia russelii venom, a medically important snake of India, by tandem mass spectrometry. Toxicon. 2015;107:266–281. doi: 10.1016/j.toxicon.2015.06.027. [DOI] [PubMed] [Google Scholar]
- Shashidharamurthy R., Jagadeesha D.K., Girish K.S., Kemparaju K. Variations in biochemical and pharmacological properties of Indian cobra (Naja Naja Naja) venom due to geographical distribution. Mol. Cell. Biochem. 2002;229(1–2):93–101. doi: 10.1023/a:1017972511272. [DOI] [PubMed] [Google Scholar]
- Tan, Hock Choo, Tan Kae Yi, Tan Nget Hong. vol. 1871. Humana Press Inc; 2019. A protein decomplexation strategy in snake venom proteomics; p. 83. (–92 in Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- Warrell D.A., Davidson N. Mod, Greenwood B.M., Ormerod L.D., Pope Helen M., Watkins Barbara J., Prentice C.R.M. Poisoning by bites of the saw-scaled or carpet viper (Echis carinatus) in Nigeria. QJM: Int. J. Med. 1977;46(1):33–62. [PubMed] [Google Scholar]
- Whitaker Romulus, Whitaker Samir. Venom, antivenom production and the medically Important snakes of India. Curr. Sci. 2012;103(6):635–643. [Google Scholar]
- Wiśniewski Jacek R., Alexandre Zougman, Nagaraj Nagarjuna, Mann Matthias. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.