Abstract
Background
Adult skin fibroblasts represent the most common starting cell type used to generate human induced pluripotent stem cells (F-hiPSC) for clinical studies. Yet, a foetal source would offer unique advantages, primarily the absence of accumulated somatic mutations. Herein, we generated hiPSC from cord blood multipotent mesenchymal stromal cells (MSC-hiPSC) and compared them with F-hiPSC. Assessment of the full activation of the pluripotency gene regulatory network (PGRN) focused on circular RNA (circRNA), recently proposed to participate in the control of pluripotency.
Methods
Reprogramming was achieved by a footprint-free strategy. Self-renewal and pluripotency of cord blood MSC-hiPSC were investigated in vitro and in vivo, compared to parental MSC, to embryonic stem cells and to F-hiPSC. High-throughput array-based approaches and bioinformatics analyses were applied to address the PGRN.
Findings
Cord blood MSC-hiPSC successfully acquired a complete pluripotent identity. Functional comparison with F-hiPSC showed no differences in terms of i) generation of mesenchymal-like derivatives, ii) their subsequent adipogenic, osteogenic and chondrogenic commitment, and iii) their hematopoietic support ability. At the transcriptional level, specific subsets of mRNA, miRNA and circRNA (n = 4,429) were evidenced, casting a further layer of complexity on the PGRN regulatory crosstalk.
Interpretation
A circRNA map of transcripts associated to naïve and primed pluripotency is provided for hiPSC of clinical-grade foetal origin, offering insights on still unreported regulatory circuits of the PGRN to consider for the optimization and development of efficient differentiation protocols for clinical translation.
Funding
This research was funded by Ricerca Corrente 2012–2018 by the Italian Ministry of Health.
Keywords: Cord blood, Multipotent mesenchymal stromal cells, hiPSC, miRNA, circRNA, PGRN
Research in context.
Evidence before study
The interest in human induced pluripotent stem cell (hiPSC) for tissue replacement approaches is at its maximum. Yet, a debate on the best cell source for hiPSC is ongoing, mainly dealing with risks associated with the use of adult fibroblasts as hiPSC cell source, such as accumulation of somatic mutations, transmission of infections and influence on differentiation properties due to memory of the tissue of origin. To this regard, the regulatory circuit controlling hiPSC pluripotency and cell fate decisions is the pluripotency gene regulatory network (PGRN), which implies a crosstalk between specific subsets of transcripts. Circular RNA (circRNA) were recently proposed as novel players of the PGRN. Our previous studies have focused on the use of cord blood as source of clinical-grade stem cells, while underpinning the crucial role of circRNA in sustaining stem cell identity.
Added value of this study
We applied a footprint-free approach to generate hiPSC from cord blood. This cell source offers unique advantaged over biopsies of adult tissue, such as no accumulation of somatic mutations, low risk of viral or environmental contamination, low immunogenicity, availability of clinical-grade, HLA-typed cord blood units at international networks of public banks. These hiPSC were functionally undistinguishable from fibroblast-derived hiPSC. A set of 4429 circRNA with potential roles in hiPSC biology were defined.
Implications of all available evidence
Cord blood hiPSC showing fully acquisition of the pluripotent identity represent a better alternative than fibroblast-derived hiPSC and could be readily implemented in translational clinical studies. Key insights on the fine tuning of primed and naïve pluripotent states by circRNA were provided, laying the ground for future studies on optimized protocols for the obtainment of hiPSC differentiated derivatives for clinical applications.
Alt-text: Unlabelled box
1. Introduction
As of 2017, the first in-human clinical application of allogeneic human induced pluripotent stem cell (hiPSC) for tissue replacement approaches has become a reality, following pioneering autologous studies [1,2]. This landmark transplantation success renewed the interest in hiPSC-based pre-clinical and clinical studies. Yet, the majority of them still rely on the use of fibroblast-derived hiPSC (F-hiPSC), starting from tissue biopsies [3]. Indeed, the issue of the best cell source to generate hiPSC is still ongoing, as well as the debate on how heavily and to what extent hiPSC retain memory of the parental cell type and tissue of origin [4], [5], [6], [7], [8], [9]. Inefficient and incomplete generation of differentiated hiPSC progeny is another bottleneck that needs to be solved before hiPSC can be successfully moved from bench to bedside. To this regard, self-renewal and cell fate decisions of pluripotent stem cells (PSC) are governed by the pluripotency gene regulatory network (PGRN). A successful reprogramming process entails the complete activation of the PGRN to induce and stably sustain the newly acquired pluripotent identity. Its most studied components are OCT4, SOX2 and NANOG core PGRN transcription factors [10]. Yet, also micro RNA (miRNA), such as those belonging to the miRNA 302/367 cluster [11] and, very recently, circular RNA (circRNA) were shown to possibly intervene in the maintenance of this regulatory circuit [12], [13], [14]. As miRNA, circRNA are a class of non-coding RNA with regulatory functions, even though the production of truncated peptides originating from the translation of a circRNA has been reported [15]. CircRNA originate from back-splicing events of RNA transcripts, leading to 3′−5′ covalent bonds and consequent circularization [16]. They can be produced from both genic and intergenic DNA regions. In the former case, they can include exonic, intronic, and 5′/3′-untranslated regions sequences. The existence of circRNA was proposed more than 20 years ago, even though they were considered functionless by-products of mRNA splicing for a long time [17,18]. Nevertheless, the highly conservation across species, the large number of endogenous circRNA identified in various tissues, at different developmental stages, suggest that circRNA have relevant functions to play in cellular biology and fate determination choices [19], [20], [21], [22], [23], [24]. The main mechanism of action of circRNA appear to be the “sponging” of miRNA thanks to complementary docking sites along their RNA primary sequences [12,20,25,26]. Other reported phenomena involve the interaction with RNA molecules via RNA-binding sites to regulate transcription [27] or with proteins via protein-binding sites to regulate pre-mRNA splicing [28] or cell cycle [29]. Up to date, a comprehensive network elucidating the precise role of circRNA in the modulation of miRNA post-transcriptional regulatory activity on stemness-associated messenger RNA (mRNA) is still missing.
Herein, we successfully generated hiPSC from cord blood multipotent mesenchymal stromal cells, a foetal cell type. The use of cord blood over other tissues presents many advantages. First, cord blood cells possess juvenile and naïve features, such as low immunogenicity [30], [31], [32] and presence of less committed progenitor cells [33,34]. In addition, unlike skin cells, cord blood cells are not exposed to UV irradiation, which leads to accumulation of somatic mutations [35], [36], [37]. Furthermore, cord blood is less exposed to risk of environmental and viral contamination, thanks to the placental maternal-foetal barrier [38], [39], [40]. Finally, cord blood units are already available for research and clinical studies at international networks of public banks [41,42]. In this work, we thoroughly addressed the acquisition of self-renewal and differentiation capabilities by state-of-the-art assays and deeper and more specific analyses at both the cellular and molecular level. For the former, we addressed differentiation and functional properties of mesenchymal-like derivatives in comparison with those by F-hiPSC. For the latter, we focused on the possible role of circRNA as novel players of the PGRN via regulation of miRNA activity. As a result, we outlined a map of interactions between pluripotency-associated mRNA and miRNA, descending from hiPSC top expressed circRNA, offering new insights on pathways possibly regulating pluripotency and cell fate decisions for the optimization of differentiation protocols to obtain hiPSC derivatives for clinical application.
2. Materials and methods
2.1. Ethics
All experiments were performed according to the amended Declaration of Helsinki. Informed consent by cord blood donors was obtained for research under resolution n° VII/18,653 by Lombardy Region, Italy, and evaluation by the Ethical Committee of Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico n° 1982, 14th January 2020.
2.2. Multipotent mesenchymal stromal cell culture
Human long-living cord blood multipotent mesenchymal stromal cells (MSC) derived from a cord blood unit of a normal term delivery of a male new-born were fully characterized in previous works by our group [43], [44], [45], [46] and were herein used, following standard culture conditions [44]. To clarify terminology: “long-living” refers to higher lifespan compared to other cord blood stromal cell types [44,45]; “multipotent mesenchymal” refers to specific immunophenotype and in vitro tri-lineage multipotent properties [43], [44], [45], [46] in accordance to ISCT guidelines [47,48]; “stromal” refers to growth in adherence and to depletion of hematopoietic cell types for MSC isolation [44]. Standard MSC medium was αMEM-GlutaMAX (32,561; Gibco, Carlsbad, CA, USA) 20% FBS (10,099,141; Gibco).
2.3. Reprogramming to pluripotency
CytoTune-iPS 2.0 Sendai Reprogramming kit (A16517; Thermo Fisher Scientific, Waltham, MA, USA) was used to generate human induced pluripotent stem cells from cord blood MSC (MSC-hiPSC). MSC cultured in standard MSC medium were transduced with CytoTune-iPS 2.0 Sendai Reprogramming vectors as follows: KOS vector, harbouring human KLF4, OCT4 and SOX2, at multiplicity of infection (MOI) = 5; M vector, harbouring human c-MYC, at MOI = 5; K vector, harbouring human KLF4, at MOI = 3. Transduced cells were incubated overnight at 37 °C, 5% CO2. The day after, the medium containing viral particles was removed and fresh standard MSC medium was added. Medium changes were performed every other day until transduced cells were detached and transferred onto PSC-qualified Matrigel (354,277; BD, Franklin Lakes, NJ, USA)-coated culture Petri dishes (Sarstedt, Nümbrecht, Germany), one week after infection. The day after, standard MSC medium was switched to StemMACS iPS-Brew XF PSC medium (130–104–368; Miltenyi Biotec, Gladbach, Germany). Next, the medium was replaced every other day and the cultures were monitored for the emergence of MSC-hiPSC colonies showing the typical epithelial-like morphology. Single MSC-hiPSC colonies were clonally subcultured by manual mechanical picking using a 150 μm diameter needle mounted on a stripper micropipette. The picking was performed under sterile conditions, using a Nikon Eclipse TS100 microscope (Nikon, Tokyo, Japan) placed inside a laminar flow hood and an external monitor. MSC-hiPSC colony clumps of 20–200 cells were seeded onto PSC-qualified Matrigel-coated culture Petri dishes in StemMACS medium to establish MSC-hiPSC lines. Bright field images of MSC-hiPSC cultures were taken with a Nikon Eclipse Ti microscope (Nikon).
2.4. Pluripotent stem cell culture
hiPSC were expanded onto PSC-qualified Matrigel-coated culture surfaces in StemMACS iPS-Brew XF chemically defined serum-free PSC medium, with medium changes performed every day. At 80% confluence, the colonies were detached by 5 mM ethylenediaminetetraacetic acid (EDTA; Gibco) non-enzymatic passaging and collected with Knock Out (KO) medium consisting of KO-DMEM (10,829–018; Gibco) supplemented with 20% KO-Serum Replacement (KO-SR, 10,828–028; Gibco), 2 mM GlutaMAX (35,050,038; Gibco), 50 μM 2-mercaptoethanol (31,350–010; Gibco), 1 mM sodium pyruvate (11,360–039; Gibco) and 1 mM non-essential amino acids (11,140–035; Gibco). hiPSC colonies were reduced to 10–50 cell clumps by gently pipetting and seeded following 1:3–1:8 split ratio depending on experimental needs. The day after passaging no fresh medium was added. Human embryonic stem cell (hESC) H9 (female) and fibroblast-derived hiPSC (F-hiPSC) lines were provided by Prof. Elvassore and were cultured under the same conditions as MSC-hiPSC and used as PSC positive control where specified. F-hiPSC were obtained either by the Sendai method (F-hiPSC 1) from human adult female normal skin fibroblasts [49] or by the modified mRNA method (F-hiPSC 2) from human new-born male normal foreskin fibroblasts [50,51].
2.5. Alkaline phosphatase activity
Direct alkaline phosphatase activity was assessed using Alkaline Phosphatase (AP) Live Stain kit (Thermo Fisher Scientific), following manufacturer's instructions. AP-positive colonies were imaged on a Nikon Eclipse Ti microscope (Nikon).
2.6. Growth properties
PSC were reduced to single cell suspension by 5 min incubation with accutase (L0950; Biowest, Nuaillé, France), the number of viable cells determined by an automated cell counter (NC-100 NucleoCounter; Chemometec, Allerod, Denmark) and seeded at 15,000 cells/cm2 in StemMACS iPS-Brew XF supplemented with 10 µM Y-27,632 Rock inhibitor (72,302; STEMCELL Technologies, Vancouver, Canada). To calculate population doubling time, single cell suspensions were seeded in 96-well E-Plates (ACEA Biosciences, San Diego, CA, USA) at 5000 cells/cm2 in StemMACS iPS-Brew XF supplemented with 10 µM Y-27,632 Rock inhibitor. Proliferation rate was determined based on the proprietary cell index parameter on xCELLigence Real Time Cell Analysis single plate system (RTCA-SP; ACEA Biosciences; RRID: SCR_014821) as already described [52].
2.7. Flow cytometry
Immunophenotyping was performed as previously described [46] on a BD FACSCanto II cytometer (RRID: SCR_018056). The following antibodies were used: HLA-ABC-FITC (555,552; BD; RRID: AB_395,935), HLA-DR-FITC (556,643; BD; RRID: AB_396,509), CD14-PerCP-C5.5 (562,692; BD; RRID: AB_2,737,726), CD31-PE (340,297; BD; RRID: AB_400,016), CD34-FITC (555,821; BD; RRID: AB_396,150), CD34-PC5 (555,823; BD; RID: AB_396,152), CD44-PE (555,479; BD; RRID: AB_395,871), CD45-APC—C7 (561,863; BD; RRID: AB_10,897,014), CD90-PC7 (561,558; BD; RRID: AB_10,714,644), CD90-PerCP-C5.5 (561,557; BD; RRID: AB_10,712,762), CD56-PC7 (557,747; BD; RRID: AB_396,853), CD73-APC (560,847; BD; RRID: AB_10,612,019), CD105-PE (560,839; BD; RRID: AB_2,033,932), CD144-AF647 (561,567; BD; RRID: AB_10,712,766), CD271-AF647 (560,326; BD; RRID: AB_1,645,403), CD271-AF647 (560,326; BD; RRID: AB_1,645,403). Passage (P) 4–6 MSC, P5–20 MSC-hiPSC, P40–50 hESC were used (n = 3 each).
2.8. Karyotype
Conventional metaphase analysis was performed on cells seeded onto amniodishes (Euroclone, Pero, Italy) by the Q-banding technique at 400 banding level. A minimum of 20 cells per sample were analysed on the IKAROS automated system (MetaSystems, Altlussheim, Germany) to exclude clonal rearrangements.
2.9. qPCR
RNA extraction, retrotranscription and amplification were performed as previously reported [43,44]. To analyse residual presence of reprogramming vectors, unreprogrammed MSC, MSC undergoing reprogramming, P1 reprogrammed colonies, early (P5–6) and late (P13–17) passage hiPSC were used and raw cycle threshold (Ct) values were considered. The primer pair was designed to target Sendai genome sequences and to amplify all reprogramming vectors (SEV), as per reprogramming kit manufacturer's instructions. Ct values above 35 were not considered indicative of successful amplification of the target. To analyse gene expression, the 2−ΔΔCt method was applied [53], using GAPDH as house-keeping gene and normalizing to the experimental control group, as specified. Validation of circRNA was performed as previously described [12]. The following primers were used (forward and reverse 5′ to 3′ sequences): CTCCTGTGATGAGCTGTCCA, CCATTCACCACGTTGTTGTC (circRNA_0034447); GGGCCATGAAGGATGAGGAG, TTGAGGGCGGCCACATC (circRNA_0008432); ATGACAACGATGGCATTCCCT, CACTGATCTCCAACCCCATC (circRNA_0034528); TGAGAGCTGCGAACTTGGTC, CAGGGCGCTGCTCCAG (circRNA_0001827); CTGGCCATGAGAGTGGAGAG, CTTGTCCGTGGAGAACATGA (circRNA_0011385); GAAATTCACAAGCGCACAGGA, TGCGGAGTCCATCATGTCAC (circRNA_0012634). Other primer sequences will be given upon request.
2.10. Pyrosequencing
DNA was extracted using the QIAamp DNA Blood Mini Kit (51,104; Qiagen), following manufacturer's instructions. Each DNA sample was treated with the EZ DNA Methylation-Gold Kit (D5005; Zymo Research, Orange, CA, USA) to obtain bisulfite converted DNA. To analyse DNA methylation, a 50 μL PCR reaction was performed with 25 μL of GoTaq Hot Start Green Master mix (M5121; Promega, Madison, WI, USA), 10 μM of forward primer, 10 μM of biotinylated reverse primer and 500 ng of bisulfite-treated DNA. Biotin-labelled primers were used to purify the final PCR product with sepharose beads: 10 μL of PCR product were bound to 1 μL of Streptavidin Sepharose HP affinity chromatography medium (Amersham Biosciences, Uppsala, Sweden) in presence of 40 μL of binding buffer (Amersham Biosciences) by 10 min incubation in agitation. Sepharose beads containing the immobilized PCR product were purified with the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Westborough, MA, USA), according to the manufacturer's instructions. Pyrosequencing primer (0.3 μM) was annealed to the purified single-stranded PCR product in presence of 15 μL of annealing buffer, during an incubation of 2 min at 85 °C. Then, pyrosequencing was performed in duplicate with the PyroMark MD System (Pyrosequencing). The percentage of methylated cytosines was calculated as the number of methylated cytosines divided by the sum of methylated and unmethylated cytosines, multiplied by 100%.
2.11. Immunofluorescence
For immunofluorescence analysis, the PSC 4-marker Immunocytochemistry Kit (A24881; Thermo Fisher Scientific) and the 3-Germ Layer Immunocytochemistry Kit (A25538; Thermo Fisher Scientific) coupled with NCAM antibody (MA1–06,801; Thermo Fisher Scientific; RRID: AB_558,237) were used, following manufacturer's instructions. Fluorescence mounting medium (DakoCytomation, Glostrup, Denmark) was used. Samples were imaged on a Nikon Eclipse 80i VideoConfocal microscope (Nikon).
2.12. Differentiation into endodermal, mesodermal and ectodermal derivatives
PSC were detached by the EDTA method and 150 µL per well of cell suspension was transferred to a low-attachment V-bottom 96-well plate (M9686; Sigma-Aldrich, St. Louis, MO, USA) in KO medium for 3–4 days to promote aggregation and allow embryoid body (EB) formation. Then, EB were transferred into low-attachment 24-well plate (Sarstedt) and maintained in suspension in KO medium for 2–3 supplementary days. Finally, EB were transferred onto 0.1% gelatine-coated (07,903; STEMCELL Technologies) chamber slides for further differentiation. The medium was replaced twice a week for 2–3 weeks. Endodermal differentiation medium was composed of DMEM (11,960,044; Gibco), 20% FBS, 2 mM L-glutamine (25,030,081; Gibco), 0.1 mM 2-mercaptoethanol and 1 mM non-essential amino acids. Mesodermal differentiation medium was further supplemented with 100 mM ascorbic acid (A4403; Sigma-Aldrich). Differentiation into ectodermal derivatives was performed following a protocol elsewhere described [54]. Differentiating cells were analysed at day 15 of the protocol, when they had already acquired an early epithelial morphology or at later stages (days 16–30) when they showed a neuronal morphology.
2.13. Animal study
Six- to 8-weeks old female SHrN hairless NOD.SCID mice, obtained from Envigo Laboratories (Huntingdon, UK), were used. Mice were maintained under specific pathogen-free conditions, housed in isolated vented cages, and handled using aseptic procedures. The Istituto di Ricerche Farmacologiche Mario Negri-IRCCS adheres to the principles set out in the following laws, regulations, and policies governing the care and use of laboratory animals: Italian Governing Law (D. lg 26/2014; authorization no.19/2008-A issued 6 March 2008 by the Ministry of Health); Mario Negri Institutional Regulations and Policies providing internal authorization for persons conducting animal experiments (Quality Management System Certificate: UNI EN ISO 9001:2015, reg. no. 6121); the National Institute of Health (NIH) Guide for the Care and Use of Laboratory Animals (2011edition) and EU directive and guidelines (European Economic Community [EEC] Council Directive 2010/63/UE). Data were reported according to ARRIVE guidelines.
2.14. Teratoma assay
Teratoma assay was performed as elsewhere described [55] at the Istituto di Ricerche Farmacologiche Mario Negri-IRCCS, under authorization n° 510/2016-PR by the Italian Ministry of Health, 24th May 2016, for subcutaneous injection of tumorigenic cells. Briefly, 1 million MSC or MSC-hiPSC were subcutaneously injected bilaterally in flanks of NOD.SCID mice (n = 2 each). Mice were monitored once a week for six months for teratoma appearance. Teratoma growth was measured with a Vernier calliper, and teratoma weight (mg = mm3) was calculated as follows: [length (mm) × width2 (mm2)]/2 and body weight was registered as index of the animal's health status. When teratoma weights reached the ethical limits (10% of mice body weight), mice were sacrificed. At sacrifice, teratomas were collected, washed in PBS and immediately fixed in neutral buffered formalin 10% to be paraffin embedded. Standard haematoxylin and eosin staining was performed on 5 μm-thick paraffin sections.
2.15. Stem cell panel PCR-array
Total RNA was isolated using TRIzol reagent (Ambion, Waltham, MA, USA), as elsewhere reported [52]. SuperScript IV VILO Master Mix for cDNA synthesis (11,756,050; Thermo Fisher Scientific), TaqMan OpenArray Real-Time PCR Master Mix (4,462,159; Thermo Fisher Scientific) and TaqMan OpenArray Human Stem Cell Panel array (4,475,390; Thermo Fisher Scientific) were used to determine mRNA expression on a QuantStudio 12 K Flex Real Time PCR System (Thermo Fisher Scientific), following manufacturer's instructions.
2.16. Directed differentiation of hiPSC toward MSC-like cells and mesenchymal derivatives
hiPSC were detached by the accutase method and seeded in PSC medium:KO medium 1:1 supplemented with 10 µM Y-27,632 Rock inhibitor at the density of 160,000 cells/well in EZSPHERE ultra-low attachment 96-well plates (Asahi Glass Corporation, Tokyo, Japan) to generate a homogeneous population of EB. After 2 days, the medium was switched to mesengenic medium, composed by KO medium supplemented with 6 µM CHIR99021 (72,052; STEMCELL Technologies) and 10 ng/mL TGFβ3 (100–36E; Peprotech, Rocky Hill, NJ, USA). After 2 days the EB were transferred onto 0.1% gelatine-coated 24-well plates in mesengenic medium:MSC medium (αMEM-GlutaMAX 10% FBS 10 ng/mL TGFβ3) 1:1. Differentiation into mesenchymal derivatives was induced as previously reported [43,44]. Primary MSC (n = 3) were used as control. Differentiation was assessed by qPCR, following the protocol described above.
2.17. Cobblestone area-forming cell assay
MSC-like cells generated from hiPSC were seeded at 30,000 cells/cm2 in 96-well plate (n = 5) onto 0.1% gelatine-coated wells in MSC medium. At 100% confluence, the cells were irradiated at 10 Gy for 10 min. The same day, CD34+ hematopoietic progenitor cells were isolated from cord blood by magnetic labelling using the Indirect CD34 MicroBead Kit (130–046–701; Miltenyi) on MS MACS separation columns (Miltenyi) following manufacturer's instructions, and seeded at 5000 cells/cm2 on top of MSC-like cells in myelocult H5100 (05,150; STEMCELL Technologies) supplemented with 10−6 hydrocortisone (H0888; Sigma-Aldrich). Co-cultures were maintained for 5 weeks with medium changes every other day and then evaluated by optical microscopy analysis on a Nikon Eclipse Ti microscope (Nikon) for the formation of typical cobblestone areas and by flow cytometry using CD45-APC—C7 (561,863; BD; RRID: AB_10,897,014) and CD34-FITC antibodies (555,821; BD; RRID: AB_396,150) to assess support of hematopoietic cells, following the protocols already described above.
2.18. miRNome PCR-array
The miRNeasy Mini Kit (217,004; Qiagen) and the RNeasy MinElute Cleanup Kit (74,204; Qiagen) were used to extract miRNA. First, cells lysed with 700 μL of QIAzol Lysis Reagent (79,306; Qiagen) were incubated with 140 μL of chloroform for 3 min at RT. Then, the samples were centrifuged for 15 min at 12,000 × g at 8 °C. The resulting upper aqueous phase was transferred to a new collection tube, and 350 μL of 70% ethanol was added. Next, the samples were transferred in RNeasy spin columns and centrifuged for 15 s at maximum speed at 20 °C. The collected flow-through was transferred to RNeasy MinElute spin columns and centrifuged for 15 s at maximum speed at 20 °C, after the addition of 450 μL of 100% ethanol. Then, 500 μL of RPE Buffer was added to the RNeasy MinElute spin columns and centrifuged for 15 s at maximum speed. A further 2 min washing step at maximum speed was performed with 500 μL of 80% ethanol, followed by an additional centrifugation at maximum speed for 5 min with open lids. To elute miRNA, the filters were incubated with 20 μL of RNase free-water at RT for 5 min and then centrifuged for 1 min at maximum speed. RNA quality and quantification were assessed on a 2100 Bioanalyzer instrument (Agilent, Santa Clara, CA, USA; RRID: SCR_018043). Cell miRNome was determined using TaqMan Advanced miRNA cDNA Synthesis Kit (Thermo Fisher Scientific), TaqMan OpenArray Real-Time PCR Master Mix (Thermo Fisher Scientific) and Real Time PCR TaqMan OpenArray Human MicroRNA Panel array (Thermo Fisher Scientific) on a QuantStudio 12 K Flex Real Time PCR System (Thermo Fisher Scientific), following manufacturer's instructions.
2.19. circRNA microarray
Sample preparation and microarray hybridization were performed as previously reported [12] according to the manufacturer's protocol (Arraystar). Briefly, total RNA was isolated from MSC-hiPSC using TRIzol reagent was treated with RNase R to remove linear RNA and enrich for circRNA. Next, circRNA was amplified and transcribed into fluorescent cRNA using the random priming method with a Super RNA Labelling Kit (Arraystar). The labelled cRNA was hybridized onto an Arraystar Human Circular RNA Microarray (Arraystar V1.0). The array was scanned with the Agilent Scanner G2505C, and raw data were extracted by Agilent Feature Extraction software (version 11.0.1.1). Comparison with MSC was performed on previously published dataset available at NCBI GEO database through series accession number GSE122178 (sample C2) [12]. Comparison with F-hiPSC [13] and hESC [19] was performed on datasets available at the indicated publications. Identification and analysis of circRNA were restricted to circBASE database available nomenclature and data.
2.20. Computational and statistical analysis
Biological pathway enrichment studies were performed interrogating GO and KEGG online databases. Pathways related to pathology were excluded. Graphical representation was done in R (R Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available online at https://www.R-project.org/). For stem cell panel analysis, all 2-fold significantly upregulated mRNA were considered. For miRNome analysis, mRNA targeted by the top 10 miRNA were considered. For circRNome analysis, mRNA targeted by miRNA sponged by the top 10 circRNA were considered. CircRNA-sponged miRNA were predicted by Arraystar proprietary algorithm and were included in the analysis only if detected in the cells of interest by the PCR-array. For the generation of the circRNA-miRNA-mRNA network by Cytoscape software (RRID: SCR_003032) [56] only mRNA detected by the PCR-array were included. All other statistical analyses were performed using Prism 6 (GraphPad Software, La Jolla, CA, USA). Details of statistical analysis are specified in the figure legends. A p-value < 0.05 was considered statistically significant.
2.21. Availability of data and material
Data of messenger (GSE144634), micro (GSE144631) and circular (GSE144629 and GSE122178) RNA are accessible through the specified series accession numbers at the GEO online public database by NCBI, which supports MIAME-compliant data submissions [57,58] (https://www.ncbi.nlm.nih.gov/geo). All other data and material are available upon request.
3. Results
3.1. Reprogramming cord blood cells to pluripotency
The protocol implemented to induce pluripotency in human cord blood multipotent mesenchymal stromal cells (MSC) is summarized in Fig. 1a. Colonies of highly-packed cells with epithelial-like morphology (n = 48) appeared as early as one week after infection with the reprogramming vectors (Supplementary fig. 1a, left panel). This typical embryonic morphology was more evident in bona fide independent MSC-hiPSC lines (n = 3) generated by feeder-free subculture of single clones (Supplementary fig. 1a, right panel).
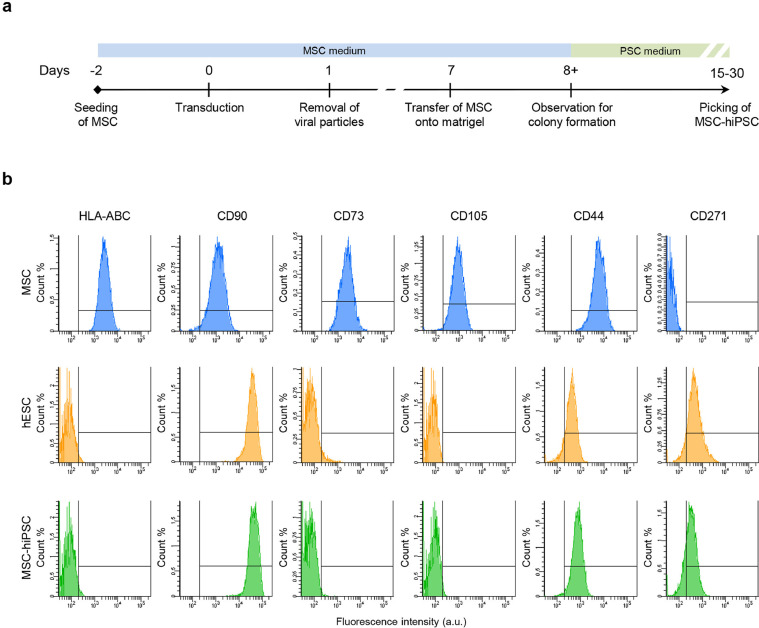
Fig. 1.
Reprogramming of MSC to pluripotency. a) Schematic of the reprogramming protocol. b) Representative flow cytometry histograms showing immunophenotype results of MSC-hiPSC compared to hESC and MSC for a selected panel of surface markers. The horizontal axis reports fluorescence intensity in logarithmic scale (arbitrary units, a.u.), the vertical axis reports event count percentage (Count%). The vertical bar gates exclude sample autofluorescence based on unstained controls.
Alkaline phosphatase (AP) activity was addressed as a rough and rapid screening for pluripotency. The AP fluorescence-based assay was performed on MSC-hiPSC colony candidates, before and after picking for MSC-hiPSC line establishment. In contrast to human fibroblasts that are usually used to generate MSC-hiPSC and show undetectable or no AP activity [59], both non reprogrammed MSC and MSC-hiPSC colony candidates stained positive, indicating AP activity (Supplementary fig. 1b). The staining was clearer in MSC-hiPSC colonies after picking, similar to that of human embryonic stem cell (hESC) colonies used as pluripotent stem cell control (Supplementary fig. 1c). The MSC-hiPSC lines were cultured for many passages in vitro (>40), resulting in a growth curve (Supplementary fig. 1d) and doubling time (Supplementary fig. 1e) during growth logarithmic phase (Supplementary fig. 1f) identical to that of hESC, maintaining a normal karyotype (Supplementary fig. 1g). Looking for alternatives to AP activity as initial pluripotency test, immunophenotyping of the MSC-hiPSC lines was performed. The flow cytometry panel targeted classic MSC positive and negative surface markers [46], which were observed to be either dramatically reduced (HLA-ABC, CD73, CD105) or increased (CD44, CD90, CD271) immediately following reprogramming, coherently with hESC immunophenotype (Fig. 1b and supplementary fig. 1h). Hematopoietic surface markers were also addressed and found negative before and after reprogramming (Supplementary fig. 1h).
3.2. Stemness identity of MSC-hiPSC
The expression of key pluripotency network-related genes was investigated by qPCR (Fig. 2a). Transcriptional levels of OCT4 (isoform A) and SOX2 in MSC-hiPSC were similar to those found in hESC, while they were undetectable in MSC parental cells. In contrast, KLF4 was more expressed in MSC (5-fold) than MSC-hiPSC and hESC, whereas cMYC was expressed at similar levels in all three cell types. More importantly, also NANOG and LIN28A were upregulated in MSC-hiPSC, matching hESC levels. These two proteins were not present in the cocktail of exogenously encoded factors shuttled by the Sendai reprogramming system. Thus, correct engagement of the endogenous pluripotency network was achieved. Additional qPCR analyses were performed to check for residual presence of the reprogramming factors at different passages (P1, P5–6 and P13–17 compared with MSC before and during reprogramming). Presence of reprogramming vectors was robustly detected only during reprogramming. Early passage MSC-hiPSC showed no or little amplification of Sendai vector sequence. Late passage MSC-hiPSC showed lack of amplification of the viral genome similarly to MSC before reprogramming, indicating loss of the reprogramming vectors (Supplementary fig. 2a).
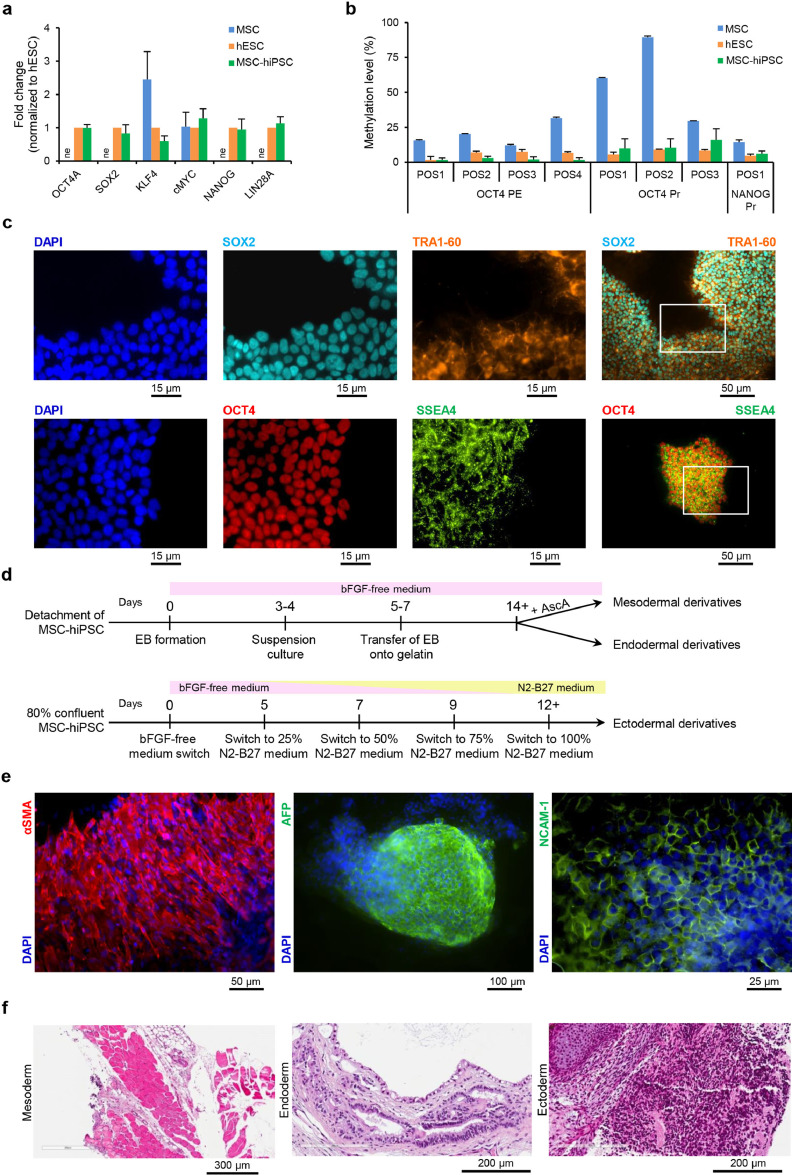
Fig. 2.
Stemness and pluripotency of MSC-hiPSC. a) Histograms showing MSC-hiPSC gene expression of pluripotency network-related genes normalized to hESC, compared to MSC. Mean with standard deviation are represented (n = 3); ne, not expressed. b) Histograms showing DNA methylation percentage of key OCT4 and NANOG gene regulatory regions for MSC, hESC and MSC-hiPSC. Mean with standard deviation are represented (n = 3); POS, position, PE, proximal enhancer, Pr, promoter. c) Representative fluorescence images showing protein expression and cellular localization of typical pluripotency markers (OCT4A, SSEA4, SOX2, TRA1–60) for MSC-hiPSC; white rectangles in images with merged fluorescence signals outline the areas selected to show single fluorescence signals at higher magnification. d) Schematics of the in vitro differentiation protocols. bFGF, basic fibroblast growth factor, EB, embryoid body, AscA, ascorbic acid. e) Representative fluorescence images showing protein expression and cellular localization of mesodermal (αSMA), endodermal (AFP) and ectodermal (NCAM-1) derivative markers for in vitro differentiated MSC-hiPSC. DAPI was used to stain nuclei in all fluorescence images. f) Representative haematoxylin-eosin histology images of cell derivatives from all the three germ layers for in vivo differentiated MSC-hiPSC. Striatal muscle cells for mesoderm, mucinous epithelium for endoderm and neuroepithelium for ectoderm are shown.
To address the contribution of epigenetic remodelling to the transcriptional shift observed in MSC-hiPSC, the methylation state of regulatory regions of OCT4 and NANOG was addressed on MSC-hiPSC bisulfite-treated DNA by pyrosequencing. The results showed dramatic methylation remodelling for CpG islands in different positions at OCT4 promoter and at OCT4 proximal enhancer sequences (Fig. 2b). Consistent demethylation at hESC levels was detected in MSC-hiPSC compared to MSC, more pronounced for OCT4 promoter. The same trend was found for NANOG promoter, further supporting a role for epigenetic modifications in the reprogramming of MSC and in the activation of the pluripotency molecular network.
Protein expression of pluripotency-associated markers was also addressed on undifferentiated MSC-hiPSC colonies by immunofluorescence (Fig. 2c). Nuclear localization of SOX2 transcription factor was observed in co-expression with TRA1–60 plasma membrane surface marker. In addition, nuclear localization of OCT4 transcription factor was observed in co-expression with SSEA4 plasma membrane surface marker. The expression level and subcellular localization of stemness markers were the same as hESC (Supplementary fig. 2b). To note, hESC, and not MSC-hiPSC, showed heterogeneous OCT4 and SOX2 signals in different cells of the same colonies, a phenomenon already reported in the literature and ascribed to physiological fluctuations in expression levels that do not impair the pluripotent identity of the cells [60], [61], [62].
3.3. Three-lineage differentiation properties of MSC-hiPSC
To challenge the differentiation potential of MSC-hiPSC, both in vitro and in vivo protocols were applied. The in vitro protocols are summarized in Fig. 2d and were different for the generation of mesodermal, endodermal and ectodermal derivatives. Differentiation of MSC-hiPSC into mesodermal and endodermal derivatives relied on embryoid body formation, which ultimately led to the generation of α-smooth muscle actin-expressing cell sheets and α-fetoprotein-expressing 3D cell clusters, respectively, assessed by immunofluorescence (Fig. 2e). Differentiation of MSC-hiPSC into ectodermal derivatives was achieved in 2D cultures in gradually increasing percentages of neural cell medium, as showed by the presence of NCAM-1-positive cells with epithelial morphology (Fig. 2e). Protein expression and subcellular localization of markers for derivatives of the three germ layers were again the same as hESC (Supplementary fig. 2c). Notably, parental MSC expressed basal levels of αSMA, consistent with their mesodermal lineage (Supplementary fig. 2d). Furthermore, the neuronal differentiation potential of MSC-hiPSC was assessed by the emergence of neuron-specific βIII-tubulin+ (TUJ1) cells showing typical neuronal morphology at later stages of the ectodermal differentiation protocol (Supplementary fig. 2e).
In addition, MSC-hiPSC in vivo differentiation properties were addressed by the teratoma assay. Cell suspensions from subconfluent cultures of MSC-hiPSC, hESC and MSC were detached and injected subcutaneously over both flanks of SCID mice. Formation of teratoma was clearly detectable as early as 8 weeks post-injection of MSC-hiPSC, hESC, but not of MSC samples, which did not show the appearance of any palpable subcutaneous cell mass (data not shown). At 12 weeks post-injection, MSC-hiPSC and hESC had generated solid teratomas (weight of 1.5 ± 1.1 g and a diameter of 14±3.4 mm). Samples from MSC-hiPSC underwent histologic analysis by haematoxylin-eosin to detect cell derivatives of all three germ layers. Striated muscle tissue, mucinous epithelium and neuroepithelium were detected, representative of mesoderm, endoderm and ectoderm, respectively (Fig. 2f). The generation of other cell types and tissues, such as chondroid matrix, smooth muscle, cartilage, squamous, pigment and ciliary epithelium was also observed (Supplementary fig. 3a-f). These latter results further confirmed fully acquisition by MSC of pluripotent properties following reprogramming to MSC-hiPSC.
3.4. Stem cell transcriptome of MSC-hiPSC
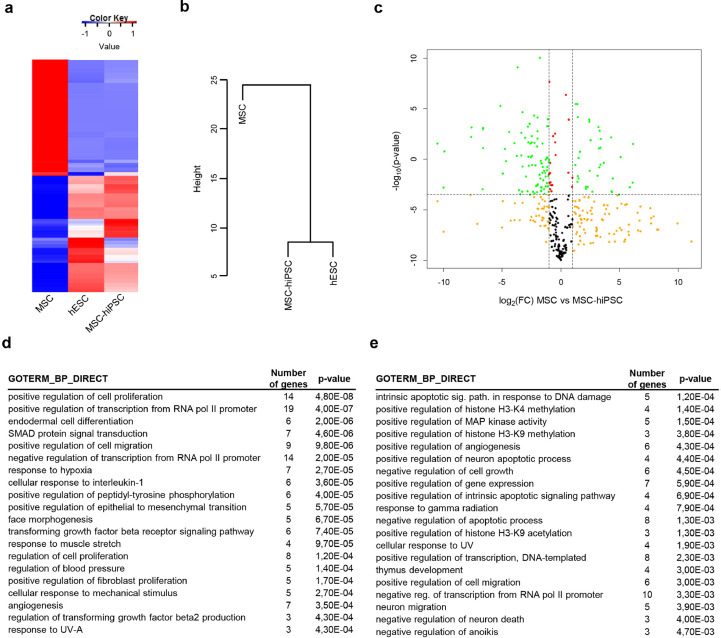
To further characterize the molecular rearrangements following reprogramming, the stem cell transcriptome of MSC-hiPSC was addressed. The reprogrammed cells showed complete change of the transcriptional profile compared to MSC, while it was similar to hESC (Fig. 3a). This was also evidenced by hierarchical clustering analysis, which grouped MSC-hiPSC and hESC under the same node (Fig. 3b). The higher gene expression similarity between MSC-hiPSC and hESC was supported by the calculation of the Pearson's correlation coefficient (Supplementary fig. 4a). Focusing on MSC and MSC-hiPSC, two groups of differentially expressed genes were selected (Fig. 3c), for later bioinformatics analysis. These results were validated on stem cell transcriptome data coming from different samples of MSC and MSC-hiPSC (Supplementary fig. 4b and c, respectively). The bioinformatics analysis of MSC upregulated genes revealed involvement of TGFβ-SMAD signalling pathway and of cellular pathways related to angiogenesis and stromal identity (Fig. 3d) or the hematopoietic niche (Supplementary fig. 4d). Conversely, MSC-hiPSC upregulated genes were found to mostly regulate response to damage, survival and epigenetic modifications (Fig. 3e), but also the pluripotency networks (Supplementary fig. 4e). Notably, pathways related to regulation of angiogenesis and to the hematopoietic niche were also preserved.
Fig. 3.
Stem cell transcriptome of MSC-hiPSC. a) Heatmap showing differentially expressed genes amongst MSC, hESC and MSC-hiPSC. Gene expression values are represented by the colour key. b) Dendrogram showing hierarchical clustering of MSC, hESC and MSC-hiPSC. c) Volcano plot showing p-value and fold change (FC) of gene expression data comparing MSC to MSC-hiPSC. Vertical dashed lines delimitate FC below and above 2, the horizontal dashed line shows p-value=0.05 [two-tailed t-test]. Colour code: FC greater than 2 reaching (green) or not (yellow) statistical significance; FC smaller than 2 reaching (red) or not (black) statistical significance. Enrichment in gene ontology terms of the biological processes category (GOTERM_BP_DIRECT) for upregulated genes of MSC (d) and MSC-hiPSC (e) is reported in the indicated tables [Fisher's exact test].
3.5. Mesenchymal potential of MSC-hiPSC
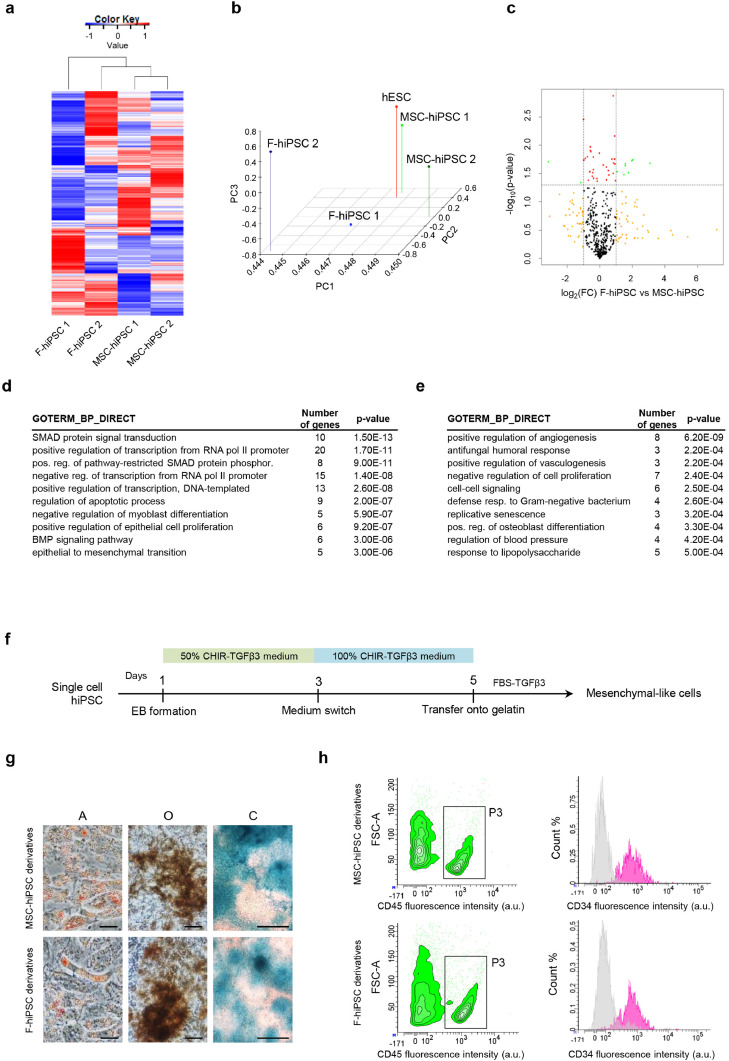
To unveil any specificity of cord blood MSC-hiPSC compared to conventional fibroblast (F)-derived hiPSC, the stem cell transcriptome of F-hiPSC was also addressed (Supplementary fig. 5a). MSC-hiPSC lines showed the higher similarity and grouped together by hierarchical clustering analysis (Fig. 4a). In contrast, F-hiPSC lines did not group under an exclusive node. For further assessment of the degree of similarity between MSC-hiPSC and F-hiPSC lines, a Principal Component Analysis (PCA) was performed including hESC as gold standard of pluripotency. PCA showed high similarity between MSC-hiPSC and hESC, whereas F-hiPSC were more distant between each other and the other PSC lines in terms of all components (PC1, PC2, PC3) (Fig. 4b). This result was supported by clustering analysis (Supplementary fig. 5b). A direct comparison of F-hiPSC with MSC-hiPSC revealed two groups of differentially expressed genes in a statistically significant fashion (Fig. 4c). Bioinformatics analysis of F-hiPSC upregulated genes showed a predominant involvement of TGFβ-SMAD signalling pathway (Fig. 4d and supplementary fig. 5c), whereas MSC-hiPSC upregulated genes were mostly involved in immune system functions and regulation of angiogenesis (Fig. 4e). Moreover, the hematopoietic niche was found amongst the most enriched pathways (Supplementary fig. 5d).
Fig. 4.
Mesenchymal potential of MSC-hiPSC. a) Heatmap showing differentially expressed genes amongst different F-hiPSC and MSC-hiPSC. Gene expression values are represented by the colour key. b) Principal Component Analysis (PCA) showing 3D visualization of Principal Component (PC) 1, PC2 and PC3 of differentially expressed genes for different F-hiPSC, MSC-hiPSC and hESC. c) Volcano plot showing p-value and fold change (FC) of gene expression data comparing F-hiPSC to MSC-hiPSC. Vertical dashed lines delimitate FC below and above 2, the horizontal dashed line shows p-value=0.05 [two-tailed t-test]. Colour code: FC greater than 2 reaching (green) or not (yellow) statistical significance; FC smaller than 2 reaching (red) or not (black) statistical significance. Enrichment in gene ontology terms of the biological processes category (GOTERM_BP_DIRECT) for upregulated genes of MSC-hiPSC (d) and F-hiPSC (e) is reported in the indicated tables [Fisher's exact test]. f) Schematic of the differentiation protocol toward MSC-like cells. g) Representative images of adipogenic (A, scale bar is 50 µm), osteogenic (O, scale bar is 50 µm) and chondrogenic (C, scale bar is 400 µm) mesenchymal derivatives. h) Left panel: representative density plot showing CD45+ hematopoietic cells (P3) gated from total cells of the cobblestone area-forming cell assay; FSC-A, forward scatter area, a.u., arbitrary units. Right panel: representative histograms showing CD34+ hematopoietic progenitor subpopulation (purple) of CD45+ cells compared to unstained control (grey); the vertical axis represents event percentage count (Count%).
To address whether these molecular traits had also a biological relevance, F-hiPSC and MSC-hiPSC were induced to differentiate into mesodermal derivatives. An in vitro differentiation protocol was implemented for the generation of MSC-like cell derivatives (Fig. 4f), to explore the MSC-hiPSC phenotype implications, if any, of TGFβ-SMAD pathway enrichment. Differentiation toward MSC-like derivatives was equally successful in both MSC-hiPSC and F-hiPSC, leading to the appearance of fibroblast-shaped cells expressing CD90 (93.8 ± 0.1 and 93.15±0.8, respectively), CD271 (31.4 ± 4.7 and 31.4 ± 0.6, respectively), CD56 (72.9 ± 23.0 and 76.3 ± 13.8, respectively) mesenchymal and stromal progenitor surface markers (Supplementary fig. 5e). These cells were negative for hematopoietic (CD34 and CD45) and endothelial (CD144 and CD31) antigens (data not shown). To address the differentiation capacity of the MSC-like derivatives, they were induced to give rise to further differentiated cells along the adipogenic, osteogenic and chondrogenic lineages. MSC-like cells from both MSC-hiPSC and F-hiPSC successfully generated fat droplets-containing cells upon adipogenic stimuli, calcium-depositing cells upon osteogenic stimuli, and glycosaminoglycans-producing cells upon chondrogenic stimuli (Fig. 4g). These results were confirmed at the transcriptional level, where upregulated expression of genes involved in the differentiation pathways was detected. Respectively, C/EBPβ for adipogenesis, RUNX2 for osteogenesis and SOX9 for chondrogenesis (Supplementary fig. 5f). To further challenge the functionality of MSC-like cells, their capacity to act as haematopoiesis-supporting stroma was addressed by the cobblestone area-forming cell (CAFC) assay. The formation of typical cobblestone areas was observed for MSC-like cells from both MSC-hiPSC and F-hiPSC within the completion of the five weeks co-culture of MSC-like cells with CD34+ cord blood hematopoietic progenitors (Supplementary fig. 5 g). The hematopoietic cell component of the CAFC assay was quantified by flow cytometry (Fig. 4h). Cells positive for CD45 were 18.3 ± 12.8% and 13.3 ± 5.7% for MSC- and F-hiPSC MSC-like derivatives, respectively. The majority of CD45+ cells were also CD34+ (81.6 ± 7.7% and 85.6 ± 10.7%, respectively). No statistical differences were found (p-value=0.8286 for CD45+, p-value=0.3429 for CD45+/CD34+ [Mann-Whitney test]).
3.6. miRNome of MSC-hiPSC
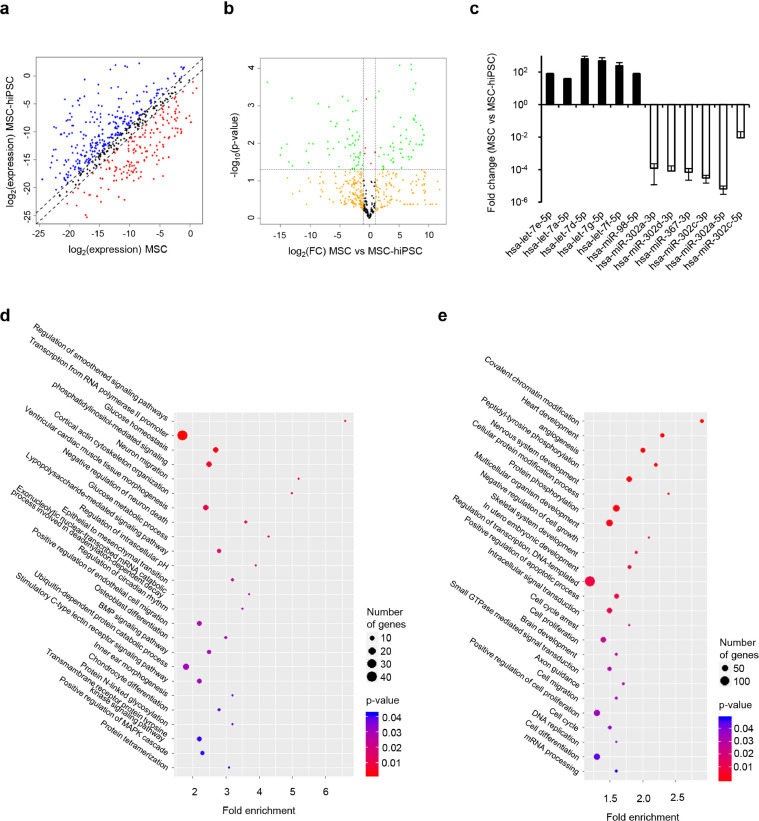
The complete miRNome of MSC-hiPSC was addressed and compared to that of MSC to further investigate molecular changes occurring upon reprogramming. The analysis revealed dramatic change of miRNA expression profile following reprogramming (Fig. 5a), where many of these miRNA were differentially expressed in a statistically significant fashion (Fig. 5b). These results were validated addressing the miRNome of different samples of MSC and MSC-hiPSC (Supplementary fig. 6a and 6b, respectively). The most evident results were the overrepresentation of the let-7 miRNA family in MSC and the overrepresentation of the miRNA 302/367 cluster in MSC-hiPSC (Fig. 5c). The function of miRNA belonging to the former is control of proliferation and development. On the contrary, miRNA belonging to the latter are involved in the control of pluripotency and exit from the naïve pluripotent state. Bioinformatics analysis of transcripts targeted by MSC upregulated miRNA showed regulation of osteochondral differentiation, mesenchymal identity, glucose metabolism (Fig. 5d) and GTPases-associated signalling pathways (Supplementary fig. 6c), whereas transcripts targeted by MSC-hiPSC upregulated miRNA were mainly involved in tissue specification, embryonic development, three-germ layer differentiation pathways (Fig. 5e) and response to mitogenic/organ growth stimuli (Supplementary fig. 6d).
Fig. 5.
miRNome of MSC-hiPSC. a) Scatter plot comparing gene expression data of MSC and MSC-hiPSC. Dashed lines delimitate fold changes (FC) of MSC-hiPSC vs MSC below (red dots) and above (blue dots) 2. b) Volcano plot showing p-value and FC of gene expression data comparing MSC to MSC-hiPSC. Vertical dashed lines delimitate FC below and above 2, the horizontal dashed line shows p-value=0.05 [two-tailed t-test]. Colour code: FC greater than 2 reaching (green) or not (yellow) statistical significance; FC smaller than 2 reaching (red) or not (black) statistical significance. c) Histogram showing expression level FC for miRNA belonging to the let-7 family (black bars) and to the miRNA302/367 cluster (white bars) comparing MSC to MSC-hiPSC. FC and standard deviation in logarithmic scale are represented. Plots showing p-value (colour key), number of genes (dot size) and fold enrichment (vertical axis) of gene ontology terms of the biological processes categories for genes targeted by MSC (d) and MSC-hiPSC (e) upregulated miRNA [Fisher's exact test].
3.7. circRNome of MSC-hiPSC
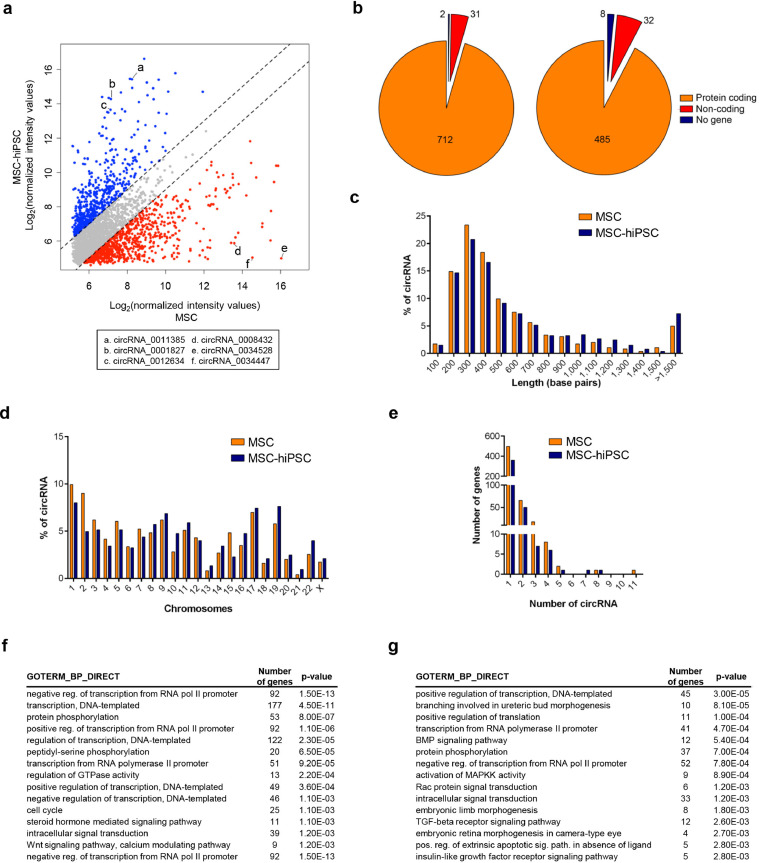
The circRNome of MSC-hiPSC was determined by means of a microarray technology and again compared to that of MSC to unveil still unreported connections between circRNA, miRNA and mRNA. Two distinct profiles of circRNA expression were obtained (Fig. 6a) and validated (Supplementary fig. 6e). MSC expressed 3290 circRNA, whereas MSC-hiPSC expressed 4429 circRNA. Differentially expressed circRNA were analysed considering 2970 shared circRNA (Supplementary fig. 6f). amongst them, 745 were found to be upregulated in MSC, whereas 525 were found to be upregulated in MSC-hiPSC, based on a 2-fold change cut-off. The majority of them were of exonic origin, whereas a minor portion was transcribed from non-coding gene regions, or more rarely from intergenic DNA (Fig. 6b). amongst non-coding gene regions, circRNA were mostly found at intronic regions and at 5′-UTR regions (Supplementary fig. 6g). CircRNA from MSC and MSC-hiPSC had similar length distribution, with the 50% of them ranging between 200 and 400 nucleotides (Fig. 6c). Likewise, chromosome localization of circRNA-generating genes showed a similar pattern (Fig. 6d). Chromosome 13 and 21 were the least rich in circRNA (≈1%) compared to an average percentage of 5% for the other chromosomes. The majority of genes harboured one circRNA, whereas some expressed from 2 up to 4 circRNA (Fig. 6e).
Fig. 6.
circRNome of MSC-hiPSC. a) Scatter plot comparing normalized intensity values of MSC and MSC-hiPSC. Dashed lines delimitate fold changes of MSC-hiPSC vs MSC below (red dots) and above (blue dots) 2. Letters on plot indicate circRNA validated by qPCR as specified by the legend. b) Pie chart representing genomic origin of circRNA for MSC (left) and MSC-hiPSC (right). Number of genes for each category is reported. c) Histograms showing size distribution of circRNA. d) Histograms showing chromosome localization of circRNA. e) Histograms showing the number of circRNA produced per gene. Enrichment in gene ontology terms of the biological processes category (GOTERM_BP_DIRECT) for genes targeted by circRNA-sponged miRNA in MSC (f) and MSC-hiPSC (g) is reported in the indicated tables [Fisher's exact test].
CircRNA-sponged miRNA were predicted by bioinformatics analysis of differentially expressed circRNA. From the resulting circRNA-sponged miRNA, enriched molecular pathways of targeted transcripts were determined. Importantly, the selected miRNA were detected in the respective cell type as evidenced by the miRNome analysis. Upregulated circRNA in MSC elicited involvement of pathways regulating signal transduction and transcription (Fig. 6f and supplementary fig. 6h). Upregulated circRNA in MSC-hiPSC were also involved in development and morphogenetic signalling pathways (Fig. 6g and supplementary fig. 6i). To further investigate the influence of cell source on PSC circRNome, MSC-hiPSC circRNA profile was compared with that of F-hiPSC [13] and hESC [19] (Supplementary fig. 6j), taking into account circRNA annotated in circBASE database. Intriguingly, MSC-hiPSC and hESC shared 1962 circRNA, which represented the 44% and 42% of their total circRNA content, respectively. This percentage was markedly reduced considering circRNA shared between F-hiPSC and hESC (32% and 8%, respectively). MSC-hiPSC and F-hiPSC expressed 367 common circRNA, which represented the 8% and 30% of their total circRNA content, respectively. The subset of circRNA shared by all PSC datasets represented the 3% of total detected circRNA.
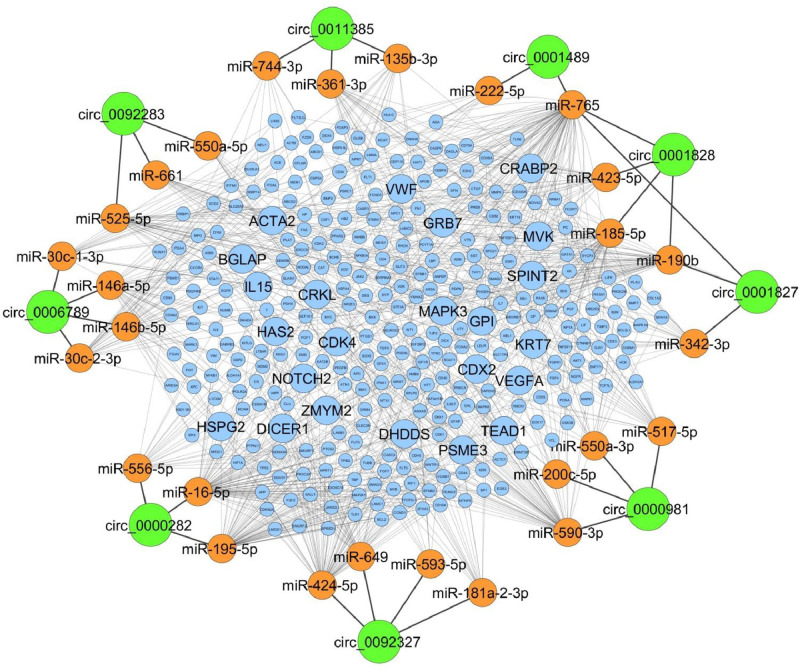
Based on data coming from the comprehensive stem cell mRNA panel and the miRNome analysis, a circRNA-guided molecular network was generated (Fig. 7). In detail, 9 out of 10 top MSC-hiPSC circRNA were predicted to interact with 27 miRNA, which were detected by the miRNA PCR-array. These miRNA targeted 310 stemness-associated mRNA expressed in MSC-hiPSC as measured by the mRNA PCR-array. Genes targeted by more than 5 miRNA by bioinformatics prediction and showing high expression by the stem cell mRNA panel were evidenced in the network. Of note, many of these genes showed preferential expression during embryogenesis or in foetal tissues, such as the placenta. Furthermore, they were mainly involved in stemness and development (i.e.: GRB7, ACTA2, TEAD1, CDX2, HSPG2, NOTCH2), in metabolic processes (i.e.: DHDDS, GPI, MVK, BGLAP) and, more intriguingly, in the regulation of naïve versus primed pluripotent sub-states (i.e.: CRKL, CRABP2, MAPK3, ZMYM2).
Fig. 7.
Map of circRNA-miRNA-mRNA network for MSC-hiPSC. The schematic shows top upregulated circRNA (green) in MSC-hiPSC, circRNA-sponged miRNA (orange) and miRNA-targeted mRNA (blue). Transcripts targeted by at least 5 miRNA are evidenced.
4. Discussion
In the wake of the compelling interest in human induced pluripotent stem cell (hiPSC) clinical applications, we successfully explored for the first time the use of cord blood multipotent mesenchymal stromal cells (MSC) as the starting cell type to obtain hiPSC. Noteworthy, other stromal cell types isolated from perinatal tissues have been reprogrammed by groups sharing the same view on the advantages of this kind of cell sources. For instance, cord blood unrestricted somatic stem cells were induced to acquire a pluripotent state by the use of an integrative retroviral approach [63]. Similarly, successful reprogramming to pluripotency was reported for cord blood endothelial cells [64], Wharton's jelly MSC [65,66] and amniotic fluid cells [67,68]. Also, non-stromal cord blood cell types were used to generate hiPSC by others [69,70]. Intriguingly, MSC showed high expression of KLF4 and cMYC compared with MSC-hiPSC. These results were also reported by others [63,71] and could be ascribed to the developmental origin of MSC and to their highly proliferative state, respectively. Being two components of the canonical reprogramming cocktail of transcription factors, KLF4 and cMYC higher transcriptional level could be addressed as a feature influencing reprogramming efficiency in ad hoc studies.
In the present work, we focused on high-throughput techniques to define coding and non-coding RNA species involved in pluripotent identity definition. At the messenger RNA (mRNA) level, we observed a molecular skew for endothelial and mesenchymal identity-associated molecular pathways in MSC-hiPSC and fibroblast (F)-hiPSC, respectively, in a context of higher similarity to human embryonic stem cells (hESC) for the former. These molecular profiles could be ascribed to preservation of parental cell traits due to epigenetic memory. The issue of source cells can be tracked down to the pioneering studies on reprogramming, which used mouse embryonic and human adult fibroblasts [72,73]. In later studies, successful reprogramming of other cell types was reported, such as keratinocytes [74], hematopoietic cells [70], pancreatic islet beta cells [75], hepatocytes [76], neural progenitors [77] and adult multipotent mesenchymal stromal cells [78]. Notwithstanding, the majority of studies still relies on the reprogramming of mesodermal derivatives, and most of them on skin fibroblasts [3]. These pivotal studies lead to the definition of reprogramming as a general biological mechanism, but also introduced the issue of epigenetic memory of the parental cell in the obtained hiPSC. Indeed, tissue of origin can heavily influence hiPSC differentiation properties. For instance, beta cell-derived hiPSC maintained open chromatin structure at key beta-cell genes and demonstrated higher differentiation ability into insulin-producing cells than isogenic non-beta cell-derived hiPSC [75]. The same phenomenon was observed for cardiac progenitor cell-derived hiPSC compared to F-hiPSC [5]. In other works, persistent and incomplete erasure of tissue-specific methylation and aberrant de novo methylation were observed in hiPSC depending on the source cell type [6,7,9]. This donor memory was found also at the transcriptional level for mRNA and micro RNA (miRNA) [79], [80], [81]. Yet, other reports underlined that this is a transitory phenomenon. Kim et al. showed that epigenetic memory could be erased by differentiation or serial reprogramming [4]. In another study, epigenetic memory was observed only at early passages after pluripotency induction and was attenuated by continuous passaging [82]. Likewise, Gao et al. evidenced minor transcriptional memory between parental cells and hiPSC, with no influence on their differentiation capacity [83]. Another study investigated erythroid differentiation properties of hiPSC derived from neural stem cells or hematopoietic stem cells. Despite persistency of epigenetic memory, no relevant differences were observed [84]. Previous works comparing MSC-hiPSC with F-hiPSC focused on cardiac differentiation [85] and transcriptional profile [86], detecting overall similarity between the two hiPSC classes. In the present study, we addressed for the first time in the literature the differentiation potential toward mesenchymal-like derivatives and their hematopoietic support ability. In line with the aforementioned reports, no relevant differences were observed.
Circular RNA (circRNA), as novel actors in the arena of non-coding functional RNA, add a layer of refinement in post-transcriptional control. To this regard, we defined the largest hiPSC circRNome ever reported, and concomitantly described their miRNome and stemness-associated mRNA. Only two very recent reports by other groups addressed the presence of circRNA in hiPSC. In one study, circBIRC6 was shown to participate in the molecular circuitry controlling pluripotency (PGRN) of hESC and reprogramming of somatic cells to hiPSC via direct sponging of miRNA targeting mRNA involved in the maintenance of the pluripotent state, thus demonstrating relevance of circRNA in hiPSC biology [14]. This study relied on expression data already available at public databases generated by RNA-seq of differentiated tissues and hESC [20,87]. In our previous study, we also reported that another circRNA, circFOXP1, is involved in the maintenance of mesenchymal stem cell identity and downregulated upon reprogramming to pluripotency [12]. To note, circBIRC6 is not amongst the most expressed circRNA by MSC-hiPSC as outlined by our microarray data. Another group described the RNA-seq-defined circRNome of F-hiPSC compared to a differentiated counterpart, detecting a subset of circRNA with cardiac-specific expression [13]. Even though they identified 614 new hiPSC-specific circRNA, the whole circRNome was constituted by only 1612 molecules. Our microarray approach instead disclosed a larger set of 4429 hiPSC-expressed circRNA, providing a more challenging ground for researchers interested in the role of these non-coding RNA in stem cell biology. Furthermore, the herein defined dataset showed higher similarity with hESC circRNA profile, although biases associated to the different methodologies applied cannot be excluded.
Herein, our comprehensive strategy allowed focusing on circRNA-miRNA interactions in hiPSC as potential novel mechanism to control the identity of pluripotent stem cells. In contrast to our expectations, top selected mRNA targeted by circRNA-sponged miRNA were not belonging to the core transcription factors of the PGRN. Yet, most of them were found to be expressed at higher levels during embryogenesis or in foetal tissues, hinting at a possible contribution in the modulation of the undifferentiated state of PSC. In particular, CRKL is an activator of the RAS signalling pathway, which was shown to be a key player in the transition from the naïve to the primed pluripotent state [88]. Likewise, MAPK3-mediated signalling was involved in the priming of naïve PSC to lineage commitment [89], albeit compatible with self-renewal at basal levels [90]. Furthermore, ZMYM2 was reported to physically interact with NANOG, one of the core actors of the PGRN, to curtail NANOG-mediated shift from the primed to the naïve pluripotent state [91]. Conversely, CRABP2 was found to be upregulated in the naïve state [92].
These intriguing results prompted us to hypothesize that circRNA may cast a further layer of molecular control over stemness and self-renewal in addition to the action of the core PGRN. In this scenario, circRNA-driven sponging of miRNA would represent a rapid post-transcriptional mechanism to achieve superior gene expression control on more refined cellular states, such as the skew toward naïve or primed pluripotency.
To conclude, we showed that MSC-hiPSC generated from cord blood units with high clinical translatability potential acquired a complete pluripotent identity. Focusing on mesodermal lineages, differentiation potential of cord blood MSC-hiPSC and functional properties of its derivatives were undistinguishable from those of F-hiPSC. Yet, deep molecular profiling revealed higher similarity to hESC, which should be carefully investigated before excluding any phenotypic peculiarities of cord blood MSC-hiPSC compared to F-hiPSC with regard to other biological processes or differentiation pathways. Furthermore, we provided new insights on unreported circRNA-miRNA-mRNA axes possibly involved in self-renewal and differentiation processes. All considered, cord blood represents a better alternative than skin fibroblasts as cell source for clinical translation of hiPSC and hiPSC-based cell derivatives, whose generation will benefit from future studies aimed at a better comprehension of how circRNA intervene in the exit from the pluripotent program.
Author contribution
Conceptualization, M.B., N.E. and L.L.; Methodology, M.B., A.C., V.B., F.G., A.G. and N.E.; Validation, M.B. and A.C.; Formal Analysis, M.B., A.C., V.P., F.G., A.G. and F.P.; Investigation, M.B., A.C., V.P., F.G., A.G. and F.P.; Resources, M.B., V.B., F.G., A.G., S.G. and N.E.; Writing – Original Draft, M.B., A.C., V.P., F.G. and L.L.; Writing – Review & Editing, M.B., A.C., V.P., N.E., V.B., F.G., A.G., C.L., S.G. and L.L.; Visualization, M.B., A.C., V.P. and F.P.; Supervision, V.B., N.E., C.L. and L.L.; Project Administration, L.L.; Funding Acquisition, L.L..
Declaration of Competing Interest
The method for isolation and reprogramming to pluripotency of long-living cord blood multipotent mesenchymal stromal cells is part of a technology developed by Dr. Lorenza Lazzari and Dr. Mario Barilani licensed by the Italian national patent agency Ufficio Italiano Brevettie Marchi (UIBM) with numbers 102017000141216 and 102017000141245 (international numbers WO2019/111197 and WO2019/111198).
Acknowledgements
This research was funded by Ricerca Corrente 2012–2018 by the Italian Ministry of Health. The funding source had no role in study design, data collection, data analysis, interpretation, and writing of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102848.
Appendix. Supplementary materials
Supplementary fig 1 Reprogramming to pluripotency of mesenchymal stem cells (MSC). a) Representative bright-field images of MSC at week 1 and week 3 after infection with reprogramming vectors. b) Representative bright-field and fluorescence images showing alkaline phosphatase (AP) activity of a representative colony (dashed line) of MSC-derived human induced pluripotent stem cells (MSC-hiPSC) and of neighbouring unreprogrammed MSC. c) Bright-field and fluorescence images of representative MSC-hiPSC (left panel) and human embryonic stem cells (hESC, right panel) colonies onto matrigel coating showing AP activity. d) Growth curves of MSC-hiPSC and hESC. Mean and standard deviation are represented (n = 3); CPD, cumulative population doublings. e) Histograms showing doubling time of MSC-hiPSC compared to hESC. Mean and standard deviation are represented (n = 3); h, hours. f) Representative xCELLigence proliferation rate curve (cell index parameter) for population doubling time calculation during the exponential growth phase (detected between the blue and red vertical lines). Mean and standard deviation of replicates are represented (n = 6); h, hours. g) Representative karyotype of MSC-hiPSC. h) Table summarizing all immunophenotyping data for MSC, hESC and MSC-hiPSC. ne, not expressed. Mean and standard deviation are reported.
Supplementary fig 2 Characterization of MSC-hiPSC. a) Table summarizing qPCR results of Sendai virus vector specific sequences for unreprogrammed MSC, MSC during reprogramming and established MSC-hiPSC lines. Ct, threshold cycle; SEV, sequence common to all Sendai virus vectors; na, not amplified; ntc, no template control. b) Representative fluorescence images showing protein expression and cellular localization of typical pluripotency markers (OCT4A, SSEA4, SOX2, TRA1–60) for hESC. c) Representative fluorescence images showing protein expression and cellular localization of mesodermal (αSMA), endodermal (AFP) and ectodermal (NCAM-1 derivative markers for in vitro differentiated hESC. d) Representative fluorescence image showing protein expression and cellular localization of αSMA mesodermal derivative marker for MSC. e) Representative fluorescence image showing protein expression and cellular localization of a typical neuronal marker (TUJ1) for in vitro differentiated MSC-hiPSC. DAPI was used to stain nuclei in all fluorescence images.
Supplementary fig 3 In vivo differentiation of MSC-hiPSC. Representative hematoxylin-eosin histology images of in vivo differentiated MSC-hiPSC. Chondroid matrix (a), smooth muscle (b), cartilage (c), squamous (d), pigment (e), and ciliary (f) epithelium are shown.
Supplementary fig 4 Stem cell transcriptome of MSC-hiPSC. a) Matrix of plots showing Pearson's correlations amongst MSC, hESC and MSC-hiPSC based on gene expression data The matrix contains density plots (matrix diagonal), scatter plots with least square reference line (below matrix diagonal) and Pearson's correlation coefficients (above matrix diagonal) of gene expression data. Scatter plots comparing gene expression data of different MSC (b) and MSC-hiPSC (c) are shown. Dashed lines delimitate fold changes of MSC 2 vs MSC 1 (b) and MSC-hiPSC 2 vs MSC-hiPSC 1 (c) below (red dots) and above (blue dots) 2. Enrichment in Kyoto encyclopaedia Genes and Genomes pathways (KEGG_PATHWAYS) and p-value for upregulated genes of MSC (d) and MSC-hiPSC (e) is reported in the indicated tables.
Supplementary fig 5 Mesenchymal potential of MSC-hiPSC. a) Scatter plot showing gene expression data of two F-hiPSC. Dashed lines delimitate fold changes of F-hiPSC 2 vs F-hiPSC 1 below (red dots) and above (blue dots) 2. b) Dendrogram showing hierarchical clustering of F- and MSC-hiPSC lines and hESC. Enrichment in Kyoto encyclopaedia Genes and Genomes pathways (KEGG_PATHWAYS) and p-value for upregulated genes of MSC-hiPSC (c) and fibroblast derived hiPSC (F-hiPSC) (d) is reported in the indicated tables. e) Left plots representative density plot showing positivity of MSC-like cells for CD90 and CD271. Right plots: representative histograms showing positivity to CD56. f) Histograms showing gene expression of the genes of interest. Mean and standard deviation are represented; n = 6 for MSC-like cells, n = 3 for primary MSC. g) Representative bright-field images of cobblestone areas. Scale bars are 100 μm.
Supplementary fig 6 miRNA and circRNA expression in MSC-hiPSC. Scatter plots comparing miRNA expression data of different MSC (a) and MSC-hiPSC (b) are shown. Dashed lines delimitate fold changes of MSC 2 vs MSC 1 (b) and MSC-hiPSC 2 vs MSC-hiPSC 1 (c) below (red dots) and above (blue dots) 2. Enrichment in Kyoto encyclopaedia Genes and Genomes pathways (KEGG_PATHWAYS), number of genes and p-value for mRNA targeted by upregulated miRNA of MSC (c) and MSC-hiPSC (d) is reported in the indicated tables. Histogram showing qPCR validations of dowregulated (black bars) and upregulated (white bars) circRNA in MSC-hiPSC compared to MSC. Mean and standard error of the mean are represented (e). Venn diagram representing shared and unique circRNA in MSC and MSC-hiPSC (f). Venn diagrams summarizing genetic origin of circRNA in percentage (g). CDS, coding DNA sequence; UTR3, 3′ untranslated region; intronic, intronic region; UTR5, 5′ untranslated region. Tables reporting KEGG_PATHWAYS, number of genes and p-value for mRNA targeted by miRNA sponged by top 10 upregulated MSC (h) and MSC-hiPSC (i) circRNA. j) Venn diagram showing shared and unique circRNA as reported in the present study for MSC-hiPSC (Barilani et al.) and in other studies for F-hiPSC (Lei et al.) and hESC (Salzman et al.).
References
- 1.Mandai M., Watanabe A., Kurimoto Y., Hirami Y., Morinaga C., Daimon T. Autologous induced stem-cell-derived retinal cells for macular degeneration. N Engl J Med. 2017;376(11):1038–1046. doi: 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]
- 2.Cyranoski D. Japanese man is first to receive ‘reprogrammed'stem cells from another person. Nature. 2017:10. doi: 10.1038/d41586-019-02597-2. [DOI] [PubMed] [Google Scholar]
- 3.Brouwer M., Zhou H., Nadif Kasri N. Choices for induction of pluripotency: recent developments in human induced pluripotent stem cell reprogramming strategies. Stem cell Rev Reports. 2016;12(1):54–72. doi: 10.1007/s12015-015-9622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez-Freire V., Lee A.S., Hu S., Abilez O.J., Liang P., Lan F. Effect of human donor cell source on differentiation and function of cardiac induced pluripotent stem cells. J Am Coll Cardiol. 2014;64(5):436–448. doi: 10.1016/j.jacc.2014.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim K., Zhao R., Doi A., Ng K., Unternaehrer J., Cahan P. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(12):1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luu P.L., Gerovska D., Scholer H.R., Arauzo-Bravo M.J. Rules governing the mechanism of epigenetic reprogramming memory. Epigenomics. 2018;10(2):149–174. doi: 10.2217/epi-2017-0098. [DOI] [PubMed] [Google Scholar]
- 8.Noguchi H., Miyagi-Shiohira C., Nakashima Y. Induced tissue-specific stem cells and epigenetic memory in induced pluripotent stem cells. Int J Mol Sci. 2018;19(4) doi: 10.3390/ijms19040930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohi Y., Qin H., Hong C., Blouin L., Polo J.M., Guo T. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 2011;13(5):541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M., Belmonte J.C. Ground rules of the pluripotency gene regulatory network. Nat Rev Genet. 2017;18(3):180–191. doi: 10.1038/nrg.2016.156. [DOI] [PubMed] [Google Scholar]
- 11.Sherstyuk V.V., Medvedev S.P., Zakian S.M. Noncoding RNAs in the Regulation of Pluripotency and Reprogramming. Stem cell Rev Reports. 2018;14(1):58–70. doi: 10.1007/s12015-017-9782-9. [DOI] [PubMed] [Google Scholar]
- 12.Cherubini A., Barilani M., Rossi R.L., Jalal M.M.K., Rusconi F., Buono G. FOXP1 circular RNA sustains mesenchymal stem cell identity via microRNA inhibition. Nucleic Acids Res. 2019;47(10):5325–5340. doi: 10.1093/nar/gkz199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lei W., Feng T., Fang X., Yu Y., Yang J., Zhao Z.A. Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Res Ther. 2018;9(1):56. doi: 10.1186/s13287-018-0793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8(1):1149. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.H. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasman Z., Been M.D., Garcia-Blanco M.A. Exon circularization in mammalian nuclear extracts. RNA (New York, NY) 1996;2(6):603–610. [PMC free article] [PubMed] [Google Scholar]
- 19.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 21.Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Shen T., Han M., Wei G., Ni T. An intriguing RNA species–perspectives of circularized RNA. Protein Cell. 2015;6(12):871–880. doi: 10.1007/s13238-015-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H., Jang H., Kim T.W., Kang B.H., Lee S.E., Jeon Y.K. Core pluripotency factors directly regulate metabolism in embryonic stem cell to maintain pluripotency. Stem Cells. 2015;33(9):2699–2711. doi: 10.1002/stem.2073. [DOI] [PubMed] [Google Scholar]
- 24.Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. 2014;9(6):e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 28.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stubbendorff M., Deuse T., Hua X., Phan T.T., Bieback K., Atkinson K. Immunological properties of extraembryonic human mesenchymal stromal cells derived from gestational tissue. Stem Cells Dev. 2013;22(19):2619–2629. doi: 10.1089/scd.2013.0043. [DOI] [PubMed] [Google Scholar]
- 31.Wang M., Yang Y., Yang D., Luo F., Liang W., Guo S. The immunomodulatory activity of human umbilical cord blood-derived mesenchymal stem cells in vitro. Immunology. 2009;126(2):220–232. doi: 10.1111/j.1365-2567.2008.02891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riordan N.H., Chan K., Marleau A.M., Ichim T.E. Cord blood in regenerative medicine: do we need immune suppression? J Transl Med. 2007;5:8. doi: 10.1186/1479-5876-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vormoor J., Lapidot T., Pflumio F., Risdon G., Patterson B., Broxmeyer H.E. Immature human cord blood progenitors engraft and proliferate to high levels in severe combined immunodeficient mice. Blood. 1994;83(9):2489–2497. [PubMed] [Google Scholar]
- 34.Cairo M.S., Wagner J.E. Placental and/or umbilical cord blood: an alternative source of hematopoietic stem cells for transplantation. Blood. 1997;90(12):4665–4678. [PubMed] [Google Scholar]
- 35.Abyzov A., Mariani J., Palejev D., Zhang Y., Haney M.S., Tomasini L. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492(7429):438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Antonio M., Benaglio P., Jakubosky D., Greenwald W.W., Matsui H., Donovan M.K.R. Insights into the mutational burden of human induced pluripotent stem cells from an integrative multi-omics approach. Cell Rep. 2018;24(4):883–894. doi: 10.1016/j.celrep.2018.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang X.X., Bernerd F., Halliday G.M. Ultraviolet A within sunlight induces mutations in the epidermal basal layer of engineered human skin. Am J Pathol. 2009;174(4):1534–1543. doi: 10.2353/ajpath.2009.080318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira L. Congenital viral infection: traversing the uterine-placental interface. Annu Rev Virol. 2018;5(1):273–299. doi: 10.1146/annurev-virology-092917-043236. [DOI] [PubMed] [Google Scholar]
- 39.Tetro N., Moushaev S., Rubinchik-Stern M., Eyal S. The placental barrier: the gate and the fate in drug distribution. Pharm Res. 2018;35(4):71. doi: 10.1007/s11095-017-2286-0. [DOI] [PubMed] [Google Scholar]
- 40.Al-Enazy S., Ali S., Albekairi N., El-Tawil M., Rytting E. Placental control of drug delivery. Adv Drug Deliv Rev. 2017;116:63–72. doi: 10.1016/j.addr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazzari L., Corsini C., Curioni C., Lecchi L., Scalamogna M., Rebulla P. The Milan cord blood bank and the italian cord blood network. J Hematother. 1996;5(2):117–122. doi: 10.1089/scd.1.1996.5.117. [DOI] [PubMed] [Google Scholar]
- 42.Bertolini F., Lazzari L., Corsini C., Lauri E., Gorini F., Sirchia G. Cord blood banking for stem cell transplant. Int J Artif Organs. 1993;16(Suppl 5):111–112. [PubMed] [Google Scholar]
- 43.Barilani M., Lavazza C., Boldrin V., Ragni E., Parazzi V., Crosti M. A chemically defined medium-based strategy to efficiently generate clinically relevant cord blood mesenchymal stromal colonies. Cell Transplant. 2016;25(8):1501–1514. doi: 10.3727/096368916X690827. [DOI] [PubMed] [Google Scholar]
- 44.Barilani M., Lavazza C., Vigano M., Montemurro T., Boldrin V., Parazzi V. Dissection of the cord blood stromal component reveals predictive parameters for culture outcome. Stem Cells Dev. 2015;24(1):104–114. doi: 10.1089/scd.2014.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barilani M., Palorini R., Votta G., Piras R., Buono G., Grassi M. Central metabolism of functionally heterogeneous mesenchymal stromal cells. Sci Rep. 2019;9(1):15420. doi: 10.1038/s41598-019-51937-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barilani M., Banfi F., Sironi S., Ragni E., Guillaumin S., Polveraccio F. Low-affinity nerve growth factor receptor (CD271) heterogeneous expression in adult and fetal mesenchymal stromal cells. Sci Rep. 2018;8(1):9321. doi: 10.1038/s41598-018-27587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy. 2006;8(4):315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 48.Horwitz E.M., Le Blanc K., Dominici M., Mueller I., Slaper-Cortenbach I., Marini F.C. Clarification of the nomenclature for MSC: the international society for cellular therapy position statement. Cytotherapy. 2005;7(5):393–395. doi: 10.1080/14653240500319234. [DOI] [PubMed] [Google Scholar]
- 49.Giobbe G.G., Michielin F., Luni C., Giulitti S., Martewicz S., Dupont S. Functional differentiation of human pluripotent stem cells on a chip. Nat Methods. 2015;12(7):637–640. doi: 10.1038/nmeth.3411. [DOI] [PubMed] [Google Scholar]
- 50.Gagliano O., Luni C., Qin W., Bertin E., Torchio E., Galvanin S. Microfluidic reprogramming to pluripotency of human somatic cells. Nat Protoc. 2019;14(3):722–737. doi: 10.1038/s41596-018-0108-4. [DOI] [PubMed] [Google Scholar]
- 51.Luni C., Giulitti S., Serena E., Ferrari L., Zambon A., Gagliano O. High-efficiency cellular reprogramming with microfluidics. Nat Methods. 2016;13(5):446–452. doi: 10.1038/nmeth.3832. [DOI] [PubMed] [Google Scholar]
- 52.Barilani M., Peli V., Cherubini A., Dossena M., Dolo V., Lazzari L. NG2 as an identity and quality marker of mesenchymal stem cell extracellular vesicles. Cells. 2019;8(12) doi: 10.3390/cells8121524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 54.Delli Carri A., Onorati M., Lelos M.J., Castiglioni V., Faedo A., Menon R. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2013;140(2):301–312. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 55.Zhang W.Y., de Almeida P.E., Wu J.C. StemBook; CambridgeMA: 2008. Teratoma formation: a tool for monitoring pluripotency in stem cell research. Harvard Stem Cell Institute Copyright: (c) 2012 Wendy Y. Zhang, Patricia E. de Almeida, and Joseph C. Wu. [PubMed] [Google Scholar]
- 56.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M. NCBI GEO: archive for functional genomics data sets—Update. Nucleic Acids Res. 2012;41(D1):D991–D9D5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodriguez-Piza I., Richaud-Patin Y., Vassena R., Gonzalez F., Barrero M.J., Veiga A. Reprogramming of human fibroblasts to induced pluripotent stem cells under xeno-free conditions. Stem Cells. 2010;28(1):36–44. doi: 10.1002/stem.248. [DOI] [PubMed] [Google Scholar]
- 60.Sustackova G., Legartova S., Kozubek S., Stixova L., Pachernik J., Bartova E. Differentiation-independent fluctuation of pluripotency-related transcription factors and other epigenetic markers in embryonic stem cell colonies. Stem Cells Dev. 2012;21(5):710–720. doi: 10.1089/scd.2011.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karwacki-Neisius V., Goke J., Osorno R., Halbritter F., Ng J.H., Weisse A.Y. Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell. 2013;12(5):531–545. doi: 10.1016/j.stem.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strebinger D., Deluz C., Friman E.T., Govindan S., Alber A.B., Suter D.M. Endogenous fluctuations of OCT4 and SOX2 bias pluripotent cell fate decisions. Mol Syst Biol. 2019;15(9):e9002. doi: 10.15252/msb.20199002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zaehres H., Kogler G., Arauzo-Bravo M.J., Bleidissel M., Santourlidis S., Weinhold S. Induction of pluripotency in human cord blood unrestricted somatic stem cells. Exp Hematol. 2010;38(9) doi: 10.1016/j.exphem.2010.05.009. 809-18, 18.e1-2. [DOI] [PubMed] [Google Scholar]
- 64.Haase A., Olmer R., Schwanke K., Wunderlich S., Merkert S., Hess C. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5(4):434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 65.Miere C., Devito L., Ilic D. Sendai virus-based reprogramming of mesenchymal stromal/stem cells from umbilical cord Wharton's Jelly into induced pluripotent stem cells. Methods Mol Biol. 2016;1357:33–44. doi: 10.1007/7651_2014_163. [DOI] [PubMed] [Google Scholar]
- 66.Devito L., Klontzas M.E., Cvoro A., Galleu A., Simon M., Hobbs C. Comparison of human isogeneic Wharton's jelly MSCs and iPSC-derived MSCs reveals differentiation-dependent metabolic responses to IFNG stimulation. Cell Death Dis. 2019;10(4):277. doi: 10.1038/s41419-019-1498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drews K., Matz P., Adjaye J. Generation of iPSC lines from primary human amniotic fluid cells. Stem Cell Res. 2015;15(3):712–714. doi: 10.1016/j.scr.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Slamecka J., Laurini J., Shirley T., Hoerstrup S.P., Weber B., Owen L. Reprogramming primary amniotic fluid and membrane cells to pluripotency in xeno-free conditions. J Vis Exp. 2017;(129) doi: 10.3791/56003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Gu Q., Hao J., Bai D., Liu L., Zhao X. Generation of induced pluripotent stem cells with high efficiency from human umbilical cord blood mononuclear cells. Genomics Proteomics Bioinformatics. 2013;11(5):304–311. doi: 10.1016/j.gpb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giorgetti A., Montserrat N., Aasen T., Gonzalez F., Rodriguez-Piza I., Vassena R. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 2009;5(4):353–357. doi: 10.1016/j.stem.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roson-Burgo B., Sanchez-Guijo F., Del Canizo C., De Las Rivas J. Transcriptomic portrait of human Mesenchymal Stromal/Stem Cells isolated from bone marrow and placenta. BMC Genomics. 2014;15:910. doi: 10.1186/1471-2164-15-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 73.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 74.Aasen T., Raya A., Barrero M.J., Garreta E., Consiglio A., Gonzalez F. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26(11):1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 75.Bar-Nur O., Russ H.A., Efrat S., Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9(1):17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 76.Liu H., Ye Z., Kim Y., Sharkis S., Jang Y.Y. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology (Baltimore, Md) 2010;51(5):1810–1819. doi: 10.1002/hep.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim J.B., Greber B., Arauzo-Bravo M.J., Meyer J., Park K.I., Zaehres H. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461(7264) doi: 10.1038/nature08436. 649-3. [DOI] [PubMed] [Google Scholar]
- 78.Oda Y., Yoshimura Y., Ohnishi H., Tadokoro M., Katsube Y., Sasao M. Induction of pluripotent stem cells from human third molar mesenchymal stromal cells. J Biol Chem. 2010;285(38):29270–29278. doi: 10.1074/jbc.M109.055889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghosh Z., Wilson K.D., Wu Y., Hu S., Quertermous T., Wu J.C. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS ONE. 2010;5(2):e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marchetto M.C., Yeo G.W., Kainohana O., Marsala M., Gage F.H., Muotri A.R. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE. 2009;4(9):e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vitaloni M., Pulecio J., Bilic J., Kuebler B., Laricchia-Robbio L., Izpisua Belmonte J.C. MicroRNAs contribute to induced pluripotent stem cell somatic donor memory. J Biol Chem. 2014;289(4):2084–2098. doi: 10.1074/jbc.M113.538702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Polo J.M., Liu S., Figueroa M.E., Kulalert W., Eminli S., Tan K.Y. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28(8):848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao S., Hou X., Jiang Y., Xu Z., Cai T., Chen J. Integrated analysis of hematopoietic differentiation outcomes and molecular characterization reveals unbiased differentiation capacity and minor transcriptional memory in HPC/HSC-iPSCs. Stem Cell Res Ther. 2017;8(1):13. doi: 10.1186/s13287-016-0466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dorn I., Klich K., Arauzo-Bravo M.J., Radstaak M., Santourlidis S., Ghanjati F. Erythroid differentiation of human induced pluripotent stem cells is independent of donor cell type of origin. Haematologica. 2015;100(1):32–41. doi: 10.3324/haematol.2014.108068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Streckfuss-Bomeke K., Wolf F., Azizian A., Stauske M., Tiburcy M., Wagner S. Comparative study of human-induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes, and skin fibroblasts. Eur Heart J. 2013;34(33):2618–2629. doi: 10.1093/eurheartj/ehs203. [DOI] [PubMed] [Google Scholar]
- 86.Pichard L., Brondelo J.M., Becker F., Desprat R., De Ceuninck F., Pastoureau P. Generation of human pluripotent stem cell lines (iPSCs) from mesenchymal stem cells (MSCs) from three elderly patients with osteoarthritis. Stem Cell Res. 2020;44 doi: 10.1016/j.scr.2020.101721. [DOI] [PubMed] [Google Scholar]
- 87.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9) doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Altshuler A., Verbuk M., Bhattacharya S., Abramovich I., Haklai R., Hanna J.H. RAS regulates the transition from naive to primed pluripotent stem cells. Stem Cell Reports. 2018;10(3):1088–1101. doi: 10.1016/j.stemcr.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134(16):2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 90.Dalton S. Signaling networks in human pluripotent stem cells. Curr Opin Cell Biol. 2013;25(2):241–246. doi: 10.1016/j.ceb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lawrence M., Theunissen T.W., Lombard P., Adams D.J., Silva J.C.R. ZMYM2 inhibits NANOG-mediated reprogramming. Wellcome Open Res. 2019;4:88. doi: 10.12688/wellcomeopenres.15250.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu Z., Robitaille A.M., Berndt J.D., Davidson K.C., Fischer K.A., Mathieu J. Wnt/beta-catenin signaling promotes self-renewal and inhibits the primed state transition in naive human embryonic stem cells. Proc Natl Acad Sci U.S.A. 2016;113(42) doi: 10.1073/pnas.1613849113. E6382-e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary fig 1 Reprogramming to pluripotency of mesenchymal stem cells (MSC). a) Representative bright-field images of MSC at week 1 and week 3 after infection with reprogramming vectors. b) Representative bright-field and fluorescence images showing alkaline phosphatase (AP) activity of a representative colony (dashed line) of MSC-derived human induced pluripotent stem cells (MSC-hiPSC) and of neighbouring unreprogrammed MSC. c) Bright-field and fluorescence images of representative MSC-hiPSC (left panel) and human embryonic stem cells (hESC, right panel) colonies onto matrigel coating showing AP activity. d) Growth curves of MSC-hiPSC and hESC. Mean and standard deviation are represented (n = 3); CPD, cumulative population doublings. e) Histograms showing doubling time of MSC-hiPSC compared to hESC. Mean and standard deviation are represented (n = 3); h, hours. f) Representative xCELLigence proliferation rate curve (cell index parameter) for population doubling time calculation during the exponential growth phase (detected between the blue and red vertical lines). Mean and standard deviation of replicates are represented (n = 6); h, hours. g) Representative karyotype of MSC-hiPSC. h) Table summarizing all immunophenotyping data for MSC, hESC and MSC-hiPSC. ne, not expressed. Mean and standard deviation are reported.
Supplementary fig 2 Characterization of MSC-hiPSC. a) Table summarizing qPCR results of Sendai virus vector specific sequences for unreprogrammed MSC, MSC during reprogramming and established MSC-hiPSC lines. Ct, threshold cycle; SEV, sequence common to all Sendai virus vectors; na, not amplified; ntc, no template control. b) Representative fluorescence images showing protein expression and cellular localization of typical pluripotency markers (OCT4A, SSEA4, SOX2, TRA1–60) for hESC. c) Representative fluorescence images showing protein expression and cellular localization of mesodermal (αSMA), endodermal (AFP) and ectodermal (NCAM-1 derivative markers for in vitro differentiated hESC. d) Representative fluorescence image showing protein expression and cellular localization of αSMA mesodermal derivative marker for MSC. e) Representative fluorescence image showing protein expression and cellular localization of a typical neuronal marker (TUJ1) for in vitro differentiated MSC-hiPSC. DAPI was used to stain nuclei in all fluorescence images.
Supplementary fig 3 In vivo differentiation of MSC-hiPSC. Representative hematoxylin-eosin histology images of in vivo differentiated MSC-hiPSC. Chondroid matrix (a), smooth muscle (b), cartilage (c), squamous (d), pigment (e), and ciliary (f) epithelium are shown.
Supplementary fig 4 Stem cell transcriptome of MSC-hiPSC. a) Matrix of plots showing Pearson's correlations amongst MSC, hESC and MSC-hiPSC based on gene expression data The matrix contains density plots (matrix diagonal), scatter plots with least square reference line (below matrix diagonal) and Pearson's correlation coefficients (above matrix diagonal) of gene expression data. Scatter plots comparing gene expression data of different MSC (b) and MSC-hiPSC (c) are shown. Dashed lines delimitate fold changes of MSC 2 vs MSC 1 (b) and MSC-hiPSC 2 vs MSC-hiPSC 1 (c) below (red dots) and above (blue dots) 2. Enrichment in Kyoto encyclopaedia Genes and Genomes pathways (KEGG_PATHWAYS) and p-value for upregulated genes of MSC (d) and MSC-hiPSC (e) is reported in the indicated tables.
Supplementary fig 5 Mesenchymal potential of MSC-hiPSC. a) Scatter plot showing gene expression data of two F-hiPSC. Dashed lines delimitate fold changes of F-hiPSC 2 vs F-hiPSC 1 below (red dots) and above (blue dots) 2. b) Dendrogram showing hierarchical clustering of F- and MSC-hiPSC lines and hESC. Enrichment in Kyoto encyclopaedia Genes and Genomes pathways (KEGG_PATHWAYS) and p-value for upregulated genes of MSC-hiPSC (c) and fibroblast derived hiPSC (F-hiPSC) (d) is reported in the indicated tables. e) Left plots representative density plot showing positivity of MSC-like cells for CD90 and CD271. Right plots: representative histograms showing positivity to CD56. f) Histograms showing gene expression of the genes of interest. Mean and standard deviation are represented; n = 6 for MSC-like cells, n = 3 for primary MSC. g) Representative bright-field images of cobblestone areas. Scale bars are 100 μm.
Supplementary fig 6 miRNA and circRNA expression in MSC-hiPSC. Scatter plots comparing miRNA expression data of different MSC (a) and MSC-hiPSC (b) are shown. Dashed lines delimitate fold changes of MSC 2 vs MSC 1 (b) and MSC-hiPSC 2 vs MSC-hiPSC 1 (c) below (red dots) and above (blue dots) 2. Enrichment in Kyoto encyclopaedia Genes and Genomes pathways (KEGG_PATHWAYS), number of genes and p-value for mRNA targeted by upregulated miRNA of MSC (c) and MSC-hiPSC (d) is reported in the indicated tables. Histogram showing qPCR validations of dowregulated (black bars) and upregulated (white bars) circRNA in MSC-hiPSC compared to MSC. Mean and standard error of the mean are represented (e). Venn diagram representing shared and unique circRNA in MSC and MSC-hiPSC (f). Venn diagrams summarizing genetic origin of circRNA in percentage (g). CDS, coding DNA sequence; UTR3, 3′ untranslated region; intronic, intronic region; UTR5, 5′ untranslated region. Tables reporting KEGG_PATHWAYS, number of genes and p-value for mRNA targeted by miRNA sponged by top 10 upregulated MSC (h) and MSC-hiPSC (i) circRNA. j) Venn diagram showing shared and unique circRNA as reported in the present study for MSC-hiPSC (Barilani et al.) and in other studies for F-hiPSC (Lei et al.) and hESC (Salzman et al.).
Data Availability Statement
Data of messenger (GSE144634), micro (GSE144631) and circular (GSE144629 and GSE122178) RNA are accessible through the specified series accession numbers at the GEO online public database by NCBI, which supports MIAME-compliant data submissions [57,58] (https://www.ncbi.nlm.nih.gov/geo). All other data and material are available upon request.