Abstract
Zaire ebolavirus (EBOV) causes Ebola virus disease (EVD), which carries a fatality rate between 25% and 90% in humans. Liver pathology is a hallmark of terminal EVD; however, little is known about temporal disease progression. We used multiplexed fluorescent immunohistochemistry and in situ hybridization in combination with whole slide imaging and image analysis (IA) to quantitatively characterize temporospatial signatures of viral and host factors as related to EBOV pathogenesis. Eighteen rhesus monkeys euthanized between 3 and 8 days post-infection, and 3 uninfected controls were enrolled in this study. Compared with semiquantitative histomorphologic ordinal scoring, quantitative IA detected subtle and progressive features of early and terminal EVD that was not feasible with routine approaches. Sinusoidal macrophages were the earliest cells to respond to infection, expressing proinflammatory cytokine interleukin 6 (IL6) mRNA, which was subsequently also observed in fibrovascular compartments. The mRNA of interferon-stimulated gene-15 (ISG-15), also known as ISG15 ubiquitin like modifier (ISG15), was observed early, with a progressive and ubiquitous hybridization signature involving mesenchymal and epithelial compartments. ISG-15 mRNA was prominent near infected cells, but not in infected cells, supporting the hypothesis that bystander cells produce a robust interferon gene response. This study contributes to our current understanding of early EVD progression and illustrates the value that digital pathology and quantitative IA serve in infectious disease research.
Zaire ebolavirus (EBOV) is a highly infectious and often lethal filovirus that causes hemorrhagic fever with a fatality rate of 25% to 90% in humans.1, 2, 3, 4 Currently, there are no therapeutics approved by the U.S. Food and Drug Administration for EBOV treatment, though several are being actively evaluated.5, 6, 7 Recently, in a landmark achievement, the European Commission (November, 2019) and Food and Drug Administration (December, 2019) granted marketing authorization to Merck's vaccine known as Ervebo (Merck, Kenilworth, NJ), which is a vesicular stomatitis virus vector vaccine system that expresses EBOV glycoprotein (GP).
Establishing efficacy of EBOV medical countermeasures in humans is inherently challenging given the unpredictable and rare nature of outbreaks, the diversity and potential for comorbidities in patient populations, as well as unknown inoculation dosage, route, and time of exposure. This emphasizes the importance of utilizing and characterizing highly controlled animal models that best recapitulate our understanding of human disease to allow us to better define the pathobiological signatures of EVD progression. Nonhuman primates (NHPs) represent the gold standard for EBOV animal model research, given their high fidelity to human disease pathophysiology.8 As in humans, elevated levels of circulating hepatic transaminases and decreased albumin in EBOV-infected NHPs suggest a combination of hepatocyte cytolysis and dysfunction.8,9 Evidence of hepatic involvement and injury as suggested by clinical chemistry values is corroborated histopathologically by hepatocyte necrosis and eosinophilic intracytoplasmic hepatocyte inclusions, which are readily observed in EBOV-infected NHPs by 5 days post-infection (DPI) as well as the rare published human autopsy reports3,4 EBOV infection also leads to activation of coagulation followed by a consumptive coagulopathy.8, 9, 10 This is reflected by increased tissue factor (TF) mRNA expression in peripheral blood mononuclear cells (PBMCs) in vitro and in EBOV-infected monkeys, in addition to thrombocytopenia and petechial hemorrhages in NHPs.10
Given the integral role the liver plays in EVD, characterizing early events of EBOV replication and dissemination within distinct anatomic compartments/cells and associated host response will serve to enhance our understanding of EVD disease progression and provide benchmarks for evaluating efficacy of medical countermeasures. Promising experimental vaccines and post-exposure medical countermeasures developed using preclinical NHP models have displayed sufficient efficacy to justify advancement into human clinical trials in the 2013 to 2016 Western Africa and ongoing EBOV outbreak in the Democratic Republic of Congo.6 The aim of this research was to evaluate the capacity for advanced tissue staining platforms combined with whole-slide imaging and quantitative image analysis (IA) to provide precise morphomolecular benchmarks of early EVD serving to shed light on the pathogenesis of this emerging infectious disease.
Materials and Methods
Virus, Animals, and Method of Infection
EBOV strain Kikwit [full designation: Zaire Ebolavirus, H.sapiens-tc/COD/1995/Kikwit-9510621, abbreviated name: EBOV/Kik (https://www.ncbi.nlm.nih.gov/nuccore, accession number MG572235.1)] was obtained from BEI Resources, NIAID, NIH (catalog number NR-50306). A total of 21, 6– to 7–year–old, Chinese origin (4.72 to 10.98 kg) rhesus macaques (8 males and 13 females) were enrolled in this study. Eighteen (n = 18) were inoculated with 1000 plaque-forming units EBOV/Kik in 1 mL of medium/animal via intramuscular injection in the left triceps muscle. Among them, three animals at each time point were humanely euthanized and necropsied at 3 to 6 DPI. Six additional animals were set as terminal disease controls and euthanized upon meeting predetermined clinical endpoint criteria. Three animals served as uninfected controls and were euthanized at 0 DPI. Additional study details not directly pertinent to the results of this paper are being compiled for publication entitled Natural Disease Progression of the Kikwit Ebola Virus Strain in the Rhesus Monkey Animal Model (D.X.L., R.S.B., J.P.L., Donna L. Perry, Timothy K. Cooper, A.M.W.H., John G. Bernbaum, R.J.H., M.A., N.I., William Dowling, Jing Qin, Ian Crozier, Peter B. Jahlring, L.E.H., unpublished data).
Tissue Inactivation, Processing, and Histopathologic Interpretation
Tissue samples were fixed for 72 hours in 10% neutral buffered formalin, after which a buffer exchanged was performed. Tissues were removed from the BSL-4 laboratory, processed in a Tissue-Tek VIP-6 automated vacuum infiltration processor (Sakura Finetek USA, Torrance, CA), followed by paraffin embedding with a Tissue-Tek model TEC unit (Sakura Finetek) resulting in formalin-fixed, paraffin-embedded blocks. Formalin-fixed, paraffin-embedded blocks were sectioned to 5 μm, transferred to positively charged slides, deparaffinized in xylene, and dehydrated in graded ethanol. Tissues sections were stained with hematoxylin and eosin for histopathology and phosphotungstic acid-hematoxylin stain for the confirmation of fibrin deposition. Qualitative and semiordinal bright-field analysis was performed blindly by a single American College of Veterinary Pathologists board-certified veterinary pathologist (N.A.C.). Individual ordinal scores for inflammation, necrosis, and fibrin, as well as a composite score, were assigned for each animal. Microscopic criteria used to assign ordinal scores are outlined in Table 1.
Table 1.
Semiquantitative Ordinal Histopathology Grading Criteria
| Hepatocellular necrosis | |
| 0, absent | No necrosis |
| 1, minimal | Rare multifocal single-cell necrosis adjacent to sinusoidal histiocyte aggregation |
| 2, mild | Multifocal necrosis of neighboring hepatocytes; more widespread |
| 3, moderate-to-marked | Multifocal coalescing areas of hepatocellular loss (absence of hepatocytes in large areas replaced by fibrin, edema, hemorrhage, and inflammatory cells composed of neutrophils and histiocytes) |
| Inflammation | |
| 0, absent | No inflammation |
| 1, mild | Rare, multifocal, localized inflammation within sinusoids consisting of aggregated histiocytes and lesser neutrophils |
| 2, moderate | Sinusoidal expansion by >5 neighboring histiocytes admixed with variable numbers of neutrophils |
| 3, severe | Inflammatory foci consisting of neutrophils admixed with histiocytes and fibrin in areas of hepatocellular loss (necrosis) |
| Fibrin deposition | |
| 0, absent | No fibrin deposition evident |
| 1, mild | Rare, multifocal occlusion of sinusoids by fibrin |
| 2, moderate | Routine, multifocal occlusion of sinusoids and veins by fibrin thrombi |
| Inclusions | |
| 0, absent | No viral inclusions observed |
| 1, present | Eosinophilic cytoplasmic viral inclusions observed |
Fluorescent mIHC
Fluorescent multiplex immunohistochemistry (mIHC) was performed using the Opal 4-color tyramide signaling amplification kit (Akoya Biosciences, Marlborough, MA). Two independent 3-plex mIHC panels were developed, optimized, and applied on liver sections from the 21 animals. Target antigens and optimization details are outlined in Table 2. Formalin-fixed, paraffin-embedded liver sections were deparaffinized and soaked in 10% formalin for 20 minutes before undergoing heat-induced antigen retrieval using AR6 buffer and a steamer set to 100°C to ensure uniform heating of liver sections. Slides were incubated in antibody diluent/blocking agent for 10 minutes to block endogenous peroxidase activity and nonspecific binding. Next, primary antibodies were applied at the concentration and in the order determined through optimization, followed by application of secondary horseradish peroxidase conjugated antibodies and tyramide signal amplification conjugated Opal fluorescent dyes. After development of each fluorochrome, heat-induced antigen retrieval was reapplied to strip the primary and secondary antibody complexes, and the process restarted in sequential fashion. Following the final round of heat-induced antigen retrieval, slides were counterstained with DAPI and mounted with ProLong Gold Antifade Mountant (Thermo Fischer Scientific). Uninfected controls served as negative controls, whereas EBOV terminal animals served as positive controls. Isotype controls were also evaluated to confirm absence of nonspecific binding of secondary antibodies.
Table 2.
Optimized Conditions for mIHC
| Panel | Primary antibody | Animal and isotype | Tissue marker | Manufacturer, catalog number, and lot number | Concentration | TSA-conjugated fluorochrome | Manufacturer, catalog number, and lot number | Concentration | AR |
|---|---|---|---|---|---|---|---|---|---|
| 1 | EBOV VP35 | Ms IgG2b | EBOV viral particles | Kerafast (Boston, MA) EMS703 090315 |
1:300 | Green (Opal 520) | PerkinElmer (Waltham, MA) FP1487A 2380385 |
1:600 | 1 |
| HepPar1 | Ms IgG1 | Hepatocytes | Thermo Fisher Scientific (Waltham, MA) MA5-12417 TD2559592A |
1:50 | Far red (Opal 690) | PerkinElmer FP1497A 2491340 |
1:50 | 2 | |
| MPO | Rb IgG | Neutrophils | Thermo Fisher Scientific PA5-16672 TE2572301G |
1:200 | Red (Opal 570) | PerkinElmer FP1488A 2496488 |
1:200 | 3 | |
| 2 | EBOV VP35 | Ms IgG2b | EBOV viral particles | Kerafast EMS703 090315 |
1:300 | Green | PerkinElmer FP1487A 2380385 |
1:600 | 1 |
| CD68 | Ms IgG1 | Monocytes/macrophages | Dako Cytomation (Glostrup, Denmark) M081401-2 20047711 |
1:50 | Far red | PerkinElmer FP1497A 2491340 |
1:50 | 2 | |
| Tissue factor (CD142) | Ms IgG1 | Extrinsic coagulation pathway | Thermo Fisher Scientific MA1-83495 TI2638538 |
1:50 | Red | PerkinElmer FP1488A 2496488 |
1:200 | 3 |
AR, antigen retrieval; mIHC, multiplexed fluorescent immunohistochemistry; MPO, myeloperoxidase; Ms, mouse; Rb, rabbit.
Fluorescent mISH
Fluorescent multiplex in situ hybridization (mISH) was conducted following the RNAscope Multiplex FL v2 User Manual (Advanced Cell Diagnostics, Newark, CA). Formalin-fixed, paraffin-embedded liver sections were deparaffinized and pretreated with hydrogen peroxide for 10 minutes to block endogenous peroxidase activity. The tissue sections underwent heat-induced antigen retrieval at 100°C for 15 minutes followed by RNAscope Protease Plus incubation in the ACD Hybez II Oven for 30 minutes at 40°C. Experimental details, including target probes and fluorochromes, are outlined in Table 3. Probes were incubated for 2 hours in the oven at 40°C followed by signal amplification and probe visualization. The rhesus macaque–specific housekeeping gene Vegfa (catalog number 487211, lot number 18143A; Advanced Cell Diagnostics) was employed as an RNA integrity–positive control, and the bacterial gene DapB of Bacilus subtilis (catalog number 310043, lot number 17264A, Advanced Cell Diagnostics) was used as a negative control probe.
Table 3.
Optimized Conditions for mISH
| Channel | Probe | mRNA marker | Manufacturer, catalog number, and lot number | HRP-conjugated fluorochrome | Manufacturer, catalog number, and lot number | Concentration |
|---|---|---|---|---|---|---|
| C1 | Mmu-ISG1501 | Interferon-stimulated gene 15 | Advanced Cell Diagnostics 540521 18136A |
Far red (Opal 690) | PerkinElmer FP1497A 2491340 |
1:350 |
| C2 | Hs-IL6-C2 | Interleukin 6 | Advanced Cell Diagnostics 310371-C2 18270A |
Red (Opal 570) | PerkinElmer FP1488A 2496488 |
1:750 |
| C3 | V-Ebola-VP35-C3 | EBOV viral protein VP35 | Advanced Cell Diagnostics 451691-C3 18116B |
Green (Opal 520) | PerkinElmer FP1487A 2380385 |
1:1500 |
HRP, horseradish peroxidase; mISH, multiplex fluorescent in situ hybridization.
Slide Scanning and Image Analysis
Fluorescently labeled slides were digitized at 200× (mIHC) or 400× (mISH) with the Vectra Polaris Automated Quantitative Pathology Imaging System (Akoya Biosciences), equipped with an LED light source and monochrome camera (ORCA-Flash 4.0; Hamamatsu Photonics, Hamamatsu, Japan). Terminal animal tissue samples, which contained abundant target protein and nucleic acid biomarkers, were used to establish exposure times for each assay via autoexposure on a region of interest (ROI). To maximize signal-to-noise ratios, 400× ISH slides were computationally spectrally unmixed using a synthetic library specific to the Opal fluorochromes used. Furthermore, an unstained liver section was used to create an autofluorescence signature that was subsequently removed from whole-slide images using InForm software version 2.4.8 (Akoya Biosciences). Whole-slide images were analyzed using IA software (HALO version 2.3; Indica Labs, Corrales, NM). Slides were manually annotated to remove tissue artifacts (ie, folds, air bubbles, fluorescent precipitate, etc.) and to select ROI for IA when applicable. Threshold values were modified in the real-time tuning window to minimize background signal and maximize specificity of target proteins and nucleic acids. For mIHC, IA was performed using the Area Quantification FL (AQ) and High-Plex FL (HP) cell phenotyping modules, whereas the AQ module was used for mISH. AQ outputs included total percentage of tissue with immunoreactivity or probe hybridization based on the total tissue area examined (μm2), as well as total percent positivity for 2 or more dyes (merged or overlapping dyes). HP outputs are based on nuclear segmentation and user defined phenotyping criteria with the output being cellular density (cells per μm2). For mIHC, AQ was applied across the entire whole-slide images, whereas for HP analysis, three 100× ROIs were selected and analyzed in HALO version 2.3 software from each animal to determine cells density per μm2. For mISH, 5 fields from each animal (1.46 × 106 μm2/per field) were stamped for batch analysis using Phenochart software version 1.0.11 (Akoya Biosciences) with subsequent multispectral unmixing via InForm software version 2.4.8 (Akoya Biosciences) with analysis conducted using the AQ module in HALO software version 2.3.
Quantitative RT-PCR of Liver Homogenates
Total RNA was isolated as described previously.11 Briefly, 70 μL of homogenized liver sample inactivated by Trizol LS was added to 280 μL of Buffer AVL (Qiagen, Germantown, MD) with carrier RNA. Samples were then extracted using the QIAamp Viral RNA Mini Kit (Qiagen) in accordance with the manufacturer's instructions, eluted in 70 μL of Buffer AVE (Qiagen), aliquoted, and then frozen. Viral RNA titer was determined using an experimental BEI Resources Critical Reagents Program EZ1 RT-qPCR kit assay in accordance with the manufacturer's instructions. Sample titers were reported as RNA genome equivalents per milliliter of sample. Temporal viral loads from homogenized liver samples are summarized in Supplemental Table S1.
Measurement of Extracellular Vesicle TF Activity
Extracellular vesicle TF activity was measured as previously described.12 Briefly, extracellular vesicles were isolated from platelet-poor plasma by centrifugation and washed to remove plasma. Extracellular vesicles were incubated with factor VIIa (2.4 nmol/l) and factor X (73.2 nmol/l) in the presence and absence of an inhibitory anti-TF antibody (HTF-1; BD Biosciences, Franklin Lakes, NJ) and 5 mmol/l calcium chloride for 2 hours at 37°C, and then the amount of factor Xa was determined using a chromogenic substrate (Pefachrome FXa 8598, 0.67 mmol/l; Enzyme Research Laboratories, South Bend, IN).
Statistical Analysis
For ordinal scoring, the nonparametric Kruskal-Wallis test and Benjamini, Krieger, and Yekutieli correction for multiple comparisons were applied given the nonparametric, noncontinuous nature of the data. For mIHC and mISH, given the quantitative and parametric nature of the data, one-way analysis of variance and post hoc Tukey correction for multiple comparisons were performed. All statistical tests were performed using GraphPad Prism version 8.2 software (GraphPad Software, La Jolla, CA) and for all tests, P < 0.05 was considered statistically significant. To investigate relationships amongst this multimodal approach, the authors utilized Pearson's correlation coefficient. Correlations (R) among the various microscopic and molecular modalities are summarized in the form of a heat map in Supplemental Figure S1, with specific P values summarized in Supplemental Table S2.
Results
Qualitative and Semiquantitative Characterization of Histomorphologic Signatures in the Liver of EBOV/Kik-Infected Rhesus Macaques
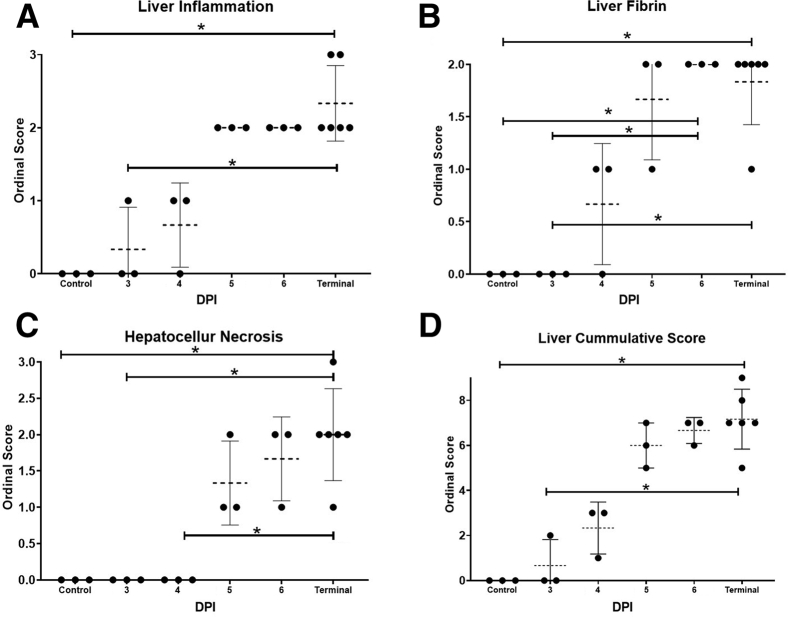
The earliest histomorphologic feature attributed to EVD was observed at 3 DPI represented by increased numbers of sinusoidal histiocytes that rarely contained eosinophilic cytoplasmic inclusions (Supplemental Figure S2). Beginning at 4 DPI, rare-to-moderate scattered intravascular fibrin thrombi (Supplemental Figure S2) were observed and persisted through terminal time points. By 5 DPI, animals began to exhibit minimal-to-moderate multifocal hepatocellular necrosis, which coincided with eosinophilic cytoplasmic inclusions involving both hepatocytes and histiocytes (Supplemental Figure S2). In the most severe instances of hepatocellular necrosis, aggregates of histiocytes were admixed with neutrophils and fibrin. In terminal control animals, inflammation (Figure 1), hepatocellular necrosis (Figure 1), and liver cumulative score (Figure 1) were statistically increased. Fibrin scores were statistically elevated in animals ≥5 DPI (Figure 1), indicating that this feature serves as a more sensitive microscopic feature in acute EVD compared with other described criteria. Cumulative scores ≤3 were only observed in animals ≤4 DPI, whereas scores ≥5 were only observed in animals ≥5 DPI. A summary of ordinal scores are summarized in Supplemental Table S3.
Figure 1.
Liver histopathologic ordinal scores increase temporally during acute infection of rhesus macaques with Zaire ebolavirus strain Kikwit (EBOV/Kik) as early as 3 days post-infection (DPI). Liver inflammation (A), fibrin (B), hepatocellular necrosis (C), and liver cumulative scores (D). For each time point, the dashed line represents the mean. Data are expressed as means ± SD. n = 21 animals (A–D). ∗P < 0.05.
Temporospatial Characterization of EBOV and Host Proteins Markers in the Liver of EBOV/Kik-Infected Rhesus Macaques
mIHC
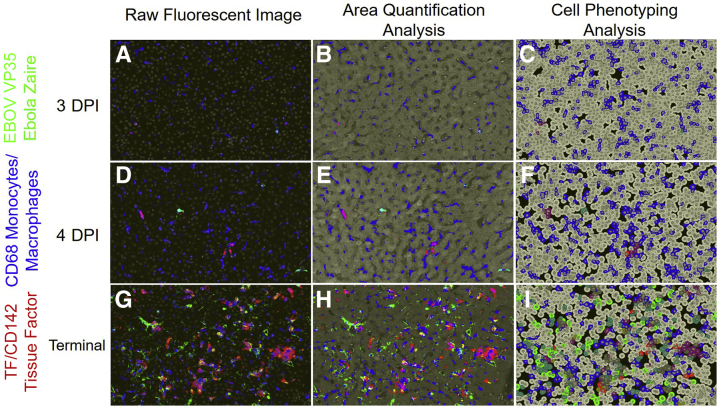
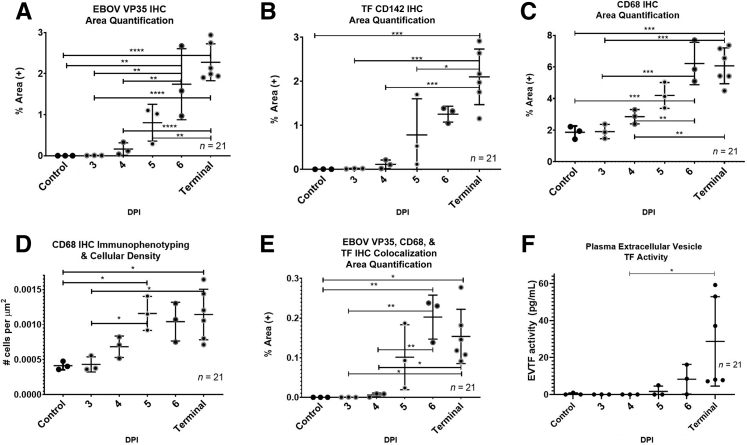
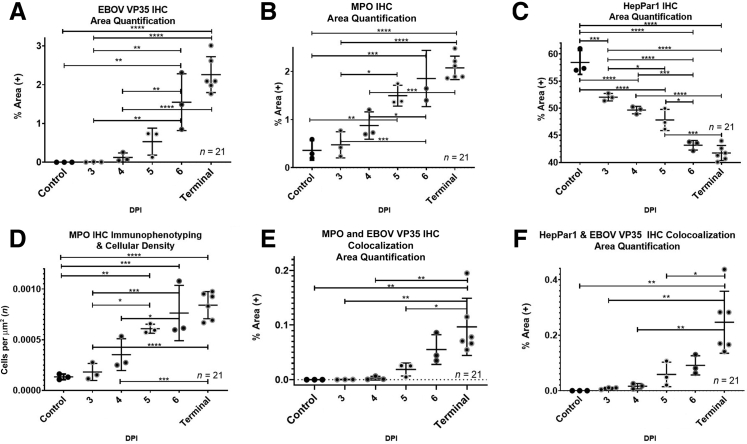
To examine the appearance and spread of EBOV alongside immune cell infiltration in response to EBOV infection, a 3-plex fluorescent immunohistochemistry cocktail was used. The authors analyzed EBOV VP35, tissue factor (TF) (also known as CD142), and CD68 expression. EBOV VP35 was selected due to its high expression during EBOV infection and involvement in repression of the host interferon responses.13 TF was selected due to its role in coagulation and up-regulation in EBOV infection.9,10 CD68 is a transmembrane GP highly expressed by monocytes and macrophages, both of which are well established as permissive cells for EBOV replication and early dissemination.9 Figure 2 shows representative mIHC images in raw area quantification (AQ) and cell phenotyping (HP) modes from three 4-DPI and terminal control animals. Quantitative IA results along with plasma extracellular vesicle TF activity are represented in Figure 3. EBOV VP35 was observed in sinusoidal histiocytes in all animals beginning at 3 DPI identifying early viral dissemination sites not observed ubiquitously with histopathology (Figure 1). CD68 immunolabeling and CD68+ cellular density (number of cells per μm2) increased starting at 4 DPI (Figure 3). For TF, the percentage of total area immunoreactivity was only statistically increased in terminal control animals (Figure 3). The temporal rise in percentage of total area immunoreactivity for TF mirrored rising TF activity in circulating extracellular vesicles (Figure 3), with a moderately positive correlation (r = 0.63, P = 3.3 × 10−3). Colocalization of EBOV VP35, CD68, and TF, were statistically increased in animals ≥6 DPI (Figure 3). Temporal increases in percentage of total area immunoreactivity for CD68 and TF were strongly positively correlated with EBOV-VP35 (r = 0.90, P = 2.7 × 10−8; r = 0.87, P = 3.6 × 10−7, respectively). Together, these results suggest significant replication of EBOV in CD68+, TF+ macrophages in the livers of infected animals.
Figure 2.
Representative images of raw fluorescent, area quantification, and cell phenotyping analysis of Zaire ebolavirus (EBOV) and host biomarkers in the liver of an experimental rhesus macaque model of EBOV strain Kikwit (EBOV/Kik). EBOV VP35 (green), CD68 monocytes/macrophages (blue), and tissue factor (CD142) (red). Three-days post-infection (DPI) raw fluorescent image (A), Area Quantification analysis (AQ) (B), and High-Plex FL (HP) cell phenotyping analysis (C). Four-DPI raw fluorescent image (D), AQ (E), and HP (F), respectively. Terminal control raw fluorescent image (G), AQ (H), and HP (I). AQ outputs include total percentage of tissue that stained positive based on the total tissue area examined, as well as total percent positivity for two or more dyes (merged or overlapping dyes). HP outputs are based on cellular segmentation using DAPI and CD68 immunoreactivity and are reported as number of CD68+ cells/μm2. Original magnification, ×100 (A–I).
Figure 3.
Zaire ebolavirus strain Kikwit (EBOV/Kik) infection in rhesus macaques results in a quantifiable temporal increase in liver monocytes/macrophages populations with concurrent increases in hepatic tissue factor (TF) and plasma extracellular TF activity. Area Quantification analysis (AQ) EBOV VP35 (A), AQ TF (CD142) (B), AQ CD68 (C), and High-Plex FL (HP) cell phenotyping analysis CD68 (D). AQ EBOV VP35, CD68, and TF colocalization (E) and plasma extracellular vesicle TF (EVTF) activity (F). Data are expressed as means ± SEM. n = 21 animals (A–C, F); n = 3 100× regions of interest per animal (D). ∗P < 0.05, ∗∗P < 0.005, ∗∗∗P < 0.0005, and ∗∗∗∗P < 0.00005. DPI, days post-infection; IHC, immunohistochemistry.
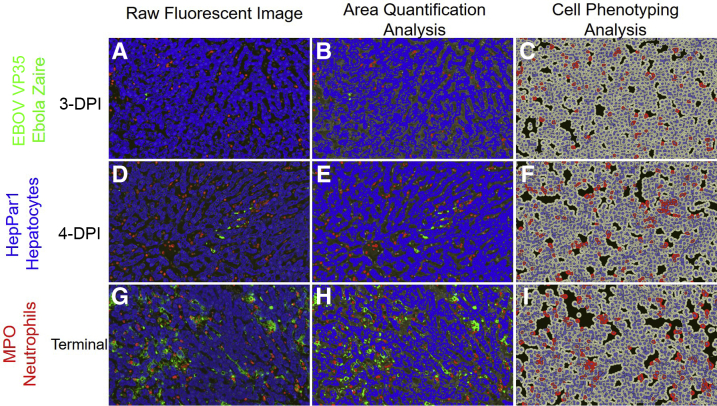
To explore the impact of EBOV infection on neutrophil infiltration and liver damage, a second mIHC cocktail was used that targeted EBOV VP35, a neutrophil marker myeloperoxidase (MPO), and a hepatocyte marker HepPar1, which served as an indirect indicator of hepatocellular dysfunction and necrosis. Representative images from three 4-DPI and terminal control animals including raw, AQ, and HP outputs are found in Figure 4, with quantitative IA results summarized in Figure 5. EBOV-VP35 percent area immunoreactivity (Figure 5) was analogous to condition 1 (mIHC results represented in Figures 2 and 3) as expected. The percentage of total area immunoreactivity for MPO (Figure 5) and neutrophil cell density (Figure 5) statistically increased in animals ≥5 DPI and was strongly positively correlated (r = 0.98, P = 1.6 × 10−15). HepPar1 percent total immunoreactivity decreased in a statistically significant and linear manner across all time points (Figure 5). The decrease in HepPar1 percentage of total area immunoreactivity was strongly negatively correlated with EBOV VP35 and MPO (r = −0.81, P = 7.8 × 10−6; r = −0.89; P = 8.8 × 10−8, respectively).
Figure 4.
Representative images of raw fluorescent, area quantification, and cell phenotyping analysis of Zaire ebolavirus (EBOV) and host biomarkers in the liver of an experimental rhesus macaque model of EBOV strain Kikwit (EBOV/Kik) infection. EBOV VP35 (green), HepPar1-hepatocytes (blue), and myeloperoxidase-neutrophils (red). Three-days post-infection (DPI) raw fluorescent image (A), Area Quantification analysis (AQ) (B), and High-Plex (HP) cell phenotyping analysis (C). Four-DPI raw fluorescent image (D), AQ (E), and HP (F). Terminal raw fluorescent image (G), AQ (H), and HP (I). AQ outputs include total percentage of tissue that stained positive based on the total tissue area examined (μm2), as well as total percent positivity for two or more dyes (merged or overlapping dyes). HP outputs are based on cellular segmentation using DAPI and myeloperoxidase (MPO) immunoreactivity and are reported as number of MPO-positive cells/μm2. Original magnification, ×100 (A–I).
Figure 5.
Zaire ebolavirus strain Kikwit (EBOV/Kik) infection in rhesus macaques results in a quantifiable temporal increase in liver neutrophils and decreased carbamoyl phosphate synthetase 1 (CPS1), a mitochondrial enzyme involved in the production of urea. Area Quantification analysis (AQ) EBOV VP35 (A), AQ myeloperoxidase neutrophils (MPO) (B), AQ HepPar1 hepatocytes (C), and High-Plex FL (HP) cell phenotyping analysis MPO (D). AQ MPO and EBOV VP35 colocalization (E) and AQ HepPar1 and EBOV VP35 colocalization (F). Data are expressed as means ± SEM. n = 21 (A–C, E, and F, animals); n = 3 (D, 100× regions of interest per animal). ∗P < 0.05, ∗∗P < 0.005, ∗∗∗P < 0.0005, and ∗∗∗∗P < 0.00005. DPI, days post-infection.
mISH
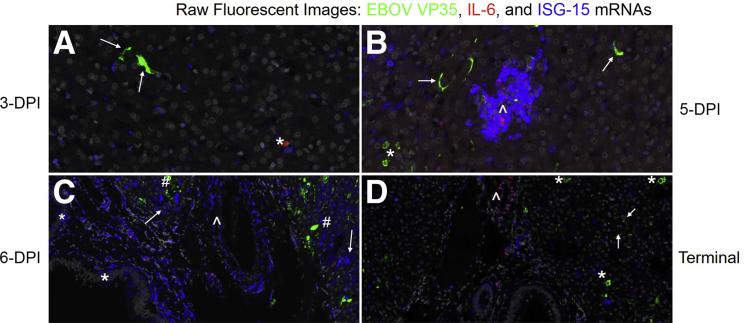
To complement studies on cellular infiltration to the liver during EBOV infection, innate immune responses were also examined by tracking the appearance of EBOV VP35, interferon-stimulated gene 15 (ISG15), and proinflammatory cytokine interleukin 6 (IL6) mRNAs. Qualitative data and representative images of fluorescent mISH are summarized in Supplemental Table S4 and Figure 6, respectively, with quantitative IA results summarized in Figure 7. EBOV VP35 mRNA (+sense) displayed hybridization patterns of either diffuse cytoplasmic and/or discrete cytoplasmic foci, with the latter interpreted to most likely indicate the presence of viral replication centers that represent multiprotein/RNA structures.14 EBOV VP35 mRNA was initially restricted to sinus histiocytes, with subsequent involvement of hepatocytes at 4 DPI, and ultimately PBMCs, fibrovascular stroma, and endothelium by 5 DPI. By 6 DPI, EBOV VP35 mRNA persisted within the cells/anatomic compartments listed above but became more prominent in the fibrovascular stroma compartment when compared with earlier time points. EBOV VP35 mRNA expression was statistically increased in animals ≥5 DPI when compared with other time points (Figure 7). Viral loads, as determined by quantitative RT-PCR on homogenized liver (Figure 7) and percent total area of EBOV VP35 mRNA hybridization, had the strongest correlation value (r = 0.80, P = 1.7 × 10−5) when comparing experimental methods employed in this study.
Figure 6.
Temporospatial characterization of Zaire ebolavirus (EBOV) and host proinflammatory and interferon-stimulating gene mRNAs in the liver of an experimental rhesus macaque model of EBOV strain Kikwit (EBOV/Kik) infection. EBOV VP35 +sense mRNA (green), interleukin 6 (IL6) mRNA (red), and interferon-stimulated gene 15 (ISG-15) mRNA, also known as ISG15 ubiquitin like modifier (ISG15) mRNA (blue). A: Three-days post-infection (DPI) rare EBOV VP35 (arrows) and IL6 mRNA hybridization (asterisk) of sinus histiocytes with mild multifocal ISG15 mRNA hybridization of neighboring sinus histiocytes and hepatocytes. B: Five-DPI multifocal EBOV VP35 mRNA hybridization of sinus histiocytes (arrow) and hepatocytes (asterisk). Regional focal aggregate of ISG15 mRNA hybridization of sinus histiocytes and hepatocytes (arrowhead), with moderate intralesional IL6 and scant EBOV VP35 mRNA hybridization. C: Six-DPI abundant ISG15 mRNA hybridization of biliary (asterisk) and hepatocyte (arrows) epithelium, blood vessels including the fibrovascular stroma and endothelium (arrowhead), and sinus histiocytes. Moderate multifocal EBOV VP35 hybridization of hepatocytes (hash mark), and sinus histiocytes. D: Terminal control, generalized decline in ISG15 mRNA hybridization. Moderate multifocal EBOV VP35 hybridization of sinus histiocytes (arrows) and hepatocytes (asterisk). Moderate perivascular fibrovascular IL6 mRNA hybridization (arrowhead). Original magnification, ×1200 (A–D).
Figure 7.
Zaire ebolavirus strain Kikwit (EBOV/Kik) infection in rhesus macaques causes a temporal increase in the mRNAs of proinflammatory cytokine interleukin 6 (IL6) and interferon-stimulated gene, also known as ISG15 ubiquitin like modifier (ISG15), within distinct anatomic compartments. A strong positive correlation of EBOV VP35 mRNA hybridization and EBOV liver viral loads is observed. Area quantification analysis (AQ) EBOV VP35 mRNA (A), Liver viral loads as determined by RT-qPCR targeting the EZ1 gene (B), AQ ISG15 mRNA (C), and AQ IL6 mRNA (D). AQ outputs include total percentage of tissue that stained positive based on the total tissue area examined (μm2) with viral loads reported as EBOV RNA copies per milligram of tissue. Data are expressed as means ± SEM. n = 21 (B, animals); n = 5 (A, C, and D, 100× regions of interest per animal). ∗P < 0.05, ∗∗P < 0.005, and ∗∗∗∗P < 0.00005. DPI, days post-infection; ISH, in situ hybridization.
IL6 mRNA hybridization was initially scant and restricted to sporadic individual sinus histiocytes. These IL-6+ histiocytes showed no EBOV VP35 mRNA hybridization. There was rare involvement of portal vasculature and central veins in both negative control and 3-DPI animals. By 4 DPI, frequency of IL6 mRNA hybridization progressed within these compartments and was also sporadically observed within the hepatic capsule. By 5 DPI, IL6 mRNA hybridization progressed to moderate severity, maintaining prominence in sinusoidal macrophages and fibrovascular stroma, but also for the first-time involved PBMCs and rarely hepatocytes.
ISG15 mRNA hybridization had a robust and diverse profile represented by distinct cytoplasmic puncta or diffuse cytoplasmic labeling. It was first observed at 3 DPI, with mild hybridization of hepatocytes and sinus histiocytes, and sporadic dense geographical aggregates affecting neighboring sinus histiocytes and subjacent hepatocytes. By 4 DPI, ISG15 mRNA was expressed in many cell types, including hepatocytes, biliary epithelium, fibrovascular stroma, endothelium, sinus histiocytes, and PBMCs. Colocalization of ISG15 mRNA within EBOV VP35 mRNA positive cells was extremely rare; however, hybridization was frequently observed near infected cells. ISG15 mRNA expression was statistically increased in animals ≥5 DPI (Figure 7). Terminal control animals showed a slight decreasing trend in overall ISG15 mRNA expression.
Discussion
The authors detected significant differences at early time points of EVD using quantitative approaches that were not evident by means of qualitative or semiquantitative traditional histomorphologic approaches. For example, trends for ordinal criteria of liver inflammation, hepatocellular necrosis, and cumulative scores increased over time, but were not significantly different for animals between 3 and 6 DPI. Although this could be in part due to the low number of animals enrolled in this study, mIHC coupled with IA algorithms detected statistically significant differences across time points in the form of percent positive immunoreactivity for EBOV VP35, CD68, TF, MPO, and HepPar1, as well as cell density for monocytes/macrophages (CD68) and neutrophils (MPO). These results suggest that an mIHC approach allows more robust detection of disease progression signatures of EVD in a highly translational animal model of EVD.
Previous studies have reported that circulating neutrophils and increased tissue recruitment of neutrophils may be associated with fatal outcomes of EBOV in humans as well as in cynomolgus and rhesus macaques.9,15,16 Neutrophil marker genes have also been shown to be up-regulated in PBMCs derived from terminal NHPs, which likely represents immature circulating neutrophils released prematurely from the bone marrow.17 Statistically increased percent total immunoreactivity of MPO and neutrophil cell density in the liver were observed at ≥5 DPI. Given the strong correlation of MPO with HepPar1 (r = −0.89, P = 8.8 × 10−8), and CD68 (r = 0.95, P = 9.4 × 10−11), it is logical to suggest that the primary driver in the recruitment of neutrophils to the liver is a combination of increased numbers of activated sinusoidal histiocytes (many of which are infected with EBOV) and hepatocellular necrosis, with the latter likely serving as a source of danger-associated molecular patterns resulting in recruitment of neutrophils to the liver.18
Colocalization of MPO and EBOV V35 was statistically increased in terminal control animals. Although neutrophils have not been conclusively shown to support EBOV replication, one report has suggested they may be susceptible to infection and could potentially modulate the innate host immune response and play a role in tissue damage and modulation of the host response.19 It cannot be definitely determined whether increased colocalization represents viral antigen within intact neutrophils, either through viral entry or phagocytosis. Additionally, this observation could be reflective of viral antigen trapped within neutrophil extracellular traps, which consist of MPO and neutrophil elastase attached to an extruded chromatin scaffolding within hepatic sinusoids. Given neutrophil extracellular traps have been affiliated with endothelium damage and activation of platelets, this provides an additional rationale for further investigating the role neutrophils play in EVD.
Although the authors did observe a statistically significant and progressive loss of HepPar1 immunoreactivity across all time points, hepatocyte necrosis was moderate in the worst-case scenario and was not evident until 5 DPI. These findings suggest that in addition to an indirect surrogate of hepatocyte necrosis, decreased HepPar1 immunoreactivity may potentially represent a clinicopathologically relevant molecular marker associated with EVD pathogenesis. HepPar1 antibody targets carbamoyl phosphate synthetase 1 (CPS1), which is a mitochondrial enzyme involved in the urea cycle, and generally displays diffuse granular cytoplasmic labeling in healthy hepatocytes.20 CPS1 deficiency, a rare autosomal recessive disorder in humans, can lead to severe life-threatening hyperammonemia that has several clinical signs that mimic EVD in humans and NHPs. These include poorly regulated breathing rate or body temperature, vomiting, anorexia, seizures, coma, and intellectual disability in surviving patients.21 To our knowledge, hyperammonemia has not been characterized in EVD, but warrants further investigation given that pharmacologic approaches are readily available (supplementation with citrulline and/or arginine and ammonia scavengers) and may be viable options for achieving metabolic stability and enhancing patient outcomes in patients with EVD.
Geisbert et al10 described significant increases in TF mRNA and protein in the peripheral blood of EBOV/Kik-infected macaques. This study’s IA results extend these findings to the liver, where colocalization of CD68, EBOV VP35, and TF immunoreactivity were significantly increased in animals ≥6 DPI. The authors acknowledge the intrinsic resolving limit of standard microscopy (ie, 200 nm); however, increasing temporal percent area of colocalization of EBOV, CD68, and TF can be explained by acknowledging that TF is expressed on the membrane of activated CD68+ cells during EBOV infection. More specifically, EBOV GP interacts with TLR4 on the macrophage cell membrane during cell entry, leading to induction of a proinflammatory response that results in increased IL-6 and TF expression.10,22,23 The authors also observed a temporal increase in levels of plasma extracellular vesicle TF activity in EBOV-infected animals, which closely mirrored TF AQ results, supported by a moderate positive correlation (r = 0.63, P = 2.0 × 10−3). This suggests extracellular vesicle TF activity could represent a promising peripheral blood biomarker serving as a surrogate for EVD-induced liver pathology.
Transcriptomic analysis of peripheral blood in both humans and in NHPs infected with EBOV has highlighted the expression of interferon-stimulated genes (ISGs) and proinflammatory cytokines as EVD develops.24, 25, 26, 27 ISG15 mRNA, has been shown to accumulate in the blood as early as 4 days before onset of fever, whereas IL6 (a proinflammatory cytokine) mRNA typically accumulates at onset of fever.25 Increasing levels of IL-6 protein in peripheral blood is a hallmark of EVD and is closely linked with severe disease and fatal outcomes in both NHPs and humans.28,29 It remains largely unknown what cellular phenotypes and their spatial signatures within EBOV-infected tissues are driving EVD transcriptomic signatures. By using mISH, the authors were able to explore the expression of these host responses in the context of early liver disease. This study’s results suggest there is little IL6 expression by hepatocytes, with some expression from sinus histiocytes, perivascular stroma cells, and less commonly, hepatic capsular fibroblasts. These findings suggest that sinus histiocytes and fibrovascular stromal cells are key contributors to IL6 gene expression in the context of the liver and could represent a significant cell population driving peripheral IL-6 cytokinemia.
ISG15 mRNA displayed the largest percentage of total area of hybridization of the three mRNAs investigated and displayed the most heterogeneity regarding cell phenotypes involved. Although ISG15 was very prominent within and around areas of EBOV VP35 hybridization, colocalization was a rare event. This supports the hypothesis that EBOV infection of certain permissive cell phenotypes inhibits the interferon host cell immune response, whereas bystander cells are activated, ultimately eliciting a robust interferon (IFN) response that is detectable in peripheral blood.17,27 It remains unknown whether this robust IFN response of predominantly uninfected cells is protective or a detriment to the host. The authors acknowledge that biological relevance in the context of protein expression would need to be confirmed by additional techniques such as IHC and/or Western blot. Furthermore, investigation of other tissues known to support early EBOV replication such as the spleen and lymph nodes would be of additional benefit to confirm if this pattern is recapitulated in other organ systems or if they carry unique gene expression signatures.
EBOV VP35 mRNA hybridization mirrored EBOV VP35 spatial and temporal immunoreactivity, with virus initially restricted to sinus histiocytes and less commonly observed within hepatocytes and detectable in all animals at ≥3 DPI. Additional involvement of the endothelium and fibrovascular stroma were evident by ≥5 DPI. This is interesting given that a separate study of EBOV-infected macaques that employed EBOV VP40 and anti-EBOV GP IHC identified widespread viral antigen deposits in fibrous interstitium and endothelium of the reproductive tract of animals only at 6 to 9 DPI, with minimal associated immune response or histopathologic changes.30 Together, this study’s findings and past research indicate that even during acute infection, EBOV is already transitioning from primary sites of replication such as macrophages and hepatocytes to anatomic compartments, such as fibroblast stromal cells, that may support viral persistence. EBOV mRNA was not evident in PBMCs (interpreted to represent monocytes) until 5 DPI, This supports previous research that described how monocytes are initially refractory to EBOV GP-mediated entry, but ultimately become permissive under conditions that promote their differentiation and up-regulation of critical EBOV entry factors.31
Implications and Future Research
This study’s data provide clear evidence that IA has the potential to facilitate more rigorous and unbiased analysis for infectious disease pathogenesis research. Integration of IA with mIHC and mISH allowed us to reliably define temporospatial host response to EBOV infection in a highly relevant translational rhesus macaque model of EVD. These signatures will serve to inform future models of EVD and aid in the evaluation of efficacy of medical countermeasures. Furthermore, these results can serve as a reference for evaluating the development of novel in vitro platforms such as single-cell (primary human hepatocytes and iPSC-derived hepatocytes), multicellular (organoids), and synthetic organ platforms (ie, vascularized microliver), to evaluate these systems' ability to accurately recapitulate EVD endpoints in translational animal models.
Acknowledgment
We thank the National Institute of Allergy and Infectious Diseases Integrated Research Facility for performing the Biosafety Level (BSL) 4 work.
Footnotes
Supported in part by NIAID interagency agreement NOR 15 003-001-0000 (L.E.H.) and under Battelle Memorial Institute contract HHSN272200700016I (L.E.H.).
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2020.03.003.
Supplemental Data
Continuous data sets derived from quantitative image analysis enables multimodal correlation analysis using Pearson's correlation coefficient (r). AQ, Area Quantification analysis; HP, High-Plex FL analysis; MPO, myeloperoxidase; TF-Tissue Factor.
Temporal histomorphologic changes in the liver of Zaire ebolavirus strain Kikwit (EBOV/Kik)-infected rhesus macaques. A: Three days post-infection (DPI), rare eosinophilic cytoplasmic inclusions are rarely observed in sinus histiocytes (black arrow). B: Four DPI, rare sinusoidal fibrin thrombi are observed occluding hepatic sinusoids (black arrow). C: Five DPI, intracytoplasmic eosinophilic inclusions affecting both hepatocytes (asterisks) and sinus histiocytes (black arrow) are routinely observed.
References
- 1.Malvy D., McElroy A.K., de Clerck H., Gunther S., van Griensven J. Ebola virus disease. Lancet. 2019;393:936–948. doi: 10.1016/S0140-6736(18)33132-5. [DOI] [PubMed] [Google Scholar]
- 2.Baseler L., Chertow D.S., Johnson K.M., Feldmann H., Morens D.M. The pathogenesis of Ebola virus disease. Annu Rev Pathol. 2017;12:387–418. doi: 10.1146/annurev-pathol-052016-100506. [DOI] [PubMed] [Google Scholar]
- 3.Zaki S.R., Goldsmith C.S. Pathologic features of filovirus infections in humans. Curr Top Microbiol Immunol. 1999;235:97–116. doi: 10.1007/978-3-642-59949-1_7. [DOI] [PubMed] [Google Scholar]
- 4.Martines R.B., Ng D.L., Greer P.W., Rollin P.E., Zaki S.R. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol. 2015;235:153–174. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- 5.Marzi A., Mire C.E. Current Ebola virus vaccine progress. BioDrugs. 2019;33:9–14. doi: 10.1007/s40259-018-0329-7. [DOI] [PubMed] [Google Scholar]
- 6.Hoenen T., Groseth A., Feldmann H. Therapeutic strategies to target the Ebola virus life cycle. Nat Rev Microbiol. 2019;17:593–606. doi: 10.1038/s41579-019-0233-2. [DOI] [PubMed] [Google Scholar]
- 7.Hayden F.G., Friede M., Bausch D.G. Experimental therapies for Ebola virus disease: what have we learned? J Infect Dis. 2017;215:167–170. doi: 10.1093/infdis/jiw496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett R.S., Huzella L.M., Jahrling P.B., Bollinger L., Olinger G.G., Jr., Hensley L.E. Nonhuman primate models of Ebola virus disease. Curr Top Microbiol Immunol. 2017;411:171–193. doi: 10.1007/82_2017_20. [DOI] [PubMed] [Google Scholar]
- 9.Geisbert T.W., Hensley L.E., Larsen T., Young H.A., Reed D.S., Geisbert J.B., Scott D.P., Kagan E., Jahrling P.B., Davis K.J. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol. 2003;163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisbert T.W., Young H.A., Jahrling P.B., Davis K.J., Kagan E., Hensley L.E. Mechanisms underlying coagulation abnormalities in Ebola hemorrhagic fever: overexpression of tissue factor in primate monocytes/macrophages is a key event. J Infect Dis. 2003;188:1618–1629. doi: 10.1086/379724. [DOI] [PubMed] [Google Scholar]
- 11.Honko A.N., Johnson J.C., Marchand J.S., Huzella L., Adams R.D., Oberlander N., Torzewski L.M., Bennett R.S., Hensley L.E., Jahrling P.B., Olinger G.G. High dose sertraline monotherapy fails to protect rhesus macaques from lethal challenge with Ebola virus Makona. Sci Rep. 2017;7:5886. doi: 10.1038/s41598-017-06179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hisada Y., Mackman N. Measurement of tissue factor activity in extracellular vesicles from human plasma samples. Res Pract Thromb Haemost. 2019;3:44–48. doi: 10.1002/rth2.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basler C.F., Amarasinghe G.K. Evasion of interferon responses by Ebola and Marburg viruses. J Interferon Cytokine Res. 2009;29:511–520. doi: 10.1089/jir.2009.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groseth A., Charton J.E., Sauerborn M., Feldmann F., Jones S.M., Hoenen T., Feldmann H. The Ebola virus ribonucleoprotein complex: a novel VP30-L interaction identified. Virus Res. 2009;140:8–14. doi: 10.1016/j.virusres.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McElroy A.K., Shrivastava-Ranjan P., Harmon J.R., Martines R.B., Silva-Flannery L., Flietstra T.D., Kraft C.S., Mehta A.K., Lyon G.M., Varkey J.B., Ribner B.S., Nichol S.T., Zaki S.R., Spiropoulou C.F. Macrophage activation marker soluble CD163 associated with fatal and severe Ebola virus disease in humans. Emerg Infect Dis. 2019;25:290–298. doi: 10.3201/eid2502.181326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Ven A.J., Netea M.G., van der Meer J.W., de Mast Q. Ebola virus disease has features of hemophagocytic lymphohistiocytosis syndrome. Front Med (Lausanne) 2015;2:4. doi: 10.3389/fmed.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Speranza E., Connor J.H. Host transcriptional response to Ebola virus infection. Vaccines (Basel) 2017;5:30. doi: 10.3390/vaccines5030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kienle K., Lammermann T. Neutrophil swarming: an essential process of the neutrophil tissue response. Immunol Rev. 2016;273:76–93. doi: 10.1111/imr.12458. [DOI] [PubMed] [Google Scholar]
- 19.Mohamadzadeh M., Coberley S.S., Olinger G.G., Kalina W.V., Ruthel G., Fuller C.L., Swenson D.L., Pratt W.D., Kuhns D.B., Schmaljohn A.L. Activation of triggering receptor expressed on myeloid cells-1 on human neutrophils by Marburg and Ebola viruses. J Virol. 2006;80:7235–7244. doi: 10.1128/JVI.00543-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler S.L., Dong H., Cardona D., Jia M., Zheng R., Zhu H., Crawford J.M., Liu C. The antigen for Hep Par 1 antibody is the urea cycle enzyme carbamoyl phosphate synthetase 1. Lab Invest. 2008;88:78–88. doi: 10.1038/labinvest.3700699. [DOI] [PubMed] [Google Scholar]
- 21.Diez-Fernandez C., Haberle J. Targeting CPS1 in the treatment of carbamoyl phosphate synthetase 1 (CPS1) deficiency, a urea cycle disorder. Expert Opin Ther Targets. 2017;21:391–399. doi: 10.1080/14728222.2017.1294685. [DOI] [PubMed] [Google Scholar]
- 22.Lai C.Y., Strange D.P., Wong T.A.S., Lehrer A.T., Verma S. Ebola virus glycoprotein induces an innate immune response in vivo via TLR4. Front Microbiol. 2017;8:1571. doi: 10.3389/fmicb.2017.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okumura A., Pitha P.M., Yoshimura A., Harty R.N. Interaction between Ebola virus glycoprotein and host toll-like receptor 4 leads to induction of proinflammatory cytokines and SOCS1. J Virol. 2010;84:27–33. doi: 10.1128/JVI.01462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Speranza E., Bixler S.L., Altamura L.A., Arnold C.E., Pratt W.D., Taylor-Howell C., Burrows C., Aguilar W., Rossi F., Shamblin J.D., Wollen S.E., Zelko J.M., Minogue T., Nagle E., Palacios G., Goff A.J., Connor J.H. A conserved transcriptional response to intranasal Ebola virus exposure in nonhuman primates prior to onset of fever. Sci Transl Med. 2018;10:eaaq1016. doi: 10.1126/scitranslmed.aaq1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X., Speranza E., Munoz-Fontela C., Haldenby S., Rickett N.Y., Garcia-Dorival I., Fang Y., Hall Y., Zekeng E.G., Ludtke A., Xia D., Kerber R., Krumkamp R., Duraffour S., Sissoko D., Kenny J., Rockliffe N., Williamson E.D., Laws T.R., N'Faly M., Matthews D.A., Gunther S., Cossins A.R., Sprecher A., Connor J.H., Carroll M.W., Hiscox J.A. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol. 2017;18:4. doi: 10.1186/s13059-016-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speranza E., Altamura L.A., Kulcsar K., Bixler S.L., Rossi C.A., Schoepp R.J., Nagle E., Aguilar W., Douglas C.E., Delp K.L., Minogue T.D., Palacios G., Goff A.J., Connor J.H. Comparison of transcriptomic platforms for analysis of whole blood from Ebola-infected cynomolgus macaques. Sci Rep. 2017;7:14756. doi: 10.1038/s41598-017-15145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caballero I.S., Honko A.N., Gire S.K., Winnicki S.M., Mele M., Gerhardinger C., Lin A.E., Rinn J.L., Sabeti P.C., Hensley L.E., Connor J.H. In vivo Ebola virus infection leads to a strong innate response in circulating immune cells. BMC Genomics. 2016;17:707. doi: 10.1186/s12864-016-3060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bixler S.L., Goff A.J. The role of cytokines and chemokines in filovirus infection. Viruses. 2015;7:5489–5507. doi: 10.3390/v7102892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchinson K.L., Rollin P.E. Cytokine and chemokine expression in humans infected with Sudan Ebola virus. J Infect Dis. 2007;196 Suppl 2:S357–S363. doi: 10.1086/520611. [DOI] [PubMed] [Google Scholar]
- 30.Perry D.L., Huzella L.M., Bernbaum J.G., Holbrook M.R., Jahrling P.B., Hagen K.R., Schnell M.J., Johnson R.F. Ebola virus localization in the macaque reproductive tract during acute Ebola virus disease. Am J Pathol. 2018;188:550–558. doi: 10.1016/j.ajpath.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez O., Johnson J.C., Honko A., Yen B., Shabman R.S., Hensley L.E., Olinger G.G., Basler C.F. Ebola virus exploits a monocyte differentiation program to promote its entry. J Virol. 2013;87:3801–3814. doi: 10.1128/JVI.02695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Continuous data sets derived from quantitative image analysis enables multimodal correlation analysis using Pearson's correlation coefficient (r). AQ, Area Quantification analysis; HP, High-Plex FL analysis; MPO, myeloperoxidase; TF-Tissue Factor.
Temporal histomorphologic changes in the liver of Zaire ebolavirus strain Kikwit (EBOV/Kik)-infected rhesus macaques. A: Three days post-infection (DPI), rare eosinophilic cytoplasmic inclusions are rarely observed in sinus histiocytes (black arrow). B: Four DPI, rare sinusoidal fibrin thrombi are observed occluding hepatic sinusoids (black arrow). C: Five DPI, intracytoplasmic eosinophilic inclusions affecting both hepatocytes (asterisks) and sinus histiocytes (black arrow) are routinely observed.