Abstract
Somatic mutations in RAS and related pathway genes such as NF1 have been strongly implicated in the development of cancer while also being implicated in a diverse group of developmental disorders named the ‘RASopathies’, including neurofibromatosis type 1 (NF1), Noonan syndrome (NS), Noonan syndrome with multiple lentigines (NSML), Costello syndrome (CS), cardiofaciocutaneous syndrome (CFC), and capillary malformation–arteriovenous syndrome (CM-AVM). It remains unclear why (i) there is little overlap in mutational subtype between Ras-driven malignancies associated with sporadic disease and those associated with the RASopathy syndromes, and (ii) RASopathy-associated cancers are usually of different histological origin to those seen with sporadic mutations of the same genes. For instance, germline variants in KRAS and NRAS are rarely found at codons 12, 13 or 61, the most common sites for somatic mutations in sporadic cancers. An exception is CS, where germline variants in codons 12 and 13 of HRAS occur relatively frequently. Given recent renewed drug interest following early clinical success of RAS G12C and farnesyl transferase inhibitors, an improved understanding of this relationship could help guide targeted therapies for both sporadic and germline cancers associated with the Ras pathway.
Key words: Costello syndrome, neurofibromatosis type 1, Noonan syndrome, RAS, RASopathy
Highlights
-
•
RAS mutations have been associated with a range of malignancies and with developmental disorders called RASopathies.
-
•
Cancers associated with sporadic and germline RAS mutations are dissimilar in histological origins and mutational subtypes.
-
•
This disparity highlights our incomplete understanding of the relationship between the RAS–MAPK pathway and cancer.
-
•
Recent advances in targeting Ras G12C, HRAS and MEK have led to renewed interest in targeted therapies in this pathway.
Introduction
RAS proteins are a family of small GTPases, critically important for cellular signalling through their interaction with a multitude of pathways, of which the RAS–MAPK pathway is best known. The RAS superfamily includes HRAS, NRAS, KRAS(4a and 4b), as well as numerous close relatives including MRAS, RRAS and TC21.1
RAS exists in two states: the active guanosine triphosphate (GTP)-bound and the inactive guanosine diphosphate (GDP)-bound conformations. These allow RAS to act as a molecular switch, controlling downstream signalling pathways. Conversion between these two states is regulated by a family of large, diverse proteins that associate with other proteins and lipids to provide tight control of RAS function.1 The switch between inactive and active state is facilitated by guanine nucleotide exchange factors (GEFs), while the switch back from the active to inactive state is facilitated by GTPase-activating proteins (GAPs).2 The relative balance of GEF and GAP activity allows strict regulation of RAS activity. It is this equilibrium that is disturbed by oncogenic RAS mutations which cause an overall decrease in RAS-mediated GTP hydrolysis, trapping RAS in its active GTP-bound state and leading to a net increase in downstream signalling independent of upstream input.
RAS proteins have numerous downstream effectors, of which RAF kinases are a major player. The downstream MAPK pathway has been shown to be vital in a number of cellular processes, including proliferation, differentiation and growth.3,4 Mutations downstream in this pathway have also been associated with malignancy. An emerging and more detailed taxonomy of class I–III RAF and MEK mutations has recently been described, augmenting our understanding of these alterations as oncogenic ‘activators’, which are independent of upstream RAS signalling, or oncogenic ‘amplifiers’ which retain some dependence on upstream processes.5
Considering the vital importance of RAS in cellular function, it is unsurprising that disruption of this pathway has been associated with numerous diseases. Somatic RAS mutations in human cancer have long been recognised, with mutations seen in ∼16% of cancers.6 More recently, it has been found that germline mutations in genes encoding components of the RAS–MAPK pathway cause a group of developmental disorders, now termed the ‘RASopathies’.
To complement a number of reports that have recently highlighted exciting clinical progress with novel RAS pathway inhibitors in cancer,7, 8, 9, 10, 11 this review examines the parallels in pathological and genetic details of RASopathies and somatic RAS-driven cancers.
RASopathies
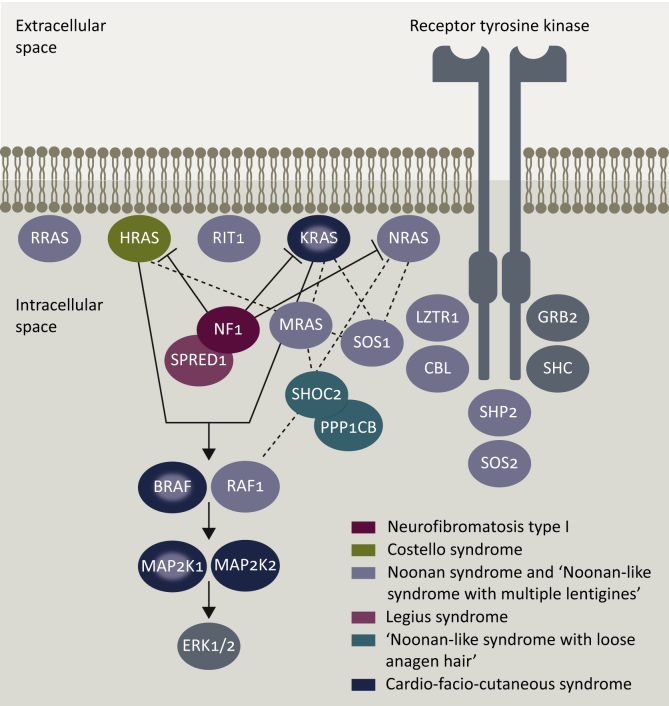
The RASopathies are a group of genetic syndromes including neurofibromatosis type 1 (NF1), Noonan syndrome (NS), Noonan syndrome with multiple lentigines (NSML), Costello syndrome (CS), Legius syndrome, cardiofaciocutaneous syndrome (CFC), capillary malformation–arteriovenous syndrome (CM–AVM) and autosomal dominant intellectual disability type 5 (Table 1). They are the product of disruption in the RAS–MAPK pathway, which can result from alteration of RAS proteins, GEFs, GAPs, scaffolding proteins, phosphatases, ubiquitin ligases and pathway inhibitors (Figure 1).12 Despite comprising a group of distinct disorders, they share several characteristics, including cardiac malformations, craniofacial dysmorphia, learning difficulties and an increased risk of cancer.13
Table 1.
Summary of genes associated with the RASopathy syndromes, including germline pathogenic RAS variants noted in these conditions
| Syndrome | Genes | Common mutations | Associated malignancies |
|---|---|---|---|
| Neurofibromatosis 1 | NF1: Ras GAP | Glioma Rhabdomyosarcoma Malignant peripheral nerve tumour JMML Breast cancer Gastrointestinal cancers |
|
| Noonan | PTPN11: Phosphatase | JMML | |
| SOS1: RasGEF | Neuroblastoma | ||
| RAF1: Kinase | Acute lymphocytic leukaemia | ||
| RIT1: GTPase | Glioma | ||
| KRAS: GTPase | V14I, Q22R, P34L, P34Q, I36M, T58I, D153V, V152G | Rhabdomyosarcoma | |
| CBL: Ubiquitin ligase | Acute myeloid leukaemia | ||
| SOS2: RasGEF | Testicular cancer | ||
| NRAS: GTPase | P34L, T50I, I24N, G60E | Non-Hodgkin lymphoma | |
| SHOC2: Scaffolding | Colon cancer | ||
| RRAS: GTPase | |||
| LZTR1: Adaptor | |||
| BRAF: Kinase | |||
| Noonan with multiple lentigines |
PTPN11: Phosphatase RAF1: Kinase BRAF: Kinase |
||
| Noonan-like syndrome with loose anagen hair | SHOC2: Scaffolding | ||
| Costello | HRAS: GTPase | G12S, G12A, G13C | Rhabdomyosarcoma, neuroblastoma, bladder cancer |
| Cardiofaciocutaneous | BRAF: Kinase | Similar to Noonan syndrome but overall risks unclear | |
| MAP2K1: Kinase | |||
| MAP2K2: Kinase | |||
| KRAS: GTPase | P34R, D153V, F156L | ||
| SHOC2: Scaffolding | |||
| Capillary malformation–arteriovenous malformation | RASA1: RasGAP | Unclear | |
| Legius | SPRED1: Negative regulator | No convincing malignancy risk |
Variant information from NSeuronet.103
JMML, juvenile myelomonocytic leukaemia.
Figure 1.
Germline disorders of the RAS–MAPK pathway.
Neurofibromatosis type 1
NF1 is an autosomal dominant disorder arising from pathogenic variants of the NF1 gene, which codes for the Ras GAP protein neurofibromin.14,15 It has a birth incidence of 1 in 2000–2700 and diagnostic prevalence of ∼1 per 4000 individuals worldwide, with around half of cases arising from de novo mutations.16, 17, 18, 19 Germline pathogenic variants in the NF1 gene are diverse: ∼2600 pathogenic variants have been reported, ranging from single-base-pair substitutions to large whole-gene deletions, most of which are predicted to result in an almost complete absence of the protein.20, 21, 22 The clinical diagnosis of NF1 is based on the presence of café-au-lait spots (hyperpigmented macules), inguinal/axillary freckling, iris hamartomas, optic gliomas, neurofibromas (peripheral nerve sheath tumours) and osseous lesions.23
Overall, the risk of cancer is ∼2.5 times higher than for the general population, primarily due to the increased risk of sarcoma and brain malignancies.24 Although most NF1 patients will have benign cutaneous and subcutaneous neurofibromas, ∼10%–15% will develop malignant peripheral nerve sheath tumours (MPNSTs).20,24 Central nervous system (CNS) tumours occur in ∼20% of NF1 patients, most of which are low-grade pilocytic astrocytomas affecting the optic nerve in childhood.25 Even discounting CNS tumours and sarcoma, NF1 patients seem to have an increased incidence of malignancy, largely due to an increased incidence of breast and gastrointestinal cancer.25 In particular, NF1 patients have an increased risk of gastrointestinal stromal tumours that are not susceptible to imatinib as they bypass PDGFRa/cKIT.26 Cancer is the most common cause of death in NF1 patients and contributes towards a 10–15-year reduced life expectancy.27,28 Even when excluding NF1-associated malignancies, neurofibromatosis patients with cancer seem to have significantly poorer 5-year survival than matched controls.25
Noonan syndrome
NS is an autosomal dominant disorder characterised by craniofacial dysmorphic features, congenital heart defects, growth impairment, bleeding disorders and neurocognitive delay.13 It affects ∼1/1000–2000 newborns, and unlike NF1 has been associated with pathogenic variants in several genes, including PTPN11, SOS1, RAF1, RIT1, KRAS, NRAS, CBL, LZTR1 and BRAF.29, 30, 31, 32, 33 There is phenotypic overlap between NF1 and NS, with NF1 patients with missense pathogenic variants in NF1 showing more distinct NS features including pulmonary stenosis.34
NS is associated with an increased risk of malignancy, with a cumulative incidence of ∼4% by age 20. The most common cancers in NS are CNS tumours, neuroblastoma, acute lymphoblastic leukaemia and rhabdomyosarcoma (RMS).35 Children with NS are also predisposed to a spectrum of myeloproliferative disorders, which may follow a benign course or an aggressive course similar to juvenile myelomonocytic leukaemia (JMML).36
The most commonly mutated gene found in NS is PTPN11, with pathogenic variants found in ∼50% cases.29 It codes for SHP2, a nonreceptor protein tyrosine phosphatase for which inhibitors are currently being tested in early phase clinical trials of cancers with somatic mutations affecting the RAS pathway.37 Mutations impair the protein's ability to switch from active to inactive conformation, causing increased RAS–MAPK signalling.
KRAS pathogenic variants rarely cause NS and CFC, and there is a clinical and mutational overlap between these conditions. Pathogenic variants have been identified at numerous locations, including within the phosphatase binding loop (codon 14), close to the switch 2 domain (codon 58), within the KRAS switch 1 domain (codons 34, 36) and in the α-5 helix of the 4B isoform (codons 152, 153 and 156).31,38, 39, 40 This is dissimilar to oncogenic RAS mutations, which are commonly located at codons 12, 13 and 61.
NRAS pathogenic variants have been identified in a small number of patients with NS at codons 24, 34, 50 and 60 which, again, are dissimilar to oncogenic NRAS mutations but are all predicted to increase NRAS signalling.41, 42, 43
CBL is a ubiquitin ligase which regulates intracellular signalling. Pathogenic variants in this gene have been reported to cause a variable Noonan-like phenotype with a predisposition to JMML.44,45
Pathogenic variants in SHOC2 have been reported to cause a distinctive phenotype, with patients displaying facial features typical of NS, along with sparse, slow growing hair, reduced growth secondary to growth hormone deficiency, cognitive deficits and hyperactive behaviour. This phenotype has been termed Noonan-like syndrome with loose anagen hair.46 SHOC2 associates with protein phosphatase 1 (PP1C) which regulates the RAS–MAPK pathway. Pathogenic variants in PPPC1B, the beta catalytic subunit of PP1C, have been reported to cause a similar phenotype to Noonan-like syndrome with loose anagen hair, but growth hormone deficiency has not been noted.47
NS with multiple lentigines (formerly LEOPARD syndrome) is a distinct, rare condition. Patients share the same craniofacial dysmorphic features as those with NS, but also have multiple skin lentigines, as well as ECG abnormalities, heart defects, growth delay and hearing loss.13 Similar to NS, NSML is associated with mutations in PTPN11, RAF1 and rarely BRAF.
Costello syndrome
CS is a rare RASopathy first described in 1991. Although difficult to accurately establish, birth prevalence in the UK has been estimated at ∼1/380 000.48,49 CS, in common with the other RASopathies, is characterised by craniofacial features including macrocephaly, epicanthal folds, downwards slanting palpebral fissures and depressed nasal bridge. Dermatological manifestations include soft skin, with excessive wrinkling and redundancy on the hands and feet.50 Patients show cardiac, musculoskeletal and ocular abnormalities as well as failure to thrive in infancy and developmental delay.13,51
CS is caused by activating mutations in HRAS, which decrease the intrinsic and/or GAP-induced GTPase activity.52 Amino acid positions 12 and 13, the most commonly mutated positions in CS, are also the most commonly mutated positions in oncogenic somatic RAS mutations.13,52
Among the RASopathies, patients with CS are at a particularly increased risk of developing malignancy, with 15% cumulative risk of cancer at age 20, compared with 4% for patients with NS. The malignancies seen in CS overlap with those in NS, the most common being RMS, neuroblastoma and bladder cancer.35,53 RAS mutations are also common in sporadic RMS, and interestingly, loss of heterozygosity at HRAS appears to be important in the development of cancer in both groups. A recent study found a high proportion of complete paternal uniparental disomy of chromosome 11 in both Costello-related and sporadic cases of RMS.54 This loss of heterozygosity may be the second hit in tumour development.
It has been suggested that the risk of malignancy in CS could correlate with mutation type, with a higher incidence of malignancy noted in patients with the uncommon G12A mutation, compared with the more common G12S mutation.55
Cardiofaciocutaneous syndrome
CFC syndrome is rare, with craniofacial features similar to those seen in NS. Prevalence has not accurately been established, but in some countries it is more common than CS. The prevalence in Japan has been estimated at 1/810 000, compared with the Japanese estimate for CS of 1/1 290 000.56 Precise estimation of birth incidence or prevalence has not yet been possible due to genetic and phenotypic heterogeneity of CFC and overlap with NS. A significant proportion of affected patients may also remain undiagnosed due to less characteristic presentation than is usually seen in CS. Cardiac anomalies commonly seen in CFC include pulmonary stenosis, atrial septal defect and hypertrophic cardiomyopathy.57 Musculoskeletal and neurological abnormalities are common, including hypotonia, epilepsy and learning disability.58 Severe developmental impairment and refractory epilepsy are seen in a significant proportion of patients. Cutaneous manifestations include sparse or very curly hair, absent eyebrows and very dry skin.
Due to the rarity of CFC, it is difficult to establish its associated risk of malignancy. However, malignancies similar to those seen in NS and CS have been observed in CFC patients, including cases of acute lymphoblastic leukaemia, non-Hodgkin lymphoma and RMS.35
CFC has been associated with pathogenic variants in BRAF, MAP2K1 (MEK1), MAP2K2 (MEK2) and KRAS (also associated with NS).12,59,60 BRAF pathogenic variants are seen in ∼75% of CFC patients.57
Capillary malformation–arteriovenous malformation syndrome
CM–AVM is an autosomal dominant disorder characterised by capillary malformations, which may be associated with arteriovenous malformations and fistulas, occurring in many tissues.13 Individuals may also have cardiovascular malformations, septal defects and valvular disorders. CM–AVM is caused by inactivating pathogenic variants in RASA1 which encodes the Ras-GAP, p120-RasGAP.61 Inactivating pathogenic variants of RASA1 cause increased signalling in the RAS–MAPK pathway.
This is not the only association between the RAS–MAPK pathway and vascular malformations. Somatic mutations in this pathway and its regulators have been associated with intracranial and extracranial ‘high-flow’ vascular abnormalities.62, 63, 64, 65 Specifically, activating KRAS mutations were found in a large proportion of a cohort of patients with sporadic arteriovenous malformations of the brain. In vitro analysis of endothelial cells expressing mutant KRAS found increased ERK activity, increased expression of genes related to angiogenesis and increased migratory behaviour.62 Zebrafish models expressing mutant BRAF and MAPK2K1 were found to develop vascular malformations, which responded to vemurafenib (a licenced BRAF inhibitor) in almost all cases.64 These studies suggest the potential for targeted medical treatment in patients with vascular malformations.
Somatic RAS mutations in cancer
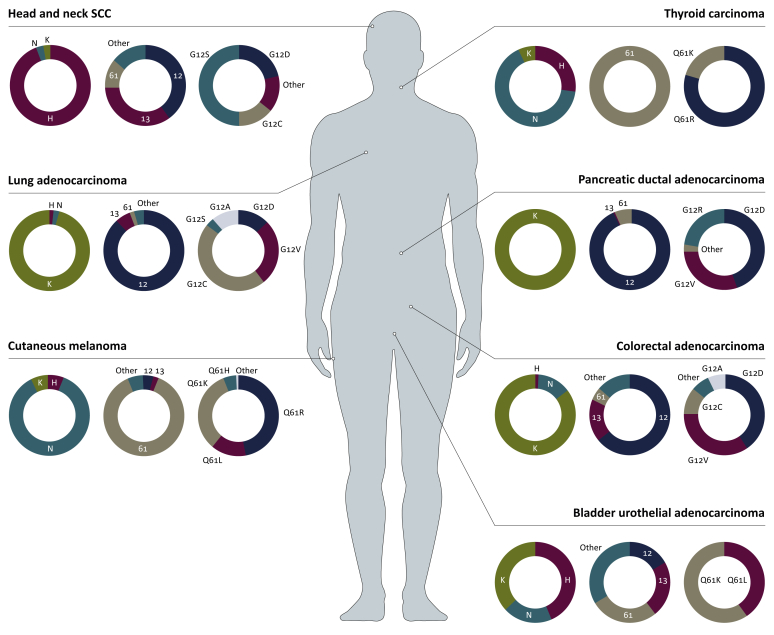
Somatic mutations in RAS genes (KRAS4a, KRAS4b, NRAS, HRAS) and related genes have been found in a wide range of malignancies (Figure 2). The vast majority of these mutations occur at codons 12, 13 or 61. Missense mutations at codons 12 and 13 are thought to limit GAPs interaction with the GTPase site of RAS proteins, preventing their hydrolysis to an inactive state.66 Glutamine 61 is part of the intrinsic GTP hydrolysis mechanism, so mutations likely prevent intrinsic and GAP-mediated hydrolysis.1 The role of RAS as an oncogene may be more nuanced than originally thought, with studies suggesting that RAS amplification through modification of copy number may play an important role in tumorigenesis. A large cohort of patients with advanced cancer found allelic imbalance in 55% of those with KRAS mutation.67
Figure 2.
RAS-mutant somatic cancers represented in terms of typically associated isoforms, codons and mutational subtypes.
H, HRAS, N, NRAS, K and KRAS. Data derived from cBioPortal.82,83 SCC, squamous cell carcinoma.
Oncogenic RAS mutations are found most frequently in KRAS (85%), followed by NRAS (11%) and HRAS (4%; Figure 2). Despite sharing common sites of mutation, the frequency of mutation in these hotspots varies between isoforms (Figure 3). For example, 83% of KRAS mutations are found at G12 followed by G13 (14%) and Q61 (2%). By contrast, Q61 is the most commonly mutated site in NRAS, whereas HRAS shows comparable frequencies between G12, G13 and Q61. The frequency of mutation sites also varies between malignancy types for a particular isoform.68 This isoform bias is striking considering that the RAS isoforms are virtually identical in structure beyond their hypervariable domain, and that these mutations occur in regions that are identical in their amino acid sequence.6
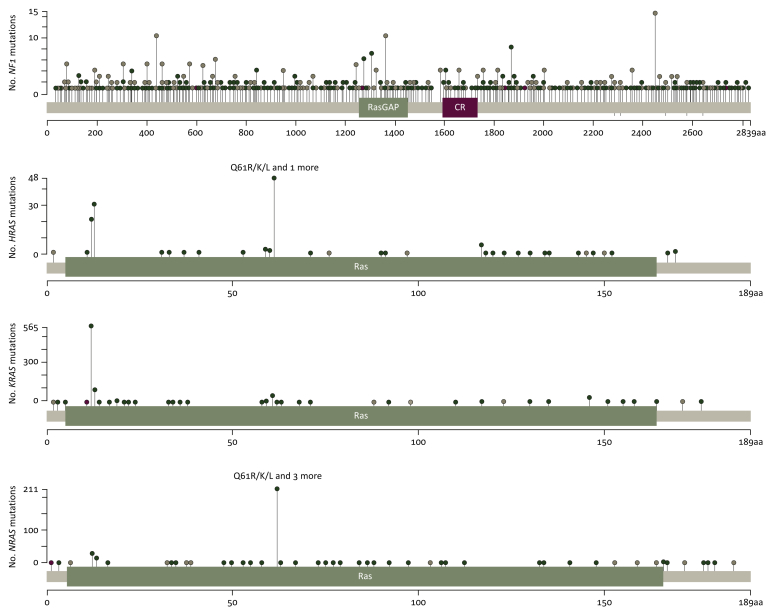
Figure 3.
Lollipop diagram representation of NF1, HRAS, NRAS and KRAS genes showing the distribution of amino acid substitutions seen in malignancy.
RAS activation in cancer can occur through other mechanisms, including disruption of GAPs (e.g. NF1) and activation of receptor tyrosine kinases.68
NF1
NF1 is a large gene found on chromosome 17, containing 61 exons and coding for a 2818 amino acid protein (of which only ∼330 code for the GAP domain).20,69 The protein is ubiquitously expressed but found in its highest levels in the CNS.70 Neurofibromin's most well-characterised function is as a GAP, promoting the conversion of GTP-bound RAS to the inactive GDP-bound RAS. Neurofibromin is also known to associate with a large number of other proteins, including tubulin, kinesin and PKA, suggesting that it is also likely to have other, as yet unidentified, functions.71
As previously mentioned, over 2600 inherited mutations in NF1 have been associated with NF1, varying massively in size and showing no localised mutation clustering in RASopathy or somatic cancer (Figure 3).20 In the germline context, NF1 mutation confers an increased risk of malignancy, particularly MPNST and gliomas.
Somatic NF1 mutations are frequently observed in melanoma, which has not been described as an associated tumour in NF1. Indeed, somatic mutations have been observed in 12%–30% of cutaneous melanomas, and up to 45%–90% of desmoplastic melanomas.20,72 Other components of the RAS–MAPK pathway have also been implicated in melanoma, with BRAF and NRAS the most commonly mutated genes.73 Mouse melanoma models have suggested that NF1 mutations work cooperatively with BRAF mutations to prevent oncogene-induced senescence and may also impair response to targeted BRAF inhibitors.74 NF1 mutations in melanoma are often seen alongside mutations in other tumour suppressor genes, however, they have also been seen in melanomas that lack both BRAF and NRAS mutations—these types of melanoma are strongly associated with UV damage.75 In one cohort, somatic NF1 mutations were identified in 46% of BRAF/RAS wild-type melanomas.72
NF1 mutations have also been identified in lung adenocarcinoma. In a recent study of 591 patients, NF1 mutations were found in 10% of cases.76 A quarter of these mutations occurred with other oncogenic mutations (e.g. in BRAF, ERBB2, KRAS, HRAS, NRAS). The NF1 mutations were found to be diverse and distributed throughout the genome, with 27/59 identified mutations predicted to result in loss of function of the NF1 protein.76 Reduced levels of neurofibromin have been associated with resistance to EGFR tyrosine kinase inhibitors, which has been overcome with MAP–ERK inhibitor treatment.77 Somatic NF1 mutations have also been identified in approximately 12% of squamous cell lung cancers.78 NF1 mutations are associated with tumours rich in transversion mutations, which are strongly associated with a history of smoking.78
Pheochromocytomas are tumours originating from chromaffin cells of the adrenal medulla. They are rarely found in the general population (incidence ∼1/100 000), but ∼0.1%–6% of neurofibromatosis patients will develop them.79 Sporadic NF1 mutations have been associated with pheochromocytoma. In a cohort of 119 cases, 25 were found to carry an inactivating NF1 mutation.80 Indeed, 56% of the somatic mutations identified in their cohort were on the NF1 gene.
Somatic NF1 mutations have also been associated with ovarian and breast carcinoma, acute myeloid leukaemia, neuroblastoma, glioblastoma, colon adenocarcinoma and bladder transitional cell carcinoma.20 In common with germline disease, somatic NF1 mutations are associated with a poorer prognosis in breast cancer.81
Despite the large number of cancers associated with somatic NF1 mutations, it remains unclear why NF1 patients appear to be predisposed only to a distinct group of malignancies, and why they do not appear to be predisposed to, for example, cutaneous melanoma or lung adenocarcinoma where somatic NF1 mutations tend to occur.
HRAS
Somatic mutations in HRAS have also been associated with malignancy, although to a much less significant degree than in KRAS and NRAS. HRAS mutations are most frequently found in cancers of the adrenal glands, thymus, head and neck and bladder.82,83 Akin to other RAS genes, oncogenic mutations occur most frequently in codons 12, 13 and 61. The Gly12Val mutation is most frequently observed in HRAS-positive cancers, and functional assays have suggested that this substitution may have the highest transformational potential.84
HRAS mutations in CS are also found at codons 12 and 13. The most common mutation is a Gly12Ser substitution in codon 12 of exon 1. Less frequent mutations include Gly12Ala at codon 12 and Gly13Cys on codon 13.48,53,85 These mutations have also been seen in somatic cancers. There have been case reports of patients with Gly12Val mutations, nearly all of whom have had severe, ultimately lethal, disease. This was first reported in 2005—the patient died at 1 year of age due to severe cardiomyopathy.52 A report published in 2007 identified a patient with congenital myopathy and excess of muscle spindles who died at the age of 3 weeks.86 Four further patients with Gly12Val mutations and severe CS have been identified, all of whom died within 6 postnatal weeks.87 Severe clinical manifestations of CS have also been associated with two rarer mutations: Gly12Asp and Gly12Cys, both of which are predicted to have a high transformational potential.88
More recently, a patient with the Gly12Val mutation and a mild clinical phenotype of CS has been identified. This mutation was found to be associated with high levels of exon 2 skipping. This suggests that the oncogenic potential of HRAS mutations may not only be related to the activity of the encoded protein, but also to the effect on splicing efficiency.89
The rarity of this mutation and its severe functional effects could suggest that some fetuses harbouring it may die in utero, or postnatally without a diagnosis being reached. However, embryonic or early lethality has not consistently been observed in mouse models: an HRASG12V-mutant model with partial expression found that mice were viable, although they did exhibit facial dysmorphism, neurological deficits and cardiomyopathies. Interestingly, these mice did not appear to be at an increased risk of malignancy.90 A later model with full HRASG12V expression demonstrated >80% mortality within 14 perinatal days, craniofacial defects and the development of papillomas and angiosarcomas.91
CS is caused by mutations not only in a single gene, but at the same codon as oncogenic HRAS mutations. This example seems to provide the greatest crossover between germline and somatic disease. It is interesting to note that germline mutations of HRAS occur at codons 12 and 13 but have not been identified at codon 61. Furthermore, RMS is the most common CS-associated malignancy, but HRAS mutations are rarely seen in sporadic RMS.
KRAS
KRAS is distinct from other RAS proteins in that it exists in two forms (KRAS4A and KRAS4B) which are the result of alternative splicing. KRAS mutation also stands out for the very high frequency at which it is associated with malignancy compared with NRAS and HRAS. KRAS mutations are most frequently found in pancreatic ductal adenocarcinoma, colorectal adenocarcinoma (CRC) and non-small-cell lung cancer.82,83
Oncogenic mutations are most commonly identified at codon 12, although this varies with cancer type. Mutations in codons 13, 146 and 117 are more common in CRC than other cancer types.92 In fact, mutations at codons 146 and 117 are almost pathognomonic for CRC compared with other cancer types.93 All oncogenic mutations in KRAS increase its intrinsic activity by increasing intrinsic hydrolysis and/or promoting nucleotide exchange.
Germline KRAS mutations have been seen at numerous locations, including K5, V14, Q22, P34, I36, T58, G60, V152, D153 and F156, and are associated with NS and CFC. Like oncogenic mutations, most of these mutations increase the levels of activated GTP-bound KRAS.94 A biochemical and functional analysis of germline KRAS mutations by Gremer et al. grouped them into five distinct classes, most of which were dissimilar from oncogenic RAS mutations, and most of which conferred a milder phenotype than cancer-associated mutations (Table 2).94 Despite resulting in accumulation of GTP-bound KRAS, it was found that most germline-mutated KRAS induced only moderately increased phosphorylation levels of downstream signalling proteins.
Table 2.
Classes of germline KRAS mutations and their effect
| Class | Mutation | Effect |
|---|---|---|
| A | K5N T58I D153V |
Higher activated state, higher downstream signalling. Mechanism not identified |
| B | V14I | Increase in intrinsic and GEF-catalysed nucleotide exchange |
| C | Q22R | Impaired GAP-stimulated hydrolysis |
| D | Q22E R156L |
Increased nucleotide exchange and resistance to GAPs |
| E | P34L P34R G60R |
Defective GAP sensitivity and strongly reduced interaction with effectors |
Adapted from Gremer et al.94
GAP, GTPase-activating proteins; GEF, guanine nucleotide exchange factor.
Class E germline KRAS alterations comprise three mutations that cause defective GAP sensitivity, locking KRAS in an activated state similar to that seen in oncogenic mutations. However, they were noted to have compromised interaction with effector proteins, accumulating in only a mild gain of function phenotype. This may not be sufficient to meet the oncogenic threshold required to cause those cancers associated with oncogenic KRAS mutations.
BRAF/MEK
Oncogenic mutations in the BRAF–MAPK pathway occur with decreasing incidence further downstream in the pathway. BRAF mutations are seen in ∼7% of cancers, and MEK in <1%.95
BRAF is a kinase effector immediately downstream of RAS, affected by somatic mutations which have been associated with various malignancies including malignant melanoma, thyroid, lung, ovarian and colorectal cancers. Germline pathogenic variants are seen in NS, and ∼75% of cases of CFC syndrome.
The most common somatic BRAF mutation found in cancer is V600E, which confers increased kinase activity. This mutation has not been noted in CFC. One study identified 11 distinct BRAF missense mutations in CFC patients, distributed more widely than seen in cancer, primarily in the cysteine-rich domain and the protein kinase domain of BRAF. Only two CFC patients were found to have mutations that have been identified in cancer and both had severe phenotypes, suggesting that germline BRAF mutations may have attenuated activity compared with somatic mutations. However, four of the 11 BRAF mutations were found to confer increased kinase activity comparable with that of V600E.96 Importantly, inhibitors of V600E-mutant BRAF have been developed and have shown efficacy in advanced BRAF-mutant melanoma.97
Mutations in MAP2K1 and MAP2K2 are associated uncommonly with CFC, and have also been seen in BRAF-mutant colorectal cancer and melanoma with acquired resistance to targeted therapy.98 MEK inhibitors have shown efficacy alone and in combination with BRAF inhibitors in BRAF-mutant melanoma.97
Conclusion
In conclusion, RAS proteins are a vital component of cellular signalling pathways that confer a wide range of effects. Their importance in normal cellular functioning is demonstrated by the diverse manifestations of germline mutations, as seen in the group of RASopathy disorders. Furthermore, the importance of strict regulation of their function can be seen from the numerous malignancies associated with oncogenic mutations in RAS and related proteins. However, it remains unclear why only a specific handful of malignancies are seen in RASopathy syndrome patients, rather than a predisposition to the full range of malignancies associated with sporadic mutations of the same genes.
Perhaps, as suggested by analyses of KRAS mutations, mutations associated with germline disease are attenuated in their effect compared with oncogenic mutations and may not reach a specific threshold required to cause cancer. Perhaps also, as suggested by the relative lack of Gly12Val mutations in patients with CS, mutations that confer a greater risk of cancer may be incompatible with life in a fetus with germline mutation. This does not account for the restricted pattern of malignancy seen in NF1—whose patients have been noted to have a very diverse range of mutations, ranging from large genomic deletions to single-base-pair substitutions, akin to oncogenic mutations of the same gene. However, somatic NF1 mutations often require co-mutations in the MAPK pathway to confer oncogenicity, possibly suggesting that mutations in this gene are generally attenuated in their effect, and may be more compatible with life in the germline context.
It is clear that there is further insight to be gained into the relationship between RAS pathway mutations and malignancy. Sporadic RAS mutations are a key driver of many malignancies and are an exciting prospect for targeted biological therapies. Early results have suggested that tipifarnib, a farnesyltransferase inhibitor, is well tolerated and may have some efficacy in HRAS-mutated head and neck cancers.99 Despite initial disappointments as a therapeutic target, there is also increasing optimism for direct inhibitors of KRAS and SHP2.100 Small molecule inhibitors of KRAS with the G12C mutation are showing early evidence of clinical efficacy.101,102 A greater understanding of RAS pathway mutations' causative relationship with cancer may help us to identify further therapeutic targets.
Acknowledgments
Funding
This work was supported by Cancer Research UK via funding to the CRUK Manchester Institute [grant number A25254] and the CRUK Lung Cancer Centre of Excellence [grant number A20465], as well as the Manchester Cancer Research Centre Town Hall programme. DGE is supported by the Manchester NIHR Biomedical Research Centre (IS-BRC-1215-20007).
Disclosure
GE: Consulting: AZ. FHB: Institutional funding as a PI: Amgen, Novartis, Pfizer; Clinical trials: AbbVie, Amgen, Ariad, AZ, BI, BMS, Celgene, Genentech, MSD, Novartis, Pfizer, Regeneron, Roche, Takeda; Consulting/advisory: AbbVie, Celgene, Cell Medica, Ipsen, Medivation, Regeneron, Takeda, Amgen, AZ. CRL: Institutional funding as a CI/PI: Roche, Amgen and BI; Consulting: CBPartners, Amgen. All remaining authors have declared no conflict of interest.
References
- 1.Simanshu D.K., Nissley D.V., McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherfils J., Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 3.Drosten M., Dhawahir A., Sum E.Y.M. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 2010;29:1091–1104. doi: 10.1038/emboj.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapiro P. Ras-MAP kinase signaling pathways and control of cell proliferation: relevance to cancer therapy. Crit Rev Clin Lab Sci. 2002;39:285–330. doi: 10.1080/10408360290795538. [DOI] [PubMed] [Google Scholar]
- 5.Yaeger R., Corcoran R.B. Targeting alterations in the RAF/MEK pathway. Cancer Discov. 2019;9:329–341. doi: 10.1158/2159-8290.CD-18-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen J., Hallin J., Engstrom L. The KRASG12C inhibitor, MRTX849, provides insight toward therapeutic susceptibility of KRAS mutant cancers in mouse models and patients. Cancer Discov. 2020;10(1):54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindsay C., Blackhall F. Direct Ras G12C inhibitors: crossing the rubicon. Br J Cancer. 2019;121(3):197–198. doi: 10.1038/s41416-019-0499-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adderley H., Blackhall F., Lindsay C. KRAS-mutant non-small cell lung cancer: converging small molecules and immune checkpoint inhibition. EBioMedicine. 2019;41:711–716. doi: 10.1016/j.ebiom.2019.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canon J., Rex K., Saiki A. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 11.Lindsay C., Jamal-Hanjani M., Forster M., Blackhall F. KRAS: reasons for optimism in lung cancer. Eur J Cancer. 2018;99:20–27. doi: 10.1016/j.ejca.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Tidyman W.E., Rauen K.A. Pathogenetics of the RASopathies. Hum Mol Genet. 2016;25:R123–R132. doi: 10.1093/hmg/ddw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauen K.A. The RASopathies. Annu Rev Genomics Hum Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett E., Thomas N., Upadhyaya M. Neurofibromatosis type 1: its association with the Ras/MAPK pathway syndromes. J Pediatr Neurol. 2009;7:105–115. [Google Scholar]
- 15.Martin G.A., Viskoohil D., Bollag G. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 16.Lammert M., Friedman J.M., Kluwe L., Mautner V.F. Prevalence of neurofibromatosis 1 in German children at elementary school enrollment. Arch Dermatol. 2005;141:71–74. doi: 10.1001/archderm.141.1.71. [DOI] [PubMed] [Google Scholar]
- 17.Huson S.M., Compston D.A., Clark P., Harper P.S. A genetic study of von Recklinghausen neurofibromatosis in south east Wales. I. Prevalence, fitness, mutation rate, and effect of parental transmission on severity. J Med Genet. 1989;26:704–711. doi: 10.1136/jmg.26.11.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans D., Howard E., Giblin C. Birth incidence and prevalance of tumour-prone syndromes: estimates from a UK family genertic register service. Am J Med Genet. 2010;152:327–332. doi: 10.1002/ajmg.a.33139. [DOI] [PubMed] [Google Scholar]
- 19.Kallionpää R., Uustilo E., Leppavirta J. Prevalence of neurofibromatosis type 1 in the Finnish population. Genet Med. 2017;20(9):1082–1086. doi: 10.1038/gim.2017.215. [DOI] [PubMed] [Google Scholar]
- 20.Philpott C., Tovell H., Frayling I.M. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics. 2017;11:13. doi: 10.1186/s40246-017-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths S., Thompson P., Frayling I., Upadhyaya M. Molecular diagnosis of neurofibromatosis type 1: 2 years experience. Fam Cancer. 2006;6:21–34. doi: 10.1007/s10689-006-9001-3. [DOI] [PubMed] [Google Scholar]
- 22.Messiaen L.M., Callens T., Mortier G. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 23.Riccardi V.M. 2nd ed. John Hopkins University Press; Baltimore, Maryland: 1992. Neurofibromatosis: Phenotype, Natural History and Pathogenesis. [Google Scholar]
- 24.Walker L., Thompson D., Easton D. A prospective study of neurofibromatosis type 1 cancer incidence in the UK. Br J Cancer. 2006;95:233–238. doi: 10.1038/sj.bjc.6603227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uusitalo E., Rantanen M., Kallionpaa R.A. Distinctive cancer associations in patients with neurofibromatosis Type 1. J Clin Oncol. 2016;34:1978–1986. doi: 10.1200/JCO.2015.65.3576. [DOI] [PubMed] [Google Scholar]
- 26.Federico Salvi P., Lorenzon L., Caterino S. Gastrointestinal stromal tumors associated with neurofibromatosis 1: a single centre experience and systematic review of the literature including 252 cases. Int J Surg Oncol. 2013;2013:398570. doi: 10.1155/2013/398570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen S.A., Yang Q., Friedman J.M. Mortality in neurofibromatosis 1: an analysis using U.S. death certificates. Am J Hum Genet. 2001;68:1110–1118. doi: 10.1086/320121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans D., O'Hara C., Wilding A. Mortality in neurofibromatosis 1: in North West England: an assessment of actuarial survival in a region of the UK since 1989. Eur J Hum Genet. 2011;19:1187–1191. doi: 10.1038/ejhg.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tartaglia M., Mehler E.L., Goldberg R. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 30.Tartaglia M., Pennacchio L.A., Zhao C. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 31.Schubbert S., Zenker M., Rowe S.L. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 32.Razzaque M.A., Nishizawa T., Komoike T. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39(8):1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 33.Aoki Y., Nilhori T., Banjo T. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am J Hum Genet. 2013;93:173–180. doi: 10.1016/j.ajhg.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Shachar S., Costantini S., Hallevi H. Increased rate of missense/in-frame mutations in individuals with NF1-related pulmonary stenosis: a novel genotype–phenotype correlation. Eur J Hum Genet. 2012;21(5):535–539. doi: 10.1038/ejhg.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kratz C.P., Rapisuwon S., Reed H. Cancer in Noonan, Costello, cardiofaciocutaneous and LEOPARD syndromes. Am J Med Genet C Semin Med Genet. 2011;157:83–89. doi: 10.1002/ajmg.c.30300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strullu M., Caye A., Lachenaud J. Juvenile myelomonocytic leukaemia and Noonan syndrome. J Med Genet. 2014;51:689–697. doi: 10.1136/jmedgenet-2014-102611. [DOI] [PubMed] [Google Scholar]
- 37.Nichols R.J., Haderk F., Stahlhut C. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol. 2018;20:1064–1073. doi: 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubbert S., Bollag G., Lyubynska N. Biochemical and functional characterization of germ line KRAS mutations. Mol Cell Biol. 2007;27:7765–7770. doi: 10.1128/MCB.00965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carta C., Pantaleoni F., Bocchinfuso G. Germline missense mutations affecting KRAS isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet. 2006;79:129–135. doi: 10.1086/504394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zenker M., Lehmann K., Schulz A.L. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J Med Genet. 2006;44:131–135. doi: 10.1136/jmg.2006.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denayer E., Peeters H., Sevenants L. NRAS mutations in Noonan syndrome. Mol Syndromol. 2012;3:34–38. doi: 10.1159/000338467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cirstea I.C., Kutsche K., Dvorsky R. A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat Genet. 2010;42:27–29. doi: 10.1038/ng.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Runtuwene V., Eekelen M., Overvoorde J. Noonan syndrome gain-of-function mutations in NRAS cause zebrafish gastrulation defects. Dis Model Mech. 2011;4:393–399. doi: 10.1242/dmm.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez B., Mechinaud F., Galamrun C. Germline mutations of the CBL gene define a new genetic syndrome with predisposition to juvenile myelomoncytic leukaemia. J Med Genet. 2010;47:686–691. doi: 10.1136/jmg.2010.076836. [DOI] [PubMed] [Google Scholar]
- 45.Martinelli S., De Luca A., Stellacci E. Heterozygous germline mutations in the CBL tumour-suppressor gene cause a Noonan syndrome-like phenotype. Am J Hum Genet. 2010;87:250–257. doi: 10.1016/j.ajhg.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cordeddu V., Di Schiavi E., Pennacchio L.A. Mutation in SHOC2 promotes abberant protein N-myristoylation and underlies Noonan-like syndrome with loose anagen hair. Nat Genet. 2010;41:1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gripp K., Aldinger K., Bennett J. A novel rasopathy caused by recurrent de novo missense mutations in PPP1CB closely resembles Noonan syndrome with loose anagen hair. Am J Med Genet. 2016;170:2237–2247. doi: 10.1002/ajmg.a.37781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rauen K.A. HRAS and the Costello syndrome. Clin Genet. 2007;71:101–108. doi: 10.1111/j.1399-0004.2007.00743.x. [DOI] [PubMed] [Google Scholar]
- 49.Giannoulatou E., McVean G., Taylor I.B. Contributions of intrinsic mutation rate and selfish selection to levels of de novo HRAS mutations in the paternal germline. Proc Natl Acad Sci U S A. 2013;110:20152–20157. doi: 10.1073/pnas.1311381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siegel D.H., Mann J.A., Krol A.L., Rauen K.A. Dermatological phenotype in Costello syndrome: consequences of Ras dysregulation in development. Br J Dermatol. 2011;166:601–607. doi: 10.1111/j.1365-2133.2011.10744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin A.E., Alexander M.E., Colan S.D. Clinical, pathological, and molecular analyses of cardiovascular abnormalities in Costello syndrome: a Ras/MAPK pathway syndrome. Am J Med Genet A. 2011;155A:486–507. doi: 10.1002/ajmg.a.33857. [DOI] [PubMed] [Google Scholar]
- 52.Aoki Y., Niihori T., Kawame H. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 53.Gripp K.W., Lin A.E., Stabley D.L. HRAS mutation analysis in Costello syndrome: genotype and phenotype correlation. Am J Med Genet A. 2006;140A:1–7. doi: 10.1002/ajmg.a.31047. [DOI] [PubMed] [Google Scholar]
- 54.Robbins K., Stabley D., Holbrook Jea. Paternal uniparental disomy with segmental loss of heterozygosity of chromosome 11 are hallmark characteristics of syndromic and sporadic embryonal rhabdomyosarcoma. Am J Med Genet. 2016;170:3197–3206. doi: 10.1002/ajmg.a.37949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerr B., Delrue M.A., Sigaudy S. Genotype-phenotype correlation in Costello syndrome: HRAS mutation analysis in 43 cases. J Med Genet. 2006;43:401–405. doi: 10.1136/jmg.2005.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abe Y., Aoki Y., Kuriyama S. Prevalence and clinical features of Costello syndrome and cardio-facio-cutaneous syndrome in Japan: Findings from a nationwide epidemiological survey. Am J Med Genet A. 2012;158A:1083–1094. doi: 10.1002/ajmg.a.35292. [DOI] [PubMed] [Google Scholar]
- 57.Allanson J.E., Anneren G., Aoki Y. Cardio-facio-cutaneous syndrome: does genotype predict phenotype? Am J Med Genet C Semin Med Genet. 2011;157:129–135. doi: 10.1002/ajmg.c.30295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoon G., Rosenberg J., Blaser S., Rauen K.A. Neurological complications of cardio-facio-cutaneous syndrome. Dev Med Child Neurol. 2007;49:894–899. doi: 10.1111/j.1469-8749.2007.00894.x. [DOI] [PubMed] [Google Scholar]
- 59.Niihori T., Aoki Y., Narumi Y. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez-Viciana P., Tetsu O., Tidyman W.E. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 61.Eerola I., Boon L.M., Mulliken J.B. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am J Hum Genet. 2003;73:1240–1249. doi: 10.1086/379793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nikolaev S., Vestiska S., Bonilla X., Boudreau E. Somatic activating KRAS mutations in arteriovenous malformations of the brain. N Engl J Med. 2018;378:1561–1562. doi: 10.1056/NEJMc1802190. [DOI] [PubMed] [Google Scholar]
- 63.Couto J.A., Huang A.Y., Konczyk D.J. Somatic MAP2K1 mutations are associated with extracranial arteriovenous malformation. Am J Hum Genet. 2017;100:546–554. doi: 10.1016/j.ajhg.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al-Olabi L., Polubothu S., Dowsett K. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J Clin Invest. 2018;128:5185. doi: 10.1172/JCI124649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ayturk U.M., Couto J.A., Hann S. Somatic activating mutations in GNAQ and GNA11 are associated with congenital hemangioma. Am J Hum Genet. 2016;98:1271. doi: 10.1016/j.ajhg.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheffzek K., Ahmadian M.R., Kabsch W. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 67.Burgess M., Hwang E., Mroue R. KRAS Allelic imbalance enhances fitness and modulates MAP kinase dependence in cancer. Cell. 2017;168:817–829. doi: 10.1016/j.cell.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hobbs G.A., Der C.J., Rossman K.L. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu G., O'Connell P., Viskochil D. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 70.Daston M.M., Ratner N. Neurofibromin, a predominantly neuronal GTPase activating protein in the adult, is ubiquitously expressed during development. Dev Dyn. 1992;195:216–226. doi: 10.1002/aja.1001950307. [DOI] [PubMed] [Google Scholar]
- 71.Peltonen S., Kallionpaa R.A., Peltonen J. Neurofibromatosis type 1 (NF1) gene: beyond cafe au lait spots and dermal neurofibromas. Exp Dermatol. 2016;26:645–648. doi: 10.1111/exd.13212. [DOI] [PubMed] [Google Scholar]
- 72.Krauthammer M., Kong Y., Bacchiocchi A. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hill V.K., Gartner J.J., Samuels Y., Goldstein A.M. The genetics of melanoma: recent advances. Annu Rev Genomics Hum Genet. 2013;14:257–279. doi: 10.1146/annurev-genom-091212-153429. [DOI] [PubMed] [Google Scholar]
- 74.Maertens O., Johnson B., Hollstein P. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3:338–349. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hodis E., Watson I.R., Kryukov G.V. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Redig A.J., Capelletti M., Dahlberg S.E. Clinical and molecular characteristics of NF1-mutant lung cancer. Clin Cancer Res. 2016;22:3148–3156. doi: 10.1158/1078-0432.CCR-15-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bruin E.C., Cowell C., Warne P.H. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov. 2014;4:606–619. doi: 10.1158/2159-8290.CD-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koch C.A., Vortmeyer A.O., Huang S.C. Genetic aspects of pheochromocytoma. Endocr Regul. 2001;35:43–52. [PubMed] [Google Scholar]
- 80.Burnichon N., Buffet A., Parfait B. Somatic NF1 inactivation is a frequent event in sporadic pheochromocytoma. Hum Mol Genet. 2012;21:5397–5405. doi: 10.1093/hmg/dds374. [DOI] [PubMed] [Google Scholar]
- 81.Dischinger P., Tovar E., Esseburg C. NF1 deficiency correlates with estrogen receptor signaling and diminished survival in breast cancer. NPJ Breast Cancer. 2018;4:29. doi: 10.1038/s41523-018-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cerami E., Gao J., Dogrusoz U. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao J., Aksoy B., Dogrusoz U. Integratie analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6 doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aoki Y., Niihori T., Narumi Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- 85.Estep A.L., Tidyman W.E., Teitell M.A. HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am J Med Genet A. 2006;140A:8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- 86.Burgt I., Kupsky W., Stassou S. Myopathy caused by HRAS germline mutations: implications for disturbed myogenic differentiation in the presence of constitutive HRas activation. J Med Genet. 2007;44:459–462. doi: 10.1136/jmg.2007.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burkitt-Wright E.M.M., Bradley L., Shorto J. Neonatal lethal Costello syndrome and unusual dinucleotide deletion/insertion mutations in HRAS predicting p.Gly12Val. Am J Med Genet A. 2012;158A:1102–1110. doi: 10.1002/ajmg.a.35296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lo I.F.M., Brewer C., Shannon N. Severe neonatal manifestations of Costello syndrome. J Med Genet. 2008;45:167–171. doi: 10.1136/jmg.2007.054411. [DOI] [PubMed] [Google Scholar]
- 89.Hartung A.M., Swensen J., Uriz I.E. The splicing efficiency of activating HRAS mutations can determine Costello syndrome phenotype and frequency in cancer. PLoS Genet. 2016;12:e1006039. doi: 10.1371/journal.pgen.1006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schuhmacher A.J., Guerra C., Sauzeau V. A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J Clin Invest. 2008;118:2169–2179. doi: 10.1172/JCI34385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen X., Mitsutake N., LaPerle K. Endogenous expression of HRASG12V induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc Natl Acad Sci U S A. 2009;106(19):7979–7984. doi: 10.1073/pnas.0900343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haigis K.M. KRAS Alleles: the devil is in the detail. Trends Cancer. 2017;3:686–697. doi: 10.1016/j.trecan.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Edkins S., O'Meara S. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gremer L., Merbitz-Zahradnik T., Dvorsky R. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum Mutat. 2011;32:33–43. doi: 10.1002/humu.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yaegar R., Corcoran R. Targeting alternations in the RAF-MEK pathway. Cancer Discov. 2019;9(3):329–341. doi: 10.1158/2159-8290.CD-18-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rodriguez-Viciana P., Tetsu O., Tidyman W. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311(5765):1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 97.Mackiewicz J., Mackiewicz A. BRAF and MEK inhibitors in the era of immunotherapy in melanoma patients. Contemp Oncol (Pozn) 2018;22:68–72. doi: 10.5114/wo.2018.73890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahronian L., Sennott E., Van Allen E.M. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 2015;5(4):358–367. doi: 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ho A.L., Chau N., Bauman J. Preliminary results from a phase II trial of tipifarnib in squamous cell carcinomas (SCCs) with HRAS mutations. Ann Oncol. 2018;29:viii373. [Google Scholar]
- 100.Novartis Pharmaceuticals An open-label, multi-center, phase i, dose finding study of oral TNO155 in adult patients with advanced solid tumors. NCT03114319. https://clinicaltrials.gov/ct2/show/NCT03114319 Available at.
- 101.Fakih M., O'Neil B., Price T.J. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J Clin Oncol. 2019;37:3003. [Google Scholar]
- 102.Mirati Therapeutics MRTX849 in patients with cancer having a KRAS G12C mutation. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03785249?term=G12C Available at.
- 103.Zenker M. NSEuroNet. https://nseuronet.com Available at.