Abstract
One notable feature of the SARS-CoV-2 genome, the spike (S) protein of SARS-CoV-2 has a polybasic furin cleavage site (FCS) at its S1-S2 boundary through the insertion of 12 nucleotides encoding four amino acid residues PRRA. Quite intriguingly, this polybasic FCS is absent in coronaviruses of the same clade as SARS-CoV-2. Thus, with currently available experimental structural data for S protein, this short article presents a set of comprehensive structural characterization of the insertion of FCS into S protein, and argues against a hypothesis of the origin of SARS-CoV-2 from purposeful manipulation: (1), the inserted FCS is spatially located at a random coil loop region, mostly distantly solvent-exposed (instead of deeply buried), with no structural proximity to the other part of the S protein; (2), the insertion of FCS itself does not alter, neither stabilize nor de-stabilize, the three-dimensional structure of S; (3), the net result here is the insertion of a furin cleavage site into S protein, whose S1 and S2 subunits will still be strongly electrostatically bonded together from a structural and biophysical point of view, even if the polybasic FCS is actually cleaved by furin protease before or after viral cell entry.
Graphical abstract

1. Introduction
The membrane of SARS-CoV-2 harbours a homotrimeric ([1]) transmembrane spike (S) glycoprotein, which is essential for the entry of virus particles into the cell. S protein contains two functional domains: a receptor binding domain (RBD) [2], and a second domain which contains sequences that mediate fusion of viral and cell membranes [[3], [4], [5], [6], [7]]. Recently, it was reported that S protein contains a potential cleavage site for furin protease [8], including four residues (Pro681 (P681), Arg682 (R682), Arg683 (R683) and Ala684 (A684)) [[9], [10], [11], [12], [13]]. Functionally, R682, R683, A684 and Arg685 (R685) constitute the minimal polybasic furin cleavage cite (FCS), i.e., RXYR, where X or Y is to be a positively charged arginine or lysine. With respect to the origin of this COVID-19 pandemic, of further interest is the fact that the S protein has a specific FCS that is absent in coronaviruses of the same clade as SARS-CoV-2 [8,[14], [15], [16]].
In the midst of this COVID-19 pandemic, FCS is reportedly linked to a natural-selection, instead of purposeful-manipulation, −based hypothesis of the origin of this COVID-19 outbreak [9,[17], [18], [19], [20], [21]]. Regardless of this COVID-19-origin hypothesis [9], to date (Thu Jun 25 09:39:392020), it still remains not clear what the actual structural consequence is of the polybasic FCS's insertion into the S protein of SARS-CoV-2. Thus, this article delves into all currently available (as of Thu Jun 25 09:39:392020) structural data [22] of S protein and aims to uncover the structural impact of the insertion of FCS into the S protein of SARS-CoV-2.
2. Materials and methods
To begin with, QHD43416.1 is the GenBank access code of the surface glycoprotein of SARS-CoV-2 [Severe acute respiratory syndrome coronavirus 2]. With QHD43416.1, the amino acid sequence of S protein was retrieved and listed below,
MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVKLHYT.
First, the sequence above was plugged into the SwissModel homology modelling [23] server in search of an experimental structural model of the S protein of SARS-CoV-2. In principle, it is expected that the experimental S protein structure is complete, i.e., with no experimentally uncharted territories (EUTs) [24]. In fact, however, the structural search led to a Cryo-EM structure (PDB ID: 6VSB) with a range of EUTs [24], representing the three-dimensional structure of the prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up [1]. From an amino acid sequence alignment (Fig. 1.2 in supplementary file supplement.pdf), it is obvious that those EUTs are widely scattered throughout the homotrimeric structure (PDB ID 6VSB) [1]. Yet, with another amino acid sequence alignment (Fig. 1.1 in supplementary file supplement.pdf), it was revealed that the sequence similarity between the Cryo-EM structure (PDB ID 6VSB) and QHD43416.1 is as high as 99.26%, making PDB ID 6VSB rather suitable to be used as a structural template for the subsequent homology structural modelling of the S protein of the SARS-CoV-2. Therefore, the Cryo-EM PDB ID 6VSB structure was used as the template by the SwissModel [23] homology modelling server to build a structural model with as less EUTs [24] as possible of the S protein of SARS-CoV-2.
Afterwards, the UCSF Chimera software [25] was employed to add hydrogen atoms to the homology homotrimeric structural model of the S protein, which was subsequently subject to a comprehensive set of electrostatic interaction analysis as described in [26] previously, and also a set of solvent accessible surface area (SASA) analysis by DSSP [27,28]. According to the DSSP-calculated SASA values, an amino acid residue of a protein can be classified as either buried or exposed according to a comparison with its standard SASA value contained in the “standard.data” file (supplementary file standard.data) available with the Naccess [29] software distribution.
3. Results
3.1. A structural electrostatic analysis of the homotrimeric structural model of the S protein of SARS-CoV-2
With SwissModel [23] and UCSF Chimera [25], a homology homotrimeric structural model (supplementary file model.pdb) of the spike protein of SARS-CoV-2 was built with only two EUTs (Fig. 1.3 in supplementary file supplement.pdf) [24], one at its N-terminal and another at its C-terminal, as compared (Fig. 1.2 in supplementary file supplement.pdf) with the widely scattered EUTs [24] inside its Cryo-EM structural template PDB ID 6VSB [1]. Subsequently, with the electrostatic analysis as described previously in [26], this short article puts forward a comprehensive set of structural electrostatic interaction analysis (supplementary file supplementary.pdf) for the homotrimeric structural model (supplementary file model.pdb) of the spike protein of SARS-CoV-2.
With a close inspection of all tables in the supplementary file supplementary.pdf,
-
1.
no salt bridge or hydrogen bond was structurally identified for the three basic residues at FCS.
-
2.
no hydrogen bond was structurally identified for Arg682 or Pro681.
-
3.
one hydrogen bond was structurally identified for Ala684 with Arg685.
-
4.
two hydrogen bonds were structurally identified for Arg683 with Thr604 and Asn679.
-
5.
four hydrogen bonds were structurally identified for Arg685 with Val687, Lys310 and Ala684.
Further details of the hydrogen bonds were included in Table 1 , where the first four (1, 2, 3 and 4 in Table 1) hydrogen bonds were formed between the side chains of the FCS residues and the main chain (backbone) oxygen atoms of the other part of the S protein, thus only affecting the backbone oxygen atoms in the structure of the S protein. In light of the double covalent bond formed between those affected backbone oxygen atoms and their neighbouring backbone carbon atoms, it is unlikely that the first four (1, 2, 3 and 4 in Table 1) hydrogen bonds are able to cause a major conformational perturbation of the structure of the S protein of SARS-CoV-2.
Table 1.
A summary of side chain and main chain hydrogen bonding analysis between the polybasic FCS and other part of the S protein of SARS-CoV-2. In this table, the residue naming scheme is Chain ID_residue name_residue number, ∠ADH represents the angle formed by acceptor (A), donor (D) and hydrogen (H) (∠ADH).
| Hydrogen bond No. | Acceptor (A) | Donor (D) | Hydrogen (H) | D-A (Å) | H-A (Å) | ∠ ADH () |
|---|---|---|---|---|---|---|
| 1 | O, A_THR_604 | NE, A_ARG_683 | HE, A_ARG_683 | 2.90 | 1.95 | 15.53 |
| 2 | O, C_ASN_679 | NH1, C_ARG_683 | HH11, C_ARG_683 | 2.73 | 1.73 | 8.46 |
| 3 | O, B_VAL_687 | NH1, B_ARG_685 | HH11, B_ARG_685 | 2.75 | 1.91 | 27.04 |
| 4 | O, B_VAL_687 | NE, B_ARG_685 | HE, B_ARG_685 | 2.86 | 1.99 | 25.09 |
| 5 | O, C_ARG_685 | NZ, C_LYS_310 | HZ3, C_LYS_310 | 2.70 | 1.83 | 24.75 |
On the other hand, however, the last (hydrogen bond No. 5 in Table 1) hydrogen bond was formed between the main chain (backbone) oxygen atom of the FCS residue (Arg685) and the positively charged side chain of Lys310, for which a comprehensive set of structural electrostatic interaction analysis is included in Table 2, Table 3 below.
Table 2.
Lys310-specific salt bridging analysis of the structure of the S protein. In this table, the residue naming scheme is Chain ID_residue name_residue number.
| Salt bridge | Residue A | Atom A | Residue B | Atom B | Distance (Å) |
|---|---|---|---|---|---|
| 1 | A_LYS_310 | NZ | A_ASP_663 | OD1 | 2.628 |
| 2 | A_LYS_310 | NZ | A_ASP_663 | OD2 | 2.667 |
| 3 | B_LYS_310 | NZ | B_ASP_663 | OD2 | 2.615 |
| 4 | B_LYS_310 | NZ | B_ASP_663 | OD1 | 2.691 |
| 5 | C_LYS_310 | NZ | C_ASP_663 | OD1 | 3.129 |
Table 3.
Lys310-specific hydrogen bonding network analysis of the structure of the S protein. In this table, the residue naming scheme is Chain ID_residue name_residue number, ∠ADH represents the angle formed by acceptor (A), donor (D) and hydrogen (H) (∠ADH).
| Hydrogen bond No. | Acceptor (A) | Donor (D) | Hydrogen (H) | D-A (Å) | H-A (Å) | ∠ ADH () |
|---|---|---|---|---|---|---|
| 1 | OD1, A_ASP_663 | NZ, A_LYS_310 | HZ2, A_LYS_310 | 2.63 | 1.74 | 22.20 |
| 2 | OD2, B_ASP_663 | NZ, B_LYS_310 | HZ3, B_LYS_310 | 2.62 | 1.67 | 16.87 |
| 3 | O, C_ARG_685 | NZ, C_LYS_310 | HZ3, C_LYS_310 | 2.70 | 1.83 | 24.75 |
| 4 | OD1, A_ASP_663 | NZ, A_LYS_310 | HZ2, A_LYS_310 | 2.63 | 1.74 | 22.20 |
| 5 | OD2, B_ASP_663 | NZ, B_LYS_310 | HZ3, B_LYS_310 | 2.62 | 1.67 | 16.87 |
From the structurally identified salt bridges and hydrogen bonds (Table 2, Table 3), it is clear that Lys310 and Asp663 forms a set of strong electrostatic interactions for all three chains (A, B and C), making it unlikely that the 5th (Table 1) hydrogen bond is able to disrupt the basic residue pair Lys310-Asp663 and induce a major conformational change for the structure of the S protein of SARS-CoV-2.
To sum up, the inserted FCS is only involved in a set of weak electrostatic interactions (five hydrogen bonds, Table 1) within the S protein of SARS-CoV-2, whose overall scaffold is not to be altered, neither stabilized nor de-stabilized, by the insertion of the polybasic FCS.
3.2. An SASA analysis of the homotrimeric structural model of the S protein of SARS-CoV-2
To delve deep into the structural impact of FCS's insertion into the S protein of SARS-CoV-2, this short article takes a close look at the spatial location of FCS in the overall scaffold (supplementary file model.pdb) of the S protein of SARS-CoV-2.
As shown in Fig. 1 , the inserted FCS is spatially located at a random coil loop region with no structural proximity to other part of the structure of the S protein. In light of the structural electrostatic analysis above, this visual observation (Fig. 1) constitutes further structural evidence that the inserted FCS does not alter, neither stabilize nor de-stabilize, the overall scaffold of the S protein of SARS-CoV-2.
Fig. 1.
An overall structure of the spike protein of SARS-CoV-2 in green cartoon. In this figure, the inserted FCS fragment from Chain B is coloured red. This figure is prepared using PyMol [30] with supplementary file model.pdb as an input. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further investigate the structural impact of the inserted FCS, a set of quantitative SASA analysis (supplementary file model.dssp) by DSSP [27,28] was conducted for all three chains (A, B and C) in the structure of the S protein (supplementary file model.pdb). Specific details are included in Table 4 for residues at FCS as below,
Table 4.
A quantitative analysis by DSSP [27] of the SASA values and the relative SASA values of the FCS residues. The relative SASAs are calculated for each amino acid in the protein by expressing the various summed residue accessible surfaces as a ratio (Value/Reference) of that observed in a ALA-X-ALA tripeptide built using the QUANTA molecular graphics software package [29].
| Residue ID | Chain ID | Residue name | Residue name | Value (Å2) | Reference (Å2) | Value/reference | Notes |
|---|---|---|---|---|---|---|---|
| 681 | A | P | PRO | 40 | 136.13 | 0.29 | Buried |
| 682 | A | R | ARG | 214 | 238.76 | 0.90 | Solvent-exposed |
| 683 | A | R | ARG | 193 | 238.76 | 0.81 | Solvent-exposed |
| 684 | A | A | ALA | 17 | 107.95 | 0.16 | Buried |
| 685 | A | R | ARG | 79 | 238.76 | 0.33 | Buried |
| 681 | B | P | PRO | 112 | 136.13 | 0.82 | Solvent-exposed |
| 682 | B | R | ARG | 175 | 238.76 | 0.73 | Solvent-exposed |
| 683 | B | R | ARG | 214 | 238.76 | 0.90 | Solvent-exposed |
| 684 | B | A | ALA | 59 | 107.95 | 0.55 | Solvent-exposed |
| 685 | B | R | ARG | 60 | 238.76 | 0.25 | Buried |
| 681 | C | P | PRO | 135 | 136.13 | 0.99 | Solvent-exposed |
| 682 | C | R | ARG | 93 | 238.76 | 0.39 | Buried |
| 683 | C | R | ARG | 168 | 238.76 | 0.70 | Solvent-exposed |
| 684 | C | A | ALA | 21 | 107.95 | 0.19 | Buried |
| 685 | C | R | ARG | 149 | 238.76 | 0.62 | Solvent-exposed |
In addition to Table 4, subsequent quantitative analysis demonstrated that the average relative accessibilities of the FCS are 56.54%, 64.55% and 58.93% for all three (A, B and C) chains in the homotrimeric [1] structure of the S protein of SARS-CoV-2, respectively. By and large, this result classifies the polybasic FCS as exposed, instead of buried, in all three chains of the homology structural model (supplementary file model.pdb). This quantitative relative accessibility analysis, along with the visual inspection of Fig. 1, indicates that the net structural consequence of FCS is the insertion of a furin cleavage site into SARS-CoV-2 S protein, leading to a question further: what if the inserted polybasic FCS is actually cleaved by furin protease?
3.3. What if the inserted polybasic FCS is actually cleaved by furin protease?
To answer this question, a similar set of electrostatic interaction analysis [26] was conducted for the trimeric [1] structural model of the S protein, with the difference here being that the whole S protein structure was splitted into two parts: the first part consists of three (chains A, B and C) S1 structural fragments, i.e., before the polybasic FCS, while the second part consists of three (chains A, B and C) S2 structural fragments, i.e., after the polybasic FCS. The results of this FCS-specific electrostatic interaction analysis are included in Table 5, Table 6, Table 7 as below.
Table 5.
FCS-specific inter-part salt bridging analysis of the S protein of SARS-CoV-2 with FCS at the boundary of its S1 and S2 units. In this table, the residue naming scheme is Chain ID_residue name_residue number. In this table, for residues A and B, one of them is located upstream (residue ID smaller than 681) of the polybasic FCS, while the other is located downstream (residue ID larger than 685) of the polybasic FCS.
| PDB File name | Residue A | Atom A | Residue B | Atom B | Distance (Å) |
|---|---|---|---|---|---|
| Model.pdb | A_ARG_319 | NH1 | B_ASP_737 | OD2 | 2.735 |
| Model.pdb | A_ARG_319 | NH1 | B_ASP_745 | OD1 | 3.587 |
| Model.pdb | A_ARG_319 | NH2 | B_ASP_737 | OD2 | 3.982 |
| Model.pdb | B_ARG_319 | NH1 | C_ASP_745 | OD1 | 2.606 |
| Model.pdb | B_ARG_319 | NH1 | C_ASP_745 | OD2 | 3.565 |
| Model.pdb | B_ARG_319 | NH2 | C_ASP_745 | OD1 | 3.325 |
| Model.pdb | B_ARG_319 | NH2 | C_ASP_745 | OD2 | 2.636 |
| Model.pdb | B_LYS_854 | NZ | A_ASP_614 | OD1 | 3.487 |
| Model.pdb | B_LYS_986 | NZ | C_ASP_427 | OD2 | 3.985 |
| Model.pdb | C_ARG_319 | NH1 | A_ASP_745 | OD1 | 3.819 |
| Model.pdb | C_ARG_646 | NH1 | A_ASP_848 | OD2 | 3.643 |
| Model.pdb | C_ARG_646 | NH2 | A_ASP_848 | OD1 | 3.695 |
| Model.pdb | C_ARG_646 | NH2 | A_ASP_848 | OD2 | 2.627 |
| Model.pdb | C_ARG_847 | NH1 | B_GLU_619 | OE1 | 3.034 |
| Model.pdb | C_LYS_854 | NZ | B_ASP_614 | OD1 | 2.686 |
Table 6.
A summary of side chain and main chain hydrogen bonding analysis between the two structural fragments generated by the hypothesized cleavage at the polybasic FCS of the S protein of SARS-CoV-2. In this table, the residue naming scheme is Chain ID_residue name_residue number, ∠ADH represents the angle formed by acceptor (A), donor (D) and hydrogen (H) (∠ADH). In this table, for residues A and B, one of them is to be located upstream (residue ID smaller than 681) of the polybasic FCS, while the other is to be located downstream (residue ID larger than 685) of the polybasic FCS.
| PDB file name | Acceptor (A) | Donor (D) | Hydrogen (H) | D-A (Å) | H-A (Å) | ∠ ADH () |
|---|---|---|---|---|---|---|
| Model.pdb | OD2, B_ASP_737 | NH1, A_ARG_319 | HH12, A_ARG_319 | 2.74 | 1.78 | 15.71 |
| Model.pdb | O, A_ILE_692 | N, A_GLU_654 | H, A_GLU_654 | 2.86 | 1.86 | 5.94 |
| Model.pdb | O, A_THR_696 | OG, A_SER_659 | HG, A_SER_659 | 2.72 | 1.91 | 26.75 |
| Model.pdb | O, B_PRO_863 | N, A_ALA_668 | H, A_ALA_668 | 2.78 | 1.79 | 9.02 |
| Model.pdb | O, B_LEU_864 | N, A_GLY_669 | H, A_GLY_669 | 2.82 | 1.95 | 24.85 |
| Model.pdb | O, A_GLU_654 | N, A_ALA_694 | H, A_ALA_694 | 2.80 | 1.83 | 12.75 |
| Model.pdb | O, A_GLU_661 | OH, A_TYR_695 | HH, A_TYR_695 | 2.62 | 1.68 | 9.06 |
| Model.pdb | O, A_GLY_669 | N, A_MET_697 | H, A_MET_697 | 2.86 | 1.87 | 9.86 |
| Model.pdb | O, A_TYR_660 | N, A_SER_698 | H, A_SER_698 | 2.75 | 1.74 | 2.22 |
| Model.pdb | O, C_ASP_614 | NE, A_ARG_847 | HE, A_ARG_847 | 2.97 | 2.12 | 27.07 |
| Model.pdb | O, C_ASP_614 | NH1, A_ARG_847 | HH11, A_ARG_847 | 2.71 | 1.79 | 19.16 |
| Model.pdb | OD2, C_ASP_614 | OG1, A_THR_859 | HG1, A_THR_859 | 2.83 | 1.95 | 19.70 |
| Model.pdb | O, C_THR_547 | ND2, A_ASN_978 | HD21, A_ASN_978 | 2.99 | 2.12 | 24.76 |
| Model.pdb | O, C_LEU_984 | NZ, B_LYS_386 | HZ3, B_LYS_386 | 2.72 | 1.89 | 28.40 |
| Model.pdb | O, B_ILE_692 | N, B_GLU_654 | H, B_GLU_654 | 2.81 | 1.82 | 9.58 |
| Model.pdb | O, B_ALA_694 | N, B_VAL_656 | H, B_VAL_656 | 2.96 | 2.02 | 17.75 |
| Model.pdb | O, B_THR_696 | OG, B_SER_659 | HG, B_SER_659 | 2.72 | 1.93 | 28.63 |
| Model.pdb | O, C_PRO_863 | N, B_ALA_668 | H, B_ALA_668 | 2.82 | 1.84 | 9.55 |
| Model.pdb | O, C_LEU_864 | N, B_GLY_669 | H, B_GLY_669 | 2.81 | 1.90 | 20.41 |
| Model.pdb | OH, B_TYR_695 | N, B_SER_673 | H, B_SER_673 | 2.96 | 2.09 | 24.82 |
| Model.pdb | O, B_GLU_654 | N, B_ALA_694 | H, B_ALA_694 | 2.77 | 1.83 | 16.90 |
| Model.pdb | O, B_GLY_669 | N, B_MET_697 | H, B_MET_697 | 2.77 | 1.77 | 5.38 |
| Model.pdb | O, B_TYR_660 | N, B_SER_698 | H, B_SER_698 | 2.65 | 1.65 | 3.25 |
| Model.pdb | OD2, A_ASP_614 | OG1, B_THR_859 | HG1, B_THR_859 | 2.98 | 2.16 | 26.02 |
| Model.pdb | O, A_ARG_983 | N, C_SER_383 | H, C_SER_383 | 2.91 | 1.90 | 2.08 |
| Model.pdb | OD2, A_ASP_745 | OG1, C_THR_549 | HG1, C_THR_549 | 2.94 | 2.12 | 25.69 |
| Model.pdb | OD2, A_ASP_848 | NH2, C_ARG_646 | HH22, C_ARG_646 | 2.63 | 1.79 | 27.55 |
| Model.pdb | O, C_ILE_692 | N, C_GLU_654 | H, C_GLU_654 | 2.81 | 1.89 | 19.55 |
| Model.pdb | O, C_THR_696 | OG, C_SER_659 | HG, C_SER_659 | 2.86 | 2.07 | 28.50 |
| Model.pdb | O, A_PRO_863 | N, C_ALA_668 | H, C_ALA_668 | 2.82 | 1.86 | 14.51 |
| Model.pdb | O, A_LEU_864 | N, C_GLY_669 | H, C_GLY_669 | 2.73 | 1.84 | 22.00 |
| Model.pdb | O, C_TYR_695 | N, C_CYS_671 | H, C_CYS_671 | 2.88 | 1.91 | 13.00 |
| Model.pdb | OG, C_SER_689 | OG1, C_THR_676 | HG1, C_THR_676 | 2.79 | 1.86 | 11.33 |
| Model.pdb | O, C_CYS_671 | N, C_TYR_695 | H, C_TYR_695 | 2.78 | 1.80 | 9.89 |
| Model.pdb | O, C_GLU_661 | OH, C_TYR_695 | HH, C_TYR_695 | 2.57 | 1.63 | 8.69 |
| Model.pdb | O, C_GLY_669 | N, C_MET_697 | H, C_MET_697 | 2.91 | 1.93 | 11.90 |
| Model.pdb | OD1, B_ASP_614 | NZ, C_LYS_854 | HZ2, C_LYS_854 | 2.69 | 1.71 | 10.87 |
| Model.pdb | O, B_THR_547 | ND2, C_ASN_978 | HD21, C_ASN_978 | 2.84 | 1.92 | 19.82 |
Table 7.
A summary of side chain hydrogen bonding analysis between the two structural fragments generated by the hypothesized cleavage at the polybasic FCS of the S protein of SARS-CoV-2. In this table, the residue naming scheme is Chain ID_residue name_residue number, ∠ADH represents the angle formed by acceptor (A), donor (D) and hydrogen (H) (∠ADH). In this table, for residues A and B, one of them is to be located upstream (residue ID scriptsizeer than 681) of the polybasic FCS, while the other is to be located downstream (residue ID larger than 685) of the polybasic FCS.
| PDB file name | Acceptor (A) | Donor (D) | Hydrogen (H) | D-A (Å) | H-A (Å) | ∠ ADH () |
|---|---|---|---|---|---|---|
| Model.pdb | OD2, B_ASP_737 | NH1, A_ARG_319 | HH12, A_ARG_319 | 2.74 | 1.78 | 15.71 |
| Model.pdb | OD2, C_ASP_614 | OG1, A_THR_859 | HG1, A_THR_859 | 2.83 | 1.95 | 19.70 |
| Model.pdb | OD2, A_ASP_614 | OG1, B_THR_859 | HG1, B_THR_859 | 2.98 | 2.16 | 26.02 |
| Model.pdb | OD2, A_ASP_745 | OG1, C_THR_549 | HG1, C_THR_549 | 2.94 | 2.12 | 25.69 |
| Model.pdb | OD2, A_ASP_848 | NH2, C_ARG_646 | HH22, C_ARG_646 | 2.63 | 1.79 | 27.55 |
| Model.pdb | OG, C_SER_689 | OG1, C_THR_676 | HG1, C_THR_676 | 2.79 | 1.86 | 11.33 |
| Model.pdb | OD1, B_ASP_614 | NZ, C_LYS_854 | HZ2, C_LYS_854 | 2.69 | 1.71 | 10.87 |
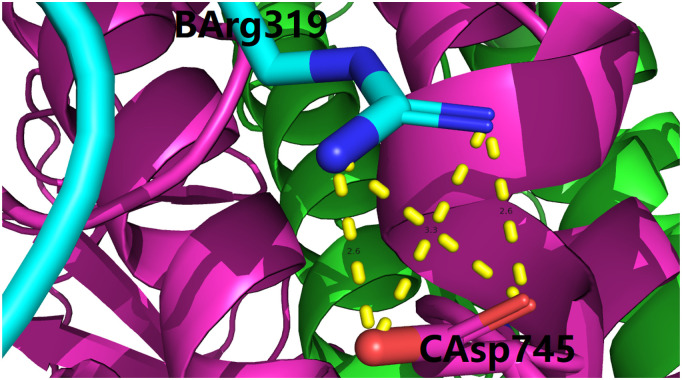
From Table 5, Table 6, Table 7, it is clear that a series of electrostatic interactions (including both salt bridges and hydrogen bonds) still exist at the interface of the two parts of the S protein generated from the hypothesized cleavage by furin proteases. For instance, a total of four inter-chain salt bridges still exist at the interface of chains B and C of the S protein, as shown in Fig. 2 . For another instance, not only does Arg319 of chain A form three inter-chain salt bridges with Asp737 and Asp745 of chain B (Table 5), Arg319 of chain A also forms a side chain hydrogen bond with Asp737 of chain B (Table 7). Among all electrostatic interactions listed in Table 5, Table 6, Table 7, these seven inter-chain salt bridges and one side chain hydrogen bond are merely two examples, which constitute a set of strong electrostatic forces towards the structural stabilization of the overall homotrimeric [1] structure of the S protein, even if the inserted polybasic FCS is actually cleaved by furin protease either before or after viral cell entry.
Fig. 2.
Four inter-chain salt bridges formed between basic residue pair B_ARG_319 and C_ASP_745 (Table 5). This figure is prepared using PyMol [30] with supplementary file model.pdb as an input. In this figure, the three (A, B and C) chains of the spike protein of SARS-CoV-2 is shown as green, cyan and purple cartoons, the residue naming scheme is Chain ID, residue name, residue number, the four salt bridges are shown here with four dotted yellow lines, with the lengths of the four inter-chain salt bridges being 2.6, 2.6, 3.3 and 3.5 Å, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusion
Incorporating currently available structural data (as of Thu Jun 25 09:39:402020) of the S protein [22], and in the hope of pushing a little bit forward the boundary of our scientific knowledge on COVID-19, this short article presents a comprehensive structural characterization of the FCS inserted into the S protein of SARS-CoV-2, and puts forward a set of structural analysis as below,
-
1.
the polybasic FCS is only involved in set of weak electrostatic interactions, and is therefore not able to alter, neither stabilize nor de-stabilize, the overall scaffold of the S protein of SARS-CoV-2.
-
2.
the polybasic FCS is spatially located at a random coil loop region, mostly distantly solvent-exposed (Fig. 1, instead of deeply buried), with no structural proximity to the other part of the S protein of SARS-CoV-2.
-
3.
the S1 and S2 subunits of the S protein will still be strongly bonded together, at least electrostatically [31] from a structural and biophysical point of view, even if the FCS is actually cleaved by furin protease.
5. Discussion
Quite recently, it has been reported that this polybasic FCS is essential for SARS-CoV-2 to infect human lung cells, and that campaigns to develop therapeutics against SARS-CoV-2 should include the evaluation of furin inhibitors [14,16]. In view of the reported in vitro functional relevance of FCS [16], it is postulated here that the polybasic FCS does need to be cleaved by furin protease, leading to a different action mechanism of SARS-CoV-2 from its siblings, where its S1 and S2 subunits (while still strongly bonded together) undergoes a major structural rearrangement before or after viral cell entry. Nevertheless, in the midst of this COVID-19 pandemic [32] with more than 5.5 million (as of Thu Jun 25 09:39:402020) confirmed cases globally, this short article puts forward a set of analysis that the net structural consequence of FCS here is the insertion of a furin cleavage site into the S protein of SARS-CoV-2, and is thus of only limited structural biophysical relevance here. Finally, along with [9], the structural biophysical analysis here makes even more unlikelier a purposeful-manipulation-based hypothesis of the origin of SARS-CoV-2.
Appendix A. Supplemetary data
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 feb;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shang J., Ye G., Shi K., Wan Y.S., Aihara H., Li F. Structure of 2019-nCoV chimeric receptor-binding domain complexed with its receptor human ACE2. Worldwide Protein Data Bank. 2020 doi: 10.2210/pdb6VW1/pdb. [DOI] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 feb;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020 feb;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A. 2020 feb. Severe acute respiratory syndrome-related coronavirus: The species and its viruses – a statement of the Coronavirus Study Group. [Google Scholar]
- 6.Jiang S., Shi Z., Shu Y., Song J., Gao G.F., Tan W. A distinct name is needed for the new coronavirus. Lancet. 2020 mar;395(10228):949. doi: 10.1016/S0140-6736(20)30419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020 feb;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian S., Huang Q., Fang Y., Wu J. FurinDB: a database of 20-residue Furin cleavage site motifs, substrates and their associated drugs. Int. J. Mol. Sci. 2011 Feb;12(2):1060–1065. doi: 10.3390/ijms12021060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020 mar;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han X., Bertzbach L.D., Veit M. Mimicking the passage of avian influenza viruses through the gastrointestinal tract of chickens. Vet. Microbiol. 2019 dec;239:108462. doi: 10.1016/j.vetmic.2019.108462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tse L.V., Hamilton A.M., Friling T., Whittaker G.R. A novel activation Mechanism of avian influenza virus H9N2 by Furin. J. Virol. 2013 nov;88(3):1673–1683. doi: 10.1128/JVI.02648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020 jan;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. 2020 feb. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020 Apr;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleine-Weber H., Elzayat M.T., Hoffmann M., Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018 Nov;8(1) doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the Spike protein of SARS-CoV-2 is essential for infection of human lung Cells. Mol. Cell. 2020 May;78(4):779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020 feb;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menachery V.D., Dinnon K.H., Yount B.L., McAnarney E.T., Gralinski L.E., Hale A. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 2019 dec;94(5) doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T., Wu Q., Zhang Z. 2020 feb. Pangolin homology associated with 2019-nCoV. [Google Scholar]
- 20.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 feb;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong M.C., Cregeen S.J.J., Ajami N.J., Petrosino J.F. 2020 feb. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. [Google Scholar]
- 22.Berman H., Henrick K., Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003 dec;10(12) doi: 10.1038/nsb1203-980. 980–980. [DOI] [PubMed] [Google Scholar]
- 23.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018 May;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W. 2020 Jan. Visualising the Experimentally Uncharted Territories of Membrane Protein Structures inside Protein Data Bank. [Google Scholar]
- 25.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Li W. How do SMA-linked mutations of SMN1 lead to structural/functional deficiency of the SMA protein? PLoS One. 2017 jun;12(6) doi: 10.1371/journal.pone.0178519. e0178519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr A. 1976;32:922–923. [Google Scholar]
- 29.Hubbard S.J., Thornton J.M. Vol. 2. Department of Biochemistry and Molecular Biology, University College London; 1993. Naccess. Computer Program. [Google Scholar]
- 30.DeLano W.L. Vol. 40. 2002. Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsletter On Protein Crystallography; pp. 82–92. [Google Scholar]
- 31.Anfinsen C.B. Principles that govern the folding of protein chains. Science. 1973 Jul;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 32.Myers E.M. 2020 Apr. Compounding Health Risks and Increased Vulnerability to SARS-CoV-2 for Racial and Ethnic Minorities and Low Socioeconomic Status Individuals in the United States. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material