Abstract
Protein aggregation is a widespread phenomenon that stems from the establishment of non-native intermolecular contacts resulting in protein precipitation. Despite its deleterious impact on fitness, protein aggregation is a generic property of polypeptide chains, indissociable from protein structure and function. Protein aggregation is behind the onset of neurodegenerative disorders and one of the serious obstacles in the production of protein-based therapeutics. The development of computational tools opened a new avenue to rationalize this phenomenon, enabling prediction of the aggregation propensity of individual proteins as well as proteome-wide analysis. These studies spotted aggregation as a major force driving protein evolution. Actual algorithms work on both protein sequences and structures, some of them accounting also for conformational fluctuations around the native state and the protein microenvironment. This toolbox allows to delineate conformation-specific routines to assist in the identification of aggregation-prone regions and to guide the optimization of more soluble and stable biotherapeutics. Here we review how the advent of predictive tools has change the way we think and address protein aggregation.
Abbreviations: A3D, AGGRESCAN3D; APRs, Aggregation-prone regions; DI, Developability index; IAPP, Islet amyloid polypeptide; IDPs, Intrinsically disordered proteins; mAbs, Monoclonal antibodies; SAP, Spatial aggregation propensity; STAP, STructural Aggregation-Prone region
Keywords: Protein aggregation, Bioinformatics, Amyloid, Protein structure, Proteomics, Evolution, Protein production
1. Introduction
Proteins are the ultimate and essential cellular players in almost all biological processes, coordinating different functions inherent to life through the establishment of molecular networks in the overcrowded cellular milieu [1]. This activity is mediated by specific inter-molecular interactions that finely regulate protein homeostasis and functioning [2]. In contrast, non-native protein–protein interactions can prompt aberrant oligomerization and ultimately protein aggregation, a process associated with the onset of a wide range of human disorders -including Alzheimer’s, Parkinson’s diseases and type II diabetes [3], [4]. Such detrimental property is not restrained to disease-related proteins but widespread and is considered a generic trait of polypeptides chains. Indeed, protein aggregation constitutes a significant bottleneck in the production of protein-based therapeutics, compromising their recombinant expression, downstream processing, and biosafety, precluding the marketing of otherwise promising biotherapeutics [5], [6]. It is therefore essential to elucidate the molecular determinants of protein aggregation, its biological connection with native functions, and its role in shaping protein evolution since such knowledge would translate into advances in biomedicine and biotechnology.
Proteins aggregate in variety of physicochemical diverse supramolecular assemblies ranging from highly ordered cross-β amyloid fibrils, formed by a repetitive array of monomers disposed orthogonally to the fibril axis, to less ordered amorphous deposits [7]. In between, an array of oligomeric and protofibrillar structures display intermediate structural properties. The same polypeptide chain can form fibrillar or amorphous assemblies depending on its microenvironment [7], [8], [9], [10]. Indeed, the molecular determinants driving the formation of fibrils in amyloidosis and less ordered aggregates during biologics production overlap significantly, making difficult to predict if the assembly of a given protein in a given condition will lead to one or other kind structure, or something in between [7]. Additionally, there exist a number of proteins, known as functional amyloids, that exploit the amyloid fold to perform their physiological activities [4], [11], [12]. For instance, functional amyloids are involved in curli-mediated biofilm formation in E. coli [13], hypersensitive response activation in plants [14], melanin polymerization in mammalian cells [15], and hormone storage in humans [16]. Functional and non-functional aggregation have evolved under selective pressures of different signs, which favored and disfavored them, respectively [17], [18], [19]. This review focus on undesired aberrant protein aggregation and how it can be predicted and modulated.
Great efforts have been devoted to the analysis and characterization of specific aggregation-prone proteins, which crystallized into a robust theoretical comprehension of the protein aggregation phenomenon [20], [21]. Yet, it remains challenging to translate these learned rules to previously uncharacterized proteins, and their study requires the dedicated and manual inspection of their sequences and folds.
Fortunately, in silico approaches have evolved hand-in-hand with the development of the field, and have become powerful platforms to systematically project our empirical and theoretical knowledge into unstudied protein sequences or structures [22], [23]. To date, more than 30 algorithms have been implemented to deal with protein aggregation, allowing to identify aggregation determinants, predict the effect of disease-related mutations, and assist in the redesign of protein solubility [23], [24]. Each of these programs relies on different principles and assumptions and face the aggregation conundrum from diverse perspectives. This diversity provides us with a versatile toolbox to orthogonally combine the outputs of conceptually different algorithms and adapt the predictive strategy to the intended purpose. Noteworthy, these predictive tools allow for the fast evaluation of extensive collections of protein variants or even complete proteomes, which has contributed substantially to illuminate the connection between protein function and aggregation while uncovering aberrant aggregation as an important constrain of protein evolution [25], [26], [27].
In this article, we review some of the most critical biocomputational advances that have contributed to our present understanding of the constraints shaping non-functional protein aggregation in living organisms, helping to provide biological context for the protein aggregation phenomenon. We define a framework for predicting protein aggregation, taking into account that function and aggregation are often two sides of the same coin. We intend to provide a comprehensive compendium of strategies that can be adapted to any specific protein of interest. We end up illustrating the potential of state-of-the-art algorithms to assist in the design and control of the solubility of proteins of biotechnological interest.
2. Proteome-wide analysis: A biological framework for protein aggregation
The implementation of predictive tools with the ability to systematically analyze extensive collections of proteins has allowed extending the analysis of aggregation to complete proteomes, resulting in a deeper understanding of the molecular determinants that govern protein aggregation while revealing crosstalk between protein evolution and aggregation [28], [29], [30]. Different computational proteome-wide analyses converged in the identification of aggregation-prone regions (APRs) similar to those identified in disease-linked proteins across all kingdoms of life [31]. The presence of these sequence stretches is not anecdotic since an average of one APR per protein was detected, independently of the considered proteome. APRs have been identified in proteins with different conformational properties: intrinsically disordered, globular, transmembrane, or oligomeric proteins. Overall, these computational studies suggest that APRs are ubiquitously present in nature and are an intrinsic trait of proteins, despite being potentially harmful. Of note, the predicted aggregation propensity of proteomes substantially decreases with increasing complexity and longevity of organisms, which seems to point to an evolutionary pressure acting against protein aggregation [26], [31]. Yet, the aggregation phenomena persist, indicating that negative selection cannot wholly abrogate it.
2.1. Defining the interplay between functional and aberrant contacts.
Cells have evolved a complex network of quality control mechanisms to mitigate protein aggregation in order to maintain protein homeostasis. Such strategies consume significant cellular energy [32]. Thus, the omnipresence of APRs in proteins does not only endorse them with the risk of sporadic aggregation but constitutively drain a substantial amount of cellular resources. It is then shocking that despite millions of years of evolution sharpening protein sequences and structures, protein aggregation has not been purged out from polypeptides. This resilience against negative natural selection has been interpreted as an indication of APRs being essential to develop certain biological functions [33]. Multiple experimental and computational studies converge to demonstrate that the physicochemical determinants of protein aggregation substantially overlap with those responsible for the establishment of native intra- and intermolecular contacts (i.e., substrate binding, protein folding, or protein–protein interactions) [34], [35], [36], [37]. This observation can be easily understood by considering that regions responsible for native contacts are usually hydrophobic and prone to establishing hydrogen-bonded networks, two features that also favor the non-native interactions that ultimately lead to protein aggregation. In globular proteins, the evolutionary suppression of APRs is often restrained by the need for a densely packed hydrophobic core to maintain their native fold [38], [39]. The interface between protein sub-units or protein complexes is also enriched in hydrophobic residues that stabilize the quaternary structure. Accordingly, protein folding, stability, and aggregation are in a close interplay in globular proteins, being governed by the same molecular features, but differently weighted [35], [40].
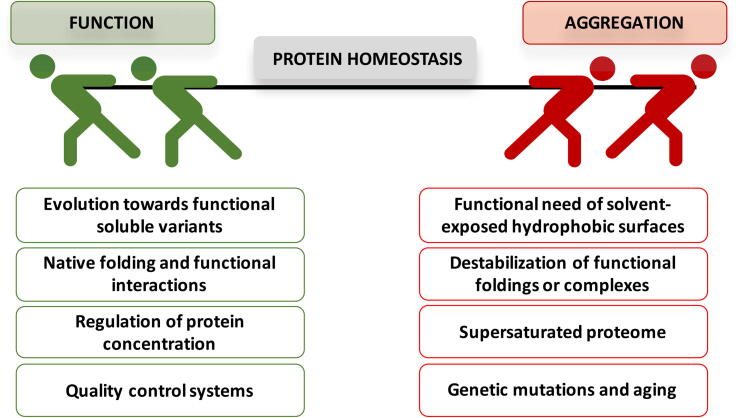
Computational algorithms have contributed substantially to clarify the function-aggregation interplay by assisting the experimental characterization of some archetypical protein examples. In this way, in the human Josephin domain, the residues with higher contributions to its predicted aggregation propensity are also fundamental for the ubiquitin-binding activity of the protein [41]. Solubilizing mutations affecting those residues lead to a concomitant loss of activity. Likewise, the aggregation of the human SUMO protein repertoire (SUMO1, SUMO2, and SUMO3) is directed by specific regions that overlap with SUMO functional interfaces [42]. All in all, protein regions accounting for the protein functionality might -under some circumstances- lead to aberrant contacts and eventually to protein aggregation, depicting an intrinsic competition between these two opposed reactions (Fig. 1).
Fig. 1.
Innate competition between functional interactions and protein non-functional aggregation. Several factors contribute to balancing this subtle equilibrium.
2.2. Computational identification of the evolutive strategies constraining protein aggregation
As discussed, almost every polypeptide is endorsed with an inherent risk to suffer aggregation and, potentially compromise cellular fitness. However, it is also true that proteins remain soluble and functional in their natural contexts, and only under certain conditions (i.e., mutations, gene duplications, or aging), a reduced set of proteins are found accumulated into insoluble deposits in vivo [4], [43]. Thus, it becomes clear that if APRs cannot be skipped from protein sequences and structures, alternative evolutionary strategies must have emerged to cope with their presence. Chaperones, co-chaperones, and degradation systems constitute the first line of defense against aggregation. In addition to this protein quality control machinery, large-scale computational analysis identified other submerged regulation mechanisms to counterbalance the unavoidable aggregation propensity of proteins [30], [31]. These strategies are adapted to the protein size, half-life, in-cell relative concentration, sub-cellular location, and translation rate.
Correlations between protein size and foldability report that longer and multi-domain proteins usually fold slower than short and single-domain polypeptides [44]. Hypothetically, if all proteins harvest equivalent aggregation loads, longer sequences would be more susceptible to aggregate as a result of more prolonged exposure of their APRs to solvent [31], [45]. Computational studies of bacterial proteomes revealed that single-domain proteins (below 20 kDa) could accommodate a higher aggregation load; the average aggregation propensity decreases with increasing protein lengths [26]. Complementarily, the principal bacterial chaperones GroEL, and DnaK bind preferentially to substrates above the 20 kDa limit [46]. The same trend has been reported for the human proteome, highlighting the conservation of a control mechanism aimed to cope with the theoretical increased risk of aggregation associated with longer proteins [25]. Aggregation propensity is also connected to protein turnover; De Baets and co-workers analyzed 611 protein sequences and their lifetimes with the aggregation predictor TANGO [47]. They show that short-living proteins can accommodate a higher aggregation load since their time window to misfold and aggregate is smaller than that of proteins with higher lifetimes.
The analysis of the aggregation propensity of proteins in different cellular compartments has documented a connection between protein location and aggregation in yeast, bacterial, and human proteomes [25], [26], [48]. Tartaglia and co-workers proposed that protein solubility correlates with the volume of the cellular compartment they populate: in smaller subcellular locations proteins display lower aggregation tendencies, likely a strategy to prevent abnormal interactions in more crowded environments [49]. Sequences of proteins that undergo secretory pathways or residing in the periplasm are, on the average, more soluble, probably because they are more exposed to extracellular stresses and have little access to protective chaperones, which are significantly depleted in those environments [25], [26].
Elevated protein expression has been traditionally linked with the formation of aberrant protein deposits, either in conformational disorders or during recombinant expression [50], [51], [52]. According to the law of mass action, the probability of establishing non-functional interactions scales with the protein concentration [53]. Certainly, in contrast to protein folding, aggregation is a second or higher-order reaction, being strongly dependent on protein abundance. Several proteomics analyses revealed that there exists an anti-correlation between gene-expression and/or protein abundance and predicted aggregation propensity in bacteria, nematodes, and humans [49], [54], [55], [56]. In essence, protein abundance in the cell is tightly regulated to attain optimal levels, sufficient for proteins to remain soluble and functional, but not more than that. Vendruscolo and co-workers have extended this hypothesis by proposing that “supersaturated” proteins -a subset of proteins living above their solubility limit- conform a metastable subproteome inherently exposed to aggregation [57], [58], [59]. They suggest that after the primary aggregation of the disease-causing amyloid proteins -i.e., Aβ42 or α-synuclein-, “supersaturated” proteins are collaterally more exposed to aggregation, expanding the dysfunction to other unrelated biological pathways and prompting a general collapse of cellular functions.
Computational analyses also revealed a link between protein aggregation and function [26], [48], [60]. Chen and co-workers demonstrated that essential proteins from three eukaryotic organisms -yeast, fly, and nematode- are the subject of a higher evolutionary pressure against protein deposition [60]. Similarly, in bacteria, operons that encode essential proteins or functions display lower aggregation loads [26]. It is not surprising that proteins in the same operon display similar aggregation propensities since they share common gene-expression regulation and thus abundance, all working in related functions.
The above-described relationships have been established by analyzing aggregation over protein sequences, without taking into account the modulation of these properties by the structural context in the folded states in which proteins spend the majority of their lifetime. Performing an equivalent structure-based proteomic analysis is not trivial since the available protein structures for a given proteome are limited, and their analysis requires significant computation time. Despite these limitations, in a recent work, we analyzed a fraction of the structurally characterized Escherichia coli proteome to explore whether, in addition to sequence properties, structural aggregation might also influence the evolution of bacterial proteins [61]. Our analysis revealed that the aggregation features of protein surfaces and interfaces in folded states are constrained according to the protein abundance, length, essentiality, subcellular location, and function. This observation indicates that protein structures would have also evolved to minimize the risk of aggregation in their natural environments.
3. Prediction of protein aggregation from different native conformations
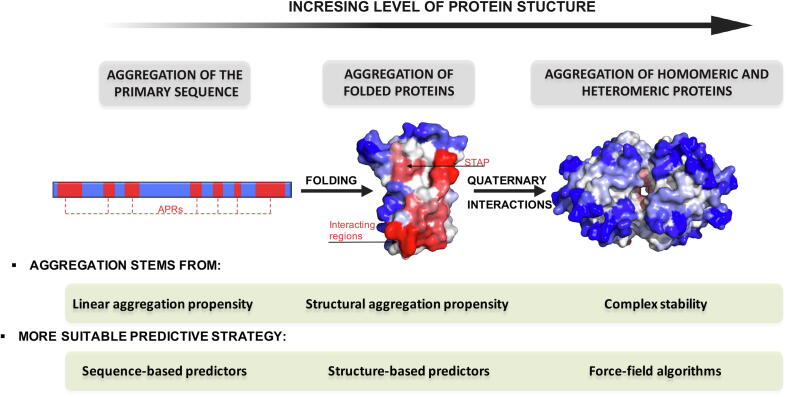
The previous section illustrates how protein aggregation cannot be understood without considering the folding, functional purpose, and cellular environment of a protein. In each conformational state, the risk of aggregation stems from different sources; globular proteins, IDPs, and oligomeric proteins pose different challenges that need to be addressed with dedicated tools. Therefore, in order to anticipate protein aggregation successfully, we need to adapt our computational scheme to the particular properties of the protein under study. Such a task can be difficult for untrained users since an in-depth knowledge of the available computational tools is needed. In this section, we apply the insights provided by proteome-wide analysis to classify and review a state-of-the-art collection of predictive tools. The aim is to establish a systematic framework for evaluating protein aggregation that can be adapted to the intended predictive purpose (Fig. 2).
Fig. 2.
Computational strategies to predict protein aggregation. In each folding state, aggregation is driven by different molecular determinants, delimiting the best-performing predictive strategy in each particular case. Aggregation-prone residues are colored in red and solubilizing amino acids in blue. APR and STAP designate Aggregation-Prone Regions and STructural Aggregation-prone Regions, respectively. PDB structures correspond to monomeric and tetrameric transthyretin (PDB: 1F41). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. Sequence-based predictors
The first generation of computational algorithms designed to predict protein aggregation is based on the identification of linear APRs across the polypeptide sequence. The conceptual pillars of these algorithms are the theoretical and experimental studies that allowed the definition of the main molecular determinants of aggregation. To date, more than 20 sequential algorithms have been developed [23], [62]. Their design exploits the evidence that protein aggregation is driven by short and well-defined sequential stretches -referred to as APRs- characterized by a high hydrophobicity, low net charge, and a remarkable preference to adopt β-sheet secondary structure [23]. However, each algorithm relies on different interpretations and weighting of the essential features driving aggregation, which allows the orthogonal combination of conceptually different algorithms to reduce method-specific biases. These algorithms can be divided into four subclasses according to their underlying principles, further described in Table 1. Briefly, the first class of algorithms, known as phenomenological, employs experimental data to define the determinants of aggregation and provide empiric aggregation scales. They include software such as AGGRESCAN, and Zyggregator, both based on the rationalization of experimentally determined factors influencing protein aggregation [63], [64], [65]. The second class of computational approaches relies on the theoretical assessment of sequence properties known to be associated with aggregation. TANGO, PASTA 2.0, FoldAmyloid, Waltz and Amyloid Mutants belong to this second class, and they evaluate the tendency of a sequence to adopt a defined β-enriched conformation, the packing density of proteins, the composition and patterning of their residues or the suitability to adopt the topologically restricted conformations that characterize amyloid-like states [66], [67], [68], [69]. A growing number of machine learning methods are being developed. They exploit the potential of neural networks to identify sequential features highly correlated with aggregation [70]. Machine learning approaches attain performances comparable -or even higher- to traditional predictors. APPNN, netCSSP or FISH Amyloid are some examples of this kind of algorithms [71], [72], [73]. Finally, a last group of software is the one based on the combination and weighting of the outputs of other predictors (either phenomenological or theoretical) into one single output. These consensus predictors assemble the different concepts behind each predictor to increase robustness while reducing method-specific biases. AMYLPRED 2 and MetAmyl exemplify this kind of software [74], [75].
Table 1.
Sequence based-prediction methods, according to the rationale behind their analysis. *Registration prior to analysis is required.
| Method* | Underlying rationale | Webserver, software or equation |
|---|---|---|
| Phenomenological methods | ||
| AGGRESCAN [63], [64] | Prediction is assayed against an aggregation propensity scale for the 20 proteinogenic amino acids derived from in vivo experiments. | http://bioinf.uab.es/aggrescan/ |
| Zyggregator [65] | Prediction of a 21-residue sliding window from an equation accounting for hydrophobicity, secondary structure propensity, and net charge built upon changing aggregation rate on mutations.It also considers the presence of gatekeeper residues or hydrophobic patches | http://www-mvsoftware.ch.cam.ac.uk/index.php/login* |
| Theoretical methods | ||
| TANGO [7], [117] | Evaluation of the population of random coil, native conformation or aggregated species from empirically and statistically derived conformational amino acidic preferences, along with physico-chemical variables. | http://tango.crg.es/* |
| PASTA 2.0 [66], [118] | Energetic function derived from high-resolution protein structures, which considers interaction potential and H-bond formation between all non-consecutive residues for parallel and anti-parallel β-pairing. | http://protein.bio.unipd.it/pasta2/ |
| FoldAmyloid[67] | A protein structure derived scale; from the notion that hydrophobic stretches exhibit higher “packing density” and H-bonding propensity. | http://bioinfo.protres.ru/fold-amyloid/ |
| WALTZ[68] | Application of a position specific matrix derived from a large group of hexapeptides, for predicting amyloid-like formation. | https://waltz.switchlab.org/ |
| Pafig [119] | Analysis of six-residue sliding window for a scale derived from machine supervised learning over 531 physicochemical properties, which led to best discrimination using 41 of them. | Code can be downloaded from their web page http://www.mobioinfor.cn/pafig/ (Requires MS Windows) |
| Betascan [120] | Evaluation of β-strand pairing propensity, obtained from probabilities of residues to be H-bonded in amphiphilic β-sheets. | http://cb.csail.mit.edu/cb/betascan/ hosts the web server and allows download of the Perl scipt. |
| GAP [121] | Discriminates amyloid-like or β-amorphous hexapeptides from position-specific pairing frequencies. | https://www.iitm.ac.in/bioinfo/GAP/ |
| 3D Profile [122] | Energetic impact on the spatial accommodation to the backbone of the fibril forming Sup35 hexapeptide is assessed. | http://services.mbi.ucla.edu/zipperdb/* |
| Machine learning methods | ||
| APPNN [71] | Machine learning approach based on the analysis of seven physicochemical and biochemical features such as β-sheet frequency, hydrophobic moment, helix termination parameters or isoelectric point. | http://cran.r-project.org/web/packages/appnn/index.html |
| NetCSSP [72] | Analysis of contact-dependent secondary structure prediction to identify hidden β-propensities. | http://cssp2.sookmyung.ac.kr/ |
| FiSH Amyloid [73] | Classification of amyloidogenic stretches based on co-ocurrence patterns in protein sequences. | http://www.comprec.pwr.wroc.pl/COMPREC_home_page.html |
| Consensus methods | ||
| AmylPred2 [74] | Generates consensus predictions over 11 algorithms but allows user-customized predictions as some methodologies can have a certain degree of redundancy, thus biasing the consensus prediction. | http://aias.biol.uoa.gr/AMYLPRED2/* |
| MetAmyl [75] | Score is obtained applying a linear combination of four predictors’ (which showed lower redundancy) outcome, weighting the individual contribution of each method. | http://metamyl.genouest.org |
This list intends to be illustrative and not to provide an extensive enumeration and description of all available methods. Programs in this list are not necessarily more accurate than those absent.
The aforementioned programs are just our lab selection among the more than 20 available algorithms that have demonstrated their efficacy in the study of disease-related proteins, allowing to discretize the experimentally relevant sequence stretches driving their aggregation [4], [23], [29], [31]. They are these tools that allowed for most of the above aforementioned proteome-wide analysis [31]. However, they do not evaluate the modulation of sequential aggregation determinants imposed by the three-dimensional conformation of the protein. This drawback defines the particular scenarios in which their predictions are particularly accurate: (i) Intrinsically disordered proteins; these proteins lack a defined three-dimensional structure and fluctuate between multiple transients unfolded or partially folded conformations. During these dynamic fluctuations, APRs are accessible to the solvent, without significant structural protection. (ii) After being synthesized in the ribosome, proteins depart from an extended conformation that transits towards the folded state. During this process, APRs are also exposed to the solvent. This situation is of particular relevance during protein overexpression, where the transient concentration of unfolded polypeptide chains increases dramatically (iii) Dynamic fluctuations or destabilization of protein structures may result in the partial unfolding, exposing previously hidden APRs.
Therefore, the straightest application of sequence-based algorithms is the prediction of IDPs aggregation. IDPs are particularly depleted in APRs since the compositional bias of disordered proteins inherently protects them from aggregation. In essence, they contain a significant proportion of protective residues (Asp, Arg, Glu, Gly, Lys, and Pro), so-called gatekeepers, that are difficult to accommodate in a β-sheet aggregated state [7], [76], [77], [78]. However, IDPs are not entirely protected against aggregation, and this stems from their innate biological functions. IDPs are generally involved in multiple protein–protein interactions, acting as signal integrators and master regulators of diverse biological processes [79]. Such activity entails the presence of short molecular recognition motifs that must be at least transiently exposed to the solvent to find their suitable binding partner. Those motifs retain an intrinsic hydrophobicity, and under pathogenic conditions, they may act as cryptic APRs, triggering aberrant interactions and finally, protein aggregation. Accordingly, computationally identified APRs in IDPs tend to overlap with interaction motifs; in a recent work, we have computationally identified and characterized the aggregation of one of such sequence stretches [80]. Aβ42, α-synuclein or IAPP are some examples of IDPs whose aggregation is associated with human disorders [4], [81]. In these proteins, different algorithms consistently predict APRs that overlap with the regions identified to drive the aggregation in vitro and lately to be part of the core that sustains the respective amyloid fibrils structure [82], [83], [84], [85].
3.2. Structure-based algorithms
Structure-based algorithms were born as a second generation of software designed to translate the predictive potential of sequence-based algorithms to globular proteins. In folded proteins, the three-dimensional arrangement of the polypeptide chain significantly reshapes the molecular determinants of aggregation, weighting the contribution of linear APRs [86]. Sequence-based algorithms are blinded to these effects, which usually result in overprediction when they are used to forecast the aggregation propensity of folded proteins. In this way, the architecture of globular proteins buries the hydrophobic residues inside the protein core, blurring the exposition of linear APRs to solvent. Thus, once a protein is folded into its compact tertiary structure, those APRs do not contribute to protein aggregation, although sequence-based algorithms would predict the contrary. Moreover, in a folded protein, neighbor residues do not need to be consecutive in sequence, and structural clustering of non-consecutive hydrophobic residues in the surface or interface of the protein might occur. Those solvent-exposed hydrophobic patches, known as STructural APRs (STAP), are usually crucial for the protein activity, as previously discussed for the Josephin domain and the SUMO proteins. This is also the case of antigen-recognition elements in antibodies that exhibit a relatively high exposed hydrophobicity required for target binding [87]. Of course, STAPs cannot be identified by linear predictors.
Structure-based algorithms use three-dimensional protein coordinates in order to evaluate the aggregation of proteins in their native fold and overcome the limitations mentioned above. Herein, we briefly describe the principles of four of the more popular structure-based algorithms. Their applications to the redesign of therapeutic proteins will be further reviewed in section 4.
SolubiS constitutes one of the most instinctive evolutions of a linear predictor to evaluate the structural context [88]. SolubiS identifies linear APRs -using the TANGO sequence-based algorithm- and applies the FOLDX force field to evaluate their contribution to global protein stability [89]. The result is an algorithm able to analyze the structural context and the relative shielding of a given APR and provide an estimation of the tendency of such APR to be solvent-exposed and thus become aggregation competent. SolubiS has successfully forecasted the behavior of model globular proteins in vitro and is an excellent tool to evaluate the impact of APR sequential variations in the solubility of the protein. Nonetheless, since SolubiS was build using a sequence-based predictor, this software is still blinded to the emergence of STAPs. SAP (spatial aggregation propensity) was the first algorithm designed with this objective, being able to identify hydrophobic patches exposed to solvent in globular proteins [90]. SAP uses a structurally corrected hydrophobicity scale as a proxy for protein aggregation and considers that the aggregation contribution of a given side chain is modulated for the residues in the vicinity. Under that premise, SAP defines a 5 Å radius sphere centered in the analyzed atom and evaluates the spatial contribution of each nearby residue as the product of the solvent-accessible area of the atoms within the sphere. In this way, SAP analyzes the local hydrophobicity of solvent-exposed residues regardless of their sequence positions. The Developability Index algorithm (DI) is an adaptation of SAP to predict the aggregation of monoclonal antibodies (mAbs) based on their structure in a faster and more accurate way [91]. DI includes the effect of electrostatic interactions, which have an essential role in solubility, counterbalancing the contribution of hydrophobicity. Aggrescan3D (A3D) is another example of structure-based algorithms, implementing the sequence-based AGGRESCAN aggregation scale to assess the aggregation of protein structures [92], [93]. A3D is conceptually similar to SAP, but it introduces an experimentally derived aggregation scale instead of using hydrophobicity. A3D calculates the aggregation propensity of each residue by computing its intrinsic aggregation propensity, which is also corrected by its solvent-exposure and properties of the side chains in the vicinity. The main particularity of A3D, when compared with other structure-based algorithms, is its capacity to assess the impact of dynamic structural fluctuations of protein structures and its effect on the exposition of APRs. The A3D “dynamic mode” uses the CABS-flex force field -based on high resolution coarse-grained molecular dynamics simulations- to reproduce the dynamism and plasticity of protein structures in their native states [94], [95]. For each particular conformer generated by CABS-flex, A3D computes its structural aggregation propensity. In this way, A3D allows the identification of dynamic aggregation-prone regions that are otherwise protected in the static PDB deposited structure [96]. Another structure-based approach has been developed by Sormanni and coworkers [97] The CamSol method applies the physicochemical principles implemented in the sequence-based predictor Zyggregator and performs structural corrections similar to those of SAP or A3D to compute the solubility of native protein structures [65].
3.3. Oligomeric proteins
The computational analysis of native oligomeric assemblies revealed that, as a general trend, protein–protein interfaces display higher aggregation propensities than the solvent-exposed surfaces of globular proteins [37]. Indeed, a remarkable overlap between interaction surfaces and APRs has been identified in oligomeric proteins associated with conformational disorders, indicating that the functional interaction of the monomeric subunits is associated with an intrinsic risk of aggregation. This exposed hydrophobicity in monomeric subunits becomes masked once they are incorporated in the quaternary structure. Accordingly, in disease-linked oligomeric proteins, pathological mutations usually impact the complex stability, favoring the dissociation of the aggregation-prone monomeric constituents. This is the case of transthyretin and SOD1, for which the disentangle of the quaternary structure is the rate-limiting step in the downhill polymerization process that drives their aggregation; once the tetramer is destabilized, aggregation becomes highly favorable [98], [99]. Besides, upon disassembly, the monomeric subunits become “supersaturated,” in comparison with the respective multimeric protein, which exacerbates the probability of spurious intermolecular contacts. For these proteins, in addition to STAP detection and scoring, accurate predictions of aggregation should also evaluate the thermodynamic stability of the assembled protein and how amino acid substitutions impact both factors. In this context, the FOLDX force field is useful for the identification of both stabilizing and destabilizing mutations associated with the aggregation of oligomeric proteins [89].
4. Aggregation in the biotechnological production of protein-based therapeutics.
The use of recombinant proteins for therapeutic applications offers a compelling alternative to small molecules. Proteins are capable of performing highly specific and intricate functions, which is impossible for small molecule drugs. The high specificity of proteins also results in less drug toxicity through interference with normal body processes. Aggregation is a significant concern in the production of protein therapeutics, and it may jeopardize the viability of the complete biotechnological process [5], [100], [101]. First, because aggregation reduces production yields, but most importantly because aggregates have the potential to trigger immunogenic responses upon administration, threatening patients' health [102], [103]. Consequently, biotech companies allocate extensive funding and resources to mitigate protein aggregation in their production pipelines; often by undertaking expensive trial/error screenings of conditions that may or may not result in a significant improvement. On top of that, aggregation can occur at every step of the process, from expression and purification to formulation and storage [104]. These aggregation-related issues stem from a simple principle: proteins are not selected to remain soluble out of their evolutionary context. In an external environment, the natural competition between functional and aberrant contacts is imbalanced by the absence of native binding partners, quality control systems, and deregulation of protein concentration, leading to uncontrolled intracellular protein deposition. Indeed, because protein-based drugs should be administered in most cases parenterally, protein concentration in the final formulation can exceed 200 mg/mL, being several orders of magnitude above their natural abundances [105]. Besides, during industrial processes, polypeptides are exposed to unnatural stresses such as pH-changes, shearing effects, or temperature fluctuations, impacting both their solubility and stability.
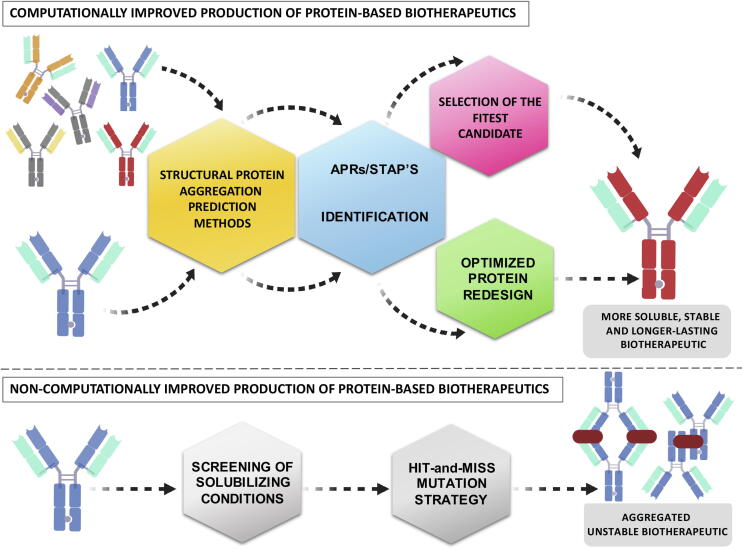
In this framework, the computational prediction of protein aggregation offers an avenue to work with pre-selected and well-characterized protein candidates producing more soluble, stable, and long-lasting therapeutic proteins (Fig. 3). Several of the algorithms cited in the previous section have been exploited to screen for more soluble protein variants and/or introduce solubilizing mutations. Human α-galactoside or Bacillus anthracis protective antigen are examples of biotherapeutic proteins whose solubility has been successfully redesigned by using SolubiS, reducing their aggregation propensity while maintaining their functional activity [106]. Such approximation has also been translated to the redesign of mAbs, which currently represent the faster-growing class of biotherapeutic molecules. SolubiS has successfully ranked a set of mAbs according to their aggregation, also being able to apply this predictive potential to the redesigning of these complex macromolecules into more soluble variants [87]. As a dedicated tool for antibody predictions, the DI algorithm also allows the screening of the aggregation propensity of mAbs in the early phases of drug discovery; thus, the development of pre-screened candidates can be prioritized attending to their solubility.
Fig. 3.
Comparison between a computationally guided pipeline for optimizing protein-based biotherapeutics and currently used strategies. The computational analysis of a candidate pool and/or the reduction of their aggregation by introducing solubilizing mutations offers a powerful alternative to expensive and blinded trial/error approaches, being cost-effective strategies to increase the success rate in the development of protein-based therapeutics.
Finally, A3D offers a user-friendly platform for the design of protein solubility by integrating structural aggregation predictions with stability prediction since, eventually, solubilizing mutations at the protein surfaces may have destabilizing effects on the native structure, even if the involved residues are fully exposed to solvent [107]. Towards the optimization of such a feature, we have recently updated A3D with an “Automated mutations” mode that automatically ranks solubilizing mutations in the protein surfaces according to their effect on aggregation and stability. This new module was tested for the redesign of a human variable heavy chain of the human antibody germline, allowing the identification of solubilizing mutations that do not perturb the stability of the antibody fragment. This routine will significantly reduce the time dedicated to the visual inspection of protein structures and manual selection of mutations [107]. Of note, A3D users can pre-exclude the complementarity-determining regions (CDRs) from the automated round of analysis. This is important because several previous phage display initiatives aimed to increase Abs solubility have resulted in variants in which the mutations cluster in or close to the hydrophobic CDRs, with the subsequent risk of hampering antigen-recognition [108]. The automated A3D selected mutations lay sequentially and structurally far from the CDRs, yet they significantly increased the solubility of the Ab of interest when this was incubated under harsh conditions. The main advantage of this upgrade is that it permits its use to academic and industrial fellows that did not have extensive training in protein redesign.
5. Modulation of the environmental conditions on the prediction of protein aggregation.
The vast majority of proteins reside and develop their functions in highly complex and overcrowded cellular microenvironments exposed to unpredictable stresses that may impact their solubility [52], [109]. However, in the absence of external stressors, cellular conditions tend to remain relatively constant and the extrinsic factors impacting protein aggregation are not expected to change significantly over time. Thus, even if the prediction of aggregation in vivo is far from being trivial, for the computational evaluation of this property, it can be considered that we face a defined and constant environment. In stark contrast, during industrial manufacturing, proteins are often exposed to very different conditions that affect their physical stability [110]. For instance, more than 65% of antibodies and related constructs are formulated at pH < 6.5 [5], [111]. Yet, only a small set of in silico tools have been designed to address the protein background, and many of them only collaterally with barely parametrized functions. The sequence-based predictors TANGO and Zyggregator can evaluate the effect of pH on protein net charge, but their performance in predicting the pH-dependent solubility of experimentally characterized proteins is relatively low; mostly because they do not compute the partial charge of the side chains [7], [65], [112], [113]. The first algorithm in which pH was implicitly considered is DI. DI can evaluate the net charge -accounting for partial ionization- of mAbs in a pH-dependent way. In such a way, DI can evaluate if mAbs with similar solubility in neutral conditions would have different solubility in a particular step of the purification, thus extending the pre-selection and optimization of candidates to a more realistic level. One of the main disadvantages of this approach and other related applications is that they only evaluate the effect of pH in terms of its modulation over protein net charge. However, it is well-known that the protonation/deprotonation of the ionizable residues also has a significant effect in the hydrophobicity, as illustrated by their shifts in the partition coefficients upon changes on the solution pH [114], [115]. On that basis, we have recently built a novel phenomenological model to predict the effect of pH on the aggregation of IDPs considering both fluctuations in hydrophobicity and global net charge [116]. Our algorithm calculates the pH-dependent hydrophobicity along the sequence at a given pH and computes the effect of the global protein net charge in order to profile the pH-dependent solubility of a given protein. Such an approach has demonstrated a higher predictive potential than mere charge-dependent approaches stressing the importance of including the changes in hydrophobicity in future predictive endeavors. We expect that this model would inspire the implementation of novel structure-based algorithms, including this feature, to develop more robust and versatile software.
6. Conclusions
The widespread nature of aggregation influences every aspect of protein function and applications either in vivo or in vitro, from disease onset to the production of biotherapeutics. Accordingly, it becomes fundamental to develop new tools to systematically address protein aggregation issues in a fast and reliable way, with the ultimate objective of transforming this arbitrary and unpredictable process into an anticipatable variable. Nowadays, in silico approximations are being progressively integrated into the routines of many laboratories, being cost-effective tools to assist and nurture experimental efforts. Likewise, it can be expected that they will also gain relevance in the biotechnological arena, replacing or complementing the actual costly and limited experimental methods used to optimize protein solubility. In the near future, we could expect that novel automated and more robust algorithms accounting from conditions extrinsic to the protein sequence and static structure would be progressively implemented in the streamlined workflow of companies as an easy and fast optimization step for protein-based therapeutics; impacting both their marketing and clinical application.
CRediT authorship contribution statement
Jaime Santos: Data curation, Visualization, Writing - original draft. Jordi Pujols: Writing - original draft. Irantzu Pallarès: Writing - original draft, Visualization, Supervision. Valentín Iglesias: Writing - original draft, Visualization, Software. Salvador Ventura: Conceptualization, Supervision, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Huttlin E.L. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamada T., Bork P. Evolution of biomolecular networks: lessons from metabolic and protein interactions. Nat Rev Mol Cell Biol. 2009;10:791–803. doi: 10.1038/nrm2787. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg D., Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiti F., Dobson C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu Rev Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 5.Roberts C.J. Therapeutic protein aggregation: mechanisms, design, and control. Trends Biotechnol. 2014;32:372–380. doi: 10.1016/j.tibtech.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts C.J. Protein aggregation and its impact on product quality. Curr Opin Biotechnol. 2014;30:211–217. doi: 10.1016/j.copbio.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rousseau F., Schymkowitz J., Serrano L. Protein aggregation and amyloidosis: confusion of the kinds? Curr Opin Struct Biol. 2006;16:118–126. doi: 10.1016/j.sbi.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Alam P., Bousset L., Melki R., Otzen D.E. alpha-synuclein oligomers and fibrils: a spectrum of species, a spectrum of toxicities. J Neurochem. 2019;150:522–534. doi: 10.1111/jnc.14808. [DOI] [PubMed] [Google Scholar]
- 9.Frare E. Characterization of oligomeric species on the aggregation pathway of human lysozyme. J Mol Biol. 2009;387:17–27. doi: 10.1016/j.jmb.2009.01.049. [DOI] [PubMed] [Google Scholar]
- 10.Vetri V. Amyloid fibrils formation and amorphous aggregation in concanavalin A. Biophys Chem. 2007;125:184–190. doi: 10.1016/j.bpc.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Avni A., Swasthi H.M., Majumdar A., Mukhopadhyay S. Intrinsically disordered proteins in the formation of functional amyloids from bacteria to humans. Prog Mol Biol Transl Sci. 2019;166:109–143. doi: 10.1016/bs.pmbts.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Otzen D. Functional amyloid: turning swords into plowshares. Prion. 2010;4:256–264. doi: 10.4161/pri.4.4.13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman M.R. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh J. Amyloidogenesis of type III-dependent harpins from plant pathogenic bacteria. J Biol Chem. 2007;282:13601–13609. doi: 10.1074/jbc.M602576200. [DOI] [PubMed] [Google Scholar]
- 15.Fowler D.M. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maji S.K. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maury C.P. The emerging concept of functional amyloid. J Intern Med. 2009;265:329–334. doi: 10.1111/j.1365-2796.2008.02068.x. [DOI] [PubMed] [Google Scholar]
- 18.Houben, B. et al. Autonomous aggregation suppression by acidic residues explains why chaperones favour basic residues. EMBO J, e102864 (2020).doi: 10.15252/embj.2019102864 [DOI] [PMC free article] [PubMed]
- 19.Sanchez de Groot, N. et al. Evolutionary selection for protein aggregation. Biochem Soc Trans 40, 1032-7 (2012).doi: 10.1042/BST20120160 [DOI] [PubMed]
- 20.Nooren I.M., Thornton J.M. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eichner T., Radford S.E. A diversity of assembly mechanisms of a generic amyloid fold. Mol Cell. 2011;43:8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Pallares I., Ventura S. Advances in the Prediction of Protein Aggregation Propensity. Curr Med Chem. 2019;26:3911–3920. doi: 10.2174/0929867324666170705121754. [DOI] [PubMed] [Google Scholar]
- 23.Ricardo Graña-Montes, J.P.-P., Carlota Gómez-Picanyol & Ventura, a.S. Prediction of Protein Aggregation and Amyloid Formation. in From Protein Structure to Function with Bioinformatics (ed. Rigden, D.J.) 205-263 (Springer, 2017).doi: 10.1007/978-94-024-1069-3_7
- 24.Santos J., Iglesias V., Ventura S. Computational prediction and redesign of aberrant protein oligomerization. Prog Mol Biol Transl Sci. 2020;169:43–83. doi: 10.1016/bs.pmbts.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Monsellier E., Ramazzotti M., Taddei N., Chiti F. Aggregation propensity of the human proteome. PLoS Comput Biol. 2008;4 doi: 10.1371/journal.pcbi.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Groot N.S., Ventura S. Protein aggregation profile of the bacterial cytosol. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0009383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallares I., Ventura S. Understanding and predicting protein misfolding and aggregation: Insights from proteomics. Proteomics. 2016;16:2570–2581. doi: 10.1002/pmic.201500529. [DOI] [PubMed] [Google Scholar]
- 28.Redler R.L. Computational approaches to understanding protein aggregation in neurodegeneration. J Mol Cell Biol. 2014;6:104–115. doi: 10.1093/jmcb/mju007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buck P.M., Kumar S., Singh S.K. On the role of aggregation prone regions in protein evolution, stability, and enzymatic catalysis: insights from diverse analyses. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monsellier E., Chiti F. Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 2007;8:737–742. doi: 10.1038/sj.embor.7401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castillo V., Grana-Montes R., Sabate R., Ventura S. Prediction of the aggregation propensity of proteins from the primary sequence: aggregation properties of proteomes. Biotechnol J. 2011;6:674–685. doi: 10.1002/biot.201000331. [DOI] [PubMed] [Google Scholar]
- 32.Sherman M.Y., Goldberg A.L. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 33.Pastore A., Temussi P.A. The two faces of Janus: functional interactions and protein aggregation. Curr Opin Struct Biol. 2012;22:30–37. doi: 10.1016/j.sbi.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J Mol Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 35.Castillo V., Ventura S. Amyloidogenic regions and interaction surfaces overlap in globular proteins related to conformational diseases. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castillo V., Chiti F., Ventura S. The N-terminal helix controls the transition between the soluble and amyloid states of an FF domain. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0058297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pechmann S., Levy E.D., Tartaglia G.G., Vendruscolo M. Physicochemical principles that regulate the competition between functional and dysfunctional association of proteins. Proc Natl Acad Sci U S A. 2009;106:10159–10164. doi: 10.1073/pnas.0812414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillo V., Espargaro A., Gordo V., Vendrell J., Ventura S. Deciphering the role of the thermodynamic and kinetic stabilities of SH3 domains on their aggregation inside bacteria. Proteomics. 2010;10:4172–4185. doi: 10.1002/pmic.201000260. [DOI] [PubMed] [Google Scholar]
- 39.Fraga H., Grana-Montes R., Illa R., Covaleda G., Ventura S. Association between foldability and aggregation propensity in small disulfide-rich proteins. Antioxid Redox Signal. 2014;21:368–383. doi: 10.1089/ars.2013.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan C., Wu F., Jernigan R.L., Dobbs D., Honavar V. Characterization of protein-protein interfaces. Protein J. 2008;27:59–70. doi: 10.1007/s10930-007-9108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masino L., Nicastro G., Calder L., Vendruscolo M., Pastore A. Functional interactions as a survival strategy against abnormal aggregation. FASEB J. 2011;25:45–54. doi: 10.1096/fj.10-161208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabate R., Espargaro A., Grana-Montes R., Reverter D., Ventura S. Native structure protects SUMO proteins from aggregation into amyloid fibrils. Biomacromolecules. 2012;13:1916–1926. doi: 10.1021/bm3004385. [DOI] [PubMed] [Google Scholar]
- 43.Stroo E., Koopman M., Nollen E.A., Mata-Cabana A. Cellular Regulation of Amyloid Formation in Aging and Disease. Front Neurosci. 2017;11:64. doi: 10.3389/fnins.2017.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ivankov D.N. Contact order revisited: influence of protein size on the folding rate. Protein Sci. 2003;12:2057–2062. doi: 10.1110/ps.0302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watters A.L. The highly cooperative folding of small naturally occurring proteins is likely the result of natural selection. Cell. 2007;128:613–624. doi: 10.1016/j.cell.2006.12.042. [DOI] [PubMed] [Google Scholar]
- 46.Ellis R.J. Chaperone substrates inside the cell. Trends Biochem Sci. 2000;25:210–212. doi: 10.1016/s0968-0004(00)01576-0. [DOI] [PubMed] [Google Scholar]
- 47.De Baets G. An evolutionary trade-off between protein turnover rate and protein aggregation favors a higher aggregation propensity in fast degrading proteins. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tartaglia, G.G. & Caflisch, A. Computational analysis of the S. cerevisiae proteome reveals the function and cellular localization of the least and most amyloidogenic proteins. Proteins 68, 273-8 (2007).doi: 10.1002/prot.21427 [DOI] [PubMed]
- 49.Tartaglia G.G., Vendruscolo M. Correlation between mRNA expression levels and protein aggregation propensities in subcellular localisations. Mol Biosyst. 2009;5:1873–1876. doi: 10.1039/b913099n. [DOI] [PubMed] [Google Scholar]
- 50.Conrad B., Antonarakis S.E. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- 51.Hardy J. Amyloid double trouble. Nat Genet. 2006;38:11–12. doi: 10.1038/ng0106-11. [DOI] [PubMed] [Google Scholar]
- 52.Wang W., Nema S., Teagarden D. Protein aggregation–pathways and influencing factors. Int J Pharm. 2010;390:89–99. doi: 10.1016/j.ijpharm.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 53.Levy E.D., De S., Teichmann S.A. Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc Natl Acad Sci U S A. 2012;109:20461–20466. doi: 10.1073/pnas.1209312109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vecchi G. Proteome-wide observation of the phenomenon of life on the edge of solubility. Proc Natl Acad Sci U S A. 2020;117:1015–1020. doi: 10.1073/pnas.1910444117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castillo V., Grana-Montes R., Ventura S. The aggregation properties of Escherichia coli proteins associated with their cellular abundance. Biotechnol J. 2011;6:752–760. doi: 10.1002/biot.201100014. [DOI] [PubMed] [Google Scholar]
- 56.Tartaglia G.G., Pechmann S., Dobson C.M., Vendruscolo M. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem Sci. 2007;32:204–206. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Ciryam P., Tartaglia G.G., Morimoto R.I., Dobson C.M., Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 2013;5:781–790. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ciryam P., Kundra R., Morimoto R.I., Dobson C.M., Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol Sci. 2015;36:72–77. doi: 10.1016/j.tips.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kundra R., Ciryam P., Morimoto R.I., Dobson C.M., Vendruscolo M. Protein homeostasis of a metastable subproteome associated with Alzheimer's disease. Proc Natl Acad Sci U S A. 2017;114:E5703–E5711. doi: 10.1073/pnas.1618417114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y., Dokholyan N.V. Natural selection against protein aggregation on self-interacting and essential proteins in yeast, fly, and worm. Mol Biol Evol. 2008;25:1530–1533. doi: 10.1093/molbev/msn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carija A., Pinheiro F., Iglesias V., Ventura S. Computational Assessment of Bacterial Protein Structures Indicates a Selection Against Aggregation. Cells. 2019;8 doi: 10.3390/cells8080856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Z, L.A. & R, M.M.B. Structure and Aggregation Mechanisms in Amyloids. Molecules 25(2020).doi: 10.3390/molecules25051195 [DOI] [PMC free article] [PubMed]
- 63.Conchillo-Sole O. AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinf. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanchez de Groot, N., Pallares, I., Aviles, F.X., Vendrell, J. & Ventura, S. Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct Biol 5, 18 (2005).doi: 10.1186/1472-6807-5-18 [DOI] [PMC free article] [PubMed]
- 65.Tartaglia G.G., Vendruscolo M. The Zyggregator method for predicting protein aggregation propensities. Chem Soc Rev. 2008;37:1395–1401. doi: 10.1039/b706784b. [DOI] [PubMed] [Google Scholar]
- 66.Walsh, I., Seno, F., Tosatto, S.C. & Trovato, A. PASTA 2.0: an improved server for protein aggregation prediction. Nucleic Acids Res 42, W301-7 (2014).doi: 10.1093/nar/gku399 [DOI] [PMC free article] [PubMed]
- 67.Garbuzynskiy S.O., Lobanov M.Y., Galzitskaya O.V. FoldAmyloid: a method of prediction of amyloidogenic regions from protein sequence. Bioinformatics. 2010;26:326–332. doi: 10.1093/bioinformatics/btp691. [DOI] [PubMed] [Google Scholar]
- 68.Maurer-Stroh S. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 69.O'Donnell, C.W. et al. A method for probing the mutational landscape of amyloid structure. Bioinformatics 27, i34-42 (2011).doi: 10.1093/bioinformatics/btr238 [DOI] [PMC free article] [PubMed]
- 70.Stanislawski, J., Kotulska, M. & Unold, O. Machine learning methods can replace 3D profile method in classification of amyloidogenic hexapeptides. BMC Bioinformatics 14, 21 (2013).doi: 10.1186/1471-2105-14-21 [DOI] [PMC free article] [PubMed]
- 71.Familia C., Dennison S.R., Quintas A., Phoenix D.A. Prediction of Peptide and Protein Propensity for Amyloid Formation. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0134679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim C., Choi J., Lee S.J., Welsh W.J., Yoon S. NetCSSP: web application for predicting chameleon sequences and amyloid fibril formation. Nucleic Acids Res. 2009;37:W469–W473. doi: 10.1093/nar/gkp351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gasior P., Kotulska M. FISH Amyloid - a new method for finding amyloidogenic segments in proteins based on site specific co-occurrence of aminoacids. BMC Bioinf. 2014;15:54. doi: 10.1186/1471-2105-15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsolis A.C., Papandreou N.C., Iconomidou V.A., Hamodrakas S.J. A consensus method for the prediction of 'aggregation-prone' peptides in globular proteins. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0054175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emily M., Talvas A., Delamarche C. MetAmyl: a METa-predictor for AMYLoid proteins. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0079722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Uversky V.N., Gillespie J.R., Fink A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 77.Dyson H.J. Making Sense of Intrinsically Disordered Proteins. Biophys J. 2016;110:1013–1016. doi: 10.1016/j.bpj.2016.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Baets G., Van Durme J., Rousseau F., Schymkowitz J. A genome-wide sequence-structure analysis suggests aggregation gatekeepers constitute an evolutionary constrained functional class. J Mol Biol. 2014;426:2405–2412. doi: 10.1016/j.jmb.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 79.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pujols J., Santos J., Pallares I., Ventura S. The Disordered C-Terminus of Yeast Hsf1 Contains a Cryptic Low-Complexity Amyloidogenic Region. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19051384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coskuner O., Uversky V.N. Intrinsically disordered proteins in various hypotheses on the pathogenesis of Alzheimer's and Parkinson's diseases. Prog Mol Biol Transl Sci. 2019;166:145–223. doi: 10.1016/bs.pmbts.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 82.Li Y. Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy. Cell Res. 2018;28:897–903. doi: 10.1038/s41422-018-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fitzpatrick A.W.P. Cryo-EM structures of tau filaments from Alzheimer's disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paravastu A.K., Leapman R.D., Yau W.M., Tycko R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fitzpatrick A.W., Saibil H.R. Cryo-EM of amyloid fibrils and cellular aggregates. Curr Opin Struct Biol. 2019;58:34–42. doi: 10.1016/j.sbi.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Navarro S., Ventura S. Computational re-design of protein structures to improve solubility. Expert Opin Drug Discov. 2019;14:1077–1088. doi: 10.1080/17460441.2019.1637413. [DOI] [PubMed] [Google Scholar]
- 87.van der Kant R. Prediction and Reduction of the Aggregation of Monoclonal Antibodies. J Mol Biol. 2017;429:1244–1261. doi: 10.1016/j.jmb.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van Durme J. Solubis: a webserver to reduce protein aggregation through mutation. Protein Eng Des Sel. 2016;29:285–289. doi: 10.1093/protein/gzw019. [DOI] [PubMed] [Google Scholar]
- 89.Schymkowitz J. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chennamsetty N., Voynov V., Kayser V., Helk B., Trout B.L. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci U S A. 2009;106:11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lauer T.M. Developability index: a rapid in silico tool for the screening of antibody aggregation propensity. J Pharm Sci. 2012;101:102–115. doi: 10.1002/jps.22758. [DOI] [PubMed] [Google Scholar]
- 92.Kuriata, A. et al. Aggrescan3D (A3D) 2.0: prediction and engineering of protein solubility. Nucleic Acids Res 47, W300-W307 (2019).doi: 10.1093/nar/gkz321 [DOI] [PMC free article] [PubMed]
- 93.Kuriata A., Iglesias V., Kurcinski M., Ventura S., Kmiecik S. Aggrescan3D standalone package for structure-based prediction of protein aggregation properties. Bioinformatics. 2019;35:3834–3835. doi: 10.1093/bioinformatics/btz143. [DOI] [PubMed] [Google Scholar]
- 94.Jamroz M., Kolinski A., Kmiecik S. CABS-flex: Server for fast simulation of protein structure fluctuations. Nucleic Acids Res. 2013;41:W427–W431. doi: 10.1093/nar/gkt332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuriata, A. et al. CABS-flex 2.0: a web server for fast simulations of flexibility of protein structures. Nucleic Acids Res 46, W338-W343 (2018).doi: 10.1093/nar/gky356 [DOI] [PMC free article] [PubMed]
- 96.Pujols J., Pena-Diaz S., Ventura S. AGGRESCAN3D: Toward the Prediction of the Aggregation Propensities of Protein Structures. Methods Mol Biol. 2018;1762:427–443. doi: 10.1007/978-1-4939-7756-7_21. [DOI] [PubMed] [Google Scholar]
- 97.Sormanni P., Aprile F.A., Vendruscolo M. The CamSol method of rational design of protein mutants with enhanced solubility. J Mol Biol. 2015;427:478–490. doi: 10.1016/j.jmb.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 98.Hurshman A.R., White J.T., Powers E.T., Kelly J.W. Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry. 2004;43:7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- 99.Nordlund A., Oliveberg M. SOD1-associated ALS: a promising system for elucidating the origin of protein-misfolding disease. HFSP J. 2008;2:354–364. doi: 10.2976/1.2995726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shah M. Commentary: New perspectives on protein aggregation during Biopharmaceutical development. Int J Pharm. 2018;552:1–6. doi: 10.1016/j.ijpharm.2018.09.049. [DOI] [PubMed] [Google Scholar]
- 101.den Engelsman J. Strategies for the assessment of protein aggregates in pharmaceutical biotech product development. Pharm Res. 2011;28:920–933. doi: 10.1007/s11095-010-0297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ratanji K.D., Derrick J.P., Dearman R.J., Kimber I. Immunogenicity of therapeutic proteins: influence of aggregation. J Immunotoxicol. 2014;11:99–109. doi: 10.3109/1547691X.2013.821564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.FDA. Guidance for Industry Immunogenicity Assessment for Therapeutic Protein Products. (2014).doi:
- 104.Cromwell M.E., Hilario E., Jacobson F. Protein aggregation and bioprocessing. AAPS J. 2006;8:E572–E579. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schermeyer M.T., Woll A.K., Kokke B., Eppink M., Hubbuch J. Characterization of highly concentrated antibody solution - A toolbox for the description of protein long-term solution stability. MAbs. 2017;9:1169–1185. doi: 10.1080/19420862.2017.1338222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ganesan A. Structural hot spots for the solubility of globular proteins. Nat Commun. 2016;7:10816. doi: 10.1038/ncomms10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gil-Garcia M. Combining Structural Aggregation Propensity and Stability Predictions To Redesign Protein Solubility. Mol Pharm. 2018;15:3846–3859. doi: 10.1021/acs.molpharmaceut.8b00341. [DOI] [PubMed] [Google Scholar]
- 108.Sidhu S.S. Phage display in pharmaceutical biotechnology. Curr Opin Biotechnol. 2000;11:610–616. doi: 10.1016/s0958-1669(00)00152-x. [DOI] [PubMed] [Google Scholar]
- 109.Breydo L., Redington J.M., Uversky V.N. Effects of Intrinsic and Extrinsic Factors on Aggregation of Physiologically Important Intrinsically Disordered Proteins. Int Rev Cell Mol Biol. 2017;329:145–185. doi: 10.1016/bs.ircmb.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 110.Zapadka, K.L., Becher, F.J., Gomes Dos Santos, A.L. & Jackson, S.E. Factors affecting the physical stability (aggregation) of peptide therapeutics. Interface Focus 7, 20170030 (2017).doi: 10.1098/rsfs.2017.0030 [DOI] [PMC free article] [PubMed]
- 111.Wang W., Singh S., Zeng D.L., King K., Nema S. Antibody structure, instability, and formulation. J Pharm Sci. 2007;96:1–26. doi: 10.1002/jps.20727. [DOI] [PubMed] [Google Scholar]
- 112.Chiti F., Stefani M., Taddei N., Ramponi G., Dobson C.M. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 113.DuBay K.F. Prediction of the absolute aggregation rates of amyloidogenic polypeptide chains. J Mol Biol. 2004;341:1317–1326. doi: 10.1016/j.jmb.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 114.Simm S., Einloft J., Mirus O., Schleiff E. 50 years of amino acid hydrophobicity scales: revisiting the capacity for peptide classification. Biol Res. 2016;49:31. doi: 10.1186/s40659-016-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.MacCallum J.L., Tieleman D.P. Hydrophobicity scales: a thermodynamic looking glass into lipid-protein interactions. Trends Biochem Sci. 2011;36:653–662. doi: 10.1016/j.tibs.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 116.Santos J. pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity. Cells. 2020;9 doi: 10.3390/cells9010145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fernandez-Escamilla A.M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 118.Trovato A., Chiti F., Maritan A., Seno F. Insight into the structure of amyloid fibrils from the analysis of globular proteins. PLoS Comput Biol. 2006;2 doi: 10.1371/journal.pcbi.0020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tian, J., Wu, N., Guo, J. & Fan, Y. Prediction of amyloid fibril-forming segments based on a support vector machine. BMC Bioinformatics 10 Suppl 1, S45 (2009).doi: 10.1186/1471-2105-10-S1-S45 [DOI] [PMC free article] [PubMed]
- 120.Bryan A.W., Jr., Menke M., Cowen L.J., Lindquist S.L., Berger B. BETASCAN: probable beta-amyloids identified by pairwise probabilistic analysis. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thangakani A.M., Kumar S., Nagarajan R., Velmurugan D., Gromiha M.M. GAP: towards almost 100 percent prediction for beta-strand-mediated aggregating peptides with distinct morphologies. Bioinformatics. 2014;30:1983–1990. doi: 10.1093/bioinformatics/btu167. [DOI] [PubMed] [Google Scholar]
- 122.Thompson M.J. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci U S A. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]