Abstract
Bacteria use two-component systems (TCSs) to sense and respond to their environments. Free-living bacteria usually contain dozens of TCSs, each of them responsible for sensing and responding to a different range of signals. Differences in the content of two-component systems are related with the capacity of the bacteria to colonize different niches or improve the efficiency to grow under the conditions of the existing niche. This review highlights differences in the TCS content between Staphylococcus aureus and Staphylococcus saprophyticus as a case study to exemplify how the ability to sense and respond to the environment is relevant for bacterial capacity to colonize and survive in/on different body surfaces.
Current Opinion in Microbiology 2020, 55:40–47
This review comes from a themed issue on Cell regulation
Edited by Charles J Dorman and Joan Geoghegan
For a complete overview see the Issue and the Editorial
Available online 19th March 2020
https://doi.org/10.1016/j.mib.2020.02.004
1369-5274/© 2020 The Author(s). Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
Bacterial genomes encode for a variable number of two-component sensor – response regulator pairs. The number of TCSs is proportional to the genome size, the diversity of environments in which organisms live, and the complexity in cellular differentiation [1]. Thus, bacteria with larger genomes, more metabolic versatility, and complex lifestyles are more likely to have a larger number of two-component systems than bacteria inhabiting relatively stable environments [2]. Acquisition of new TCSs occurs through mechanisms of lateral gene transfer or gene duplication and subsequent accumulation of mutations that insulate the new pathways from the existing two-component pathways [3]. In few cases, the newly introduced genes will improve the efficiency to grow under the conditions of the existing niche and consequently will be fixed in the genome. In most of the cases, their presence will interfere with existing TCSs and they will be eliminated from the genome and thus, no longer present in extant species. Comparative analysis of the collection of TCSs present in two closely related bacterial species can be useful to explain why one bacterial species can colonize a wide range of tissues and cause many different types of infections, while the other is far more restricted in its distribution and pathogenicity [1,4].
Staphylococcus aureus is a highly versatile opportunistic pathogen able to adapt to very different types of environments. It can live freely outside the host or exist either as a commensal external colonizer or as a pathogen in both humans and animals [5]. The anterior nares are the main ecological niche for S. aureus [6]. However, multiple other sites in the human body such as the skin, axillae, vagina, and gastrointestinal tract can also be colonized by this bacterium. The core genome of S. aureus contains 16 TCSs (http://mistdb.com, http://www.ncbi.nlm.nih.gov/Complete_Genomes/SignalCensus.html, http://www.p2cs.org) [7,8,9•]. Among the sixteen TCSs, only WalRK, is essential for bacterial growth [10]. All the other TCSs are dispensable, and they can be deleted individually [11] or simultaneously in the same strain without affecting cell viability [12••]. Using S. aureus mutant strains deprived of its complete non-essential sensorial TCS network, Villanueva et al. [12••] showed that complementation with unique TCSs was sufficient to restore the capacity to grow under different environmental conditions such as low pH (GraRS) and low temperature (SrrBA) and to reduce nitrate to nitrite (NreCB) or to resist to Triton X-100 (VraRS), experimentally validating the widely offered idea that TCSs are self-sufficient, autonomous entities able to confer the capacity to sense and respond to a particular environmental condition. This study also showed that sensor histidine kinases exhibit strong preference for their cognate response regulators (RR), though in some cases, cross-regulation between non-cognate sensor-RR pairs can occur in vivo.
The set of 16 TCSs of S. aureus are conserved in other closely related coagulase negative staphylococcal species such as Staphylococcus epidermidis and Staphylococcus haemolyticus (https://mistdb.co) [9•]. However, Staphylococcus saprophyticus, a coagulase negative staphylococcus whose genome is just 0.3 Mb smaller than that of S. aureus, contains only 11 TCSs (Table 1) [13]. This lack of correlation between the genome size and the number of TCSs is very likely due to various environmental factors. S. saprophyticus is a common inhabitant of the urinary tract, perineum, rectum, urethra, cervix, and gastrointestinal tract [14]. It is the second most common cause of community-acquired urinary tract infections (UTI) in young and middle-aged female outpatients, after Escherichia coli, without the involvement of indwelling catheters [15,16]. The narrow niche of tissues that S. saprophyticus colonizes compared to S. aureus is very likely related with the reduced number of TCSs in S. saprophyticus and consequently to the capacity of the bacterium to adapt to the environmental conditions encountered in the different tissues [17•]. In this review, we summarize and discuss our current knowledge about the TCSs that are missing in the S. saprophyticus genome compared to S. aureus and the consequences that their lack has for the bacterium.
Table 1.
Two-component systems in S. aureus and S. saprophyticus
| Two-component system | S. aureus | S. saprophyticus | Function |
|---|---|---|---|
| walRK | MW0018 | SSP0021 | Cell wall maintenance, cell viability |
| MW0019 | SSP0022 | ||
| hptSR | MW0198 | Intracellular survival, uptake of hexose phosphate | |
| MW0199 | |||
| lytSR | MW0236 | SSP0463 | Autolysis, eDNA release, biofilm |
| MW0237 | SSP0464 | ||
| graRS | MW0621 | SSP2061 | AMPs resistance, growth at low pH |
| MW0622 | SSP2062 | ||
| saeSR | MW0667 | Virulence factors regulation (toxins, exoenzymes…) | |
| MW0668 | |||
| tcs7SR | MW1208 | SSP1446 | Uncharacterized function |
| MW1209 | SSP1547 | ||
| arlSR | MW1304 | SSP1323 | Pathogenesis mechanisms: autolysis, adhesion, biofilm… |
| MW1305 | SSP1324 | ||
| srrBA | MW1445 | SSP1260 | Anaerobic respiration metabolism, growth at low temperature |
| MW1446 | SSP1261 | ||
| phoRP | MW1636 | SSP1073 | Phosphate uptake and homeostasis |
| MW1637 | SSP1074 | ||
| airRS | MW1789 | SSP0946 | Oxidative stress response |
| MW1790 | SSP0947 | ||
| vraRS | MW1824 | SSP0908 | Cell wall-affecting antibiotic resistance, cell wall biosynthesis |
| MW1825 | SSP0909 | ||
| agrCA | MW1962 | SSP0839 | Quorum sensing control of adhesion and virulence factors |
| MW1963 | SSP0840 | ||
| kdpDE | MW2002 | Potassium homeostasis regulation | |
| MW2003 | |||
| hssRS | MW2282 | SSP0540 | Heme metabolism regulation |
| MW2283 | SSP0541 | ||
| nreCB | MW2313 | Response to low oxygen, nitrate reduction | |
| MW2314 | |||
| braSR | MW2544 | Antimicrobial peptide resistance | |
| MW2545 |
KdpDE
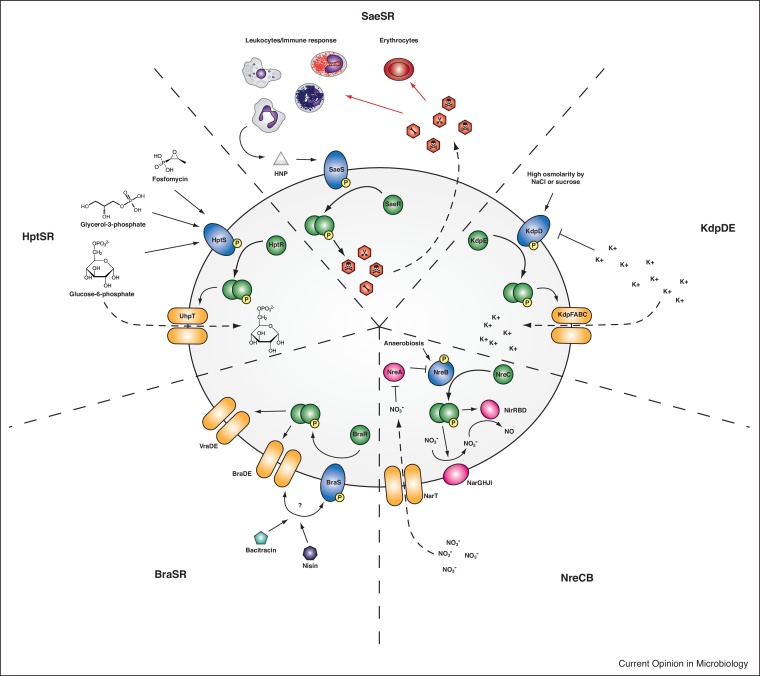
In general S. aureus can tolerate a high concentration of salt and low water activity for a non-halophilic bacterium [18]. It is assumed that this osmotolerance supports bacterial growth on a high-salt environment such as the human skin. Potassium is the major monovalent cation in cells and plays an essential role for all living organisms. Within bacterial cells, potassium is required for the maintenance of a constant pH, membrane potential and osmotic pressure. S. aureus maintains high intracellular potassium concentrations of 0.5–1.5 M, even in the absence of a high osmolarity environment, thanks to two specific potassium uptake systems, the inducible Kdp and the constitutively expressed Ktr [19]. The activation of Kdp requires the presence of the functional KdpDE TCS which is induced by high osmolarity and inhibited by cyclic di-AMP [20]. Once activated, the most highly induced genes by the KdpDE TCS are the constituents of the KdpFABC transport machinery involved in uptake of K+, as well as genes involved in other compatible solute and sugar uptake; capsule biosynthesis; and amino acid and central metabolism (Figure 1). The KdpFABC system plays a physiological role under very low K+ conditions [21]. At high K+ concentrations, a lower-affinity and constitutively expressed Ktr ion transporter is responsible for K+ transport using energy generated by electrochemical ion gradients [22]. The analysis of mutants in Kdp and Ktr systems revealed that the Ktr system is the major K+ uptake system in S. aureus and the function of Kdp is required primarily during times of K+ starvation and/or fluctuating ionic conditions. However, acquisition of potassium ions in a potassium-limited environment seems to be important during S. aureus infection [23]. Accordingly, some methicillin resistant S. aureus strains carry a second KdpDE homologous TCS in the SCCmec mobile element [24]. Similarly, a KdpDE paralog has also been described in some types of the arginine catabolic mobile element (ACME) of S. epidermidis [25••,26]. In some S. epidermidis strains containing the KdpDE system of the ACME element, the chromosomal KdpDE copy has been lost very likely to avoid cross-talk between both systems. S. saprophyticus has to cope with the highly variable ion content of urine without the contribution of the KdpDE system. In this bacterium, osmotolerance relies on Ktr and other osmoprotectant transport systems (proline/betaine, glycine betaine/choline transporter, proline permease) (Figure 2) [13]. Because urine contains large amounts of potassium, it seems that the Ktr system is sufficient to import enough K+ in the absence of the high affinity uptake system. Another interesting peculiarity regarding the cellular osmotic tolerance of S. saprophyticus and not S. aureus is the presence of plasmids carrying the aquaporin gene (aqpZ). Aquaporins are water channels that mediate rapid entry or exit of water in response to changes in osmolarity, and consequently, it has been proposed that they may also aid to osmotic balancing [13]. However, this hypothesis remains to be tested.
Figure 1.
TCSs absent in S. saprophyticus and their main functional roles. Five of the 16 TCSs in S. aureus are absent in S. saprophyticus. The HptSR TCS responds to extracellular phosphates and activates the glucose-6-phosphate transporter UhpT but can also facilitate the uptake of the antibiotic Fosfomycin. SaeSR controls the expression of secreted toxins and immune evasion factors involved in erythrocyte and leukocyte lysis as well as modifying several immune evasion pathways. SaeSR responds to antimicrobial peptides such as the human neutrophil peptide (HNP) secreted by neutrophils. The KdpDE TCS is involved in the control of a highly specific potassium uptake system (KdpFABC) required for growth under highly potassium restricting conditions. The TCS is repressed by potassium ions and cyclic di-AMP and can be activated by high osmotic conditions. Activation of KdpDE also leads to the expression of genes involved in the uptake of compatible solutes and sugars, capsule synthesis as well as amino acid biosynthesis and central metabolism. The NreCB TCS controls the reduction of nitrate to nitrite to nitric oxide. The TCS is inhibited by NreA in the absence of nitrate and induced under anaerobic conditions. The BraSR TCS is involved in the response to antimicrobial peptides such as bacitracin and nisin and involves the BraDE transporter in sensing these peptides. It also activates the expression of the GraRS TCS-associated VraDE transporter.
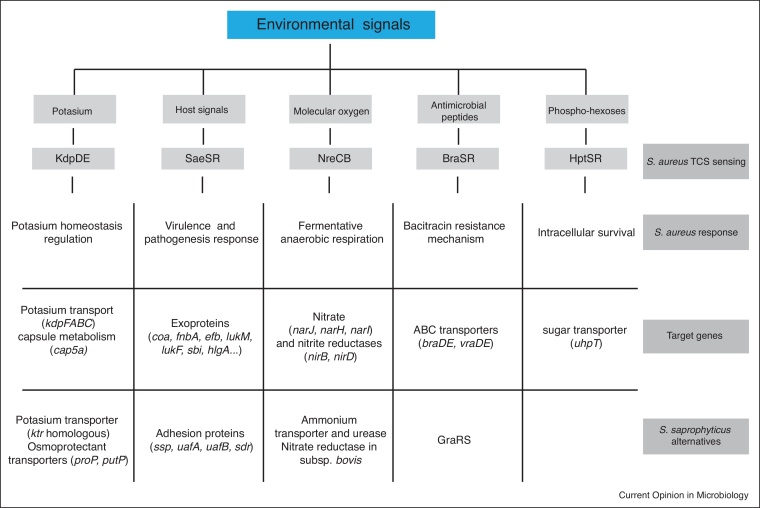
Figure 2.
Summary of the activity of the TCSs absent in S. saprophyticus. HptSR, SaeSR, KdpDE, NreCB, and BraSR are the five TCSs present in S. aureus but missing in S. saprophyticus. S. aureus colonises different niches to S. saprophyticus and consequently has to adapt to different environmental conditions. The five TCSs specific to S. aureus allow it to sense relevant environmental signals and allow it to respond by activating the expression of different target genes. Thus, S. aureus uses its larger arsenal of TCSs to adapt its physiology to the new environments. The genome of S. saprophyticus encodes genes that may compensate for the loss of genes and pathways regulated by the TCSs absent in this staphylococcal species. HptSR and its function in the uptake of extracellular hexose-6-phosphates is the only TCS-regulated adaptation mechanism for which S. saprophyticus does not encode any alternative.
SaeSR
The staphylococcal accessory element (sae) TCS has been previously characterized as a positive regulator of many secreted toxins, exoenzymes, and immunomodulatory proteins involved in staphylococcal pathogenesis (Figure 1). Loss of SaeSR abolishes the secretion of many of these proteins including exotoxins, immune evasion factors, superantigens, adhesins, staphylocoagulase, and proteases [27]. All these factors have a direct causative link with S. aureus pathogenicity and play a role once bacteria have crossed the epithelial barrier by inhibiting complement activation, neutrophil recruitment, as well as blocking opsonization by immunoglobulins [28,29]. It is therefore not surprising that mutants lacking saeSR result in lower mortality in systemic and intraperitoneal mouse infection models and show reduced ability to adhere to lung epithelial cells [30,31]. S. saprophyticus infections affect almost exclusively the urinary tract, without crossing the epithelial barrier and consequently, most of the typical virulence factors regulated by SaeSR in S. aureus are not needed in S. saprophyticus’ lifestyle. Instead, S. saprophyticus needs to maintain tight adherence to the bladder and ureter epithelium. For that, it produces different types of adhesins such as the highly conserved Aas hemagglutinin, the surface-associated lipase, Ssp, that forms fimbria-like surface appendages, and cell wall-anchored proteins (UafA, UafB, SssF and SdrI) [13,25••] (Figure 2). UafA is a chromosome-encoded adhesin that mediates hemagglutination and adherence to human bladder cells. This adhesin is exclusive of S. saprophyticus strains and it is absent in other staphylococci. UafB is a plasmid encoded serine-rich glycoprotein that binds fibronectin, fibrinogen, and human bladder-epithelial cells. SssF is another plasmid-encoded cell-wall associated protein that is highly prevalent in clinical isolates. It is associated with resistance to linoleic acid, a component of the human skin innate immune defense system. It has been proposed that SssF may aid to the survival on the skin during non-UTI periods [32•]. SdrI is a plasmid-encoded cell wall-associated serine-aspartate-rich protein that binds collagen and plays a role in acute UTI and persistent kidney infections [33]. Another specific feature of uropathogenicity found in S. saprophyticus is the presence of a d-serine deaminase responsible for degrading high concentrations of d-serine present in the urine [34]. Regulation of the expression of all these colonization factors in S. saprophyticus is not integrated under a single TCS. Instead, S. saprophyticus has more specific regulatory genes that may act individually for the modulation of such adhesion factors and metabolic enzymes for a prompt and individual response in metabolite-rich urine.
NreCB
Under conditions of low oxygen tension, S. aureus uses nitrate and nitrite as its final oxygen acceptors. NreCB is an oxygen sensing system that activates the expression of the cluster of genes needed for nitrate reduction (narGHJI) and nitrite reduction (nirRBD) [35,36] (Figure 1). Activation of NreCB is controlled by the nitrate-sensing NreA protein, which inhibits NreB autophosphorylation in the absence of nitrate (via binding to NreB) [36]. Inactivation of NreCB abrogates the ability of S. aureus to reduce nitrate, forcing the bacterium to upregulate fermentative pathways for survival [5]. The nreCB genes are transcribed together with the nitrate reductase genes (narGHJI operon) which is located downstream the genes encoding the nitrite reductase. Also located within the vicinity of nreCB is the nitrate transporter encoding gene (narT). This genome organization strongly suggests that nreCB belongs to a gene cluster that has been acquired by horizontal gene transfer. S. saprophyticus cannot use nitrate and nitrite as final oxygen acceptors because of the lack of respiratory nitrate reductase NarGHJI and assimilatory nitrite reductase NirRBD as well as the corresponding NreCB TCS on the genome. An exception to this is S. saprophyticus subsp. bovis, a regular colonizer of bovine nostrils that contains the NreCB TCS and also the nitrate but not the nitrite reductase genes on what appears to be a transposable element [37]. Another consequence of the nitrate and nitrite reductase activities is the generation of ammonia that allows nitrogen to be converted to an organic form. S. saprophyticus produces a potent urease that can obtain ammonia from urea (Figure 2). The urease activity of S. saprophyticus is significantly higher than other staphylococcal species and it is required for persistent infection in the urinary tract [38]. Besides, the urease activity is responsible for raising the pH of human urine, which allows precipitation of normally soluble polyvalent ions to carbonate apatite. These compounds aggregate around bacteria and very often are the cause of the formation of urinary stones.
BraSR
BraSR (bacitracin resistance associated) is associated with resistance to the antimicrobial peptides, nisin and bacitracin [39, 40, 41, 42]. BraSR forms a module with the ABC transporter BraDE. The ABC transporter is involved in sensing the signal and participates by mechanisms that still are not completely understood in the phosphotransfer between BraS to BraR [43]. In this system, the ABC transporter and the TCS have an absolute mutual requirement for each other in both sensing and responding [44••]. BraSR also activates the expression of VraDE, which encodes for the cognate ABC transporter associated with GraRS (Figure 1).
BraSR is absent in several staphylococcal species including S. saprophyticus, Staphylococcus xylosus, Staphylococcus arlettae, Staphylococcus intermedius, and Staphylococcus pseudointermedius [44••]. The absence of BraSR in S. saprophyticus is likely compensated by the presence of its orthologous TCS, GraRS. Indeed, experimental evolution of S. saprophyticus in the presence of increasing concentration of nisin, confirmed the role of GraRS in nisin resistance mechanisms [44••]. In S. aureus, activation of GraRS induces the expression mprF and the dltABCD operon. The dltABCD operon contributes to the net positive surface charge by covalently incorporating d-alanine to the cell wall linked teichoic acids whereas MprF adds positively charged lysine residues to phosphatidyl glycerol within the cell membrane [45]. It is hypothesized that GraRS in S. saprophyticus would also induce the expression of mprF and the dltABCD operon to confer resistance to AMPs (Figure 2). However, this hypothesis has not been experimentally tested.
HptSR
HptSR is required to sense extracellular hexose phosphates and to activate the transcription of uhpT, a gene located directly downstream hptSR that encodes the unique glucose 6-phosphate transporter of S. aureus [46] (Figure 1). UhpT is medically relevant because it can mediate the uptake of the antibiotic Fosfomycin [47]. S. aureus is currently regarded as a facultative intracellular pathogen [48]. Host cell invasion and intracellular survival is used by S. aureus to infect macrophages, spread to secondary points of infection, evade immune recognition, and avoid exposure to last-resort antibiotics [49,50]. To survive and multiply within host cells, S. aureus, as many other intracellular pathogens [51], needs to adapt to the available nutrients and other physiological conditions (pH, temperature, oxygen). Since hexose phosphates are abundant carbon sources within the host cell cytosol, the HptSR system is important for intracellular survival and multiplication of S. aureus within host cells (Figure 2). Indeed, S. aureus strains deficient in hptSR show impaired survival/multiplication within mammalian cells [46]. S. saprophyticus is an extracellular pathogen that shows strong adhesion to various epithelial cell lines. Internalization of S. saprophyticus has been described in human bladder carcinoma cell lines [46], but the relevance to UTI infection has not been documented. Even if S. saprophyticus might get internalized by some epithelial cells during its life cycle, the absence of a system to take up hexose phosphates would impair the replication and multiplication of the bacterium in the cell’s cytoplasm. Alternatively, it cannot be excluded that the absence of HptSR might be replaced by a different pathway for the uptake of glucose 6-phosphate or that other nutrients are important for intracellular survival of S. saprophyticus. The HptSR system involves four genes that are located together in the S. aureus genome. Thus, complementation of S. saprophyticus with the whole system would be the direct approach to determine whether HptSR is sufficient to confer S. saprophyticus the capacity to replicate in the cytosol of the host cell.
Conclusion and future directions
S. aureus and S. saprophyticus have been classified as high and medium-pathogenic staphylococci, respectively. Medium-pathogenic staphylococci are more specialized in their infective strategies and cause a narrow spectrum of diseases [52]. The reduced number of TCSs is not by itself the reason for a lower pathogenic capacity because many other medium-pathogenic staphylococci (S. epidermidis, S. lugdunensis, S. haemolyticus and S. pseudointermedius) have at least the same number of TCSs as S. aureus. Instead, the number of TCSs correlates with the presence/absence of those genes encoding for metabolic pathways necessary to adapt bacterial growth to the hostile environment of the host and to harmonize their expression. A question that remains open is whether the common staphylococcal ancestor contained the sixteen TCSs and S. saprophyticus suffered genome reduction events during evolution or alternatively, the staphylococcal ancestor contained a lower number of TCSs and S. saprophyticus, contrary to other staphylococcal species, did not gain additional TCSs. The mechanisms of TCS acquisition/loss during bacterial evolution have been theoretically predicted but they have not been experimentally addressed. Hence, an effort that considers long-term experiments, in which bacteria gain a selective advantage through the acquisition/loss of a TCS would provide insights into how adaptation to different niches was established over evolutionary time.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
CRediT authorship contribution statement
Beatriz Rapun-Araiz: Data curation, Investigation, Methodology, Writing - review & editing. Andreas F Haag: Data curation, Investigation, Methodology, Writing - review & editing. Cristina Solano: Funding acquisition, Writing - review & editing. Iñigo Lasa: Conceptualization, Data curation, Investigation, Methodology, Funding acquisition, Writing - original draft, Writing - review & editing.
Acknowledgements
B.R is recipient of a PhD grant from Universidad Pública de Navarra. Work in the Laboratory of Microbial Pathogenesis is funded by the Spanish Ministry of Science, Innovation and Universities grant BIO2017-83035-R Agencia Española de Investigación/Fondo Europeo de Desarrollo Regional, European Union. A.F.H. is supported by the European Research Council ERC under the European Union’s Horizon 2020 research and innovation program Grant Agreement ERC-ADG-2014 Proposal n° 670932 Dut-signal from EU awarded to José R. Penadés) and was the recipient of a Tenovus Project Grant (S16-12).
References
- 1.Galperin M.Y. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulrich L.E., Zhulin I.B. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2010;38:D401–407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capra E.J., Laub M.T. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salazar M.E., Laub M.T. Temporal and evolutionary dynamics of two-component signaling pathways. Curr Opin Microbiol. 2015;24C:7–14. doi: 10.1016/j.mib.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian D., Harper L., Shopsin B., Torres V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog Dis. 2017;75 doi: 10.1093/femspd/ftx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krismer B., Weidenmaier C., Zipperer A., Peschel A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat Rev Microbiol. 2017;15:675–687. doi: 10.1038/nrmicro.2017.104. [DOI] [PubMed] [Google Scholar]
- 7.Barakat M., Ortet P., Jourlin-Castelli C., Ansaldi M., Méjean V., Whitworth D.E. P2CS: a two-component system resource for prokaryotic signal transduction research. BMC Genomics. 2009;10:315. doi: 10.1186/1471-2164-10-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galperin M.Y., Higdon R., Kolker E. Interplay of heritage and habitat in the distribution of bacterial signal transduction systems. Mol BioSyst. 2010;6:721–728. doi: 10.1039/b908047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Gumerov V.M., Ortega D.R., Adebali O., Ulrich L.E., Zhulin I.B. MiST 3.0: an updated microbial signal transduction database with an emphasis on chemosensory systems. Nucleic Acids Res. 2020;48:D459–D464. doi: 10.1093/nar/gkz988. [DOI] [PMC free article] [PubMed] [Google Scholar]; An updated microbial signal transduction database that facilitates theoretical and experimental studies on signal transduction and gene regulation.
- 10.Dubrac S., Boneca I.G., Poupel O., Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189:8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgui S., Gil C., Solano C., Lasa I., Valle J. A systematic evaluation of the two-component systems network reveals that ArlRS is a key regulator of catheter colonization by Staphylococcus aureus. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00342. 6069–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Villanueva M., García B., Valle J., Rapun B., Ruiz de Los Mozos I., Solano C., Marti M., Penadés J.R., Toledo-Arana A., Lasa I. Sensory deprivation in Staphylococcus aureus. Nat Commun. 2018;9 doi: 10.1038/s41467-018-02949-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that S. aureus containing a unique TCS, WalKR, is viable in laboratory conditions, confirming that WalKR is the only TCS strictly required for growth. Importantly, it also shows that TCSs are self-sufficient autonomous entities.
- 13.Kuroda M., Yamashita A., Hirakawa H., Kumano M., Morikawa K., Higashide M., Maruyama A., Inose Y., Matoba K., Toh H. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc Natl Acad Sci U S A. 2005;102:13272–13277. doi: 10.1073/pnas.0502950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Argemi X., Hansmann Y., Prola K., Prévost G. Coagulase-negative staphylococci pathogenomics. IJMS. 2019;20 doi: 10.3390/ijms20051215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kline K.A., Lewis A.L. Gram-positive uropathogens, polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.UTI-0012-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores-Mireles A.L., Walker J.N., Caparon M., Hultgren S.J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Haag A.F., Bagnoli F. The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr Top Microbiol Immunol. 2017:145–198. doi: 10.1007/82_2015_5019. [DOI] [PubMed] [Google Scholar]; This study provides a careful overview of the current knowledge regarding the function of the TCSs of S. aureus.
- 18.Graham J.E., Wilkinson B.J. Staphylococcus aureus osmoregulation: roles for choline, glycine betaine, proline, and taurine. J Bacteriol. 1992;174:2711–2716. doi: 10.1128/jb.174.8.2711-2716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gründling A. Potassium uptake systems in Staphylococcus aureus: new stories about ancient systems. mBio. 2013;4:e00784–13. doi: 10.1128/mBio.00784-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moscoso J.A., Schramke H., Zhang Y., Tosi T., Dehbi A., Jung K., Gründling A. Binding of cyclic di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the universal stress protein domain and downregulates the expression of the Kdp potassium transporter. J Bacteriol. 2016;198:98–110. doi: 10.1128/JB.00480-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price-Whelan A., Poon C.K., Benson M.A., Eidem T.T., Roux C.M., Boyd J.M., Dunman P.M., Torres V.J., Krulwich T.A. Transcriptional profiling of Staphylococcus aureus during growth in 2 M NaCl leads to clarification of physiological roles for Kdp and Ktr K+ uptake systems. mBio. 2013;4:398. doi: 10.1128/mBio.00407-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gries C.M., Bose J.L., Nuxoll A.S., Fey P.D., Bayles K.W. The Ktr potassium transport system in Staphylococcus aureus and its role in cell physiology, antimicrobial resistance and pathogenesis. Mol Microbiol. 2013;89:760–773. doi: 10.1111/mmi.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeman Z.N., Dorus S., Waterfield N.R. The KdpD/KdpE two-component system: integrating K+ homeostasis and virulence. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003201.t003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David M.Z., Daum R.S. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.O’Connor A.M., McManus B.A., Coleman D.C. First description of novel arginine catabolic mobile elements (ACMEs) types IV and V harboring a kdp operon in Staphylococcus epidermidis characterized by whole genome sequencing. Infect Genet Evol. 2018;61:60–66. doi: 10.1016/j.meegid.2018.03.012. [DOI] [PubMed] [Google Scholar]; This study identifies S. epidermidis strains in which the chromosomal copy of the kdpDE TCS has been replaced by a KdpDE system contained in a new type of mobile element (ACME element).
- 26.McManus B.A., O’Connor A.M., Egan S.A., Flanagan P.R., Coleman D.C. First description of arginine catabolic mobile element (ACME) type VI harboring the kdp operon only in Staphylococcus epidermidis using short and long read whole genome sequencing: further evidence of ACME diversity. Infect Genet Evol. 2019;71:51–53. doi: 10.1016/j.meegid.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Cue D., Junecko J.M., Lei M.G., Blevins J.S., Smeltzer M.S., Lee C.Y. SaeRS-Dependent inhibition of biofilm formation in Staphylococcus aureus newman. PLoS One. 2015;10:e0123027–20. doi: 10.1371/journal.pone.0123027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foster T.J., Geoghegan J.A., Ganesh V.K., Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown A.F., Leech J.M., Rogers T.R., McLoughlin R.M. Staphylococcus aureus colonization: modulation of host immune response and impact on human vaccine design. Front Immunol. 2014;4:507. doi: 10.3389/fimmu.2013.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giraudo A.T., Calzolari A., Cataldi A.A., Bogni C., Nagel R. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett. 1999;177:15–22. doi: 10.1111/j.1574-6968.1999.tb13707.x. [DOI] [PubMed] [Google Scholar]
- 31.Liang X., Yu C., Sun J., Liu H., Landwehr C., Holmes D., Ji Y. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun. 2006;74:4655–4665. doi: 10.1128/IAI.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.de Paiva-Santos W., de Sousa V.S., Giambiagi-deMarval M. Occurrence of virulence-associated genes among Staphylococcus saprophyticus isolated from different sources. Microb Pathog. 2018;119:9–11. doi: 10.1016/j.micpath.2018.03.054. [DOI] [PubMed] [Google Scholar]; This study investigates the distribution of virulence-associated genes and analyzes the pathogenic potential of S. saprophyticus strains from different sources.
- 33.Kline K.A., Ingersoll M.A., Nielsen H.V., Sakinc T., Henriques-Normark B., Gatermann S., Caparon M.G., Hultgren S.J. Characterization of a novel murine model of Staphylococcus saprophyticus urinary tract infection reveals roles for Ssp and SdrI in virulence. Infect Immun. 2010;78:1943–1951. doi: 10.1128/IAI.01235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakinc T., Michalski N., Kleine B., Gatermann S.G. The uropathogenic species Staphylococcus saprophyticus tolerates a high concentration of d-serine. FEMS Microbiol Lett. 2009;299:60–64. doi: 10.1111/j.1574-6968.2009.01731.x. [DOI] [PubMed] [Google Scholar]
- 35.Schlag S., Fuchs S., Nerz C., Gaupp R., Engelmann S., Liebeke M., Lalk M., Hecker M., Gotz F. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J Bacteriol. 2008;190:7847–7858. doi: 10.1128/JB.00905-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilkens S., Koch-Singenstreu M., Niemann V., Götz F., Stehle T., Unden G. Nitrate/oxygen co-sensing by an NreA/NreB sensor complex of Staphylococcus carnosus. Mol Microbiol. 2014;91:381–393. doi: 10.1111/mmi.12464. [DOI] [PubMed] [Google Scholar]
- 37.Hajek V., Meugnier H., Bes M., Brun Y., Fiedler F., Chmela Z., Lasne Y., Fleurette J., Freney J. Staphylococcus saprophyticus subsp, bovis subsp nov, isolated from bovine nostrils. Int J Syst Bacteriol. 1996;46:792–796. doi: 10.1099/00207713-46-3-792. [DOI] [PubMed] [Google Scholar]
- 38.Gatermann S., John J., Marre R. Staphylococcus saprophyticus urease: characterization and contribution to uropathogenicity in unobstructed urinary tract infection of rats. Infect Immun. 1989;57:110–116. doi: 10.1128/iai.57.1.110-116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo M., Kato F., Oogai Y., Kawai T., Sugai M., Komatsuzawa H. Distinct two-component systems in methicillin-resistant Staphylococcus aureus can change the susceptibility to antimicrobial agents. J Antimicrob Chemother. 2010;65:1536–1537. doi: 10.1093/jac/dkq141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blake K.L., Randall C.P., O’Neill A.J. In vitro studies indicate a high resistance potential for the lantibiotic nisin in Staphylococcus aureus and define a genetic basis for nisin resistance. Antimicrob Agents Chemother. 2011;55:2362–2368. doi: 10.1128/AAC.01077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiron A., Falord M., Valle J., Débarbouillé M., Msadek T. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol Microbiol. 2011;81:602–622. doi: 10.1111/j.1365-2958.2011.07735.x. [DOI] [PubMed] [Google Scholar]
- 42.Falord M., Mäder U., Hiron A., Débarbouillé M., Msadek T. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Randall C.P., Gupta A., Utley-Drew B., Lee S.Y., Morrison-Williams G., O’Neill A.J. Acquired nisin resistance in Staphylococcus aureus involves constitutive activation of an intrinsic peptide antibiotic detoxification module. mSphere. 2018;3 doi: 10.1128/mSphereDirect.00633-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Coates-Brown R., Moran J.C., Pongchaikul P., Darby A.C., Horsburgh M.J. Comparative genomics of Staphylococcus reveals determinants of speciation and diversification of antimicrobial defense. Front Microbiol. 2018;9:2753. doi: 10.3389/fmicb.2018.02753. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using experimental evolution of representative braSR-positive and braSR-negative species with nisin selection, this study shows a differential selection of BraSR and GraSR to produce resistance to these antimicrobial peptides.
- 45.Yang S.-J., Bayer A.S., Mishra N.N., Meehl M., Ledala N., Yeaman M.R., Xiong Y.Q., Cheung A.L. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect Immun. 2012;80:74–81. doi: 10.1128/IAI.05669-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J.Y., Kim J.W., Moon B.Y., Lee J., Fortin Y.J., Austin F.W., Yang S.-J., Seo K.S. Characterization of a novel two-component regulatory system, HptRS, the regulator for the hexose phosphate transport system in Staphylococcus aureus. Infect Immun. 2015;83:1620–1628. doi: 10.1128/IAI.03109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu S., Fu Z., Zhou Y., Liu Y., Xu X., Wang M. Mutations of the transporter proteins GlpT and UhpT confer Fosfomycin resistance in Staphylococcus aureus. Front Microbiol. 2017;8:914. doi: 10.3389/fmicb.2017.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rollin G., Tan X., Tros F., Dupuis M., Nassif X., Charbit A., Coureuil M. Intracellular survival of Staphylococcus aureus in endothelial cells: a matter of growth or persistence. Front Microbiol. 2017;8:1354. doi: 10.3389/fmicb.2017.01354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jorch S.K., Surewaard B.G., Hossain M., Peiseler M., Deppermann C., Deng J., Bogoslowski A., van der Wal F., Omri A., Hickey M.J. Peritoneal GATA6+ macrophages function as a portal for Staphylococcus aureus dissemination. J Clin Invest. 2019;129:4643–4656. doi: 10.1172/JCI127286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowe S.E., Wagner N.J., Li L., Beam J.E., Wilkinson A.D., Radlinski L.C., Zhang Q., Miao E.A., Conlon B.P. Reactive oxygen species induce antibiotic tolerance during systemic Staphylococcus aureus infection. Nat Microbiol. 2019;339:520–529. doi: 10.1038/s41564-019-0627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chico-Calero I., Suárez M., Gonzalez-Zorn B., Scortti M., Slaghuis J., Goebel W., Vázquez-Boland J.A. European listeria genome consortium: Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci U S A. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenstein R., Götz F. What distinguishes highly pathogenic Staphylococci from medium-and non-pathogenic? In: Rosenstein R., Götz F., Svanborg C., editors. Between Pathogenicity and Commensalism. Springer Science & Business Media; 2014. pp. 33–91. [DOI] [PubMed] [Google Scholar]