Abstract
PIK3CA mutation frequency varies among breast cancer (BC) subtypes. Recent evidence suggests combination therapy with the PI3K inhibitor (PI3Ki) alpelisib and endocrine therapy (ET) improves response rates and progression-free survival (PFS) in PIK3CA-mutant, hormone receptor positive (HR+) BC versus ET alone; thus, better understanding the clinical and epidemiologic elements of these mutations is warranted. This systematic review characterizes the PIK3CA mutation epidemiology, type of testing approaches (e.g., liquid or tissue tumor biopsy), and stability/concordance (e.g., consistency in results by liquid versus solid tumor sample, by the same method over time) in patients with HR+/HER2– advanced (locally unresectable) or metastatic disease (HR+/HER2– mBC) and explores performance (e.g., pairwise concordance, sensitivity, specificity, or predictive value) of respective mutation findings. A comprehensive search of PubMed/MEDLINE, EMBASE, Cochrane Central, and select conference abstracts (i.e., AACR, ASCO, SABCS, ECCO, and ESMO conferences between 2014 and 2017) identified 39 studies of patients with HR+, HER2– mBC. The median prevalence of PIK3CA mutation was 36% (range: 13.3% to 61.5%); identified testing approaches more commonly used tissue over liquid biopsies and primarily utilized next-generation sequencing (NGS), polymerase chain reaction (PCR), or Sanger sequencing. There was concordance and stability between tissues (range: 70.4% to 94%) based on limited data. Given the clinical benefit of the PI3Ki alpelisib in patients with PIK3CA mutant HR+/HER2– mBC, determination of tumor PIK3CA mutation status is of importance in managing patients with HR+/HER2– mBC. Prevalence of this mutation and utility of test methodologies likely warrants PIK3CA mutation testing in all patients with this breast cancer subtype via definitive assessment of PIK3CA mutational status.
1. Introduction
With an estimated 271,270 new cases in 2019, breast cancer (BC) is the most common nonskin cancer in women in the United States (US) [1]. Although most BC cases are diagnosed in the early stages, approximately 10 to 41% of patients develop metastatic or advanced (locally unresectable; stage 3 or 4) disease, depending on tumor characteristics and presentation [2]. The BC subtype known as hormone receptor positive, human epidermal growth factor receptor-2 negative (HR+/HER2-) represents 70% of cases [3]. The phosphoinositide 3-kinase (PI3K) pathway is the most frequently altered pathway in HR+ BC and is associated with tumor development, disease progression, and endocrine resistance [4]. The impact of PIK3CA mutation status on BC progression (e.g., localized to metastatic disease) is uncertain [5]. Current treatment options for postmenopausal HR+/HER2- advanced BC include endocrine therapy (ET) +/- a CDK 4/6 inhibitor, an mTOR inhibitor, or chemotherapy (CT) [6]. However, ET or TT+ET rather than chemotherapy constitutes the initial therapy usually administered for women with HR+ advanced BC; TT+ET has more manageable safety profiles than CT [7]. The National Comprehensive Cancer Network (NCCN) guidelines recommend that CT can be used [8] for patients where no clinical benefit is observed after 3 consecutive endocrine-based therapies (including ET and TT+ET) or for patients with symptomatic visceral disease. A growing body of research suggests that use of a phosphoinositide 3-kinase inhibitor (PI3Ki) in conjunction with ET may improve response rates and progression-free survival in PIK3CA-mutant, estrogen receptor positive (ER+) BC relative to ET alone [7, 9–12], precluding or delaying the need for CT. Additionally, the recent results of the SOLAR-1 Phase III trial provided evidence that the PI3Ki alpelisib given with fulvestrant, as opposed to placebo plus fulvestrant, improved PFS among patients with PIK3CA-mutated HR+/HER2- mBC who had received endocrine therapy previously [13], leading to FDA approval of alpelisib. The majority of patients in this trial had metastatic disease.
The frequency of PIK3CA mutations varies across different BC molecular subgroups [5]. One study found a 41.1% frequency of PIK3CA mutation in HR+/HER2- breast cancer compared to 12.5% of patients with triple negative breast cancer [14]. Previous evidence indicates that PI3Ki are active in postmenopausal women with PIK3CA-mutant HR+/HER2- advanced or metastatic breast cancer [12, 15, 16]; thus, detection of these PIK3CA mutations in tumors is important in identifying those patients most likely to benefit from treatment using a PI3Ki. Until recently, clinical guidelines did not recommend PIK3CA mutation testing as a part of standard testing (such as HR and HER2 status) [17, 18]. As such, the majority of testing has been performed by commercial next-generation sequencing (NGS) platforms and at institutions where in-house gene panels have been developed [19]. To date, the diagnostic yield (i.e., the proportion of patients in whom the testing technique yields a definitive diagnosis) of BC PIK3CA mutation testing has been challenging to measure, given the variability in prevalence of PIK3CA mutations throughout BC subtypes and lack of guidelines for testing in clinical practice [20]. Furthermore, a systematic understanding is lacking regarding the prevalence of PIK3CA mutations in HR+/HER2- advanced/unresectable or metastatic breast cancer or within clinically relevant molecular BC subgroups. Understanding the prevalence is important to support quantifying the size of the patient pool that may benefit from receiving PIK3CA testing.
Various biopsy and analytical testing approaches exist to detect PIK3CA mutations. However, evidence is lacking with respect to the real-world generalizability and applicability across tests due to the variations in test performance across approaches. Namely, tests for PIK3CA mutation have not been routinely performed in the clinical setting among patients with HR+/HER2- mBC, and concordance between testing methods (e.g., NGS vs. PCR), test location (e.g., primary site vs. metastasis), type of biopsy (e.g., liquid vs. tissue), or retest concordance (i.e., stability over time) are not well documented. Patients may undergo multiple tumor biopsies over time, particularly if the disease progresses during a specific treatment, and consequently, the test results will influence clinician decisions regarding subsequent therapy. Also, the tumor biopsy site may change over time. For example, tissue may be tested initially using archived tumor obtained at time of diagnosis, and due to a lack of a convenient site for new tissue biopsy, a liquid biopsy may be performed at a later time during therapy for metastatic disease [20, 21]. Demonstrating concordance (i.e., agreement in test results among testing methods) and stability (i.e., consistency in test results) between tests over time will reduce the clinical burden for both patients and providers by minimizing the need for repeat and/or invasive testing. Further, high concordance between tissue and liquid biopsy test results could lead to better convenience in patient care based on the availability of different testing technologies in various global clinical settings. Discordance between samples analyzed for gene mutations using distinct methods from the same or other sources of tumor DNA could be minimized if differences in sensitivities among testing methods are identified/understood.
The purpose of this systematic review was to describe the epidemiology of PIK3CA mutations and variation across respective PIK3CA testing methods among patients with PIK3CA-mutant or wild-type HR+/HER2- advanced or metastatic breast cancer. Additionally, this review is aimed at (1) describing the type of biopsy and analytical approaches for PIK3CA mutation testing and (2) assessing the performance (e.g., pairwise concordance, sensitivity, specificity, or predictive value between test types) and stability (e.g., consistency of result over time) of PIK3CA mutation findings.
2. Methods
2.1. Study Design
This review followed the Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) guidelines [22].
2.2. Participants, Interventions, and Comparators
At the level of titles and abstracts, we screened the following inclusion criteria: conducted with human subjects or human tissue, included a population or subpopulation with HR+/HER2- mBC, and reported information on the presence of the PIK3CA mutation. Status of cases as advanced or metastatic was accepted based on study authors' determination, but definitions for advanced cases were not universally available. Unless otherwise defined, advanced refers to those cases that are locally unresectable. Following title and abstract-level screenings, full-text articles and posters were reviewed for the following inclusion criteria: included patients with HR+/HER2- mBC, included either tissue or liquid biopsies, and reported PIK3CA mutation status among the HR+/HER2- mBC subgroup.
2.3. Systematic Review Protocol
The search strategy followed an a priori review protocol developed internally.
2.4. Search Strategy
A comprehensive search of PubMed/MEDLINE, EMBASE, Cochrane Central, and select conference abstracts (i.e., AACR, ASCO, SABCS, ECCO, and ESMO conferences between 2014 and 2019) was performed including, but not limited to, keywords: “breast neoplasm,” “PIK3CA protein,” “hormone receptor positive,” and “metastases” including all the search terms: text words (free text), subject index headings (e.g., MeSH), and the relationship between the search terms (e.g., Boolean). Full search strategy is available in Appendix A. Databases were searched on August 28, 2017, for English-language publications of patients with HR+, HER2– mBC published between January 1993 and April 2019 (with the last search occurring April 2019). The initial search performed in 2017 was updated in 2019 using both the comprehensive search terms described in Appendix A plus hand searching of new conference abstracts as well as the acronyms for clinical trials ongoing as of 2017 that were identified in the initial search (e.g., BELLE-3).
2.5. Data Sources, Study Sections, and Data Extraction
Data extraction methods followed Cochrane guidelines for systematic literature reviews [23]. Two independent researchers screened titles and abstracts resulting from all searches to identify potentially eligible studies. A full-text review was then performed by two independent researchers; any discrepancies were settled by an independent third researcher. Data extraction from full publication texts was performed by two independent researchers. The following data were compiled in a standardized table and validated by one or more other authors: study design, study country, number of participants and samples, participant demographics, testing methods, analytical approach and corresponding manufacturer data, mutations detected using corresponding analytical approach, primary tumor or metastasis location and size, frequency of mutation, concordance between testing sites or testing method as reported, and stability over time (Table 1). Where multiple reports were published based on the same study, frequency data were not duplicated but, rather, extracted from the most comprehensive publication. No contact with study authors was necessary.
Table 1.
Study characteristics, prevalence of PIK3CA mutations, and reported hotspots.
| Authors | Year | Country | Number of participants with HR+/HER2- BC | Age1 | Type of biopsy | HR+/HER2- PIK3CA wild type | HR+/HER2- PIK3CA mutants | Prevalence of PIK3CA mutants | HR+/HER2- PIK3CA hotspots2 |
|---|---|---|---|---|---|---|---|---|---|
| n | Median (range) | n | n | % | (n) | ||||
| Abramson et al. [44] | 2014 | US only | 110 | 52 | Tissue | 71 | 39 | 35.5% | |
| Andre et a1. [13] | 2019 | US including international | 572 | 63 (25-92) | Tissue | 231 | 341 | NA3 | E542K (60); E545X (105); H1047 (193); C420 (6); Q546 (5)3 |
| Rugo et al. [33]a | 2018 | 284 | Liquid | 115 | 169 | NA3 | |||
| Ahmad et al. [45] | 2016 | International excluding US | 89 | 50 | Tissue | 67 | 22 | 24.7% | H1047R (13); H1047L (1); E542K (2); E545A (1); E545G (1) |
| Arthur et al. [43] | 2014 | International excluding US | 50 | 68 (25-94) | Tissue | 24 | 26 | 52.0% | |
| Baselga et al. [9] | 2017 | US including international | 1147 | 62 | Liquid | 387 | 200 | 34.1% | |
| Tissue | 584 | 276 | 32.1% | ||||||
| Campone et al. [31]b | 2018 | 1147 | |||||||
| Basho et al. [46] | 2016 | US only | 343 | 47 (23-74) | Tissue | 236 | 107 | 31.2% | |
| Baird et al. [47] | 2016 | International excluding US | 30 | 53 (35-81) | Liquid | 26 | 4 | 13.3% | H1047R (3); E545K (1) |
| Basu et al. [48] | 2017 | US only | 11 | n/a | Tissue | 7 | 4 | 36.4% | E545K (2) H1047R (2) |
| Beelen et al. [49] | 2014 | International excluding US | 41 | n/a | Tissue | 308 | 153 | 33.2% | Exon 20 (82), exon 9 (71) |
| Bertucci et al. [29] | 2016 | International excluding US | 9 | 41 (33-72) | Tissue | 4 | 5 | 55.6% | N345K (1); C420R (1); E542K (2) |
| Blackwell et al. [50] | 2015 | US including international | 72 | n/a | Tissue | 37 | 18 | 32.7% | E545K (1); Q546K (1); H1047R (10); H1047L (2) |
| Board et al. [51] | 2010 | International excluding US | 64 | n/a | Liquid, tissue | 33 | 10 | 28.00% (L) 23.26% (T) |
H1047R (4); H1047L (2); E545K (6); E542K (1) |
| Christgen et al. [52] | 2013 | International excluding US | 46 | n/a | Tissue | 22 | 24 | 52.2% | |
| Dickler [30] | 2016 | US including international | 60 | 62 (31-82) | Liquid | 25 | 22 | 46.8% | |
| Tissue | 29 | 31 | 51.7% | ||||||
| Di Leo et al. [10] | 2018 | US including international | 432 | 61 | Tissue | 204 | 109 | 34.0% | |
| Filipenko et al. [53] | 2017 | International excluding US | 263 | n/a | Tissue | 10 | 32 | 76.2% | E542K (19); E545K (2); H1047R (8); H1047L (3) |
| Fleming et al. [54] | 2012 | US only | 21 | n/a | Tissue | 18 | 5 | 21.7% | |
| Gasch et al. [55] | 2016 | International excluding US | 23 | n/a | Liquid | 13 | 10 | 43.5% | |
| Gonzalez-Angulo et al. [56] | 2011 | US only | 37 | 48 (30-83) | Tissue | 19 | 15 | 44.1% | |
| Henderson et al. [57] | 2016 | US only | 237 | n/a | Tissue | 129 | 108 | 45.6% | H1047A (37); Glu542Lys (27); GLu545Lys (26); other (18) |
| Lefebvre et al. [58] | 2016 | International excluding US | 143 | 55 (26-82) | Tissue | 90 | 53 | 37.1% | |
| Liu et al. [59] | 2015 | US including international | 44 | 56 | Tissue | 30 | 14 | 32.5% | |
| Mayer et al. [60] | 2017 | US only | 26 | 53 | Tissue | 10 | 16 | 61.5% | |
| Mayer et al. [61] | 2014 | US only | 51 | 55 (34-77) | Tissue | 35 | 16 | 31.4% | |
| Moynahan et al. [28] | 2017 | US including international | 550 | 61 (54-68) | Liquid | 312 | 238 | 43.3% | H1047R (138); E545K (61); E542K (39) |
| Muller et al. [62] | 2016 | US only | 13 | 61 | Tissue | 8 | 5 | 38.5% | |
| Oliveira et al. [63] | 2016 | US including international | 91 | 56 | Tissue | 65 | 26 | 28.6% | |
| Roy-Chowdhuri et al. [64] | 2015 | US only | 132 | n/a | Tissue | 82 | 50 | 38.0% | |
| Sakr et al. [65] | 2014 | US only | 31 | n/a | Tissue | 16 | 8 | 33.3% | |
| Soucier-Ernst et al. [66] | 2015 | US only | 28 | 56 (31-78) | Tissue | 14 | 14 | 50.0% | |
| Vetter [67] | 2014 | US including international | 618 | n/a | Tissue | 441 | 177 | 28.6% | |
| Wang et al. [68] | 2015 | US only | 22 | 57 (32-79) | Tissue | 19 | 5 | 20.8% | |
| Welt et al. [69] | 2013 | US including international | 73 | 63 | Tissue | 13 | 4 | 23.5% | |
| Williams et al. [70] | 2014 | US only | 39 | n/a | Tissue | 23 | 8 | 26.0% | |
| Yuan et al. [71] | 2016 | US only | 16 | 54.5 (34-78) | Tissue | 7 | 9 | 56.3% | |
| Yuan et al. [35] | 2015 | International excluding US | 376 | 49 | Tissue | 261 | 115 | 30.6% | |
| Zhang et al. [72] | 2014 | International excluding US | 90 | 47 (25-71) | Tissue | 60 | 30 | 33.3% | H1047R (13); H1047L (4); E545K (9); E542K (4) |
1Median age and range reported where available; 2hotspots as reported; not all studies assessed by hotspot or reported hotspot by ER+/HER2- status; 3Participants with confirmed PIK3CA mutations were intentionally overselected for this trial. US: United States; HR+/HER2- BC: hormone receptor positive/human epidermal growth factor receptor-2 negative breast cancer; PIK3CA: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; L: liquid; T: tissue. aTwo reports on participants in the SOLAR-1 phase 3 trial. bTwo reports on participants in the BELLE-2 phase 3 trial.
Among full-text reports or posters considered for inclusion, the following criteria were used to determine if PIK3CA prevalence was reported: proportion of HR+/HER2- samples with PIK3CA mutation, number of samples tested for PIK3CA hotspot mutation, or performed a subgroup analysis by mutation status. To perform subanalyses relating to the prevalence of the PIK3CA mutation, the testing types utilized, and concordance between tests, studies meeting the initial inclusion criteria were subjected to additional specific inclusion criteria (e.g., relative to subanalysis) and categorized accordingly.
Concordance data related to the following factors were extracted: between analytical approaches (i.e., PCR vs. NGS vs. Sanger sequencing), between tissue samples (presence of mutations in primary tumors versus those from metastatic lesions), and between biopsy methods (i.e., tumor tissue biopsy versus liquid biopsy) when the assessment was done at a variety of time points, including time of initial diagnosis and time at disease progression. When available, we sought to compare results obtained using different modalities of testing obtained at the same time during a patient's oncologic course, the same modality of testing obtained over different times during a patient's oncologic course, and different modalities of testing obtained at different times over a patient's oncologic course.
2.6. Data Analysis
Included studies were assessed using content analysis, a systematic technique for describing data and outcomes using qualitative methods [24] and reports of prevalence. Descriptive statistics on the overall prevalence as well as prevalence for these subgroups considered clinically important by the clinical experts on this study team were computed: geographic location (US vs. international), study design (clinical trial vs. observational research), testing approaches (including type and location of sample tested and type of analytical testing method employed), and the manufacturer of the test. Among these subgroups, independent group t-tests were performed to measure difference in the mean prevalence of the PIK3CA mutation. Considering the wide range of study types and study designs included in the review, no meta-analysis was conducted.
The risk of bias for each study included for full-text review was independently completed by two authors using the Mixed Methods Appraisal Tool (MMAT, [25], Table 2). This tool was selected given the variety of study types, using both clinical and observational data, considered in this report. The risk of bias results were used to contextualize the relative strength of each included publication but did not influence inclusion or exclusion of studies. The MMAT has been previously evaluated for content validity and methodological quality [26, 27].
Table 2.
Risk of bias assessment results using the Mixed Methods Appraisal Tool (MMAT).
| Study | Study design | MMAT criteria met1 | Risk of bias score2 | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| Quantitative descriptive | ||||||
| Abramson et al. (2014) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Andre et al. (2018)3 | Clinical | N | Y | Y | Y | ∗∗∗ |
| Ahmad et al. (2016) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Arthur et al. (2014) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Basho et al. (2016) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Basu et al. (2016)3 | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Beelen et al. (2014) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Bertucci et al. (2016) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Board et al. (2010) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Campone et al. (2018) | Clinical | Y | N | Y | Y | ∗∗∗ |
| Christgen et al. (2013) | Observational | Y | Y | Y | N | ∗∗∗ |
| Dickler et al. (SABCS, 2016)3 | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Filipenko et al. (2017) | Observational | N | N | Y | Y | ∗∗ |
| Fleming et al. (2012) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Gasch et al. (2016) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Gonzalez-Angulo et al. (2011)3 | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Fitzgerald et al. (2016) | Observational | Y | Y | Y | N | ∗∗∗ |
| Lefebvre et al. (2016) | Observational | Y | N | Y | Y | ∗∗∗ |
| Liu et al. (2015) | Observational | N | Y | Y | Y | ∗∗∗ |
| Mayer et al. (2014) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Mayer et al. (2016) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Moynahan et al. (2017) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Muller et al. (2016) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Oliveira et al. (2016)3 | Observational | N | N | Y | N | ∗ |
| Roy-Chowdhuri et al. (2015) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Rugo et al. (2019)3 | Clinical | N | Y | Y | Y | ∗∗∗ |
| Sakr et al. (2014) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Soucier-Ernst et al. (2015)3 | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Vetter et al. (2014)3 | Observational | Y | N | Y | N | ∗∗ |
| Wang et al. (2015)3 | Observational | N | N | Y | N | ∗ |
| Welt et al. (2013) | Observational | Y | Y | Y | Y | ∗∗∗∗ |
| Williams et al. (2014)3 | Observational | Y | N | Y | N | ∗∗ |
| Yuan et al. (2015) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Yuan et al. (2016) | Clinical | Y | N | Y | N | ∗∗ |
| Zhang et al. (2014) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Quantitative randomized controlled | ||||||
| Baselga et al. (2017) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Di Leo et al. (2018) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Quantitative nonrandomized | ||||||
| Baird et al. (2016)3 | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Blackwell et al. (2015) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
| Dickler et al. (ASCO, 2016) | Clinical | Y | Y | Y | Y | ∗∗∗∗ |
1Mixed Methods Appraisal Tool (MMAT) criteria available in Appendix B (corresponding with questions 1-4 for respective study type). 2Each ∗ indicates percentage (25%, 50%, 75%, or 100%) of criteria met where ∗ (25%) corresponds to a high risk of bias and ∗∗∗∗ (100%) corresponds to a low risk of bias. 3Study data reported in a poster or conference abstract. Y: yes; N: no.
3. Results
Of 3068 articles and conference abstracts identified, 572 met the inclusion criteria and were included in a full-text review, of which 39 were included. Full study selection process is shown in Figure 1.
Figure 1.
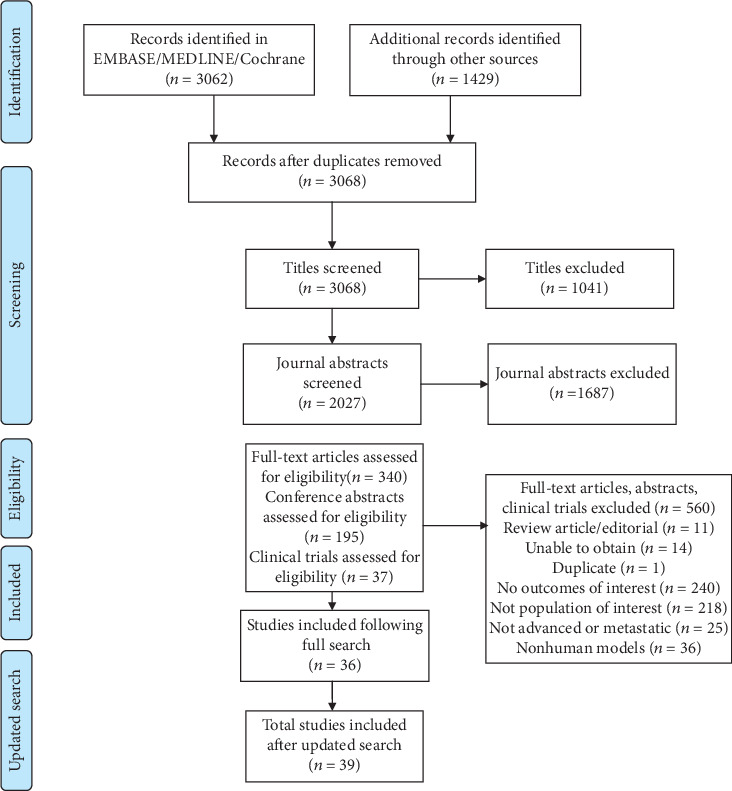
PRISMA flow diagram of article eligibility for inclusion.
3.1. Study Selection and Characteristics
Prevalence of the PIK3CA mutation among HR+/HER2- mBC was reported in 37 of 39 studies that met the inclusion criteria (Table 1). Two studies (both presenting results of the SOLAR-1 Phase 3 trial) were enriched for patients with PIK3CA-mutated tumors and were excluded from prevalence estimation. Eighteen studies were observational in nature, and fifteen were performed in the US; twelve studies were conducted internationally, and twelve included both US and international populations. Across studies, a total of 6825 nonduplicated individual samples were tested for genetic mutations including PIK3CA (inclusive of samples from SOLAR-1, n = 572). The majority of included studies performed genetic testing using tissue biopsy samples (n = 36/39; 92.3%); tissue samples accounted for 73.1% of all samples across all studies (n = 4992). Liquid biopsies were performed in 9 studies, accounting for 26.7% of all samples (n = 1829). Six studies (four trials) reported both tissue and liquid biopsy data (Andre et al., 2019; Baselga et al., 2017; Board et al., 2010; Campone et al., 2018; Dickler et al., 2018; and Rugo et al., 2019). Among 15 studies that reported tumor histology, most samples were from ductal carcinomas (range: 51.5% to 96.1%). Across nine studies that reported tumor histology by PIK3CA mutation status, most were also from ductal carcinomas (range: 63.2% to 99%). The majority (n = 25) of studies included molecular data while 14 studies included data from 11 clinical trials. The sample sizes tested ranged from 9 to 618. Most studies did not specify which samples came from advanced versus metastatic cases so it was not possible to describe identified studies on these strata. At the level of study inclusion, cases were considered advanced based on study authors' determination though definitions for this term were not generally provided. Our baseline assumption for advanced BC was cases that were locally unresectable.
3.2. Prevalence
The overall reported prevalence of the PIK3CA mutation among all HR+/HER2-mBC samples ranged from 13.3% to 61.5% (median = 36.4%, IQR = 31.2%‐45.6%). More specifically, PIK3CA mutation prevalence ranged from 20.8% to 61.5% among tissue biopsies in this subgroup (n = 33 studies) and 43.4% to 46.8% among liquid biopsies (n = 6 studies). No statistically significant differences were observed in reported PIK3CA mutation prevalence by study location (i.e., U.S.-based vs. international) (mean difference = 0.035, p = 0.225), study type (i.e., clinical trial vs. observational) (mean difference = 0.0095, p = 0.836), or biopsy type (i.e., liquid or tissue) (mean difference = −0.033, p = 0.567); none of these variables varied by more than 3% between subgroups. Data from the SOLAR-1 phase 3 trial were excluded from the described prevalence estimates because it selectively enrolled a PIK3CA-mutant cohort and a PIK3CA wild-type cohort that did not reflect the likelihood of the mutation in the overall HR+/HER2- mBC population.
3.3. Hotspot Mutations
Hotspot mutations were reported in six of 39 studies; the most commonly reported among the HR+/HER2- mBC subgroup were H1047R and E545K. Frequency of the H1047R hotspot mutation ranged from 22% to 75% among identified PIK3CA mutations while the E545K mutation ranged from 11.1% to 50%. Other hotspot mutations identified were Q546K, H1047L, E542K, C420R, and N345. Of note, data from SOLAR-1 reported the following frequencies of hotspot mutations in their oversampled PIK3CA-mutant population: H1047X 57%; E542K 18%; E545K 31%. Although several additional studies reported the presence of hotspot mutations, they were not reported for the HR+/HER2- mBC subgroup specifically.
3.4. Testing Methodology and Analysis
The most common source of tumor DNA for identifying PIK3CA mutations among the HR+/HER2-mBC subgroup was tumor tissue biopsies (n = 36). Formalin-fixed paraffin-embedded (FFPE) tissue samples were used in most studies (n = 30) relative to other tissue or liquid sample types. Among liquid biopsies, two studies used circulating tumor cells (CTC), two studies used cell-free DNA (cfDNA), and seven studies (five trials) reported using circulating tumor DNA (ctDNA). Most studies reported using PCR for analysis (n = 28). Additional techniques included NGS (n = 11), Sanger sequencing (n = 5), and liquid chip technology (n = 2). Among clinical trials specifically (n = 12), PCR (n = 9), NGS (n = 1), and NGS plus Sanger sequencing (n = 2) were used. Four studies used multiple methods for analysis (Basho et al., 2016; Blackwell et al., 2015; Board et al., 2010; and Di Leo et al., 2017). Among studies using the NGS technique (n = 10), seven were cross-sectional and 3 were clinical trials. Additionally, ten of the eleven (90.1%) studies using NGS were published between 2015 and 2017.
3.5. Concordance
Among the 39 studies that met the inclusion and exclusion criteria, only six reported data on concordance specific to the HR+/HER2-mBC subgroup (Figure 2). Of these six studies, concordance of PIK3CA mutations between tissue and liquid biopsies ranged from 70.4% to 94%. Studies generally did not report timing of sample collection (e.g., fresh or archival tissues), although some [10, 28, 29] specified whether samples from archival tissue were from primary or metastatic tumor, and one [9] reported that 3% of samples were from fresh biopsy. One study reported that subjects were enrolled according to local PIK3CA mutation testing; however, central testing for PIK3CA mutation was performed but did not report the concordance between the test results [30]. One study (n = 9 participants) reported 100% concordance between the mutation status of primary breast tumors and metastatic lesions [29]. No analysis of stability over time was performed due to insufficient information. For example, in the BOLERO-2, BELLE-2, and BELLE-3 trials, cfDNA or ctDNA was collected at randomization for PIK3CA mutation analysis and compared to results from archival tissue; however, only one trial reported concordance by tissue source [9, 10, 28]. In BELLE-2, 21% of patients who had PIK3CA-wild-type tumor tissue at randomization had evolved to PIK3CA-mutant status via ctDNA collected at a later time point, though the reference period is not reported [31]. In BOLERO-2, concordance of ctDNA against tissue biopsy was numerically higher among metastatic (n = 49) lesions (81.6%) relative to using all tissue samples (either primary or metastatic lesions) (70.4%) [28]. Variations in modalities of testing obtained at one or more time points in a patient's oncologic course were not reported in any of the six studies describing concordance.
Figure 2.
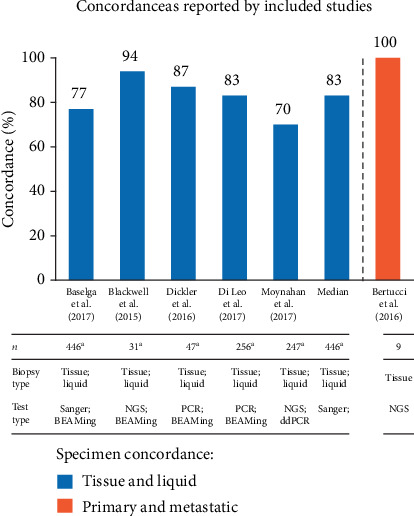
Concordance of PIK3CA mutation status as reported by included studies of HR+/HER2-mBC in clinical or observational trial patients.
3.6. Risk of Bias
The risk of bias for each included publication was stratified by study type per the MMAT (Table 2). Ten publications were rated as moderate to high risk of bias (i.e., did not meet all criteria), with 26 publications rated as low risk of bias (i.e., met all criteria). The risk of bias was performed at the publication rather than at the study level; hence, for those publications (e.g., posters and abstracts) where reported information was limited and a reduced MMAT score was assigned, poor study design (bias) is not necessarily implied. Including gray literature introduces additional bias given that such publications are not peer reviewed. Further, this tool does not account for potential publication bias nor selective reporting across studies that could influence the cumulative evidence given the inclusion of clinical trials.
4. Discussion
Increasingly, systemic therapy in patients with metastatic cancer is being driven by actionable mutations and rearrangements present within tumor tissue. In patients with metastatic non-small-cell lung cancer, for example, testing for EGFR mutations and ALK rearrangements, among other alterations, is critical for selecting the appropriate systemic therapy [32]. Among patients with mBC, systemic treatment has historically been guided by HR and HER2 status; the role of tumor profiling beyond HR/HER2 was less clear. More recently, however, the importance of testing for BRCA and PIK3CA in selected patients with breast cancer has been established by the utility of PARP inhibitors and the PI3Ki alpelisib in selected subpopulations, respectively. In the case of alpelisib, the SOLAR-1 study established improved PFS with alpelisib given with fulvestrant, as opposed to placebo plus fulvestrant, in patients with metastatic, PIK3CA-mutated, HR-positive, HER2-negative advanced or metastatic breast cancer [33], leading to FDA approval of the drug. The results of SOLAR-1 highlight the tangible benefit of testing for PIK3CA mutations in patients with HR+/HER2- mBC, an approach recommended by the NCCN consensus guidelines as well [8]. Various clinical and molecular studies have considered the epidemiological prevalence and clinical significance of PIK3CA mutation in BC using a variety of testing methods. The presence of a PIK3CA mutation is common in BC; however, the associated prognostic and predictive value is uncertain in the current clinical context due to variations in the source of tumor DNA, times when samples are obtained for testing during the disease course, and analytical methodologies and their respective sensitivity/specificity, as well as which hotspots are detected by the test. Our systematic review reports on 5701 advanced/locally unresectable or metastatic BC samples tested for PIK3CA mutation, primarily derived from retrospective analyses of tissue from archived samples, acquired as a part of observational or clinical trials. There is an increasing trend towards conducting clinical trials that test for the presence of genetic mutations up front (at initial diagnosis), yet this typically requires fresh tissue biopsies. However, published data regarding PIK3CAmutations in HR+/HER2– tumors obtained at diagnosis of metastatic disease is limited. Most samples were analyzed from tissue archived from primary tumor and a minority from metastatic tissue biopsies. Tumor DNA from liquid biopsies in some studies was collected with a paired tissue sample, though collection may have occurred at a different time. Since tumor PIK3CA mutation status has been shown to change over time in response to various lines of treatment, better understanding of the prevalence of PIK3CA mutation timeline is warranted, particularly in HR+/HER2– tumors [34, 35].
The median prevalence (36.4%) of PIK3CA mutation among patients with HR+/HER2– mBC, based on included literature, was similar to an estimated rate of 30% among all BC patients [5]. This estimated prevalence among the population of interest varied across testing techniques (i.e., tissue vs. liquid biopsy methods), although there was heterogeneity of reporting methods across studies. Differences in PIK3CA mutation prevalence by study site, biopsy type, and study type were not statistically significant. However, PIK3CA mutations and mutation hotspots (specifically H1047R and E545K) frequently occur in HR+/HER2– mBC. Given the potential prognostic and predictive value of PIK3CA mutation for both courses of treatment and overall survival, future research on the mutation is warranted for both this and other BC subtypes. Also, given differences in outcomes among intrinsic molecular subtypes of HR+ tumors (e.g., luminal A and luminal B), the implications of PIK3CA mutation with regard to these populations warrant consideration [36].
Most hotspot mutations among those with HR+/HER2– mBC were reported in H1047R and E545K and exist in exons 20 and 9, respectively. This finding is consistent with prior literature suggesting that PIK3CA mutations in these two exons are functionally important as oncogenes in breast cancer [37]. Specifically, exon 9 mutations are situated in the helical domain of p110α enabling it to avoid inhibitory regulation by p85, and thus, PI3K becomes an active kinase catalyzing conversion of PIP2 to PIP3. Consequently, subsequent cell signaling leads to increased growth, antiapoptosis, cell-cycle progression, and translation [5, 38]. Exon 20 mutations are situated in the kinase domain; however, the mechanism by which they confer PI3K signaling is not well understood [37]. A study by Lai et al. demonstrated that PIK3CA mutations were present in 26% of patients with invasive ductal breast carcinomas, with more than half of those occurring in exon 20. They further concluded that mutations in exon 20 were an independent risk factor of poor prognosis although any association with BC stage is not reported [39]. Our findings suggest that PIK3CA mutations in these hotspots remain present among the HR+/HER2- mBC subtype, which could have important prognostic value. However, literature reporting PIK3CA hotspot mutations among this important subgroup is lacking, and the sample sizes of those included studies reporting hotspot frequencies were mostly small.
This review reflects the most up-to-date summary of testing methods used to identify PIK3CA mutations associated with HR+/HER2- mBC. Despite the introduction and availability of newer, more precise analytical technologies such as NGS, the majority of studies still utilized PCR to test for PIK3CA mutation, using both tissue samples and liquid biopsies. However, NGS methodology was used more frequently in recent publications from both clinical and molecular trials, suggesting an overall trend towards the newer sequencing method. This discrepancy between PCR and NGS may be problematic for the prevalence of PIK3CA mutation comparisons in the population of interest given that the relative concordance between the two is not fully understood. Given the better suitability of NGS for high-throughput testing requirements with large clinical trials and commercial genomic profiling, PIK3CA mutation testing using PCR or Sanger sequencing may become less frequent [40]. However, recent clinical trials have used PCR to determine PIK3CA mutation status [10, 41]. While data were limited for measures of concordance across time and type of analysis, available evidence suggested a high level of concordance across sampling methods. Both clinicians and research investigators lack consistent approaches regarding if or how to retest for PIK3CA mutations at different time points or between different testable materials. This lack of protocol represents a significant gap in knowledge for the clinical applicability of PIK3CA testing for HR+/HER2– mBC patients. Additional evidence is needed to increase study generalizability as well as assess other possible confounding factors, such as differential timing between tumor tissue and liquid biopsies.
4.1. Limitations
This systematic review had several limitations. The included studies were limited to those published in English and those published by 2019. The included studies were further limited to those that specifically reported on cases of HR+/HER2- mBC and therefore do not necessarily represent all tests being implemented in PIK3CA research nor are they necessarily generalizable to other BC subtypes. Most studies only reported tests at a single time point or were cross-sectional samples of a larger study and did not necessarily describe when the tests were performed, if tests were performed more than once, or report on the reliability of the tests. In regard to analyses of concordance, the number of included studies was limited by search criteria used to estimate general prevalence of PIK3CA mutation, increasing subjection to reporting bias, and uncertainty around study findings. Heterogeneity in outcome reporting across studies limited consistency in the data presented.
5. Conclusions
This review emphasizes the significance of PIK3CA prevalence and mutation testing methodologies of HR+/HER2– mBC and the need for clinician familiarity with the relative value of available molecular tests in the context of targeted therapy. Routine PIK3CA testing from tissue or ctDNA isolated from plasma samples from patients with HR+/HER2– advanced/mBC is now recommended, as patients with PIK3CA-mutated breast cancer may be appropriate candidates for alpelisib, a recently FDA approved selective inhibitor of the α isoform of PI3Ki [42]. The use of alpelisib in combination with fulvestrant improved progression-free survival in patients with tumors harboring a PIK3CA mutation, which is associated with eventual endocrine resistance that occurs in advanced/metastatic disease [13]. Determinations of tumor PIK3CA mutation status may have additional implications for future research or treatment strategies [43].
Acknowledgments
The authors would like to thank Aditya Bardia, MD, and Alejandra Aguilar, PharmD, for their contributions to this paper. This project was supported by Novartis.
Appendix
A. Search Strategy
A.1. MEDLINE, MEDLINE (R) In-Process, EMBASE CDSR, DARE, and CENTRAL (Ovid)
Tables 3 and 4 describe the search strategy used for databases and selected conference abstracts, respectively.
Table 3.
| No. | Search String |
|---|---|
| 1 | exp breast neoplasms/ or exp breast cancer/ |
| 2 | (breast$ adj3 (cancer$ or neoplas$ or oncolog$ or tumo?r$ or malignanc$ or carcinoma$ or adenocarcinoma$ or sarcoma$)).ti,ab. |
| 3 | (mammar$ adj3 (cancer$ or neoplas$ or oncolog$ or tumo?r$ or malignanc$ or carcinoma$ or adenocarcinoma$ or sarcoma$)).ti,ab. |
| 4 | (metasta$ or advance$ or second$ or recurren$ or inoperab$ or disseminat$ or incur$).ti,ab,sh. |
| 5 | (1 or 2 or 3) and 4 |
| 6 | exp Breast/ and exp Neoplasm Metastasis/ |
| 7 | (breast$ adj3 (metasta$ or advance$ or second$ or recurren$ or inoperab$ or disseminat$ or incur$)).ti,ab. |
| 8 | (mammar$ adj3 (metasta$ or advance$ or second$ or recurren$ or inoperab$ or disseminat$ or incur$)).ti,ab. |
| 9 | (breast$ or mammar$).ti,ab,sh. |
| 10 | ((stage or grade or type) adj2 ("3" or III or "c" or "4" or "IV" or d)).ti,ab. |
| 11 | (N1 or N2$ or N3$ or pN1$ or pN2$ or pN3$).ti,ab,sh. |
| 12 | 9 and (10 or 11) |
| 13 | OR/5-8,12 |
| 14 | (Antineoplastic Agents). ti,ab,rn,kw. |
| 15 | (selective estrogen receptor modulators). ti,ab,rn,kw. |
| 16 | (estrogen antagonists). ti,ab,rn,kw. |
| 17 | (drug therapy). ti,ab,rn,kw. |
| 18 | (letrozole or Femara or CGS 20267 or CGS-20267 or 112809-51-5). ti,ab,rn,kw |
| 19 | (anastrozole or Arimidex or ZD1033 or ZD-1033 or ICI D1033 or 120511-73-1). ti,ab,rn,kw. |
| 20 | (exemestane or examestane or Aromasin or Aromasine or Aromasil or FCE 24304 or FCE-24304 or 107868-30-4). ti,ab,rn,kw. |
| 21 | (tamoxifen or Nolvadex or Novaldex or Soltamox or Tomaxithen or Zitazonium or ICI 46474 or ICI-46474 or ICI 47699 or ICI-47699 or 10540-29-1). ti,ab,rn,kw |
| 22 | (fulvestrant or Faslodex or ICI 182780 or ICI-182780 or ZM 182780 or ZM-182780 or 129453-61-8). ti,ab,rn,kw. |
| 23 | (palbociclib or Ibrance or PD 0332991 or PD-0332991 or 571190-30-2). ti,ab,rn,kw. |
| 24 | (everolimus or Afinitor or Certican or RAD001 or RAD 001 or SDZ RAD or SDZ-RAD or 159351-69-6).ti,ab,rn,kw. |
| 25 | (LEE011 or LEE-011 or Ribociclib or 1211441-98-3). ti,ab,rn,kw. |
| 26 | (abemaciclib or LY2835219 or LY2835210 or 1231929-97-7).ti,ab,rn,kw. |
| 27 | (capecitabine or Xeloda or 154361-50-9).ti,ab,rn,kw. |
| 28 | (doxorubicin or Adriamycin or Doxil or Adriablastin or Adriablastine or Adriblastin or Adriblastina or Adriblastine or Adrimedac or Doxolem or Doxorubicin or Doxotec or Farmiblastina or Myocet or Onkodox or Ribodoxo or Rubex 23214-92-8).ti,ab,rn,kw. |
| 29 | (paclitaxel or Abraxane or Paxene or NSC-125973 or NSC125973 or Anzatax or Onxol or Praxel or Taxol or 33069-62-4).ti,ab,rn,kw. |
| 30 | (docetaxel or Taxotere or Docefrez or RP 56976 or RP-56976 or 114977-28-5).ti,ab,rn,kw. |
| 31 | (cyclophosphamide or cytophosphane or Cytoxan or Endoxan or NSC 26271 or B 518 or B-518 or Cyclophosphane or Cytophosphan or Neosar or Procytox or NSC-26271 or 50-18-0).ti,ab,rn,kw. |
| 32 | (eribulin or Halaven or NSC 707389 or NSC-707389 or B 1793 or B 1939 or B-1793 or B-1939 or E 7389 or E-7389 or ER 086526 or ER-086526 or ER-86526 or ER086526 or eribulin or Halaven or 253128-41-5).ti,ab,rn,kw. |
| 33 | (ethinyl estradiol).ti,ab,rn,kw. |
| 34 | (megestrol acetate or Megace or Mestrel or Maygace or Megostat).ti,ab,rn,kw. |
| 35 | (methotrexate or Amethopterin or Trexall).ti,ab,rn,kw. |
| 36 | (Fluorouracil or 5FU or 5-FU or 5-Fluorouracil or 5 Fluorouracil or Fluoruracil or Adrucil or Carac or Efudix or Efudex or Fluoroplex or Flurodex or Fluracedyl).ti,ab,rn,kw. |
| 37 | (toremifene or Fareston or FC-1157a or FC 1157a or FC1157a).ti,ab,rn,kw. |
| 38 | (gemcitabine or Gemzar or LY 188011 or LY-188011).ti,ab,rn,kw. |
| 39 | (vinorelbine or Navelbine or KW 2307 or KW-2307).ti,ab,rn,kw. |
| 40 | (abraxane or Albumin Bound Paclitaxel or ABI007 or ABI-007 or ABI 007).ti,ab,rn,kw. |
| 41 | (ixabepilone or azaepothilone B or BMS247550 or BMS 247550 or BMS-247550 or Ixempra).ti,ab,rn,kw. |
| 42 | (cisplatin or cis-platinum or NSC-119875 or Platino or Platinol or Platidiam).ti,ab,rn,kw. |
| 43 | (carboplatin or Carbosin or Carbotec or Paraplatin or Carboplat or NSC-241240 or NSC 241240 or NSC241240).ti,ab,rn,kw. |
| 44 | (Fluoxymesterone or Stenox or Halotestin or fluoximesterone).ti,ab,rn,kw. |
| 45 | OR/14-44 |
| 46 | (animals not humans).sh. |
| 47 | (comment or editorial or editorial or book or practice-guideline or letter or journal correspondence).pt. |
| 48 | 45 not (46 or 47) |
| 49 | 13 and 45 and 48 |
| 50 | (Phosphatidylinositol 3-Kinases or “PIK3CA protein, human”) |
| 51 | 49 and 50 |
| 52 | limit 51 to english |
| 53 4 | remove duplicates from 52 |
Table 4.
| American Association for Cancer Research (AACR) (2014-2019) | “hormone receptor positive” OR “HR+” OR “ER+” OR “PR+” OR “estrogen receptor positive” |
| American Society of Clinical Oncology (ASCO) Breast Cancer Symposium (2014-2018) | breast cancer AND (“hormone receptor positive” OR “HR+” OR “ER+” OR “PR+” OR “estrogen receptor positive”) |
| San Antonio Breast Cancer Symposium (2014-2018) | breast cancer AND (“hormone receptor positive” OR “HR+” OR “ER+” OR “PR+” OR “estrogen receptor positive”) |
| European CanCer Organisation (ECCO) (2015-2016) | breast cancer AND (“hormone receptor positive” OR “HR+” OR “ER+” OR “PR+” OR “estrogen receptor positive”) Abstract title: breast cancer |
| European Society for Medical Oncology (ESMO) (2014, 2016) | breast cancer AND (“hormone receptor positive” OR “HR+” OR “ER+” OR “PR+” OR “estrogen receptor positive”) Abstract title: breast cancer |
A.2. ClinicalTrials.gov
(letrozole OR Femara) AND (breast OR mammar∗)
(anastrozole OR Arimidex) AND (breast OR mammar∗)
(exemestane or Aromasin or Aromasil) AND (breast OR mammar∗)
(tamoxifen OR Nolvadex OR Soltamox) AND (breast OR mammar∗)
(fulvestrant OR Faslodex) AND (breast OR mammar∗)
(palbociclib OR Ibrance) AND (breast OR mammar∗)
(everolimus OR afinitor) AND (breast OR mammar∗)
(LEE011 OR LEE-011 OR Ribociclib) AND (breast OR mammar∗)
(abemaciclib) AND (breast OR mammar∗)
(capecitabine OR Xeloda) AND (breast OR mammar∗)
(doxorubicin OR Adriamycin OR Doxil) AND (breast OR mammar∗)
(paclitaxel OR Abraxane) AND (breast OR mammar∗)
(ethinyl estradiol) AND (breast OR mammar∗)
((Fluoxymesterone OR Stenox OR Halotestin OR fluoximesterone) AND (breast OR mammar∗)
(megestrol acetate OR Megace OR Mestrel OR Maygace OR Megostat) AND (breast OR mammar∗)
(docetaxel OR Taxotere OR Docefrez) AND (breast OR mammar∗)
(cyclophosphamide OR cytophosphane OR Cytoxan OR Endoxan) AND (breast OR mammar∗)
(eribulin OR Halaven) AND (breast OR mammar∗)
(methotrexate OR Trexall) AND (breast OR mammar∗)
(fluorouracil OR Adrucil OR Fluorouracil Novaplus OR PremierPro Rx Fluorouracil) AND (breast OR mammar∗)
(toremifene OR Fareston) AND (breast OR mammar∗)
(gemcitabine OR Gemcitabine Novaplus OR Gemzar OR PremierPro Rx Gemcitabine) AND (breast OR mammar∗)
(vinorelbine OR Navelbine OR Vinorelbine Novaplus) AND (breast OR mammar∗)
(paclitaxel OR paclitaxel protein bound OR Taxol OR Abraxane) AND (breast OR mammar∗)
(ixabepilone OR Ixempra) AND (breast OR mammar∗)
(cisplatin OR Platinol-AQ) AND (breast OR mammar∗)
(carboplatin OR Paraplatin OR Paraplatin NovaPlus OR) AND (breast OR mammar∗)
A.3. Selected Conference Abstracts
B. Abridged Mixed Methods Appraisal Tool (2011) Criteria Used for Systematic Review
Table 5 notes the decision-making criteria for assignment of a quality score per the Mixed Methods Appraisal Tool.
Table 5.
| Quantitative descriptive |
| Is the sampling strategy relevant to address the quantitative research question (quantitative aspect of the mixed methods question)? |
| Is the sample representative of the population understudy? |
| Are measurements appropriate (clear origin, or validity known, or standard instrument)? |
| Is there an acceptable response rate (60% or above)? |
|
|
| Quantitative randomized controlled (trials) |
| Is there a clear description of the randomization (or an appropriate sequence generation)? |
| Is there a clear description of the allocation concealment (or blinding when applicable)? |
| Are there complete outcome data (80% or above)? Is there low withdrawal/drop-out (below 20%)? |
| Is there low withdrawal/drop-out (below 20%)? |
|
|
| Quantitative nonrandomized |
| Are participants (organizations) recruited in a way that minimizes selection bias? |
| Are measurements appropriate (clear origin, or validity known, or standard instrument; and absence of contamination between groups when appropriate) regarding the exposure/intervention and outcomes? |
| In the groups being compared (exposed vs. nonexposed; with intervention vs. without; cases vs. controls), are the participants comparable, or do researchers take into account (control for) the difference between these groups? |
| Are there complete outcome data (80% or above), and, when applicable, an acceptable response rate (60% or above), or an acceptable follow-up rate for cohort studies (depending on the duration of follow-up)? |
Data Availability
The systematic review search strategy is available for replication.
Disclosure
This project was presented, in part, as posters at the 2018 Meeting of the American Association for Cancer Research.
Conflicts of Interest
The authors declare that this study received funding from Novartis. The funder had the following involvement with the study: study design, writing of the article, and the decision to submit for publication. EJA, LM, and JD received research funds from Novartis to support this study. EP is an employee of Novartis. At the time of the study, DT was an employee of Novartis. AA receives research funding from Varian Medical Systems unrelated to the current project. LD and TW have no conflicts to disclose.
Authors' Contributions
DT, EP, and TW conceived the research project and oversaw development of the literature search. EJA, LM, and JD performed the search, data extraction, and data analysis. EJA, LM, and LD led the writing of this article. LD and AA provided subject matter expertise and feedback.
References
- 1.American Cancer Society. Cancer Facts & Figures 2019. Atlanta: American Cancer Society; 2019. [Google Scholar]
- 2.Pan H., Gray R., Braybrooke J., et al. 20-year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. New England Journal of Medicine. 2017;377(19):1836–1846. doi: 10.1056/NEJMoa1701830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howlader N., Altekruse S. F., Li C. I., et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. JNCI: Journal of the National Cancer Institute. 2014;106(5) doi: 10.1093/jnci/dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuels Y., Ericson K. Oncogenic PI3K and its role in cancer. Current Opinion in Oncology. 2006;18(1):77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 5.Mukohara T. PI3K mutations in breast cancer: prognostic and therapeutic implications. Breast Cancer: Targets and Therapy. 2015;7:p. 111. doi: 10.2147/bctt.s60696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Cancer Institute. SEER stat fact sheets: female breast cancer. http://seer.cancer.gov/statfacts/html/breast.html.
- 7.Miller T. W., Balko J. M., Arteaga C. L. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. Journal of Clinical Oncology. 2011;29(33):4452–4461. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Breast Cancer Version 3. Fort Washington, MD, USA: National Comprehensive Cancer Network; 2014. [Google Scholar]
- 9.Baselga J., Im S. A., Iwata H., et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. The Lancet Oncology. 2017;18(7):904–916. doi: 10.1016/S1470-2045(17)30376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Leo A., Johnston S., Lee K. S., et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor- positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): a randomised, double-blind, placebo-controlled, phase 3 trial. The Lancet Oncology. 2018;19(1):87–100. doi: 10.1016/S1470-2045(17)30688-5. [DOI] [PubMed] [Google Scholar]
- 11.Massacesi C., di Tomaso E., Urban P., et al. PI3K inhibitors as new cancer therapeutics: implications for clinical trial design. OncoTargets and Therapy. 2016;9:p. 203. doi: 10.2147/OTT.S89967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller T. W., Balko J. M., Fox E. M., et al. ERα-dependent E2F transcription can mediate resistance to estrogen deprivation in human breast cancer. Cancer Discovery. 2011;1(4):338–351. doi: 10.1158/2159-8290.CD-11-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andre F., Ciruelos E., Rubovszky G. Alpelisib+ fulvestrant for HR+, HER2-advanced breast cancer: results of the phase III SOLAR-1 trial. European Society for Medical Oncology (ESMO) 2018 Congress (Abstract LBA3_PR) on October. 2018.
- 14.Cizkova M., Susini A., Vacher S., et al. PIK3CA mutation impact on survival in breast cancer patients and in ERα, PR and ERBB2-based subgroups. Breast Cancer Research. 2012;14(1, article R28) doi: 10.1186/bcr3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paplomata E., O’Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Therapeutic Advances in Medical Oncology. 2014;6(4):154–166. doi: 10.1177/1758834014530023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stemke-Hale K., Gonzalez-Angulo A. M., Lluch A., et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Research. 2008;68(15):6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller T. W., Rexer B. N., Garrett J. T., Arteaga C. L. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Research. 2011;13(6):p. 224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Poznak C., Harris L. N., Somerfield M. R. Use of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American Society of Clinical Oncology clinical practice guideline. Journal of Oncology Practice. 2015;11(6):514–516. doi: 10.1200/JOP.2015.005215. [DOI] [PubMed] [Google Scholar]
- 19.Davis A. A., McKee A. E., Kibbe W. A., Villaflor V. M. Complexity of delivering precision medicine: opportunities and challenges. American Society of Clinical Oncology Educational Book. 2018;38:998–1007. doi: 10.1200/EDBK_200279. [DOI] [PubMed] [Google Scholar]
- 20.Hagemann I. S. Molecular testing in breast cancer: a guide to current practices. Archives of Pathology & Laboratory Medicine. 2016;140(8):815–824. doi: 10.5858/arpa.2016-0051-RA. [DOI] [PubMed] [Google Scholar]
- 21.Desmedt C., Voet T., Sotiriou C., Campbell P. J. Next-generation sequencing in breast cancer. Current Opinion in Oncology. 2012;24(6):597–604. doi: 10.1097/CCO.0b013e328359554e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moher D., Liberati A., Tetzlaff J., Altman D. G., The PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Medicine. 2009;6(7, article e1000097) doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Tulder M., Furlan A., Bombardier C., Bouter L., the Editorial Board of the Cochrane Collaboration Back Review Group Updated method guidelines for systematic reviews in the Cochrane collaboration back review group. Spine. 2003;28(12):1290–1299. doi: 10.1097/01.BRS.0000065484.95996.AF. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh H.-F., Shannon S. E. Three approaches to qualitative content analysis. Qualitative Health Research. 2016;15(9):1277–1288. doi: 10.1177/1049732305276687. [DOI] [PubMed] [Google Scholar]
- 25.Pluye P., Robert E., Cargo M., et al. Proposal: a mixed methods appraisal tool for systematic mixed studies reviews. Vol. 2. Montréal: McGill University; 2011. [Google Scholar]
- 26.Pace R., Pluye P., Bartlett G., et al. Testing the reliability and efficiency of the pilot mixed methods appraisal tool (MMAT) for systematic mixed studies review. International Journal of Nursing Studies. 2012;49(1):47–53. doi: 10.1016/j.ijnurstu.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Souto R. Q., Khanassov V., Hong Q. N., Bush P. L., Vedel I., Pluye P. Systematic mixed studies reviews: updating results on the reliability and efficiency of the mixed methods appraisal tool. International Journal of Nursing Studies. 2015;52(1):500–501. doi: 10.1016/j.ijnurstu.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Moynahan M. E., Chen D., He W., et al. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR+, HER2 − advanced breast cancer: results from BOLERO-2. British Journal of Cancer. 2017;116(6):726–730. doi: 10.1038/bjc.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertucci F., Finetti P., Guille A., et al. Comparative genomic analysis of primary tumors and metastases in breast cancer. Oncotarget. 2016;7(19):27208–27219. doi: 10.18632/oncotarget.8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickler M. N. Phase II study of taselisib (GDC-0032) plus fulvestrant in HER2-negative, hormone receptor-positive advanced breast cancer: analysis by PIK3CA and ESR1 mutation status from circulating tumor DNA. SABCS; 2016. [Google Scholar]
- 31.Campone M., Im S.-A., Iwata H., et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant for postmenopausal, hormone receptor-positive, human epidermal growth factor receptor 2-negative, advanced breast cancer: overall survival results from BELLE-2. European Journal of Cancer. 2018;103:147–154. doi: 10.1016/j.ejca.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Ettinger D. S., Wood D. E., Aisner D. L., et al. Non–small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. Journal of the National Comprehensive Cancer Network. 2017;15(4):504–535. doi: 10.6004/jnccn.2017.0050. [DOI] [PubMed] [Google Scholar]
- 33.Rugo H. S., Mayer I., Conte P., et al. Abstract CT142: prevalence of PIK3CAmutations in patients with hormone receptor-positive, human epidermal growth factor-2-negative advanced breast cancer from the SOLAR-1 trial. AACR; 2019. [Google Scholar]
- 34.Takeshita T., Yamamoto Y., Yamamoto-Ibusuki M., et al. Clinical significance of plasma cell-free DNA mutations in PIK3CA, AKT1, and ESR1 gene according to treatment lines in ER-positive breast cancer. Molecular Cancer. 2018;17(1):p. 67. doi: 10.1186/s12943-018-0808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan H., Chen J., Liu Y., et al. Association of PIK3CA mutation status before and after neoadjuvant chemotherapy with response to chemotherapy in women with breast cancer. Clinical Cancer Research. 2015;21(19):4365–4372. doi: 10.1158/1078-0432.CCR-14-3354. [DOI] [PubMed] [Google Scholar]
- 36.Prat A., Cheang M. C. U., Galván P., et al. Prognostic value of intrinsic subtypes in hormone receptor–positive metastatic breast cancer treated with letrozole with or without lapatinib. JAMA Oncology. 2016;2(10):1287–1294. doi: 10.1001/jamaoncol.2016.0922. [DOI] [PubMed] [Google Scholar]
- 37.Samuels Y., Wang Z., Bardelli A., et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):p. 554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 38.Liu P., Cheng H., Roberts T. M., Zhao J. J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nature Reviews Drug Discovery. 2009;8(8):627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai Y.-L., Mau B.-L., Cheng W.-H., Chen H.-M., Chiu H.-H., Tzen C.-Y. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Annals of Surgical Oncology. 2008;15(4):1064–1069. doi: 10.1245/s10434-007-9751-7. [DOI] [PubMed] [Google Scholar]
- 40.LE M. C. Existing and emerging technologies for tumor genomic profiling. Journal of Clinical Oncology. 2013;31(15):1815–1824. doi: 10.1200/JCO.2012.46.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krop I. E., Mayer I. A., Ganju V., et al. Pictilisib for oestrogen receptor-positive, aromatase inhibitor-resistant, advanced or metastatic breast cancer (FERGI): a randomised, double-blind, placebo-controlled, phase 2 trial. The Lancet Oncology. 2016;17(6):811–821. doi: 10.1016/S1470-2045(16)00106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.NCCN. Clinical Practice Guidelines in Oncology. Breast Cancer. National Comprehensive Cancer Network; 2019. [DOI] [PubMed] [Google Scholar]
- 43.Arthur L. M., Turnbull A. K., Renshaw L., et al. Changes in PIK3CA mutation status are not associated with recurrence, metastatic disease or progression in endocrine-treated breast cancer. Breast Cancer Research and Treatment. 2014;147(1):211–219. doi: 10.1007/s10549-014-3080-x. [DOI] [PubMed] [Google Scholar]
- 44.Abramson V. G., Cooper Lloyd M., Ballinger T., et al. Characterization of breast cancers with PI3K mutations in an academic practice setting using SNaPshot profiling. Breast Cancer Research and Treatment. 2014;145(2):389–399. doi: 10.1007/s10549-014-2945-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahmad F., Badwe A., Verma G., Bhatia S., Das B. R. Molecular evaluation of PIK3CA gene mutation in breast cancer: determination of frequency, distribution pattern and its association with clinicopathological findings in Indian patients. Medical Oncology. 2016;33(7):p. 74. doi: 10.1007/s12032-016-0788-y. [DOI] [PubMed] [Google Scholar]
- 46.Basho R. K., Gagliato D. M., Ueno N. T., et al. Clinical outcomes based on multigene profiling in metastatic breast cancer patients. Oncotarget. 2016;7(47):76362–76373. doi: 10.18632/oncotarget.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baird R. D., Van Rossum A., Oliveira M., et al. POSEIDON trial phase 1b results: safety and preliminary efficacy of the isoform selective PI3K inhibitor taselisib (GDC-0032) combined with tamoxifen in hormone receptor (HR) positive, HER2-negative metastatic breast cancer (MBC) patients (pts)-including response monitoring by plasma circulating tumor (ct) DNA. American Society of Clinical Oncology; 2016. [DOI] [PubMed] [Google Scholar]
- 48.Basu G. D., White T., LoBello J. R., et al. Assessment of ESR1 and ERBB2 mutations in estrogen receptor positive (ER+) metastatic breast cancers (MBC) American Society of Clinical Oncology; 2017. [Google Scholar]
- 49.Beelen K., Opdam M., Severson T. M., et al. PIK3CA mutations, phosphatase and tensin homolog, human epidermal growth factor receptor 2, and insulin-like growth factor 1 receptor and adjuvant tamoxifen resistance in postmenopausal breast cancer patients. Breast Cancer Research. 2014;16(1, article R13) doi: 10.1186/bcr3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blackwell K., Burris H., Gomez P., et al. Phase I/II dose-escalation study of PI3K inhibitors pilaralisib or voxtalisib in combination with letrozole in patients with hormone-receptor-positive and HER2-negative metastatic breast cancer refractory to a non-steroidal aromatase inhibitor. Breast Cancer Research and Treatment. 2015;154(2):287–297. doi: 10.1007/s10549-015-3615-9. [DOI] [PubMed] [Google Scholar]
- 51.Board R. E., Wardley A. M., Dixon J. M., et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Research and Treatment. 2010;120(2):461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 52.Christgen M., Noskowicz M., Schipper E., et al. Oncogenic PIK3CA mutations in lobular breast cancer progression. Genes, Chromosomes and Cancer. 2013;52(1):69–80. doi: 10.1002/gcc.22007. [DOI] [PubMed] [Google Scholar]
- 53.Filipenko M. L., Os’kina N. A., Oskorbin I. A., et al. Association between the prevalence of somatic mutations in PIK3CA gene in tumors and clinical and morphological characteristics of breast cancer patients. Bulletin of Experimental Biology and Medicine. 2017;163(2):250–254. doi: 10.1007/s10517-017-3777-z. [DOI] [PubMed] [Google Scholar]
- 54.Fleming G. F., Ma C. X., Huo D., et al. Phase II trial of temsirolimus in patients with metastatic breast cancer. Breast Cancer Research and Treatment. 2012;136(2):355–363. doi: 10.1007/s10549-011-1910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasch C., Oldopp T., Mauermann O., et al. Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2-negative metastatic breast cancer. Molecular Oncology. 2016;10(8):1330–1343. doi: 10.1016/j.molonc.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalez-Angulo A. M., Ferrer-Lozano J., Stemke-Hale K., et al. PI3K pathway mutations and PTEN levels in primary and metastatic breast cancer. Molecular Cancer Therapeutics. 2011;10(6):1093–1101. doi: 10.1158/1535-7163.MCT-10-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fitzgerald D. M., Henderson L. E., Isakoff S. J., et al. Abstract P1-12-03: Association between tumor genotype and development of brain metastases in patients with hormone receptor positive (HR+)/HER2- metastatic breast cancer. Cancer Research. 2017;77(4) doi: 10.1158/1538-7445.SABCS16-P1-12-03. [DOI] [Google Scholar]
- 58.Lefebvre C., Bachelot T., Filleron T., et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Medicine. 2016;13(12, article e1002201) doi: 10.1371/journal.pmed.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S., Wang H., Zhang L., et al. Rapid detection of genetic mutations in individual breast cancer patients by next-generation DNA sequencing. Human Genomics. 2015;9(1):p. 2. doi: 10.1186/s40246-015-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mayer I. A., Abramson V. G., Formisano L., et al. A phase Ib study of alpelisib (BYL719), a PI3Kα-specific inhibitor, with letrozole in ER+/HER2− metastatic breast cancer. Clinical Cancer Research. 2017;23(1):26–34. doi: 10.1158/1078-0432.CCR-16-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mayer I. A., Abramson V. G., Isakoff S. J., et al. Stand up to cancer phase Ib study of pan-phosphoinositide-3-kinase inhibitor buparlisib with letrozole in estrogen receptor-positive/human epidermal growth factor receptor 2-negative metastatic breast cancer. Journal of Clinical Oncology. 2014;32(12):1202–1209. doi: 10.1200/JCO.2013.54.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muller K. E., Marotti J. D., de Abreu F. B., et al. Targeted next-generation sequencing detects a high frequency of potentially actionable mutations in metastatic breast cancers. Experimental and Molecular Pathology. 2016;100(3):421–425. doi: 10.1016/j.yexmp.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 63.Oliveira M., Dienstmann R., Bellet M., et al. Clonality of PIK3CA mutations (mut) and efficacy of PI3K/AKT/mTOR inhibitors (PAMi) in patients (pts) with metastatic breast cancer (MBC) American Society of Clinical Oncology; 2016. [Google Scholar]
- 64.Roy-Chowdhuri S., de Melo Gagliato D., Routbort M. J., et al. Multigene clinical mutational profiling of breast carcinoma using next-generation sequencing. American Journal of Clinical Pathology. 2015;144(5):713–721. doi: 10.1309/AJCPWDEQYCYC92JQ. [DOI] [PubMed] [Google Scholar]
- 65.Sakr R. A., Weigelt B., Chandarlapaty S., et al. PI3K pathway activation in high-grade ductal carcinoma in situ—implications for progression to invasive breast carcinoma. Clinical Cancer Research. 2014;20(9):2326–2337. doi: 10.1158/1078-0432.CCR-13-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soucier-Ernst D., Colameco C., Troxel A. B., et al. Abstract P6-07-05: Mutational spectrum and tumor response in metastatic breast cancer. Cancer Research. 2016;76(4) doi: 10.1158/1538-7445.SABCS15-P6-07-05. [DOI] [Google Scholar]
- 67.Vetter M. Prevalence of PIK3CA mutations in tumor tissue of a consecutive cohort of breast cancer patients. 2014.
- 68.Wang K., Ali S. M., Khaira D., et al. Abstract P6-03-12: Comprehensive genomic profiling of clinically advanced mucinous carcinoma of the breast. Cancer Research. 2016;76(4) doi: 10.1158/1538-7445.SABCS15-P6-03-12. [DOI] [PubMed] [Google Scholar]
- 69.Welt A., Tewes M., Aktas B., et al. Preemptive tumor profiling for biomarker-stratified early clinical drug development in metastatic breast cancer patients. Breast Cancer Research and Treatment. 2013;142(1):81–88. doi: 10.1007/s10549-013-2718-4. [DOI] [PubMed] [Google Scholar]
- 70.Williams C. B., De P., Dey N., et al. New treatment options for metastatic breast cancer revealed by reverse-phase protein microarray and genomic profiling. Cancer Research. 2015;75(9 Supplement) [Google Scholar]
- 71.Yuan Y., Yost S., Yuan Y.-C., et al. Abstract P6-16-08: The impact of genomic mutation on metastatic breast cancer treatment: A retrospective clinical trial. Cancer Research. 2017;77(4) doi: 10.1158/1538-7445.SABCS16-P6-16-08. [DOI] [Google Scholar]
- 72.Zhang Y., Liu M., Yang H., et al. PIK3CA mutations are a predictor of docetaxel plus epirubicin neoadjuvant chemotherapy clinical efficacy in breast cancer. Neoplasma. 2014;61(4):461–467. doi: 10.4149/neo_2014_057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The systematic review search strategy is available for replication.


