Abstract
Hedgehog (Hh) signaling has been shown to regulate multiple developmental processes, however, it is unclear how it regulates lipid metabolism. Here, we demonstrate that Hh signaling exhibits potent activity in Drosophila fat body, which is induced by both locally expressed and midgut-derived Hh proteins. Inactivation of Hh signaling increases, whereas activation of Hh signaling decreases lipid accumulation. The major lipase Brummer (Bmm) acts downstream of Smoothened (Smo) in Hh signaling to promote lipolysis, therefore, the breakdown of triacylglycerol (TAG). We identify a critical Ci binding site in bmm promoter that is responsible to mediate Bmm expression induced by Hh signaling. Genomic mutation of the Ci binding site significantly reduces the expression of Bmm and dramatically decreases the responsiveness to Ci overexpression. Together, our findings provide a model for lipolysis to be regulated by Hh signaling, raising the possibility for Hh signaling to be involved in lipid metabolic and/or lipid storage diseases.
Keywords: Hh, Smo, Ci, Bmm, ATGL, Lipolysis, Lipid droplet, Fat body
1. Introduction
Hh signaling is a highly conserved pathway that controls organ development and tissue homeostasis. Dysregulation of Hh signaling is implicated in many human disorders, including several types of cancer (Briscoe and Therond, 2013; Jiang and Hui, 2008). Smo, an atypical G protein-coupled receptor (GPCR) family member, acts as a key regulator of the pathway for both insects and vertebrates. Abnormal Smo activation results in basal cell carcinoma (BCC) and medulloblastoma, therefore, Smo is an attractive therapeutic target (Guha, 2012; Sekulic and Von Hoff, 2016) that can, however, acquire drug resistance (resistance to GDC-0449 that was approved by FDA in 2012) through a single amino acid mutation (Rudin et al., 2009; Yauch et al., 2009). Besides the knowledge of Hh/Smo signaling in cancer, little is known whether Hh/Smo signaling is involved in metabolic disorders.
In Drosophila, the Hh signal is transduced through a reception system at the plasma membrane, which includes the receptor complexes Patched (Ptc)-Ihog and the signal transducer Smo (Jia and Jiang, 2006; Lum and Beachy, 2004; Zheng et al., 2010). Binding of Hh to Ptc-Ihog relieves the inhibition of Smo by Ptc and allows Smo to activate the Ci/Gli family of Zn-finger transcription factors to induce the expression of Hh target genes, including ptc, decapentaplegic (dpp), and engrailed (en) (Hooper and Scott, 2005; Jia and Jiang, 2006). In the absence of Hh, Ptc inhibits Smo. Unphosphorylated Smo is ubiquitinated and degraded through the proteasome and lysosome (Li et al., 2012; Xia et al., 2012). Ci is processed into a truncated repressor form that enters into the nucleus to block target gene expression (Hooper and Scott, 2005). Gli1 is a transcription factor and also a Hh target gene in vertebrate Hh signaling (Jiang and Hui, 2008). In addition to these classical target genes, some other genes involved in neural tube patterning and axon guidance are also shown to be regulated by Hh signaling (Briscoe and Therond, 2013); however, it is unknown whether Hh signaling directly regulates gene(s) from the metabolic pathways.
The Drosophila fat body has emerged as a model tissue to determine the roles of Hh signaling in metabolism. The fat body mimics vertebrate liver and adipose tissue and functions in metabolizing nutrients and storing glycogen and triacylglycerol (TAG), but also in sensing nutritional conditions and responding with synthesis and release of energy (Droujinine and Perrimon, 2016). Hh functions as an organizer of tissue morphogenesis, as well as an endocrine hormone, circulating to the fat body to orchestrate the nutrient response during fat body development (Pospisilik et al., 2010; Rodenfels et al., 2014). Hh signaling inhibits fat formation and blocks adipogenesis in the fat body (Suh et al., 2006), therefore, Hh has been shown to be a key modulator of fat body formation in Drosophila (Pospisilik et al., 2010) and plays a key role in brown versus white adipocyte determination in mice (Nosavanh et al., 2015; Pospisilik et al., 2010). Hh also activates Smo-AMP kinase (AMPK) to promote a Warburg-like glycolytic metabolism and glucose uptake (Teperino et al., 2012, 2014). Consistently, Smo inactivation results in liver steatosis and significantly increases lipid droplet accumulation (Matz-Soja et al., 2016); whereas Hh specifically activated in the fat leads to lean mice with a severe reduction in fat tissue (Pospisilik et al., 2010). These studies suggest a role for Hh signaling in adipogenesis regulation, however, the underlying molecular mechanisms are unclear. In addition, it is unknown whether Hh signaling regulates lipolysis.
In this study, we found that Hh signaling upregulated the transcription of a specific lipase with a subsequent increase in lipolysis. We found that Hh signaling was highly activated in fat body, demonstrated by the elevated Hh target gene expression, which was significantly reduced by the inactivation of Smo and Ci. We further found that both Hh protein circulated from gut and Hh protein locally expressed contributed to the high activity of Hh signaling in fat body. We further found that the transcription of lipase Bmm, the Drosophila Adipose Triglyceride Lipase (ATGL) (Gronke et al., 2005), was significantly inhibited by the inactivation of Smo or Ci. Analysis of bmm promoter identified critical Ci binding sites. Mutating a Ci binding site adjacent to transcription start significantly reduced the transcription and Hh responsiveness of Bmm. Finally, we found that, in Drosophila fat body, genomic mutation of the Ci binding site in bmm promoter significantly reduced the ability for Ci to regulate Bmm-mediated lipolysis. These findings indicate that Hh signaling directly regulates lipolysis, raising the possibility for Hh signaling to be involved in metabolic disease(s).
2. Results
2.1. Hh signaling inhibits lipid droplet accumulation in Drosophila fat body and in cultured adipocyte
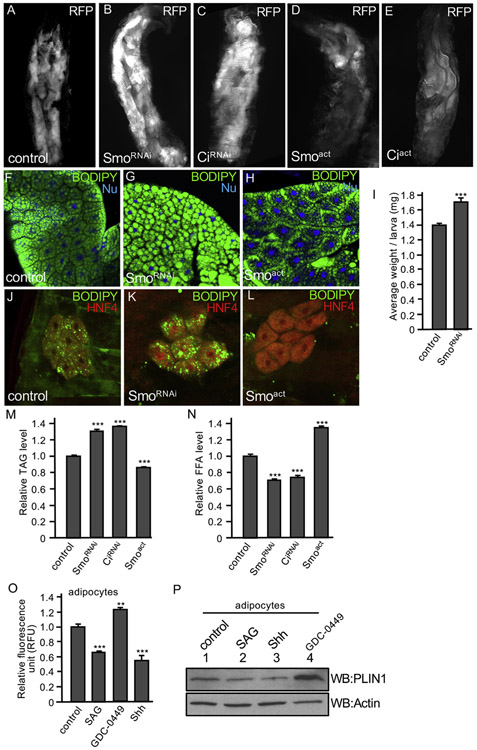
In Drosophila, during larva development, TAG and glycogen are highly accumulated as nutrient storage in the larval fat bodies. By the stage of late 3rd instar larva, lipid droplets in the fat body, accounting for about 30% of the larval body weight, can be metabolized to provide energy for locomotor activity. Later during development, the fat body undergoes morphological remodeling to provide energy for pupae and newly enclosed adults, which raises a good model to study lipid metabolism (Nelliot et al., 2006). To determine whether Hh signaling plays a role in regulating lipid accumulation, we examined the fat body size in late 3rd instar larvae when Smo and Ci were either overexpressed or inactivated. We found that RNAi of either Smo or Ci by the fat body-specific cg-Gal4 increased fat body sizes in late 3rd instar larvae (Fig. 1B-C, compared to WT in Fig. 1A). In contrast, the overexpression of Smoact, a constitutively active form of Smo in which Ser/Thr residues are substituted by Asp to mimic phosphorylation (Fan et al., 2012; Jia et al., 2004), or Ciact, a constitutively active form of Ci with phosphorylation clusters mutated to block Ci processing (Jia et al., 2010), dramatically reduced the fat body sizes (Fig. 1D-E). The phenotypes in the increase or decrease of fat body size were highly penetrant, and the change was not due to nutrition intake because no change in food content was seen upon spectrophotometric measurement of extracts from flies grown on food with 1% Brilliant Blue.
Fig. 1. Hh signaling regulates fat body size and lipid accumulation.
(A) A 3rd instar larva expressing RFP by the fat body-specific cg-Gal4, indicating the fat bodies marked by RFP. (B–E) A larva expressing SmoRNAi, CiRNAi or the constitutively active Smoact, Ciact by the cg-Gal4. Fat bodies are marked by RFP. Male and female larvae have similar pattern in this experiment. (F) A fat body from larvae expressing the fat body-specific ppl-Gal4 in w1118 wild-type background was immunostained for BODIPY (green) and nuclear dye Hoechst 33342 (Nu, blue). (G–H) Fat bodies from larvae expressing SmoRNAi or Ciact by the ppl-Gal4 were stained with BODIPY and nuclear dye. Male and female larvae have similar pattern in this experiment. (I) Larvae from ppl-w1118 (control) or ppl-SmoRNAi were collected and analyzed for their weight. 16 larvae (8 males and 8 females) for each group, with 15 repeats. *** indicates a p < 0.001, versus control (Student’s t-test). (J) Oenocyte from larvae with oen-Gal4-w1118 (control) were immunostained with BODIPY (green) and the anti-HNF4 antibody that marks the oenocyte cells (red). (K-L) Oenocyte from larvae expressing SmoRNAi or Smoact by oen-Gal4 were stained with BODIPY and HNF4. Male and female larvae exhibit similar phenotypes. (M–N) Early 3rd instar larvae expressing Smoact, SmoRNAi, or CiRNAi by ppl-Gal4 combined with tub-Gal80ts were shifted to 29 °C (non-permissive temperature) to inhibit Gal80 expression for 16 h and collected for the analysis of TAG and FFA. Concentration of TAG and FFA was normalized by protein. ppl-Gal4-w1118 served as control. 20 larvae (10 males and 10 females) for each group were collected and the assays were performed with 3 repeats. *** indicates a p < 0.001, versus control in the first column in each assay (Student’s t-test). (O–P) Lipid accumulation regulated by Hh signaling in adipocyte. 3T3-L1 adipocytes were treated with DMSO (control), 200 nM SAG, 100 nM GDC-0449, or 5 nM Shh peptide, followed by staining with BODIPY and subjected to fluorescence activity measurement. RFU: relative fluorescence units. ** indicates a p < 0.01 and *** indicates a p < 0.001, versus control in the first column in each assay (Student’s t-test). 3T3-L1 adipocytes with the same set of treatments were also analyzed by western blots with the anti-PLIN1. Western blot with the anti-Actin antibody serves as loading control.
We further examined the lipid droplets in the fat body and found that RNAi of Smo by larval fat body-specific ppl-Gal4 increased, whereas overexpression of Smoact by ppl-Gal4 decreased lipid droplets in the fat body (Fig. 1G-H, compared to WT in Fig. 1F). Although the wing disc is not considered the best model to study lipid metabolism, it can be used to examine lipid accumulation under some circumstances. We found that the overexpression of Ciact and Smoact by the dorsal compartment specific ap-Gal4 reduced lipid droplet accumulation in the wing disc (Fig. S1), which is consistent with the finding in fat body. In another separate experiment, we found that RNAi of Smo by ppl-Gal4 in the fat body significantly increased larval body weight (Fig. 1I). We further analyzed the effect of Hh signaling in lipid accumulation in the oenocyte, which are specialized hepatocyte-like cells laying beneath the epidermis in apparent cell clusters (Owusu-Ansah and Perrimon, 2014). We found that RNAi of Smo by the oenocyte-specific oen-Gal4 increased, whereas Smoact expression by the oen-Gal4 decreased lipid accumulation in the oenocyte (Fig. 1K-L, compared to Fig. 1J), suggesting a cell-autonomous effect of Hh signaling in lipid accumulation.
The increase in fat body size and the accumulation of lipid droplets are likely due to elevated adipogenesis or blocked lipolysis, or both. We therefore analyzed lipolysis activity by examining the levels of TAG and free fatty acid (FFA) in Drosophila, using the inducible Gal80ts system to activate or inactivate Hh signaling from the early to late 3rd instar larvae stage. We found that the expression of Smoact by ppl-Gal4 significantly decreased, whereas Smo RNAi or Ci RNAi by the ppl-Gal4 significantly increased the levels of TAG in fat body (Fig. 1M). Consistently, the levels of FFA were significantly increased by the expression of Smoact, however decreased by RNAi of Smo or Ci (Fig. 1N). We also examined TAG levels in adult flies and found that the expression of Smo RNAi by the adult fat body-specific yolk-Gal4 significantly increased, whereas the expression of Smoact by the yolk-Gal4 significantly decreased the levels of TAG in adult flies (Fig. S2A). Taken together, these data suggest that Hh signaling inhibits lipid droplet accumulation through promoting lipolysis in Drosophila late stages of development.
3T3-L1 cells recapitulate signaling events and can be differentiated into primary adipocytes (Green and Kehinde, 1975), which have Hh signaling responsiveness. We found that, in 3T3-L1 adipocytes, the activation Hh signaling by the treatment of either SAG (a Smo agonist) or Shh peptide significantly reduced fluorescence intensities of BODIPY that labeled lipid in the adipocytes (Fig. 1O). In contrast, the inhibition of Hh signaling by the treatment of GDC-0449 (a potent Smo inhibitor) significantly increased fluorescence intensities (Fig. 1O). Perilipin 1 (PLIN1), an adipocyte-specific protein associated with lipid droplet (Beller et al., 2010; Greenberg et al., 1991; Yao et al., 2018), is often used to monitor lipid droplet accumulation. We found that the level of PLIN1 was decreased by the treatment of SAG or Shh, indicating a decrease in lipid accumulation in these adipocytes (Fig. 1P). In contrast, PLIN1 was increased by the treatment of GDC-0449, suggesting an increase in lipid accumulation (Fig. 1P). Taken together, the observations in both Drosophila tissue and cultured adipocytes suggest that Hh signaling inhibits lipid droplet accumulation by promoting lipolysis.
2.2. Hh signaling is highly active in Drosophila fat body
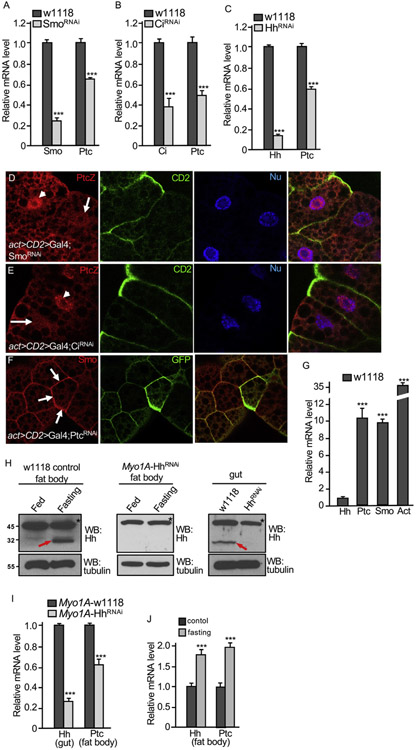
The inactivation of Smo and Ci caused dramatic increase in lipid accumulation, raising the possibility for Hh signaling to be very actively involved in lipolysis. To test this hypothesis, we first examined the expression of ptc, one of the Hh target gene, in late 3rd instar larvae and found that RNAi of Smo or Ci by the ppl-Gal4 significantly reduced ptc mRNA levels (Fig. 2A-B). In addition, RNAi of Hh by the ppl-Gal4 also reduced the level ptc mRNA (Fig. 2C), which was consistent with a previous finding that RNAi of Hh produced phenotype in Drosophila fat body (Pospisilik et al., 2010). To further examine the activity of Hh signaling in fat body, we generated clones using the act > CD2 > Gal4 to express Smo RNAi or Ci RNAi and monitored the reporter ptc-lacZ expression. We found that ptc-lacZ expression was blocked in cells expressing Smo RNAi or Ci RNAi (Fig. 2D-E). Hh signaling activity can be monitored by expression of different responsive genes, with dpp induction indicating low level, whereas ptc induction indicating high level of Hh signaling (Fan et al., 2012). Thus, our findings suggest that Hh signaling is highly active in fat body.
Fig. 2. Fat body exhibits high levels of Hh signaling activity.
(A–C) Fat bodies were collected from late 3rd instar larvae carrying individual genotype of ppl-w1118 (control), ppl-SmoRNAi, ppl-CiRNAi, and ppl-HhRNAi and examined the levels of Smo, Ci, and Hh mRNA (for RNAi efficiency), and Ptc mRNA (for Hh signaling activity). *** indicates a p < 0.001, versus control in each assay (Student’s t-test). (D–E) Fat bodies expressing SmoRNAi or CiRNAi by the act > CD2 > Gal4 were stained for ptc-lacZ and CD2. The lack of CD2 expression marks the RNAi cells. Arrows indicate ptc-lacZ expression down-regulated and arrowheads indicate ptc-lacZ expression in WT cells. (F) PtcRNAi was expressed by the act > CD2 > Gal4, and fat bodies were stained for Smo and GFP. GFP expression marks the RNAi cell. Arrows indicate the accumulation of Smo on the cell surface. (G) Fat bodies from late 3rd instar larvae of w1118 strain were analyzed for their expression of Hh, Ptc, Smo and Act. Hh expression was at relatively low level. *** indicates a p < 0.001, versus Hh expression in the first column (Student’s t-test). (H) Late 3rd instar larvae from Myo1A-w1118 or Myo1A-HhRNAi were collected and grown on normal medium or fasting on 1.0% agar for 8 h. Fat bodies were collected from each group and subjected to Western blot for Hh (left and middle panels). Guts from larvae grown on normal medium and subjected to Western blot for Hh to monitor the RNAi efficiency in the gut (right panel). Western blot with the anti-tubulin antibody serves as loading control. Asterisks indicate a non-specific band that also serves as a loading control. Red arrows indicate Hh protein. (I) Fat bodies and guts from late 3rd instar larvae of Myo1A-w1118 or Myo1A-HhRNAi were collected. mRNA of each group was isolated and used for RT-PCR analysis to examine Hh expression in the gut and Ptc expression in the fat body. *** indicates a p < 0.001, versus w1118 control in each assay (Student’s t-test). (J) Late 3rd instar larvae of the w1118 strain were cultured with normal food or fasting condition with 1.0% agar for 8 h. Fat bodies were collected and mRNA from each group were isolated and subjected to RT-PCR analysis for Ptc and Hh expression. *** indicates a p < 0.001, versus control (Student’s t-test).
To further examine Hh signal transduction in fat body, we expressed Ptc RNAi in the same clonal analysis using the act > CD2 > Gal4 in fat body and found that RNAi of Ptc dramatically increased the accumulation of Smo on the cell surface (Fig. 2F), a mechanism for Smo activation in Hh signaling (Jia et al., 2004), suggesting that Hh signal is transduced in the same fashion as that in the wing disc.
We wondered how Hh signaling adopt a such high level of activity in the fat body. By analyzing the relative mRNA level, we found that both Ptc and Smo mRNA exhibited very high levels in WT fat body; however, Hh itself expression was relatively low (Fig. 2G). It is possible that Hh signaling may also be activated by Hh protein circulated from the gut (Rodenfels et al., 2014). To test this, we examined the levels of Hh protein in the fat body when Hh expression is inhibited in the gut. Consistent with the previous findings (Rodenfels et al., 2014), fasting condition increased the level of Hh protein in fat body, which was blocked by Hh RNAi by the enterocyte cell-specific Myo1A-Gal4 in the whole midgut (Fig. 2H, compare the middle to the left column). In support of our hypothesis, RNAi of Hh in the gut decreased the level of Ptc mRNA in the fat body (Fig. 2I), suggesting the circulated Hh protein contributes to part of the Hh signaling activity in fat body. We further analyzed the expression of a Hh-lacZ reporter and found that the Hh-lacZ was expressed at higher levels in the gut and wing disc, compared to that in the fat body (Fig. S3). Furthermore, we found that, consistent to the fasting induced Hh protein circulation (Fig. 2H), fasting indeed increased the level of Ptc mRNA in fat body (Fig. 2J). We also found that fasting condition also increased the expression of Hh (Fig. 2J). Taken together, these findings suggest that both the circulated Hh protein and the local expression of Hh contribute to the high level of Hh signaling in the fat body.
2.3. Hh signaling promotes TAG production through regulating Bmm/ATGL transcription
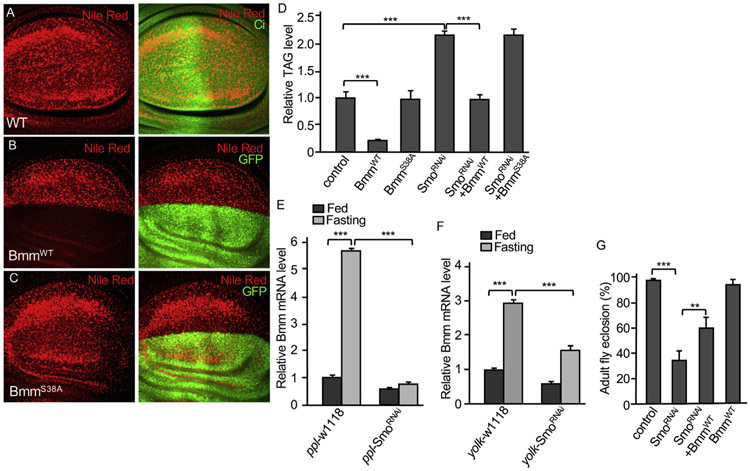
Lipid droplet accumulation decreased by Hh signaling is possibly caused a decrease in adipogenesis (Pospisilik et al., 2010; Suh et al., 2006), or an increase in lipolysis, or both. To determine whether Hh signaling regulates lipolysis, we examined Bmm, a major lipase in Drosophila. Wing disc can be used to access the activity of the lipase. We found that the overexpression of wild type Bmm (BmmWT) by the ap-Gal4 blocked, whereas the expression of an enzyme dead form Bmm (BmmS38A) (Gronke et al., 2005) by the ap-Gal4 failed to inhibit lipid accumulation in wing disc (Fig. 3B-C, compared to Fig. 3A), indicating that Bmm enzymatic activity was lost by S38A mutation. Consistently, the level of TAG was significantly decreased by the expression of BmmWT, but not the expression of BmmS38A, using ppl-Gal4 in the fat body (Fig. 3D). RNAi of Smo by the ppl-Gal4 significantly increased the level of TAG in fat body (Figs. 1M and 3D). Interestingly, TAG level increased by Smo RNAi was significantly decreased by the expression of BmmWT, but not BmmS38A (Fig. 3D). Similar to the increased levels of Ptc and Smo mRNA (Fig. 2G), fasting condition increased Bmm transcription (Fig. 3E). Interestingly, the increased Bmm transcription was blocked by Smo RNAi (Fig. 3E), suggesting Smo is required for fasting-induced Bmm expression. The same phenotypes were also observed in adult flies with fasting condition in combination with Smo RNAi expression by the yolk-Gal4 (Fig. 3F). Consistent with this finding, fasting condition significantly decreased the level of TAG in WT adult flies but not in adult flies expressing Smo RNAi using the yolk-Gal4 (Fig. S2B). The fat body-specific ppl-Gal4 expresses Gal4 starting from 1st instar larval stage to early adult stage. By examining adult fly eclosion, we found that adult fly eclosion was significantly decreased by Smo RNAi, which was recovered by BmmWT expression (Fig. 3G). Taken together, these data suggest that Bmm acts downstream of Smo to regulate lipolysis.
Fig. 3. Smo acts upstream of Bmm to regulate lipolysis.
(A) A WT wing disc was stained with Nile Red and Ci. (B–C) Wing discs expressing BmmWT or BmmS38A by the dorsal compartment-specific ap-Gal4 were stained with Nile Red and GFP. GFP indicates the Gal4 expression domain. (D) Late 3rd instar larvae expressing individual or in combination of BmmWT, BmmS38A, and SmoRNAi, by ppl-Gal4 were collected and subjected for TAG analysis, which was normalized by protein. ppl-Gal4-w1118 served as control. 16 larvae (8 males and 8 females) for each group were collected and the assays were performed with 3 repeats. *** indicates a p < 0.001 (Student’s t-test). (E) Fat bodies from late 3rd instar larvae expressing SmoRNAi by the ppl-Gal4 were treated under fed or fasting conditions, followed by RT-PCR to examine the expression of Bmm. ppl-Gal4-w1118 served as control. (F) 15-day female adults expressing SmoRNAi by the yolk-Gal4 (yolk-Gal4 is specifically expressed in adult female fat body) were treated with normal fed or fasting conditions, followed by RT-PCR to examine the levels of Bmm mRNA. yolk-Gal4-w1118 females served as control. 16 female adults were collected from each group and the assay was performed with 3 repeats. (G) Rate of adult eclosion were examined for the indicated genotypes and ppl-Gal4 were used to drive SmoRNAi and BmmWT expression. ppl-Gal4-w1118 served as control. In Figure E–G, ** indicates a p < 0.01, and *** indicates a p < 0.001 (Student’s t-test).
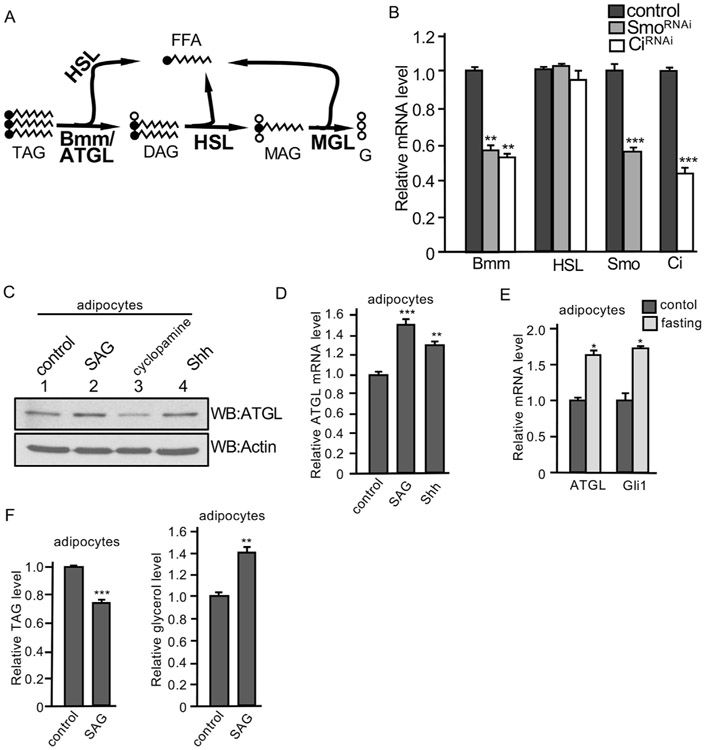
Bmm/ATGL hydrolyzes TAG and releases the first FFA from the glycerol backbone (Fig. 4A). This enzyme exhibits high substrate specificity for TAG. Both ATGL and hormone-sensitive lipase (HSL) can catalyze the reaction for TAG to generate FFA and diacylglycerol (DAG). DAG is the preferred substrate for HSL, leading to the formation of FFA and monoacylglycerol (MAG). MAG lipase (MGL) catabolizes MAG to generate fatty acid and glycerol (Fig. 4A). We examined the expression of Bmm in larva fat body and found that Smo RNAi or Ci RNAi by the ppl-Gal4 significantly decreased Bmm mRNA levels, but did not change the expression of HSL (Fig. 4B), suggesting Hh signaling specifically regulates Bmm. We further examined the expression of ATGL in 3T3-L1 adipocytes and found that the treatment with SAG or Shh increased, whereas the treatment with cyclopamine (a Smo antagonist) decreased the level of ATGL protein (Fig. 4C). In addition, the treatment of SAG or Shh significantly increased the levels of ATGL mRNA (Fig. 4D). Furthermore, similar to Bmm and Ptc expression induced by fasting condition (Figs. 2J and 3E), ATGL and Gli1 (a Hh target gene in vertebrate) transcription were induced by fasting in adipocytes (Fig. 4E). Finally, the treatment of SAG decreased the level of TAG and increased the level of glycerol in adipocytes (Fig. 4F), which was consistent with the findings in Drosophila fat body (Fig. 1M and N). Taken together, these data suggest that Hh signaling promotes the transcription of Bmm/ATGL in adipose tissue.
Fig. 4. Hh signaling upregulates Bmm/ATGL transcription.
(A) Scheme of the conversion of TAG, DAG, and MAG in the lipolytic pathway, with FFA and glycerol (G) indicated. Critical enzymes for these reactions are indicated in bold. (B) Fat bodies from late 3rd instar larvae expressing HhRNAi or SmoRNAi by the ppl-Gal4 were collected and mRNA was extracted for RT-PCR analysis to monitor the levels of gene transcription. Fat bodies from ppl-Gal4-w1118 served as control. The right two groups of columns indicate the RNAi efficiency. ** indicates a p < 0.01, and *** indicates a p < 0.001, versus control in the first column in each assay (Student’s t-test). (C) Differentiated 3T3-L1 adipocytes were treated with SAG (200 nM), cyclopamine (10 μM), or Shh (5 nM). Cell lysates were collected and subjected to Western blot with the anti-ATGL antibody. Western blots with the anti-Actin antibody served as loading control. (D) Differentiated 3T3-L1 adipocytes were treated with SAG (200 nM) or Shh (5 nM) and mRNA was isolated for RT-PCR assay to examine the level of ATGL expression. ** indicates a p < 0.01, and *** indicates a p < 0.001, versus control in the first column. (E) RT-PCR assay to examine the transcription of ATGL and Gli1 in differentiated 3T3-L1 adipocytes cultured with either fasting medium containing normal glucose and 1% FBS or control regular medium for 12 h * indicates a p < 0.05, versus control in the first column of each group. (F) Differentiated 3T3-L1 adipocytes were treated with SAG or DMSO (control) and analyzed for their levels of TAG in the cell and secreted free glycerol in the medium. ** indicates a p < 0.01, and *** indicates a p < 0.001, versus control in the first column of each panel.
2.4. Ci promotes Bmm transcription through direct interaction with bmm promoter
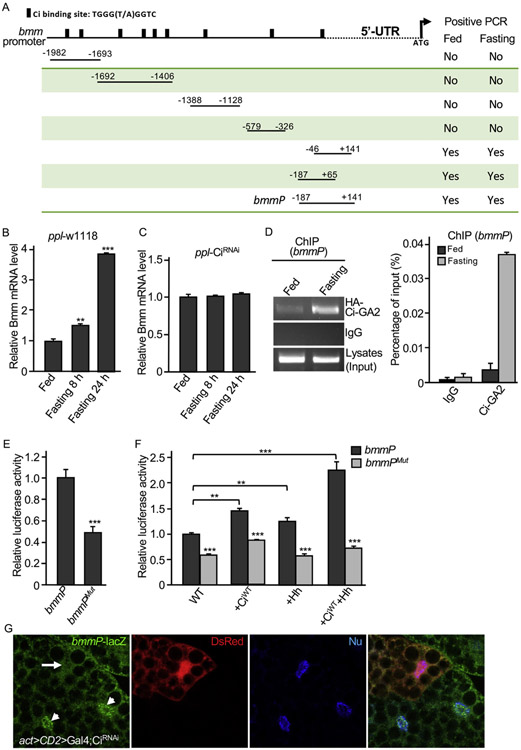
Ci binding site has been very well characterized in previous studies (Alexandre et al., 1996; Muller and Basler, 2000; Von Ohlen et al., 1997). By analyzing the bmm promoter, we identified 9 potential Ci binding sites (Fig. 5A). To determine whether Ci regulates Bmm/ATGL expression, we first tested Ci loss of function in regulating Bmm transcription. We found that the levels of Bmm mRNA were significantly increased by fasting condition in a time dependent manner in WT adult flies (Fig. 5B); however, levels of Bmm mRNA were not changed in flies expressing Ci RNAi (Fig. 5C), suggesting that Ci is required for fasting-induced Bmm transcription, in addition to Bmm transcriptional regulation by Ci under normal conditions. We thus move forward to examine the activity of bmm promoter fragments containing different Ci binding site(s) (Fig. 5A). We carried out ChIP analysis using adult flies expressing Ci-GA2 that contained the Zn finger Ci DNA-binding domain fused to the Gal4 activation domain (Jiang et al., 2014), in combination with fed or fasting conditions. Fasting condition increased the interaction between Ci-GA2 and bmm promoter regions of +141 to −46 and + 65 to −187 (Fig. S4), which contains a Ci binding site adjacent to the 5′-UTR (Fig. 5A). We verified the binding between Ci-GA2 and the +141 to −187 fragment (named bmmP) in a second round ChIP analysis, which consistently showed that fasting condition increased the interaction between Ci-GA2 and bmm promoter (Fig. 5D, left panel), which was further confirmed a quantitative analysis (Fig. 5D, right panel), suggesting that the binding site adjacent to the 5′-UTR is critical for the recruitment of Ci to the bmm promoter in response to fasting condition.
Fig. 5. Characterization of Ci binding sites in bmm promoter.
(A) A schematic drawing of the bmm promoter region with Ci binding sites indicated. (B–C) Adult male flies from ppl-Gal4-w1118 or ppl-Gal4-CiRNAi were cultured in either normal food (Fed) or 1% agar (Fasting) for 8 or 24 h. RT-PCR with samples of adult whole bodies were performed to examine the levels of Bmm mRNA. ** indicates a p < 0.01, and *** indicates a p < 0.001, versus fed flies in the first column. (D) Shown here is a ChIP analysis using adult flies expressing Ci-GA2 driven by ppl-Gal4 in fat body in combination with the primers covering the bmmP region indicated in A. For the ChIP analysis, male adults expressing HA-Ci-GA2 by the ppl-Gal4 were cultured in fed or fasting conditions for 24 h. Flies were collected, lysed, and immunoprecipitated with the anti-HA antibody, followed by PCR with primers covering fragments in bmm promoter. Only the primer combinations covering the potential Ci binding site adjacent to the 5′-UTR gave rise to elevated signals by fasting (See Fig. S4). IgG served as a negative control in these experiments. A Quantitative PCR analysis was performed and shown in the right panel. (E) bmmP-luc or bmmPMut-luc reporter construct was transfected into S2 cells in combination with tub-Ci followed by luciferase reporter activity analysis. ** indicates a p < 0.01. (F) S2 cells were transfected with tub-Ci and bmmP-luc or bmmPMut-luc construct, in combination with Flag-CiWT, Hh, or both, and analyzed for luciferase reporter activity. ** indicates a p < 0.01, and *** indicates a p < 0.001. (G) A fat body from late 3rd instar larvae expressing CiRNAi by the act > CD2 > Gal4 was immunostained for DsRed and bmmP-lacZ. Arrow indicates the decreased bmmP-lacZ expression in a cell expressing CiRNAi. Arrowheads indicate bmmP-lacZ expression in WT cells.
To further characterize the Ci binding site in bmmP, we generated luciferase reporter constructs containing either bmmP WT or bmmPMut in which eight nucleotides (AGGTGGTA) of the Ci binding site were deleted. We found that bmmPMut had a significant reduction in the luciferase reporter assay (Fig. 5E). We further found that bmmP reporter activity was significantly increased by CiWT expression; however, bmmPMut has significant low level of luciferase activity even in the presence of CiWT (Fig. 5F, the 2nd pair of columns). Consistently, the treatment with Hh-conditioned medium enhanced the reporter activity of bmmP but not bmmPMut (Fig. 5F, the 3rd pair of columns). We further found that the combination of CiWT and Hh treatment had potent activity to induce the highest levels of bmmP luciferase activity but not the activity of bmmPMut luciferase reporter (Fig. 5F, the last pair of columns). Finally, we constructed a bmmP-lacZ reporter and generated transgenic lines with this reporter construct. We found that, in fat body from late 3rd instar larvae, the inactivation of Ci by RNAi using the act > CD2 > Gal4 dramatically reduced bmmP-lacZ expression (Fig. 5G). Taken together, these data suggest that the Ci binding site in bmmP region is required for Ci to induce Bmm transcription.
2.5. Bmm mutation abolishes Ci regulation in vivo
To further characterize Bmm transcription regulated by Ci in vivo, we took advantage of the CRISPR-Cas9 technique and generated bmmDel and bmmMut mutants (see Methods) that contain genomic deletion of the whole promoter and genomic deletion of the eight nucleotides of Ci binding site adjacent to the transcription start, respectively (Fig. 6A). These mutants exhibited ~2 days delay in adult fly eclosion, otherwise are viable and fertile. By analyzing late 3rd instar larvae fat bodies, we found that the levels of Bmm mRNA from bmmDel and bmmMut larvae were significantly decreased compared to the level of Bmm mRNA in WT larvae (Fig. 6B). Consistently, bmmDel and bmmMut adult flies expressed Bmm mRNA at significantly low levels (Fig. 6C). Furthermore, the levels of TAG were significantly increased in bmmDel and bmmMut adult flies fed with normal food (Fig. 6D). Although fasting condition consistently decreased the levels of TAG, bmmDel and bmmMut adult flies also had much higher levels of TAG compared to WT flies under fasting conditions (Fig. 6D), suggesting that fasting induced lipolysis relies on, at least in part of, the Hh signaling. Finally, we found that the expression of Ciact significantly decreased the level of TAG in WT larvae; however, the ability for Ciact to decrease the level of TAG was significantly interfered by bmmMut mutation (Fig. 6E), suggesting that Ci requires the binding site in bmm promoter to promote lipolysis, and that mutating Ci binding site in bmm promoter significantly interferes with Bmm transcription. Together with the findings above, our observations indicate that Bmm transcription is regulated by Ci through Ci direct interaction with the bmm promoter.
Fig. 6. Analysis of bmm mutants generated by CRISPR.
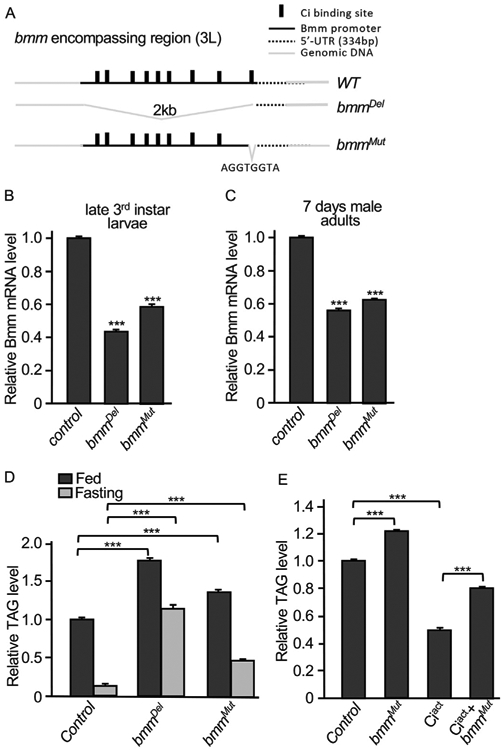
(A) A schematic drawing of the bmm promoter region, with Ci binding sites and genomic deletions, bmmDel and bmmMut, indicated. AGGTGGTA Ci-binding nucleotides were deleted in the genome of bmmMut. (B–C) RT-PCR analysis was performed using fat bodies from late 3rd instar larvae or using whole bodies from 7-day old male adults to examine Bmm mRNA levels in bmmDel and bmmMut. yw strain was used as control because both bmmDel and bmmMut were back-crossed to yw strain. *** indicates a p < 0.001, versus control in the first column. (D) 5–7 days male adult flies from the indicated genotypes were fed with normal food or fasted in 1% agar for 16 h, followed by the examination of TAG. yw strain was used as control. Each group had 20 flies and the experiment was performed with 3 repeats. *** indicates a p < 0.001, versus control in the first column, for both fed and fasting groups. (E) Ciact was expressed by yolk-Gal4 in either WT or bmmMut homozygous background. 7 days adult females were examined for the levels of TAG. 7 days adult females of the yolk-Gal4 flies under the same condition were used as control. *** indicates a p < 0.001.
3. Discussion
Hh signaling has been shown to play critical roles in animal development and also shown to be the driver for many types of cancer. Recent published studies have indicated that Hh signaling regulates adipogenesis and is likely involved in brown versus white adipocyte determination. However, it is unknown whether Hh signaling is involved in lipolysis in order to regulate lipid accumulation. In this study, we have uncovered two mechanisms for Hh to regulate lipid metabolism. First, we describe that Hh signaling has potent activity in Drosophila fat body. This high level of activity in the fat body are likely contributed by both Hh protein locally expressed and Hh protein circulated from the gut. Second, we identified bmm as a Hh signaling target gene directly regulated by Ci transcription factor that mediates Hh response in adipose tissue. Using the in vivo models of Drosophila larva, fat body, oenocyte, gut, and adult fly, we have collected evidence to support the idea that Hh signaling promotes the transcription of Bmm lipase (Fig. 7), uncovering the first lipolysis gene directly regulated by Hh signaling. We found that Hh signaling possesses potent activity in the fat body to induce lipolysis, therefore, provides energy for increased locomotor activity in larva stage and energy needed for metamorphosis during puparium formation and adult enclosion. The mechanisms in this study are likely conserved among species because the observations are replicated in differentiated adipocytes.
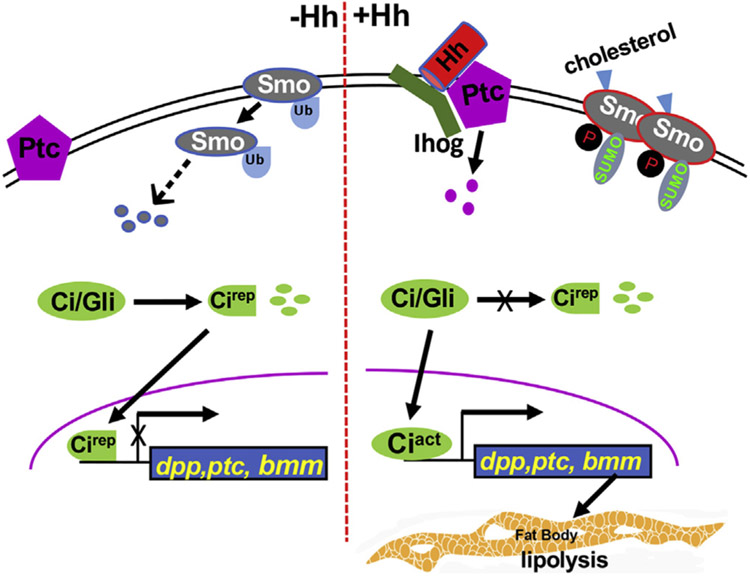
Fig. 7. A model of Hh signaling and its connection with lipolysis.
In the absence of Hh, Ptc inhibits Smo. Smo undergoes endocytosis that is mediated by ubiquitination. Ci/Gli is processed into a truncated repressor form that enters into the nucleus to block the expression of target genes, such as dpp and ptc. In the presence of Hh, the Ptc inhibition on Smo is relieved, causing increased Smo phosphorylation and accumulation on the cell surface. Phosphorylation and sumoylation counteracts ubiquitination to activate Smo. Upon Hh stimulation, full-length Ci is activated to turn on target gene expression. In this study, Bmm is identified as a direct target of Hh signaling. Hh signaling promotes lipolysis through Ci binding to the bmm promoter, therefore, elevates Bmm transcription.
The Hh protein locally expressed in the fat body and remotely circulated from the gut may contribute to high levels, but might not be all parts, of the Hh signaling activation. Recent structural studies have shown that Smo can be activated by cholesterol interaction with both the cysteine rich and the transmembrane domains of Smo (Byrne et al., 2017; Huang et al., 2016, 2018; Rana et al., 2013). Interestingly, extracellular sodium and the transmembrane sodium gradient may also regulate Smo activation (Blassberg and Briscoe, 2018; Myers et al., 2017). Lipid droplets, mainly found in adipose tissue, are composed of a neutral lipid core consisting of TAG and cholesteryl esters and serve as a reservoir for cholesterol (Farese and Walther, 2009; Olzmann and Carvalho, 2019), raising the possibility of Smo activation by cholesterol in fat body, although there has not been report on whether Smo can be activated by rich cholesterol in the fat body independent of Hh stimulation. It might also be possible for a Smo independent activation of Ci in a noncanonical fashion (Rodenfels et al., 2014). Taking consideration of these possibilities, the high levels of Hh signaling activity may be contributed by various mechanisms. In support of this idea, high levels of Hh signaling in the fat body is also evidenced by the status of Ci activation. It has been suggested that Ci processing is blocked in the fat body (Rodenfels et al., 2014). Our unpublished data indicate that the Ci processing component proteins, such as Slimb, are expressed in the fat body, indicating the high level of Hh signaling activity (both canonical and noncanonical) in the fat body is sufficient to block Ci processing.
In this study, we show that Hh signaling promotes lipolysis and decreases the levels of TAG in larvae and adult flies. One might think of the possibility that the inhibition of adipogenesis by Hh signaling might also contribute to the overall decrease in TAG; however, we examined the fat body at relatively later stages (3rd instar larvae and adult flies) and examined the size of the fat body and the accumulation of lipid droplets in the already developed fat body. The late 3rd instar larva and adult fly have been shown as good models to examine lipolysis, thus the use of these models allowed us to draw the conclusion that Hh signaling regulates lipolysis. In addition, we monitored the levels of TAG, FFA and/or glycerol, indicating the specificity of lipolysis regulated in vivo. Moreover, we have identified Ci binding site responsible for Hh signaling to regulate the expression of a specific lipase. Finally, the use of differentiated adipocyte also allowed us to specifically examine the lipolysis monitored at the level of TAG. All the evidence collected suggest that Hh signaling promotes lipolysis in adipose tissue.
3.1. Experimental procedures
3.1.1. Constructs, transgenes, and Gal4 lines
The bmmP-luciferase (bmmP-luc) reporter construct was generated by PCR from w1118 wild-type strain genomic DNA, and the 328bp fragment from −187 to +141 of Bmm transcription start site was inserted into pGL4 vector by XhoI and BglII sites. bmmPMut-luciferase (bmmPMut-luc) construct was generated based on bmmP-luc but with deletion of nucleotides AGGTGGTA (Ci binding site) near the Bmm transcription start site. bmmP-lacZ was constructed by fusing lacZ into the downstream of bmmP and inserted into attB-UASTΔUAS vector, in which UAS binding sites were deleted in the attB-UAST backbone (Fan et al., 2012). Transgenic line of bmmP-lacZ was made using the 75B1 attP locus. Transgenic lines of UAS-Myc-SmoWT, UAS-Myc-Smoact (previously named SmoSD123, a mutant form of Smo in which three clusters of PKA and CK1 sites were mutated to Asp to mimic phosphorylation and thus the activation of Smo) (Fan et al., 2012; Jia et al., 2004), UAS-HA-Ciact (previously named Ci−3P, a mutant form of Ci in which three PKA sites were mutated to Ala to abolish PKA phosphorylation and thus Ci processing) (Jia et al., 2010), and UAS-HA-Ci-GA2 (Jiang et al., 2014) have been described. UAS-BmmWT and UAS-BmmS38A transgenic lines (gifts from Dr. Ronald Kuhnlein) have been described (Gronke et al., 2005). UAS-PtcRNAi (BSC#28795) and UAS-SmoRNAi (BSC#27037) were obtained from Bloomington Stock Center (BSC). Gal80ts flies were from BSC (#7017). Hh-lzcZ has been described (Jia et al., 2005). UAS-HhRNAi (v1402) was obtained from Vienna Drosophila Resource Center (VDRC) and characterized in our previous study (Jiang et al., 2018). The wing dorsal-compartment-specific ap-Gal4 have been described in our previous studies (Jiang et al., 2018). act > CD2 > Gal4-DsRed strain (gifted from Dr. Duojia Pan) has been described (Liu et al., 2016). Other Gal4 lines are: cg-Gal4 (Ghosh et al., 2014), ppl-Gal4 (Zinke et al., 1999), oen-Gal4 (Li et al., 2016), and yolk-Gal4 (Georgel et al., 2001). yw or w1118 strains were used as controls.
3.1.2. Generating bmm mutants using CRISPR-Cas9
bmmDel and bmmMut mutant flies were generated according to Fly-CRISPR Scarless Gene Editing (Gratz et al., 2014, 2015; Li et al., 2015). See supplemental information for details.
3.1.3. Examination of lipid accumulation and ATGL expression in adipocyte
Generation of 3T3-L1 differentiated adipocyte was performed as previously described (Zebisch et al., 2012). Briefly, 3T3-L1 pre-adipocytes were cultured in DMEM (Sigma, D6429) with 10% newborn calf serum. Upon reaching confluence (about 48 h after seeding), differentiation was initiated with DMEM containing 0.5 mM iso-butylmethylxanthine (IBMX), 0.25 μM dexamethasone, 1 μg/mL insulin and 2 μM rosiglitazone. 48 h later, medium was changed to DMEM with 10% FBS and 1 μg/mL insulin, cells were maintained in this medium for additional 7–14 days before testing assays. To examine lipid accumulation, 3T3-L1 adipocytes were seeded into 96 well plate with 1 × 105 cell per well. 48 h later, cells were incubated in DMEM with desired compounds for additional 24 h. Half cells were then stained with BODIPY for 1 h, followed by fluorescence activity measurement taken by Molecular Devices Spectra Max M5 with excitation at 485 nm and emission at 538 nm, cutoff at 515 nm. For data analysis, relative fluorescence unit (RFU) of each group was obtained by the deduction of BODIPY stained cell with unstained cell. Each group has 8 repeats. Compounds used: SAG (Sigma, 200 nM); Vismodegib (GDC-0449, Selleckchem, 100 nM); Shh (R&D Systems, 5 nM). For Western blot analysis, 3T3-L1 adipocytes were starved overnight in serum free medium, then treated with SAG (200 nM), Shh (5 nM), or Cyclopamine (Selleckchem, 10 μM) for 24 h. Washed twice with PBS, cells were lysed in lysis buffer (50 mM Tris-HCl (pH8.0), 100 mM NaCl, 1.5 mM EDTA, 10% (v/v) glycerol, 1% (v/v) NP-40, 1 mM DTT and protease inhibitor tablet (Roche)) for 30 min on ice. Western bolt was performed by using primary antibody rabbit anti-ATGL (Cell Signaling, 1:1000), anti-PLIN1(Thermo Fisher, 1:2000), and anti-Actin (Cell Signaling, 1:10000).
3.1.4. Chromatin immunoprecipitation (ChIP)
About 50 5–7 days ppl-Gal4-HA-Ci-GA2 adult male fasted or fed flies were fixed with DSG (Thermo Fisher Scientific) at 2 mM for 45 min at room temperature, washed 3 times with PBS, then cross-linked with 1% (v/v) fresh prepared formaldehyde for 15 min. Reaction was stopped by 125 nM Glycine. Flies were then homogenized in 900 μL L1 buffer containing 50 mM Tris-HCl, pH 8.0; 2mM EDTA; 0.1% IGEPAL 630 (Sigma-Aldrich); 10% glycerol; 1 mM dithiothreitol (DTT); supplemented with 1 × protease inhibitor cocktail (Sigma-Aldrich) and 1 mM PMSF, for 30 min on ice. Nuclei were precipitated by centrifuged at 1200×g for 5 min and resuspended in 500 μL SDS lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS). Chromatin was sonicated 10 s for 8 times with 10 s break on ice (90% confluence, 50% power). Samples were centrifuged at 5000×g for 5min. Soluble chromatin was measured by absorbance at 260 nm by NanoDrop™ 2000/2000c Spectrophotometers, and equivalent amounts of input DNA were used for immunoprecipitation. 4 μg mouse anti-HA antibody (Santa Cruz sc-7392) was used for precipitation of samples. PCR analysis was performed using primers covering different regions of bmm promoter. Quantitative PCR analysis was performed to determine the percentage of bmmP region precipitated with the anti-HA antibody against PCR product from the input DNA. IgG served as a negative control in these experiments.
3.1.5. RT-PCR analysis
RNA from fat bodies or adult flies were isolated using TRIzol® Reagent (Invitrogen™ #15596018). Equivalent amounts purified RNA were reverse transcribed into cDNAs by High Capacity cDNA Reverse Transcription Kit (AB Applied Biosystems, #4368814) according to manufacturer’s instruction. Quantitative Real-Time PCR reactions were carried out using SYBR Green qPCR Master Mix (Thermo Scientific), and signals were detected by QuantStudio 3 (Applied Biosystems). L32 was used as reference gene and assays were performed with 3 repeats. Statistical analysis of the mRNA level was obtained using the (2−ΔΔCt) method.
3.1.6. Immunostaining of Drosophila tissues
Wing discs were dissected in PBS and fixed with 4% formaldehyde in PBS for 20 min at room temperature. Fat bodies were dissected in PBS and fixed with 4% formaldehyde for 30 min at 37 °C. After permeabilization with PBST (PBS supplemented with 1% Triton X100), tissues were blocked in 5% goat serum for 30 min then incubated with primary antibodies for 3 h and the appropriate secondary antibodies for 1 h, and washed three times with PBST after each incubation. Overexpression or RNAi clones in fat bodies were generated with the act > CD2 > Gal4 in combination with heat shock at 72 h after egg laying (AEL). Oenocytes on the basal surface of the lateral epidermis from 3rd instar larvae with specific genotypes were dissected and stained with the methods we described (Li et al., 2016), which was modified from a previous study (Burns et al., 2012). Briefly, larvae were dechorionated in 80% bleach for 8 min, rinsed with H2O twice, fixed with 8% formaldehyde/heptane for 30 min with continuous shaking, followed by washing with heptane/methanol twice, washing with methanol twice, washing with methanol/0.1% PBST twice, and washing with 0.2% PBST twice. Then samples were dissected and stained desired antibody in 0.2% PBST. Primary antibodies used were: mouse anti-SmoN (DSHB); rabbit anti-β-Gal (Cappel), anti-GFP (Clontech); rat anti-Ci (2A1, DSHB), anti-HNF4 (gift from Dr. B. Gebelein). Affinity-purified secondary antibodies (Jackson ImmunoResearch) for multiple labeling were used. For lipid staining, BODIPY (493/503, Invitrogen, D3922) or Nile Red (Sigma) was used during secondary antibody staining. Hoechst 33342 (Invitrogen) was used for nuclear staining. Fluorescence signals were acquired on a Nikon A1+ confocal microscope and images processed with Nikon Elements software. About 15 imaginal discs, fat bodies, or oenocytes were scored and 3–5 images were taken for each genotype. About 100% wing discs, 80% fat bodies, and 90% oenocytes exhibited the immunostaining phenotypes.
3.1.7. TAG, glycerol, and free fatty acid measurement
Free fatty acid was measured according to Free Fatty Acid Quantitation Assay (Sigma, # MAK044). Briefly, 10 male and 10 female larvae were weighed and homogenized in 50 μL PBST (1 × PBS + 1% Triton X-100), added chloroform to final volume of 30 μL/mg tissue, extracted by shaking the vial for 5 min, then centrifuged at 13000×g for 10 min. 300 μL of organic phase (lower phase) were collected and air dried at 50 °C, then vacuum dried for another 15 min. 200 μL Fatty Acid Assay Buffer were added to each sample to dissolve dried lipids. 10 μL liquid samples was mixed with 40 μL Fatty Acid Assay Buffer and 2 μL ACS Reagent, incubated for 30 min at 37 °C. After that, 50 μL Master Reaction Mix was added into each well, then incubated for another 30 min at 37 °C, protected from light. Measurement was taken at 570 nm by Spetra MR DYNEX Technologies. A set of gradient diluted palmitic acid was used as standard.
TAG assay was performed according to previous described (Tennessen et al., 2014). Briefly, 10 male and 10 female larvae were homogenized in 50 μL PBST (1 × PBS + 0.05% Tween 20, filtered before use) buffer/larva, then 50 μL of each sample were subjected for protein assay. The rests were heated at 70 °C for 10 min and 100 μL of each sample was mixed with equal volume of triglyceride reagent, incubated at 37 °C for 60 min, centrifuged at 12000×g for 5 min. 30 μL of each supernatant was added 100 μL Free Glycerol Reagent and incubated at 37 °C for additional 10 min, measurement was taken at 540 nm by Spetra MR DYNEX Technologies. Each sample was performed with 3 repeats. For glycerol assay in 3T3-L1 cells, medium samples were collected, heated at 70 °C for 10 min, centrifuged at 12000×g for 5 min then mixed with 100 μL Free Glycerol Reagent, incubate at 37 °C for 10 min before measurement.
In this study, when we collect fat bodies, we do not separate male from female, because fat bodies are thought to be identical. When we examine TAG levels in the larvae, we use half male and half female (normally 8 for each) as a pool. When we examine adult flies, we normally use male flies because eggs carried in female flies might cause variation.
Supplementary Material
Acknowledgements
We are grateful to Dr. Duojia Pan for the act > CD2 > Gal4-DsRed strain; Dr. Ronald Kuhnlein for the UAS-BmmWT and UAS-BmmS38A transgenic lines; Dr. Savraj S. Grewal for the cg-Gal4 line; Vienna Drosophila Resource Center (VDRC) and TRiP at Harvard Medical School for fly stocks; Dr. Brian Gebelein for the anti-HNF4 antibody; Dr. Tom Kornberg for the anti-Hh antibody; and the Developmental Studies Hybridoma Bank (DSHB) for the anti-Ci and anti-β-tubulin antibodies. We also thank the Redox Metabolism Shared Resource Facilities of the University of Kentucky Markey Cancer Center (supported by National Cancer Institute grant P30 CA177558, USA). We also thank the Imaging Core Facility of the COBRE (P20 GM121327, USA) for providing the imaging systems. This study was supported by grants from the National Institute of Health (NIH), USA (R01GM079684 and R35GM131807) to J. J.
Funding
This study was supported by the National Institutes of Health grants (R01GM079684 and R35GM131807), and was also supported by the Shared Resource Facilities of the University of Kentucky Markey Cancer Center (P30CA177558), and the Imaging Core and Pilot funds from the COBRE (P20GM121327).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ydbio.2019.09.009.
References
- Alexandre C, Jacinto A, Ingham PW, 1996. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 10, 2003–2013. [DOI] [PubMed] [Google Scholar]
- Beller M, Bulankina AV, Hsiao HH, Urlaub H, Jackle H, Kuhnlein RP, 2010. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metabol. 12, 521–532. [DOI] [PubMed] [Google Scholar]
- Blassberg R, Briscoe J, 2018. Smoothened sensor places sodium and sterols center stage. Dev. Cell 44, 3–4. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Therond PP, 2013. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol 14, 416–429. [DOI] [PubMed] [Google Scholar]
- Burns KA, Gutzwiller LM, Tomoyasu Y, Gebelein B, 2012. Oenocyte development in the red flour beetle Tribolium castaneum. Dev. Gene. Evol 222, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne EF, Luchetti G, Rohatgi R, Siebold C, 2017. Multiple ligand binding sites regulate the Hedgehog signal transducer Smoothened in vertebrates. Curr. Opin. Cell Biol 51, 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droujinine IA, Perrimon N, 2016. Interorgan communication pathways in Physiology: focus on Drosophila. Annu. Rev. Genet 50, 539–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Liu Y, Jia J, 2012. Hh-induced Smoothened conformational switch is mediated by differential phosphorylation at its C-terminal tail in a dose- and position-dependent manner. Dev. Biol 366, 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese RV Jr., Walther TC, 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA, 2001. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 1, 503–514. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Rideout EJ, Grewal SS, 2014. TIF-IA-dependent regulation of ribosome synthesis in drosophila muscle is required to maintain systemic insulin signaling and larval growth. PLoS Genet. 10, e1004750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Rubinstein CD, Harrison MM, Wildonger J, O’Connor-Giles KM, 2015. CRISPR-Cas9 genome editing in Drosophila. Curr. Protoc. Mol. Biol 111, 31 32 31–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM, 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Kehinde O, 1975. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5, 19–27. [DOI] [PubMed] [Google Scholar]
- Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C, 1991. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem 266, 11341–11346. [PubMed] [Google Scholar]
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP, 2005. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metabol. 1, 323–330. [DOI] [PubMed] [Google Scholar]
- Guha M, 2012. Hedgehog inhibitor gets landmark skin cancer approval, but questions remain for wider potential. Nat. Rev. Drug Discov 11, 257–258. [DOI] [PubMed] [Google Scholar]
- Hooper JE, Scott MP, 2005. Communicating with hedgehogs. Nat. Rev. Mol. Cell Biol 6, 306–317. [DOI] [PubMed] [Google Scholar]
- Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, Salic A, 2016. Cellular cholesterol directly activates smoothened in hedgehog signaling. Cell 166, 1176–1187 e1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Zheng S, Wierbowski BM, Kim Y, Nedelcu D, Aravena L, Liu J, Kruse AC, Salic A, 2018. Structural basis of smoothened activation in hedgehog signaling. Cell 175, 295–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Liu Y, Xia R, Tong C, Yue T, Jiang J, Jia J, 2010. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. J. Biol. Chem 285, 37218–37226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Jiang J, 2006. Decoding the Hedgehog signal in animal development. Cell. Mol. Life Sci 63, 1249–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J, 2004. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature 432, 1045–1050. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, Amanai K, Jiang J, 2005. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for slimb/beta-TRCP-mediated proteolytic processing. Dev. Cell 9, 819–830. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC, 2008. Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Liu Y, Fan J, Epperly G, Gao T, Jiang J, Jia J, 2014. Hedgehog-regulated atypical PKC promotes phosphorylation and activation of Smoothened and Cubitus interruptus in Drosophila. Proc. Natl. Acad. Sci. U.S.A 111, E4842–E4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Liu Y, Zhang J, Jia J, 2018. An intracellular activation of Smoothened that is independent of Hedgehog stimulation in Drosophila. J. Cell Sci 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Song J, Zaytseva YY, Liu Y, Rychahou P, Jiang K, Starr ME, Kim JT, Harris JW, Yiannikouris FB, Katz WS, Nilsson PM, Orho-Melander M, Chen J, Zhu H, Fahrenholz T, Higashi RM, Gao T, Morris AJ, Cassis LA, Fan TW, Weiss HL, Dobner PR, Melander O, Jia J, Evers BM, 2016. An obligatory role for neurotensin in high-fat-diet-induced obesity. Nature 533, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barish S, Okuwa S, Volkan PC, 2015. Examination of endogenous rotund expression and function in developing Drosophila olfactory system using CRISPR-cas9-mediated protein tagging. G3 (Bethesda) 5, 2809–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chen Y, Shi Q, Yue T, Wang B, Jiang J, 2012. Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS Biol. 10, e1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zheng Y, Yin F, Yu J, Silverman N, Pan D, 2016. Toll receptor-mediated hippo signaling controls innate immunity in Drosophila. Cell 164, 406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Beachy PA, 2004. The Hedgehog response network: sensors, switches, and routers. Science 304, 1755–1759. [DOI] [PubMed] [Google Scholar]
- Matz-Soja M, Rennert C, Schonefeld K, Aleithe S, Boettger J, Schmidt-Heck W, Weiss TS, Hovhannisyan A, Zellmer S, Kloting N, Schulz A, Kratzsch J, Guthke R, Gebhardt R, 2016. Hedgehog signaling is a potent regulator of liver lipid metabolism and reveals a GLI-code associated with steatosis. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B, Basler K, 2000. The repressor and activator forms of Cubitus interruptus control Hedgehog target genes through common generic gli-binding sites. Development 127, 2999–3007. [DOI] [PubMed] [Google Scholar]
- Myers BR, Neahring L, Zhang Y, Roberts KJ, Beachy PA, 2017. Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc. Natl. Acad. Sci. U.S.A 114, E11141–E11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelliot A, Bond N, Hoshizaki DK, 2006. Fat-body remodeling in Drosophila melanogaster. Genesis 44, 396–400. [DOI] [PubMed] [Google Scholar]
- Nosavanh L, Yu DH, Jaehnig EJ, Tong Q, Shen L, Chen MH, 2015. Cell-autonomous activation of Hedgehog signaling inhibits brown adipose tissue development. Proc. Natl. Acad. Sci. U.S.A 112, 5069–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Carvalho P, 2019. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol 20, 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Perrimon N, 2014. Modeling metabolic homeostasis and nutrient sensing in Drosophila: implications for aging and metabolic diseases. Dis. Model Mech 7, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, Bayer M, Haschemi A, Puviindran V, Tar K, Orthofer M, Neely GG, Dietzl G, Manoukian A, Funovics M, Prager G, Wagner O, Ferrandon D, Aberger F, Hui CC, Esterbauer H, Penninger JM, 2010. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell 140, 148–160. [DOI] [PubMed] [Google Scholar]
- Rana R, Carroll CE, Lee HJ, Bao J, Marada S, Grace CR, Guibao CD, Ogden SK, Zheng JJ, 2013. Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling. Nat. Commun 4, 2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenfels J, Lavrynenko O, Ayciriex S, Sampaio JL, Carvalho M, Shevchenko A, Eaton S, 2014. Production of systemically circulating Hedgehog by the intestine couples nutrition to growth and development. Genes Dev. 28, 2636–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, LoRusso PM, Von Hoff DD, de Sauvage FJ, Low JA, 2009. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med 361, 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic A, Von Hoff D, 2016. Hedgehog pathway inhibition. Cell 164, 831. [DOI] [PubMed] [Google Scholar]
- Suh JM, Gao X, McKay J, McKay R, Salo Z, Graff JM, 2006. Hedgehog signaling plays a conserved role in inhibiting fat formation. Cell Metabol. 3, 25–34. [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Barry WE, Cox J, Thummel CS, 2014. Methods for studying metabolism in Drosophila. Methods 68, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R, Aberger F, Esterbauer H, Riobo N, Pospisilik JA, 2014. Canonical and non-canonical Hedgehog signalling and the control of metabolism. Semin. Cell Dev. Biol 33, 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R, Amann S, Bayer M, McGee SL, Loipetzberger A, Connor T, Jaeger C, Kammerer B, Winter L, Wiche G, Dalgaard K, Selvaraj M, Gaster M, Lee-Young RS, Febbraio MA, Knauf C, Cani PD, Aberger F, Penninger JM, Pospisilik JA, Esterbauer H, 2012. Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell 151, 414–426. [DOI] [PubMed] [Google Scholar]
- Von Ohlen T, Lessing D, Nusse R, Hooper JE, 1997. Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc. Natl. Acad. Sci. U.S.A 94, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Jia H, Fan J, Liu Y, Jia J, 2012. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS Biol. 10, e1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Li X, Wang W, Liu Z, Chen J, Ding M, Huang X, 2018. MRT, functioning with NURF complex, regulates lipid droplet size. Cell Rep. 24, 2972–2984. [DOI] [PubMed] [Google Scholar]
- Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, Pujara K, Stinson J, Callahan CA, Tang T, Bazan JF, Kan Z, Seshagiri S, Hann CL, Gould SE, Low JA, Rudin CM, de Sauvage FJ, 2009. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 326, 572–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebisch K, Voigt V, Wabitsch M, Brandsch M, 2012. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal. Biochem. 425, 88–90. [DOI] [PubMed] [Google Scholar]
- Zheng X, Mann RK, Sever N, Beachy PA, 2010. Genetic and biochemical definition of the Hedgehog receptor. Genes Dev. 24, 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I, Kirchner C, Chao LC, Tetzlaff MT, Pankratz MJ, 1999. Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development 126, 5275–5284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.