Abstract
Mass spectrometry (MS) has made enormous contributions to comprehensive protein identification and quantification in proteomics. MS is also gaining momentum for structural biology in a variety of ways, complementing conventional structural biology techniques. Here, we will review how MS-based techniques, such as hydrogen/deuterium exchange, covalent labeling, and chemical cross-linking, enable the characterization of protein structure, dynamics, and interactions, especially from a perspective of their data analyses. Structural information encoded by chemical probes in intact proteins is decoded by interpreting MS data at a peptide level, i.e., revealing conformational and dynamic changes in local regions of proteins. The structural MS data are not amenable to data analyses in traditional proteomics workflow, requiring dedicated software for each type of data. We first provide basic principles of data interpretation, including isotopic distribution and peptide sequencing. We then focus particularly on computational methods for structural MS data analyses and discuss outstanding challenges in a proteome-wide large scale analysis.
Keywords: Structural biology, Mass spectrometry, Hydrogen/deuterium exchange, Covalent labeling, Chemical cross-linking, Computational proteomics, Bioinformatics software
1. Introduction
Protein function is regulated by changes in its three-dimensional structure and enzymatic activity in response to protein modifications, ligand binding, or protein–protein interactions. Elucidating the three-dimensional structures of proteins, therefore, is an important step toward understanding their role in molecular functional biology. For decades, the structural models of proteins have been studied at atomic resolution using X-ray crystallography [1], nuclear magnetic resonance (NMR) spectroscopy [2], and (cryo-)electron microscopy (EM) [3]. However, these approaches are not applicable to all proteins and protein complexes owing to their limited amount, low solubility, large size, or unavailable crystals. To bridge the gap and study dynamic protein complexes, there are needs for complementary techniques taking advantages of 1) analyzing proteins in native solution condition; 2) dealing with limited sample amounts; 3) analyzing protein complexes or interactions; and 4) providing information for active dynamic structural changes of a protein under various biological conditions. In that respect, mass spectrometry (MS) has become increasingly popular in the field of structural biology [4]. Over the past few decades, MS has made substantial contributions to proteomics, from the identification and quantification of individual proteins, to high throughput analyses of whole proteome and the studies of protein localization and post-translational modifications [5], [6], [7]. Recent high-performance liquid chromatography (HPLC) and high MS acquisition speed enabled the detection of tens of thousands of peptides in an hour [8], the detection of over 10,000 proteins from human cell lines [9], and the exploration of the human interactome consisting of ~ 56,000 interactions [10]. MS approaches have also been applied to various structural biology studies ranging from epitope mapping and protein–ligand interactions to probing structures of membrane proteins [11], [12], [13], [14], [15], [16], [17], [18]. MS coupled with chemistry could infer global, local and site-specific structural information with high sensitivity and accuracy even for low concentration proteins.
The unique strength of MS is its ability to detect changes in each individual amino acid level as well as in protein and peptide levels. MS-based structural proteomics can be divided into two main streams, 1) peptide-centric and 2) protein-centric, depending on what analyte MS actually analyzes to obtain the structural information [19], [20], [21]. In peptide-centric methods, more popular than protein-centric methods, the concept is that 1) chemical reagents introduce modifications into native proteins or protein complexes in solution, 2) such modifications encode some structural information about which local regions are exposed/buried and which residues are in close spatial proximity (mediating protein–protein interactions) in native proteins or complexes, 3) MS-based proteomics identifies and localizes the modifications at the peptide level to infer the structural information. In a protein-centric method, called native MS, MS directly analyzes intact biomolecules that include all subunits and cofactors that make up the functional complex. It is possible only when the process of native MS can maintain weak noncovalent interactions between protein subunits and associated biomolecules such as DNA, cofactors, and ligands. The mass and charge state distribution detected by native MS allows for the determination of protein complex stoichiometry, cofactor content, and conformation. Native MS has also been coupled with other hybrid techniques such as ion mobility spectrometry, hydrogen/deuterium exchange, and top-down proteomics. Ion mobility spectrometry enables an additional separation by the overall shape of protein complexes [22], hydrogen/deuterium exchange reveals the conformational dynamics of intact proteins [23], and top-down proteomics, sequencing intact proteins, provides an additional level of information about specific proteoforms arising from sequence variations and post-translational modifications [24]. Although MS-based methods provide opportunities to probe the protein structure, dynamics, and interactions in native environments, the individual pieces of data from different methods typically do not provide sufficient information to derive a structural model of a protein or complex by itself. The structural features and constraints monitored by MS-based methods can be used complementary to conventional techniques such as X-ray crystallography, EM, and NMR spectroscopy. The improved accuracy and completeness of a model will be achieved by simultaneous use of all such information from multiple sources of data [25], [26], [27].
This review will focus on computational methods and discuss challenges in three MS-based peptide-centric methods, 1) hydrogen/deuterium exchange - exchanging peptide amide hydrogen atoms with deuterium atoms in solution, 2) covalent labeling - introducing irreversible modifications to amino acid residues, and 3) chemical cross-linking - covalently linking two spatially proximal amino acid residues. The three approaches generate different type of MS data, each requiring dedicated software to decode the structural information. The MS-based identification of disulfide bonds involved in protein tertiary structures will be also discussed under chemical cross-linking category owing to their similarity.
2. Basics of mass spectrometry
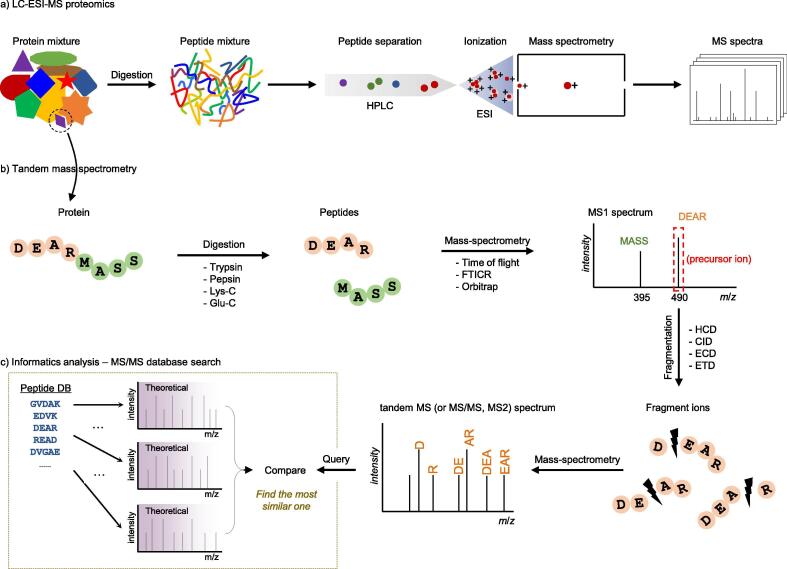
MS has become a powerful tool for analyzing complex protein mixtures [28]. Protein mixtures are digested by residue-specific enzymes, and the digested peptides are separated by liquid chromatography (LC), thereby reducing the complexity of the peptide mixture prior to MS. The separated peptides are then subjected to electrospray ionization (ESI) and introduced into a mass spectrometer (Fig. 1a). The instrument generates a mass spectrum (MS1), in which a peptide ion is detected and recorded as a peak with the intensity at its mass-to-charge ratio (m/z) value measured by a mass analyzer. The measurement accuracy is achieved at a level of a few parts per million (ppm) using high-resolution mass analyzers such as Orbitrap and Fourier transform ion cyclotron resonance (FTICR).
Fig. 1.
Conventional MS-based proteomics experiments for peptide and protein identification. a) Overview of MS-based proteomics. A protein mixture from a biological source is digested into peptides (usually by trypsin). The peptides are separated by one or more steps of high-performance liquid chromatography (HPLC) column and are ionized by electrospray ionization (ESI) at the end of the column. The resulting peptides enter the mass spectrometer and the peptides eluting at the time point are recorded in a mass spectrum (MS1). The peptides can also be ionized using matrix-assisted laser desorption/ionization (MALDI), where the peptides are ionized out of a dry, crystalline matrix via laser pulses. b) Besides a mass list of peptides in MS1 spectra, some prioritized peptides (precursor ions) are fragmented by energetic collision with gas, and the products are recorded in the tandem or MS/MS spectrum. (This figure is the conceptual illustration for a single protein. All peptides from a protein mixture shown in a) are analyzed together in single MS run). c) Peptides are most commonly identified using a database search approach, where an experimental MS/MS spectrum is compared with theoretical spectra predicted for peptides from a protein sequence database.
Tandem mass spectrometry (MS/MS or MS2) is used to obtain peptide sequence information (Fig. 1b). The isolated peptide, a precursor ion, is split into fragment ions by low-energy collision-induced dissociation (CID) and their peak (a pair of m/z and intensity) information is recorded in MS/MS spectra. Series of these fragment ions detected at MS/MS readily reveal the amino acid sequence of the precursor ion. To interpret an experimentally obtained MS/MS spectrum as a peptide, the most popular approach is to compare the experimental MS/MS spectrum with theoretical MS/MS spectra, generated from candidate peptides stored in a protein sequence database, using database search software [29], [30], [31] (Fig. 1c). The software retrieves from the database, candidate peptides whose masses are within a specified mass tolerance of a precursor ion mass. The validity of peptide-spectrum matches can be assessed by target-decoy strategy [32]. For a comprehensive review of the peptide and protein identification, see refs [33], [34].
3. Hydrogen/deuterium exchange
Surface labeling is based on the concept that solvent-exposed regions in proteins will react quickly with labeling reagents and therefore will be labeled/modified, while buried regions will be labeled/modified slowly or not at all [35]. Protein conformational changes or protein–ligand binding can affect the degree of solvent exposure for certain protein regions, and the changes in labeled/modified degree by labeling reagents indicate which regions are undergoing a structural change or forming an interface with an interacting partner. The best-known and most widely used strategy for surface labeling is hydrogen–deuterium exchange (HDX) that monitors the exchange of backbone amide hydrogens with deuteriums [36], [37], [38]. The exchange rates are sensitive to changes in hydrogen bonding, secondary structure, solvent accessibility and dynamics. The general HDX-MS workflow is depicted in Fig. 2a. The target protein(s) are incubated with D2O to exchange accessible hydrogen atoms with deuterium atoms. The exchange reaction is quenched at different time points to plot deuteration rate as a function of exchange time (from seconds to days). The deuterated proteins are subjected to proteolytic digestion followed by LC-MS. MS measures mass increases of peptides by deuterium incorporation. The amount of deuterium incorporation is usually determined using only peptide masses without MS/MS fragmentation due to H/D scrambling under conventional collisional activation (intramolecular H/D rearrangement). Electron-mediated fragmentation techniques such as electron capture and transfer dissociation (ECD/ETD) can be employed to avoid such scrambling and measure the exchange at the individual amino acid level [39]. In fact, the digestion and LC-MS workflow leads to back exchange [36]. To minimize the back exchange effect, in most applications of HDX, the deuterated proteins are digested using pepsin at low temperature and low pH (with a minimum at ~ 2.5) and the peptide mixtures are separated through chromatography columns cooled to temperatures close to 0 °C. Back exchange may be corrected on the basis of deuterium incorporation in a completely deuterated sample [37]. Most HDX analyses, however, measure relative rather than absolute deuterium incorporation and studies have shown that back exchange correction does not affect relative measurement.
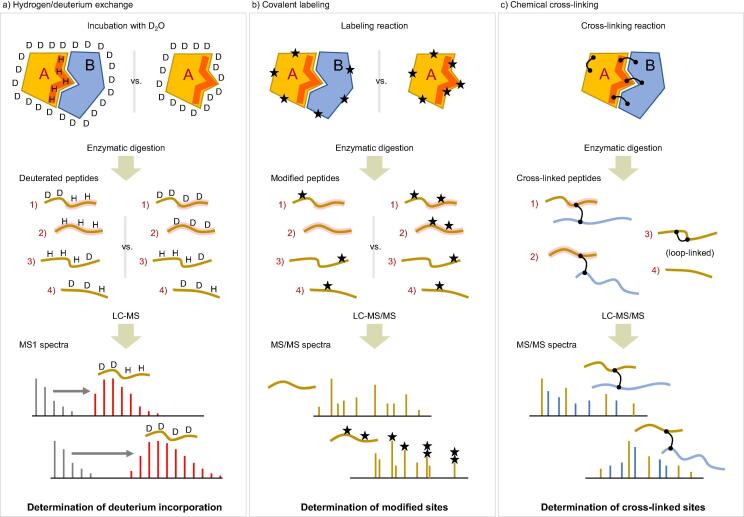
Fig. 2.
Schematic representation for revealing binding interfaces in protein–protein interaction. For the sake of simplicity, this example focuses on the analysis of protein A (the same goes for the analysis of protein B). Each MS-based method shows how to probe protein surface topology and reveal specific protein–protein interaction sites. a) Hydrogen/deuterium exchange MS. The exchange is rapidly progressed for solvent accessible regions, while slower for protected regions by ligand binding, protein–protein interactions, or stabilization of secondary structure. The changes in exchange rates under two different conditions can reveal protein surfaces involved in transient interactions. For example, after D2O incubation of native proteins and proteolytic digestion, peptides 1 and 2 of protein A were less deuterated under the left condition than under the right condition, indicating that peptides 1 and 2 are from the protected (red) region, while peptides 3 and 4 have no difference in their deuteration. The deuterium incorporation of these peptides can be measured by monitoring their isotopic distributions by LC-MS (shown at the bottom). The high precision and sensitivity of mass instruments enable the detection of such subtle changes. b) The concept of covalent labeling MS is similar to that of HDX-MS. The difference is that covalent labeling introduces irreversible modifications (marked by stars in this figure) to solvent accessible regions. The protected red region of protein A was rarely modified when interacting with protein B. The covalent modifications are highly stable during sample handling and thus modified sites as well as modification types on the same peptides can be distinguished by MS/MS as shown at the bottom. c) Chemical cross-linking MS covalently connects two residues in close spatial proximity between two proteins or within a protein. The constraints obtained by the chemical cross-links narrow down the possible location of the binding interface. Since the link of two peptides is covalent, the analysis is amenable to MS/MS. In the MS/MS spectra, fragment ions from two peptides are present at the same time. In this figure, curvy lines represent peptides. Lines or parts in lines shaded in red means that their sequences belong to the red region in protein A. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
HDX is a non-covalent labeling approach, while covalent labeling measures the solvent accessibility of side chains by introducing modifications to side chains. Covalent labeling is discussed in Section 4.
3.1. Fundamentals of data interpretation
HDX data analysis generates a list of peptides and quantifies the deuterium incorporated to each peptide at each labeling time point. The peptide list is usually compiled from database search-driven identification of a separated MS/MS experiment for an unlabeled sample. To increase the protein sequence coverage, additional peptides from in silico digestion (using protease specificity) can be considered if they can be assigned to peaks in MS1 spectra. It is assumed that the chromatographic properties of deuterated peptides are identical to those of natural peptides and thus the isotopic distributions of deuterated peptides can be annotated using the retention time of natural peptides. Since there are small differences in retention time between the two, the deuterated isotopic distributions are annotated within a small window around the retention time of natural peptides.
The exchange of hydrogens in a peptide backbone with deuteriums causes a mass increase of the peptide, which depends on the number of incorporated deuterium atoms (approximately 1.006 Da per D atom). Although such mass increases can be detected by MS, they are not readily calculated since deuterated peptides are represented as complicated forms in MS1 spectra due to natural isotopes and partial deuteration. Because of isotopes existing in nature such as 13C, 15N, 18O, and 34S, a peptide is not represented as a single peak in an MS1 spectrum, but represented by an isotopic cluster of peaks spaced by 1.00235 Da (i.e., average isotope spacing) (see natural isotope distribution of Fig. 3). Furthermore, deuterium incorporation to a peptide is a gradual process and the resulting partial deuteration can result in multiple species of deuterated peptides with different deuteration levels. Thus the deuterated isotopic distribution is the union of isotopic distributions of differently deuterated peptides and spans a wider m/z range than the corresponding natural isotopic distribution (compare natural and deuterated isotope distributions of Fig. 3).
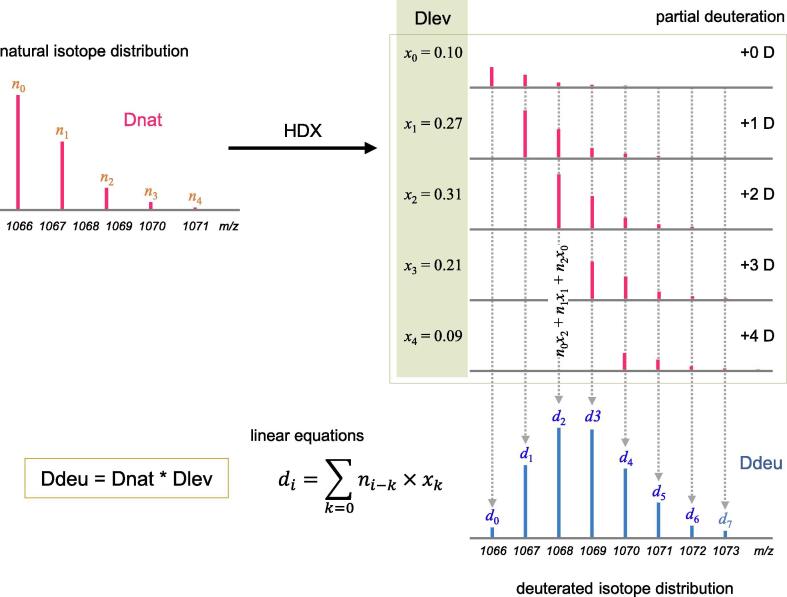
Fig. 3.
Three distributions in HDX-MS data analysis are shown. The observed, deuterated isotopic distribution, Ddeu is represented as the convolution of natural isotopic distribution (Dnat) and deuterium incorporation distribution (Dlev), the parameter values of which we want to solve. The HDX process would produce various partially deuterated peptides from a single peptide and thus the abundance of each deuterium number (#), x# values in this figure, should be determined. Many studies assumed that the distribution of x# conforms to the binomial distribution. After all, Ddeu is a linear combination of mass-shifted natural isotope distributions, yielding a set of linear equations of variable x#, for example, . In the figure, the average deuterium incorporation can be determined as two.
Deuterium incorporation into a peptide can be analyzed mainly in two different ways, 'centroid' or 'theoretical fitting' methods [40]. The centroid methods simply calculate the difference between the weighted average mass of the deuterated isotopic distribution and its natural (non-deuterated) isotopic distribution. Theoretical fitting methods determine the number of incorporated deuterium by fitting theoretical deuterated isotopic distributions with the observed isotopic distribution.
3.2. The determination of deuterium incorporation
Many software tools are available for automated data analysis and have made HDX-MS more widely used [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56]. Theoretical fitting methods are computationally more expensive, but are often more accurate than centroid methods. As deuterium is gradually incorporated, the number of incorporated deuterium is not determined as a single value, but as a distribution of incorporated deuterium numbers. More specifically, theoretical fit method is to solve linear equations, where the (partially) deuterated isotopic distribution is defined as convolution of its natural isotopic distribution and the deuterium incorporation distribution of the peptide.
In many studies, the deuterium incorporation distribution of a peptide was assumed a statistical model such as the binomial distribution [49], [50], [51]. The methods involve three relevant distributions as shown in Fig. 3: two observed isotopic distributions- 1) natural isotopic distribution, 'Dnat'; 2) deuterated isotopic distribution, 'Ddeu', and one distribution of deuterium incorporation to Ddeu, 'Dlev'. Dlev is expected to conform to the binomial distribution B(n, d/n), where n is the number of exchangeable hydrogens in a peptide and d, of interest in HDX-MS analysis, is the average deuterium incorporation. The convolution of Dnat and Dlev results in Ddeu, which spans a wider m/z range than Dnat since Dlev is not a single value but a distribution. Given Dnat and Ddeu of a peptide, the estimation of d is performed as follows: for every possible d, theoretical Dlev(d) is generated and theoretical Ddeu(d) can also be generated by convoluting Dlev(d) with Dnat (Let Dlev(#) and Ddeu(#) be the distributions based on deuterium number #). The convolution can be calculated using the fast Fourier transform (FFT) [49]. Then d can be determined by fitting any theoretical Ddeu(d) with the observed Ddeu.
The best match between the two distributions was estimated by various error functions such as least-squares, chi-squares, and asymmetric least squares. The core part in theoretical fitting methods is the implementation of error functions, which should be robust against noise or interfering peaks in experimental MS data since deuterated isotopic distributions frequently overlap with other isotopic distributions due to their wide span. HX-Express2 [50] implemented asymmetric least squares regression so that an error contribution gets greater for peaks of lower intensity than the calculated distributions, thereby favoring the calculated distribution to fit to uncontaminated peaks. Contrary to minimizing errors, deMix [51] proposed a measure, referred as to 'Matched Peak Count', which counts matching peaks between the two distributions, whose intensities are comparable within an intensity-proportional tolerance. In particular, the measure was very robust against high-intensity interfering peaks by minimizing their effect.
As an alternative, the observed deuterated isotopic distribution could directly be deconvoluted using the natural isotopic distribution, where Ddeu is a linear combination of mass-shifted natural isotope distributions (i.e., there is no assumption about Dlev in Fig. 3). The best fit to all of the linear equations was achieved using least-squares method (LSQ) [52] or maximum entropy method (MEM) [53]. The LSQ-based method is simple but not desirable when the number of exchangeable hydrogens is big or signal-to-noise ratio is poor. The MEM‐based method was more robust to the noise than LSQ and could result in a smoothed deuteration distribution at the expense of longer computation time. Hexicon 2 [54] employed Gold’s iterative deconvolution algorithm that assigns positive distribution coefficients using a gradient descent and favors smooth solutions similar to maximum entropy regularization. DEX [55] applied a Fourier deconvolution method and the deconvolution step resulted in the noise reduction, because the noise is averaged by Fourier transforms. These approaches are interested in the deuterium incorporation distribution itself for a peptide (allowing variations in extent of deuterium incorporation) as well as the average deuterium incorporation, while the binomial fitting-based approaches relatively emphasize the average deuterium incorporation for a peptide.
3.3. Deconvolution of bimodal behavior
One of the difficulties in determining deuterium incorporation is that often a deuterated isotopic distribution is observed as a bimodal form, not unimodal, which arises from EX1 behavior or heterogeneous conformational populations [57]. The kinetics for HDX has two limiting regimes, referred to as EX2 and EX1 [38], [58]. In EX1 condition, as a portion of the proteins unfold, the deuterium incorporation takes place on a number of residues simultaneously before returning to their ‘closed’ state. As a result, there are two populations represented as a bimodal form: 1) a lower mass population that has never undergone the slow unfolding transition and 2) a higher mass population that has undergone the transition at least once. With increasing D2O labeling time, the higher mass population would gradually grow while the lower mass population shrink. In EX2 condition (much more common under native conditions), the rate constant of the closing reaction is faster than the intrinsic rate of exchange and a progressive mass shift in single deuterated distribution is observed with increasing D2O labeling time. Most interestingly, the coexistence of two protein conformations may lead to simultaneous, progressive mass shifts in two distributions with increasing D2O labeling time. Some software tools such as ExMS [48], ExMS2 [56], HX-Express v2 [50], deMix [51], and Hexicon 2 [54] have been proposed to analyze a bimodal exchange pattern.
deMix [51] first performs an unimodal analysis via binomial fitting for an observed deuterated distribution and decides whether to perform bimodal analysis without any human intervention based on the unimodal analysis results. In the bimodal analysis, Dlev in Fig. 3 was assumed as a mixture of two binomial distributions. deMix used 'Matched Peak Count' for distribution fitting (as mentioned in the previous section), which not only was robust over random noises in comparing two distributions but also had strength in fast dissection of bimodal deuterated distributions by efficaciously fitting non-overlapped area. ExMS2 [56] provides various fitting options in multimodal analysis, where the number (up to 3) and shape (binomial, Gaussian, or reference) of component distributions can be set.
4. Covalent labeling
Covalent labeling is a powerful tool for monitoring the solvent accessibility of side chains in proteins or protein complexes by introducing irreversible modifications to reactive side chains [59], [60], [61], [62], [63], [64]. This is conceptually similar to HDX-MS analysis except that the labeling is restricted to one or a few amino acids. The most common targets have been primary amines (Lys and N-termini), carboxyl groups (Asp/Glu and C-termini), or thiols (Cys) [59]. The general covalent labeling MS workflow is depicted in Fig. 2b. In brief, subsequent labeling of proteins, the modified proteins are subjected to proteolytic digestion followed by LC-MS/MS. Since the modifications by covalent labeling reagents are highly stable and irreversible during sample handling, the downstream analyses to calculate the extent of modification are relatively flexible and MS/MS is usually applied to determine modification types as well as modified sites in peptides. On top of the restricted labeling to a few amino acids, diethyl pyrocarbonate (DEPC) reagent can modify nucleophilic side chains (Cys, His, Lys, Thr, Tyr and Ser) and N-termini via nucleophilic substitution reactions [60]. Oxidative labeling with hydroxyl radicals introduces simultaneous modifications into many different amino acids and has been widely used in many studies [61], [62], [63], [64].
The strength of covalent labeling is that the samples can be analyzed using conventional proteomics workflows, including sample handling techniques, LC-MS/MS, and data analysis. In HDX, optimized sample handling techniques are needed to minimize back exchange and scrambling due to reversible and labile nature of deuterium incorporation. In chemical cross-linking in Section 5, two or more peptides are connected and thus the data interpretation requires dedicated software.
4.1. Fundamentals of data interpretation
A list of unmodified and expectedly modified peptides from in silico digestion of target proteins are assigned to peptide peaks detected by MS (peptide mass fingerprinting). For confirmed peptides by MS, the peptide modification extents are calculated based on Extracted Ion Chromatograms (XIC) [65], [66]. The fraction of unmodified or modified peptide is derived from the ratio of XIC areas under the unmodified/modified species to total XIC areas of all the species (including unmodified and modified). Plotting the extent of modification relative to labeling reaction time can tell which protein regions are the most solvent-exposed. Peptide mass fingerprinting relying on accurate mass is sometimes insufficient to determine modified peptides. MS/MS peptide sequencing is necessary in cases of labeling many amino acids or studying unspecific proteins (i.e., a large search space). MS/MS enables the identification of specific amino acids undergoing modifications; however many modifiable amino acids give rise to a combinatorial issue in identifying modified sites using MS/MS (we discuss this issue in the next section).
4.2. MS/MS-based identification of modified residues
MS/MS allows site resolution for modifications, which can be detected by modification-related diagnostic mass shifts of fragment ions in MS/MS spectra as shown in Fig. 4. MS/MS has been the most powerful tool to identify numerous types of modifications on peptides and a large number of software tools have been developed to search for modified peptides [67]. For residue-specific labeling [59], standard database search tools [29], [30], [31] could detect the modified sites by allowing the corresponding modification on targeted residues during search. However, if the labeling reagent can target several residues or unspecific residues [60], dedicated software to modification searches might be needed. Oxidative labeling studies with hydroxyl radicals have reported that in practice every amino acid with the exception of Gly could be modified in oxidative labeling and various mass shifts such as + 16, +14, and + 32 Da were also observed [68]. In general, modification searches are performed by matching all possible modified forms of each peptide from a protein sequence database with MS/MS spectra. Given a list of possible modifications, the number of possible modified forms for a peptide of length n is expressed as , where Vi is the number of possible modifications at i-th residue of the peptide. For a large scale analysis, the modification search speed and sensitivity are overwhelmed by the huge search space that follows from enumerating all possible modified forms of peptides derived from a large database. What matters more than the much longer search time, is the fact that more false positives require a more stringent threshold to maintain an acceptable error rate, resulting in the loss of true identifications [69].
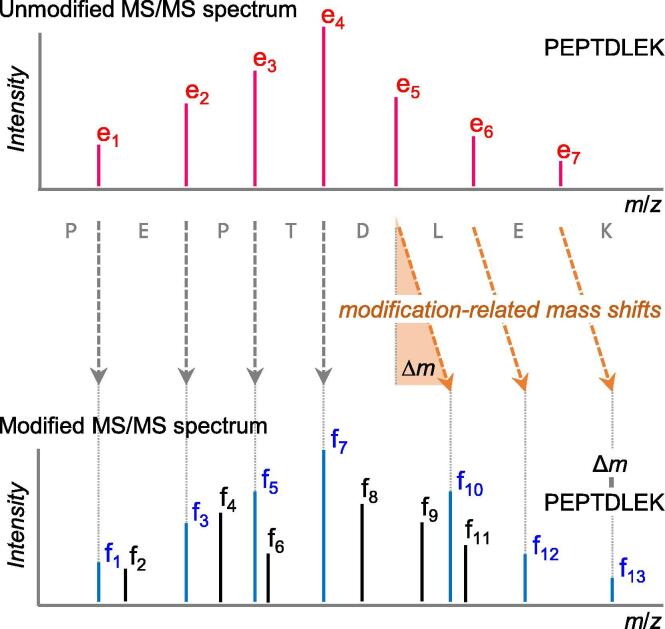
Fig. 4.
MS/MS spectra of unmodified (top) and modified (bottom) forms of a peptide PEPTDLEK are shown. In the modified spectrum, modification-related mass shifts (f10, f12 and f13 relative to e5, e6 and e7, respectively) are detected, starting with the modified site (i.e., the fifth residue D). Even with the same modification, peptides modified at different sites generate different spectra, but very similar. Theoretically, differently modified spectra can always be distinguished and thus database search tools can work for modified peptides. Experimental spectra typically contain lots of noise (shown as black peaks) that were not aligned in the bottom spectrum and matching such noise peaks might lead to incorrect assignments.
A number of computational methods and strategies have been developed to identify modified peptides while taking into account more modification types [70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94]. Recent developments for modification searches have mainly focused on open search, often referred to as 'blind' search, a strategy that searches MS/MS spectra for modifications of arbitrary masses [67]. The open searches take into account all peptides in a database as candidates of MS/MS spectra and a modification mass is calculated as the mass difference between an MS/MS precursor ion and a candidate peptide. MSFragger [91] could identify modified peptides in ultrafast fashion by pre-indexing the masses of all possible peptides from a database and even their fragment ions. However, most open searches usually allowed for only one unexpected modification per peptide because their accuracy and performance tend to degrade rapidly with increasing numbers of modified sites per peptide.
For oxidative labeling studies, the capacity to identify multiple modifications on a peptide is absolutely necessary to find the most solvent-exposed region since many amino acids in the region could be simultaneously modified by several type of modifications. Byonic [92] was applied to a study for probing in vivo structural dynamics of membrane proteins [95]. Byonic improved the search speed based on Lookup Peaks algorithm [93] that can fast extract candidate peptides from a database by utilizing likely b- and y-ion peaks in a spectrum. To narrow down the search space for multiple modifications, some approaches [74], [75], [76], [77] take advantage of sequence tags which are partial sequences derived from fragment ions of a peptide in an MS/MS spectrum, e.g., a sequence tag 'LE' can be derived using fragment ions f10-f12-f13 in the bottom spectrum of Fig. 4. On one hand, the sequence tags enable fast extraction of candidate peptides from a database. At the same time, they can localize modified regions within a peptide and thus make it possible to consider modifications only against shorter regions instead of the whole peptide’s extent. MODplus [94] further subdivides the regions into even shorter ones by simultaneously using multiple sequence tags. A dynamic programming algorithm, guided by the sequence tags, rapidly localizes modified regions in possibly modified peptides and then unexplained delta masses of the modified regions are interpreted with known modifications. In analyses of human proteome MS/MS datasets, MODplus worked well with the whole human database and ~ 1,000 modification types.
5. Chemical cross-linking
In cross-linking MS, cross-linking reagents covalently link two spatially proximal amino acid residues in native proteins or protein complexes, and MS/MS is applied to identify the linked amino acid residues. The two linked residues provide a powerful means to study the three-dimensional structure of proteins and protein complexes [96], [97], [98]. The general cross-linking MS workflow is depicted in Fig. 2c. In brief, proteins or protein assemblies under denaturing conditions are first reacted with bifunctional cross-linking reagents whose spacer-arm length confers a distance constraint on the two linked residues. The enzymatic digestion of the cross-linked proteins produces cross-linked peptides, followed by LC-MS/MS analysis. Subsequent informatics analysis of cross-linked MS data identifies cross-linked peptides and their linkage sites. Finally, distance constraints of the linkage sites are imposed on structure modeling. Owing to the usefulness of cross-linking MS, a large number of cross-linking reagents have been developed. Their underlying chemistry and reaction mechanisms are beyond the scope of this review. For more detailed discussion, see refs [99], [100].
5.1. Fundamentals of data interpretation
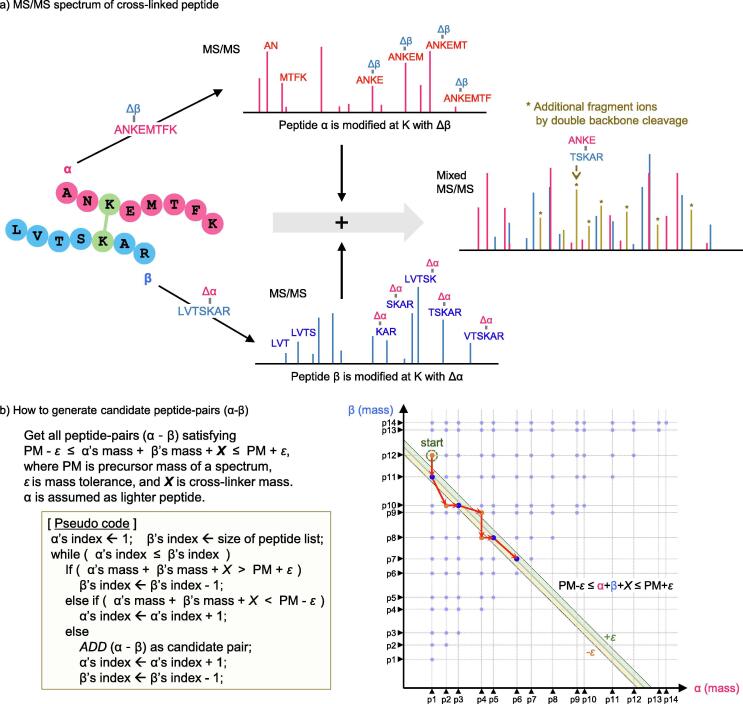
Algorithms to identify cross-linked peptides would follow a strategy similar to conventional database searches for linear peptides, comparing an experimental MS/MS spectrum with theoretical spectra of candidate peptides in a database. The key difference is retrieving peptide-pairs as candidates, a computationally challenging aspect of interpreting MS/MS spectra of cross-linked peptides. A candidate peptide-pair can be determined if the sum of the masses of two peptides in a pair and the cross-linker spacer arm is matched with the precursor mass of an MS/MS spectrum within a mass tolerance. Assuming that a cross-linked peptide involves only two peptides, the number of all possible peptide-pairs as candidates is equal to the binomial coefficient n+1C2, approximately n2 / 2, where n is the number of peptides from a database. Since the search space grows quadratically as the number of peptides in a database increases, the database size is critical to search performance. The theoretical MS/MS spectrum of a candidate peptide-pair (α-β) can be generated as a multiplexed spectrum including all fragment ions from modified peptides, αΔβ and βΔα, of the two peptides, where each peptide is modified by the mass of the other peptide and the cross-linker on the reactive site (Fig. 5a). Then, theoretical spectra of all candidate peptide-pairs were compared with an experimental MS/MS spectrum and the best-scoring peptide-pair is assigned as a cross-linked peptide.
Fig. 5.
a) MS/MS spectrum of a cross-linked peptide (α-β) can conceptually be regarded as multiplexed by the two spectra generated from the two modified peptides, αΔβ and βΔα, where each peptide is modified by the mass of the other peptide (although actually two peptides in a cross-linked peptide are simultaneously fragmented). The real spectrum, however, becomes more complex than a simply multiplexed form due to additional ions (* peaks) originating from double backbone cleavage. This characteristic makes the interpretation very complicated due to ambiguous peak assignments. b) Pseudo code for retrieving candidate peptide-pairs matched with a query mass (i.e., precursor ion mass, PM in the figure). Basically, the computational complexity is nC2 or O(n2). Using the sorted peptide list by their masses, the complexity gets down to O(n), where peptides that have been checked are never checked again, given a spectrum. For the schematic representation of the algorithm, let us assume a two-dimensional coordinate plane consisting of α-axis and β-axis representing peptide masses (right bottom in the figure). The triangles on each axis represent all (linear) peptides from a database at coordinates corresponding to their masses and then the light blue dots on the αβ plane are all possible peptide-pairs (105 pairs for 14 peptides in the figure, calculated as 14+1C2). Given a PM, candidate peptide-pairs are dots within the area satisfying an inequality PM-ε ≤ α + β + X ≤ PM + ε on the αβ plane, where ε is a mass tolerance and X is a cross-linker mass, (e.g., blue dots in green and yellow diagonal bands). To find candidate peptide-pairs, the pseudo code algorithm moves exactly along the red arrows and checks only eight dots (4 orange and 4 blue dots) while taking into account p12 ~ p7 as β and p1 ~ p6 as α, respectively (p13 and p14 were excluded from pairing since they themselves were heavier than PM + ε). Consequently, the entire peptide list is retrieved once. Due to the mass tolerance ε, the real implementation was a little different because one peptide could be paired with each of several peptides whose masses were almost the same (i.e., the mass difference was within ε). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The molecule types generated from cross-linking MS are mono-linked (or dead end), loop-linked (a peptide contains a linker spanning two reactive sites), and cross-linked peptides [98]. Mono-linked and loop-linked types involve only single peptide and they can be identified as modified peptides by standard database searches allowing the cross-linker as a variable modification. Although mono-linked and loop-linked types also provide useful information on protein structure, this review will focus on cross-linked types in which identifying peptide-pairs gives rise to significant computational issues.
5.2. Computational methods for cross-linked peptides
In its early days, some efforts have been made to make use of the existing database search tools designed for linear peptide identification to identify cross-linked peptides. It involves the generation of a chimeric database, XL-DB, containing linear (concatenated together) sequences of all possible peptide pairs [101], [102]. Then, the database search was performed against the XL-DB while allowing the cross-linker as a variable modification. The method works reasonably well, but the scoring schemes for linear peptides were not optimal for cross-linked peptides. The search never consider all possible fragment ion types from cross-linked peptides (α-β) simultaneously, since the linearized sequences, αβ and βα, were separately written in XL-DB and thus were matched to an experimental MS/MS spectrum one at a time during the search, i.e., utilizing only half fragment ions to identify cross-linked peptides. In a sense, the XL-DB approach eliminated the demand for dedicated software for identification of cross-linked peptides, but it faces the quadratic growth problem in the database size, with suboptimal fragment ion matching.
With the developments of open searches (details in MS/MS-based identification of modified residues in covalent labeling section), there also have been efforts to apply open search algorithms to the identification of cross-linked peptides [103], [104], [105], [106], [107]. The rationale is that a cross-linked peptide may be considered as one peptide with a large variable mass modification (1,000 ~ 4,000 Da) corresponding to the mass of the other peptide, i.e., αΔβ is a modified form of peptide α with the modification mass of peptide β in the example in Fig. 5a. This open search-based approach, first, identifies a modified peptide αΔm (m is an arbitrary mass) that best matches an MS/MS spectrum and then, finds the second peptide β corresponding to the mass m. If there were many second peptide candidates within a specified mass tolerance, additional validation is required. This approach, however, finds constituent peptides of a cross-linked peptide separately, and thus cannot interpret an MS/MS spectrum simultaneously with two constituent peptides, similar to XL-DB approach. Several software tools such as CXMS pipeline [103], Protein Prospector [104], Kojak [105], pLink [106], and MetaMorpheus [107] have adopted the open search strategy. CXMS pipeline that employs open search Popitam [108] reduced the search space of open search by considering only MS/MS spectra of high-charge state precursors unidentified from linear peptide search. As a recent automated approach, Kojak [105] finds top 250 modified peptides in the first pass via open search allowing modifications on linkable residues and then searches for cross-linked peptides via pairing the 250 peptides, in which two peptide masses plus the cross-linker mass is equal to the precursor ion mass. pLink [106] used similar strategy to Kojak, but in pairing peptides identified from open search, the heavier peptide in a pair is selected from top 50 peptides while top 250 peptides for the lighter peptide, assuming that the heavier (or longer) peptides would be identified more confidently from open search.
Various tools have been developed to take into account all possible fragment ions of cross-linked peptides at once during search [109], [110], [111], [112], [113]. The searches, however, have been limited to a database of only a few proteins or small purified protein complexes due to the huge search space. As an effort to reduce the search space, SIM-XL [112] created a dynamic database that consisted of all possible pairs containing at least one linear peptide identified with a dead-end via a preliminary search. Nevertheless, the underlying n-square problem originating from all peptide-peptide combinations could not be readily resolved. The expansion of cross-linking studies to a large database such as a complete human proteome has become feasible with the advent of isotopically coded cross-linkers and MS-cleavable cross-linkers [100], [114], [115]. For isotopically coded cross-linkers, cross-linked peptides by isotopically coded light and heavy linkers are mixed and analyzed together by MS. Isotopic pairs of chemically identical peptides could be recognized owing to the mass difference between light and heavy linkers. In MS/MS spectra of the two isotopic pairs, there are common fragment ions with identical masses, i.e., independent of cross-linkers. The software, xQuest [114], utilized such common ions to fast query candidate peptides to fragment-ion index generated from all peptides in the database. The queried peptides are compared with MS/MS spectra and only the combinations of the best matching peptides are evaluated as candidate cross-linked peptides. On one hand, MS-cleavable cross-linkers contain a preferential cleavage site (or sites) in the spacer arm and thus produces signature fragment ion peaks that indicate the masses of component peptides in a cross-linked peptide. XlinkX [115] software was designed to take advantage of MS-cleavable cross-linkers. XlinkX extracts the mass of each component peptide using signature peaks spaced by a unique mass difference (referred to as Δm principle) derived from the cross-linker. If at least one component peptide mass is observed with the Δm principle in an MS/MS spectrum, the spectrum is regarded as generated from a cross-linked peptide and all deduced peptide-pairs are submitted for further peptide sequence analysis. The extraction of component peptide masses reduced the search space from O(n2) to O(2n) by eliminating the combination of all peptides in a database. While xQuest and XlinkX were designed for specific cross-linkers, they also supported ‘enumeration mode’ that took into account all the peptide pair combinations against a small database. In other approaches, the signature peaks from cross-linkers were utilized for more sophisticated scoring of cross-linked peptides [116]. As a kind of MS-cleavable cross-linker, protein interaction reporter (PIR) containing two labile bonds in the cross-link can be specifically cleaved in situ to release a signature reporter ion and two intact peptide ions. X-links [117] supported identification of PIR-labeled products.
5.3. Disulfide bond
A disulfide bond is a post-translational modification that covalently links the sulfur atoms of two cysteine residues in close spatial proximity so that proteins are internally or externally cross-linked. Disulfide bonds are vital for the stabilization of final protein structures, and at the same time known to regulate mediation of various signaling pathways in a cell [118], [119]. Thus the determination of their presence and location in proteins can provide invaluable insight into protein folds and functions. Similar to chemical cross-linking MS strategy, information on the disulfide bonds can be obtained by analyzing MS/MS spectra of intact disulfide-linked peptides, resulting from enzymatic digestion of native proteins under non-reducing condition [120], [121]. The formation of nonnative disulfide bonds is prevented during sample preparation by controlling temperature, pH, and free cysteine [121]. The nature of disulfide-linked peptides is the same as chemically cross-linked peptides. Thus, algorithms for cross-linked peptides can be equally applied to the identification of disulfide-linked peptides by simply adjusting the linkable residues to Cysteine and the mass of cross-linker to −2.01565 Da of didehydro.
Dedicated software such as DBond [122], [123], SlinkS [124] and MassMatrix [125] has been introduced for the analysis of the MS/MS spectra of disulfide-linked peptides. The software tools were designed to recognize diagnostic fragment ion series from disulfide bonds that indicate the presence of disulfide bonds (not detected in linear peptides) and help assess the correctness of all the component peptides. SlinkS [124] utilized the abundance of fragment ions from disulfide bond-specific cleavages as well as peptide backbone fragments under electron transfer higher energy dissociation (EThcD) condition and improved the identification performance of disulfide-linked peptides. The identification of disulfide-linked peptides requires the enumeration of all combinations of two peptides containing Cys. DBond [122] implemented a fast algorithm for enumeration of peptide combination by sorting all peptides in a database based on their masses (Fig. 5b). In the sorted list, all possible peptide-pairs can be obtained by retrieving the entire list once while keeping the sum of masses of lighter and heavier peptides closest to the query precursors mass. The algorithm could reduce the quadratic search space to a linear search space and DBond worked well with a whole human database without any filtration step. Disulfide-linked peptides with more than one disulfide bond can be analyzed by informatics methods, but their poor fragmentation would make the identification less confident. MassMatrix [125] supported MS/MS-based identification of disulfide-linked peptides with up to 2 disulfide bonds.
5.4. Confidence and false positives
The confidence of cross-linked peptide assignments involves the confidence of each component peptides. The assignment of cross-linked peptides (α-β) would be confident if distinguishing fragment ions are evenly observed from both αΔβ and βΔα (i.e., both peptides are identified with very high confidence). There are ambiguous assignments where one peptide is identified with very high confidence but the other identification is poor (if the peptide is very short or if its fragment ions are rarely observed). The complete matching score of cross-linked peptides to MS/MS spectra is unable to distinguish such ambiguous assignments since their scores could be comparable to those in which both peptides are correctly identified. It should be noted that most false positive identifications in cross-linking MS arise from cases where one peptide is correctly identified but the second peptide is incorrect. In particular, the proportion of such cases increases if one peptide is very short [126]. To compensate for the problem, XLSearch [127] implemented a data-driven scoring scheme that independently estimates the probability of correctly identifying each individual peptide given knowledge of the correct or incorrect identification of the other peptide. Protein Prospector [128] reported the uneven fragmentation efficiency of the two component peptides and suggested to use metrics reflecting the quality of the spectral match to the less confident peptide, resulting in the most discriminatory power between correct and incorrect assignments. Machine-learning techniques were also applied to improve the classification. xProphet [129] applied linear discriminant analysis to optimally combine multiple scores so that the combined score maximized the separation of true positive and false positive hits.
False Discovery Rate (FDR) estimation using target-decoy strategy [32] is more complicated for cross-linked peptides than linear peptides. Because two peptides are involved, three labels exist, 1) target-target (TT), 2) target-decoy or decoy-target (TD), and 3) decoy-decoy (DD). False positives could be estimated using those with at least one decoy match. The most commonly used formula for false positives is #TD - #DD [130]. Some corrected formulas have been introduced as the types of cross-linkers (e.g., directional, non-directional and heterobifunctional) [131]. Finally, FDRs were calculated separately for mono-linked, loop-linked, and cross-linked peptides (that can be further separated into intraprotein and interprotein) because their a priori probabilities for matching as well as score distributions are different [129]. The separate FDR calculation showed an improvement in the validation when compared to global FDR calculation [128].
6. Conclusions
MS-based proteomics has been a versatile tool to study many aspects of proteins. Here, we reviewed state-of-the-art MS-based techniques and computational tools for structural analyses of proteins and protein complexes. The peptide-centric structural proteomics have limitations as well as particular advantages. It necessarily includes two procedures to introduce modifications into native proteins and digest the modified proteins into peptides. First of all, the protein digestion causes major limitations. If sequence coverage of a protein is observed to be low by recovered peptides, a significant amount of structural information is lost. If a peptide sequence is repeated within a protein or complex, it cannot be unambiguously assigned. For protein isoforms, assembly of recovered peptides can hardly determine exact proteoforms. Second, the excessive labeling may cause unwanted structural perturbations of native proteins. Third, the comparison between unmodified and modified samples assumes the same properties of unmodified and modified peptides in MS experiments. The detectability of the two, however, might be different because modifications might prevent enzymatic cleavage or change chromatographic retention time and ionization efficiency, making their quantitative comparison complicated. Finally, unexpected modifications that existed in native proteins or are introduced during MS experiments might hinder the identification of peptides of interest. For the success of MS-based techniques, the experimental conditions should be carefully controlled throughout the experiment and the choice of labeling or cross-linking reagents and proteases should be carefully made by considering the distribution of amino acids in the protein sequence. A combination of different reagents or techniques may also be considered to increase the number of restraints if target amino acids for labeling are not sufficient in regions of interest.
MS-based techniques are extending the scope toward endogenous biomolecular or proteome-wide studies. It can be expected that the commensurate advances in data analysis tools will enable large-scale analyses, at the same time reduce false positives, and in the end increase the quality of constraints on protein or network modeling. The recent focus has been on integrative approaches that combine information from different types of experiments to compute structural models. MS-based techniques can work as complements to emerging in vivo and in situ technologies such as live-cell imaging and in-/on-cell NMR. Improvements would be necessary for integrative software to best combine a variety of structural data from different methods.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2017R1E1A1A01077412, 2019M3E5D3073568). S.N. was supported by BK21 Plus project.
References
- 1.Ilari A., Savino C. Protein structure determination by x-ray crystallography. Methods Mol Biol. 2008;452:63–87. doi: 10.1007/978-1-60327-159-2_3. [DOI] [PubMed] [Google Scholar]
- 2.Tugarinov V., Hwang P.M., Kay L.E. Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu Rev Biochem. 2004;73:107–146. doi: 10.1146/annurev.biochem.73.011303.074004. [DOI] [PubMed] [Google Scholar]
- 3.Bai X.C., McMullan G., Scheres S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Lössl P., van de Waterbeemd M., Heck A.J. The diverse and expanding role of mass spectrometry in structural and molecular biology. EMBO J. 2016;35:2634–2657. doi: 10.15252/embj.201694818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 6.Ong S.E., Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 7.Mann M., Jensen O.N. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 8.Hebert A.S., Richards A.L., Bailey D.J., Ulbrich A., Coughlin E.E., Westphall M.S. The one hour yeast proteome. Mol Cell Proteomics. 2014;13:339–347. doi: 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geiger T., Wehner A., Schaab C., Cox J., Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11(M111) doi: 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 12.Greber B.J., Boehringer D., Leibundgut M., Bieri P., Leitner A., Schmitz N. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;515:283–286. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- 13.Staals R.H., Zhu Y., Taylor D.W., Kornfeld J.E., Sharma K., Barendregt A. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Wei H., Krystek S.R., Jr, Bond D., Brender T.M., Cohen D. Mapping the energetic epitope of an Antibody/Interleukin-23 interaction with hydrogen/deuterium exchange, fast photochemical oxidation of proteins mass spectrometry, and alanine shave mutagenesis. Anal Chem. 2017;89:2250–2258. doi: 10.1021/acs.analchem.6b03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limpikirati P., Hale J.E., Hazelbaker M., Huang Y., Jia Z., Yazdani M. Covalent labeling and mass spectrometry reveal subtle higher order structural changes for antibody therapeutics. MAbs. 2019;11:463–476. doi: 10.1080/19420862.2019.1565748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Arcy S., Martin K.W., Panchenko T., Chen X., Bergeron S., Stargell L.A. Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A–H2B in an unconventional tetrameric form. Mol Cell. 2013;51:662–677. doi: 10.1016/j.molcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rostislavleva K., Soler N., Ohashi Y., Zhang L., Pardon E., Burke J.E. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350:aac7365. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urnavicius L., Zhang K., Diamant A.G., Motz C., Schlager M.A., Yu M. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leitner A. Cross-linking and other structural proteomics techniques: how chemistry is enabling mass spectrometry applications in structural biology. Chem Sci. 2016;7:4792–4803. doi: 10.1039/c5sc04196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokmina-Lukaszewska M., Patterson A., Berry L., Scott L., Balasubramanian N., Bothner B. The role of mass spectrometry in structural studies of flavin-based electron bifurcating enzymes. Front Microbiol. 2018;9:1397. doi: 10.3389/fmicb.2018.01397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinz A., Arlt C., Chorev D., Sharon M. Chemical cross-linking and native mass spectrometry: A fruitful combination for structural biology. Protein Sci. 2015;24:1193–1209. doi: 10.1002/pro.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcoux J., Wang S.C., Politis A., Reading E., Ma J., Biggin P.C. Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proc Natl Acad Sci U S A. 2013;110:9704–9709. doi: 10.1073/pnas.1303888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan J., Zhang S., Borchers C.H. Comparative higher-order structure analysis of antibody biosimilars using combined bottom-up and top-down hydrogen-deuterium exchange mass spectrometry. Biochim Biophys Acta. 2016;1864:1801–1808. doi: 10.1016/j.bbapap.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 24.Li H., Nguyen H.H., Ogorzalek Loo R.R., Campuzano I.D.G., Loo J.A. An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes. Nat Chem. 2018;10:139–148. doi: 10.1038/nchem.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward A.B., Sali A., Wilson I.A. Biochemistry. Integrative structural biology. Science. 2013;339:913–915. doi: 10.1126/science.1228565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faini M., Stengel F., Aebersold R. The evolving contribution of mass spectrometry to integrative structural biology. J Am Soc Mass Spectrom. 2016;27:966–974. doi: 10.1007/s13361-016-1382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaptein R., Wagner G. Integrative methods in structural biology. J Biomol NMR. 2019;73:261–263. doi: 10.1007/s10858-019-00267-z. [DOI] [PubMed] [Google Scholar]
- 28.Steen H., Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 29.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 30.Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 31.Kim S., Pevzner P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 33.Nesvizhskii A.I., Vitek O., Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods. 2007;4:787–797. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 34.Nesvizhskii A.I. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L., Chance M.R. Protein footprinting comes of age: mass spectrometry for biophysical structure assessment. Mol Cell Proteomics. 2017;16:706–716. doi: 10.1074/mcp.O116.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konermann L., Pan J., Liu Y.H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 37.Brown K.A., Wilson D.J. Bottom-up hydrogen deuterium exchange mass spectrometry: data analysis and interpretation. Analyst. 2017;142:2874–2886. doi: 10.1039/c7an00662d. [DOI] [PubMed] [Google Scholar]
- 38.Percy A.J., Rey M., Burns K.M., Schriemer D.C. Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry-a review. Anal Chim Acta. 2012;721:7–21. doi: 10.1016/j.aca.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 39.Rand K.D., Zehl M., Jørgensen T.J. Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling. Acc Chem Res. 2014;47:3018–3027. doi: 10.1021/ar500194w. [DOI] [PubMed] [Google Scholar]
- 40.Claesen J., Burzykowski T. Computational methods and challenges in hydrogen/deuterium exchange mass spectrometry. Mass Spectrom Rev. 2017;36:649–667. doi: 10.1002/mas.21519. [DOI] [PubMed] [Google Scholar]
- 41.Pascal B.D., Chalmers M.J., Busby S.A., Mader C.C., Southern M.R., Tsinoremas N.F. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinf. 2007;8:156. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pascal B.D., Chalmers M.J., Busby S.A., Griffin P.R. HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J Am Soc Mass Spectrom. 2009;20:601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pascal B.D., Willis S., Lauer J.L., Landgraf R.R., West G.M., Marciano D. HDX workbench: software for the analysis of H/D exchange MS data. J Am Soc Mass Spectrom. 2012;23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slysz G.W., Baker C.A., Bozsa B.M., Dang A., Percy A.J., Bennett M. Hydra: software for tailored processing of H/D exchange data from MS or tandem MS analyses. BMC Bioinf. 2009;10:162. doi: 10.1186/1471-2105-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S., Liu L., Uzuner U., Zhou X., Gu M., Shi W. HDX-analyzer: a novel package for statistical analysis of protein structure dynamics. BMC Bioinf. 2011;12:S43. doi: 10.1186/1471-2105-12-S1-S43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller D.E., Prasannan C.B., Villar M.T., Fenton A.W., Artigues A. HDXFinder: automated analysis and data reporting of deuterium/hydrogen exchange mass spectrometry. J Am Soc Mass Spectrom. 2012;23:425–429. doi: 10.1007/s13361-011-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kan Z.Y., Walters B.T., Mayne L., Englander S.W. Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis. Proc Natl Acad Sci U S A. 2013;110:16438–16443. doi: 10.1073/pnas.1315532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kan Z.Y., Mayne L., Chetty P.S., Englander S.W. ExMS: data analysis for HX-MS experiments. J Am Soc Mass Spectrom. 2011;22:1906–1915. doi: 10.1007/s13361-011-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmblad M., Buijs J., Håkansson P. Automatic analysis of hydrogen/deuterium exchange mass spectra of peptides and proteins using calculations of isotopic distributions. J Am Soc Mass Spectrom. 2001;12:1153–1162. doi: 10.1016/S1044-0305(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 50.Guttman M., Weis D.D., Engen J.R., Lee K.K. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J Am Soc Mass Spectrom. 2013;24:1906–1912. doi: 10.1007/s13361-013-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Na S., Lee J.J., Joo J.W.J., Lee K.J., Paek E. deMix: decoding deuterated distributions from heterogeneous protein states via HDX-MS. Sci Rep. 2019;9:3176. doi: 10.1038/s41598-019-39512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chik J.K., Vande Graaf J.L., Schriemer D.C. Quantitating the statistical distribution of deuterium incorporation to extend the utility of H/D exchange MS data. Anal Chem. 2006;78:207–214. doi: 10.1021/ac050988l. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z., Guan S., Marshall A.G. Enhancement of the effective resolution of mass spectra of high-mass biomolecules by maximum entropy-based deconvolution to eliminate the isotopic natural abundance distribution. J Am Soc Mass Spectrom. 1997;8:659–670. [Google Scholar]
- 54.Lindner R., Lou X., Reinstein J., Shoeman R.L., Hamprecht F.A., Winkler A. Hexicon 2: automated processing of hydrogen-deuterium exchange mass spectrometry data with improved deuteration distribution estimation. J Am Soc Mass Spectrom. 2014;25:1018–1028. doi: 10.1007/s13361-014-0850-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hotchko M., Anand G.S., Komives E.A., Ten Eyck L.F. Automated extraction of backbone deuteration levels from amide H/2H mass spectrometry experiments. Protein Sci. 2006;15:583–601. doi: 10.1110/ps.051774906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kan Z.Y., Ye X., Skinner J.J., Mayne L., Englander S.W. ExMS2: An integrated solution for hydrogen-deuterium exchange mass spectrometry data analysis. Anal Chem. 2019;91:7474–7481. doi: 10.1021/acs.analchem.9b01682. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J., Ramachandran P., Kumar R., Gross M.L. H/D exchange centroid monitoring is insufficient to show differences in the behavior of protein states. J Am Soc Mass Spectrom. 2013;24:450–453. doi: 10.1007/s13361-012-0555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weis D.D., Wales T.E., Engen J.R., Hotchko M., Ten Eyck L.F. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J Am Soc Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 59.Mendoza V.L., Vachet R.W. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Limpikirati P., Liu T., Vachet R.W. Covalent labeling-mass spectrometry with non-specific reagents for studying protein structure and interactions. Methods. 2018;144:79–93. doi: 10.1016/j.ymeth.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konermann L., Stocks B.B., Pan Y., Tong X. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom Rev. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 62.Johnson D.T., Di Stefano L.H., Jones L.M. Fast photochemical oxidation of proteins (FPOP): A powerful mass spectrometry-based structural proteomics tool. J Biol Chem. 2019;294:11969–11979. doi: 10.1074/jbc.REV119.006218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bavro V.N., Gupta S., Ralston C. Oxidative footprinting in the study of structure and function of membrane proteins: current state and perspectives. Biochem Soc Trans. 2015;43:983–994. doi: 10.1042/BST20150130. [DOI] [PubMed] [Google Scholar]
- 64.Garcia N.K., Deperalta G., Wecksler A.T. Current trends in biotherapeutic higher order structure characterization by irreversible covalent footprinting mass spectrometry. Protein Pept Lett. 2019;26:35–43. doi: 10.2174/0929866526666181128141953. [DOI] [PubMed] [Google Scholar]
- 65.Ziemianowicz D.S., Sarpe V., Schriemer D.C. Quantitative analysis of protein covalent labeling mass spectrometry data in the Mass Spec Studio. Anal Chem. 2019;91:8492–8499. doi: 10.1021/acs.analchem.9b01625. [DOI] [PubMed] [Google Scholar]
- 66.Bellamy-Carter J., Oldham N.J. PepFoot: a software package for semiautomated processing of protein footprinting data. J Proteome Res. 2019;18:2925–2930. doi: 10.1021/acs.jproteome.9b00238. [DOI] [PubMed] [Google Scholar]
- 67.Na S., Paek E. Software eyes for protein post-translational modifications. Mass Spectrom Rev. 2015;34:133–147. doi: 10.1002/mas.21425. [DOI] [PubMed] [Google Scholar]
- 68.Xu G., Chance M.R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 69.Ahrné E., Müller M., Lisacek F. Unrestricted identification of modified proteins using MS/MS. Proteomics. 2010;10:671–686. doi: 10.1002/pmic.200900502. [DOI] [PubMed] [Google Scholar]
- 70.Tsur D., Tanner S., Zandi E., Bafna V., Pevzner P.A. Identification of post-translational modifications by blind search of mass spectra. Nat Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y., Chen W., Cobb M.H., Zhao Y. PTMap–a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc Natl Acad Sci U S A. 2009;106:761–766. doi: 10.1073/pnas.0811739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chalkley R.J., Baker P.R., Medzihradszky K.F., Lynn A.J., Burlingame A.L. In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol Cell Proteomics. 2008;7:2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han X., He L., Xin L., Shan B., Ma B. PeaksPTM: mass spectrometry-based identification of peptides with unspecified modifications. J Proteome Res. 2011;10:2930–2936. doi: 10.1021/pr200153k. [DOI] [PubMed] [Google Scholar]
- 74.Searle B.C., Dasari S., Wilmarth P.A., Turner M., Reddy A.P., David L.L. Identification of protein modifications using MS/MS de novo sequencing and the OpenSea alignment algorithm. J Proteome Res. 2005;4:546–554. doi: 10.1021/pr049781j. [DOI] [PubMed] [Google Scholar]
- 75.Dasari S., Chambers M.C., Slebos R.J., Zimmerman L.J., Ham A.J., Tabb D.L. TagRecon: high-throughput mutation identification through sequence tagging. J Proteome Res. 2010;9:1716–1726. doi: 10.1021/pr900850m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Na S., Jeong J., Park H., Lee K.J., Paek E. Unrestrictive identification of multiple post-translational modifications from tandem mass spectrometry using an error-tolerant algorithm based on an extended sequence tag approach. Mol Cell Proteomics. 2008;7:2452–2463. doi: 10.1074/mcp.M800101-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Na S., Bandeira N., Paek E. Fast multi-blind modification search through tandem mass spectrometry. Mol Cell Proteomics. 2012;11(M111) doi: 10.1074/mcp.M111.010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Savitski M.M., Nielsen M.L., Zubarev R.A. ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol Cell Proteomics. 2006;5:935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- 79.Bandeira N., Tsur D., Frank A., Pevzner P.A. Protein identification by spectral networks analysis. Proc Natl Acad Sci U S A. 2007;104:6140–6145. doi: 10.1073/pnas.0701130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Falkner J.A., Falkner J.W., Yocum A.K., Andrews P.C. A spectral clustering approach to MS/MS identification of post-translational modifications. J Proteome Res. 2008;7:4614–4622. doi: 10.1021/pr800226w. [DOI] [PubMed] [Google Scholar]
- 81.Na S., Payne S.H., Bandeira N. Multi-species identification of polymorphic peptide variants via propagation in spectral networks. Mol Cell Proteomics. 2016;15:3501–3512. doi: 10.1074/mcp.O116.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.David M., Fertin G., Rogniaux H., Tessier D. SpecOMS: a full open modification search method performing all-to-all spectra comparisons within minutes. J Proteome Res. 2017;16:3030–3038. doi: 10.1021/acs.jproteome.7b00308. [DOI] [PubMed] [Google Scholar]
- 83.Ye D., Fu Y., Sun R.X., Wang H.P., Yuan Z.F., Chi H. Open MS/MS spectral library search to identify unanticipated post-translational modifications and increase spectral identification rate. Bioinformatics. 2010;26:i399–i406. doi: 10.1093/bioinformatics/btq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma C.W., Lam H. Hunting for unexpected post-translational modifications by spectral library searching with tier-wise scoring. J Proteome Res. 2014;13:2262–2271. doi: 10.1021/pr401006g. [DOI] [PubMed] [Google Scholar]
- 85.Burke M.C., Mirokhin Y.A., Tchekhovskoi D.V., Markey S.P., Heidbrink Thompson J., Larkin C. The Hybrid Search: a mass spectral library search method for discovery of modifications in proteomics. J Proteome Res. 2017;16:1924–1935. doi: 10.1021/acs.jproteome.6b00988. [DOI] [PubMed] [Google Scholar]
- 86.Chick J.M., Kolippakkam D., Nusinow D.P., Zhai B., Rad R., Huttlin E.L. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat Biotechnol. 2015;33:743–749. doi: 10.1038/nbt.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baliban R.C., DiMaggio P.A., Plazas-Mayorca M.D., Young N.L., Garcia B.A., Floudas C.A. A novel approach for untargeted post-translational modification identification using integer linear optimization and tandem mass spectrometry. Mol Cell Proteomics. 2010;9:764–779. doi: 10.1074/mcp.M900487-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fu Y., Xiu L.Y., Jia W., Ye D., Sun R.X., Qian X.H. DeltAMT: a statistical algorithm for fast detection of protein modifications from LC-MS/MS data. Mol Cell Proteomics. 2011;10(M110) doi: 10.1074/mcp.M110.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chi H., He K., Yang B., Chen Z., Sun R.X., Fan S.B. pFind-Alioth: A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. J Proteomics. 2015;125:89–97. doi: 10.1016/j.jprot.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 90.Solntsev S.K., Shortreed M.R., Frey B.L., Smith L.M. Enhanced global post-translational modification discovery with MetaMorpheus. J Proteome Res. 2018;17:1844–1851. doi: 10.1021/acs.jproteome.7b00873. [DOI] [PubMed] [Google Scholar]
- 91.Kong A.T., Leprevost F.V., Avtonomov D.M., Mellacheruvu D., Nesvizhskii A.I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods. 2017;14:513–520. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Current Protocols in Bioinformatics. 2012;40(1) doi: 10.1002/0471250953.2012.40.issue-110.1002/0471250953.bi1320s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bern M., Cai Y., Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal Chem. 2007;79:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- 94.Na S., Kim J., Paek E. MODplus: Robust and unrestrictive identification of post-translational modifications using mass spectrometry. Anal Chem. 2019;91:11324–11333. doi: 10.1021/acs.analchem.9b02445. [DOI] [PubMed] [Google Scholar]
- 95.Zhu Y., Guo T., Park J.E., Li X., Meng W., Datta A. Elucidating in vivo structural dynamics in integral membrane protein by hydroxyl radical footprinting. Mol Cell Proteomics. 2009;8:1999–2010. doi: 10.1074/mcp.M900081-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leitner A., Faini M., Stengel F., Aebersold R. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem Sci. 2016;41:20–32. doi: 10.1016/j.tibs.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 97.Yu C., Huang L. Cross-linking mass spectrometry: an emerging technology for interactomics and structural biology. Anal Chem. 2018;90:144–165. doi: 10.1021/acs.analchem.7b04431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Holding A.N. XL-MS: Protein cross-linking coupled with mass spectrometry. Methods. 2015;89:54–63. doi: 10.1016/j.ymeth.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 99.Petrotchenko E.V., Borchers C.H. Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom Rev. 2010;29:862–876. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- 100.Paramelle D., Miralles G., Subra G., Martinez J. Chemical cross-linkers for protein structure studies by mass spectrometry. Proteomics. 2013;13:438–456. doi: 10.1002/pmic.201200305. [DOI] [PubMed] [Google Scholar]
- 101.Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics. 2007;6:2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 102.Panchaud A., Singh P., Shaffer S.A., Goodlett D.R. xComb: a cross-linked peptide database approach to protein-protein interaction analysis. J Proteome Res. 2010;9:2508–2515. doi: 10.1021/pr9011816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Singh P., Shaffer S.A., Scherl A., Holman C., Pfuetzner R.A., Larson Freeman T.J. Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal Chem. 2008;80:8799–8806. doi: 10.1021/ac801646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chu F., Baker P.R., Burlingame A.L., Chalkley R.J. Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. Mol Cell Proteomics. 2010;9:25–31. doi: 10.1074/mcp.M800555-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoopmann M.R., Zelter A., Johnson R.S., Riffle M., MacCoss M.J., Davis T.N. Kojak: efficient analysis of chemically cross-linked protein complexes. J Proteome Res. 2015;14:2190–2198. doi: 10.1021/pr501321h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang B., Wu Y.J., Zhu M., Fan S.B., Lin J., Zhang K. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 107.Lu L., Millikin R.J., Solntsev S.K., Rolfs Z., Scalf M., Shortreed M.R. Identification of MS-cleavable and noncleavable chemically cross-linked peptides with MetaMorpheus. J Proteome Res. 2018;17:2370–2376. doi: 10.1021/acs.jproteome.8b00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hernandez P., Gras R., Frey J., Appel R.D. Popitam: towards new heuristic strategies to improve protein identification from tandem mass spectrometry data. Proteomics. 2003;3:870–878. doi: 10.1002/pmic.200300402. [DOI] [PubMed] [Google Scholar]
- 109.Lee Y.J., Lackner L.L., Nunnari J.M., Phinney B.S. Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J Proteome Res. 2007;6:3908–3917. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- 110.McIlwain S., Draghicescu P., Singh P., Goodlett D.R., Noble W.S. Detecting cross-linked peptides by searching against a database of cross-linked peptide pairs. J Proteome Res. 2010;9:2488–2495. doi: 10.1021/pr901163d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Du X., Chowdhury S.M., Manes N.P., Wu S., Mayer M.U., Adkins J.N. Xlink-identifier: an automated data analysis platform for confident identifications of chemically cross-linked peptides using tandem mass spectrometry. J Proteome Res. 2011;10:923–931. doi: 10.1021/pr100848a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lima D.B., de Lima T.B., Balbuena T.S., Neves-Ferreira A.G.C., Barbosa V.C., Gozzo F.C. SIM-XL: A powerful and user-friendly tool for peptide cross-linking analysis. J Proteomics. 2015;129:51–55. doi: 10.1016/j.jprot.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 113.Yılmaz Ş., Drepper F., Hulstaert N., Černič M., Gevaert K., Economou A. Xilmass: A new approach toward the identification of cross-linked peptides. Anal Chem. 2016;88:9949–9957. doi: 10.1021/acs.analchem.6b01585. [DOI] [PubMed] [Google Scholar]
- 114.Rinner O., Seebacher J., Walzthoeni T., Mueller L.N., Beck M., Schmidt A. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu F., Rijkers D.T., Post H., Heck A.J. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods. 2015;12:1179–1184. doi: 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- 116.Götze M., Pettelkau J., Fritzsche R., Ihling C.H., Schäfer M., Sinz A. Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J Am Soc Mass Spectrom. 2015;26:83–97. doi: 10.1007/s13361-014-1001-1. [DOI] [PubMed] [Google Scholar]
- 117.Anderson G.A., Tolic N., Tang X., Zheng C., Bruce J.E. Informatics strategies for large-scale novel cross-linking analysis. J Proteome Res. 2007;6:3412–3421. doi: 10.1021/pr070035z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sevier C.S., Kaiser C.A. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 119.Bulleid N.J., Ellgaard L. Multiple ways to make disulfides. Trends Biochem Sci. 2011;36:485–492. doi: 10.1016/j.tibs.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 120.Gorman J.J., Wallis T.P., Pitt J.J. Protein disulfide bond determination by mass spectrometry. Mass Spectrom Rev. 2002;21:183–216. doi: 10.1002/mas.10025. [DOI] [PubMed] [Google Scholar]
- 121.Lakbub J.C., Shipman J.T., Desaire H. Recent mass spectrometry-based techniques and considerations for disulfide bond characterization in proteins. Anal Bioanal Chem. 2018;410:2467–2484. doi: 10.1007/s00216-017-0772-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Choi S., Jeong J., Na S., Lee H.S., Kim H.Y., Lee K.J. New algorithm for the identification of intact disulfide linkages based on fragmentation characteristics in tandem mass spectra. J Proteome Res. 2010;9:626–635. doi: 10.1021/pr900771r. [DOI] [PubMed] [Google Scholar]
- 123.Na S., Paek E., Choi J.S., Kim D., Lee S.J., Kwon J. Characterization of disulfide bonds by planned digestion and tandem mass spectrometry. Mol Biosyst. 2015;11:1156–1164. doi: 10.1039/c4mb00688g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu F., van Breukelen B., Heck A.J. Facilitating protein disulfide mapping by a combination of pepsin digestion, electron transfer higher energy dissociation (EThcD), and a dedicated search algorithm SlinkS. Mol Cell Proteomics. 2014;13:2776–2786. doi: 10.1074/mcp.O114.039057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu H., Zhang L., Freitas M.A. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J Proteome Res. 2008;7:138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ji C., Li S., Reilly J.P., Radivojac P., Tang H. XLSearch: a Probabilistic Database Search Algorithm for Identifying Cross-Linked Peptides. J Proteome Res. 2016;15:1830–1841. doi: 10.1021/acs.jproteome.6b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Trnka M.J., Baker P.R., Robinson P.J., Burlingame A.L., Chalkley R.J. Matching cross-linked peptide spectra: only as good as the worse identification. Mol Cell Proteomics. 2014;13:420–434. doi: 10.1074/mcp.M113.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Walzthoeni T., Claassen M., Leitner A., Herzog F., Bohn S., Förster F. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9:901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- 130.Fischer L., Rappsilber J. Quirks of error estimation in cross-linking/mass spectrometry. Anal Chem. 2017;89:3829–3833. doi: 10.1021/acs.analchem.6b03745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fischer L., Rappsilber J. False discovery rate estimation and heterobifunctional cross-linkers. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0196672. [DOI] [PMC free article] [PubMed] [Google Scholar]